Abstract
In vertebrates, olfaction is mediated by several families of G protein–coupled receptors (GPCRs) including odorant receptors (ORs). In this study, we investigated the antiquity of OR genes by searching for amino acid motifs found in chordate ORs among the protein predictions from 12 nonchordate species. Our search uncovered a novel group of genes in the cnidarian Nematostella vectensis. Phylogenetic analysis that included representatives from the other major lineages of rhodopsin-like GPCRs showed that the cnidarian genes, the cephalochordate and vertebrate ORs, and a family of genes from the echinoderm, Strongylocentrotus purpuratus, form a monophyletic clade. The taxonomic distribution of these genes indicates that the formation of this clade and therefore the diversification of the rhodopsin-like GPCR family began at least 700 million years ago, prior to the divergence of cnidarians and bilaterians. ORs and other rhodopsin-like GPCRs have roles in cell migration, axon guidance, and neurite growth; therefore, duplication and divergence in this family may have played a key role in the evolution of cell type diversity (including the emergence of complex nervous systems) and in the evolution of metazoan body plan diversity.
Keywords: Nematostella vectensis, odorant receptors, rhodopsin-like GPCR, metazoa, cnidaria
Vertebrate olfaction involves several families of G protein–coupled receptors (GPCRs), including the odorant receptors (ORs) (Buck and Axel 1991), the trace amine–associated receptors (TAARs) (Liberles and Buck 2006), the type 1 (Dulac and Axel 1995) and type 2 vomeronasal receptors (Herrada and Dulac 1997; Matsunami and Buck 1997; Ryba and Tirindelli 1997), and the formyl peptide receptor–like proteins (FPRs) (Rivière et al. 2009). In this study, we focused on the ORs which are GPCRs that belong to the rhodopsin-like gene family (Fredriksson and Schiöth 2005). Like other GPCRs, genes in the rhodopsin-like family encode seven-transmembrane (TM) domain receptors that convert external stimuli into intracellular biochemical signals. ORs, TAARs, and FPRs belong to the rhodopsin-like gene family, whereas the type 1 and type 2 vomeronasal receptors are not rhodopsin-like (reviewed in Bargmann 2006). Rhodopsin-like GPCRs are present in placozoans (Srivastava et al. 2008) and sponges (Srivastava et al. 2010), and the first representatives of this family are believed to have appeared between 580 and 800 million years ago (Römpler et al. 2007). The rhodopsin-like GPCRs found in humans can be divided into four subgroups: the α subgroup contains genes such as the amine receptors, melatonin receptors, TAARs, and opsins; the β subgroup contains peptide receptors (e.g., gonadotropin-releasing hormone receptors and oxytocin receptors); the γ subgroup includes chemokine, somatostatin, and opioid receptors; and the δ subgroup includes ORs, purinergic receptors, FPRs, and leucine-rich repeat containing GPCRs (Fredriksson et al. 2003).
The OR repertoires in several mammals, fish, amphibians, and cephalochordates have been described, yet orthologs of chordate ORs do not appear to occur in protostomes (reviewed in Kaupp 2010). Though chemosensory receptors have been described in protostomes (e.g., insects and nematode worms), these genes are unrelated to the ORs found in chordates. The apparent absence of chordate ORs in the protostomes does not mean that they are a deuterostome innovation. To explore the origin of ORs relative to the emergence of early metazoan lineages, we used amino acid motifs (table 1) that are commonly found in cephalochordate and vertebrate ORs (Churcher and Taylor 2009) to query the protein predictions of 12 nonchordate species. Sequences possessing these motifs were then used as queries in Blastp searches for paralogous genes. Motif-possessing sequences and Blastp hits were then used in two phylogenetic analyses.
Table 1.
Occurrence of Chordate OR Motifs in the Protein Predictions from 12 Nonchordate Species
| No. | Motifb | Speciesa |
|||||||||||
| Monosiga brevicollis | Trichoplax adhaerens | Hydra magnipapillata | Nematostella vectensis | Helobdella robusta | Lottia gigantea | Capitella sp I | Caenorhabditis elegans | Drosophila melanogaster | Anopheles gambiae | Ciona intestinalis | Strongylocentrotus purpuratus | ||
| 1 | L..P.Y.L…L...D........P | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 |
| 2 | L..P.Y.....L…D........P | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 |
| 3 | MA.D.Y…C.PL.Y | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | MA…Y...C.PL.Y | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | M....Y…C.PL.Y | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
See methods for the numbers of proteins for each species.
Positions denoted with '.' represent any amino acid.
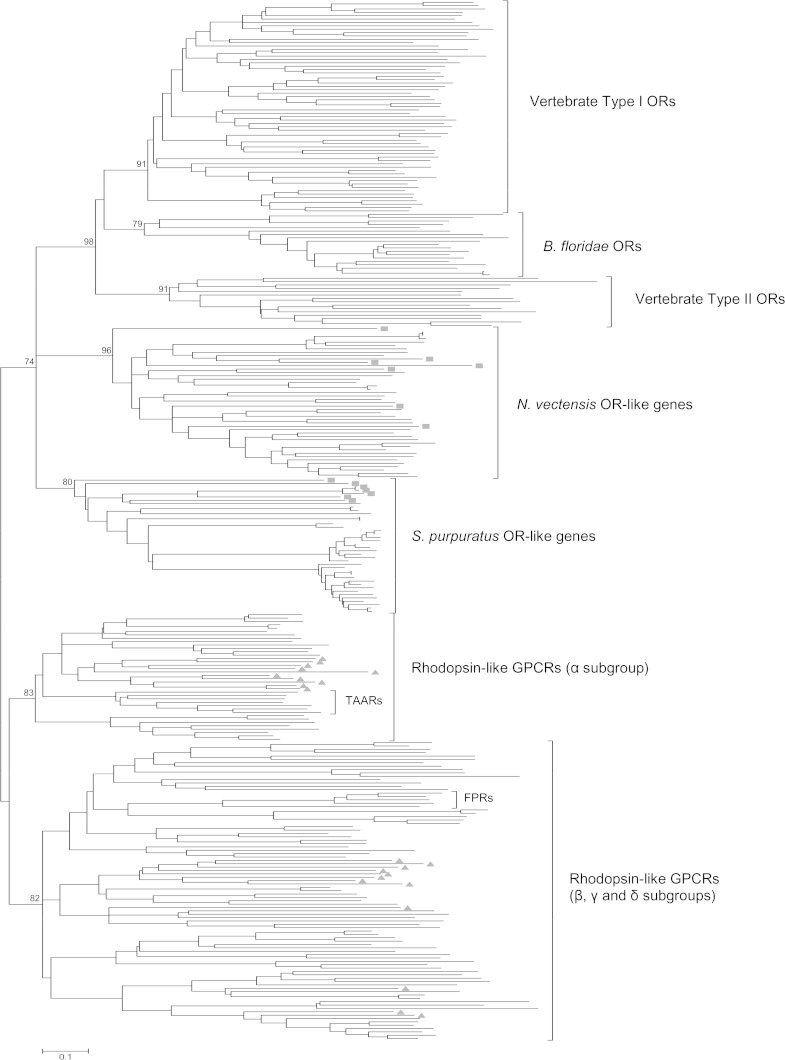
Using this approach, we uncovered a family of genes from the cnidarian, Nematostella vectensis that belongs to a monophyletic clade that includes echinoderm, cephalochordate, and vertebrate ORs (fig. 1, supplementary fig. S1, Supplementary Material online). This N. vectensis gene family includes 35 full-length and 11 partial OR-like genes (supplementary table S1, Supplementary Material online) and contains six of the seven sequences with motif 1 and seven of the nine sequences with motif 2 (table 1). Sequence identity among the full-length OR-like genes over the TM-spanning domains ranges from 14–98%. Like the chordate ORs, many of these genes have only one exon and are tandemly arrayed in the N. vectensis genome (supplementary table S1, Supplementary Material online). Our data suggest that chordate ORs evolved from a distinct clade of rhodopsin-like genes that was present in the ancestor of cnidarians and bilaterians and that is approximately 700 million years (My) old (Putnam et al. 2007).
FIG. 1.—
Phylogenetic analysis of Nematostella vectensis and Strongylocentrotus purpuratus OR-like genes, cephalochordate, and vertebrate ORs, and a diversity rhodopsin-like GPCRs that includes the TAARs and the FPRs. The unrooted NJ tree is based on approximately 200 amino acid positions and Poisson-corrected distances. The tree was constructed using the pairwise deletion option, and support for nodes was estimated using 1,000 bootstrap replicates. The bootstrap values for major clades are shown and the scale bar below indicates distance. Non-OR rhodopsin-like GPCRs were included based on the α, β, γ, and δ subgroup classification (Fredriksson et al. 2003). A list of these genes is included in supplementary table S2 (Supplementary Material online). Type I and II vertebrate OR designations are based on the classification in Niimura and Nei (2005). Motif-containing sequences are marked with “▪” and non-OR rhodopsin-like GPCRs from cnidarians are marked with “▴.”
Our search also uncovered 27 full-length OR-like genes in the echinoderm, Strongylocentrotus purpuratus that, with the N. vectensis OR-like genes, form a monophyletic clade with the chordate ORs (fig. 1, supplementary fig. S1 and table S1, Supplementary Material online). Most of the 27 S. purpuratus genes are single exon genes that are tandemly arrayed in the sea urchin genome (supplementary table S1, Supplementary Material online). The 27 S. purpuratus genes belong to a family of at least 40 OR-like genes that were characterized by Burke et al. (2006) and Raible et al. (2006) (fig. 1, supplementary table S2, Supplementary Material online). Although 13 of the 40 genes do not possess our query motifs and were not Blastp hits to motif-containing sequences, they are single exon genes and most are linked to genes that were detected in our survey. We therefore suspected that the 13 genes may be related to the genes from this survey. We added them to our alignment and our phylogenetic analysis and our suspicions were confirmed; the 13 additional genes form a monophyletic clade with the 27 genes from this survey. More importantly, this new analysis clarifies the position of the S. purpuratus OR-like gene family relative to chordate ORs and other rhodopsin-like GPCRs.
Three other proteins in our survey contained a query motif. Two were from N. vectensis and one was from the gastropod snail Lottia gigantea. One of the N. vectensis genes has motif 2 (table 1). Phylogenetic analysis showed that this gene, and eight genes that were Blastp hits to this gene, form a monophyletic clade with the α subgroup of rhodopsin-like GPCRs (data not shown). The other N. vectensis gene has motif 1 and 2 (table 1) and it had a single Blastp hit. Although these two genes are clearly related to each other, we were unable to assign them to a rhodopsin-like GPCR clade based on the amino acid positions used in our phylogenetic analysis. The L. gigantea protein has motif 5 (table 1) and retrieved a single hit in our Blastp search for paralogs. Both of the L. gigantea genes are most closely related to receptors in the α subgroup of rhodopsin-like GPCRs (data not shown).
Consistent with previous genome surveys, we did not uncover orthologs of chordate ORs in the protostomes surveyed. Therefore, chordate ORs join the list of genes that are present in N. vectensis and vertebrates but appear to have been lost in flies and nematode worms (Putnam et al. 2007). In mammals, fish, and cephalochordates, the OR genes appear to have arisen from only a few ancestral genes (Niimura and Nei 2005; Niimura 2009; Churcher and Taylor 2009) and therefore the apparent lack of chordate ORs in the protostomes may be the result of the loss of one or a few genes in the common ancestor of protostomes. Although our survey included representatives from both the ecdysozoa and lophotrochozoa, it remains possible that OR orthologs occur in unsurveyed protostomes.
Although our survey uncovered orthologs of chordate ORs in N. vectensis, we did not find representatives of this clade in the freshwater hydrozoan Hydra magnipapillata. This result is not surprising given that the amino acid substitution rates in H. magnipapillata proteins relative to their human orthologs are typically greater than the substitution rates found in N. vectensis and that the divergence time between anthozoans and H. magnipapillata is estimated to be 540 My (Chapman et al. 2010). It is possible that OR-like genes are present in H. magnipapillata and other hydrozoans but that these genes lack our query motifs. Many of the N. vectensis genes described above lack the motifs as did many of the S. purpuratus genes. And, although the motifs are common in chordate ORs, not all of them have the motifs. Therefore, these motifs can uncover OR orthologs in very distantly related metazoans, but the lack of motif-possessing sequences in a given genome does not rule out the possibility that OR orthologs are present.
Although the cnidarian and echinoderm genes are more closely related to the chordate ORs than any other rhodopsin-like GPCRs, the function of these genes is not yet clear. Expression data suggest that the amphioxus genes in this clade function as ORs (Satoh 2005), but without experimental data from cnidarians and echinoderms, we cannot attribute the same role to these genes. The conservation of motif 1 over time, however, combined with the results of our phylogenetic analysis makes these N. vectensis and S. purpuratus OR-like genes excellent candidates for expression and functional analysis.
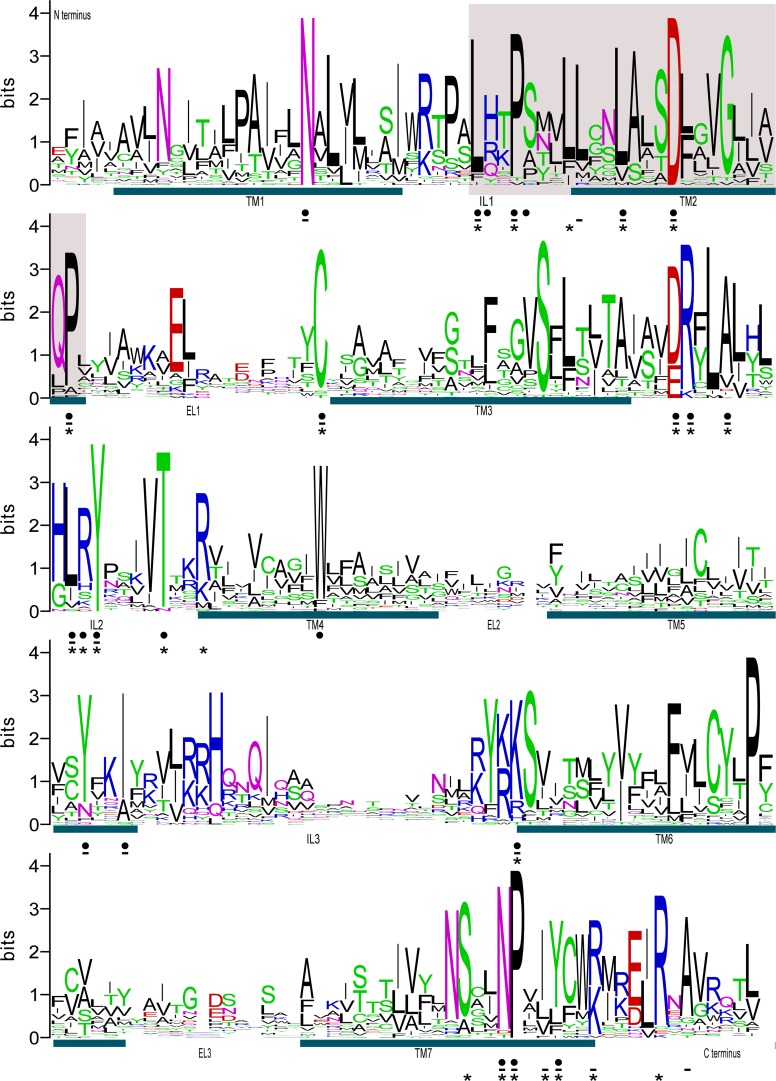
Several amino acid positions in the N. vectensis OR-like genes (fig. 2) are also conserved in cephalochordate (Churcher and Taylor 2009) and vertebrate ORs (Alioto and Ngai 2005). In more closely related sequences (e.g., mouse or rat ORs), these sites are not obvious because residues at other positions can appear highly conserved simply because they occur in a large number of genes produced by recent duplication events. The discovery of orthologs of chordate ORs in cnidarians, however, allows us to compare genes among lineages that diverged a long time ago and therefore exposes sites that are likely to be functionally significant.
FIG. 2.—
Conserved amino acid residues among Nematostella vectensis OR-like genes. The Weblogo (Crooks et al. 2004) is based on an alignment of 35 full-length N. vectensis OR-like genes from which the N and C termini have been removed. The seven TM-spanning domains (TM1-7) are labeled with blue bars and the intracellular (IL1-3) and extracellular (EL1-3) loops are also labeled. Amino acid positions that are common to fish ORs (Alioto and Ngai 2005) are indicated with “•.” Positions that are common to mouse ORs (Alioto and Ngai 2005) are shown with “-“ and those commonly found in cephalochordate ORs (Churcher and Taylor 2009) are shown by “*.” The region of the alignment that contains motifs 1 and 2 is highlighted in light gray.
Conserved amino acid positions found in N. vectensis and chordate ORs include an asparagine (N) in transmembrane domain one (TM1) and several leucines (L), a proline (P), and an aspartate (D) in intracellular loop one (IL1) and TM2 (fig. 2). These residues were part of our query motifs (motifs 1 and 2) that also include a tyrosine (Y) residue. Although the tyrosine residue is present in a subset of N. vectensis OR-like genes, it appears to have been lost in the majority. This residue is also absent in some cephalochordate ORs (Churcher and Taylor 2009) and zebrafish ORs (Alioto and Ngai 2005). In chordate ORs, the KAxxTxxxH (where x represents a variable amino acid position) motif in IL3 occurs frequently (Churcher and Taylor 2009). In N. vectensis OR-like genes, however, only the lysine (K) and the threonine (T) residues are common.
The N. vectensis OR-like genes also have several amino acid residues that are common to rhodopsin-like GPCRs and that are believed to have roles in receptor activation. These include the D/ERY motif at the boundary between TM3 and IL2, a tryptophan (W) residue in TM4 and the NPxxY motif in TM7. The arginine (R) in the D/ERY motif and the NPxxY motif are believed to function in receptor activation (reviewed in Nygaard et al. 2009).
Most of the conserved residues occur in or very close to the intracellular loops of the protein (fig. 2). In other GPCRs, the intracellular loops interact with G proteins and other proteins on the cytosolic side of the cell (reviewed in Ritter and Hall 2009). In the mouse odorant receptor, mOR-EG, conserved positions within the intracellular loops are required for receptor function (Kato et al. 2008). It is therefore possible that signal transduction in cnidarians and chordates is regulated in a similar fashion.
Our phylogenetic analysis combined with the data presented by Anctil (2009) shows that there was a diversity of rhodopsin-like GPCRs in the cnidarian–bilaterian ancestor; cnidarians have representatives of at least three of the four main subgroups of rhodopsin-like GPCRs found in humans (fig. 1,supplementary table S2, Supplementary Material online). Because the receptors involved in olfaction represent much of the diversity within the rhodopsin-like gene family (e.g., ORs, TAARs and FPRs) and receptors for environmental sensing are likely to have evolved prior to those for cell–cell communication, sensory receptors such as the ORs may be appropriate outgroups for understanding the evolution of rhodopsin-like GPCRs that have other functions.
We suggest that the evolution of rhodopsin-like GPCRs, including ORs, played a key role in metazoan evolution. These genes may have been associated with the emergence of cells that could migrate from one location to another and cells that form connections with distant cells. This hypothesis is based on the observation that several rhodopsin-like GPCRs have roles in cellular development. For example, the δ subgroup includes genes that influence axon path-finding (Mombaerts et al. 1996; Wang et al. 1998; Bozza et al. 2002); the γ subgroup includes genes that regulate the formation of nerve cell projections and cell migration (Li and Ransohoff 2008; Tiveron and Cremer 2008); and the α subgroup contains genes involved in neurite growth (Gaspar et al. 2003; Jordan et al. 2005; McKenna et al. 2008; Bhide 2009; Galve-Roperh et al. 2009). This hypothesis may explain why organisms with unorthodox body plans and sensory organs such as the echinoderms have unusual repertoires of rhodopsin-like GPCRs (described in Raible et al. 2006). Thus, the duplication and divergence of rhodopsin-like GPCRs in early metazoans may have been a prerequisite for the cellular and neuronal flexibility that led to the evolution of complex body plans.
Methods
Identification of OR-Like Genes
The protein predictions from Monosiga brevicollis (n = 9196, genome assembly version 1.0; King et al. 2008), Capitella sp. I (n = 32,415, genome assembly version 1.0, unpublished data), Helobdella robusta (n = 23,432, genome assembly version 1.0, unpublished data), L. gigantea (n = 23,851, genome assembly version 1.0, unpublished data), Trichoplax adhaerens (n = 11,520, genome assembly version 1.0; Srivastava et al. 2008), N. vectensis (n = 27,273, genome assembly version 1.0; Putnam et al. 2007), and Ciona intestinalis (n = 14,002, genome assembly version 2.0; Dehal et al. 2002) were downloaded from the US Department of Energy Joint Genome Institute (http://www.jgi.doe.gov). The sea urchin, S. purpuratus, proteins (n = 28,944, assembly version 0.5; Sea Urchin Genome Sequencing Consortium 2006) were downloaded from the Human Genome Sequencing Center at Baylor College of Medicine (ftp://ftp.hgsc.bcm.tmc.edu/pub/data). The Caenorhabditis elegans proteins (n = 39,620, release WS190; C. elegans Sequencing Consortium 1998) were downloaded from WormBase (http://www.wormbase.org). The Drosophila melanogaster (n = 20,815, release 54; Adams et al. 2000) and Anopheles gambiae (n = 13,133, release 54; Holt et al. 2002) proteins were downloaded from Ensembl (www.ensembl.org). The H. magnipapillata proteins (n = 17,398, assembly version 1.0; Chapman et al. 2010) were downloaded from the National Center for Biotechnology Information (ftp://ftp.ncbi.nih.gov). Protein sets were used to construct 12 MySQL databases, which were searched using five amino acid motifs (table 1) that are present in less than 1% of non-OR rhodopsin-like GPCRs (Churcher and Taylor 2009). These motifs were derived from a diverse set of lamprey, fish, mammalian, and cephalochordate ORs and were selected because of their ability to discriminate between ORs and non-ORs from the rhodopsin-like GPCR family. Proteins containing a query motif were used in Blastp (Altschul et al. 1997) searches for paralogs. Sequences that were at least 40% identical to the query sequence over a minimum of 100 amino acids were used in iterative Blastp searches until no more genes meeting our search criteria could be identified. Only hits to at least part of any of the TM domains were retained; however, three sequences were excluded because they do not appear to be GPCRs (S. purpuratus protein 028230, N. vectensis proteins 205877 and 212306). Proteins with all seven TM domains were considered full-length sequences; all others were considered partial sequences.
Phylogenetic Analysis
Motif-containing sequences and Blastp hits were aligned to vertebrate and cephalochordate ORs using ClustalW (Thompson et al. 1994) and the alignment was adjusted by hand in BioEdit (Hall 1999). The alignment file is included as supplementary figure S2 (Supplementary Material online). The Neighbor-Joining (NJ) tree was constructed in MEGA version 4.0 (Tamura et al. 2007) using the pairwise deletion option. The unrooted NJ tree is based on approximately 200 amino acid positions and Poisson-corrected distances. Support for nodes was estimated using 1,000 bootstrap replicates. Outgroup sequences from the protostomes, deuterostomes, cnidarians, and a placozoan were carefully selected to ensure that, where possible, representatives of the α, β, γ, and δ subgroups of rhodopsin-like GPCRs were included. The list of sequences can be found in supplementary table S2 (Supplementary Material online). Nematostella vectensis protein (214496) is very short and was excluded from the NJ tree to allow pairwise distances to be calculated from the alignment. Therefore, figure 1 includes 45 of the 46 N. vectensis OR-like genes. Tree topology was confirmed using PHYML (Guindon and Gascuel 2003) and the tree is included in supplementary figure S1 (Supplementary Material online).
Identification of Conserved Amino Acid Residues
To highlight conserved amino acid residues we constructed a WebLogo (Crooks et al. 2004) from the alignment of 35 full-length N. vectensis OR-like genes (fig. 2). Amino acid residues that are commonly found in fish (Alioto and Ngai 2005), mouse (Alioto and Ngai 2005), and cephalochordate ORs (Churcher and Taylor 2009) are marked in the figure.
Supplementary Material
Supplementary tables S1and S2 and supplementary figures S1 and S2 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
We are grateful to George O. Mackie for his contribution and to Christine E. Churcher for her editorial support. This work was supported by grants from the Canadian Foundation for Innovation (9639 to J.S.T.), the British Columbia Knowledge Development Fund, and the Natural Sciences and Engineering Research Council of Canada (262783 to J.S.T. and OGP0001427 to G.O.M.).
References
- Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Alioto TS, Ngai J. The odorant receptor repertoire of teleost fish. BMC Genomics. 2005;6:173. doi: 10.1186/1471-2164-6-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anctil M. Chemical transmission in the sea anemone Nematostella vectensis: a genomic perspective. Comp Biochem Physiol D. 2009;4:268–289. doi: 10.1016/j.cbd.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Bargmann CI. Comparative chemosensation from receptors to ecology. Nature. 2006;444:295–301. doi: 10.1038/nature05402. [DOI] [PubMed] [Google Scholar]
- Bhide PG. Dopamine, cocaine and the development of cerebral cortical cytoarchitecture: a review of current concepts. Semin Cell Dev Biol. 2009;20:395–402. doi: 10.1016/j.semcdb.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Bozza T, Feinstein P, Zheng C, Mombaerts P. Odorant receptor expression defines functional units in the mouse olfactory system. J Neurosci. 2002;22:3033–3043. doi: 10.1523/JNEUROSCI.22-08-03033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Burke RD, et al. A genomic view of the sea urchin nervous system. Dev Biol. 2006;300:434–460. doi: 10.1016/j.ydbio.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Chapman JA, et al. The dynamic genome of Hydra. Nature. 2010;464:592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churcher AM, Taylor JS. Amphioxus (Branchiostoma floridae) has orthologs of vertebrate odorant receptors. BMC Evol Biol. 2009;9:242. doi: 10.1186/1471-2148-9-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Lagerström MC, Lundin LG, Schiöth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Schiöth HB. The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol Pharmacol. 2005;67:1414–1425. doi: 10.1124/mol.104.009001. [DOI] [PubMed] [Google Scholar]
- Galve-Roperh I, Palazuelos J, Aguado T, Guzmán M. The endocannabinoid system and the regulation of neural development: potential implications in psychiatric disorders. Eur Arch Psy Clin N. 2009;259:371–382. doi: 10.1007/s00406-009-0028-y. [DOI] [PubMed] [Google Scholar]
- Gaspar P, Cases O, Maroteaux L. The developmental role of serotonin: news from mouse molecular genetics. Nat Rev Neurosci. 2003;4:1002–1012. doi: 10.1038/nrn1256. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;53:686–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Herrada G, Dulac C. A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell. 1997;90:763–773. doi: 10.1016/s0092-8674(00)80536-x. [DOI] [PubMed] [Google Scholar]
- Holt RA, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Jordan JD, et al. Cannabinoid receptor-induced neurite outgrowth is mediated by Rap1 activation through Gαo/i-triggered proteasomal degradation of Rap1GAPII. J Biol Chem. 2005;280:11413–11421. doi: 10.1074/jbc.M411521200. [DOI] [PubMed] [Google Scholar]
- Kato A, Katada S, Touhara K. Amino acids involved in conformational dynamics and G protein coupling of an odorant receptor: targeting gain-of-function mutation. J Neurochem. 2008;107:1261–1270. doi: 10.1111/j.1471-4159.2008.05693.x. [DOI] [PubMed] [Google Scholar]
- Kaupp UB. Olfactory signalling in vertebrates and insects: differences and commonalities. Nat Rev Neurosci. 2010;11:188–200. doi: 10.1038/nrn2789. [DOI] [PubMed] [Google Scholar]
- King N, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Ransohoff RM. Multiple roles of chemokine CXCL12 in the central nervous system: a migration from immunology to neurobiology. Prog Neurobiol. 2008;84:116–131. doi: 10.1016/j.pneurobio.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442:645–650. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
- Matsunami H, Buck LB. A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell. 1997;90:775–784. doi: 10.1016/s0092-8674(00)80537-1. [DOI] [PubMed] [Google Scholar]
- McKenna WL, et al. Netrin-1-independent adenosine A2b receptor activation regulates the response of axons to netrin-1 by controlling cell surface levels of UNC5A receptors. J Neurochem. 2008;104:1081–1090. doi: 10.1111/j.1471-4159.2007.05040.x. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, et al. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Niimura Y. On the origin and evolution of vertebrate olfactory receptor genes: comparative genome analysis among 23 chordate species. Genome Biol Evol. 2009;1:34–44. doi: 10.1093/gbe/evp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc Natl Acad Sci U S A. 2005;102:6039–6044. doi: 10.1073/pnas.0501922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard R, Frimurer TM, Holst B, Rosenkilde MM, Schwartz TW. Ligand binding and micro-switches in 7tm receptor structures. Trends Pharmacol Sci. 2009;30:249–259. doi: 10.1016/j.tips.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Raible F, et al. Opsins and clusters of sensory G-protein-coupled receptors in the sea urchin genome. Dev Biol. 2006;300:461–475. doi: 10.1016/j.ydbio.2006.08.070. [DOI] [PubMed] [Google Scholar]
- Ritter SL, Hall RA. Fine-tuning of GPCR activity by receptor-interacting proteins. Nat Rev Mol Cell Biol. 2009;10:819–830. doi: 10.1038/nrm2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière S, Challet L, Fluegge D, Spehr M, Rodriguez I. Formyl peptide receptor-like proteins are a novel family of vomeronasal chemosensors. Nature. 2009;459:574–577. doi: 10.1038/nature08029. [DOI] [PubMed] [Google Scholar]
- Römpler H, et al. G protein-coupled time travel. Mol Interv. 2007;7:17–25. doi: 10.1124/mi.7.1.5. [DOI] [PubMed] [Google Scholar]
- Ryba NJP, Tirindelli R. A new multigene family of putative pheromone receptors. Neuron. 1997;19:371–379. doi: 10.1016/s0896-6273(00)80946-0. [DOI] [PubMed] [Google Scholar]
- Satoh G. Characterization of novel GPCR gene coding locus in amphioxus genome: gene structure, expression, and phylogenetic analysis with implications for its involvement in chemoreception. Genesis. 2005;41:47–57. doi: 10.1002/gene.20082. [DOI] [PubMed] [Google Scholar]
- Sea Urchin Genome Sequencing Consortium. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–961. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- Srivastava M, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–727. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiveron MC, Cremer H. CXCL12/CXCR4 signalling in neuronal cell migration. Curr Opin Neurobiol. 2008;18:237–244. doi: 10.1016/j.conb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Wang F, Nemes A, Mendelsohn M, Axel R. Odorant receptors govern the formation of a precise topographic map. Cell. 1998;93:47–60. doi: 10.1016/s0092-8674(00)81145-9. [DOI] [PubMed] [Google Scholar]