Abstract
Label-free biosensors offer integrated, kinetic and multi-parametric measures of receptor biology and ligand pharmacology in whole cells. Being highly sensitive and pathway-unbiased, label-free receptor assays can be used to probe the systems cell biology including pleiotropic signaling of receptors, and to characterize the functional selectivity and phenotypic pharmacology of ligand molecules. These assays provide a new dimension for elucidating receptor biology and for facilitating drug discovery.
Introduction
Historically, receptor signaling was viewed as a linear cascade [1]. As a result, point-of-contact based measurements, such as alterations in intracellular cyclic AMP (cAMP), Ca2+ mobilization, protein trafficking and phosphorylation of downstream kinases, have become the most commonly used strategy in receptor biology research and drug discovery campaigns [2, 3]. The quest to discover the full complement of cell signaling components has led to identification of various activators, effectors, enzymes, and substrates involved in receptor signaling, many (if not all) of which work in concert to guide pivotal intracellular processes [4]. The integration of distinct signaling pathways dedicates how signals propagate within the cell and ultimately how the cell responds to environmental cues [5, 6]. Pharmacological assays now in development, such as label-free receptor assays, are making it possible to pathway-unbiasedly monitor cell signaling both spatially and temporally [7, 8]. As cells use sophisticated regulatory mechanisms to ensure an appropriate integrative response upon stimulation, assays based on integrated readouts as promised by label-free receptor assays are naturally suited to sense the complexity of receptor signaling and ligand pharmacology. Together with high sensitivity, the pathway unbiased nature permits the label-free receptor assay a single platform for recording a wide array of receptor signaling in native cells including primary cells.
Label-free receptor assays
Biosensor systems
Central to label-free receptor assays is the biosensor that is capable of converting a cellular response into a quantifiable signal [9]. Label-free biosensors including surface plasmon resonance [10] and its derivatives [11] were initially developed as an analytical tool for biomolecular interaction analysis. Soon after birth these biosensors were also explored to sense cellular activity; but early attempts were mostly limited to cell adhesion [12, 13] and cytotoxicity [14]. With sophisticated engineering and advanced assay designs, the new generation biosensor systems such as Cellkey™ (MDS Analytical Technologies), xCELLigence (Acea Biosciences), BIND (SRU Biosystems) and Epic® (Corning Inc) systems have made it possible to robustly probe receptor signaling, particularly in the microtiter plate format which is the standard footprint for large scale research and drug discovery. The detailed descriptions of these systems can be found in the websites of these vendors (see the Links) and are not reviewed here.
Both CellKey™ [15] and xCELLigence [16] systems exploit a microelectrode array electrical biosensor to probe changes in impedance of a cell layer. Sinusoidal voltages that are swept through a range of frequencies in continuous wave mode are used to generate electric fields between the electrodes, which are impeded by the presence of cells.
Both SRU BIND [17] and Epic® [18] systems use a nano-grating waveguide biosensor to characterize cellular responses. Lights that cover a range of wavelengths are used to generate a diffraction grating coupled waveguide resonance, thus leading to a surface-bound electromagnetic wave, which is perturbed by the presence of cells [18].
Biosensor signals
Electrical biosensor uses a sophisticated algorithm to determine and separate two types of electrical currents, extracellular and transcellular currents [15]. This is due to the fact that the cellular plasma membrane acts as an insulating barrier directing the current to flow between or beneath the cells. The flow of both currents is termed impedance. The cellular impedance is a measure of changes in the electrical conductivity or permeability of the cell layer, and the extracellular current is more robust than the transcellular current. Since the impedance is mostly a function of ionic environment surrounding or within the cells, the electric biosensor is largely sensitive to, and thus biased towards, changes in cellular morphology and ionic redistribution.
Optical biosensor mostly tracks changes in the central wavelength or angle (i.e., resonance wavelength or angle, respectively) of the biosensor resonant spectrum [8, 15]. For resonant waveguide grating (RWG) systems such as Epic® and SRU BIND, the resonance wavelength is a function of the local refractive index at or near the sensor surface, which is mostly proportional to the mass density and distribution of biomaterials within the cells. Receptor signaling often involves protein trafficking, microfilament remodeling, cell adhesion alterations, and morphological changes of cells, all of which can lead to significant dynamic mass redistribution (DMR) [8]. Such a redistribution is not random; instead, it is tightly regulated and is often dynamic both spatially and temporally. The biosensor simply acts a non-invasive monitor to record the DMR in real time. Since most optical biosensors use long-wavelength light for illumination, these biosensors are believed to be, or close to be, truly non-invasive. However, since most optical biosensors including SPR and RWG have limited detection volume due to the short penetration depth (~200nm) of the surface-bound evanescent wave, the cellular responses measured are often biased towards cellular changes within the bottom portion of cells [8].
Assay formats
Cell signaling propagates in several sequential waves, from the early responses including receptor activation, second messengers and protein trafficking, to the intermediate responses including de novo synthesis, exocytosis and gene expression alterations, and to the late responses such as cell proliferation and survival. Label-free receptor assays are non-invasive in general, thus permitting multiple assay formats to track the entire course of cell signaling in real time. The two common assay formats are co-stimulation and sequential stimulation assays [19]. Most label-free receptor assays are to probe the early processes of cell signaling.
Label-free receptor assays for receptor biology
Cell signaling by membrane receptors is a coordinated relay of messages derived from environmental cues to intracellular effectors. It is evident that sophisticated regulatory mechanisms are utilized for cells to encode, process, and integrate the information received. Common to label-free receptor assays is to record an integrated signal in real time of whole cells. Such integrated measurements often reflect the complexity of receptor signaling, permitting the study of the systems cell biology of receptors, the crosstalks between receptors, and the integration of receptor signaling.
Systems cell biology of receptors
The quest to discover the full complement of receptor signaling components has led to identification of various activators, effectors, enzymes, and substrates. Pharmacological tools including small molecules and interference RNA have become widely available, thus making it possible to selectively perturb the cellular activities of many signaling proteins downstream of a receptor. Assays that measure point-of-contacts in the complexed signaling network are also useful for orthogonal confirmation. Thus, it is feasible to study the systems cell biology of receptors using an integrated and complex readout of whole cells as promised by label-free receptor assays.
The DMR assay using Epic® system was the first label-free receptor assay used to map the signaling and its network interactions of several receptors including epidermal growth factor (EGF) receptor [20]. The EGFR is one of the most well studied receptor tyrosine kinases. EGF binds to the receptor and stimulates its intrinsic protein-tyrosine kinase activity, initiating signal transduction that principally involves the MAPK cascades, Akt signaling, STAT activation and the PLCγ pathway. Using the EGF DMR signal in A431 cells as a readout, we found that EGFR signaling is cellular status dependent–quiescent cells respond more robustly to EGF than proliferating cells. The modulation profiles of an array of known modulators also linked several targets and cellular processes to the EGFR signaling. The EGFR signaling was found to require its intrinsic tyrosine kinase activity and to be mostly originated from the internalized receptors. The EGFR signaling also led to actin remodeling, dynamin and clathrin dependent receptor internalization, and MEK pathway-mediated cell detachment (possibly via FAK).
A recent cellular impedance study for the EGFR in Cos7 cell, using the xCELLigence system, confirmed the ability of label-free receptor assays to probe the systems cell biology of the EGFR [21]. In this study, the authors claimed that the impedance signal is a direct result of growth factor-induced morphological changes, and can be used as a quantitative readout for receptor tyrosine kinase activity in COS7 cells. Both EGF and insulin-induced impedance signals were found to be correlated with the morphological changes of cells and levels of receptor autophosphorylation measured using fluorescent microscopy and enzyme-linked immunosorbent assay, respectively.
However, since the EGFR signaling consists of hundreds (if not thousands) of components which often display different temporal dynamics and spatial gradients [4, 5], it will be naturally difficult to map out the full complement of signaling components using label-free receptor assays, and thus to fully comprehend the origin of the biosensor signal of the receptor. For this reason, label-free receptor assays are often referred to “black box” [22, 23]. However, the abovementioned studies suggest that it is quite promising for these assays to identify many important nodes in the receptor signaling network (Fig. 1).
Figure 1.
Schematic depiction of the ability of label-free receptor assays to detect critical nodes in the signaling network of a receptor. Cells employ regulatory and compensatory pathways to fine tune their decision making process. It is highly possible for label-free receptor assays to identify at least some of these important signaling nodes. For example, the binding of agonist 1 to the receptor A leads to two major pathways: the first one consists effectors E1a, E1b, E2, E3, leading to response R1, while another consists of E4, E5a, E5b, E6, and E7, leading to responses R2 and R3. The integration of the responses R1, R2 and R3 when occurred within the sensing zone of the biosensor leads to the DMR1. The effectors E1a and E1b form the critical Node N1, while E5a and E5b form another critical node N2. Similarly, the activation of the receptor B by ligand 2 leads to activation of two pathways: the first one consists effectors E8a, E8b, E8c, E7, and E9, leading to the responses R2 and R3, while the second one consists of the effectors E10 and E11, leading to the response R4. The integration of R3 and R4 when occurred within the sensing zone of the biosensor leads to the DMR2. E8a, E8b and E8c form a critical node N3 for the receptor B signaling.
Pleiotropic signaling of receptors
G protein-coupled receptors (GPCRs) have been and continue to be one of the richest families of drug targets. There are at least two key drivers for this. The first driver is the increasing numbers of orphan receptors being deorphanized, some of which have implications for human diseases. Examples are GPR3 for Alzheimer’s disease [24] and GPR40 for diabetes [25]. The second driver is associated with the recent realization that GPCRs are competent to elicit a rich array of cell signaling pathways (i.e., pleiotropic signaling) [7, 26], and ligands may give operational biases to activate the receptor [27]. These pathway biased ligands may open new revenues for drug discovery.
Label-free offers new means in discovering new receptor behaviors. Our recent DMR assays for endogenous bradykinin B2 receptor in A431 cells showed that the receptor mediates signaling via at least dual pathways – Gαs- and Gαq-mediated signaling; and remarkably, the two signaling pathways counter-regulate each other [28]. Kostenis and her colleagues [29] have found, using DMR assays and classical endpoint assays, that CRTH2 receptor utilizes its C-tail domain to silence its own signaling, and the C terminus dose not encode G protein specificity determinants. A CRTH2 mutant receptor lacking this domain displays paradoxically enhanced Gαi and ERK1/2 activation.
Signaling compartmentalization and crosstalks
Cell signaling specificity requires that signaling molecules encounter their intracellular interactants in the right place and at the right time. Cell surface receptors including GPCRs may preferentially locate within some cholesterol and sphingolipid-enriched microdomains. For example, the endogenous B2 receptor in A431 may be associated with such microdomains, since its signaling was found to be sensitive to cholesterol depletion [28].
Compartmentalization of enzymes in proximity to substrates is another means of spatially restricting cell signaling events [30]. As a result, distinct signaling pathways may undertake spatially different routes. For this reason, the activation of different pathways could lead to distinct biosensor signals, and the biosensor signals obtained can be used as “signatures” to identify pathways being activated [15, 16, 31]. Using the DMR assays, we [32] found that the activation of endogenous Gq-coupled histamine H1 receptor in A431 led to a Gq-type DMR, while the activation of endogenous Gs-coupled β2-adrenergic receptor in the same cell led to a Gs-type DMR. The co-activation of both receptors by epinephrine and histamine led to a DMR signal that is close to the sum of both histamine and epinephrine DMR signals. This result suggests that both Gq and Gs mediated signals undergo spatially distinct routes at the complex pathway level.
Cells use various regulatory and compensatory pathways to fine tune cellular responses. It is common for distinct receptors to cross-talk at multiple levels, including receptor oligomerization [33], interactions of intracellular signal transduction cascades [34, 35], and phosphorylation of receptors and regulatory proteins by kinases [36]. The cross-talk ensures the information exchange between pathways and allows the cells to make appropriate responses. The recent DMR study of A431 cells [32] showed that histamine slightly attenuated the epinephrine response, while epinephrine partially attenuated the histamine response. This result indicates that there are cross-talks between the epinephrine- and histamine-mediated signaling. Label-free receptor desensitization assays [37] showed that in A431 cells, the forskolin pretreatment heterologously desensitized Gαs signaling, partially attenuated Gαq signaling, but had complicate impacts on Gαi signaling. This result suggests that adenylyl cyclase acts as an effective integrator for GPCR signaling.
Label-free receptor assays for ligand pharmacology
Central to drug development is the pharmacological characterization of drug molecules. Because of the rich behavior of receptors and the heterogeneity of signaling components in different cell types, drug molecules can have complicated pharmacology, including pathway biased activity (i.e., functional selectivity), phenotypic pharmacology and polypharmacology. The possibility to have multiple efficacies for a given ligand makes it difficult in practice to systematically represent the ligand efficacy using pathway-biased approaches, due to the large number of possible signaling outputs [2, 3]. The ability to detect the integrated response of whole cells by endogenous receptors empowers the label-free assays to classify drug molecules in new dimensions [7, 31, 38].
Ligand-directed functional selectivity
The quest to fully characterize the pharmacological activity of drug molecules with a wide spectrum of point-of-contact and phenotypic assays has led to the discovery of novel pathway biased activity of many ligands for increasing numbers of GPCRs. A classical example is the beta blocker propranolol. Propranolol was recently identified as an inverse agonist for Gαs pathway, and also a β-arrestin dependent extracellular signal regulated kinase (ERK) agonist [39]. These pathway-biased activities may contribute to the complex therapeutic profiles of drug molecules.
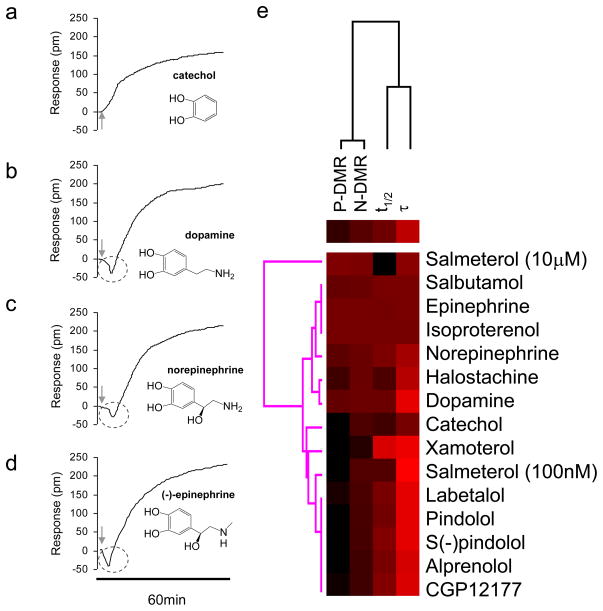
Label-free receptor assays allow a greater array of changes in the receptor to be detected. Using the DMR assays, a panel of β2-adrenergic receptor (β2-AR) ligands was characterized in quiescent A431 cells [38]. Multi-parameter analysis revealed unique patterns in the characteristics of their corresponding DMR signals (Fig. 2). Full agonists such as epinephrine and isoproterenol gave rise to a DMR with maximum amplitudes, fast transition time but slow kinetics for the P-DMR event. In comparison, partial agonists such as catechol and halostachine led to a DMR signal with smaller amplitudes, slightly slower transition time but faster kinetics for the P-DMR event. Similarity analysis suggests that these parameters can be used to categorize the agonism activity of these molecules (Fi.2). Interestingly, the partial agonists catechol and pindolol bind to different portions of the binding pocket of the β2-AR, triggering largely independent signaling. This is consistent with results obtained using both theoretical and biophysical approaches. Protein ensemble theory predicts a range of receptor active states, each of which could have its own signaling preference [40]. Biophysical studies suggest that for the β2AR ligand binding and receptor activation is a kinetically and conformationally complex process, and agonist binding and receptor conformational changes occur through a sequence of conformational intermediates [41, 42].
Figure 2.
Characteristics of β2-AR agonist DMR signals of quiescent A431 cells. (a–d) Representative DMR signals and structures of a panel of β2-AR agonists: (a) catechol of 500 μM; (b) dopamine of 32μM; (c) norepinephrine of 100nM; and (d) (−)epinephrine of 8nM; each at the saturating concentration. (e) The heat map classification of β2-AR agonist pharmacology based on the characteristics of their corresponding DMR signals. Data used were from ref. 36. The heat map was generated using the Euclidean hierarchical cluster analysis (ref. 55), after all DMR parameters were normalized to the epinephrine response. Data suggests that the first subgroup consists of full agonists and strong partial agonists including isoproterenol, epinephrine, norepinephrine, and salbutamol, while the second group consists of partial agonists including halostachine and dopamine. The third group consists of the beta-blockers with weak partial agonism activity, including labetalol, pindolol, S(−)pindolol, alprenolol, CGP12177, and to certain extent, salmeterol of 100nM. The weak agonist catechol and the partial agonist xamoterol are between the second and third group. Salmeterol of 10μM leads to very unique DMR that is similar but not identical to the full agonists.
The most notable finding from this study is the dual efficacies of the long-acting agonist salmeterol. Salmeterol resulted in a biphasic dose-dependent response, leading to two well-separated EC50 values. Salmeterol not only binds to the active site (Ser-204, Ser-207 and Asp-113) of the β2AR but also to its exosite, and results in a slow internalization of the β2AR [43]. Salmeterol was known to have dual efficacies: a very weak partial agonist for producing an effective interaction between the receptor and β-arrestin 2, and a full agonist of cAMP accumulation in C2C12 cells stably expressing the β2AR [44]. In human kidney embryonic 293 cells, despite stimulating GRK-mediated receptor phosphorylation after 30 min to an extent similar to those observed with agonists of high intrinsic efficacy such as epinephrine, salmeterol of 50nM did not induce significant β2AR internalization and was incapable of stimulating the translocation of green fluorescent protein-arrestin 2 chimera proteins to the cell surface [45].
Allosteric modulators that edit the behavior of the receptor towards agonists can also provide basis for functional selectivity. For example, neurokinin normally triggers Gs and Gq signaling via NK1 receptor. The co-binding with neurokinin of allosteric modulator LP1805 to the receptor led to enhanced Gq response and antagonism of Gs activation [46]. Label-free receptor assays have also been used to characterize allosteric modulators for metabotropic glutamate receptor 7 [47], and to examine a novel class of ligands that can simultaneously bind to both allosteric and conserved orthosteric sites of the same muscarinic receptor [48].
Protease activator receptor-1 (PAR1) plays important roles in regulating the permeability of the endothelial cell barrier at the blood–tissue interface. Thrombin activates PAR1 by cleavage of its amino terminus to unmask a tethered ligand [49]. Synthetic PAR-activating peptides (PAR-APs), corresponding to the first five or six amino acids of the tethered ligand sequences, can directly activate PAR1 [50]. Using an electric biosensor cellular assay, Hamm and her colleagues [51] had found that thrombin increased the permeability of the monolayer of a human dermal microvascular endothelial cell line HMEC-1 primarily via Rho kinase pathway. Furthermore, thrombin gave rise to relatively lower potency in activating Ca2+ mobilization. However, the opposite order of activation was observed for the two PAR-AP agonists, SFLLRN-amide and TFLLRNKPDK. A mathematic numerical analysis suggests that the PAR-AP agonists alter receptor/G protein binding to favor Gαq activation over Gα12/13 by ~800-fold.
Phenotypic pharmacology
The cellular environment can influence, sometime determine, the functional responses of receptors, thus to large extent, define drug actions. The cellular context dependent drug actions are also viewed as phenotypic pharmacology. The cellular factors that could influence drug actions include the receptor expression level, the diversity in receptor conformations and organizations, and the heterogeneity in expression and organization of cytosolic interactants. There are many examples in literature that a given receptor may prefer different signaling pathways when presented in different cellular backgrounds [52, 53]. Recently, Peters and Scott [54] used cellular impedance assays to characterize several receptors in different cell backgrounds, and found that melanocortin-4 receptor (MC4R) can trigger either Gαs or Gαq coupling in a cellular context dependent manner. In HEK-MC4R cells MC4R preferred Gαs coupling, while in CHO-MC4R cells it favored Gαq coupling.
Conclusion
The evolution of pharmacological assays has led to the discovery of a wide array of cellular behavior of receptors and drug molecules. Label-free receptor assays are emerging as a powerful assay platform to study receptor signaling, and to elucidate critical nodes of receptor signaling networks. Label-free receptors also offer new dimensions to characterize and categorize drug molecules. With recent advancements in biosensor instrumentation such as Epic® system, high throughput screening becomes a reality for the activity assessments of drug molecules acting on native cells including primary cells. The next generation biosensor systems will have higher sensitivity and spatial resolution in directions perpendicular and/or parallel to the sensor surface [55, 56]. It will be possible to characterize receptor biology and ligand pharmacology in single cells, and in mixed populations of cells such as tissue cells, reprogrammed cells, and unpurified primary cells. Together with smarter assay design and development of novel methodologies, label-free receptor assays will provide new perspectives in cell biology and drug pharmacology.
Links
For Epic® system: http://www.corning.com/lifesciences/us_canada/en/whats_new/epic_system.aspx
For CellKey™ system: http://www.mdssciex.com/products/instruments/cellkey_new/default.asp?s=1
For xCELLigence system: http://www.aceabio.com/Company/index.htm
For SRU BIND system: http://srubiosystems.com/
Related articles
- Fang Y, et al. Resonant waveguide grating biosensor for living cell sensing. Biophys J. 2006;91:1925–1940. doi: 10.1529/biophysj.105.077818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, et al. Label-free cell assays for GPCR screening. Comb Chem High Throughput Screen. 2008;11:357–369. doi: 10.2174/138620708784534789. [DOI] [PubMed] [Google Scholar]
- Kenakin T. Cellular assays as portals to seven-transmembrane receptor-based drug discovery. Nat Rev Drug Discov. 2009;8:617–626. doi: 10.1038/nrd2838. [DOI] [PubMed] [Google Scholar]
- McGuinness R. Impedance-based cellular assay technologies: recent advances, future promise. Curr Opin Pharmacol. 2007;7:535–540. doi: 10.1016/j.coph.2007.08.004. [DOI] [PubMed] [Google Scholar]
Acknowledgments
This work is supported partially by National Institutes of Health Grant 5U54MH084691.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kenakin TP. New concepts in drug discovery: ccollateral efficacy and permissive antagonism. Nat Rev Drug Discov. 2005;4:919–927. doi: 10.1038/nrd1875. [DOI] [PubMed] [Google Scholar]
- 2.Galandrin S, et al. The evasive nature of drug efficacy: implications for drug discovery. Trends Pharmacol Sci. 2007;8:423–430. doi: 10.1016/j.tips.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Fang Y, et al. Label-free cell assays for GPCR screening. Comb Chem High Throughput Screen. 2008;11:357–369. doi: 10.2174/138620708784534789. [DOI] [PubMed] [Google Scholar]
- 4.Oda K, et al. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol. 2005;1:0010. doi: 10.1038/msb4100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jordan JD, et al. Signaling networks: the origins of cellular multitasking. Cell. 2000;103:193–200. doi: 10.1016/s0092-8674(00)00112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kholodenko BN. Four-dimensional organization of protein kinase signaling cascades: the roles of diffusion, endocytosis and molecular motors. J Exper Biol. 2003;206:2073–2082. doi: 10.1242/jeb.00298. [DOI] [PubMed] [Google Scholar]
- 7.Kenakin T. Cellular assays as portals to seven-transmembrane receptor-based drug discovery. Nat Rev Drug Discov. 2009;8:617–626. doi: 10.1038/nrd2838. [DOI] [PubMed] [Google Scholar]
- 8.Fang Y, et al. Resonant waveguide grating biosensor for living cell sensing. Biophys J. 2006;91:1925–1940. doi: 10.1529/biophysj.105.077818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang Y. Label-free cell-based assays with optical biosensors in drug discovery. Assays Drug Dev Technol. 2006;4:583–595. doi: 10.1089/adt.2006.4.583. [DOI] [PubMed] [Google Scholar]
- 10.Leatherbarrow RJ, Edwards PR. Analysis of molecular recognition using optical biosensors. Curr Opin Chem Biol. 1999;3:544–547. doi: 10.1016/s1367-5931(99)00006-x. [DOI] [PubMed] [Google Scholar]
- 11.Tollin G, et al. Techniques: plasmon-waveguide resonance (PWR) spectroscopy as a tool to study ligand-GPCR interactions. Trends Pharmacol Sci. 2003;24:655–659. doi: 10.1016/j.tips.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 12.Tiruppathi C, et al. Electrical method for detection of endothelial cell shape change in real time: assessment of endothelial barrier function. Proc Natl Acad Sci USA. 1992;89:7919–7923. doi: 10.1073/pnas.89.17.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramsden JJ, et al. Kinetics of adhesion and spreading of animal cells. Biotechnol Bioeng. 1994;43:939–945. doi: 10.1002/bit.260431007. [DOI] [PubMed] [Google Scholar]
- 14.Xiao C, et al. Assessment of cytotoxicity using electric cell-substrate impedance sensing: concentration and time response function approach. Anal Chem. 2002;74:5748–5753. doi: 10.1021/ac025848f. [DOI] [PubMed] [Google Scholar]
- 15.McGuinness R. Impedance-based cellular assay technologies: recent advances, future promise. Curr Opin Pharmacol. 2007;7:535–540. doi: 10.1016/j.coph.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Yu N, et al. Real-time monitoring of morphological changes in living cells by electronic cell sensor arrays: an approach to study G protein-coupled receptors. Anal Chem. 2006;78:35–43. doi: 10.1021/ac051695v. [DOI] [PubMed] [Google Scholar]
- 17.Li P, et al. Label-Free Assays on the BIND System. J Biomol Screen. 2004;9:481–490. doi: 10.1177/1087057104267604. [DOI] [PubMed] [Google Scholar]
- 18.Fang Y. Non-invasive optical biosensor for probing cell signaling. Sensors. 2007;7:2316–2329. doi: 10.3390/s7102316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang Y, et al. Resonant waveguide grating biosensor for whole cell GPCR assays. In: Leifert WR, editor. G protein-coupled receptors in Drug Discovery. Humana Press; New York: 2009. pp. 239–252. [DOI] [PubMed] [Google Scholar]
- 20.Fang Y, et al. Characteristics of dynamic mass redistribution of EGF receptor signaling in living cells measured with label free optical biosensors. Anal Chem. 2005;77:5720–5725. doi: 10.1021/ac050887n. [DOI] [PubMed] [Google Scholar]
- 21.Atienza JM, et al. Label-free and real-time cell-based kinase assay for screening selective and potent receptor tyrosine kinase inhibitors using microelectronic sensor array. J Biomol Screen. 2006;11:634–643. doi: 10.1177/1087057106289334. [DOI] [PubMed] [Google Scholar]
- 22.Rocheville M, Jerman JC. 7TM pharmacology measured by label-free: a holistic approach to cell signaling. Curr Opin Pharmacol. 2009;9:643–649. doi: 10.1016/j.coph.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 23.Eisenstein M. GPCRs: insane in the membrane. Nat Methods. 2009;6:929–933. [Google Scholar]
- 24.Thathiah A, et al. The orphan G protein–coupled receptor 3 modulates amyloid-beta peptide generation in neurons. Science. 2009;323:946–951. doi: 10.1126/science.1160649. [DOI] [PubMed] [Google Scholar]
- 25.Alquier T, Poitout V. GPR40: good cop, bad cop? Diabetes. 2009;58:1035–1036. doi: 10.2337/db09-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hermans E. Biochemical and pharmacological control of the multiplicity of coupling at G-protein-coupled receptors. Pharmacol Ther. 2003;99:25–44. doi: 10.1016/s0163-7258(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 27.Mailman RB. Ligand-selective signaling and high content for GPCR drugs. Trends Pharmacol Sci. 2007;8:390–396. doi: 10.1016/j.tips.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang Y, et al. Optical biosensor provides insights for bradykinin B2 receptor signaling in A431 cells. FEBS Lett. 2005;579:6365–6374. doi: 10.1016/j.febslet.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 29.Schroeder R, et al. The C-terminal tail of CRTH2 is a key molecular determinant that constrains Gαi- and downstream-signaling cascade activation. J Biol Chem. 2009;284:1324–1336. doi: 10.1074/jbc.M806867200. [DOI] [PubMed] [Google Scholar]
- 30.Willoughby D, Cooper DM. Organization and Ca2+ regulation of adenylyl cyclases in cAMP microdomains. Physiol Rev. 2007;87:965–1010. doi: 10.1152/physrev.00049.2006. [DOI] [PubMed] [Google Scholar]
- 31.Fang Y, et al. Non-invasive optical biosensor for assaying endogenous G protein-coupled receptors in adherent cells. J Pharmacol Toxicol Methods. 2007;55:314–322. doi: 10.1016/j.vascn.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 32.Tran E, Fang Y. Duplexed label-free G protein-coupled receptor assays for high throughput screening. J Biomol Screen. 2008;13:975–985. doi: 10.1177/1087057108326141. [DOI] [PubMed] [Google Scholar]
- 33.Bouvier M. Oligomerization of G-protein-coupled transmitter receptors. Nat Rev Neuroscience. 2001;2:274–286. doi: 10.1038/35067575. [DOI] [PubMed] [Google Scholar]
- 34.Houslay MD, Kolch W. Cell-type specific integration of cross-talk between extracellular signal-regulated kinase and cAMP signaling. Mol Pharmacol. 2000;58:659–668. [PubMed] [Google Scholar]
- 35.Chuang TT, et al. G protein-coupled receptors: heterologous regulation of homologous desensitization and its implications. Trends Pharmacol Sci. 1996;17:416–421. doi: 10.1016/s0165-6147(96)10048-1. [DOI] [PubMed] [Google Scholar]
- 36.Tobin AB. G-protein-coupled receptor phosphorylation: where, when and by whom. Br J Pharmacol. 2008;153(S1):S167–S176. doi: 10.1038/sj.bjp.0707662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tran E, Fang Y. Label-free optical biosensor for probing integrative role of adenylyl cyclase in G protein-coupled receptor signaling. J Receptors Signal Transduction. 2009;29:154–162. doi: 10.1080/10799890903052544. [DOI] [PubMed] [Google Scholar]
- 38.Fang Y, Ferrie AM. Label-free optical biosensor for ligand-directed functional selectivity acting on β2 adrenoceptor in living cells. FEBS Lett. 2008;582:558–564. doi: 10.1016/j.febslet.2008.01.021. [DOI] [PubMed] [Google Scholar]
- 39.Azzi M, et al. β-arrestin-mediated activation of MAPK by inverse agonists reveals distinct active conformations for G protein-coupled receptors. Proc Natl Acad Sci USA. 2003;100:11406–11411. doi: 10.1073/pnas.1936664100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kenakin TP, Morgan PH. The theoretical effects of single and multiple transducer receptor coupling proteins on estimates of the relative potency of agonists. Mol Pharmacol. 1989;35:214–222. [PubMed] [Google Scholar]
- 41.Swaminath G, et al. Sequential binding of agonists to the β2 adrenoceptor: kinetic evidence for intermediate conformational states. J Biol Chem. 2004;279:686–691. doi: 10.1074/jbc.M310888200. [DOI] [PubMed] [Google Scholar]
- 42.Swaminath G, et al. Probing the β2 adrenoceptor binding site with catechol reveals differences in binding and activation by agonists and partial agonists. J Biol Chem. 2005;280:22165–22171. doi: 10.1074/jbc.M502352200. [DOI] [PubMed] [Google Scholar]
- 43.Green SA, et al. Sustained activation of a G protein coupled receptor via “anchored” agonist binding: molecular localization of the salmeterol exosite within the β2-adrenergic receptor. J Biol Chem. 1996;271:24029–24035. doi: 10.1074/jbc.271.39.24029. [DOI] [PubMed] [Google Scholar]
- 44.Carter AA, Hill SJ. Characterization of isoprenaline- and salmeterol-stimulated interactions between β2-adrenergic receptors and β-arrestin 2 using β-galactosidase complementation in C2C12 cells. J Pharmacol Exp Ther. 2005;315:839–848. doi: 10.1124/jpet.105.088914. [DOI] [PubMed] [Google Scholar]
- 45.Moore RH, et al. Salmeterol stimulation dissociates β2-adrenergic receptor phosphorylation and internalization. Am J Respir Cell Mol Biol. 2007;36:254–261. doi: 10.1165/rcmb.2006-0158OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maillet EL, et al. A novel, conformation-specific allosteric inhibitor of the tachykinin NK2 receptor (NK2R) with functionally selective properties. FASEB J. 2007;21:2124–2134. doi: 10.1096/fj.06-7683com. [DOI] [PubMed] [Google Scholar]
- 47.Niswender CM, et al. Context-dependent pharmacology exhibited by negative allosteric modulators of metabotropic glutamate receptor 7. Mol Pharmacol. 2010 doi: 10.1124/mol.109.058768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Antony J, et al. Dulasteric GPCR targeting: a novel route to binding and signaling pathway selectivity. FASEB J. 2009;23:442–450. doi: 10.1096/fj.08-114751. [DOI] [PubMed] [Google Scholar]
- 49.Vu TK, et al. Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell. 1991;64:1057–1068. doi: 10.1016/0092-8674(91)90261-v. [DOI] [PubMed] [Google Scholar]
- 50.Macfarlane SR, et al. Proteinase-activated receptors. Pharmacol Rev. 2001;53:245–282. [PubMed] [Google Scholar]
- 51.McLaughlin JN, et al. Functional selectivity of G protein signaling by agonist peptides and thrombin for the protease-activated receptor-1. J Biol Chem. 2005;280:25048–25059. doi: 10.1074/jbc.M414090200. [DOI] [PubMed] [Google Scholar]
- 52.Watson C, et al. The use of stimulus-biased assay systems to detect agonist-specific receptor active states: Implications for the trafficking of receptor stimulus by agonists. Mol Pharmacol. 2000;58:1230–1238. doi: 10.1124/mol.58.6.1230. [DOI] [PubMed] [Google Scholar]
- 53.Kinzer-Ursem TL, Linderman JJ. Both ligand- and cell-specific parameters control ligand agonism in a kinetic model of G protein-coupled receptor signaling. PLOS Comput Biol. 2007;3:e6. doi: 10.1371/journal.pcbi.0030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peters MF, Scott CW. Evaluating cellular impedance assays for detection of GPCR pleiotropic signaling and functional selectivity. J Biomol Screen. 2009;14:246–255. doi: 10.1177/1087057108330115. [DOI] [PubMed] [Google Scholar]
- 55.Horvath R, et al. Mutlidepth screening of living cells using optical waveguide. Biosensors Bioelectronics. 2008;24:799–804. doi: 10.1016/j.bios.2008.06.059. [DOI] [PubMed] [Google Scholar]
- 56.Ziblat R, et al. Infrared surface plasmon resonance: a novel tool for real time sensing of variations in living cells. Biophys J. 2006;90:2592–2599. doi: 10.1529/biophysj.105.072090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eisen MB, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]