Abstract
Genome-wide association studies (GWAS) have replicably identified multiple loci associated with population-based plasma lipid concentrations1-5. Common genetic variants at these loci together explain <10% of the total variation of each lipid trait4,5. Rare variants of individually large effect may contribute additionally to the “missing heritability” of lipid traits6,7, however it remains to be shown to what extent rare variants will affect lipid phenotypes. Here, we demonstrate a significant accumulation of rare variants in GWAS-identified genes in patients with an extreme phenotype of abnormal plasma triglyceride (TG) metabolism. A GWAS of hypertriglyceridemia (HTG) patients revealed that common variants in APOA5, GCKR, LPL and APOB genes were associated with the HTG phenotype at genome-wide significance. We subsequently resequenced protein coding regions of these genes and found a significant burden of 154 rare missense or nonsense variants in 438 HTG patients, in contrast to 53 variants in 327 controls (P=6.2X10-8); this corresponds to a carrier frequency of 28.1% of HTG patients and 15.3% of controls (P=2.6X10-5). Many rare variants were predicted in silico to have compromised function; additionally some had previously demonstrated dysfunctionality in vitro. Rare variants in these 4 genes explained 1.1% of total variation in HTG diagnoses. Our study demonstrates a marked mutation skew that likely contributes to disease pathophysiology in patients with HTG.
Genome-wide association studies (GWAS) have replicably identified novel and known loci associated with population-based plasma lipid concentrations1-5. Despite the robustness of these associations, the proportion of variability explained by GWAS-identified loci is relatively modest, <10% in most studies4,5. While vastly expanded study sample sizes continue to reveal new associations, each newly associated variant has an incrementally smaller effect size and contributes only marginally to the cumulative variation of each lipid phenotype6. This suggests that GWAS of population-based subjects may be reaching their limits to explain genetic variation of complex traits. A question that has arisen is whether additional forms of genetic variation, such as rare variants of individually large effect, could contribute to the “missing heritability” of complex traits such as plasma lipid concentrations6,7. While the mechanistic basis for the association between lipid traits and most common variants discovered in GWAS is still largely unknown, it remains possible that rare variants in GWAS-identified genes may contribute significantly to lipid phenotypes.
Studying subjects at the extremes of a quantitative phenotype distribution has proven useful to identify functional rare variants8-12. Using missense-accumulation analysis in genes defined a priori as likely to contain rare variants, a burden of mutations can be quantified statistically in subjects with severe phenotypes, prior to functional assessment of each variant. Primary hypertriglyceridemia (HTG) is one such complex polygenic disease broadly defined by fasting plasma TG concentrations >95th percentile13. Resequencing of TG-modulating candidate genes has implicated both common and rare variants in HTG disease pathophysiology9,14-16, however the majority of phenotypic variation underlying severe HTG remains unattributed17. Our objectives were: 1) to perform a non-biased GWAS of patients with HTG to identify common variants associated with HTG; and 2) to resequence coding regions of candidate genes in loci reaching genome-wide significance to evaluate the burden of rare variants in HTG patients compared with controls. Here, we demonstrate that loci found to be associated with HTG by GWAS using common variants also harbour a significant excess of rare variants.
In total, 555 HTG patients and 1319 controls were included in two cohorts of the study: the GWAS cohort included 463 HTG patients and 1197 controls and the sequencing cohort included 438 HTG patients and 327 controls. HTG patients were unrelated subjects diagnosed with Fredrickson hyperlipoproteinemia (HLP) phenotypes 2B (MIM 144250), 3 (MIM 107741), 4 (MIM 144600) or 5 (MIM 144650), ascertained primarily from a single tertiary referral lipid clinic. The mean plasma TG concentration of HTG patients was 14.3 mmol/L. Controls had maximum recorded fasting plasma TG concentrations <2.3 mmol/L to exclude undiagnosed HTG. All study subjects were of self-declared European ancestry; subjects deviating from European ancestry as determined by multi-dimensional scaling using whole-genome SNP data were removed from sequencing analysis (Supplemental Fig. 1). As expected, clinical characteristics of HTG patients were less favorable than controls, with worse lipid profiles and an increased prevalence of type 2 diabetes (Table 1).
Table 1.
Baseline clinical attributes of the study sample.
| GWAS cohort | Resequencing cohort | |||
|---|---|---|---|---|
| | ||||
| HTG | Controls | HTG | Controls | |
| Number | 463 | 1197 | 438 | 327 |
| Female | 30.7% | 40.4% | 33.2% | 50.5% |
| Diabetes | 25.7% | 0.4% | 28.1% | 4.1% |
| Age (years) | 50.9 ± 13.0 | 47.8 ± 11.1 | 51.3 ± 13.1 | 49.9 ± 15.1 |
| Body mass index (kg/m2) | 29.9 ± 4.9 | 26.4 ± 4.6 | 30.0 ± 4.9 | 26.8 ± 4.5 |
| Plasma total cholesterol (mmol/L) | 8.2 ± 3.9 | 5.3 ± 1.3 | 8.7 ± 4.3 | 4.9 ± 0.8 |
| Plasma HDL cholesterol (mmol/L) | 0.9 ± 0.3 | 1.4 ± 0.4 | 0.9 ± 0.3 | 1.3 ± 0.4 |
| Plasma LDL cholesterol (mmol/L) | - | 3.4 ± 1.2 | - | 3.2 ± 0.9 |
| Plasma triglycerides (mmol/L) | 14.3 ± 19.8 | 1.1 ± 0.7 | 14.2 ± 19.0 | 1.2 ± 0.4 |
GWAS, genome-wide association study; HDL, high-density lipoprotein; HTG, hypertriglyceridemia; LDL, low-density lipoprotein. There are 346 HTG patients and 205 low triglyceride controls common between both cohorts. Lipid measurements were conducted after a 12-h fasting period. Values are mean ± standard deviation. LDL cholesterol is not accurately calculated using the Friedewald equation for HTG patients when plasma triglyceride concentration exceeds 4.5 mmol/L.
The HTG phenotype was tested for association with >2.1 million single nucleotide polymorphisms (SNPs) using an additive multivariate logistic regression model (Supplemental Fig. 2). This model appropriately adjusted for sex, body mass index, diabetes status and 10 principal components of ancestry (Supplemental Fig. 3). Four loci were significantly associated with HTG at genome-wide significance: APOA5, GCKR, LPL and APOB (Table 2). Most associations with HTG were mediated by the same genomic loci associated with fasting plasma TG concentration in population-based GWAS5: APOA5 and GCKR were associated at the same lead SNP, and LPL was associated with the same haplotype block. Conversely, the HTG-associated SNPs in APOB were ~123-kb upstream of the gene, perhaps consistent with the involvement of regulatory elements in the over-expression of TG-rich lipoproteins in HTG pathophysiology. Investigation of sub-threshold association signals did not provide any additional insight into novel HTG-associated genes.
Table 2.
Genetic loci associated with HTG.
| Locus | SNP | CHR | Position | Minor Allele | HTG MAF | Control MAF | OR (95% CI) | P-value |
|---|---|---|---|---|---|---|---|---|
| APOA5 | rs964184 | 11 | 116.2 | G | 0.33 | 0.14 | 3.28 (2.61-4.14) | 5.4 × 10-24 |
| GCKR | rs1260326 | 2 | 2.8 | T | 0.52 | 0.41 | 1.75 (1.45-2.12) | 6.5 × 10-9 |
| LPL | rs7016880 | 8 | 19.9 | C | 0.03 | 0.10 | 0.32 (0.21-0.49) | 2.0 × 10-7 |
| APOB | rs4635554 | 2 | 21.2 | G | 0.39 | 0.31 | 1.67 (1.38-2.02) | 2.0 × 10-7 |
| | ||||||||
| MLXIPL | rs714052 | 7 | 72.5 | G | 0.07 | 0.13 | 0.44 (0.31-0.62) | 0.000003 |
| TRIB1 | rs2954029 | 8 | 126.6 | T | 0.37 | 0.46 | 0.71 (0.59-0.86) | 0.0004 |
| ANGPTL3 | rs10889353 | 1 | 62.9 | C | 0.27 | 0.32 | 0.73 (0.59-0.89) | 0.002 |
| NCAN | rs17216525 | 19 | 19.5 | T | 0.07 | 0.09 | 0.71 (0.50-1.00) | 0.05 |
| FADS | rs174547 | 11 | 61.3 | C | 0.40 | 0.33 | 1.20 (0.99-1.44) | 0.07 |
| XKR6 | rs7819412 | 8 | 11.1 | G | 0.46 | 0.50 | 0.87 (0.72-1.05) | 0.14 |
| PLTP | rs7679 | 20 | 44.0 | C | 0.20 | 0.19 | 1.17 (0.94-1.47) | 0.16 |
CHR, chromosome; CI, confidence interval; HTG, hypertriglyceridemia; MAF, minor allele frequency; OR, odds ratio; SNP, single nucleotide polymorphism. Association was tested using an additive multivariate logistic regression model, entering sex, body mass index, diabetes status, and 10 principal components of ancestry as covariates. The top four loci surpassed a pre-specified threshold for genome-wide significance of P<5×10-7. Remaining loci were replicated from GWAS meta-analysis of population-based TG concentrations5, using a Bonferroni corrected significance threshold of P<0.005.
Subsequently, we tested the hypothesis that common genetic variants in remaining known TG-associated loci are similarly associated with HTG5. Only three loci were replicated at a Bonferroni-corrected significance threshold of P<0.005: MLXIPL, TRIB1, and ANGPTL3 (Table 2). Positive replication of these TG-associated loci, combined with trends towards significance at FADS (P=0.05) and NCAN (P=0.07), suggest that additional TG-modulating loci may also be involved in HTG pathophysiology, however smaller effect sizes likely limit their detection.
We next hypothesized that HTG-associated genes would harbour rare variants related to HTG disease causation. The protein-coding sequences of APOA5, GCKR, LPL and exons 26 and 29 (67.8%) of APOB were resequenced in individual subjects as regions most likely to harbour protein-compromising mutations. Across the 4 genes, 80 distinct rare variants were identified with minor allele frequencies <1% in controls (Fig. 1, Supplemental Table 1). A significant accumulation of rare variants was identified in HTG patients (Table 3), including 154 total variants in 438 HTG diploid genomes compared to 53 total variants in 327 control diploid genomes (P=6.2×10-8), corresponding to a significantly increased carrier frequency of 28.1% in HTG patients versus 15.3% in controls (P=2.6×10-5). A more restricted analysis of rare variants found exclusively in either HTG patients or controls, deliberately removing all reported variants without demonstrated functional compromise, similarly revealed a significant burden of 47 variants in HTG patients compared to 9 variants in controls (P=2.4×10-5); this corresponds to a significantly increased carrier frequency of 10.3% in HTG patients compared to 2.8% in controls (P=4.4×10-5). HTG carriers’ fasting plasma TG concentrations ranged from 3.10–88.5 mmol/L, whereas control carriers’ fasting plasma TG concentrations ranged from 0.45–1.93 mmol/L. No discernable patterns were observed between such attributes as the gene, mutation type or mutation position with plasma TG concentration or HTG phenotype.
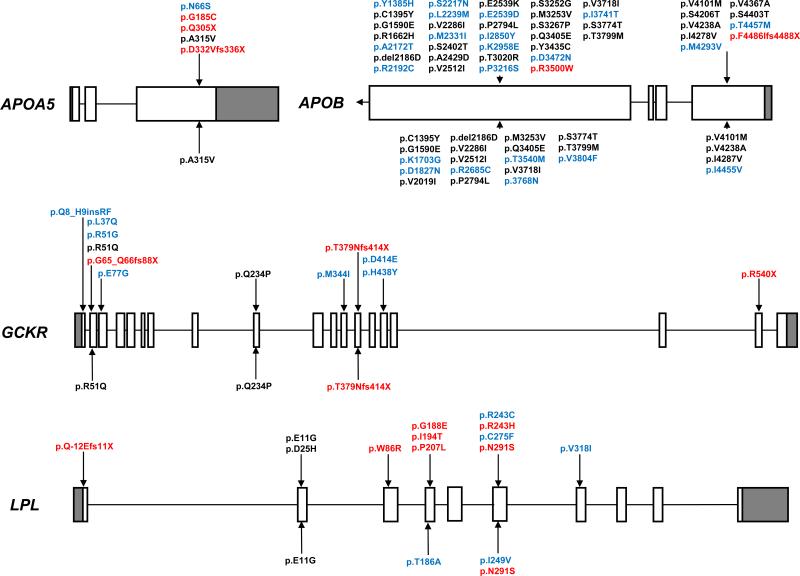
Figure 1. Rare variants identified by resequencing GWAS-identified genes in HTG patients and controls.
Variants above gene maps were identified in HTG patients and variants below gene maps were identified in controls. Rare variants are coloured according to their identification in control subjects or previous identification in subjects of unknown clinical status (black), exclusivity to HTG patients or controls (blue), or proven biological dysfunction or truncation (red). Nomenclature refers to functional protein sequences. Only exons 26 and 29 were resequenced in APOB. Gene maps are roughly to scale, although differ in scale between genes. GWAS, genome-wide association study; HTG, hypertriglyceridemia; TG, triglyceride.
Table 3.
Rare variant accumulation identified by resequencing GWAS-identified genes in HTG patients and controls.
| All mutations | Missense/Indels | Nonsense | |||||
|---|---|---|---|---|---|---|---|
| HTG | Controls | HTG | Controls | HTG | Controls | ||
| Total alleles | 876 | 654 | 876 | 654 | 876 | 654 | |
| All Variants | APOA5 | 5 | 1 | 3 | 1 | 2 | 0 |
| GCKR | 20 | 5 | 14 | 4 | 6 | 1 | |
| LPL | 44 | 8 | 43 | 8 | 1 | 0 | |
| APOB | 85 |
39 |
84 |
39 |
1 |
0 |
|
| Total | 154 | 53 | 146 | 52 | 9 | 1 | |
| P = 6.2 × 10-8 | P = 3.2 × 10-7 | P = 0.051 | |||||
| | |||||||
| Exclusive Variants | APOA5 | 4 | 0 | 2 | 0 | 2 | 0 |
| GCKR | 9 | 0 | 7 | 0 | 2 | 0 | |
| LPL | 19 | 2 | 18 | 2 | 1 | 0 | |
| APOB | 15 |
7 |
14 |
7 |
1 |
0 |
|
| Total | 47 | 9 | 42 | 9 | 5 | 0 | |
| P = 2.4 × 10-5 | P = 1.4 × 10-4 | P = 0.075 | |||||
GWAS, genome-wide association study; HTG, hypertriglyceridemia. Exclusive variants refer to rare variants found exclusively in HTG cases or low triglyceride controls; previously reported variants without characterized functional compromise are deliberately excluded. Fisher's exact test was used to calculate the significance of rare variant accumulation in HTG patients, defining nominal statistical significance as a two-sided P < 0.05. Mutation counts and annotations are found in Supplemental Table 1.
The strength of association between HTG and genomic loci did not predict the mutation accumulation observed in the resequenced genes. LPL harboured the largest relative proportion of rare variants, followed by GCKR, APOB and APOA5, with 30.9, 10.7, 9.3, and 4.5 rare variants per kilobase of coding sequence in HTG patients, whereas the same genes harboured 5.6, 2.7, 4.3 and 0.9 rare variants per kilobase of coding sequence in controls. The burden of rare variants found in HTG patients is highly suggestive of phenotype causation, supported by several truncation mutations, in silico predictions of deleterious effects, and bonafide characterized deleterious mutations (Supplemental Table 1). The majority of subjects carried only 1 rare variant, however subjects with multiple rare variants were also significantly over-represented in HTG patients (6.6% HTG carriers versus 0.9% control carriers; P=3.7×10-5). Not all rare variants in HTG patients are necessarily sufficient to cause HTG, however they likely contribute to the biochemical heterogeneity observed between patients. For instance, the APOB R3500W variant causes hypercholesterolemia18, but was found in a HTG patient with Fredrickson HLP phenotype 2B, defined by both plasma TG and total cholesterol in excess of the 95th percentile. For this individual case, APOB R3500W is more likely contributing to the elevated total cholesterol phenotype, but the mutation is part of the genetic background of this patient that led to his ascertainment through the lipid clinic. This patient exemplifies our working model that both common and rare genetic determinants in TG-associated genes together contribute to the phenotypic heterogeneity underlying HTG.
Finally, we assessed the contribution of genetic and clinical variables to the total variation in HTG diagnosis, in subjects common between GWAS and sequencing cohorts. A comprehensive logistic regression model including clinical variables and both common and rare genetic variants explained 41.6% of total variation in HTG diagnosis: clinical variables explained 19.7%, common genetic variants in 7 HTG-associated loci explained 20.8% and rare genetic variants in 4 HTG-associated loci explained 1.1%. These data suggest that rare variants found in 4 GWAS-identified genes incrementally contribute to the unexplained variation underlying HTG pathophysiology.
In summary, we performed a GWAS and resequencing of HTG-associated genes and found a significant accumulation of missense and nonsense mutations that contribute to the unexplained genetic component of HTG. Our results suggest that a complex genetic architecture of both common and rare variants in a spectrum of TG-associated genes is responsible for HTG. Future studies using high-throughput next generation sequencing are required to determine whether these associations extend to additional HTG-associated genes, including MLXIPL, TRIB1, and ANGPTL3, and TG-associated genes identified by epidemiological-scale GWAS of population-based samples. It also remains possible that rare variants in TG-modulating genes that have not yielded signals on GWAS, such as GPIHBP1 or LMF1, will further contribute to HTG phenotypes19,20. Functional analyses may more accurately define the extent of dysfunction of rare variants identified in HTG patients and their role in disease causation, while higher level analyses including gene-gene and gene-environment interactions will determine the combined impact of multiple genetic variants on plasma TG concentration in patients with HTG. Our study demonstrates that an accumulation of rare variants is present in GWAS-identified genes and that these contribute to the missing heritability of complex traits among individuals at the extreme of a lipid phenotype.
Methods Summary
Study Subjects
HTG patients were ascertained through tertiary referral lipid clinics. Controls were obtained from population-based studies including the Study of Health Assessment and Risk in Ethnic Groups21 and the Myocardial Infarction Genetics Consortium22 (Supplemental Methods). Biochemical analyses were conducted separately in each cohort, as previously described14,21,22.
GWAS
All subjects were genotyped using Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA) according to protocols of the London Regional Genomics Centre (www.lrgc.ca) or the Broad Institute (http://www.broadinstitute.org/). Imputation was conducted using HapMap CEU phased haplotypes in MACH23. All genotypes were filtered for minor allele frequency >1%, Hardy Weinberg P>0.0001, and 95% call rate or imputation quality r2>0.4. Identity-by-state calculations, multi-dimensional scaling, and association testing were conducted in PLINK24. Genome-wide significance was pre-specified as P<5X10-7; nominal significance for replication of known TG-associated SNPs was a Bonferroni-corrected threshold P<0.005. Covariates entered into all analyses included sex, body mass index, diabetes status and 10 principal components of ancestry as generated by Eigenstrat25,26.
Sequencing and Mutation Accumulation
All genes were bidirectionally sequenced in individual samples using an ABI 3730 Automated DNA Sequencer and called using automated software (Applied Biosystems, Foster City, CA). Rare variants were manually curated, confirmed by repeat analysis, and annotated in silico for functional effects using Polyphen (http://genetics.bwh.harvard.edu/pph/). Only rare variants <1% in controls causing missense or nonsense mutations were counted towards mutation accumulation. Rare variant accumulation was compared between HTG patients and controls using Fisher's exact test, defining nominal significance as a 2-sided P<0.05 (Supplemental Methods).
Explained Variation
Total explained variation was calculated from the residuals of multivariate logistic regression model including age, sex, body mass index, diabetes status, and HTG-associated common and rare variants as independent variables, using a SAS v9.2 (SAS Institute, Cary, NJ) macro written for this purpose27 (Supplemental Methods).
Supplementary Material
Acknowledgements
We thank the London Regional Genomics Centre (David Carter, Gerry Barbe, and Kevin Kang) for their dedication to this project. We also thank the Myocardial Infarction Genetics Consortium (MIGen) study for the use of their genotype data as control data in our study. The MIGen study was funded by the U.S. National Institutes of Health and National Heart, Lung, and Blood Institute's STAMPEED genomics research program (R01 HL087676) and a grant from the National Center for Research Resources (U54 RR020278). This work was made possible by the facilities of the Shared Hierarchical Academic Research Computing Network (SHARCNET; www.sharcnet.ca). CTJ is supported by a CIHR Banting and Best Canada Graduate Scholarship, a Heart and Stroke Foundation Program Grant, and a CIHR Vascular Research Fellowship. VS was supported by the Sigrid Juselius Foundation and by the Finnish Academy (grant #129494). SSA is supported by the Michael G. DeGroote Heart and Stroke Foundation of Ontario Chair and the Eli Lilly May Cohen Chair in Women's Health Research at McMaster University. RAH is supported by the Jacob J. Wolfe Distinguished Medical Research Chair, the Edith Schulich Vinet Canada Research Chair in Human Genetics (Tier I), the Martha G. Blackburn Chair in Cardiovascular Research, and operating grants from the Canadian Institutes for Health Research (MOP-13430, MOP-79523, CTP-79853), the Heart and Stroke Foundation of Ontario (NA-6059, T-6018, PRG-4854), the Pfizer Jean Davignon Distinguished Cardiovascular and Metabolic Research Award, and Genome Canada through the Ontario Genomics Institute.
References
- 1.Kathiresan S, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–97. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Willer CJ, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–9. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sabatti C, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aulchenko YS, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kathiresan S, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–39. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plomin R, Haworth CM, Davis OS. Common disorders are quantitative traits. Nat Rev Genet. 2009;10:872–8. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, et al. Resequencing genomic DNA of patients with severe hypertriglyceridemia (MIM 144650). Arterioscler Thromb Vasc Biol. 2007;27:2450–5. doi: 10.1161/ATVBAHA.107.150680. [DOI] [PubMed] [Google Scholar]
- 10.Nejentsev S, et al. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–9. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen JC, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–72. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 12.Romeo S, et al. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet. 2007;39:513–6. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan G, Al-Shali KZ, Hegele RA. Hypertriglyceridemia: its etiology, effects and treatment. CMAJ. 2007;176:1113–20. doi: 10.1503/cmaj.060963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegele RA, et al. A polygenic basis for four classical Fredrickson hyperlipoproteinemia phenotypes that are characterized by hypertriglyceridemia. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, et al. Polygenic determinants of severe hypertriglyceridemia. Hum Mol Genet. 2008;17:2894–9. doi: 10.1093/hmg/ddn188. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, et al. APOA5 genetic variants are markers for classic hyperlipoproteinemia phenotypes and hypertriglyceridemia. Nat Clin Pract Cardiovasc Med. 2008;5:730–7. doi: 10.1038/ncpcardio1326. [DOI] [PubMed] [Google Scholar]
- 17.Hegele RA. Plasma lipoproteins: genetic influences and clinical implications. Nat Rev Genet. 2009;10:109–21. doi: 10.1038/nrg2481. [DOI] [PubMed] [Google Scholar]
- 18.Gaffney D, et al. Independent mutations at codon 3500 of the apolipoprotein B gene are associated with hyperlipidemia. Arterioscler Thromb Vasc Biol. 1995;15:1025–9. doi: 10.1161/01.atv.15.8.1025. [DOI] [PubMed] [Google Scholar]
- 19.Beigneux AP, et al. Chylomicronemia with a mutant GPIHBP1 (Q115P) that cannot bind lipoprotein lipase. Arterioscler Thromb Vasc Biol. 2009;29:956–62. doi: 10.1161/ATVBAHA.109.186577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterfy M, et al. Mutations in LMF1 cause combined lipase deficiency and severe hypertriglyceridemia. Nat Genet. 2007;39:1483–7. doi: 10.1038/ng.2007.24. [DOI] [PubMed] [Google Scholar]
- 21.Anand SS, et al. Differences in risk factors, atherosclerosis, and cardiovascular disease between ethnic groups in Canada: the Study of Health Assessment and Risk in Ethnic groups (SHARE). Lancet. 2000;356:279–84. doi: 10.1016/s0140-6736(00)02502-2. [DOI] [PubMed] [Google Scholar]
- 22.Kathiresan S, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–41. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Abecasis GR. Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- 24.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 27.Mittlbock M, Schemper M. Computing measures of explained variation for logistic regression models. Comput Methods Programs Biomed. 1999;58:17–24. doi: 10.1016/s0169-2607(98)00061-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.