Abstract
We describe studies in molecular profiling and biological pathway analysis that use sparse latent factor and regression models for microarray gene expression data. We discuss breast cancer applications and key aspects of the modeling and computational methodology. Our case studies aim to investigate and characterize heterogeneity of structure related to specific oncogenic pathways, as well as links between aggregate patterns in gene expression profiles and clinical biomarkers. Based on the metaphor of statistically derived “factors” as representing biological “subpathway” structure, we explore the decomposition of fitted sparse factor models into pathway subcomponents and investigate how these components overlay multiple aspects of known biological activity. Our methodology is based on sparsity modeling of multivariate regression, ANOVA, and latent factor models, as well as a class of models that combines all components. Hierarchical sparsity priors address questions of dimension reduction and multiple comparisons, as well as scalability of the methodology. The models include practically relevant non-Gaussian/nonparametric components for latent structure, underlying often quite complex non-Gaussianity in multivariate expression patterns. Model search and fitting are addressed through stochastic simulation and evolutionary stochastic search methods that are exemplified in the oncogenic pathway studies. Supplementary supporting material provides more details of the applications, as well as examples of the use of freely available software tools for implementing the methodology.
Keywords: Biological pathways, Breast cancer genomics, Decomposing gene expression patterns, Dirichlet process factor model, Evolutionary stochastic search, Factor regression, Gene expression analysis, Gene expression profiling, Gene networks, Non-Gaussian multivariate analysis, Sparse factor models, Sparsity priors
1. INTRODUCTION
Gene expression assays of human cancer tissues provide data that reflect the heterogeneity characteristic of oncogenic processes. The studies described herein use gene expression data from human breast cancer tissue samples and aim to improve our understanding of aspects of key cancer-related molecular mechanisms. In mammals, the complex Rb/E2F network of intersecting molecular pathways is fundamental to the control of the cell cycle, links the activity of cellular proliferation processes with the determination of cell fate, and is subject to many aspects of deregulation related to the development of human cancers (Nevins 1998). Some of our studies aim to better characterize the state and nature of a tumor based on expression patterns associated with this network and also to link this characterization to potential prognostic uses of expression profiles.
Our studies use the framework of sparse multivariate latent factor models of gene expression data, with extensions for regression and ANOVA components based on explanatory variables, as well as predictive regression components for measured responses. Our approach builds on the framework introduced by West (2003). In modeling dependencies among many variables, we use latent factor models in which the factor loadings matrix is sparse; that is, each factor is related to only a relatively small number of variables, representing a sparse, parsimonious structure underlying the associations among genes. With genes related to a set of interacting pathways, one key idea is that recovered factors overlay the known biological structure and that genes appearing to be linked to any specific “pathway characterizing” factor may be known or otherwise putatively linked to function in that pathway. The modeling approach provides for the infusion of biological information into the model in various ways, but also critically serves as an exploratory analysis approach to enrich the existing biological pathway representations. This analysis is enabled through the use of a flexible class of sparsity-inducing priors that allow the introduction of arbitrary patterns of zeros in sets of factor loadings and regression parameters, so that data can inform on the sparsity structure.
One other key methodological development is the use of non-parametric model components for the distributions of latent factors. This allows for flexible adaption to the often radically non-Gaussian structure in multiple aspects of the high-dimensional distributions of gene expression outcomes, reflecting aspects of experimental/technological noise, as well as the more important non-Gaussianity that relates to biological heterogeneity.
The statistical methodology involves computation using evolutionary stochastic search and Marcov chain Monte Carlo (MCMC) methods, as we describe and illustrate in our examples. The evolutionary component uses the theory underlying MCMC methods for our sparse latent factor models to generate a variable selection method that is useful in enriching an existing model with new variables (here genes) that appear to relate to the factor structure identified by an existing set of genes already modeled. In the pathway study context, model-based analysis of genes linked to a known biological pathway naturally recommends beginning with genes (variables) of known relevance and then gradually exploring beyond these initial variables to include others showing an apparent association so as to “evolve” the model specification to higher dimensions. This method meshes with MCMC analysis in the sparse factor models on a given set of genes. Our examples focus on the Rb/E2F signalling pathway and also on hormonal pathways to illustrate the methodology as an approach to exploring, evaluating, and defining molecular phenotypes of subpath-way characteristics—for both characterization and prediction— in this important disease context. We include comments about software for these analyses, as well as on open issues and current research directions.
2. GENERAL FACTOR REGRESSION MODEL FRAMEWORK
The framework combines latent factor modeling of a high-dimensional vector x with regression of response variables z, while allowing for additional regression and/or ANOVA effects of other known covariates, h, on both x and z. In our gene expression case studies, x represents a column vector of gene expression measures on a set of genes in one sample; z represents a set of outcomes or characteristics, such as survival time after surgery or a hormonal protein assay measure; and h may represent clinical or treatment variables or normalization covariates relevant as correction factors for technical errors or “assay artifacts” (see Lucas et al. 2006; app. E).
2.1 Basic Factor Regression Model Structure
Observations are made on a p-dimensional random quantity x with the ith sample modeled as a regression on independent variables, combined with a latent factor structure for patterns of covariation among the elements of xi not explained by the regression, that is,
| (1) |
or, elementwise,
| (2) |
for g = 1 : p (variables/genes) and i = 1 : n (samples), where μ = (μ1,…, μp)′ is the p-vector of intercept terms; B is the p × r matrix of regression parameters βg,j(g = 1 : p, j = 1 : r) with rows ; A is the p × k matrix of factor loadings αg,j (g = 1 : p, j = 1 : k) with rows ; hi = (h1,i,…, hr,i)′ is the r-vector of known covariates or design factors for sample i; λi = (λ1,i,…, λk,i)′ is the latent factor k-vector for sample i; and νi = (ν1,i…, νp,i)′ is a p-dimensional vector of independent, idiosyncratic noise terms, with νi ~ N(0, Ψ), where Ψ = diag(ψ1,…, ψp).
Variation in xg,i not predicted by the regression is defined by the common factors through whereas ψg is the unexplained component of xg,i, representing natural variation and technical and measurement error idiosyncratic to that variable. We use the usual zero upper-triangular parameterization of A to define identifiable models, the parameterization in which the first k variables have distinguished status (Aquilar and West 2000; Lopes and West 2003; West 2003). In addition, for identification purposes, here αg,g > 0 for g = 1,…, k, and αg,j = 0 for factors j = g + 1,…, k and g = 1,…, k – 1. The choice of these k lead variables is then a key modeling decision, and one of the questions addressed in our development of evolutionary model search in Section 4. We refer to the lead, ordered k variables as the founders of the factors.
The factors λi are assumed to be independently drawn from a latent factor distribution F(·). Traditionally, F(λi) = N(λi|0, I), where 0 and I are the zero vector and identity matrix (used generically); the zero mean and unit variance matrix are identifying assumptions. A key methodological development, discussed later, introduces nonparametric factor models based on a Dirichlet process extension of this traditional latent factor distribution.
2.2 General Predictive Factor Regression Models
The foregoing model for x combines with regression for response variables z in an overall multivariate model for (z, x). This extends the work of West (2003) to incorporate the view that predictions of z from x may be influenced in part by the latent factors λ underlying x, as well by additional aspects of x. These potential “additional aspects” of x are additional latent factors shown as response factors.
To be specific, suppose that z is q-vector with ith observation zi = (z1,i,…, zq,i)′ and redefine xi to now be the (p + q) × 1 vector . The general model is then as in (1) with this extended dimension; elementwise,
| (3) |
for g = 1 : (p + q) and i = 1 : n, and with the additional following changes:
μ = (μ1, …, μp+q)′ is the extended vector of intercepts for both xi and zi vectors.
B is the extended (p + q) × r matrix of regression parameters of xi and zi on the regressor variables in hi; now B has elements βg,j (g = 1 : (p + q), j = 1 : r) with rows .
A is the extended (in both rows and columns) (p + q) × (k + q) matrix of factor loadings αg,j (g = 1 : (p + q), j = 1 : (k + q)) with rows .
λi = (λ1,i,…, λk+q,i)′ is the extended (k + q)-vector of latent factors, where the additional q are introduced as response factors.
νi = (ν1,i,…, νp+q,i)′ is the extended idiosyncratic noise or error vector, with the additional q elements now related to zi; the variance matrix is extended accordingly.
Beyond notation, the key extension is the introduction of additional potential latent factors, the final q in the revised λi vectors, each linked to a specific response variable in zi. The structure of the extended factor loadings matrix A reflects this; each of the q response variables serves to define an additional latent factor (i.e., serves as a a founder of a factor), whereas the first k of the x variables in the order specified serve (as originally) to define the k factors in the latent model component reflecting inherent structure in x. Thus the structure of A is
where both Ax and Az have the structure as described in the initial model of x alone (i.e., the traditional zero upper-triangular parameterization); that is, the structural constraints on A = {αg,j} have two components. First, as in the initial model for x alone, the p × k matrix Ax has αg,g > 0 for g = 1,…, k, and αg,j = 0 for j = g + 1,…, k and g = 1,…, k – 1. Second, the square response factor loadings matrix Az is lower triangular with positive diagonal elements, that is, αp+g, p+g > 0 for g = 1,…, q, and αp+g, p+j = 0 for j = g + 1,…, q and g = 1,…, q – 1.
Different scales of response variables can be corrected so that all variables lie on the same scale. This simplifies specification of prior distributions over the elements of A, B, and Ψ. Otherwise, the differing nature of the response variables and expression data implies the need for flexibility to specify different priors over the loadings and regression coefficients. This is incorporated in our analysis and the BFRM software used (Wang, Carvalho, Lucas, and West 2007); see Appendix D. Additional considerations relate to specification of values or priors for the variance terms in Ψ, some of which arise in connection with non-Gaussian responses.
2.3 Non-Gaussian, Nonparametric Factor Modeling
A relaxation of the Gaussian assumption for latent factors is of interest in expression studies and other areas of application. Figure 4 (in Sec. 5.4) displays scatterplots of estimated factors from the analysis of a sample of expression profiles from breast tumors in the first study; the two factors are labelled as representing key biological growth factor pathways. The scatterplot reflects something like three overlapping groups of tumors that can be identified as distinct biological subtypes of breast cancer. The known biology underlying this structure is discussed in Section 5.
Figure 4.
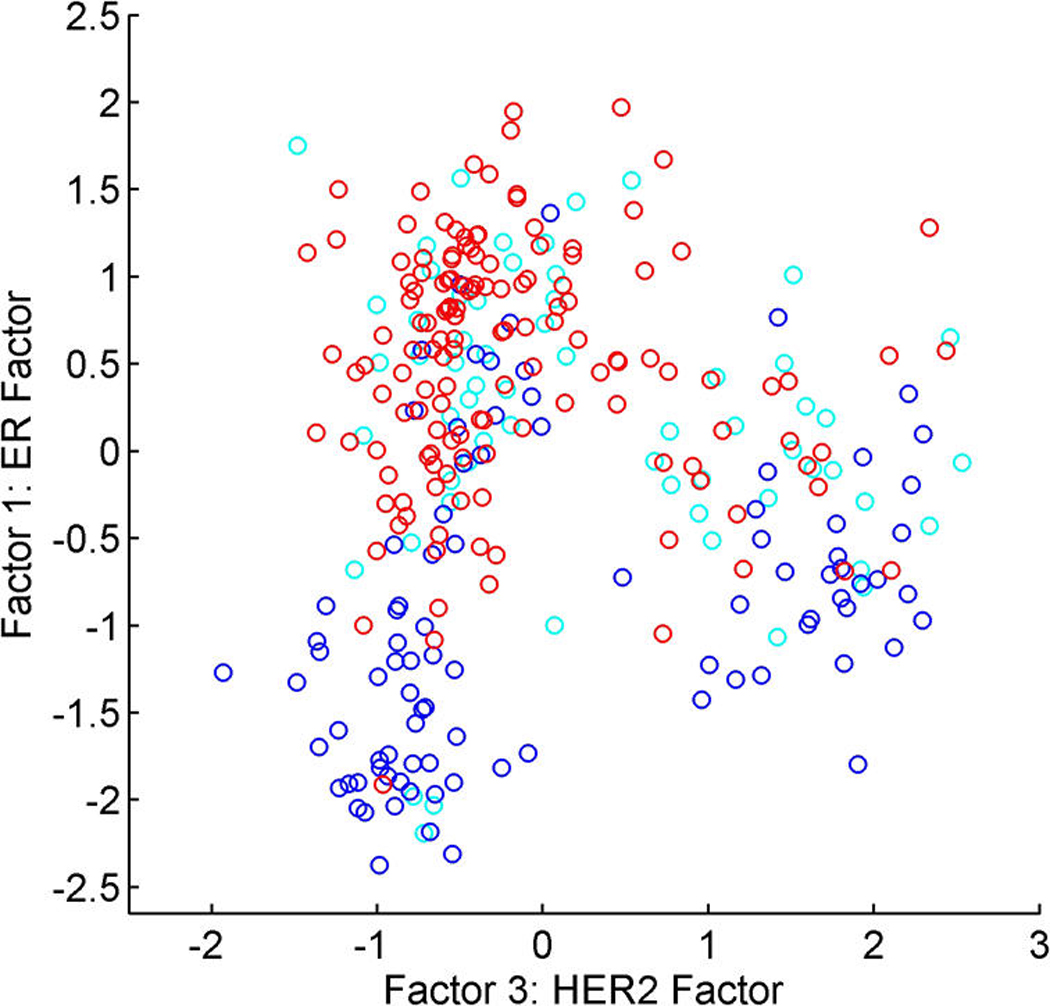
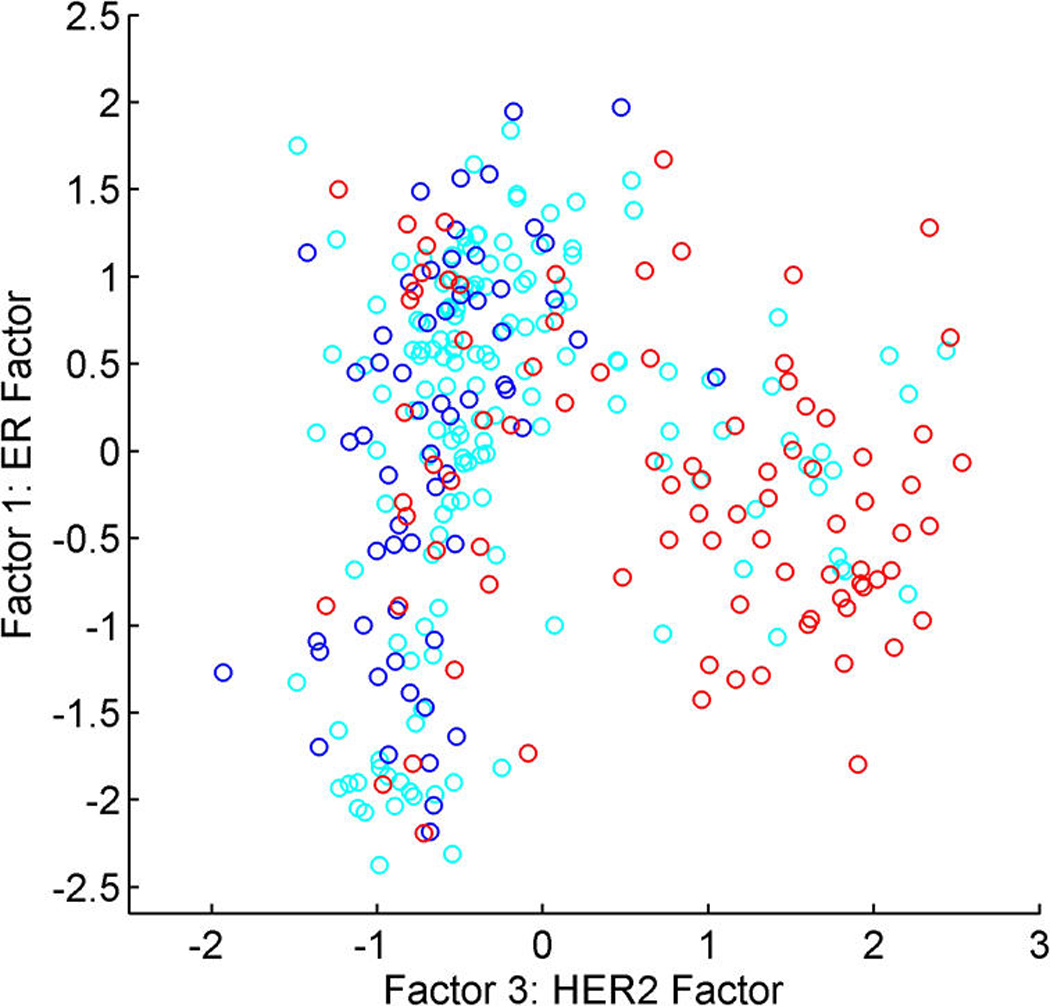
Breast cancer hormonal pathways. Scatterplots of the posterior means of designated ER factor 1 and HER2/ERB–B2 factor 3. Color coding indicates the global measurement of protein level from IHC assays. (a) Red, ER+; blue, ER−; cyan, missing/indeterminate. (b) Red, HER2+; blue, HER2−; cyan, missing/indeterminate.
A first step toward nonparametric modeling of the latent factor distribution F(λi) is to use the widely used Dirichlet process (DP) framework (West, Müller, and Escobar 1994; Escobar and West 1995, 1998; MacEachern and Müller 1998). Direct relaxation of the standard normal model simply embeds the normal distribution as a prior expectation of a DP over what is now considered an uncertain k-variate distribution function F(λi). In standard notation, F ~ Dir(α F0), a DP prior with base measure α F0 for some total mass or precision parameter, α > 0, and prior expectation F0(λ) = N(λ|0, I). Write λ1:n = {λ1, …, λn}, and for each i = 1 : n, let λ−i denote the set of n − 1 factor vectors with λi removed. A key feature of the DP model is the set of implied complete conditionals for λi (marginalizing over the uncertain F). These are given by
| (4) |
where δλ(·) is the Dirac delta function, representing a distribution degenerate at λ, and an−1 = α/(α + n − 1). Conditional on λ−i, the vector λi comes from the prior normal with probability an−1; otherwise, it takes the same value as one of the existing λr’s, with those n – 1 values having equal probability. A sample of n factor vectors then reduces to k ≤ n distinct values, and the samples are configured across that number of “clusters” in factor space; of course, the latency means that we will never know the configuration or number, and all inferences average over the implied posterior distributions. Full details and supporting theory can be found in the aforementioned references. For our purposes here, the key is the utility of the DP model as a flexible and robust nonparametric approach that will adapt to non-Gaussian structure evident in data. Concentration of factor realizations on common values also aids in, for example, allowing representation of “inactive” and “up-regulated” biological pathways across a number of samples, while also permitting variation in levels of activity of a pathway across other samples. In many cases, expression patterns are consistent with Gaussianity, and the DP model naturally “cuts back” to reflect that. Little additional model specification complexity or computational cost is incurred in moving to the DP model, whereas flexibility—to respond automatically to observed non-Gaussian structure if and when it is seen—and robustness are gained.
3. SPARSITY MODELING
A basic perspective is that of sparsity in the factor loadings matrix. Any given gene may associate with one or a few factors but is unlikely to be related to (or implicated in) all factors. Any single factor will link to a number of genes, generally a relatively (to p) small number; that is, in problems with large p, the factor loadings, matrix A will be expected to have many zero elements, although the pattern of nonzero values is unknown and must be estimated. A priori, each (of the unconstrained) αg,j may be 0 or take some nonzero value, so that relevant priors should mix point masses at 0 with distributions over nonzero values as in standard Bayesian “variable selection” analyses in regression and other areas (George and McCulloch 1993; Raftery, Madigan, and Hotelling 1997; Clyde and George 2004). This was initiated in factor models of West (2003), and is used in other models, including large p regression (Rich et al. 2005; Dressman et al. 2006; Hans, Dobra, and West 2007) and graphical models (Dobra, Jones, Hans, Nevins, and West 2004; Jones et al. 2005). The standard mixture priors (sometimes referred to as “slab and spike” priors) have been used effectively in ANOVA and related models for gene expression (Broet, Richardson, and Radvanyi 2002; Ishwaran and Rao 2003, 2005; Lee, Sha, Dougherty, Vannucci, and Mallick 2003; Do, Müller, and Tang 2005). Our extensions here represent generalizations of the standard methods for multivariate regression and ANOVA, as well as extensions of the original sparse factor regression model versions of West (2003). We focus our discussion here on the factor loadings matrix A, but the methodology implemented applies the same ideas, and resulting sparsity prior distributional models, to B as well.
A sparsity prior assigns each element αg,j of A a probability, πg,j, of taking a nonzero value. The model for these sparsity probabilities introduced by Lucas et al. (2006) has the form
| (5) |
independently over g, where δ0(·) is the Dirac delta function at 0. This states that variables have individual probabilities of association with any factor, πg,j, for variable g and factor j, and that nonzero loadings on factor j are drawn from a normal prior with variance τj. A slight modification is required for the case of diagonal elements, because they are constrained to be positive to ensure identifiability; thus the normal component of (5) is adapted to N(αg,g|0, τj)I(αg,g > 0) for g = 1,…, k and g = p + 1,…, p + q, where I(·) is the indicator function.
The usual variable selection prior model adopts πg,j = πj, a common likelihood (“base rate”) of nonzero loading on factor j for all variables, and estimates this base rate πj under a prior that heavily favors very small values. One problem is that with larger p, a very informative prior on πj favoring very small values is required, resulting in posterior probabilities for αg,j ≠ 0 that are spread out quite widely on the unit interval. Although generally consistent with smaller values of πj, this leads to a counterintuitively high level of uncertainty concerning whether or not αg,j = 0 for a nontrivial fraction of the variables. This was clearly illustrated by West (2003) and has been demonstrated in other models with the use of these standard priors (Lucas et al. 2006).
The more general model (5) addresses this problem by adding an hierarchical component for the loading probabilities πg,j. Sparsity indicates that many of these probabilities will be small or 0 and a small number will be high; this is reflected in the model of Lucas et al. (2006),
| (6) |
where Be(·|am, a(1 – m)) is a beta distribution with mean m and precision parameter a > 0. Each ρj has a prior that favors very small values, such as Be(ρj|sr, s(1 – r)), where s > 0 is large (e.g., s = p + q) and r is a small prior probability of nonzero values, usually taken as r0/(p + q) for some small integer r0 (e.g., r0 = 5). The beta prior on nonzero values of πg,j is fairly diffuse while favoring relatively larger probabilities, such as defined by aj = 10 and mj = .75. On integrating out the variable-specific probabilities πg,j from the prior for αg,j in (5), we obtain a similar distribution, but now with πg,j replaced by E(πg,j|ρj) = ρjmj. This is precisely the traditional variable selection prior discussed earlier, with the common base rate of nonzero factor loadings set at ρjmj. Insertion of an additional layer of uncertainty between the base- rate and the new πg,j now reflects the view that many (as represented by a high value of ρj) of the loadings will be 0 for sure and permits the separation of significant factor loadings from the remainder. The practical evidence of this is that in many examples that we have studied, the posterior expectations of the πg,j’s generally have a large fraction concentrated heavily at or near 0, a smaller number at very high values, and only a few in regions of higher uncertainty within the unit interval. In contrast, the standard variable selection prior leads to posterior probabilities on αg,j = 0 that are overly diffused on the unit interval. More discussion and examples in regression variable selection in ANOVA models have been given by Lucas et al. (2006). This generally better isolates nonzero effects and induces increased shrinkage toward 0 for many insignificant loadings. Key elements for the assessment of sparsity are the posterior probabilities π̂g, j = Pr(αg,j ≠ 0|x1:n).
4. EVOLUTIONARY STOCHASTIC MODEL SEARCH
In the case studies, models are defined by a process of evolutionary refinement developed to address variable (gene) selection, choice/limitation on the number of factors, and specification of the order of the first k founding variables in the model. This model search method is inspired by interest in evaluating patterns of expression of genes linked to a particular pathway and has been a key discovery tool in some of our recent studies. The method is of general utility in other application areas (Carvalho 2006), although here we describe it in the pathway exploration context.
Directly fitting models with large numbers of variables and factors is a challenge both statistically and computationally. In applied contexts, such as biological pathway exploration, attempting to fit models to all of the available variables (genes) would be misguided scientifically in any case. Our pathway studies focus on genes that play roles in chosen cancer pathways. We aim to develop an understanding of gene expression patterns among genes already assumed to be featured in a pathway by identifying additional genes and factors linked to that known biology. This “pathway-focused” view mandates beginning with an initial small set of biologically relevant genes and then expanding the model by adding new genes that appear to be linked to the factors identified in the initial model. This might be followed by refitting the model, allowing more factors if the new genes suggest additional structure. Repeating this process to iteratively refine the model underlies our evolutionary model search.
The technical key is to note that, given an initial set of p0 variables and a model denoted by M0 with k0 latent factors, we can view the model as being embedded in a larger model on all p ≫ p0 variables and k > k0 factors in which the extended matrix of loadings probabilities has πg,j = 0 for g > p0 and k > k0. Within this “full” overarching model, consider any of these variables g > p0 and evaluate whether it should be added to the current model with a single nonzero factor loading on, say, latent factor j ∈ 1 : k0. Based on model parameters fixed at their posterior means for the current model, we then can compute approximately the conditional posterior probability of inclusion, that is, just π̃g,j = Pr(αg,j ≠ 0|x1:n, M0), where M0 in conditioning simply represents the current model and estimated parameters. (Note that we use π̃g, j rather than π̂g, j to denote these inclusion probabilities for variables currently not included in the set to which the model is fitted.) Variables g with high values of π̃g, j are candidates for inclusion. These are variables showing significant associations with one or more of the currently estimated factors and so provide directions for model expansion around the currently identified latent structure. We then can rank and choose some of these variables—perhaps those for which π̃g, j > θ for some threshold or, more parsimoniously, a specified small number of them—and refit the model.
Expanding the set of variables may identify other aspects of common association that suggest additional latent factors; enriching the sample space allows a broader exploration of the complexity of associations around the initial model neighborhood. This promotes exploration of an expanded model M1 on the new p1 > p0 variables and with k1 = k0 + 1 latent factors for which the first k0 variables remain ordered as under M0. The factor founders in M0 are those of the first k0 factors in M1. We then can refit M1 and continue. This raises the question of the choice of the variable k1 as the founder of the new potential factor. We address this by fitting the model with some choice of this variable, perhaps just a random selection from the p1 variables in M1; from this model, we generate the posterior probabilities π̂g, j and choose that variable with highest loading on the new factor j = k1. We then refit model M1 with this variable as founder of the new factor, assuming that these probabilities are appreciable for more than one or two variables.
Algorithmically, the evolutionary analysis proceeds as follows. Initialize a model M0 and i = 0. For i = 0, 1,…, do the following:
Compute approximate variable inclusion probabilities, π̃g, j, for variables g not in Mi and relative to factors j = 1 : ki in Mi. Rank and select at most r variables with highest inclusion probabilities subject to π̃g, j > θ for some threshold. Stop if no additional variables are significant at this threshold.
Set i = i + 1 and refit the expanded model Mi on the new pi variables with ki = ki–1 + 1 latent factors. First, fit the model by MCMC with a randomly chosen founder of the new factor, and then choose that variable with highest estimated π̂g,ki as the founder. Refit the model and recompute all posterior summaries, including revised π̂g, j. Reject the factor model increase if fewer than some small prespecified number of variables have π̂g, j > θ, then cut back to ki–1 factors. Otherwise, accept the expanded model and continue to iterate the model evolutionary search at stage i + 1.
Stop if the foregoing process does not include additional variables or factors, or if the numbers exceed some pre-specified targets on the number of variables included in the model and/or the number of factors.
This analysis has been developed and evaluated across a number of studies, and it offers an effective way of iteratively refining a factor model based on a primary initial set of variables of interest—the nucleating variables. Computational efficiencies can be realized by starting each new model MCMC analysis using information from the previously fitted model to define initial values. Control parameters include thresholds θ on inclusion probabilities for both variables and additional factors at each step, a threshold to define the minimum number of significant variables “required” to add a new latent factor, and overall targets to control the dimension of the final fitted model—a specified maximum number of variables to include out of the overall (large) p and (possibly) a specified maximum number of latent factors. A number of simulated and real examples that investigate the efficacy of this procedure have been presented by Carvalho (2006). The main conclusions from these examples is that the evolutionary search is able to identify variables associated with a latent component and to correctly identify relevant values of k.
5. STUDY 1: HORMONAL PATHWAYS
5.1 Goals, Context, and Data
The first study uses a large, heterogenous data set that combines summary robust multichip average (RMA) measures of expression from Affymetrix u95av2 microarray profiles on three sets of breast cancer samples: 138 tumor samples from the Taiwanese–U.S. CODEx study (Huang, West, and Nevins 2002; Huang et al. 2003; Nevins et al. 2003; Pittman et al. 2004), 74 additional samples from the same center collected a year or two later, and 83 samples on breast cancer patients collected in 2000–2004 at the Duke University Medical Center. The combined set of n = 295 samples was processed using the standard RMA code from Bioconductor (www.bioconductor.org) and screened to identify 5,671 genes showing nontrivial variation (median RMA, >6.5; range, >1 across samples). We use “genes” and “probesets” interchangeably when referring to the Affymetrix data; each “gene” is really a single oligonucleotide sequence—a probeset—representing a gene, and some genes have multiple, distinct probesets.
We aim to explore expression patterns related to the two key biological growth factor pathways, the estrogen receptor (ER) pathway and the HER2/ERB–B2 pathway, which are central to the pathogenesis of breast cancer. One interest relates to how mRNA signatures of biological variation in these key pathways relate to the global and cruder designations of ER positive or negative based on the IHC assays. Discordance between expression and protein measures arises from many factors, not the least of which is the geographical variation in expression (of both genes and proteins) throughout a tumor. For each of the two binary response variables, ER+ versus ER− and HER+ versus HER−, there are quite a few missing or indeterminate outcomes, so that the analysis imputes a good fraction of the response values. The numbers are 143 ER+, 91 ER−, 61 ER missing or uncertain, and 86 HER2+, 60 HER2−, 149 HER2 missing or uncertain. We discuss summaries of analysis of a “final” set of 250 genes. We began with 10 genes known to be regulated by or co-regulated with ER, that is, key genes in the ER pathway. We then included four genes similarly related to HER/ERB–B2. We then ran the evolutionary analysis, adding in at most 10 genes per step at a thresholded inclusion probability of .75 and stopping when the total reached 250. Based on multiple reanalyses, the final MCMC sampler was run to generate 20,000 iterates, with a burn-in of 2,000.
5.2 Exploring Variable-Factor Associations and Sparsity Patterns
High values of π̂g, j = Pr(αg,j ≠ 0|x1:n) define significant gene–factor relationships. Figure 1 provides a visual summary from a model with k = 10 latent factors and q = 2 response factors. Frame (a) is a “skeleton” of the fitted model, displaying the indicator of π̂g, j > θ, where θ = .99 for this figure. Frame (b) displays the posterior estimates of loadings for those gene–factor pairs that pass this threshold, that is, α̂g, j = E(αg,j|αg,j ≠ 0, x1:n)I(π̂g, j > θ). These figures give a useful general impression of the relative sparsity/density of factors, as well as the cross-talk in terms of genes significantly linked to multiple factors.
Figure 1.
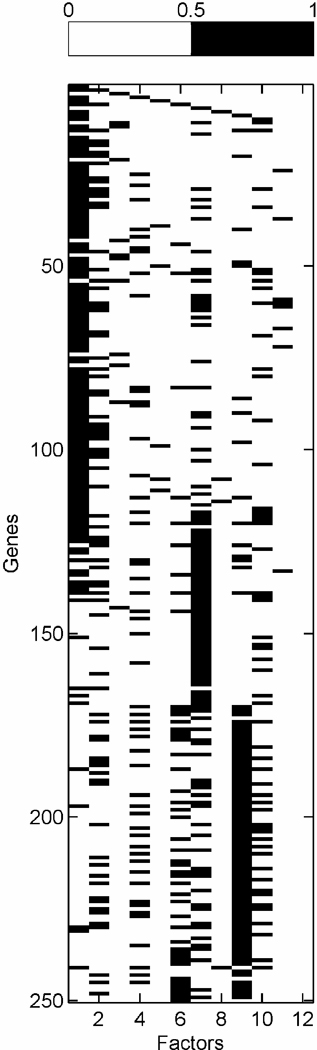
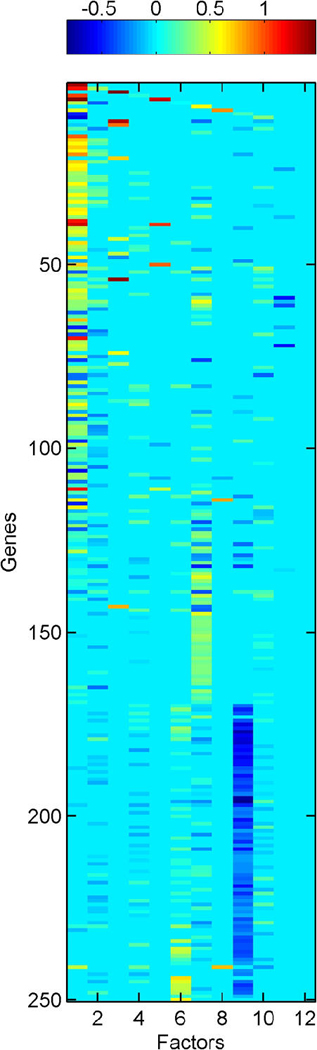
Breast cancer hormonal pathways. Skeleton of the fitted model for the 250 selected genes and 12 factors. (a) (Binary) heatmap of thresholded approximate posterior loading probabilities, I(π̂ g, j > .99). (b) Heatmap of approximate posterior means of significant gene-factor loadings, α̂ g, j
Inferred latent factors labelled 1, 2, 4, and 5 are founded by known ER-related genes and have a number of genes known to be linked to the ER pathways with significant loadings. Factor 1 is a primary ER factor strongly associated with the protein assay for ER status (see Fig. 4); factors 2, 4, and 5 contain highly loaded genes known to be related to the ER gene pathways but do not seem to be directly related to the IHC measure. Factor 3 is founded by the primary sequence probe on the Affymetrix array for HER2/ERB–B2. The array has three separate probesets with DNA oligonucleotides representing different sections of this gene, which historically has been referred to as both ERB–B2 and HER2. This factor picks up co-variation in these three genes along with a small number of other genes (12 at the threshold of π̂g,i > .99) defining a HER2 factor. Table 1 lists a few “top genes” on some selected factors. All genes listed are known to be regulated by, co-regulated with, interactive/synergistic with, or (based on previous prior experimental studies) co-expressed with ER for factors 1 and 5 and with HER2 for factor 3 (Spang et al. 2001; West et al. 2001; Huang et al. 2003). Factor 5 is loaded on a very small number of ER-related genes, led by the transcription factor TFF3 that is known to be estrogen-responsive or associated with ER status. The emergence of this additional ER-related factor indicates potential connections in the TFF3-related signalling pathway. In the list for factor 1, we include some of these that have lower loadings but are still significant. Some of these arise in factor 8; this factor has very few significantly loaded genes, and the top three here are all sequences from the cyclin D1 gene. The Affymetrix array has three separate probesets with DNA oligonucleotides representing different sections of cyclin D1, and this factor picks up co-variation in the three along with a small number of other genes, defining a cyclin D1 factor. Later we return to the cyclin D1 factor, discussing its biological connections and how it highlights some of the discovery utility of this modeling approach.
Table 1.
Breast cancer hormonal pathways: Some genes significantly loaded on latent factors 1, 3, 5, and 8 in the ER/HER2 breast cancer data analysis
| α̂g,j | Gene | Gene symbol | |
|---|---|---|---|
| Factor 1 | 1.5 | Intestinal trefoil factor | TFF3 |
| 1.4 | Carbonic anhydrase precursor | CA12 | |
| 1.3 | Clone AA314825:EST186646 | – | |
| 1.1 | Secreted cement gland protein XAG-2 homologue | AGR2 | |
| 1.1 | Hepatocyte nuclear factor-3 alpha (HNF-3α) | FOXA1 | |
| 1.1 | Trans-acting T-cell specific transcription factor | GATA-3 | |
| 1.1 | Clone AL050025:DKFZp564D066 | – | |
| 1.0 | Breast cancer, estrogen regulated LIV-1 protein … |
LIV-1 | |
| .71 | Myeloblastosis viral oncogene homolog | C–MYB | |
| .47 | Human epidermal growth factor receptor | HER3 | |
| .46 | Human epidermal growth factor receptor | HER3 | |
| .44 | BCL-2 | BCL-2 | |
| .42 | Androgen receptor … |
AR | |
| .54 | PRAD1 (cyclin D) | CCND1 | |
| .53 | BCL-1 (cyclin D) | CCND1 | |
| .45 | CYCD1 (cyclin D) | CCND1 | |
| Factor 3 | 1.5 | c-Erb-B2 | ERB–B2 |
| 1.4 | Human tyrosine kinase-type receptor (HER2) | HER2b | |
| 1.4 | Human tyrosine kinase-type receptor (HER2) | HER2 | |
| .93 | Growth factor receptor-bound protein 7 | GRB7 | |
| .78 | CAB1 | STARD3 | |
| Factor 5 | 1.3 | Intestinal trefoil factor | TFF3 |
| 1.1 | Clone AA314825:EST186646 | – | |
| .97 | Clone AI985964:wr79d08.x1 | – | |
| .45 | Secreted cement gland protein XAG-2 homolog | AGR2 | |
| .18 | Cytochrome b5 | CYB5 | |
| .16 | Cytochrome b5 | CYB5 | |
| Factor 8 | .83 | PRAD1 (cyclin D) | CCND1 |
| .81 | BCL-1 (cyclin D) | CCND1 | |
| .73 | CYCD1 (cyclin D) | CCND1 | |
| .15 | Cytochrome b5 | CYB5 |
5.3 Factor Variation, Decompositions, and Interactions
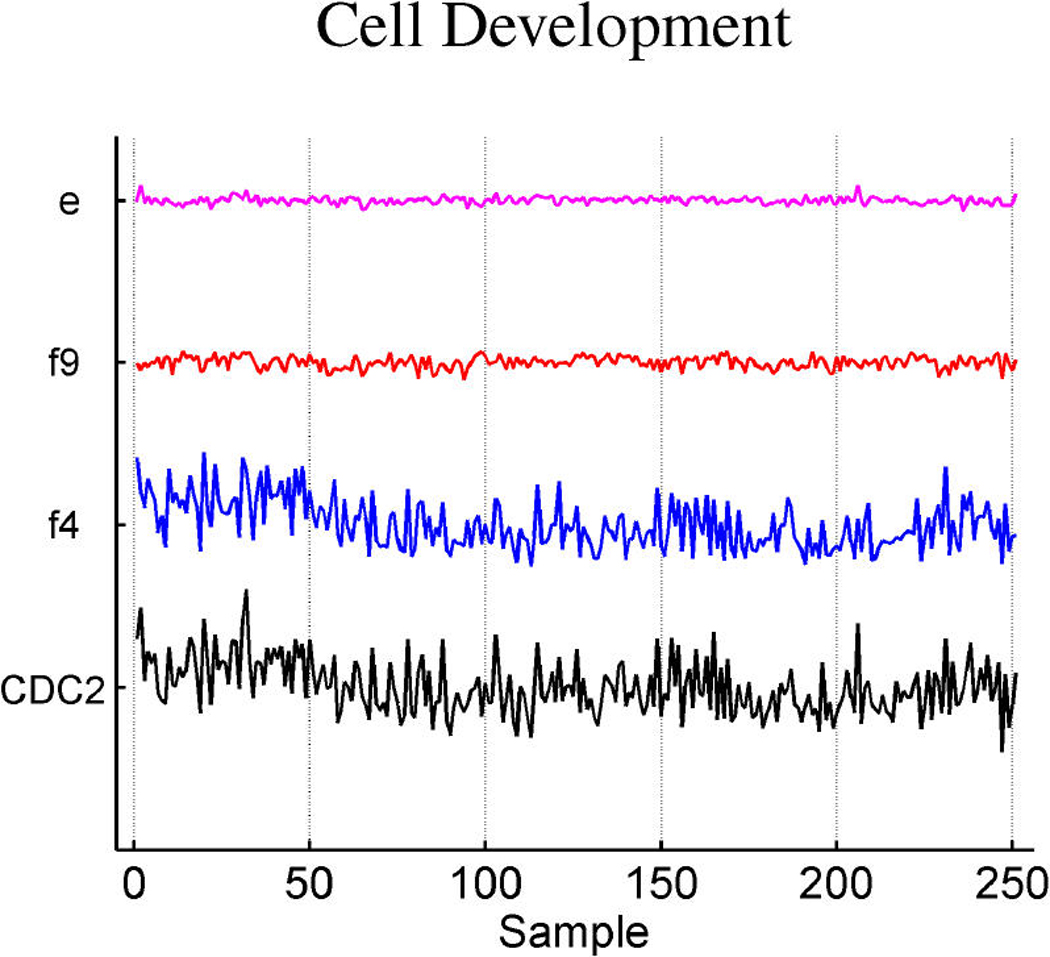
Exploring plots of estimated latent factors across samples can provide useful insights into the nature of the contributions of the factors to patterns of variation in expression gene-by-gene as well as relationships across genes. Figure 2 and Figure 3 add to the discussion of the ER factor structure and the utility of this form of analysis in revealing interacting pathways. Factors are plotted only in cases of significant gene–factor association (π̂g, j > .99). Figure 2(a) and (b) represent two versions of cyclin D1. The corresponding estimates of gene–factor loadings, α̂g, j, are approximately as follows: for gene PRAD1, loadings of .53 on the ER factor 1 and .83 on the cyclin D1 factor 8; for gene BCL-1, .54 on the ER factor and .81 on the cyclin D1 factor. The agreement is clear: Cyclin D1 expression fluctuations are–up to residual noise and the assay artifact correction components (App. E), labelled c3 and c2–described by these two factors in an approximate 5 : 8 ratio. This is not only a nice example of the agreement between factor model decompositions for what by design should be highly related expression profiles, but also is consonant with known biology. Cyclin D1 is a regulatory component of the protein kinase Cdk4, which together mediate the phosphorylation and inactivation of the Rb protein. Thus its activity is required for cell cycle transitions and control of growth and proliferation. ER binds to the CCND1 gene that encodes the cyclin D1 protein (Sabbah, Courilleau, Mester, and Redeuilh 1999) and thus can promote cell proliferation. The relationship has feedback through the regulation of ER itself by cyclin D1; for example, cyclin D1 also acts to antagonize BRCA1 repression of ER (McMahon, Suthiphongchai, DiRenzo, and Ewen 1999; Wang et al. 2005). There are further experimentally defined interactions between cyclin D1 and ER with consequences for the resulting levels of activation of each of the two pathways, as reviewed by Fu, Wang, Li, Sakamaki, and Pestell (2004). Thus the description of cyclin D1 expression fluctuations though a non–ER-related cell cycle component (factor 8), together with a significant ER-related component, is consonant with known regulatory interactions between the cell cycle/cyclin D1 pathway and the ER pathway. The factor analysis reveals and quantifies these interactions.
Figure 2.
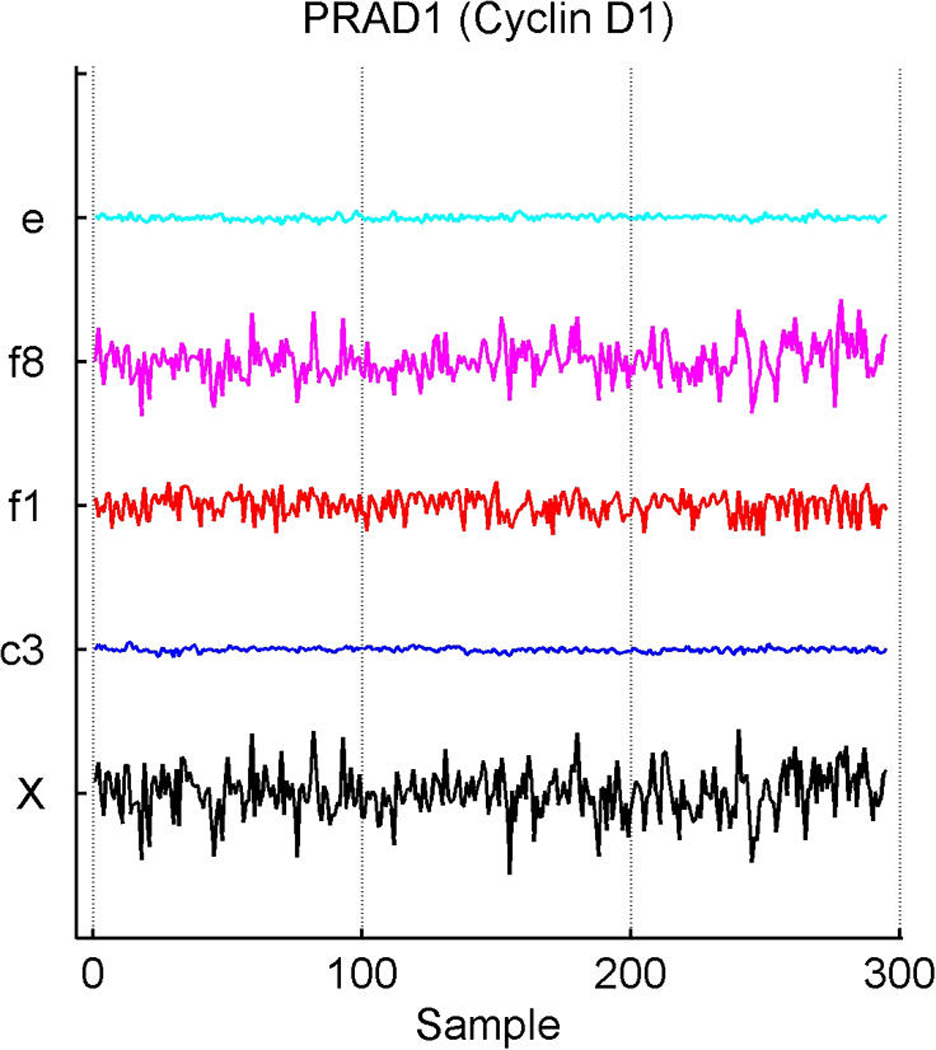
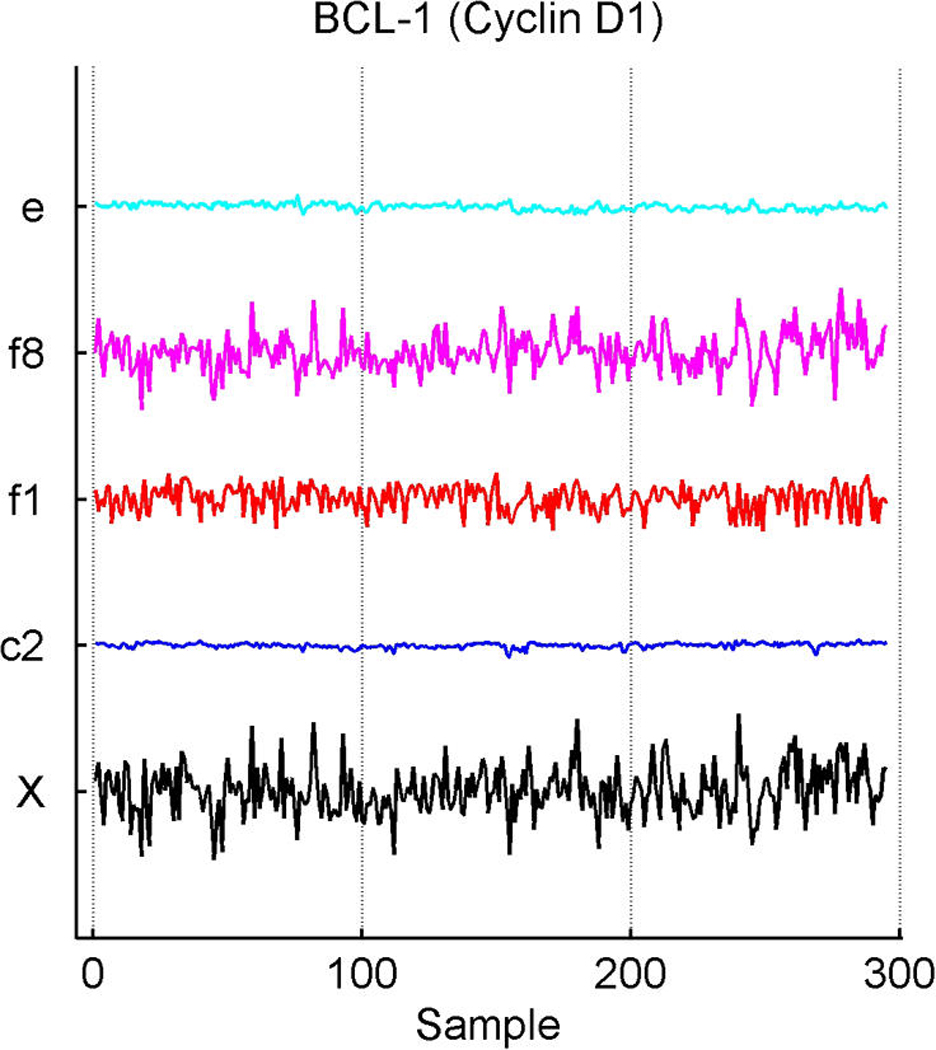
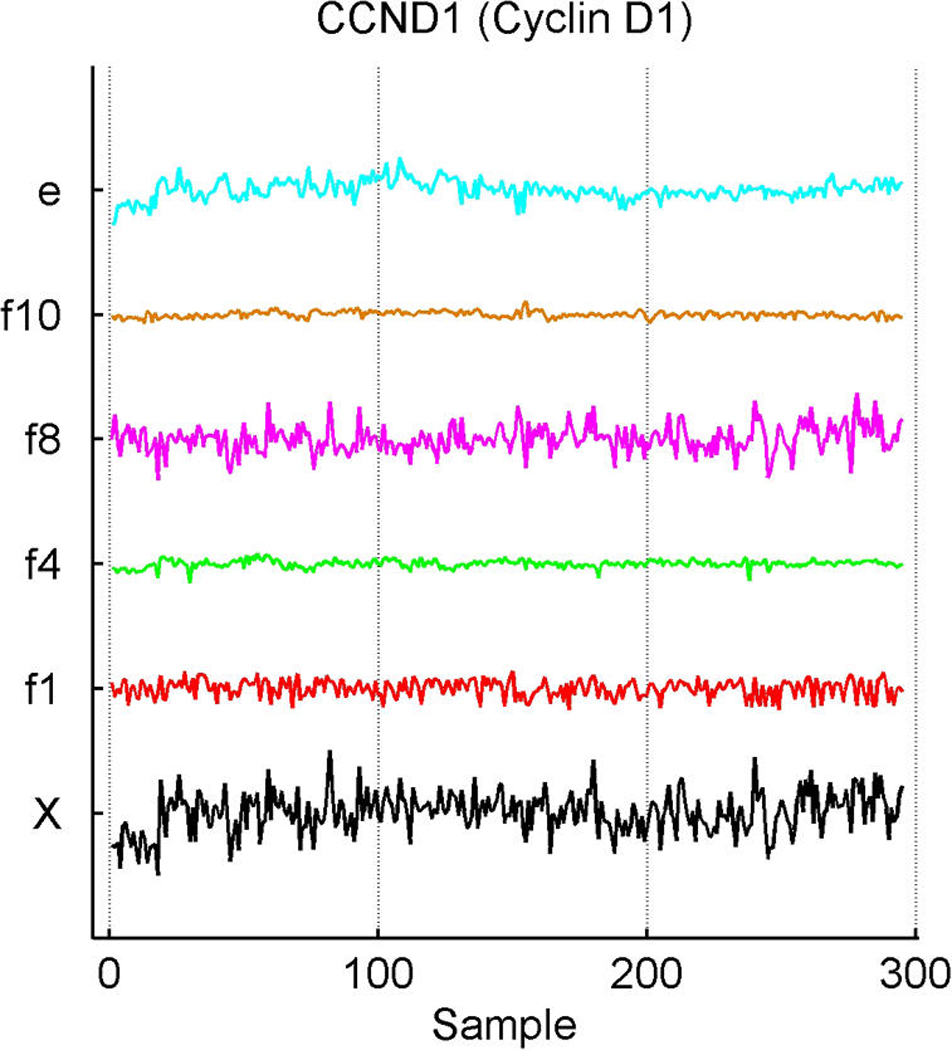
Breast cancer hormonal pathways. Plot across breast tumour samples of levels of expression (X) of the gene Cyclin D1. (a) The PRAD1/CCND1 probeset on the Affymetrix u95av2 microarray, one of the three probe sets for cyclin D1 on this array. (b) The BCL-1/CCND1 probeset. (c) The primary CCND1 probeset. Factors labelled “f” are primary latent factors, “c” indicates assay artifact covariates, and “e” represents the fitted residuals. In each of three frames, the plotted gene expression, factor, and residual levels are on the same vertical scale within the frame, so indicating the breakdown of the expression fluctuations for cyclin D1 gene probesets according to contributions from the factors. Factor 1 is the primary ER factor, and factor 8 a factor defined by the three probesets for cyclin D1, as discussed in the text.
Figure 3.
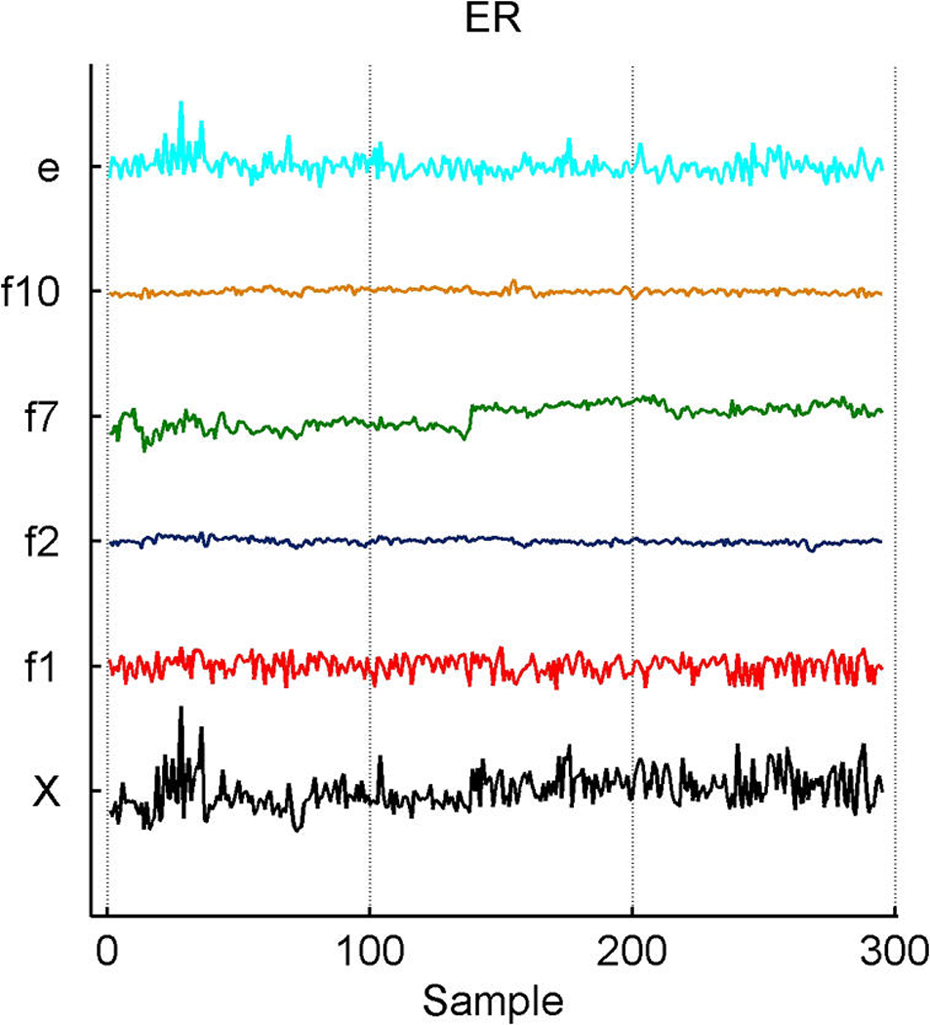
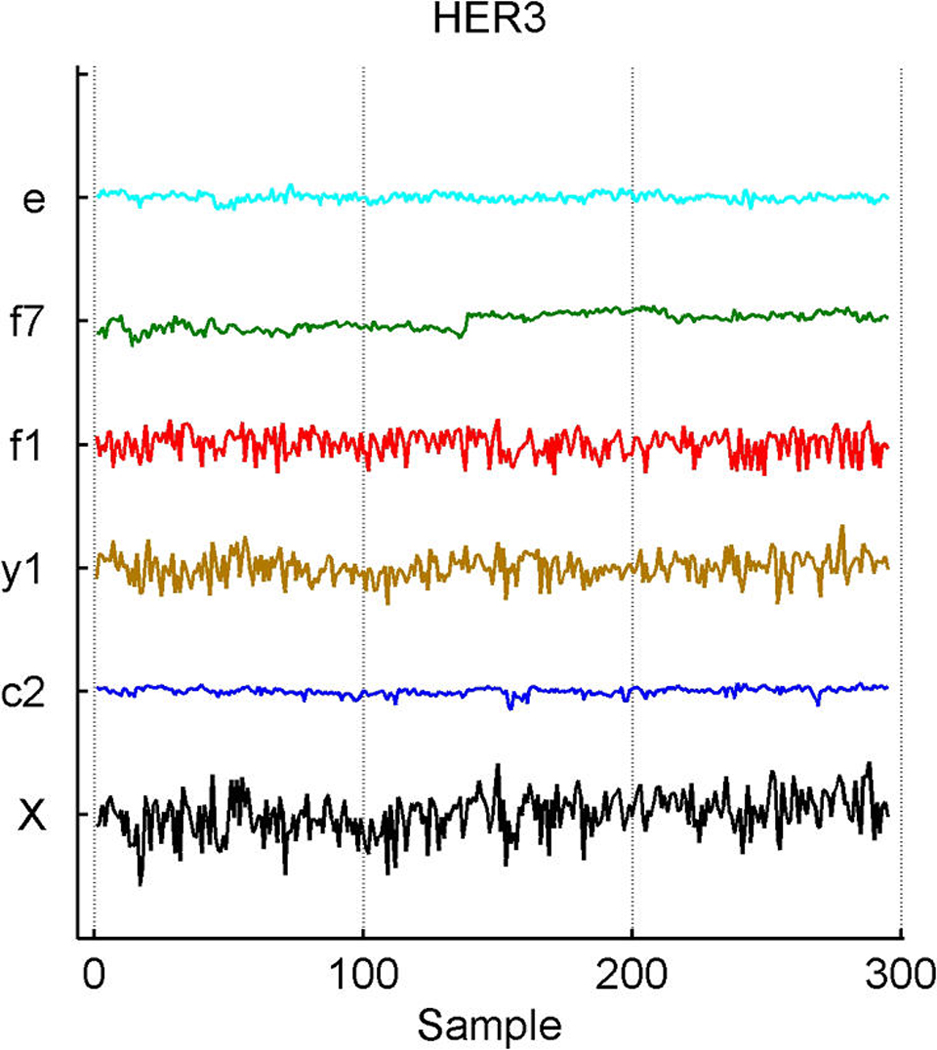
Breast cancer hormonal pathways. Plot across breast tumor samples of levels of expression (X) of the ER gene (a) and of the HER3 epidermal growth factor receptor tyrosine kinase (b), together with the estimates of factors contributing significantly to their expression fluctuations. Factors labelled “f” are primary latent factors, “y” indicates response factors, “c” indicates assay artifact covariates, and “e” represents the fitted residuals; other layout details are as in Figure 2. Note that f7 picks up what is clear artifact related to the different substudies generating the data, and also that some residual structure remains evident in the residual plot that appears to be batch-related (e.g., an early burst of positively correlated cases).
In Figure 2(c), the third cyclin D1 gene probeset, CCND1, shows substantial association with the ER and cell cycle cyclin D1 factors, as expected. The estimated loadings are reduced relative to those of the other two probesets, at about .45 for the ER factor and .73 for the cell-cycle factor relative to the .5/.8 levels of the other two probesets. CCND1 shows an additional significant association with latent factor j = 4, with an estimated coefficient of .24. Although apparently not related to the ER IHC response (unlike factor 1), factor 4 is loaded on genes that include several ER-related genes and other cyclins. The founder for factor 4 is the LIV-1 gene, which also scores highly on the ER factor 1. LIV-1 is regulated by estrogen and co-regulated with estrogen receptors in some breast cancers, although not in some other cancers. Factor 4 may reflect more complexity of the interactions between the ER and early cell-cycle pathways. The CCND1 gene probeset shows a significant association with this factor, although the practical contribution of factor 4 to expression levels of CCND1 is relatively small compared with that of the others.
This example highlights differences in data measured in different ways on a single gene, as well as the need to explore data quality issues. To highlight this, Figure 2(c) indicates some concern about the measurements for CCND1 in the early samples, transferred to the residuals for this probeset. One strength of the model is the realistic attribution of substantial levels of variation in expression data to residual, unexplained terms. In many cases, purely experimental artifact and noise can be evident concordantly across multiple genes; the sparse factor and regression model can then help protect the estimation of biological effects from such contamination.
5.4 Response Factors and Expression Signatures of Hormonal Status
Figure 4 scatters the samples on the estimated values of ER factor 1 and HER2 factor 3, with color coding by the measured immunohistocompatability (IHC) assays for ER and HER2. This shows biologically interpretable groupings into ER+/HER2−, ER−/HER2−, and HER2+ as designated by the broad IHC-based protein assay for hormonal status. These two primary latent factors are capable of refining the ER and HER2 scales and placing each tumor on the biologically relevant continuum.
The model includes the binary ER and HER2 responses and two response factors for them. The model has p + 2 entries in xi, the final two being the linear predictors in probit regressions for ER and HER2. Figure 5 illustrates the overall signatures of ER and HER2 in terms of the probit transforms of the posterior means of the linear predictors. The posterior turns out to strongly favor only rather modest additional predictive values in the gene expression data beyond those captured by the k = 10 latent factors; that is, the posteriors for the response factor loadings elements αg,j for g = p + 1, p + 2 and j = 11, 12 are almost all very concentrated at 0. A few genes contribute significantly to the ER response prediction over and above the ER latent factors (eight genes at π̂g, j > .99), but none do so for HER2 prediction; this can be seen in the images in Figure 1. For ER, it is notable that a further key signal receptor gene is significant and most highly loaded on the ER response factor; this is the HER3 gene, known to play roles in the development of more highly proliferative cellular states in breast cancers (Holbro et al. 2003), as well as biochemically partnering with HER2 in promoting cellular transformation. The top two genes loaded on the ER response factor are the two probesets for HER3 on the Affymetrix array. One of these is displayed in Figure 3(b), where the significant association with the primary latent ER factor 1 along with the ER response factor (labelled y1) is clear. The posterior estimates α̂g, j for the ER gene on the factors f1, f2, f7, and f10 are approximately .3, .09, .35, and –.09; those for the HER3 gene on factors y1, f1, and f7 are −.5, .46, and .3. Note that the probeset for HER3 also loads significantly on the likely artifactual factor 7, as does the ER gene and the assay artifact covariate c2. Although it is not displayed, the second probeset for HER3 has a fitted decomposition that is almost precisely the same in terms of the split between contributions from f1 and y1, but apparently is not significantly linked to the artifactual factors. As with cyclin D1, this is an example of different probesets for one gene (here HER3) that can behave somewhat differently in terms of expression readouts. The model analysis nevertheless identifies and extracts the commonalities. The posterior estimates α̂g, j for the two HER3 probes on the ER+/− response factor 1 are each approximately −.51, and those on the primary ER latent factor 1 are approximately .46 and .47. Thus the sparse factor model analysis cleans up the artifacts to find and quantify the relevant associations with biologically interpretable and predictive factors.
Figure 5.
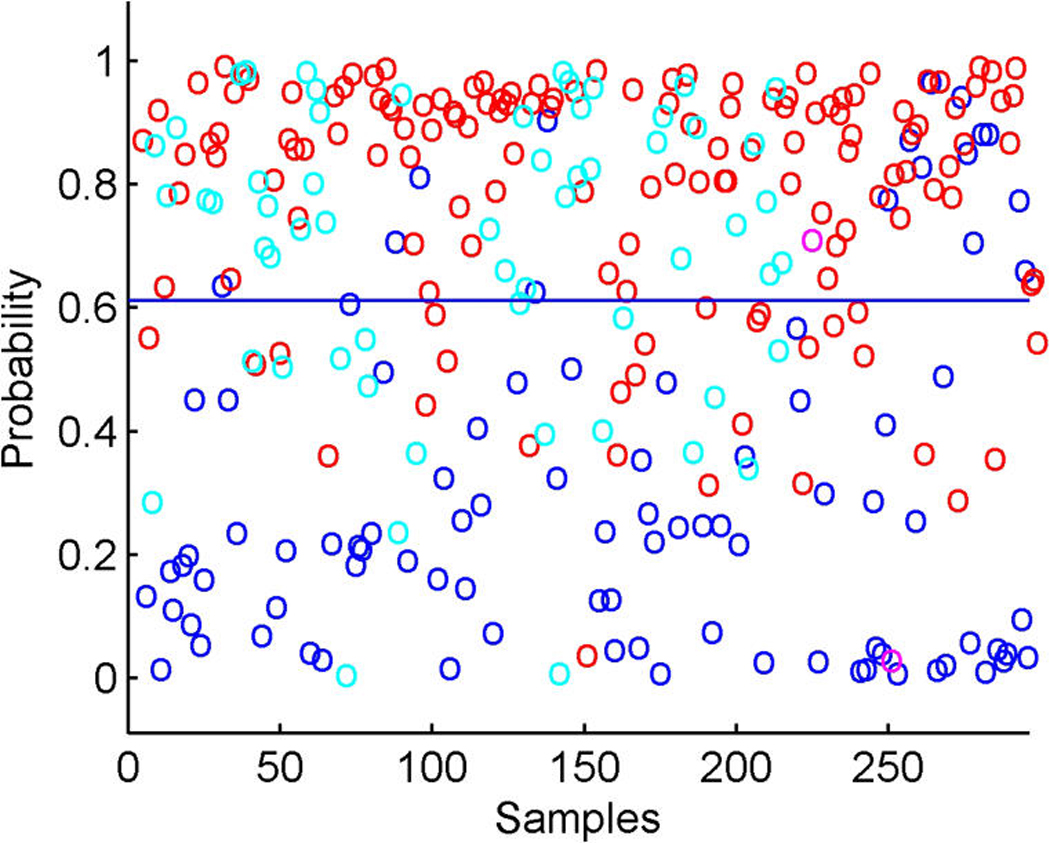
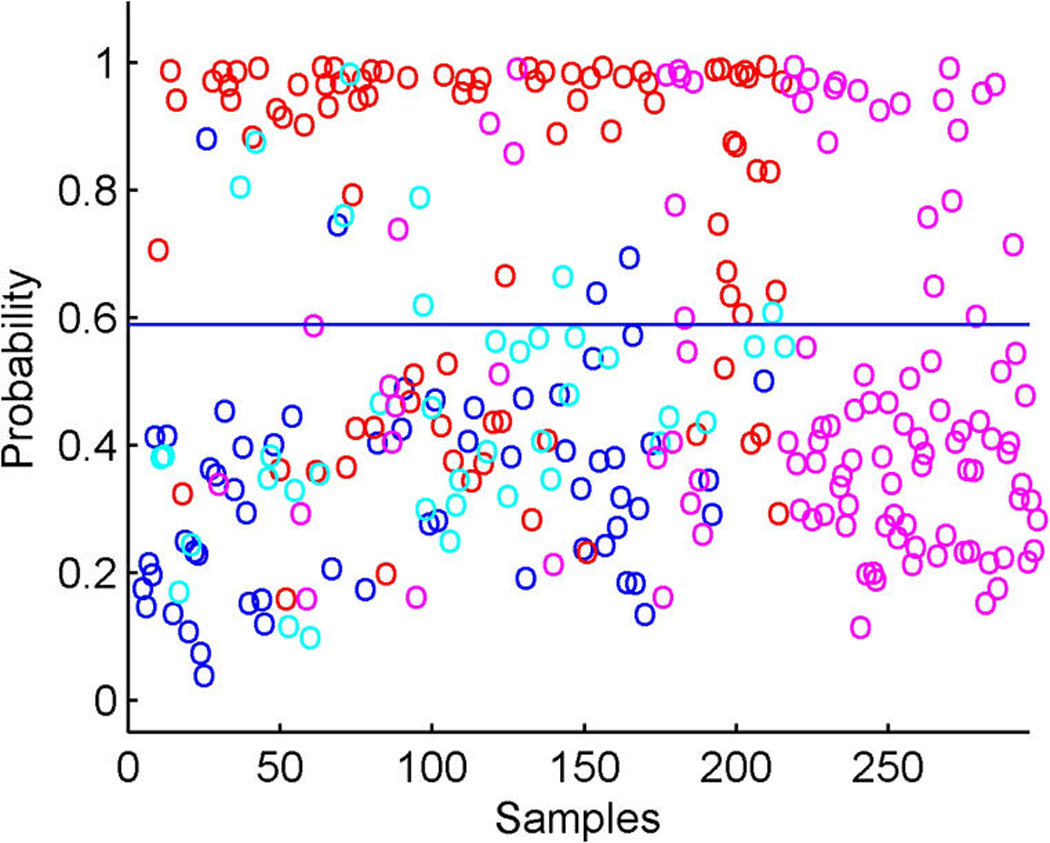
Breast cancer hormonal pathways. Scatterplots of fitted probabilities of ER+ (a) and HER2+ (b) from the overall factor regression model that includes probit components for these two binary responses. Color coding indicates hormonal receptor status; in each case, red, positive; blue, negative; magenta, missing; cyan, indeterminate.
Finally, we point out that this identification of expression predictors of the ER and HER2 variables is viewed as an exploratory component of the overall analysis that is focused on increasing our understanding of structure related to these biological phenomena. We have not set out to generate predictive models for ER and HER2 alone, but aim to include those components to aid in the identification of factor patterns underlying related genes. A direct prognostic development could be overlaid, but that is not the primary goal here.
5.5 Non-Gaussian Factor Structure Linked to Biology
Non-Gaussianity is apparent in Figure 4. Other scatterplots suggest elliptical structure for some factor dimensions, although the full joint distribution is evidently highly non-Gaussian. The biologically interpretable groupings are identified by using the nonparametric model that is designed to flexibly adapt to what can be a quite marked non-Gaussian structure.
Non-Gaussianity in the factor model naturally feeds through from the observed non-Gaussian structure in expression of many genes. The posterior distribution for the DP model for latent factors is easily simulated, so that we can simulate from the posterior predictive distribution of a future latent factor vector λn+1; this leads to simulation of the approximate predictive distributions for future outcomes xn+1 by fixing model parameters in the loadings and noise variance matrixes at posterior estimates. Suppose, based on the posterior from the model fitted to x1:n, that a specific gene g clearly is not associated with the regression component; that is, the posterior for the regression parameters βg is highly concentrated around 0. For such a gene, all of the action is in the latent factor component, so that simulating the posterior predictive distribution for λn+1 translates, through the addition of simulated noise terms νg,n+1, directly to predictions for xg,n+1.
Figure 6 shows two of the bivariate margins involving four genes highly associated with one or more factors, but not with regressors. These are the HER2/ERB–B2 gene and the ER-related FOXA1 in (a), and the two genes TFF3 and CA12, which are highly related to ER, in (b). The predictive simulation generates large samples of the full joint distribution of all genes in the model, and the samples on these two selected bivariate margins are contoured here. The data on these genes are scattered over the contours, and the concordance is some reflection of model adequacy, at least in these dimensions. Sequencing through many such plots provides a useful global assessment of overall model structure and at least some guide to genes for which the model may be lacking. These plots also highlight the relevance of the non-Gaussian factor model structure that feeds through to represent the observed non-Gaussianity of expression gene by gene.
Figure 6.
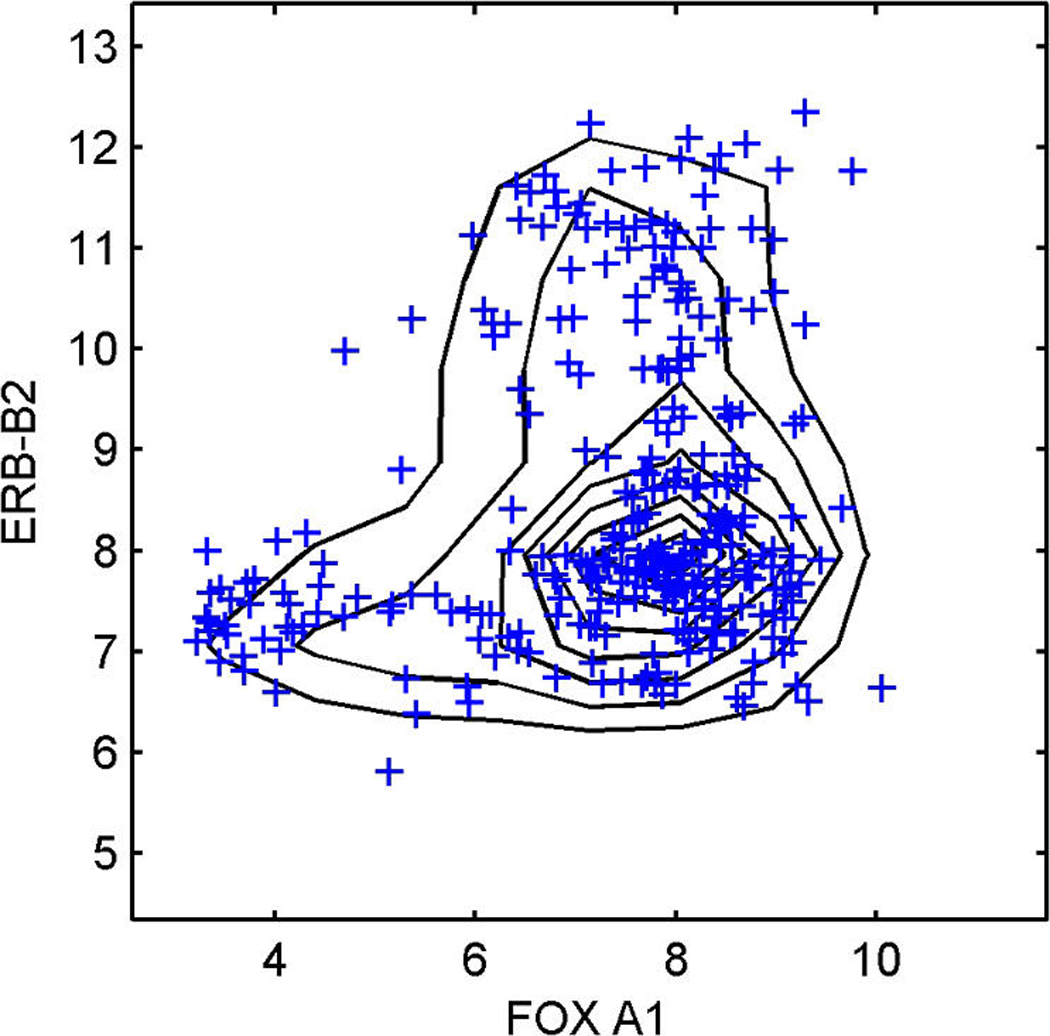
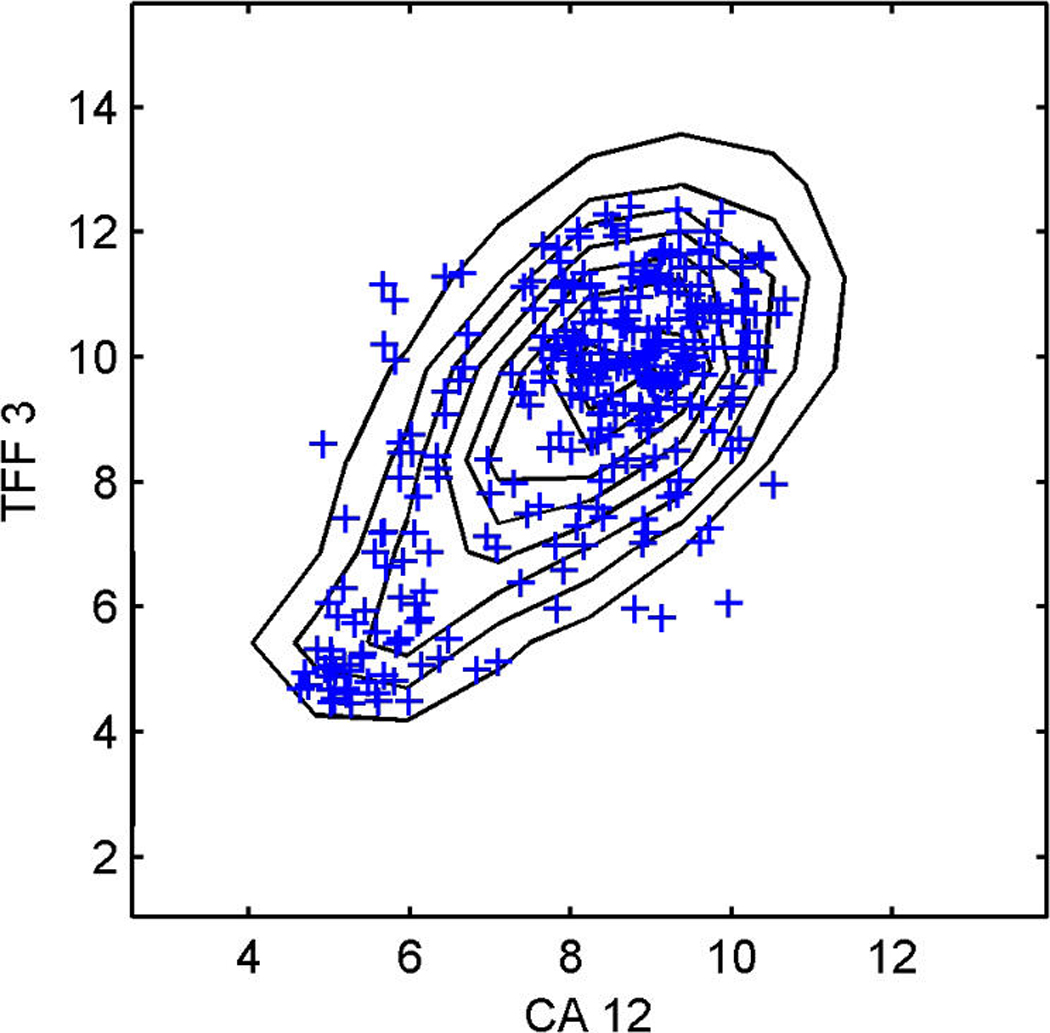
Breast cancer hormonal pathways. The plots display the approximate predictive density contours and observed data for two selected bivariate margins on four genes, HER2/ERB–B2 (a) and the ER-related FOXA1, TFF3, and CA12 (b), with the observed data marked as crosses.
6. STUDY 2: THE P53 PATHWAY AND CLINICAL OUTCOME
6.1 Goals, Context, and Data
A second application in breast cancer genomics explores gene expression data from a study of primary breast tumors described by Miller et al. (2005). One original focus of this study was the patterns of tumor-derived gene expression potentially related to mutation of the p53 gene, and we explore this as well as broader questions of pathway characterization and links between expression factors and cancer recurrence risk. The p53 transcription factor is a potent tumor suppressor that responds to DNA damage and oncogenic activity. The latter is seen in the connection of the p53 pathway to the primary Rb/E2F cell signalling pathway. The consequences of p53 activation, either by oncogenic events or DNA damage, is an arrest of the cell cycle or an induction of cell death (apoptosis). Discovery of structure in expression patterns that may relate to known, putative, or novel connections between these pathways is certainly of current interest. Mutations in p53 occur in roughly 50% of human cancers. Multiple direct mutations lead to deregulation of key aspects of the p53 pathway and thus play roles in increasing the risk and aggressiveness of cancer due to the inability to properly program cell death. Various current anti-cancer therapies target the p53 pathway as a result. The limited efficacy of such therapies is another motivation for studies that enrich our understanding of the biological interactions in the Rb/E2F/p53 network and that aid in characterizing functional interactions of signalling mechanisms central to the control of cell proliferation and oncogenic processes.
This data set (Miller et al. 2005) contains expression data on n = 251 primary breast tumors. The profiles were created on Affymetrix u133a+ microarrays, which, after RMA processing and screening to identify genes (probesets) showing nontrivial variation across samples, generates about p = 30,000 genes. Coupled with the expression data are clinical and genetic information on each patient. In each patient the p53 gene was sequenced for mutations at a number of loci, thus generating one initial key binary variable: p53 mutant versus wild-type. (No other mutational information was made available.) Clinical and pathological information includes recurrence survival and the usual binary ER status from IHC assays. Miller et al. (2005) aimed to identify a gene expression signature associated with p53 mutational status; toward this end, they started by filtering the data set to find genes that were correlated with p53 status through a series of univariate logistic regressions. Genes with p value >.001 were excluded from the analysis. From the remaining genes, they identified a 32-gene signature by evaluating a collection of supervised learning methods including diagonal linear discriminant analysis, k nearest neighbors, and support vector machines. The concept under investigation was that such a signature would better represent perturbations in the p53 pathway and could be used as a refined clinical risk predictor in the same way that expression signatures of ER, HER2, and so forth will provide improvements over IHC assays. With this in mind, we have developed a detailed sparse factor regression analysis to thoroughly explore gene expression patterns linked to the broader p53 pathway and its neighborhood connections to other pathways. Our final analysis seems to effectively dissect p53 activation into latent factors that represent core aspects of known underlying biology, identifies p53-related factors that are unrelated to mutations as well as others that are, suggests new pathway connections that tie into cell developmental activities in the Rb/E2F pathways and also contributes, through the factor regression component, to more accurate recurrence risk prediction.
6.2 Factor Model Analysis and Latent Structure Linked to p53
We begin with 25 genes known to participate in the p53 pathway (Sherr and McCormick 2002), none of which were included in Miller’s final 32-gene signature. The model includes three response variables: the binary p53 mutational status, the binary ER status, and the continuous, right-censored log of time to death. The MCMC analysis easily incorporates censoring of the continuous response, imputing the censored survival times from relevant conditional distributions at each iteration (see App. A). The evolutionary model search allows the model to evolve and sequentially include genes related to the factors in any current model—beginning with this known nucleating set of p53-related genes—as well as genes associated with the response factors in the current model. Thus the analysis can simultaneously explore subbranches of the p53 pathway while identifying its connections to the outcomes of interest; that is, the regression variable selection process is part of the evolutionary analysis. Using thresholds of θ = .75 for both variable and factor inclusion probabilities, and constraining such that a minimum of 3 genes are required to exceed this threshold on a factor to include that factor in an expanded model, the analysis terminated with a model on 1,010 genes distributed across k = 12 latent factors and q = 3 response factors.
Exploration of genes loaded on each factor provides some annotation of common biological function, as well as potential pathway interpretations of factors. All factors have “top genes” known to be associated with cell cycle and oncogenic activity. Table 2 summarizes some of the top gene–factor pairings, with a brief biological annotation and their association with the binary response p53 mutant/wild-type.
Table 2.
Breast cancer p53 study: Biological annotation for the latent factors defined by the model analysis
| Factor | Top genes | Function | p53 status |
|---|---|---|---|
| 1 | CA12, TFF3, GATA3, SPDEF, FOXA1 |
ER | Yes |
| 2 | PERP, E2F3, EP300, BAX, RB1 |
Cell development, apoptosis |
No |
| 3 | ESR1, SCUBE2, NAT1, BCMP11, MAPT |
ER | Yes |
| 4 | TOP2A, ASPM, CDC2, RRM2, BUB1B |
Cell development, apoptosis |
Yes |
| 5 | ASPN, COL5A2, COL10A1, COl3A1, COL6A2 |
No | |
| 6 | CCL5, CXCL9, CXCL13, LTB, TRGC2 |
Immunoregulation | Yes |
| 7 | FOS, JUN, EGR1, EGR3, ATF3 |
Cell development, apoptosis |
Yes |
| 8 | KRT14, KRT17, KRT5, KRT6B, SFRP1 |
No | |
| 9 | COL12A1, LTB4DH, CSPG2 MYC, RRM1 |
No | |
| 10 | CDKN2A, SFN, SCUBE2, CXCL10, BCL2 |
Cell development, apoptosis |
No |
| 11 | AFF3, VTCN1, CPEB2 ENO2, PERP |
No | |
| 12 | CAV1, GPR116, TGFBR2 CAV2, PLVAP |
No |
NOTE: The p53 status column simply refers to the direct association between p53 mutational status and the posterior mean of the factor scores in a univariate model.
Of the genes in the p53 expression signature of Miller et al. (2005), our factor model identifies all but two, and they all have significant loadings on the p53 factors 1, 3, and 4. Exploring the p53 pathway guided by the mutational status combined with a set of canonical p53 genes also gives us the opportunity to identify other instances of the pathway that are not affected by mutations. This expands our understanding of alternative ways, other than mutation, in which p53 activity may be affected in cancer processes. To illustrate this, we select three genes known to be key players in the p53 pathway: BAX, PERP, and SFN. Miller et al. (2005) observed that the expression profiles of these genes do not relate to p53 mutations, raising the question that this might represent cross-talk between p53 and other pathways. These genes are significantly loaded on factors 2 and 10; these two factors are not directly associated with p53 mutation status, nor do they contribute significantly to p53 binary regression prediction in the analysis. They are composed primarily of genes that participate in cell development and apoptosis, thus reflecting substructure in the p53 pathway that could not be identified simply through mutational status alone.
6.3 p53, ER, and Cancer Recurrence
Table 3 summarizes the estimated probabilities and coefficients of the most highly weighted latent and response factors in the linear predictors of the three response variables. Additional predictive value is generated by the included response factors that link in a number of genes to elaborate on the predictions from the expression factors themselves. Table 4 provides some information on a few of the top genes of the response factors. It is of interest to explore the genes that best define the factors that are implicated in prediction. Miller et al. (2005) noted that p53 wild-type and mutant tumors can be distinguished by molecular differences heavily influenced by three major gene clusters comprising genes involved in immune response, proliferation and estrogen response, respectively (fig. 5 of supplemental material of Miller et al.). All of the genes listed in these published clusters appear in our model in factors 1 and 3 (which we denote as ER factors), factor 4 (which we denote as a proliferation/p53 factor) and factor 6 (which we denote as an immunologic response factor). Each of these listed factors is directly associated with p53 status, as displayed in Table 2. A few examples of relevant genes in each of these factors, using the gene-factor decomposition format presented earlier, are displayed in Figure 7.
Table 3.
Breast cancer p53 study: Coefficient probabilities and estimates for factors contributing to the linear predictors for the three response variables
| Linear predictor | Factor | π̂g, j | α̂g, j |
|---|---|---|---|
| 1: p53 status | Response 1 | 1.000 | .111 |
| Factor 3 | .926 | −.347 | |
| Factor 4 | 1.000 | .617 | |
| 2: ER | Response 2 | 1.000 | .104 |
| Factor 1 | .953 | −.359 | |
| Factor 3 | .910 | .3867 | |
| 3: Survival | Response 3 | 1.000 | .089 |
| Factor 4 | .830 | −.345 | |
| Factor 6 | .882 | .261 |
Table 4.
Breast cancer p53 study: Some of the top genes and their estimated loadings on each of the three response factors
| Response factor | Top genes | α̂g, j |
|---|---|---|
| p53 status | CSPG2 | .721 |
| ASPN | .486 | |
| COL3A1 | −.468 | |
| COL3A1 | −.452 | |
| COL1A1 | −.434 | |
| ER | EGR1 | −.226 |
| NPDC1 | .180 | |
| EFEMP2 | .1607 | |
| MCM4 | .156 | |
| RBM5 | .968 | |
| Survival | COL5A2 | .380 |
| VIL2 | .342 | |
| TRPS1 | .281 | |
| TOP2A | .272 | |
| YY1 | .183 |
Figure 7.
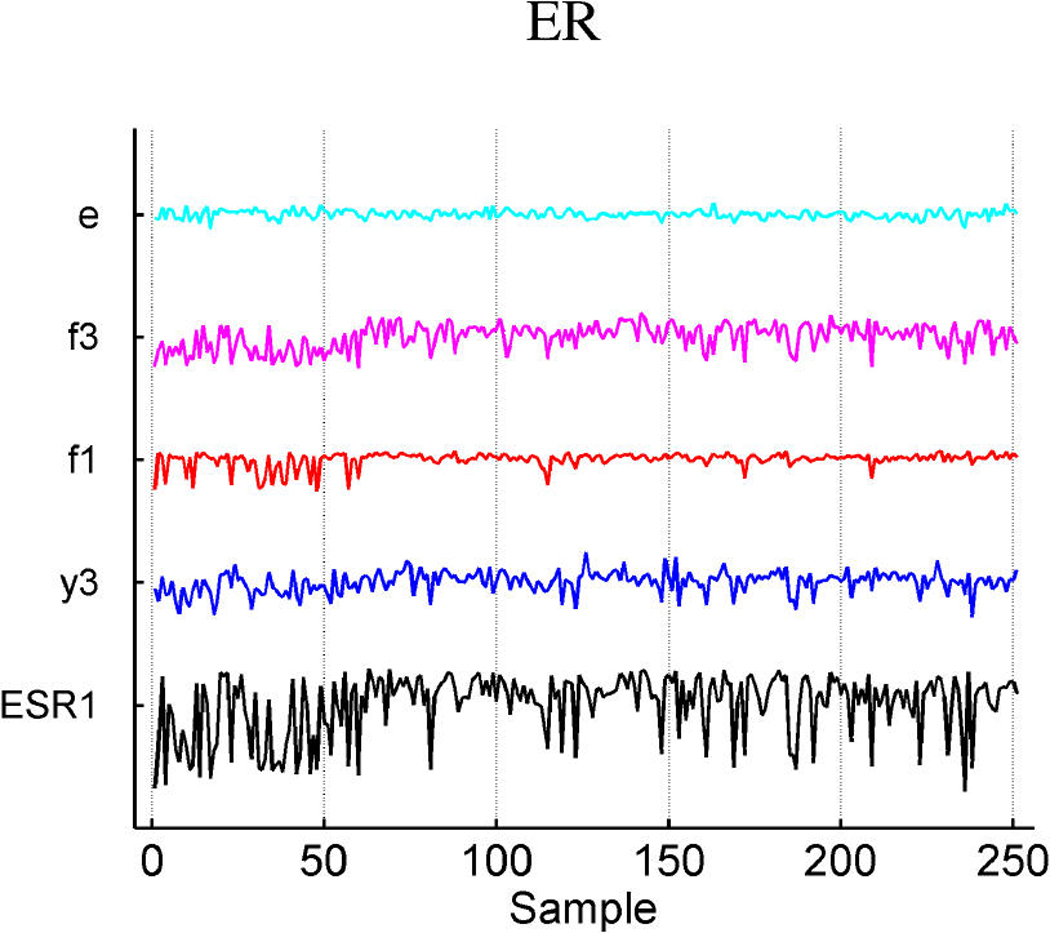
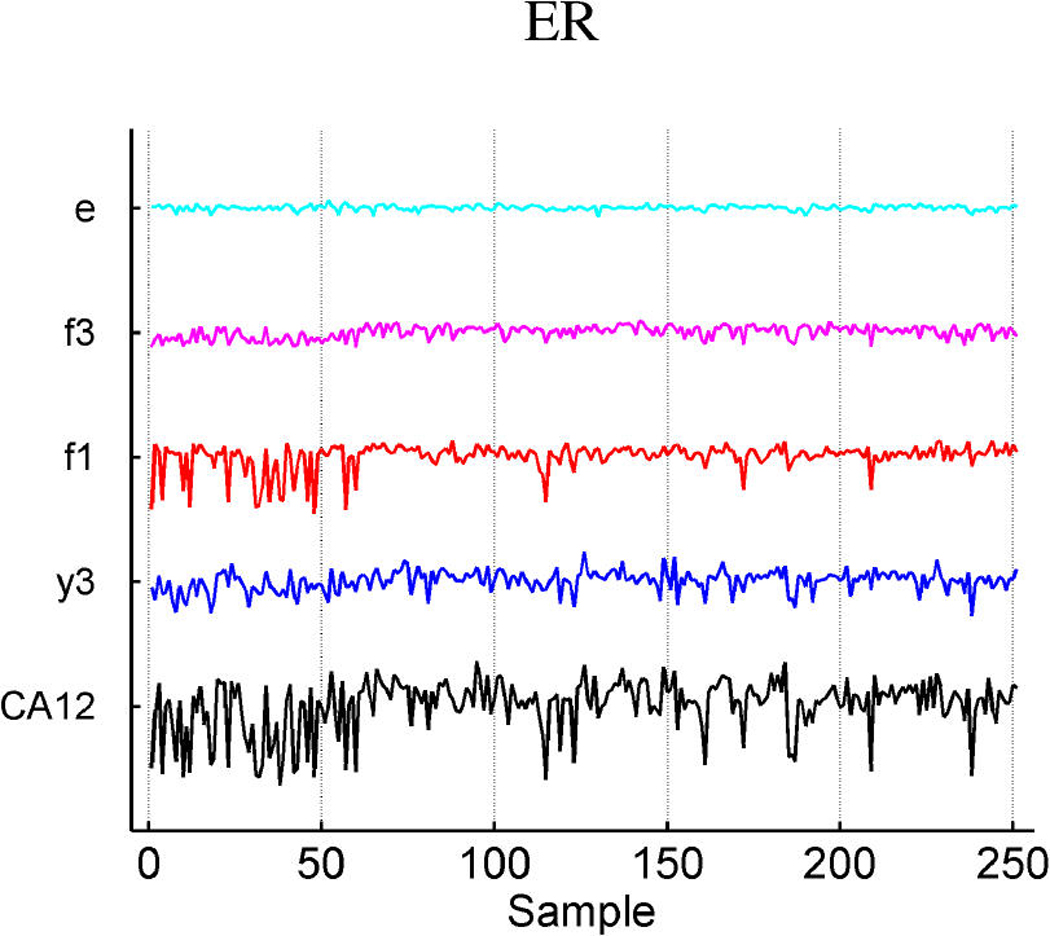
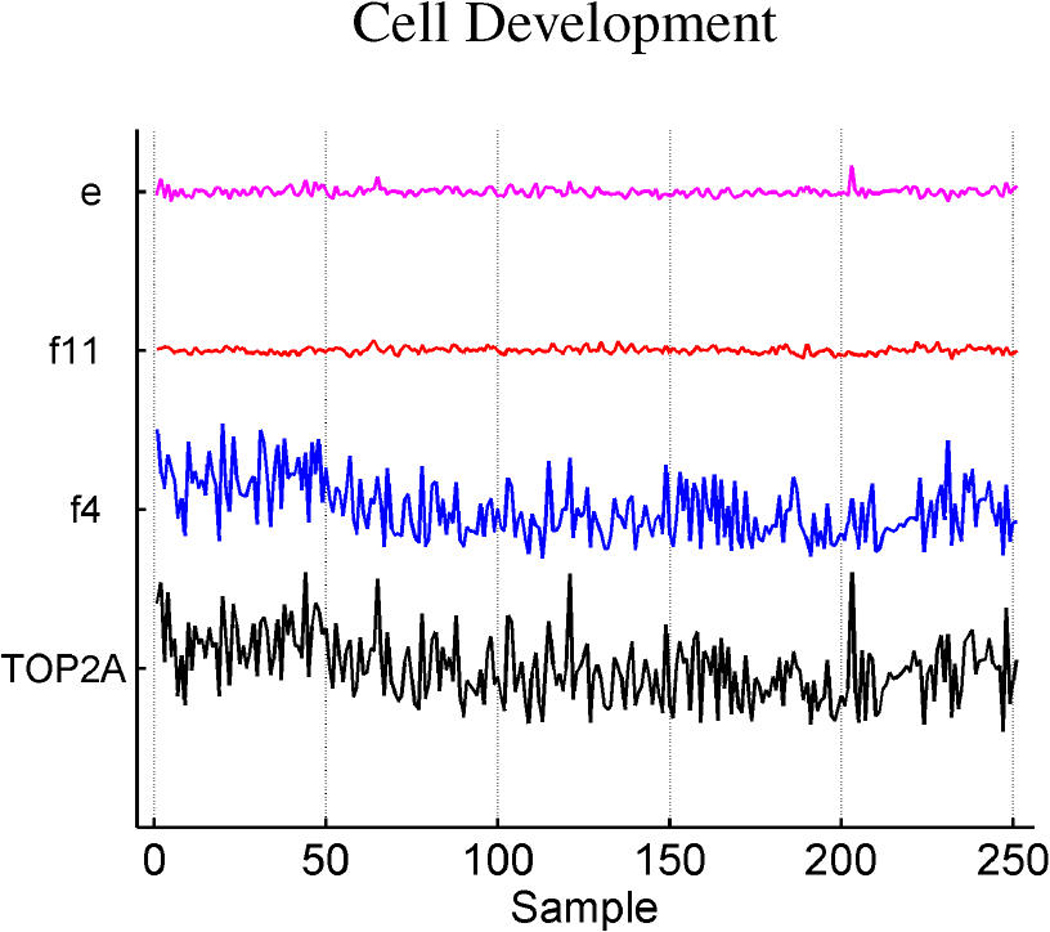
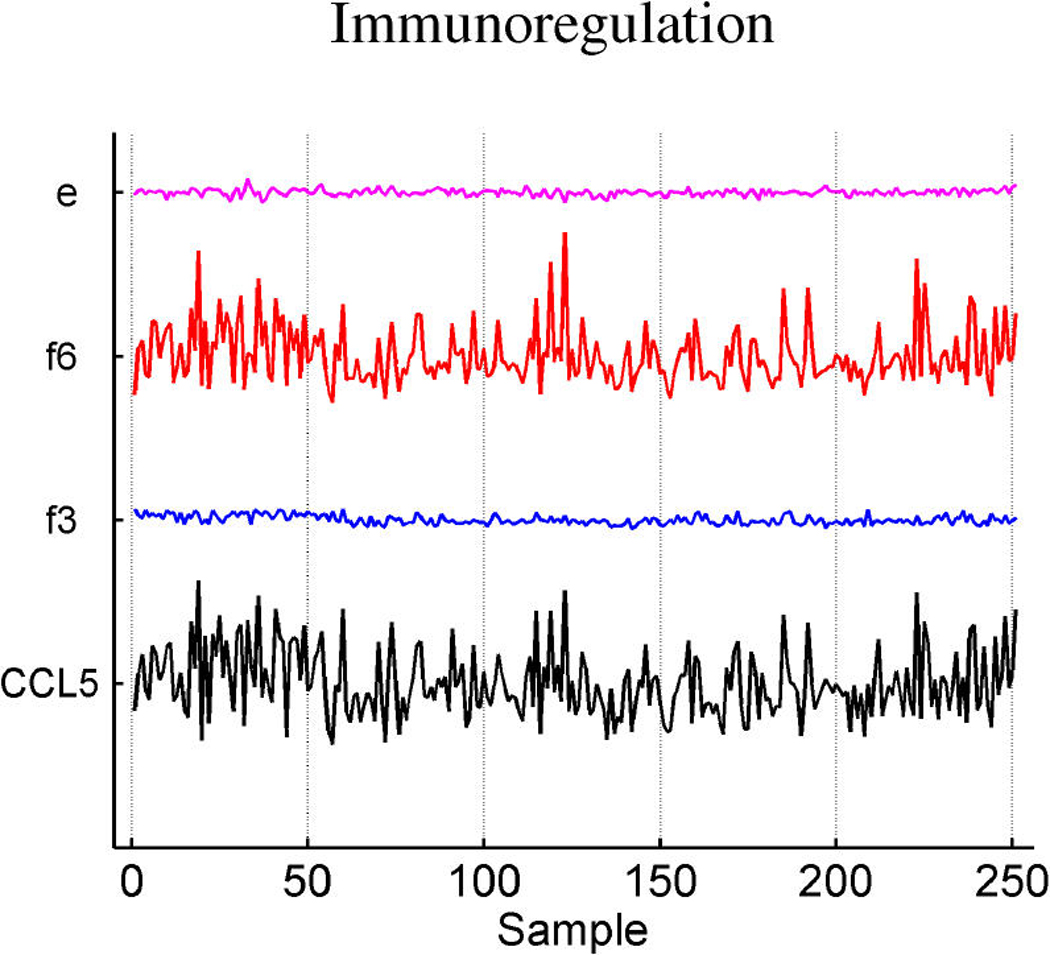
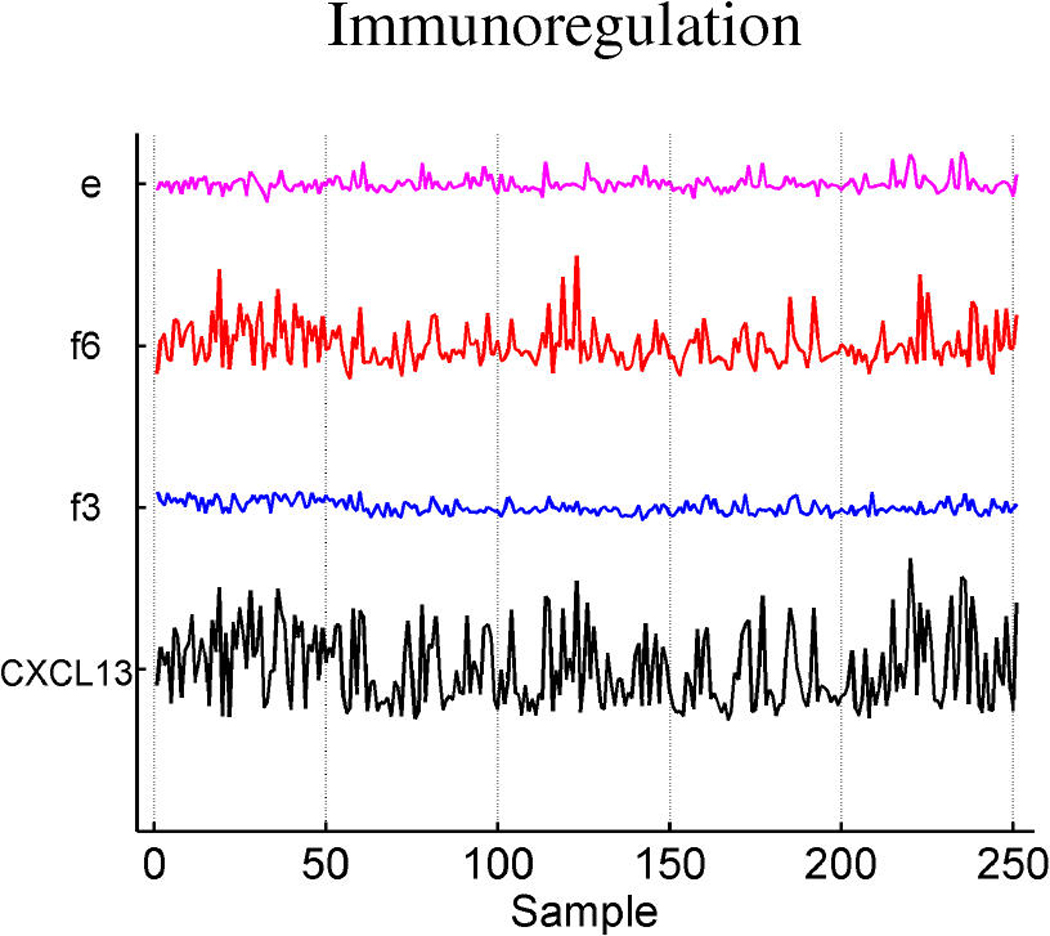
Decomposition of expression over samples of several top genes in the annotated ER factors 1 and 3 (a), cell development factor 4 (b), and immunoregulatory factor 6 (c).
The regression component of the model, and some informal predictive evaluations, are highlighted in Figure 8 and Figure 9. The model analysis summarized is based on fitting to a randomly selected 201 samples as training data, treating the remaining 50 as test or validation samples to be predicted. Figure 8 provides some indication of the within-sample discrimination for the two binary responses, p53 and ER, together with the out-of-sample predictive discrimination. For p53 status, we achieve a predictive accuracy (around 86%) very similar to the classification method of Miller et al. (2005). This observation also holds in the test set in which both methods misclassified the same samples (8 out of 50). As for ER, the in-sample accuracy of the model is approximately 85%, with 88% in the test set. The approach of Miller et al. (2005) does not allow for multiple outcome variables, so predictions of ER status are not provided. Evidently, the combined factor regression model is capable of quite accurate prediction of both ER and p53 mutational status; these predictions are based on the integration over a few latent factors rather than a single direct signature, a point that the clinical genomics community perhaps has often undervalued in studies to develop genomic prognostics (Huang et al. 2003; Pittman et al. 2004; West, Huang, Ginsberg, and Nevins 2006).
Figure 8.
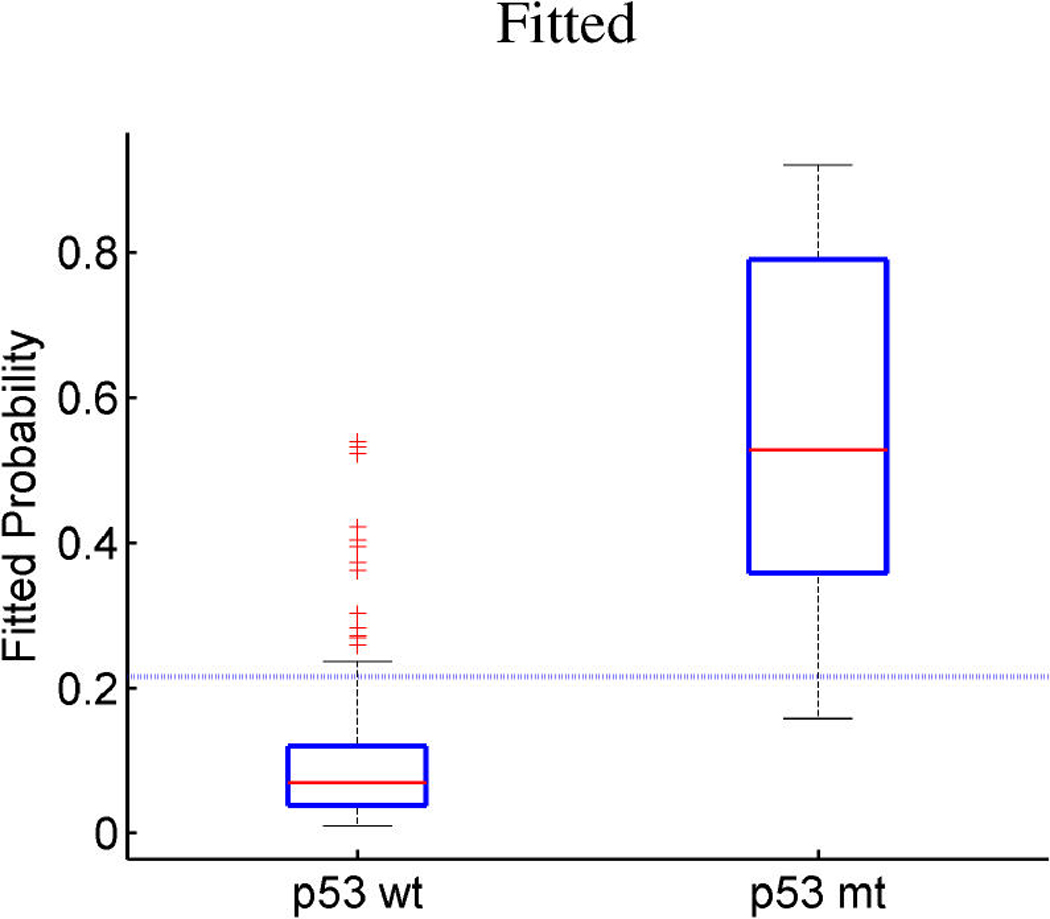
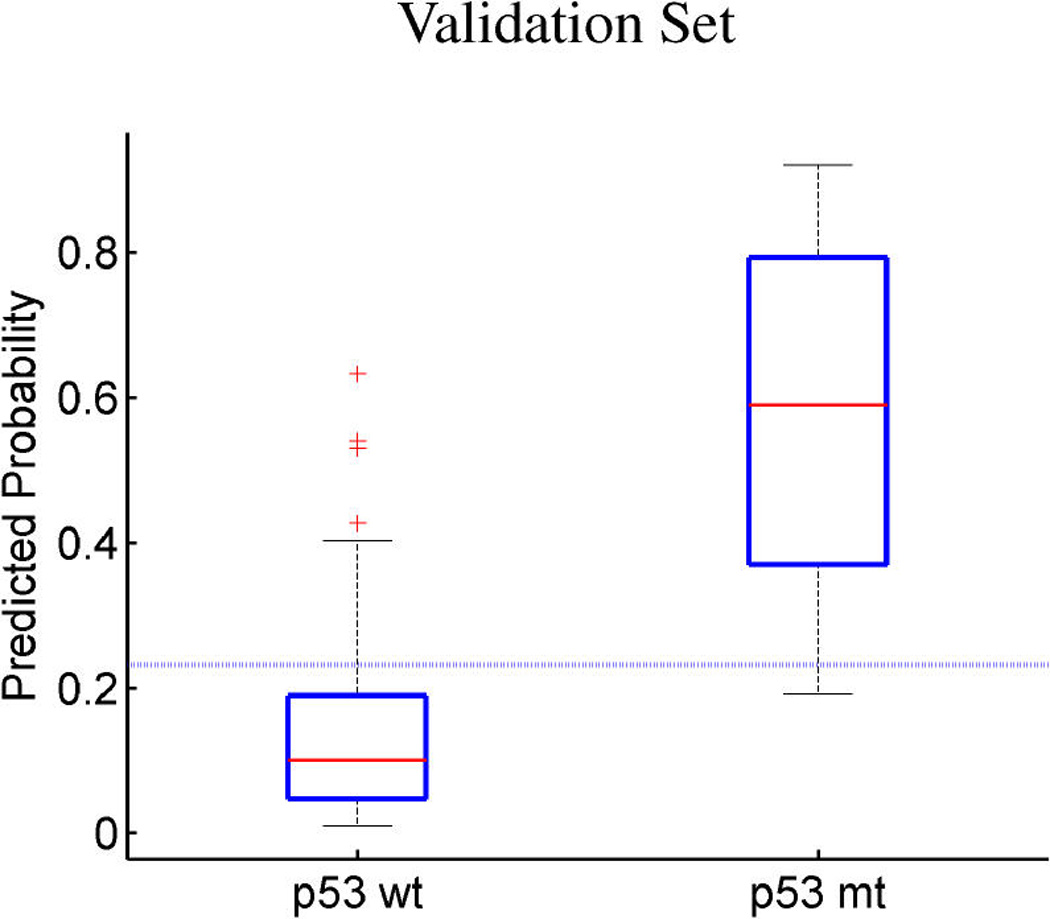
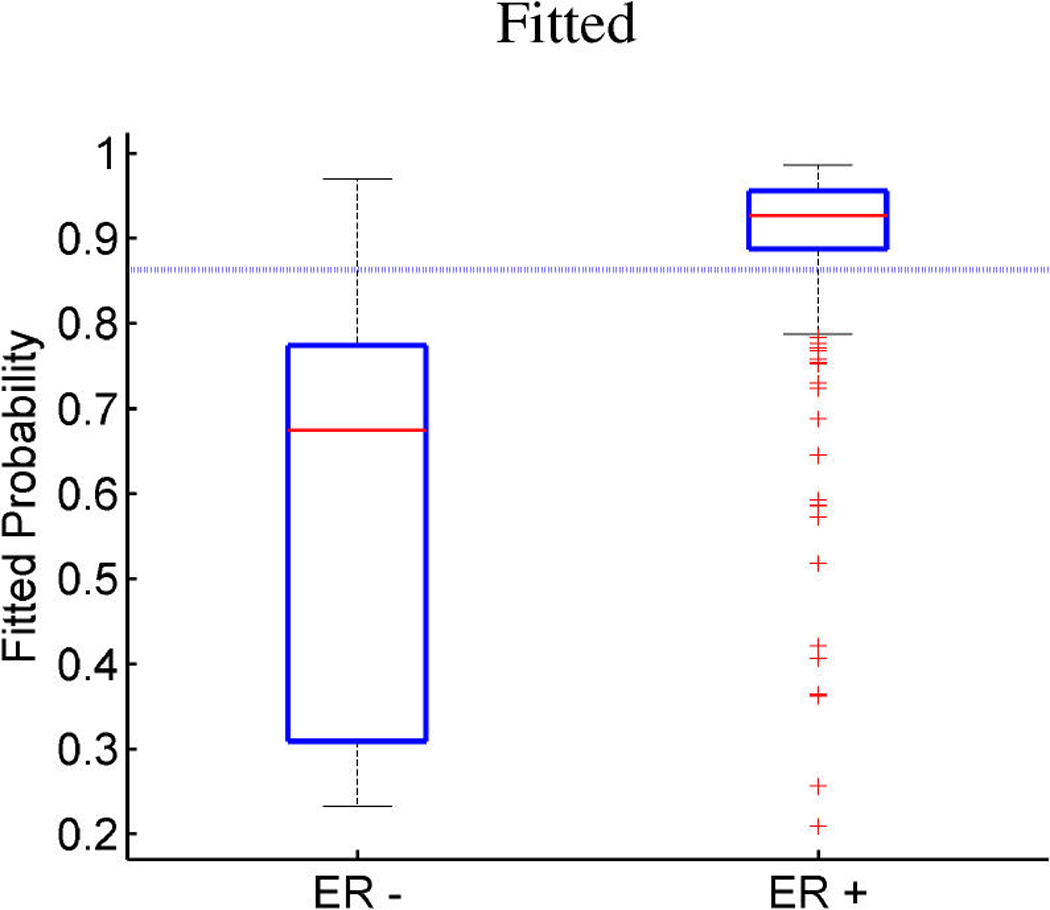
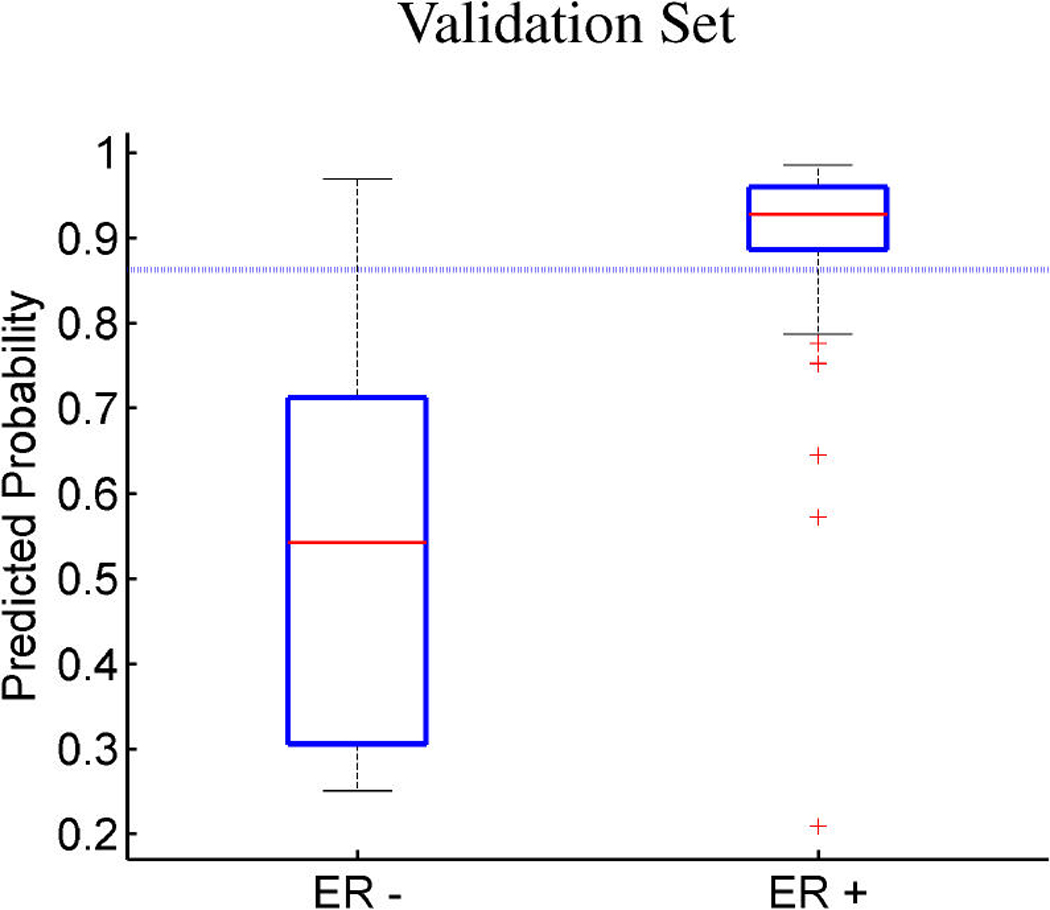
Breast cancer p53 study. Boxplots of fitted (a, c) and out-of-sample predicted (b, d) probabilities of p53 mutant versus wild-type (a, b) and ER positive versus negative (c, d).
Figure 9.
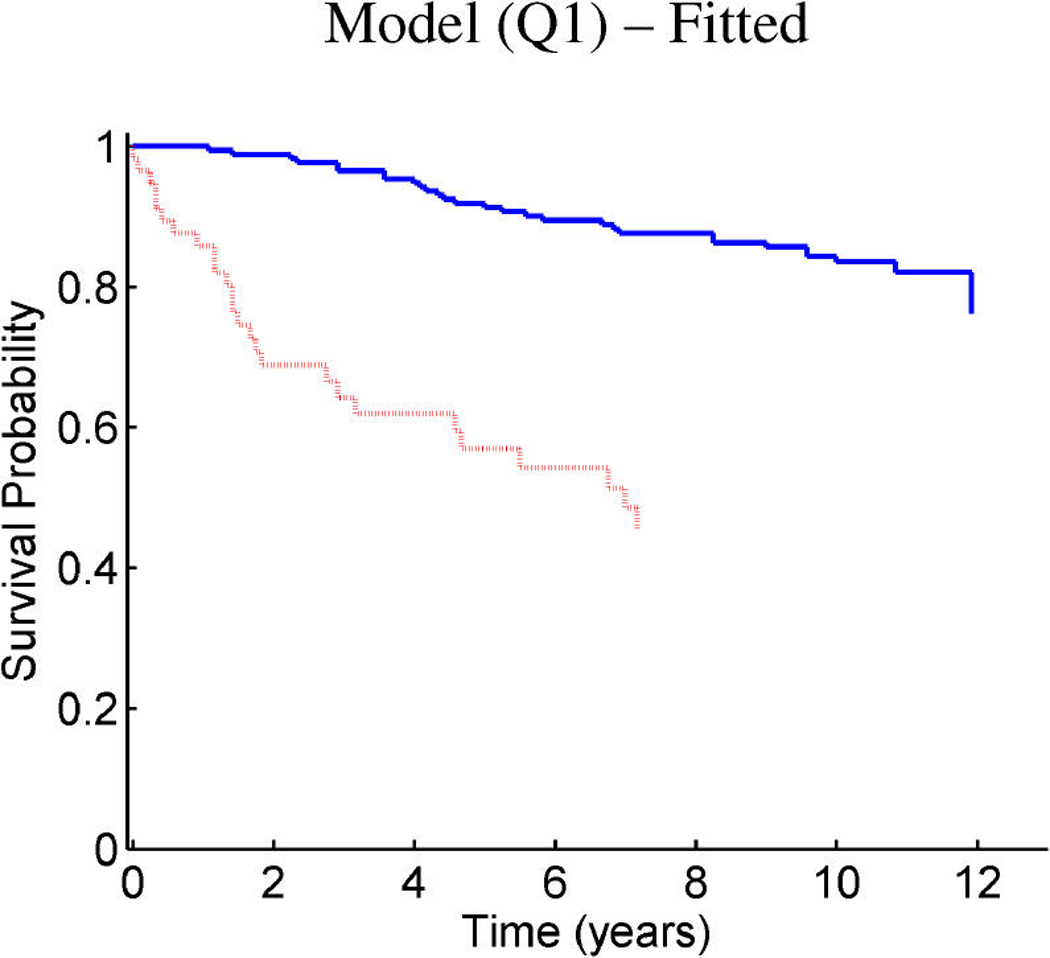
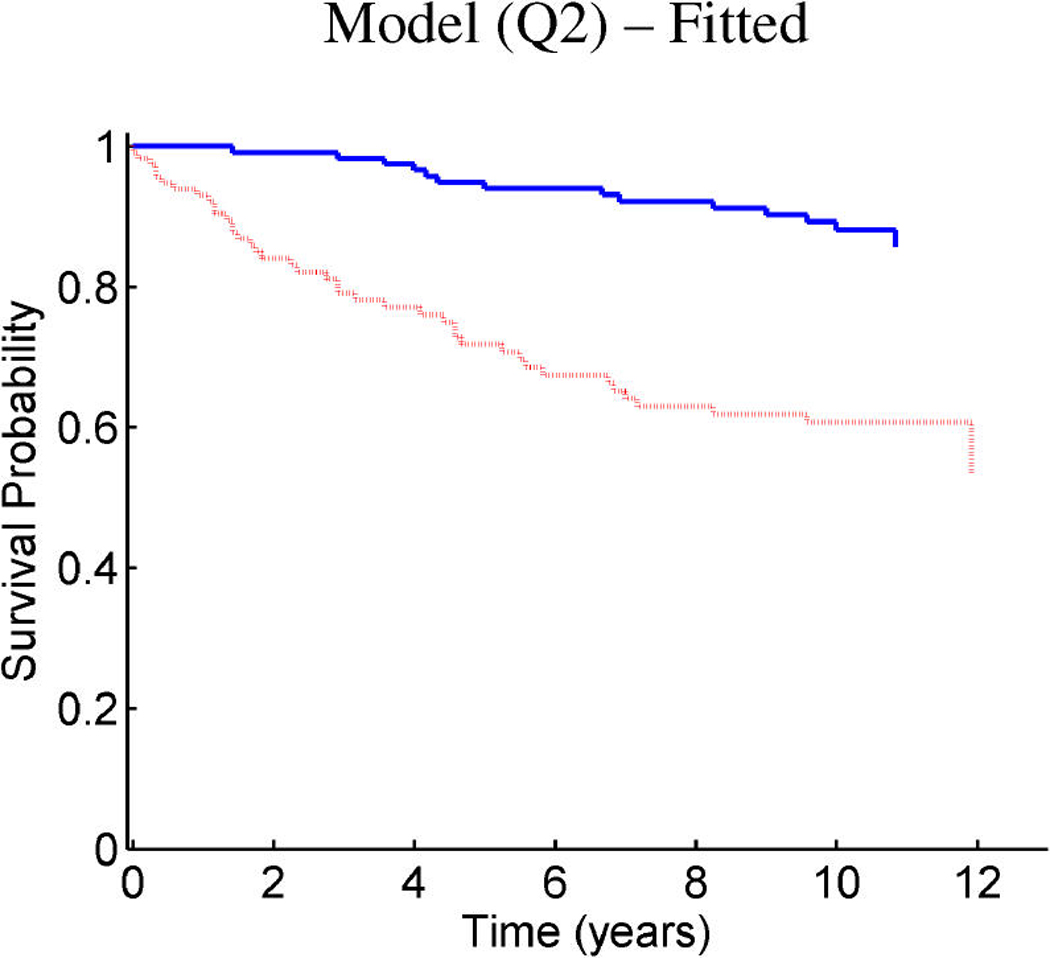
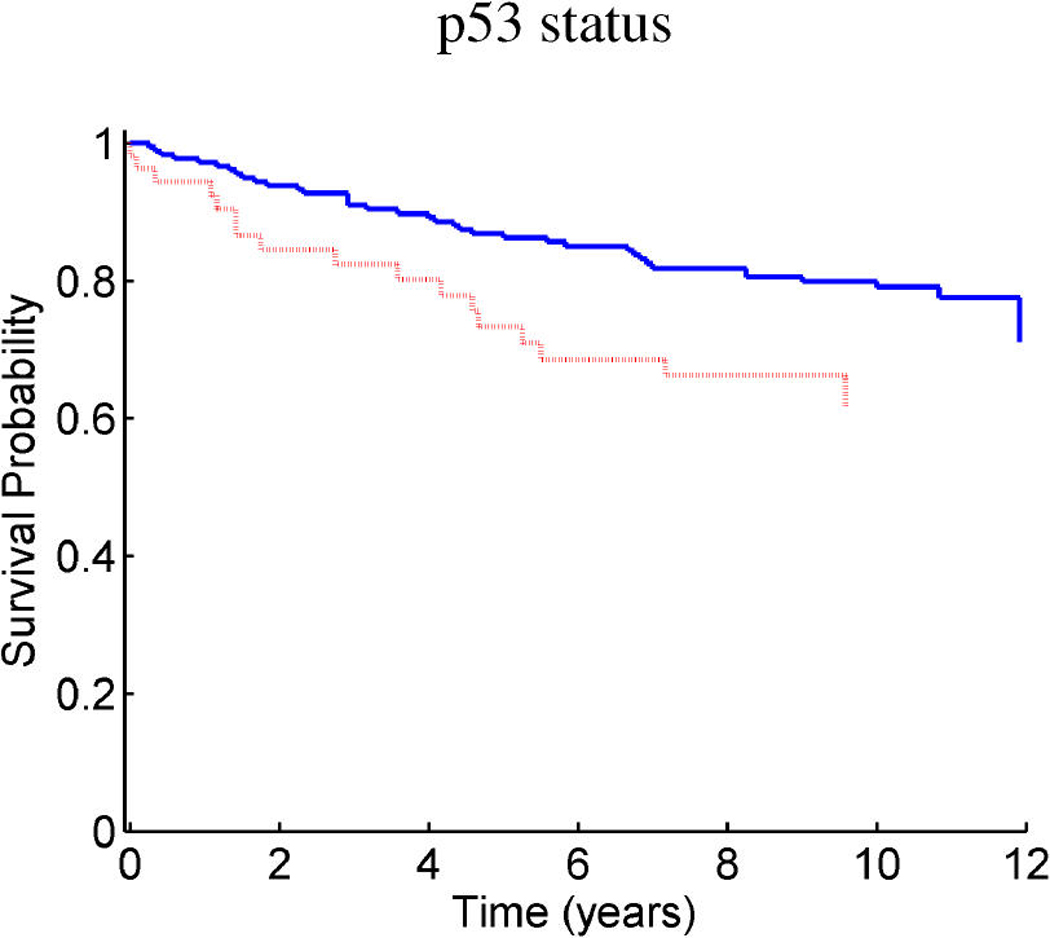
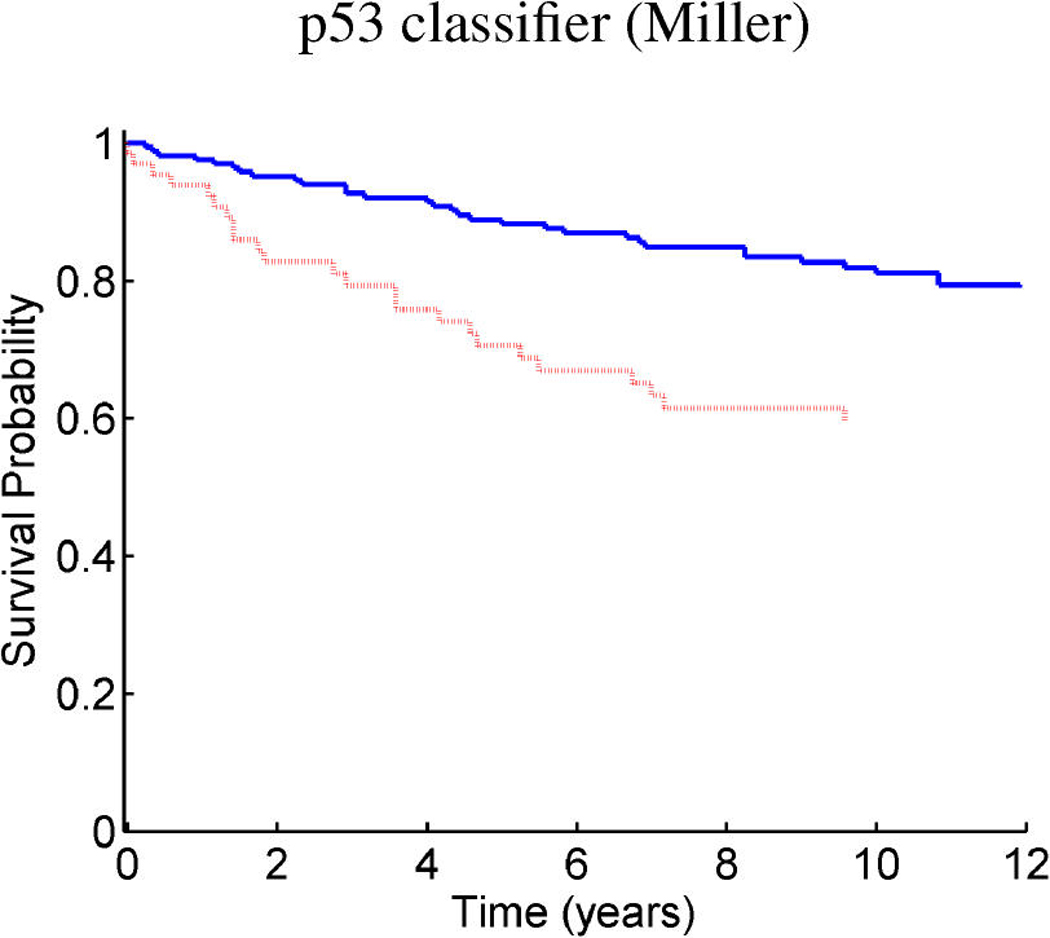
Breast cancer p53 study: Kaplan–Meier survival curves for the training samples (n = 201) split according to the indicated thresholds. (a) Q1 represents thresholding at the first quartile of the fitted linear predictor. (b) Q2 represents thresholding at the median. (c) Stratification simply on p53 wild-type versus mutant. (d) Stratification based on the p53 classification proposed by Miller et al. (2005).
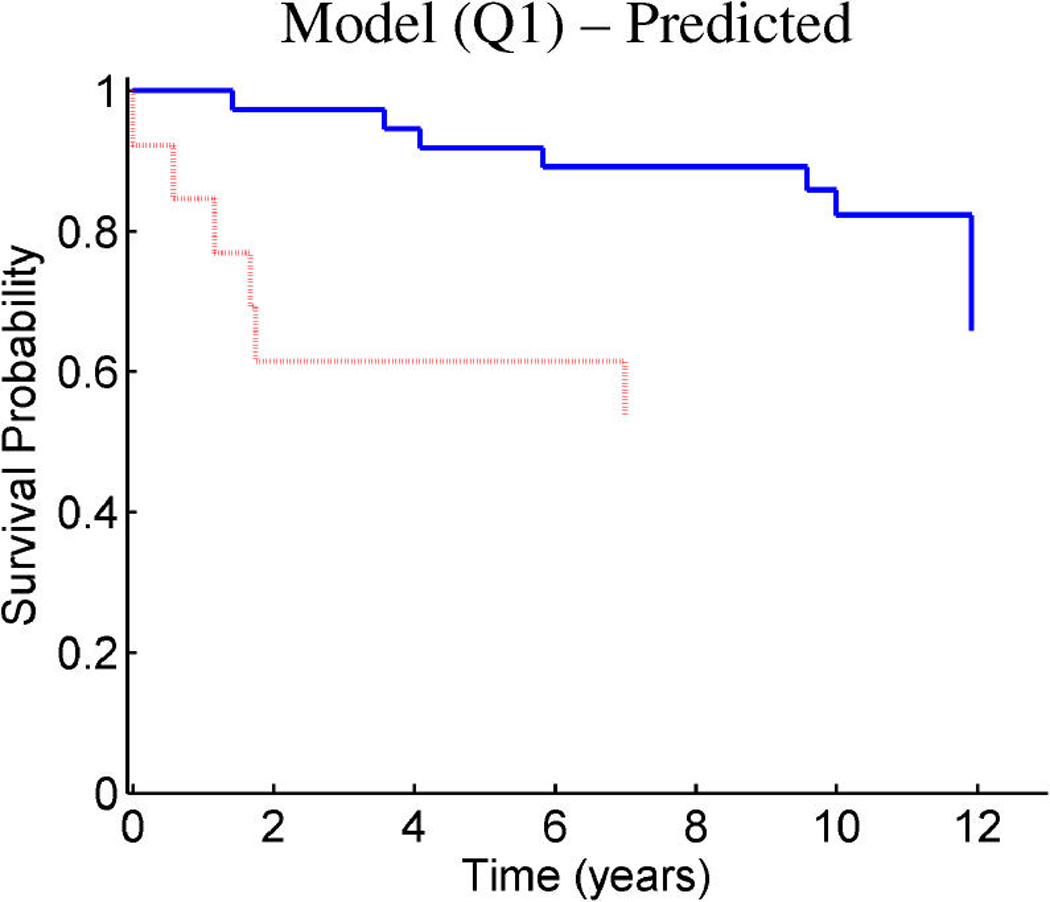
Figure 9 and Figure 10 provide similar insight into the nature of the predictions for cancer recurrence, displayed in a format consistent with the use of expression signatures to indicate patient stratification into risk groups (Huang et al. 2003; Pittman et al. 2004). Here both the test and validation samples are split by thresholds on the linear predictor of the survival regression on factors. The concordance between the resulting displays for test and training data is excellent and supports the statistical validity of these model predictions. For comparison purposes, we also present the stratification generated by the p53 classification based on the 32-gene signature proposed by Miller et al. (2005) in both test and training data. The potential practical relevance lies in the fact that these predictions are more accurate than those based on stratification purely by p53 status (because they improve on the predictions of Miller et al., which improve on p53) as is currently commonly used in clinical practice. Compared with the signature of Miller et al., our contribution goes beyond the improvements in the stratification of subpopulations as, more importantly, further investigation of the factors involved in predicting survival allows for a deeper understanding of the biological mechanism underlying the aggressiveness of the disease.
Figure 10.
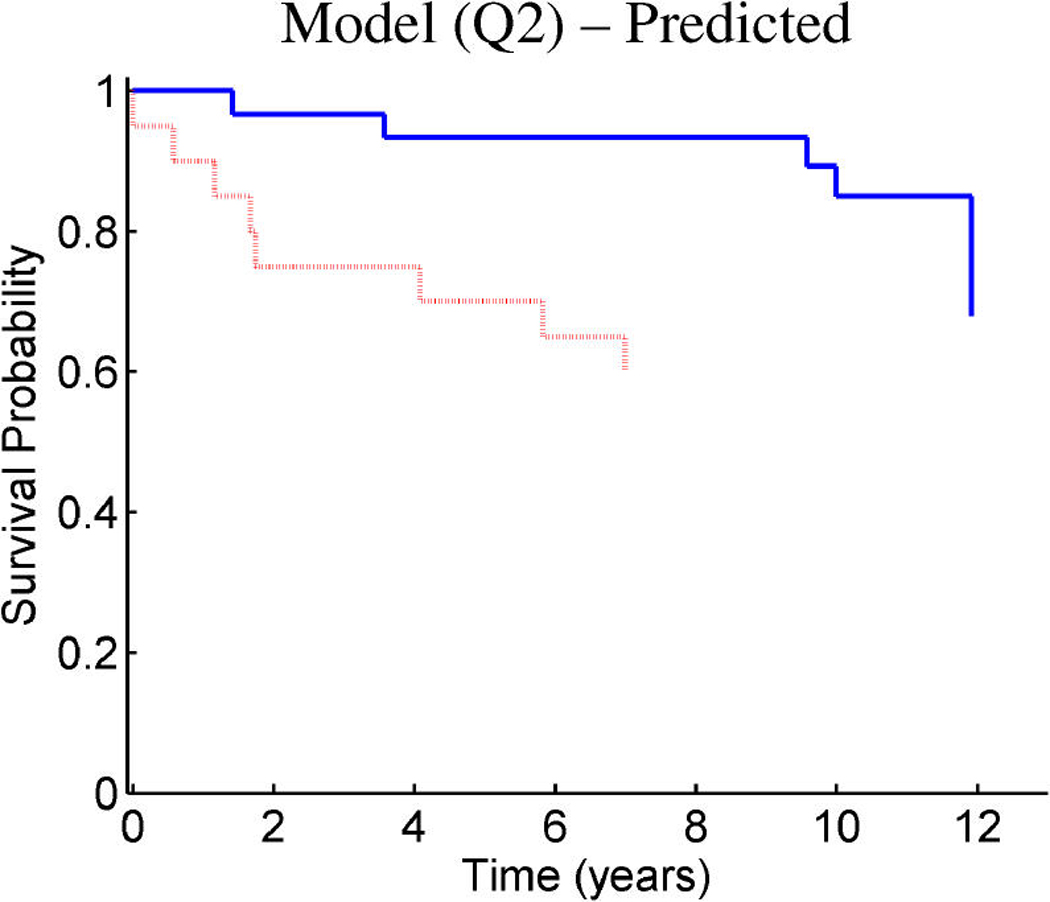
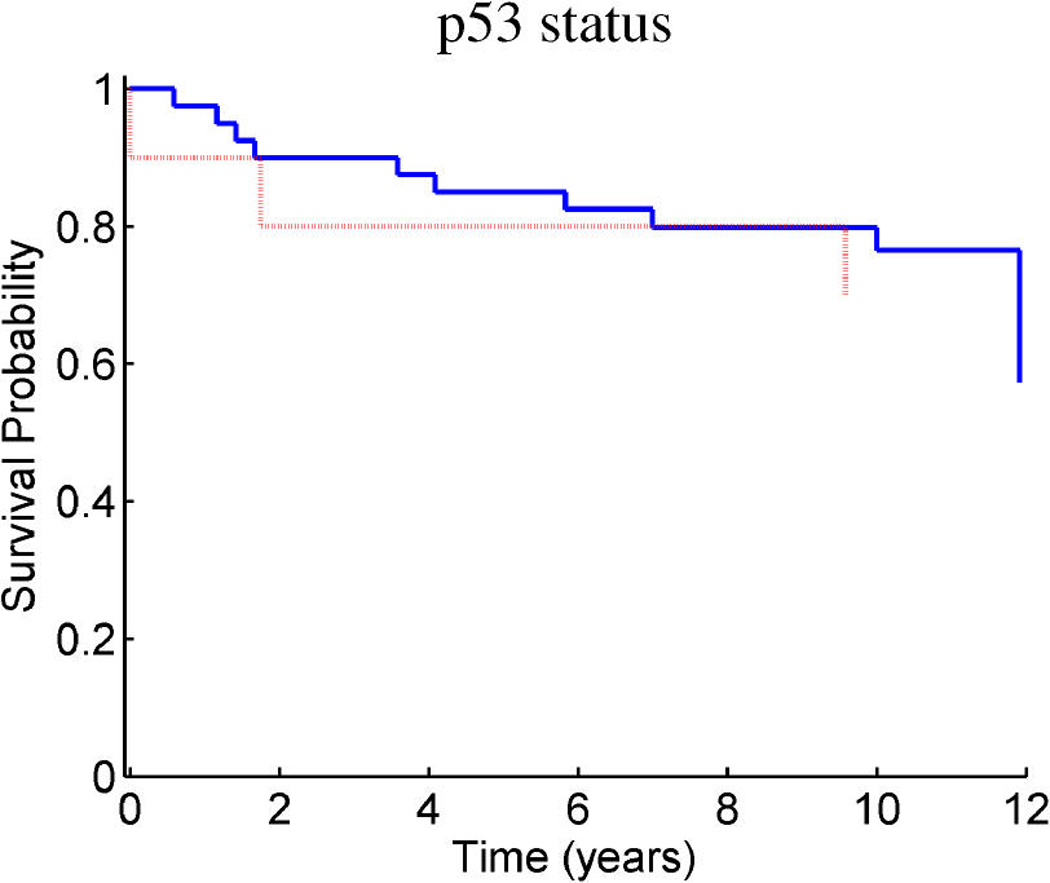
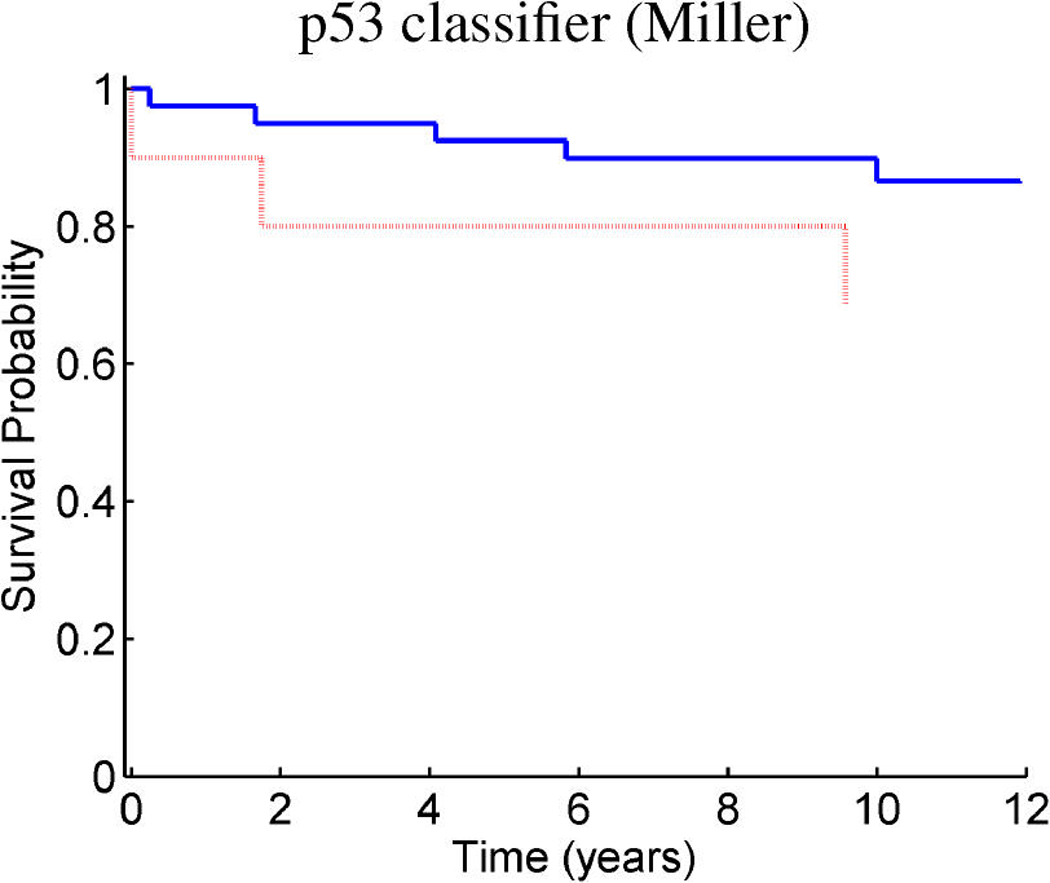
Breast cancer p53 study: Kaplan–Meier survival curves for the test samples (n = 50) split according to the indicated thresholds. (a) Q1 represents thresholding at the first quartile of the predicted linear predictor. (b) Q2 represents thresholding at the median. (c) Stratification simply on p53 wild-type versus mutant. (d) Stratification based on the p53 classification proposed by Miller et al. 2005.
Exploration of the p53 pathway guided by both mutational status and transcriptional activity of genes known to be associated in the biological process of interest is a key strategy in the generation of new biological hypotheses. The predictive power of the identified subpathway components provides additional evidence that the insights generated by the models are worth investigating. Learning methods, such as those used by Miller et al. (2005), are able to generate good predictions of outcomes, but they suffer from lack of interpretability and biological characterization and thus are unable to achieve the final goal of our studies. As an example, in this analysis, one of the factors with direct association with p53 (factor 7), designated a cell development factor, had a series of genes related to the RAS pathway through oncogenes FOS and JUN; the RAS/FOS–JUN pathways are known to link into the Rb/E2F network. This discovery of significant gene expression factor structure linked to the complex p53 pathway suggests a connection between two very important branches of the major cell signalling network, raising questions to be explored and highlighting the potential discovery uses of this analysis.
7. CLOSING COMMENTS
Sparsity of model structures and parameterizations is fundamental to the scaling of scientific models to the higher-dimensional problems that are becoming common in many areas. Gene expression genomics is one such active arena. Models of multivariate distributions in high dimensions and regression for prediction when there are many candidate predictors yield practicable methodologies only if the effective dimension is explicitly or implicitly reduced. Sparsity—in terms of low-dimensional relationships underlying high-dimensional patterns of association of many variables and defined though parametric and conditional independence constraints—is key to this reduction. We have demonstrated some of the utility of sparse factor models in these applications and are using this approach in a number of related studies in cancer genomics, as well as noncancer areas.
The breast cancer genomics applications here illustrate a range of uses of the sparse factor modeling framework. Key elements of the model framework include the use of new sparsity-inducing prior distributions over factor loadings and regression coefficients alike, with the ability to more adequately screen out insignificant variable–factor pairings and highlight (with quantitative probabilistic assessments) associations of interest; the isolation of idiosyncratic noise terms; the coupling of response prediction with factor analysis in an overall framework; the ability to handle missing or censored responses and missing data in the multivariate outcomes x space itself; and the integration of non-Gaussian, nonparametric factor components that are practically relevant in reflecting structure in common underlying patterns and their implications for non-Gaussian marginal data configurations in x space, among others. Key elements of the model fitting and analysis framework include implementation of efficient MCMC methods for analysis of a specified model, the use of evolutionary stochastic search methods for model extension based on an initial specified set of variables, and the investigation of model implications through evaluation and visualization of variable decompositions, among others.
It is clear that this modeling framework is of broader utility beyond the extensive development of applied studies in a number of genomics applications and may be considered for applications in such areas as large-scale financial time series, where it represents a natural extension of existing factor modeling (Aquilar and West 2000). In terms of methodology, the work opens up some challenging questions related to computational developments. One important question is the convergence characteristics of MCMC in the sparse factor models that couple sparsity priors with latent variables under a DP model. Our work has involved extensive repeated analyses of these and many other data sets, as well as experimentation with simulated data sets (Carvalho 2006), and choices of Monte Carlo sample sizes and other control parameters have been based on this cumulated experience. These samplers can be quite “sticky,” and there is certainly interest in exploring modifications of the basic Gibbs samplers that we use here to investigate the potential for improving mixing and convergence properties from a practical standpoint. This is one of our current research areas, and we know that this work has generated interest in these questions among other research groups that are actively developing MCMC methods for highly structured stochastic systems. A linked topic for further research is to a deeper investigation of the connections between the evolutionary stochastic model search approach and related search methods, including projection pursuit methods and the shotgun stochastic search approach for regression variable selection (Hans et al. 2007) that has proven successful in graphical modeling (Dabra et al. 2004; Jones et al. 2005) and a range of prognostic applications in “large p” regression variable uncertainty problems in gene expression genomics (Rich et al. 2005; Dressman et al. 2006). Additional topical investigations include extensions of the DP latent factor model, investigations of nonlinear variants of the overall framework using kernel regression methods, and related studies of theoretical aspects of this approach to joint distributional modeling in prediction problems that link to the use of unlabelled data (Liang, Mukherjee, and West 2007). Further development and refinement of the software used for all analyses here, which is available to interested readers (see App. B), also are in process.
Supplementary Material
Acknowledgments
The authors are grateful for the constructive comments of the editor, associate editor, and three anonymous referees on the original version of this manuscript. Support was provided by National Science Foundation (NSF) grants DMS-0102227 and DMS-0342172 and National Institutes of Health (NIH) grants NHLBI P01-HL-73042-02 and NCI U54-CA-112952-01. Any opinions, findings, and conclusions or recommendations expressed in this work are those of the authors and do not necessarily reflect the views of the NSF or NIH.
APPENDIX A: NON–GAUSSIAN RESPONSE VARIABLES
Model extensions to allow for binary, categorical, and censored data (such as survival time data) involve the following additional response-defining latent variables:
Binary probit responses. Interpret z1,i as the unobserved, underlying latent variable such that an observable response y1,i = 1 if and only if, z1,i > 0, and fix the variance ψ1 = 1 accordingly. Modifications to logistic and other link functions can be incorporated using standard methods (Albert and Johnson 1999).
Categorical responses. An observable response variable y1,i taking the value 0, 1, or 2 is modeled through two underlying latent variables, z1,i and z2,i—now two elements of zi in the factor regression—such that (a) zi,1 ≤ 0 implies that y1,i = 0, (b) zi,1 > 0 and zi,2 ≤ 0 imply that y1,i = 1, and (c) zi,1 > 0 and zi,2 > 0 imply that y1,i = 1. The hierarchical/triangular structure of the factor model for zi makes this construction for categorical data most natural.
Right-censored survival responses. One useful model includes outcome data that are logged values of survival times, in which case z1,i represents the mean of the normal on the log scale for case i. For observed times, z1,i is observed; for a case right-censored at time ci, we learn only that zi,1 ≥ ci.
In each case the uncertain elements of zi—whether due to the inherent latent structure of binary and categorical variables or to the censored data in survival analysis—are included in MCMC analyses with all model parameters and latent factors. This standard strategy also applies to cases of missing data when some elements of zi are simply missing at random, as well in predictive assessment and validation analysis when we hold out the response values of some (randomly) selected samples to be predicted based on the model fitted to the remaining data.
APPENDIX B: PRIOR TO POSTERIOR ANALYSIS MCMC COMPUTATION
Assume sparsity priors specified independently for each of the columns of A and B. Model completion then requires specification of priors for the variance components in Ψ and the τj of the sparsity priors. This involves consideration of context and ranges of variation of noise/error components. The priors for ψp+1,…, ψp+q will be response variable specific, although some values may be fixed, as noted in the foregoing discussion of binary and categorical variables. Inverse gamma priors are conditionally conjugate and are used for the Ψg and τj parameters. For the former, substantial prior information exists from previous experience with DNA microarrays across multiple experiments and observational contexts, and should be used to at least define the location of proper priors. Finally, the hyperparameters of the sparsity priors on factor loadings are to be specified; we discussed general considerations earlier.
MCMC analysis is implemented in a Gibbs sampling format. The component conditional distributions are noted here, although full details are omitted, because most are standard. This comment also applies to the conditional posteriors for the latent factor vectors arising as a result of the nonparametric DP structure; in other model contexts, this is a routinely used model component, and MCMC is well developed and understood. Some specifics of the MCMC components related to the sparsity priors are developed. Importantly, much of the computation at each iteration can be done as a parallel calculation by exploiting conditional independencies in certain complete conditionals of the posterior distribution.
Write x1:n for the set of n observations on the (p + q)-dimensional outcomes and λ1:n for the corresponding set of n (p + k)-dimensional latent factor vectors. For any quantity Δ (i.e., any subset of the full set of parameters, latent factors, and variables), let p(Δ|−) denote the complete conditional posterior of Δ given the data x1:n and all other parameters and variables. Then the sequence of conditional posteriors to sample is a follows:
Sample the conditional posterior over latent factors, p(λ1:n|−). Under the DP structure, this generates a set of some dn≤n distinct vectors and assigns each of the λi to one of these vectors (Escobar and West 1995, 1998). The inherent stochastic clustering underlying this assignment is algorithmically defined using the standard configuration sampling of DP mixture models. We simply note that, conditional on the data and all other model parameters, the model (3) can be reexpressed as a linear regression of each “residual” vector xi – μ – Bhi on Aλi, with the matrix A and the variance matrix Ψ of the regression errors known at the current values at each MCMC iterate. This then falls under the general regression and hierarchical model framework of Dirichlet mixtures as used by West et al. (1994) and MacEachern and Müller (1998). We then have access to the standard and efficient configuration sampling analysis for resampling the λ1:n at each MCMC step, as described in these references. For convenience, additional brief details are given here.
For all j = 1, …, q, the use of inverse-gamma priors for the τj leads to conditionally independent inverse-gamma complete conditionals p(τj|–). These are trivially simulated.
For all g’s for which ψg is not specified, the use of inverse-gamma priors implies that the complete conditionals p(ψg|−) are similarly inverse-gamma and so are easily simulated.
-
A novel MCMC component arises in the conditional posteriors for αg, j and βg,j together with their sparsity-governing probabilities, πg,j. The structure for resampling entries in B is completely analogous to that for A, so here we discuss only the latter. For given factor index j, this focuses on the complete conditional posterior for the full (p + q)– jth column of A, namely aj = (α 1,j,…,αp+q,j)′.
An efficient strategy is to sample the bivariate conditional posterior distribution for each pair {αg,j, πg,,j} through composition, sampling p(αg, j|−) followed by p(πg, j|αg, j, −). The model is such that for a fixed factor index j, these pairs of parameters (as g varies from g = j,…, p) are conditionally independent, so that this sampling may be performed in parallel with respect to variable index g.
The first step is to draw αg, j from its conditional posterior marginalized over πg,j. This is proportional to the conditional prior of eq. (5) but, as discussed earlier, with πg, j substituted by its prior mean ρjmj and then multiplied by the relevant conditional likelihood function; here it easily follows that this likelihood component contributes a term proportional to a normal density for αg, j. This defines a posterior that is a point mass at 0 mixed with a normal for αg, j in the case of unrestricted parameters. The computation is more complicated for the diagonal elements because of the constraint to positivity. Simulation of this is still standard and accessible using either direct calculation or accept/reject methods.
The second step is to sample the conditional posterior p(πg, j | αg, j, −), as follows: (a) if αg, j ≠ 0, then πg, j ~ Be(ajmj + 1, aj(1 − mj)); (b) if αg, j = 0, then set πg, j = 0 with probability 1 − ρ̃j, where ρ̃j = ρj(1 − mj)/(1 − ρjmj), and otherwise draw πg, j from Be(ajmj, aj(1 − mj) + 1).
Finally, draw each ρj independently from p(ρj|−) = Be(sr + σj, s(1 − r) + p + q − j − σj), where σj = #{πg, j ≠ 0:g = j + 1, …, p+q}.
APPENDIX C: ELEMENTS OF CONFIGURATION SAMPLING FOR FACTORS
Conditional on the data and all other model parameters, set ei = xi − μ − Bhi, so that the model (1) can be reexpressed as a linear regression ei = Aλi + νi, where the matrix A and the variance matrix Ψ of the errors νi are fixed at the current values at each MCMC iterate. With λi ~ F independent and F ~ Dir(α0F0), this is special case of the regression and hierarchical model framework of Dirichlet mixtures as used by West et al. (1994) and MacEachern and Müller (1998). The standard and efficient configuration sampling analysis for resampling the λ1:n uses the following steps at each of the MCMC iterates:
Step 1: Resampling configuration indicators. For each i = 1,…, n in sequence, do the following:
Remove λi from the set of currently assigned factor vectors, leaving the set of n − 1 vectors in λ−i. This current set of simulated factor vectors, λ−i, is configured into some s ≤ n − 1 groups, with a common value within each group. Denote the s distinct factor vectors by θ1:s = {θ1,…,θs} and the configuration indicators by cr = j to indicate that λr = θj for r = 1, …, i – 1, i + 1,…, n. Write nj for the number of occurrences of θj in λ−i, that is, .
- The complete conditional posterior for λi is the mixture
so that λi equals θj with probability qi, j; otherwise, it is sampled anew from N(·|mi, M) with probability qi, 0. These moments and probabilities are as follows: mi = MA′ Ψ−1ei; M−1 = I + A′ Ψ−1A; qi,0 ∝ αN(ei|0, AA′ + Ψ); and, for j = 1, s, qi, j ∝ njN(ei|Aθj, Ψ), where the notation N denotes the evaluated multivariate normal densities. Draw a new configuration indicator ci from 0: s using the probabilities qi, 0:s. If ci = 0, then sample a new value, λi ~ N(λi |mi, M).
Step 2: Resample unique factor vectors. Following step 1, the full set of resampled configuration indicators defines a set of (some final number) s conditionally independent linear regressions. For each group j = 1: s, the “data” in group j is the set of nj observations ei ~ N(ei|Aθj, Ψ) such that ci = j. Resample the unique factor vector θj of each group j = 1: s from the implied conditional posterior N(θj |tj, Tj), where tj = TjA′ Ψ−1 Σi:ci = ei and .
APPENDIX D: SOFTWARE
Efficient software implementing the MCMC and evolutionary stochastic search for the full class of sparse Bayesian factor and regression models is available to interested readers. The BFRM code implements the analysis in the framework of sparse latent factor models coupled with sparse regression and ANOVA for multivariate data, relevant in many exploratory and predictive problems with high-dimensional multivariate observations, as well as in the type of biological pathway studies of the applications here. The software also includes model components that allow for missing and censored data; binary, categorical, and continuous responses; hold-out analyses for predictive validation; and customization to gene expression studies to include automatic handling of data issues (with the generalized normalization and assay artifact correction examples here as cases in point) that arise in all expression studies that combine data on microarrays across experimental conditions or laboratories. Interested readers can download the executable BFRM code and review instructions and examples from links available on the JASA website. Examples include studies of complex networks of intersecting biological pathways in cancer genomics, as in the studies reported here, with complete details for replicating the analyses reported.
APPENDIX E: ARTIFACT CORRECTION REGRESSORS
As mentioned earlier the studies use covariates based on so-called “control genes” on microarrays to aid in identifying variation that is purely experimentally derived rather than representing biological variation of interest. We have previously introduced sparse regression terms for housekeeping/control gene data (Lucas et al. 2006). With Affymetrix arrays, we use the principal components of sets of 60–100 housekeeping gene probesets as readouts of such assay artifacts. These measures are designed to produce mRNA expression levels that show little or no biological or hybridization variation across samples, so that concordant patterns in these genes that define systematic variation through dominant principal components are potential artifact correction terms. Experience across multiple studies has demonstrated that indeed, substantial assay artifacts can be identified in this way, and that typically variation over samples in some of the dominant housekeeping correction factors can be reflected in multiple genes of interest. Contamination by assay artifact is usually sporadic, affecting multiple genes but by no means all genes, and thus the immediate relevance of the sparse regression components of the model. All of the examples given herein use this method, including corresponding assay artifact regressors in the design matrix H.
Experimental artifacts and induced variation across samples reflected in multiple genes also can be picked up by latent factors. Systematic variation that can be linked back to batch effects (e.g., sets of samples processed in different labs or under slightly different conditions at different times) often can be quite substantial and affect many genes in complex ways. Analysis that allows inclusion of latent factors in the model because collections of genes show evidence of common components of structure across samples has the ability to soak up non-biological variation of this kind. This is a strength of the sparse factor modeling approach: it can confer robustness, protecting the estimation of biologically interesting structures.
Contributor Information
Carlos M. Carvalho, Assistant Professor of Econometrics and Statistics, The University of Chicago, Graduate School of Business, Chicago, IL 60637, carlos.carvalho@chicagogsb.edu
Jeffrey Chang, Postdoctoral Fellow, Institute for Genome Sciences and Policy, Duke University, Durham, NC 27710.
Joseph E. Lucas, Assistant Research Professor, Institute for Genome Sciences and Policy, Duke University, Durham, NC 27710, and Assistant Research Professor, Department of Statistical Science, Duke University, Durham, NC 27708
Joseph R. Nevins, Barbara Levine University Professor of Breast Cancer Genomics, Department of Molecular Genetics & Microbiology, Duke University Medical Center, and Director of the Center for Applied Genomics & Technology, Institute for Genome Sciences and Policy, Duke University, Durham, NC 27710
Quanli Wang, Senior Bioinformation, Institute for Genome Sciences and Policy, Duke University, Durham, NC 27710.
Mike West, The Arts & Sciences Professor of Statistical Science, Department of Statistical Science, Duke University, Durham, NC 27708.
REFERENCES
- Aguilar O, West M. Bayesian Dynamic Factor Models and Portfolio Allocation. Journal of Business & Economic Statistics. 2000;18:338–357. [Google Scholar]
- Albert J, Johnson V. Ordinal Data Models. New York: Springer-Verlag; 1999. [Google Scholar]
- Broet P, Richardson S, Radvanyi F. Bayesian Hierarchical Model for Identifying Changes in Gene Expression From Microarray Experiments. Journal of Computational Biology. 2002;9:671–683. doi: 10.1089/106652702760277381. [DOI] [PubMed] [Google Scholar]
- Carvalho C. unpublished doctoral thesis. Duke University, ISDS; 2006. Structure and Sparsity in High-Dimensional Multivariate Analysis. available at http://stat.duke.edu/people/theses/carlos.html. [Google Scholar]
- Clyde M, George E. Model Uncertainty. Statistical Science. 2004;19:81–94. [Google Scholar]
- Do K, Müller P, Tang F. A Bayesian Mixture Model for Differential Gene Expression. Journal of the Royal Statistical Society. 2005;54:627–644. Ser. C. [Google Scholar]
- Dobra A, Jones B, Hans C, Nevins JR, West M. Sparse Graphical Models for Exploring Gene Expression Data. Journal of Multivariate Analysis. 2004;90:196–212. [Google Scholar]
- Dressman HK, Hans C, Bild A, Olsen J, Rosen E, Marcom PK, Liotcheva V, Jones E, Vujaskovic Z, Marks JR, Dewhirst MW, West M, Nevins JR, Blackwell K. Gene Expression Profiles of Multiple Breast Cancer Phenotypes and Response to Neoadjuvant Therapy. Clinical Cancer Research. 2006;12 doi: 10.1158/1078-0432.CCR-05-1447. 819-216. [DOI] [PubMed] [Google Scholar]
- Escobar M, West M. Bayesian Density Estimation and Inference Using Mixtures. Journal of the American Statistical Association. 1995;90:577–588. [Google Scholar]
- Escobar M, West M. Computing Nonparametric Hierarchical Models. In: Müller P, Dey D, Sinha D, editors. Practical Non- and Semiparametric Bayesian Statistics. New York: Springer-Verlag; 1998. pp. 1–16. [Google Scholar]
- Fu M, Wang C, Li Z, Sakamaki T, Pestell R. Minireview. Cyclin D1. Normal and Abnormal Functions. Endocrinology. 2004;145:5439–5447. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- George E, McCulloch R. Variable Selection via Gibbs Sampling. Journal of the American Statistical Association. 1993;88:881–889. [Google Scholar]
- Hans C, Dobra A, West M. Shotgun Stochastic Search in Regression With Many Predictors. Journal of the American Statistical Association. 2007;102:507–516. [Google Scholar]
- Holbro T, Beerli R, Maurer F, Koziczak M, Barbas CR, Hynes N. The ErbB2/ErbB3 Heterodimer Functions as an Oncogenic Unit: ErbB2 Requires ErbB3 to Drive Breast Tumor Cell Proliferation. Proceedings of the National Academy of Sciences. 2003;100:8933–8938. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang E, Chen S, Dressman HK, Pittman J, Tsou MH, Horng CF, Bild A, Iversen ES, Liao M, Chen CM, West M, Nevins JR, Huang AT. Gene Expression Predictors of Breast Cancer Outcomes. The Lancet. 2003;361:1590–1596. doi: 10.1016/S0140-6736(03)13308-9. [DOI] [PubMed] [Google Scholar]
- Huang E, West M, Nevins JR. Gene Expression Profiles and Predicting Clinical Characteristics of Breast Cancer. Hormone Research. 2002;58:55–73. doi: 10.1210/rp.58.1.55. [DOI] [PubMed] [Google Scholar]
- Ishwaran H, Rao J. Detecting Differentially Expressed Genes in Microarrays Using Bayesian Model Selection. Journal of the American Statistical Association. 2003;98:438–455. [Google Scholar]
- Ishwaran H, Rao J. Spike and Slab Gene Selection for Multigroup Microarray Data. Journal of the American Statistical Association. 2005;100:764–780. [Google Scholar]
- Jones B, Dobra A, Carvalho C, Hans C, Carter C, West M. Experiments in Stochastic Computation for High-Dimensional Graphical Models. Statistical Science. 2005;20:388–400. [Google Scholar]
- Lee K, Sha N, Dougherty E, Vannucci M, Mallick B. Gene Selection: A Bayesian Variable Selection Approach. Bioinformatics. 2003;19:90–97. doi: 10.1093/bioinformatics/19.1.90. [DOI] [PubMed] [Google Scholar]
- Liang F, Mukherjee S, West M. Understanding the Use of Un-labelled Data in Predictive Modelling. Statistical Science. 2007;22:189–205. [Google Scholar]
- Lopes H, West M. Bayesian Model Assessment in Factor Analysis. Statistica Sinica. 2003;14:41–67. [Google Scholar]
- Lucas J, Carvalho C, Wang Q, Bild A, Nevins JR, West M. Sparse Statistical Modelling in Gene Expression Genomics. In: Müller P, Do K, Vannucci M, editors. Bayesian Inference for Gene Expression and Proteomics. Cambridge, U.K.: Cambridge University Press; 2006. pp. 155–176. [Google Scholar]
- MacEachern SN, Müller P. Estimating Mixture of Dirichlet Process Models. Journal of Computational and Graphical Statistics. 1998;7:223–238. [Google Scholar]
- McMahon C, Suthiphongchai T, DiRenzo J, Ewen M. P/CAF Associates With Cyclin D1 and Potentiates Its Activation of the Estrogen Receptor. Proceedings of the National Academy of Sciences. 1999;96:5382–5387. doi: 10.1073/pnas.96.10.5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L, Smeds J, George J, Vega V, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu E, Bergh J. An Expression Signature for p53 Status in Human Breast Cancer Predicts Mutation Status, Transcriptional Effects, and Patient Survival. Proceedings of the National Academy of Sciences. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. Toward an Understanding of the Functional Complexity of the E2F and Retinoblastoma Families. Cell Growth and Differentiation. 1998;9:585–593. [PubMed] [Google Scholar]
- Nevins JR, Huang ES, Dressman H, Pittman J, Huang AT, West M. Towards Integrated Clinico-Genomic Models for Personalized Medicine: Combining Gene Expression Signatures and Clinical Factors in Breast Cancer Outcomes Prediction. Human Molecular Genetics. 2003;12:153–157. doi: 10.1093/hmg/ddg287. [DOI] [PubMed] [Google Scholar]
- Pittman J, Huang E, Dressman H, Horng CF, Cheng SH, Tsou MH, Chen CM, Bild A, Iversen ES, Huang AT, Nevins JR, West M. Integrated Modeling of Clinical and Gene Expression Information for Personalized Prediction of Disease Outcomes. Proceedings of the National Academy of Sciences. 2004;101:8431–8436. doi: 10.1073/pnas.0401736101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftery A, Madigan D, Hoeting J. Bayesian Model Averaging for Linear Regression Models. Journal of the American Statistical Association. 1997;92:1197–1208. [Google Scholar]
- Rich J, Jones B, Hans C, Iversen ES, McClendon R, Rasheed A, Bigner D, Dobra A, Dressman HK, Nevins JR, West M. Gene Expression Profiling and Genetic Markers in Glioblastoma Survival. Cancer Research. 2005;65:4051–4058. doi: 10.1158/0008-5472.CAN-04-3936. [DOI] [PubMed] [Google Scholar]
- Sabbah M, Courilleau D, Mester J, Redeuilh G. Estrogen Induction of the Cyclin D1 Promoter: Involvement of a Camp Response-Like Element. Proceedings of the National Academy of Sciences. 1999;96:11217–11222. doi: 10.1073/pnas.96.20.11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C, McCormick F. The Rb and p53 Pathway in Cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- Spang R, Zuzan H, West M, Nevins JR, Blanchette C, Marks JR. Prediction and Uncertainty in the Analysis of Gene Expression Profiles. In Silico Biology. 2001;2:369–381. [PubMed] [Google Scholar]
- Wang C, Fan S, Li Z, Fu M, Rao M, Ma Y, Lisanti M, Albanese C, Katzenellenbogen B, Kushner P, Weber B, Rosen E, Pestell R. Cyclin D1 Antagonizes BRCA1 Repression of Estrogen Receptor Alpha Activity. Cancer Research. 2005;65:6557–6567. doi: 10.1158/0008-5472.CAN-05-0486. [DOI] [PubMed] [Google Scholar]
- Wang Q, Carvalho C, Lucas J, West M. BFRM: Bayesian Factor Regression Modelling. Bulletin of the International Society for Bayesian Analysis. 2007;14:4–5. [Google Scholar]
- West M. Bayesian Factor Regression Models in the Large p, Small n Paradigm. In: Bernardo J, et al., editors. Bayesian Statistics. Vol. 7. Oxford U.K.: Oxford University Press; 2003. pp. 723–732. [Google Scholar]
- West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, Zuzan H, Marks JR, Nevins JR. Predicting the Clinical Status of Human Breast Cancer Utilizing Gene Expression Profiles. Proceedings of the National Academy of Sciences. 2001;98:11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M, Huang AT, Ginsberg G, Nevins JR. Embracing the Complexity of Genomic Data for Personalized Medicine. Genome Research. 2006;16:559–566. doi: 10.1101/gr.3851306. [DOI] [PubMed] [Google Scholar]
- West M, Müller P, Escobar MD. Hierarchical Priors and Mixture Models, With Application in Regression and Density Estimation. In: Smith A, Freeman P, editors. Aspects of Uncertainty: A Tribute to D.V. Lindley. London: Wiley; 1994. pp. 363–386. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


