Abstract
Nucleomorphs are the remnant nuclei of algal endosymbionts that were engulfed by nonphotosynthetic host eukaryotes. These peculiar organelles are found in cryptomonad and chlorarachniophyte algae, where they evolved from red and green algal endosymbionts, respectively. Despite their independent origins, cryptomonad and chlorarachniophyte nucleomorph genomes are similar in size and structure: they are both <1 million base pairs in size (the smallest nuclear genomes known), comprised three chromosomes, and possess subtelomeric ribosomal DNA operons. Here, we report the complete sequence of one of the smallest cryptomonad nucleomorph genomes known, that of the secondarily nonphotosynthetic cryptomonad Cryptomonas paramecium. The genome is 486 kbp in size and contains 518 predicted genes, 466 of which are protein coding. Although C. paramecium lacks photosynthetic ability, its nucleomorph genome still encodes 18 plastid-associated proteins. More than 90% of the “conserved” protein genes in C. paramecium (i.e., those with clear homologs in other eukaryotes) are also present in the nucleomorph genomes of the cryptomonads Guillardia theta and Hemiselmis andersenii. In contrast, 143 of 466 predicted C. paramecium proteins (30.7%) showed no obvious similarity to proteins encoded in any other genome, including G. theta and H. andersenii. Significantly, however, many of these “nucleomorph ORFans” are conserved in position and size between the three genomes, suggesting that they are in fact homologous to one another. Finally, our analyses reveal an unexpected degree of overlap in the genes present in the independently evolved chlorarachniophyte and cryptomonad nucleomorph genomes: ∼80% of a set of 120 conserved nucleomorph genes in the chlorarachniophyte Bigelowiella natans were also present in all three cryptomonad nucleomorph genomes. This result suggests that similar reductive processes have taken place in unrelated lineages of nucleomorph-containing algae.
Keywords: nucleomorph, cryptomonads, chlorarachniophytes, genome reduction, endosymbiosis
Introduction
Genome reduction is a well known but generally poorly understood phenomenon most often seen in organisms that have adopted a symbiotic, endosymbiotic, or parasitic lifestyle (Martin and Herrmann 1998; Martin et al. 2002; Keeling and Slamovits 2005; Nakabachi et al. 2006; McCutcheon et al. 2009; Moran et al. 2009). The most extreme examples of highly reduced genomes are those of plastids (chloroplasts) and mitochondria, which are organelles derived from cyanobacterial and alphaproteobacterial endosymbionts, respectively (Gray et al. 1999; Dolezal et al. 2006; Reyes-Prieto et al. 2007; Gould et al. 2008; Kim and Archibald 2008). Modern-day plastid genomes range between ∼70 and 200 kbp in size and possess at most ∼200 genes, whereas those of mitochondria are typically 15–350 kbp (not considering higher plants), significantly smaller than those of even the smallest free-living bacteria (Kaneko and Tabata 1997; Martin and Herrmann 1998; Gray et al. 1999; Martin et al. 2002; Timmis et al. 2004; Nakabachi et al. 2006). Within bacteria, the smallest known genomes are those of symbionts living in association with insects, such as the ∼144 kbp genome of Hodgkinia cicadicola (McCutcheon et al. 2009) and the 420–650 kbp genomes of Buchnera species (Nikoh et al. 2010). Mycoplasma species, which are important obligate parasites and human pathogens, also have significantly reduced genomes in the range of 0.6–1.4 Mbp and with ∼480 to 1,000 genes (Sasaki et al. 2002).
At less than 1 Mbp in size, the “nucleomorph” genomes of chlorarachniophyte and cryptomonad algae are far and away the most reduced and compact nuclear genomes known. Nucleomorphs are the residual nuclei of eukaryotic photosynthetic endosymbionts that evolved into fully integrated cellular organelles in the context of nonphotosynthetic eukaryotic hosts (Cavalier-Smith 2002; Archibald and Lane 2009; Moore and Archibald 2009). Unlike the “primary” endosymbiotic origin of plastids from cyanobacterial endosymbionts, cryptomonad and chlorarachniophyte nucleomorphs—and the plastids with which they are intimately associated—are the product of “secondary” endosymbiosis (Gilson and McFadden 2002; Bhattacharya et al. 2004; Keeling 2004; Archibald 2007). This process has generated a large fraction of algal biodiversity, but the cryptomonads and the chlorarachniophytes are unusual in their shared retention of the algal endosymbiont nucleus, which has been lost in all other secondary plastid-bearing algae, such as diatoms and haptophytes (Archibald 2009a, 2009b). Genomic diversity studies of cryptomonads and chlorarachniophytes have revealed that their nucleomorph genomes are similar in size (∼485 to 845 kbp in cryptomonads and ∼330 to 610 kbp in chlorarachniophytes) and structures, with both consisting of three chromosomes and subtelomeric ribosomal DNA (rDNA) operons (Rensing et al. 1994; Lane and Archibald 2006; Lane et al. 2006; Silver et al. 2007; Phipps et al. 2008; Tanifuji et al. 2010). These similarities are intriguing given that the nucleomorphs in the two groups are of independent origin. The cryptomonad nucleomorph and plastid are derived from a red algal endosymbiont (Douglas et al. 1990; Cavalier-Smith et al. 1996; Douglas and Penny 1999), whereas in chlorarachniophytes, the endosymbiont is of green algal ancestry (McFadden et al. 1995; Ishida et al. 1997, 1999; Rogers et al. 2007).
Complete nucleomorph genome sequences have been published for two cryptomonads, Guillardia theta (Douglas et al. 2001) and Hemiselmis andersenii (Lane et al. 2007), as well as a single chlorarachniophyte, Bigelowiella natans (Gilson et al. 2006). The G. theta and H. andersenii genomes are 551 and 571 kbp in size, respectively, and are extremely gene dense, with 487 and 472 protein-coding genes each. Most of the evolutionarily conserved genes in both genomes are housekeeping in nature (e.g., translation, transcription, and protein folding/degradation). The G. theta and H. andersenii nucleomorph genomes also share an identical set of 30 genes for plastid-associated proteins. The two genomes are, however, significantly different in the presence/absence of introns: G. theta has 17 spliceosomal introns and RNA and protein genes for splicing, whereas the H. andersenii nucleomorph genome has no introns or genes for spliceosomal components. Furthermore, the average length of both genes/proteins and intergenic spacer regions are smaller in G. theta than in H. andersenii, a feature that was attributed to the higher degree of genomic compaction seen in G. theta (Lane et al. 2007).
The nucleomorph genome of the chlorarachniophyte B. natans, completely sequenced by Gilson et al. (2006), is a mere 323 kbp in size and possesses 331 protein-coding genes. As in cryptomonads, a large proportion of B. natans nucleomorph genes are involved in core housekeeping processes. A remarkable difference between the B. natans and the cryptomonad nucleomorphs is that the B. natans genome contains 852 very short introns (18–21 bp) and more genes for spliceosomal components than does G. theta, despite being smaller in size. More interestingly, only three of 17 plastid-associated genes in the B. natans nucleomorph genome (cpn60 and two clpP isoforms) overlap with the 30 retained in cryptomonads. A long-standing and as yet unresolved question in nucleomorph genome biology is whether they are still undergoing reductive evolution or are “evolutionary endpoints” (Archibald and Lane 2009).
Despite lacking photosynthesis, the secondarily nonphotosynthetic cryptomonad Cryptomonas paramecium still possesses a plastid and nucleomorph. The plastid genome of C. paramecium was recently sequenced and shown to be approximately half the size of the genome of photosynthetic species, lacking many photosynthesis-related genes such as members of the psa and psb gene families (Douglas and Penny 1999; Khan et al. 2007; Donaher et al. 2009). Here, we present the complete C. paramecium nucleomorph genome sequence and compare its structure and gene content with other nucleomorph genomes. Our results provide insight into the biology of this fascinating organism, expanding our knowledge of the set of proteins still functioning in its nonphotosynthetic plastid. They also reveal unexpected overlap between the gene sets present in the independently evolved nucleomorph genomes of cryptomonads and chlorarachniophytes. Similar evolutionary forces may have driven the reduction of the ancestral nucleomorph genomes in these two unrelated algal lineages.
Material and Methods
Cell Culture and Isolation of Nucleomorph DNA
Cryptomonas paramecium strain CCAP977/2A was obtained from the Culture Collection of Algae and Protozoa (CCAP) and maintained in the laboratory at room temperature as described previously (Donaher et al. 2009). Approximately, 10 mg of total cellular DNA was extracted from a total of 50 l of 3-day-old culture (∼10 × 1010 cells) as described previously and subjected to Hoechst dye-cesium chloride density gradient centrifugation to purify nucleomorph DNA. Three distinct bands were isolated, purified, and eluted in 50 μl of Tris–ethylenediaminetetraacetic acid buffer. Semiquantitative polymerase chain reaction (PCR) was used to assess the purity of each of the isolated fractions using gene-specific primers encoding plastid rbcL, mitochondrial coxI, nucleomorph small subunit ribosomal RNA (SSU rRNA), and nuclear actin as follows: rbcL_C.para-F1 (5′-GAACTTCCGTGTCATTTGTAAGTGGATGCG-3′), rbcL_C.para-R1 (5′-GCCTGTATACCATCAGGGTGCCCAAT-3′), cox1_C.para-F1 (5′-GAATGGAACTAGCTGGTCCTGGTGTTCA-3′), cox1_C.para-R1 (5′-ACCACCTGGATGTCCAGAGATACTACTTAA-3′), SSUrDNA_C.para-F1 (5′-CCAGCTATCGAGAGAAGTCTATCCTG-3′), SSUrDNA_C.para-R1 (5′-AAAGGCCTACGATCGTTATTTTCTGTCG-3′), Actin_C.para-F1 (5′-TCGTGCGCGACATCAAGGAGAAGCT-3′), and Actin_C.para-R1 (5′-GCGCTGATCTCCTTCTGCATGCG-3′). Approximately 4 μg of nucleomorph DNA was purified with significant mitochondrial DNA contamination (∼50%) and minor plastid DNA contamination (∼1%).
Genome Sequencing and Assembly
Genome sequencing and initial genome assembly were performed at the McGill University and Génome Québec Innovation Center using a 454 GS FLX pyrosequencer and titanium reagents (Roche Diagnostics). A 3/4-plate run generated ∼716,000 reads (with an average read length of 343 bp) and ∼230 Mbp of raw sequence data. 10.1% of the reads were successfully assembled into contigs 500 bp or larger. Fifteen nucleomorph-derived contigs between 3.5 and 124 kbp were produced, each with ∼30× coverage. These 15 contigs were refined manually and assembled into seven larger contigs. The remaining gaps were bridged using PCR: amplicons were purified, cloned into the pGEM-T Easy vector (Promega), and Sanger sequenced on a Beckman-Coulter CEQ 8000 capillary DNA sequencer (Beckman Coulter, Inc.). Approximately 150 ambiguous regions of the assembly (e.g., with potential frameshifts or stop codons within open reading frames [ORFs]) were PCR amplified using Platinum Taq polymerase High Fidelity (Invitrogen). Amplicons were directly sequenced or cloned into Topo XL or Topo 2.1 cloning vectors (Invitrogen). To verify the C. paramecium nucleomorph telomere sequence, telomere-containing clones were screened from a nucleomorph and mitochondrial DNA–enriched fosmid library made using the CopyControl Fosmid Library Production Kit (EPICENTRE Biotechnologies). Isolated clones were sequenced on a Beckman-Coulter CEQ 8000 capillary DNA sequencer using the pCC1/pEpiFOS (EPICENTRE Biotechnologies) sequencing primer (5′-GGATGTGCTGCAAGGCGATTAAGTTGG-3′) and a primer designed to the 5S rDNA locus of the C. paramecium nucleomorph genome (Cp_5SrDNA primer; 5′-CGCAACTTAAGCGCACGTAGGC-3′). Sequencher 4.7 (GeneCodes Inc.) was used to combine 454 contigs with Sanger sequence data.
Genome Annotation
ORFs larger than 50 amino acids were identified and annotated using Artemis 8.0 (Rutherford et al. 2000). ORFs were searched against the non-redundant protein sequence (nr) database using BlastP (Blast ver. 2.2.18, Altschul et al. 1997) and HMMER3 (ver. 3.0, Eddy 1998; http://hmmer.org). Additional support for remote homologs was attained using Pfam searches (ver. 24.0, Finn et al. 2010).
For comparative purposes, C. paramecium ORFs were assigned to one of three general categories. ORFs with annotated homologs (e value < 0.001) in nucleomorph genomes as well as other nuclear genomes were designated “conserved ORFs.” ORFs with no homology to annotated eukaryotic proteins but with significant hits to either hypothetical proteins in nr (e value < 0.001) or known Pfam families (e value < 1 × 10−10) were labeled “ambiguous.” Genes showing similarity only to known genes in distantly related organisms were also put in this category because their orthology was uncertain. Finally, C. paramecium ORFs sharing no similarities with ORFs in any other genome (ORFans) or showing similarities only to other cryptomonad nucleomorph ORFs (Blast e value < 0.001 or e values < 0.02 with additional support from synteny) were designated nucleomorph ORFans (nORFans). Functional categorization of genes/proteins followed Douglas et al. (2001), Lane et al. (2007), and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa et al. 2010; http://www.kegg.jp/ja/).
Transfer RNAs (tRNAs) were identified with trnaScan-SE (Lowe and Eddy 1997; http://lowelab.ucsc.edu/tRNAscan-SE/). rRNA genes were identified by BlastN. To search for small nuclear RNAs (snRNAs), a local PatScan search was employed with consensus sequences of known snRNAs used as a guide (Guthrie and Patterson 1988). The C. paramecium nucleomorph genome sequence has been deposited in GenBank under accession number CP002172 (chromosome 1), CP002173 (chromosome 2), and CP002174 (chromosome 3).
Protein Length and Intergenic Spacer Size Calculations
The average lengths of proteins encoded in the C. paramecium, G. theta, and H. andersenii genomes were calculated based on all protein genes (n = 466, n = 486, and n = 470, respectively), a set of 266 genes present in all three cryptomonads (240 conserved ORFs, including plastid-associated genes, spliceosomal genes, and multiple copy genes, plus three ambiguous genes and 23 nORFans) and ORFan genes (n = 143, n = 160, and n = 127, respectively). For estimation of average intergenic spacer size, 96 syntenic regions found in C. paramecium, G. theta, and H. andersenii were examined, as was the average spacer size for each genome individually (n = 516, n = 511, and n = 522, respectively). Statistical significance of size differences was determined using ANOVA4 (http://www.hju.ac.jp/∼kiriki/anova4/) for both one-way analysis of variance (ANOVA) and multiple comparisons. A P value of 0.01 was used as a significance level.
Results and Discussion
Chromosome and Genome Structure
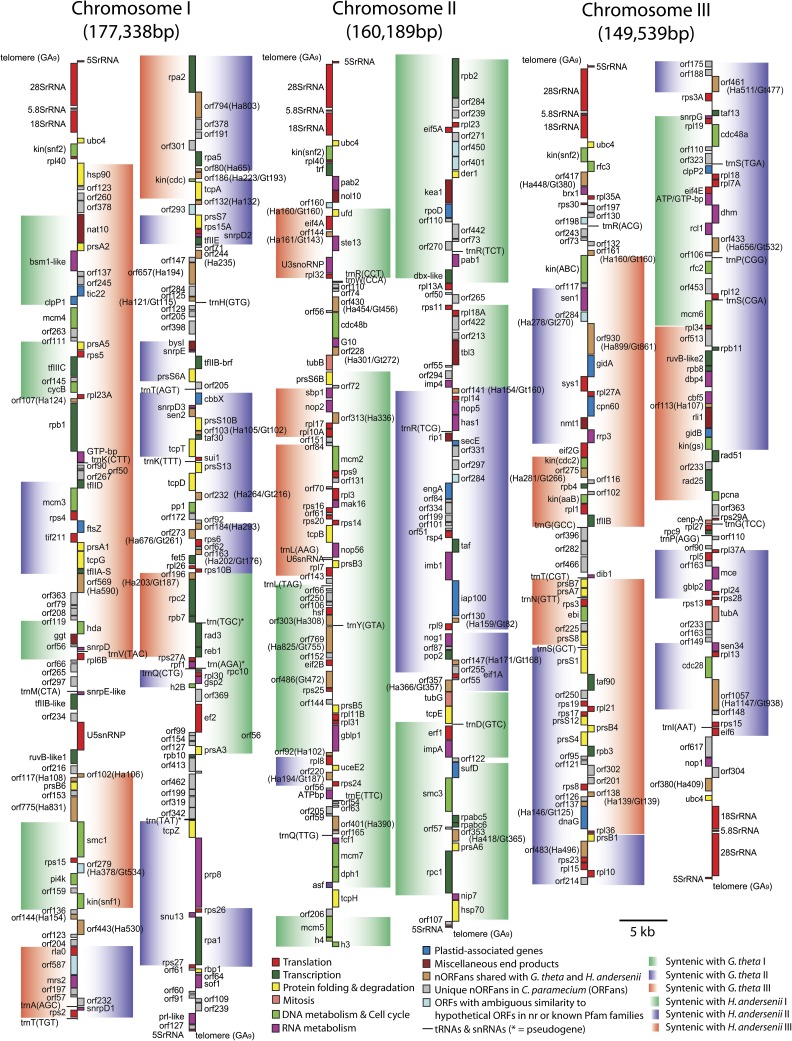
The complete nucleomorph genome of C. paramecium CCAP977/2a was 454 pyrosequenced to ∼30× coverage, assembled into seven large contigs, and finished and polished using PCR and traditional Sanger sequencing techniques. Three chromosomes were sequenced telomere-to-telomere (177.3, 160.2, and 149.5 kbp in size), consistent with previous karyotype analyses (e.g., Rensing et al. 1994), resulting in a total genome size of 487,066 bp (fig. 1). Subtelomeric rDNA regions consisting of an 18S–5.8S–28S rDNA operon and associated 5S gene are present on both ends of chromosome 3 and one end each of chromosomes 1 and 2. The remaining two chromosome ends possess stand-alone 5S rDNA loci (fig. 1). Telomere sequences comprised ten or more GA9 repeats (the exact number of repeats on each of the six chromosome ends was not determined). Reduced genomes such as those of plastids, mitochondria, and obligate endosymbionts are well known to have low G + C content (Nakabachi et al. 2006; Moran et al. 2009; Smith 2009), and nucleomorph genomes are no exception. The G + C content of the C. paramecium nucleomorph genome is 26.05%, slightly higher than that of the larger H. andersenii genome (25.18%) but lower than G. theta (26.43%; table 1).
FIG. 1.—
Physical map of the Cryptomonas paramecium nucleomorph genome. The genome is ∼487 kbp in size with three chromosomes, shown artificially broken at their midpoint. Colors correspond to predicted functional categories, and shaded bars indicate regions of synteny with the nucleomorph genome of Guillardia theta (left) or Hemiselmis andersenii (right). Gray boxes show nORFan genes (see main text). Cryptomonas paramecium ORFs with clear homologs of unknown function in H. andersenii (Ha) and/or G. theta (Gt) are shown in brown. ORFs with low sequence similarity to known genes and/or with functional motifs are shown as light blue boxes. Genes mapped on the left side of each chromosome are transcribed bottom to top and those on the right, top to bottom.
Table 1.
Overview of Nucleomorph Genome Sequences for Three Cryptomonads
| Species | Cryptomonas paramecium | Guillardia theta | Hemiselmis andersenii |
| Genome size (kbp)a | Total 485.9 | Total 550.5 | Total 571.4 |
| chr.1 177.0 | chr.1 195.9 | chr.1 207.3 | |
| chr.2 159.7 | chr.2 180.6 | chr.2 184.6 | |
| chr.3 149.1 | chr.3 173.9 | chr.3 179.4 | |
| G + C content (%) | 26.05 | 26.43 | 25.18 |
| Number of genes (protein-coding genes/total) | 466 (519) | 487 (548)b | 472 (525)c |
| Amino acid length (AAs) (all ORFs/shared ORFs/ORFans) | 289.39/333.37/187.97 | 311.66/330.94/267.89 | 338.41/351.14/294.28 |
| Intergenic spacer length (bp) (syntenic/total region) | 65.39/103.49 | 43.74/94.89 | 87.28/132.14 |
| Number of predicted spliceosomal introns | 2 | 17 | 0 |
| Telomere | GA9 | (AG)7AAG6A | GA17 |
Telomere sequences were excluded from total genome size.
Data taken from current GenBank database plus nonannotated rps30 gene in the genome (Williams et al. 2005) and one pseudo-rpl24 gene. Numbers vary from the original publication (Douglas et al. 2001) due to updated analyses.
Data taken from current GenBank plus two pseudogenes (nip7 and Yrpl24).
A comparison of gene order conservation between C. paramecium and the previously sequenced G. theta and H. andersenii nucleomorph genomes revealed the presence of large blocks of synteny (fig. 1). Twenty-eight syntenic blocks composed of four or more genes exist between C. paramecium and G. theta, whereas 20 regions of synteny were apparent between C. paramecium and H. andersenii (subtelomeric rDNA operons were not considered in this analysis, and “nORFans” [see below] and structural RNA genes were not considered interruptions of a syntenic block). The largest C. paramecium–G. theta and C. paramecium–H. andersenii syntenic blocks were 26 and 46 kbp, respectively. Several large blocks of gene order conservation (e.g., from mcm5 to the dbx-like gene on chromosome 2) were found among all three cryptomonads (fig. 1). Overall, the structure of the C. paramecium nucleomorph genome is more like that of H. andersenii than G. theta: Syntenic blocks between C. paramecium and H. andersenii were fewer and larger than those shared between C. paramecium and G. theta. Also, the telomere sequence of C. paramecium (GA9) is more similar to H. andersenii (GA17) than G. theta ((AG)7AAG6A). Despite extensive phylogenetic analyses (Deane et al. 2002; Hoef-Emden et al. 2002; Hoef-Emden 2008), the relationship between the three genera is still unclear. Nucleomorph genes are often highly divergent in nature and thus difficult to accurately place in phylogenetic trees (Hoef-Emden et al. 2002; Lane et al. 2006; Phipps et al. 2008). More extensive analyses using multiple loci will be necessary to provide a better phylogenetic framework for determining whether the higher degree of synteny between C. paramecium and H. andersenii is due to common ancestry or an increased rate of genome rearrangement in G. theta relative to the other two species.
Nucleomorph Genome Reduction and Compaction in C. paramecium
Cryptomonas paramecium has one of the smallest cryptomonad nucleomorph genomes characterized thus far (Tanifuji et al. 2010). We compared its structural features with those of the larger G. theta (Douglas et al. 2001) and H. andersenii (Lane et al. 2007) genomes to explore the relationship between total nucleomorph genome size and degree of genome reduction/compaction. Table 1 summarizes the salient features of this three-way comparison. Excluding telomere sequences, the 485.9 kbp C. paramecium nucleomorph genome is 64.6 and 85.5 kbp smaller than the G. theta and H. andersenii nucleomorph genomes, respectively. The total number of protein-coding genes in C. paramecium is 466, 21 fewer than in G. theta (table 1). Given an average gene length of ∼1 kbp, this difference in protein gene number accounts for ∼21 kbp of the genome size difference between C. paramecium and G. theta. In contrast, the C. paramecium genome has only five fewer genes than does H. andersenii despite being ∼86 kbp smaller. Furthermore, the G. theta nucleomorph genome has 487 predicted protein genes (548 genes in total), 15 more than in H. andersenii whose genome is 20 kbp larger. Therefore, nucleomorph genome coding capacity does not strictly correlate with genome size for the three species examined.
With respect to gene density, the average intergenic spacer length for C. paramecium is 103.49 bp for the genome as a whole and 65.39 bp when a set of 96 spacers shared between the three cryptomonad genomes are examined in isolation. Unexpectedly, despite the smaller size of the C. paramecium genome, the 65.39 bp average for homologous spacers was significantly larger than that of G. theta (43.74 bp, P = 0.001). The whole-genome average for C. paramecium (103.49 bp) is also larger than G. theta (94.89 bp), although this difference is not significant by ANOVA (P = 0.360). Hemiselmis andersenii has significantly larger intergenic spacers than both C. paramecium and G. theta in syntenic regions and for the genome as a whole. The smaller C. paramecium genome is thus not the most compact when intergenic spacer length is considered in isolation. Overall, 11.0% of the C. paramecium genome is noncoding compared with 8.8% in G. theta and 12.1% in H. andersenii. In addition, although 33 instances of overlapping genes are found in the C. paramecium genome, this number is fewer than that of G. theta (44 in total). Differences in intron size and abundance between C. paramecium, which has two predicted spliceosomal introns (rfc2 and orf80, which are 62 and 100 bp, respectively) plus five predicted tRNA introns (7–20 bp), and G. theta (17 spliceosomal introns between 42 and 52 bp plus 13 tRNA introns between 1 and 24 bp) are negligible. In sum, the amount of noncoding DNA in cryptomonad nucleomorph genomes does not correlate with genome size.
Lane et al. (2007) compared the average length of shared genes and syntenic spacer regions between G. theta and H. andersenii and showed that the nucleomorph genome of G. theta, which is smaller than that of H. andersenii, had significantly shorter ORFs and intergenic spacer regions. Here, we compared the C. paramecium nucleomorph proteome with those of G. theta and H. andersenii in a similar fashion. In isolation, the average lengths of C. paramecium proteins based on 1) total number of protein genes, 2) 266 genes shared between all three genomes (i.e., 240 conserved ORFs, including plastid-associated genes, spliceosomal genes, and multiple copy genes, plus three ambiguous genes and 23 nORFans), and 3) ORFs showing no homology to any other genome (true ORFans) are 289.39, 333.37, and 187.97 amino acids, respectively (table 1). The average ORFan gene length for C. paramecium (187.97 amino acids) is significantly smaller than that of G. theta (267.89 amino acids, P = 0.001) and H. andersenii (294.28 amino acids, P < 0.001). ORFan gene lengths account for ∼47 of the 65 kbp of the genome size difference between C. paramecium and G. theta, with the remainder corresponding to differences in other protein genes and structural RNA genes. This result suggests that average gene/protein length, especially among ORFans, significantly affects nucleomorph genome size. However, the average length of shared protein genes is not significantly different between C. paramecium and G. theta (P = 0.318). One explanation is that because these conserved (shared) genes are presumably necessary for gene expression and maintenance of the nucleomorph, further protein size reduction is no longer possible. In addition, intergenic spacer lengths based on syntenic regions were shorter than those of the whole genome (table 1). This is consistent with the notion that closely spaced genes should preserve their synteny for longer periods of time than genes that are further apart due to the reduced frequency of intergenic recombination (Archibald and Lane 2009). It is also possible that for unknown reasons, the syntenic/conserved regions of nucleomorph chromosomes have been subjected to stronger reductive pressures relative to more recombinant areas.
Gene Content: Conserved and Cryptomonad-Specific Genes
The C. paramecium nucleomorph genome contains 519 predicted genes: 466 putative protein genes, one snRNA (U6 snRNA), 34 tRNAs, and 18 rDNAs (table 1 and supplementary fig. S1, Supplementary Material online). Two hundred and sixty-nine protein genes (including multiple copy loci; kin(snf2) × 3, rpl40 × 2, ubc4 × 4) have clear homologs with known or predicted functions in other nuclear genomes and, in many cases, nucleomorphs (supplementary fig. S1, Supplementary Material online). These were considered conserved ORFs. Most of these genes are “housekeeping” in nature, with predicted roles in gene expression, protein folding/degradation, etc., and only 18 genes were plastid-associated genes of cyanobacterial origin. Eleven protein genes were considered ambiguous; although they showed obvious similarity to known genes or protein families, orthology was difficult to determine with confidence.
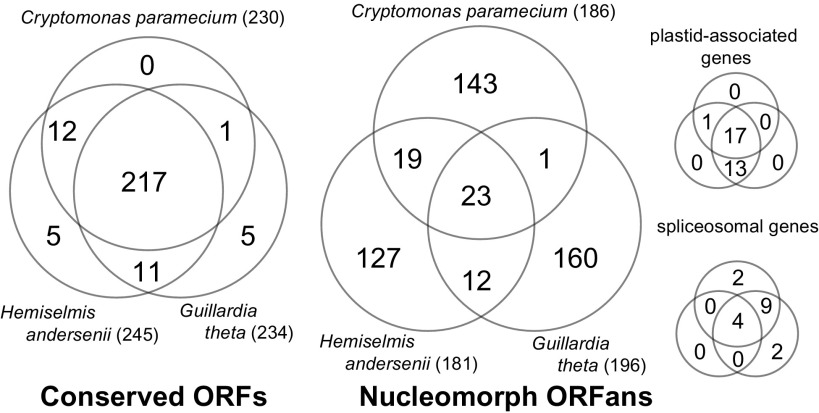
We examined how many conserved C. paramecium ORFs were shared among all three cryptomonad nucleomorphs. Because C. paramecium lacks photosynthesis, plastid-associated genes were compared separately, as were spliceosome-related genes (see below). This left a total of 230 conserved C. paramecium proteins to be compared with 234 from G. theta and 245 in H. andersenii. 94.3% (217 of 230) of the conserved ORFs in the C. paramecium nucleomorph genome were present in all three cryptomonad nucleomorph genomes (fig. 2). In fact, C. paramecium does not possess a single conserved ORF that is not present in the G. theta and/or H. andersenii genomes. These 217 conserved ORFs would appear to be essential “core” genes, that is, those that still remain after the massive reduction of the endosymbiont nuclear genome in the common ancestor of these three cryptomonads.
FIG. 2.—
Gene content comparison of three cryptomonad nucleomorph genomes. The Venn diagrams show the number of shared and/or unique genes in four categories: conserved ORFs (left), nucleomorph ORFans (middle), plastid-associated genes (right top), and spliceosomal genes (right bottom). The numbers beside species names are the total gene numbers under consideration.
Remarkably, 186 ORFs in the C. paramecium nucleomorph genome either show no similarity whatsoever to sequences in any other genome or have a detectible homolog only in the G. theta and/or H. andersenii genomes. These 186 genes were designated nORFans. In stark contrast to the pattern seen for the conserved ORFs, an analysis of cryptomonad nORFans (186, 196, and 181 genes in C. paramecium, G. theta, and H. andersenii, respectively) revealed that only 23 were shared among the three species. The majority of the nORFans in each genome showed no detectible similarity to ORFs in the other two and (by definition) to ORFs in any other genomes. This amounts to 143 genes in C. paramecium, 160 in G. theta, and 127 in H. andersenii (fig. 2). The overall proportions of the cryptomonad nucleomorph genome-specific nORFans per genome are 30.7% for C. paramecium, 32.9% for G. theta, and 26.9% for H. andersenii.
Lane et al. (2007) showed that many of the nORFans in the H. andersenii genome are contained within syntenic blocks and in the same position as G. theta nORFans (syntenic ORFs). Furthermore, nORFans in the same location in the two genomes are usually very similar in size. These syntenic ORFs were thus considered likely to be homologs of one another but with such rapid rates of evolution that sequence similarity is no longer detectible (Lane et al. 2007). We compared the precise locations of the C. paramecium, G. theta, and H. andersenii nORFans within syntenic blocks and found a similar pattern: 75 of 91 (82.4%; C. paramecium vs. H. andersenii) and 48 of 65 (73.8%; C. paramecium vs. G. theta) C. paramecium nORFans can be considered syntenic ORFs (data not shown). This result lends further support to the hypothesis that the syntenic ORFs in cryptomonad nucleomorph genomes are indeed homologous to one another, effectively eliminating the possibility that, as a whole, the class of genes we have designated nORFans are not real genes. Indeed, it is significant that roughly half of the G. theta nORFans have expressed sequence tag support (http://www.jgi.doe.gov/sequencing/why/50026.html), indicating that they are at least transcribed if not translated into protein.
What are the functions of nORFans and why do they persist in cryptomonad nucleomorph genomes despite retaining little or no primary sequence similarity? As in H. andersenii and G. theta (Lane et al. 2007; Archibald and Lane 2009), the C. paramecium nORFans encode proteins that are significantly enriched in amino acids encoded by A + T-rich codons (phenylalanine, isoleucine, asparagine, lysine, and tyrosine). This particular combination of amino acids is consistent with the possibility that nORFans encode membrane interacting/transmembrane proteins (Deber et al. 1999; Archibald and Lane 2009), a hypothesis that can and should be tested experimentally. Regardless, the fact that ∼30% of the genes in each of the three cryptomonad nucleomorph genomes sequenced thus far fall into this category is intriguing. The abundance of “ORFans” in highly reduced genomes varies. For reference, a comparison of the plastid genomes of C. paramecium, G. theta, and Rhodomonas salina revealed a total of only six “orphan” genes: four of 71 ORFs in C. paramecium and two of 147 in R. salina (Douglas and Penny 1999; Khan et al. 2007; Donaher et al. 2009). In the case of the aphid bacterial endosymbiont Buchnera sp. APS, only seven of 575 protein genes could not be assigned to clusters of orthologous group of proteins (COGs) (Shigenobu et al. 2000). In contrast, 20–40% of the genes in different strains of Mycoplasma could not be placed into COGs (Sasaki et al. 2002). In any given genome, the designation of an ORF as “unique” depends on the search criteria used and the genomes available for comparison at the time, and so it is difficult to compare such percentages directly. Overall, however, the presence and stability of both a rapidly evolving nORFan gene set and a highly conserved core (the conserved ORFs) in the three nucleomorph genomes investigated here is worthy of further investigation. The 23 nORFans conserved in all three cryptomonad nucleomorph genomes (fig. 2) might also be considered core ORFs whose evolutionary origins and predicted functions will hopefully be elucidated when more red algal genomes become available for comparison.
Convergent Gene Content in Cryptomonad and Chlorarachniophyte Nucleomorph Genomes
Given that endosymbiont nuclei have completely disappeared in secondary plastid-containing algae, such as haptophytes, stramenopiles, and euglenids (Bhattacharya et al. 2004), a central question in nucleomorph genome biology is whether cryptomonad and chlorarachniophyte nucleomorphs represent an intermediate state or an endpoint beyond which no further reduction is possible. In an attempt to answer this question, the three cryptomonad genomes examined above were compared and contrasted with the nucleomorph genome of the chlorarachniophyte B. natans (Gilson et al. 2006). A significant difference between cryptomonad and B. natans nucleomorphs is the number and size of spliceosomal introns. The B. natans nucleomorph genome is ∼373 kbp, significantly smaller than those of cryptomonads, yet it possesses 852 tiny (18–21 bp) spliceosomal introns (Gilson et al. 2006). Cryptomonas paramecium and G. theta possess only two and 17 predicted spliceosomal introns, respectively, whereas H. andersenii has no introns at all. For this reason, we omitted spliceosome-related genes in our comparison of nucleomorph gene content between the two groups.
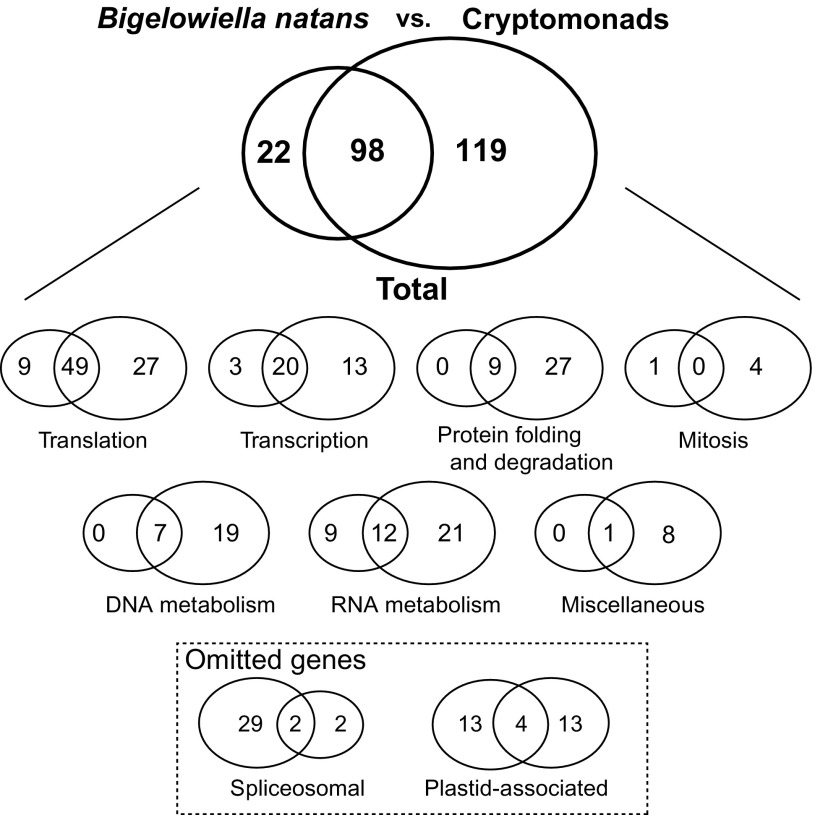
One hundred and twenty of 331 protein genes in the B. natans nucleomorph genome (i.e., those for which orthology could confidently be ascribed) were compared with the 217 core genes from cryptomonads as described above. Ninety-eight of these 120 B. natans genes (81.7%) were contained in the cryptomonad core set (fig. 3). In terms of functional category, these genes can be broken down as follows: 49 of 58 genes in translation, 20 of 23 genes in transcription, all nine genes in protein folding and degradation, all seven genes in DNA metabolism, and 12 of 21 genes in RNA metabolism. There were no shared genes in the mitosis category (0 of 1). In sum, although cryptomonads and chlorarachniophytes engulfed different endosymbionts (red and green algae, respectively), their nucleomorph genomes possess an intriguingly similar “basal set” of housekeeping genes.
FIG. 3.—
Comparison of nucleomorph gene overlap between Cryptomonas paramecium, Guillardia theta, and Hemiselmis andersenii (cryptomonads) and the chlorarachniophyte Bigelowiella natans. The top Venn diagram shows the total number of shared and unique genes, whereas those in the middle are broken down by functional category. The two categories shown at the bottom were omitted from the total gene number comparisons (see text).
Core eukaryotic translation genes have been classified into three categories in the KEGG database: 79 ribosomal genes, 32 translation factor genes, and 25 aminoacyl-tRNA biogenesis genes (Katinka et al. 2001; http://www.genome.jp/kegg/). Cryptomonad and chlorarachniophyte nucleomorph genomes share not only a similar set of ribosomal protein genes but also the exact same aminoacyl tRNA synthetase gene (for the amino acid serine), the only one known to be retained in nucleomorph genomes thus far, as noted by Gilson et al. (2006) (supplementary table S1, Supplementary Material online). Each of the five translation factors in the chlorarachniophyte nucleomorph genome is found in cryptomonads and for transcription, an identical (but compared with other eukaryotes, incomplete) set of 13 RNA polymerase I, II, and III subunit genes is present in both lineages (supplementary table S2, Supplementary Material online). These observations appear inconsistent with a pattern of random retention of nucleomorph genes in the two lineages from presumably “unreduced” green algal and red algal nuclear genomes in chlorarachniophytes and cryptomonads, respectively. These results strongly suggest that similar reductive pressures have led to convergence upon a core set of eukaryotic cellular machineries functioning in the remnant cytoplasmic compartments surrounding the plastids in the two lineages (i.e., their “periplastidial” compartments).
On the one hand, our analyses have revealed a significant overlap between the gene set present in B. natans and the core set in cryptomonads. And yet the B. natans nucleomorph gene set (and the genome itself) is significantly smaller than that of any of the sequenced cryptomonad nucleomorphs (Gilson et al. 2006). There is no obvious reason why the 119 genes that are currently present in the cryptomonad nucleomorph but absent in B. natans could not, in principle, be lost or transferred to the cryptomonad nuclear genome. The same is true of the 22 genes that are conserved in B. natans but absent in cryptomonads. It should be noted that it is unknown whether the 120 analyzed genes of B. natans are representative of the chlorarachniophyte nucleomorph core set because only one nucleomorph genome from this lineage is available at present. Additional chlorarachniophyte nucleomorph genome sequences will allow further elucidation of this core set and, in turn, more meaningful comparisons between cryptomonads and chlorarachniophytes.
The reason(s) why genome reduction in the chlorarachniophyte nucleomorph genome is more advanced is/are still unknown, but a possible slower progression of genome reduction of cryptomonad nucleomorphs is consistent with the inference of a slower rate of sequence evolution relative to the chlorarachniophyte nucleomorph (Patron et al. 2006). Regardless, determining the extent to which the housekeeping machineries functioning in the cryptomonad and chlorarachniophyte periplastidial compartments are supplemented by nucleus-encoded proteins is an important next step. The nuclear genomes of B. natans and G. theta have been sequenced by the Joint Genome Institute and should soon provide this crucial data (http://www.jgi.doe.gov/sequencing/why/50026.html). For the time being, it seems significant that in cases where a specific transcription or translation factor is not universally present in red and green algae, this same factor is almost always absent from both the cryptomonad and the chlorarachniophyte nucleomorphs (supplementary table S2, Supplementary Material online). Should the cryptomonad and chlorarachniophyte nucleomorph proteomes prove not to be supplemented to a great extent by nucleus-encoded gene products, nucleomorphs could serve as a valuable model for elucidating the minimal protein components required to maintain fundamental eukaryotic cellular processes.
Nucleomorph Genes for Plastid Proteins
The C. paramecium nucleomorph genome harbors 18 genes for plastid-associated proteins (fig. 2 and supplementary fig. S1, Supplementary Material online), whereas G. theta and H. andersenii share a total of 30 (Douglas et al. 2001; Lane et al. 2007). Thirteen of these 18 C. paramecium genes are present in all three genomes (fig. 2). Because C. paramecium is a nonphotosynthetic organism, the loss of photosynthesis-related genes such as cpeT-like, hcf136, hlip, and rub, each of which are found in both G. theta and H. andersenii, is not surprising. However, another four nonphotosynthesis plastid protein genes (gyrA, gyrB, tha4, and met) and five unknown ORFs (designated ORFs 173, 235, 237, 263, and 337 in H. andersenii) have also been lost in the C. paramecium nucleomorph genome. Furthermore, we found a novel plastid-associated gene, gidB, shared between C. paramecium and H. andersenii but absent in G. theta. Overall, these results suggest that plastid-associated genes, including those not directly involved in photosynthesis, are not strictly conserved in cryptomonad nucleomorphs.
Genes encoding plastid-targeted proteins have previously been considered to be the most evolutionarily significant genes in cryptomonad and chlorarachniophyte nucleomorph genomes (Gilson et al. 2006). This is because if all of these genes are lost or relocated to the host nucleus, then in principle, all of the nucleomorph genes encoding the housekeeping machinery required to express them can also be lost. Comparing the B. natans and G. theta nucleomorph genomes, Gilson et al. (2006) found that only a minor proportion of plastid-associated genes in G. theta and B. natans were shared. These authors took this as evidence in favor of the hypothesis that the overlap of plastid-associated genes in the two groups is essentially random and that nucleomorphs “may yet disappear.”
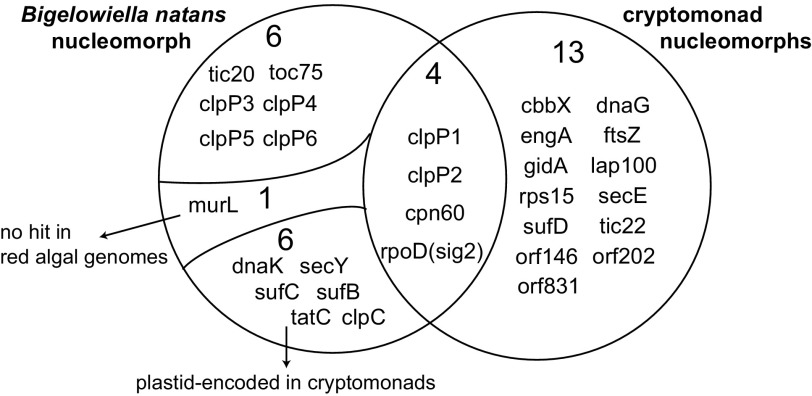
To further assess the significance of the apparent lack of overlap of plastid-associated genes in nucleomorph genomes, we compared the plastid gene sets in the three cryptomonads with those of B. natans (fig. 4). We determined that only four plastid-associated genes are in fact shared between cryptomonads and B. natans. This result is similar to the observations of Gilson et al. (2006), except they considered rpoD in cryptomonads and sig2 of chlorarachniophytes to be different genes. We consider these two genes to be orthologous because both are similar to one another in Blast searches and the same functional domains (sigma-70 domains 2, 3, and 4) were found using HMMER searches (data not shown). Furthermore, the evidence suggests that the six unique plastid genes in chlorarachniophytes (dnaK, secY, sufB, sufC, tatC, and clpC) were not encoded in the ancestral cryptomonad nucleomorph. These six genes are still present in the plastid genomes of red algae, cryptomonads, haptophytes, and stramenopiles (Donaher et al. 2009), and thus strictly speaking, should not factor in discussions of differential loss of plastid-associated genes from the cryptomonad and chlorarachniophyte nucleomorph genomes. In addition, murL is not found in the genomes of the red algae Cyanidioschyzon merolae and Galdieria sulphuraria and thus may not have been present in the ancestral cryptomonad nucleomorph genome. In sum, given that only two envelope protein translocases (tic20 and toc75) and four clp protease subunit genes 1) are present in the B. natans nucleomorph genome, 2) were demonstrably present in the ancestral cryptomonad nucleomorph, and 3) are now missing in the cryptomonad genomes thus far investigated, it is difficult to assess whether retention of plastid-associated genes in the two lineages is truly random. Nevertheless, given that eight of the 13 plastid-associated genes present in the cryptomonad nucleomorph genomes but absent in B. natans can be found in the host nuclear genome of the haptophyte Emiliania huxleyi (Patron et al. 2007; Burki et al. 2008), there is no obvious reason why these genes could not be transferred in the future. Elucidation of the tempo and mode of plastid- and nucleomorph-to-host nucleus gene transfer in cryptomonads and chlorarachniophytes should allow us to better understand why nucleomorphs persist.
FIG. 4.—
Shared and unique nucleomorph genes for plastid-targeted proteins between the chlorarachniophyte Bigelowiella natans and the cryptomonads Cryptomonas paramecium, Guillardia theta, and Hemiselmis andersenii. Six of 13 unique genes in B. natans were found in red algal nuclear genomes, whereas murL was not. Another six B. natans genes are located in cryptomonad plastid genomes.
Supplementary Material
Supplementary figure S1 and tables S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank C. Lane for providing H. andersenii nucleomorph genome data, M. Gray, D. Spencer, and S. Murray for assistance with nucleomorph DNA isolation and snRNA gene prediction, Y. Sasaki for interesting discussion about Mycoplasma genomes, T. Jones for providing guidance on methods for remote homology search with HMMER and Pfam, and two anonymous reviewers for helpful comments on an earlier version of this manuscript. J. Rainey and S. Bearne are thanked for discussions about the possible functions of nORFan proteins. K. Dewar and his colleagues at the McGill University and Génome Québec Innovation Center are also thanked for help assembling 454 sequence data. This work was supported by an operating grant (MOP-85016) from the Canadian Institutes for Health Research. G.T. is supported by a postdoctoral fellowship from the Tula Foundation and the Centre for Comparative Genomics and Evolutionary Bioinformatics at Dalhousie University. J.M.A. is a Fellow of the Canadian Institute for Advanced Research, Program in Integrated Microbial Biodiversity, and holder of a New Investigator Award from the Canadian Institutes of Health Research.
References
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald JM. Nucleomorph genomes: structure, function, origin and evolution. BioEssays. 2007;29(4):392–402. doi: 10.1002/bies.20551. [DOI] [PubMed] [Google Scholar]
- Archibald JM. The origin and spread of eukaryotic photosynthesis: evolving views in light of genomics. Bot Mar. 2009a;52(2):95–103. [Google Scholar]
- Archibald JM. The puzzle of plastid evolution. Curr Biol. 2009b;19(2):R81–R88. doi: 10.1016/j.cub.2008.11.067. [DOI] [PubMed] [Google Scholar]
- Archibald JM, Lane CE. Going, going, not quite gone: nucleomorphs as a case study in nuclear genome reduction. J Hered. 2009;100(5):582–590. doi: 10.1093/jhered/esp055. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Yoon HS, Hackett JD. Photosynthetic eukaryotes unite: endosymbiosis connects the dots. BioEssays. 2004;26(1):50–60. doi: 10.1002/bies.10376. [DOI] [PubMed] [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Pawlowski J. Phylogenomics reveals a new ‘megagroup’ including most photosynthetic eukaryotes. Biol Lett. 2008;4(4):366–369. doi: 10.1098/rsbl.2008.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Nucleomorphs: enslaved algal nuclei. Curr Opin Microbiol. 2002;5(6):612–619. doi: 10.1016/s1369-5274(02)00373-9. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, et al. Cryptomonad nuclear and nucleomorph 18S rRNA phylogeny. Eur J Phycol. 1996;31(4):315–328. [Google Scholar]
- Deane JA, et al. Cryptomonad evolution: nuclear 18SrDNA phycologeny versus cell morphology and pigmentation. J Phycol. 2002;38:1236–1244. [Google Scholar]
- Deber CM, Liu LP, Wang C. Perspective: peptides as mimics of transmembrane segments in proteins. J Pept Res. 1999;54:200–205. doi: 10.1034/j.1399-3011.1999.00118.x. [DOI] [PubMed] [Google Scholar]
- Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313(5785):314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- Donaher N, et al. The complete plastid genome sequence of the secondarily nonphotosynthetic alga Cryptomonas paramecium: reduction, compaction, and accelerated evolutionary rate. Genome Biol Evol. 2009;1:439–448. doi: 10.1093/gbe/evp047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas SE, Durnford DG, Morden CW. Nucleotide-sequence of the gene for the large subunit of ribulose-1,5-bisphosphate carboxylase oxygenase from Cryptomonas phi—evidence supporting the polyphyletic origin of plastids. J Phycol. 1990;26(3):500–508. [Google Scholar]
- Douglas SE, Penny SL. The plastid genome of the cryptophyte alga, Guillardia theta: complete sequence and conserved synteny groups confirm its common ancestry with red algae. J Mol Evol. 1999;48(2):236–244. doi: 10.1007/pl00006462. [DOI] [PubMed] [Google Scholar]
- Douglas S, et al. The highly reduced genome of an enslaved algal nucleus. Nature. 2001;410(6832):1091–1096. doi: 10.1038/35074092. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14(9):755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson PR, McFadden GI. Jam packed genomes—a preliminary, comparative analysis of nucleomorphs. Genetica. 2002;115(1):13–28. doi: 10.1023/a:1016011812442. [DOI] [PubMed] [Google Scholar]
- Gilson PR, et al. Complete nucleotide sequence of the chlorarachniophyte nucleomorph: nature's smallest nucleus. Proc Natl Acad Sci U S A. 2006;103(25):9566–9571. doi: 10.1073/pnas.0600707103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SB, Waller RR, McFadden GI. Plastid evolution. Annu Rev Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283(5407):1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Hoef-Emden K. Molecular phylogeny of phycocyanin-containing cryptophytes: evolution of biliproteins and geographical distribution. J Phycol. 2008;44(4):985–993. doi: 10.1111/j.1529-8817.2008.00530.x. [DOI] [PubMed] [Google Scholar]
- Hoef-Emden K, Marin B, Melkonian M. Nuclear and nucleomorph SSU rDNA phylogeny in the cryptophyta and the evolution of cryptophyte diversity. J Mol Evol. 2002;55(2):161–179. doi: 10.1007/s00239-002-2313-5. [DOI] [PubMed] [Google Scholar]
- Ishida K, Cao Y, Hasegawa M, Okada N, Hara Y. The origin of chlorarachniophyte plastids, as inferred from phylogenetic comparisons of amino acid sequences of EF-Tu. J Mol Evol. 1997;45(6):682–687. doi: 10.1007/pl00006272. [DOI] [PubMed] [Google Scholar]
- Ishida K, Green BR, Cavalier-Smith T. Diversification of a chimaeric algal group, the chlorarachniophytes: phylogeny of nuclear and nucleomorph small-subunit rRNA genes. Mol Biol Evol. 1999;16(3):321–331. [Google Scholar]
- Kaneko T, Tabata S. Complete genome structure of the unicellular cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol. 1997;38(11):1171–1176. doi: 10.1093/oxfordjournals.pcp.a029103. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katinka MD, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414(6862):450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- Keeling PJ. Diversity and evolutionary history of plastids and their hosts. Am J Bot. 2004;91(10):1481–1493. doi: 10.3732/ajb.91.10.1481. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Slamovits CH. Causes and effects of nuclear genome reduction. Curr Opin Genet Dev. 2005;15(6):601–608. doi: 10.1016/j.gde.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Khan H, Parks N, Kozera C, Curtis BA, Parsons BJ, Bowman S, Archibald JM. Plastid genome sequence of the cryptophyte alga Rhodomonas salina CCMP1319: Lateral transfer of putative DNA replication machinery and a test of chromist plastid Phylogeny. Mol Biol Evol. 2007;24(8):1832–1842. doi: 10.1093/molbev/msm101. [DOI] [PubMed] [Google Scholar]
- Kim E, Archibald JM. Diversity and evolution of plastids and their genomes. In: Aronsson H, Sandelius AS, editors. The chloroplast—interactions with the environment. Berlin (Germany): Springer-Verlag; 2008. pp. 1–39. [Google Scholar]
- Lane CE, Archibald JM. Novel nucleomorph genome architecture in the cryptomonad genus Hemiselmis. J Eukaryot Microbiol. 2006;53(6):515–521. doi: 10.1111/j.1550-7408.2006.00135.x. [DOI] [PubMed] [Google Scholar]
- Lane CE, et al. Insight into the diversity and evolution of the cryptomonad nucleomorph genome. Mol Biol Evol. 2006;23(5):856–865. doi: 10.1093/molbev/msj066. [DOI] [PubMed] [Google Scholar]
- Lane CE, et al. Nucleomorph genome of Hemiselmis andersenii reveals complete intron loss and compaction as a driver of protein structure and function. Proc Natl Acad Sci U S A. 2007;104(50):19908–19913. doi: 10.1073/pnas.0707419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Herrmann RG. Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol. 1998;118(1):9–17. doi: 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci U S A. 2002;99(19):12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, McDonald BR, Moran NA. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PloS Genet. 2009;5(7):e1000565. doi: 10.1371/journal.pgen.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI, Gilson PR, Waller RF. Molecular phylogeny of Chlorarachniophytes based on plastid ribosomal-RNA and rbcl sequences. Arch Protistenkunde. 1995;145(3–4):231–239. [Google Scholar]
- Moore CE, Archibald JM. Nucleomorph genomes. Annu Rev Genet. 2009;43:251–264. doi: 10.1146/annurev-genet-102108-134809. [DOI] [PubMed] [Google Scholar]
- Moran NA, McLaughlin HJ, Sorek R. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science. 2009;323(5912):379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, et al. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314(5797):267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- Nikoh N, et al. Bacterial genes in the aphid genome—absence of functional gene transfer from Buchnera to its host. PLoS Genet. 2010;6(2):e1000827. doi: 10.1371/journal.pgen.1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patron NJ, Inagaki Y, Keeling PJ. Multiple gene phylogenies support the monophyly of cryptomonad and haptophyte host lineages. Curr Biol. 2007;17(10):887–891. doi: 10.1016/j.cub.2007.03.069. [DOI] [PubMed] [Google Scholar]
- Patron NJ, Rogers MB, Keeling PJ. Comparative rates of evolution in endosymbiotic nuclear genomes. BMC Evol Biol. 2006 doi: 10.1186/1471-2148-6-46. doi: 10.1186/1471-2148-6-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps KD, Donaher NA, Lane CE, Archibald JM. Nucleomorph karyotype diversity in the freshwater cryptophyte genus Cryptomonas. J Phycol. 2008;44(1):11–14. doi: 10.1111/j.1529-8817.2007.00434.x. [DOI] [PubMed] [Google Scholar]
- Rensing SA, Goddemeier M, Hofmann CJB, Maier UG. The presence of a nucleomorph hsp70 gene is a common feature of Cryptophyta and Chlorarachniophyta. Curr Genet. 1994;26(5–6):451–455. doi: 10.1007/BF00309933. [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A, Weber APM, Bhattacharya D. The origin and establishment of the plastid in algae and plants. Annu Rev Genet. 2007;41:147–168. doi: 10.1146/annurev.genet.41.110306.130134. [DOI] [PubMed] [Google Scholar]
- Rogers MB, Gilson PR, Su V, McFadden GI, Keeling PJ. The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol Biol Evol. 2007;24(1):54–62. doi: 10.1093/molbev/msl129. [DOI] [PubMed] [Google Scholar]
- Rutherford K, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16(10):944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, et al. The complete genomic sequence of Mycoplasma penetrans, an intracellular bacterial pathogen in humans. Nucleic Acids Res. 2002;30(23):5293–5300. doi: 10.1093/nar/gkf667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407(6800):81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- Silver TD, et al. Phylogeny and nucleomorph karyotype diversity of chlorarachniophyte algae. J Eukaryot Microbiol. 2007;54(5):403–410. doi: 10.1111/j.1550-7408.2007.00279.x. [DOI] [PubMed] [Google Scholar]
- Smith DR. Unparalleled GC content in the plastid DNA of Selaginella. Plant Mol Biol. 2009;71(6):627–639. doi: 10.1007/s11103-009-9545-3. [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5(2):123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- Tanifuji G, Onodera NT, Hara Y. Nucleomorph genome diversity and its phylogenetic implications in cryptomonad algae. Phycol Res. 2010;58(3):230–237. [Google Scholar]
- Williams BAP, Slamovits CH, Patron NJ, Fast NM, Keeling PJ. A high frequency of overlapping gene expression in compacted eukaryotic genomes. Proc Natl Acad Sci U S A. 2005;102(31):10936–10941. doi: 10.1073/pnas.0501321102. [DOI] [PMC free article] [PubMed] [Google Scholar]