Abstract
Iron oxide nanoparticles (NPs) with a diameter 21.6 nm were coated with poly(maleic acid-alt-1-octadecene) (PMAcOD) modified with grafted 5,000 Da poly(ethyelene glycol) (PEG) or short ethylene glycol (EG) tails. The coating procedure utilizes hydrophobic interactions of octadecene and oleic acid tails, while the hydrolysis of maleic anhydride moieties as well as the presence of hydrophilic PEG (EG) tails allows the NP hydrophilicity. The success of the NP coating was found to be independent of the degree of grafting which was varied between 20 and 80% of the –MacOD-units, but depended on the length of the grafted tail. The NP coating and hydrophilization did not occur when the modified copolymer contained 750 Da PEG tails independently of the grafting degree. To explain this phenomenon the micellization of the modified PMAcOD copolymers in water was analyzed by small angle x-ray scattering (SAXS). The PMAcOD molecules with the grafted 750 Da PEG tails form compact non-interacting disk-like micelles, whose stability apparently allows for no interactions with the NP hydrophobic shells. The PMAcOD containing the 5,000 Da PEG and EG tails form much larger aggregates capable of an efficient coating of the NPs. The coated NPs were characterized using transmission electron microscopy, dynamic light scattering, ζ-potential measurements, and thermal gravimetry analysis. The latter method demonstrated that the presence of long PEG tails in modified PMAcOD allows the attachment of fewer macromolecules (by a factor of ~20) compared to the case of non-modified or EG modified PMAcOD, emphasizing the importance of PEG tails in NP hydrophilization. The NPs coated with PMAcOD modified with 60% (towards all –MAcOD- units) of the 5,000 PEG tails bear a significant negative charge and display good stability in buffers. Such NPs can be useful as magnetic cores for virus-like particle formation.
Keywords: monodisperse nanoparticles, amphiphilic, alternating copolymer, hydrophilic, modification, small angle X-ray scattering
1. Introduction
Magnetic nanoparticles (NPs) with hydrophilic shells have received considerable attention because of their promising bioapplications such as biosensors,1,2 contrast enhancement agents for magnetic resonance imaging,3–7 bioprobes,8–11 etc. In the majority of these applications it is important to use NPs with a narrow particle size distribution because magnetic properties are size dependent.12–15 Iron oxide NPs are commonly recommended for bioapplications because they are easily metabolized or degraded in vivo.16,17 Monodisperse iron oxide NPs can be prepared by thermal decomposition of iron acetylacetonates18–20 or carboxylates20–24 in high-boiling solvents in the presence of such surfactants as oleic, palmitic and other fatty acids and/or oleylamine. In all these methods the NPs are coated with fatty acids or amines and soluble only in a few organic solvents. On the other hand, hydrophilicity and biocompatibility are required for all biomedical applications.
There are several methods to impart water solubility (hydrophilicity) and biocompatibility. They are a place (ligand) exchange reaction,25–29 placement of polymer chains on a nanoparticle surface,30,31 NP growth in the presence of polymeric surfactants,32 or coating of NPs with amphiphilic molecules due to formation of hydrophobic double layers.33–35 The last method was shown to be particularly facile for the NP hydrophilization.33–35 The encapsulation of monodisperse iron oxide NPs by PEGylated phospholipids was described in our preceding papers.36,37 This coating yields stable hydrophilic magnetic NPs of different sizes and shapes which are also well compatible with various buffers and physiological solutions. The only downside of PEGylated phopsholipids as coating agents to yield hydrophilic NPs is high cost which hampers their application.
Alternating amphiphilic copolymers showed promise for NP functionalization.38,39 The encapsulation of hydrophobic NPs with short (7,300 Da) poly(maleic anhydride-alt-1-tetradecene) was described in ref.38 To stabilize the polymer coating, the authors used crosslinking of the alternating copolymer anhydride groups with bis(6-aminohexyl)amine. In a preceding paper we reported a successful encapsulation of even large magnetic NPs (16–21 nm in diameter) (the larger the particles from the same material, the stronger the magnetic attraction) using poly(maleic anhydride-alt-1-octadecene) (PMAOD), with a molecular weight of 30,000–50,000 Da. The longer copolymer compared to that reported in ref.38 provides a stable NP shell without additional crosslinking accompanying with the loss of carboxyl groups (decreased negative charge), while the longer hydrophobic tail (C16 vs C12 in the previously studied copolymer38) allows the more stable hydrophobic double layer. Interestingly, in a recent paper by Di Corato et al.40 the same PMAOD was used as well for NP coating, however, the authors again employed crosslinking with an amine for coating stabilization which we proved to be unnecessary. In water, PMAOD is easily hydrolyzed yielding poly(maleic acid-alt-1-octadecene), PMAcOD, where maleic acid units are highly hydrophilic and negatively charged.41
In our preceding paper were reported PMAcOD self-assembling in water and structure of NPs coated with PMAcOD.41 It was demonstrated that NPs with PMAcOD coating are stable for many months without changes of their characteristics or aggregation. However, these NPs were only stable in water while addition of salt immediately resulted in NP aggregation because the NP stabilization here is purely electrostatic and it is screened when a salt is added. To impart stability in buffers and physiological solutions to NPs, steric stabilization is required. In the present paper we report coating of iron oxide NPs with a diameter 21.6 nm with altered PMAODs containing grafted poly(ethylene glycol) (PEG) or short ethylene glycol (EG) brushes. This type of alteration was described in ref.39 using 6,000 Da PEG with an amino or hydroxyl terminal group or a number of PEGs with different molecular weights in the following paper of the same authors.42 However, although in the ref.42 PEG-NH2 with molecular weights 6,000, 9,900, and 19,300 Da and PEG-OH with molecular weights 550, 750, and 2,000 Da were mentioned for PMAOD alteration it was not specified anywhere in the paper that these copolymers were equally successful in NP coating.
In the present work we studied the dependence of the coating on the density of the grafted brush and on the PEG chain length. Discovery of the surprising phenomenon that PMAOD-PEG with a 750 Da PEG tail does not coat NPs, while PMAOD-PEG with 5,000 Da PEG and PMAOD-EG do, prompted a synchrotron small-angle X-ray scattering (SAXS) study of the micellization of the modified PMAcOD in water. Using the advanced SAXS data analysis methods it became for the first time possible to establish a link between the properties of micellar solutions and the ability of an alternating copolymer to efficiently coat the NPs. The NPs coated with modified PMAcODs were characterized using transmission electron microscopy (TEM), dynamic light scattering (DLS), ζ-potential measurements, and thermal gravimetric analysis (TGA).
2. Experimental
2.1 Materials
Hexanes (85%), ethanol (95%), and acetone (99.78%) were purchased from EMD and used as received. Chloroform (Mallinckrodt, 100%), FeCl3.6H2O (98%, Aldrich), docosane (99%, Aldrich), oleic acid (OA, 90%, Aldrich), oleic acid sodium salt (97%, TCI), and TBE buffer (1.3 M Tris, 450 mM boric acid, 25 mM EDTA·Na2 in H2O, Fluka) were used without purification. PMAOD (30 000 – 50 000 Da, Aldrich) was used as received. O-(2-aminoethyl)-o’-methyl polyethylene glycols with MW5000 and MW 750 (PEG-NH2) and (±)-3-amino-1,2-propanediol (98.0%) (EG) were purchased from Fluka and used as received. Water was purified with a Milli-Q (Millipore) water purification system (18 μS).
2.2. Synthetic procedures
2.2.1. Synthesis of Iron Oxide Nanoparticles
For the detailed procedure see our preceding paper.43 The synthesis of iron oleate was carried out according to the published procedure.22 Iron oleate was then purified and dried for 24 h at room temperature and 20 h at 70 °C in the vacuum oven. Spherical iron oxide nanoparticles of 21.6 nm in diameter were prepared through thermal decomposition of iron oleate in the presence of oleic acid in docosane as solvent.43 The round bottom three-neck flask was charged with 1.91 g of iron oleate, 7.02 g of docosane, and 0.93 mL of oleic acid. The flask was evacuated three times and filled with argon afterwards. Then the flask was heated to 70 °C to melt docosane and then a vigorous stirring was set. The reaction mixture was heated up to ~370 °C with a heating rate of 3.3 °C/min and let stirring at this temperature for 5 min. Then the reaction mixture was allowed to cool down to 60 °C and was placed in a vial while it is a liquid. At room temperature the reaction solution solidifies, allowing for long term storage of NPs. When needed, waxy NP solid is melted with a heat gun and precipitated with a mixture of acetone and hexane (5:1), followed by two consecutive washings with mixtures of acetone and hexane with the volume ratios 3:1 and 1:1. The NP solution is centrifuged after each wash to remove side products. The NPs are easily resuspended in chloroform after the final wash. The yield is 84%. The TEM image of these NPs is shown in the Supporting Information (SI, Figure S1).
2.2.2. Synthesis of PMAOD-PEG(EG) copolymers
In order to encapsulate the iron oxide NPs in PMAOD-PEG(EG), first modified PEG polymers were synthesized. Notations “P”, “S”, and “G” are reserved for PMAOD-PEG(5,000), PMAOD-PEG(750) and PMAOD-EG, respectively. Mole ratios of 1:20 (P1, S1, G1), 1:30 (P2, S2, G2), 1:60 (P3, S3, G3), and 1:80 (P4) of PMAOD to PEG or EG were used. The stock solutions of these samples were prepared in chloroform. The PMAOD stock solution (10.0 mg/mL) was also prepared in chloroform. For a typical modified stock solution with a mole ratio PMAOD:PEG-NH2=20:1 (P1, see Table S1, the SI, for notations), 37.5 mg of 5000 Da PEG-NH2 was dissolved in 1 mL of CHCl3 and sonicated for 10 minutes. Upon completion, 1.5 mL of the PMAOD stock was added and the solution was allowed to stir overnight. Table S1 (the SI) summarizes the amounts of reagents for the modified PMAOD synthesis and the yields of the copolymers.
2.2.3. Encapsulation of Iron Oxide Nanoparticles with PMAOD-PEG(EG) copolymers
For nanoparticle encapsulation, a 1:1 mass ratio between iron oxide NPs and PMAOD was maintained, except for the EG stock solutions, where the 1:1.5 ratio was employed. In a typical experiment, 1 mg of NPs is combined with 0.17 mL of the P1 stock solution. The mixture is allowed to stir for 1 h. Chloroform is then evaporated under vacuum and 2 mL of 20% TBE buffer were added. After sonicating for 10 minutes, the solution was heated at 60 °C for additional 10 minutes. The mixture was allowed to stir overnight. The following day, the polymer excess was removed by ultracentrifugation (1 h, 90 000 rpm/440 kg). The supernatant was discarded and NP pellet resuspended in water. Aggregates were removed by low speed centrifugation. The isolated yield was not taken because of the reaction small scale.
2.2.4. Hydrolysis of modified PMAOD
The solutions of modified PMAOD (2 mL each; P3: 7.84 mg/mL, S3: 6.4 mg/ml, and G3: 7.4 mg/mL) in CHCl3 were evaporated in a vacuum oven. To the dried samples 2 mL of 20% TBE buffer was added and the solutions were stirred for 24h.
2.3. Characterization
Dynamic light scattering (DLS) and ζ-potential measurements were performed using Malvern Zetasizer Nano ZS). For the DLS measurements, typically, the diluted sample in water (concentration was in the range 0.05–0.15 mg/mL) underwent sonication for about 10–20 min and filtration with a 0.2 μm syringe filter before the measurement. Measurement duration was set to be determined automatically, and data were averaged from at least three runs. Intensity and volume distributions of the particle sizes were recorded.
ζ-potential potential was measured at pH 7.4. Data was processed using the absorption of bulk iron oxide, the indices of refraction of iron oxide and solvent, and the viscosity of the pure water. The Smoluchowski approximation was used to convert the electrophoretic mobility to a ζ-potential.
Electron-transparent specimens for TEM were prepared by placing a drop of a dilute solution onto a carbon-coated Cu grid. Images were acquired at an accelerating voltage of 80 kV on a JEOL JEM1010 transmission electron microscope. Images were analyzed with ImageJ software package to estimate NP diameters. Normally 150 to 300 NPs were used for analysis.
Thermal gravimetric analysis (TGA) was performed on TGAQ5000 IR manufactured by TA Instruments. The TGA samples were prepared in the following way. An aqueous solution of the NP sample coated with a copolymer was evaporated and the NPs were resuspended in the minimum amount of chloroform. The solution was completely transferred into a 100 μL platinum pan, by filling the pan and allowing chloroform to evaporate. The experiments were carried upon heating to 700 °C with a rate 10.0 °C/min.
The composition of modified PMAOD was studied by NMR. All NMR samples were dissolved in CDCl3 and all spectra were recorded on a Varian UNITY INOVA 500MHz spectrometer at 25 °C, unless stated otherwise. 1H, 13C, 13C DEPT (distortionless enhancement by polarization transfer), 1H DQF-COSY (double-quantum-filtered correlation spectroscopy), and 13C-1H HSQC (heteronuclear single quantum correlation) experiments were performed. The 1H and 13C spectral assignment was made based on the results of these experiments and the spectral simulation results using ACD HNMR and CNMR Predictor 9.0 software. Spin-lattice relaxation times (T1) were measured for these samples using inversion recovery method. The T1 values determined for each 13C peak of these samples were in the range of 0.8 to 5.5 seconds. These values were used to optimize NMR parameters for quantitative 13C measurements. The inverse gated decoupling 13C{1H} NMR experiments, with acquisition time of 0.8 s, pulse delay of 7 s, and pulse flip angle 30°, were performed for the quantitative measurements.
The synchrotron radiation X-ray scattering measurements were done in the European Molecular Biology Laboratory (EMBL) on the storage ring DORIS III of the Deutsches Elektronen Synchrotron (DESY, Hamburg) on the X33 camera with a MAR Image plate detector.44 The scattering was recorded in the range of the momentum transfer 0.07 < s < 5.0 nm−1, where s = (4πsinθ)/λ, 2θ is the scattering angle, and λ= 0.15 nm is the X-ray wavelength. All measurements were carried out in a vacuum cuvette to diminish the parasitic scattering, and the samples were exposed in four 30 seconds frames to monitor for possible radiation damage. Three sets of modified PMAcOD samples with four different concentrations in solution were studied by SAXS: (1) P3 with the solution concentrations of 1, 2, 3 and 7.84 mg/mL, (2) S3 with the solution concentrations of 1, 2, 3 and 6.4 mg/ml, and (3) G3 with the solution concentrations of 1, 2, 3 and 7.4 mg/mL. The scattering profiles were corrected for the background scattering from distilled water and processed using standard procedures.45
The distance distribution functions p(r) of the self-assembled nanoparticles of the modified PMAcODs were calculated using an indirect transform program GNOM.46 Low-resolution shapes and internal structure of the micelles of the grafted copolymer were reconstructed ab initio from the scattering patterns using the program DAMMIN.47 The SAXS patterns from the modeled structures of the modified PMAcODs in solution were calculated using the program CRYSOL48 and compared with the experimental scattering curves.
3. Results and Discussion
3.1. Synthesis and Characterization of Modified PMAOD
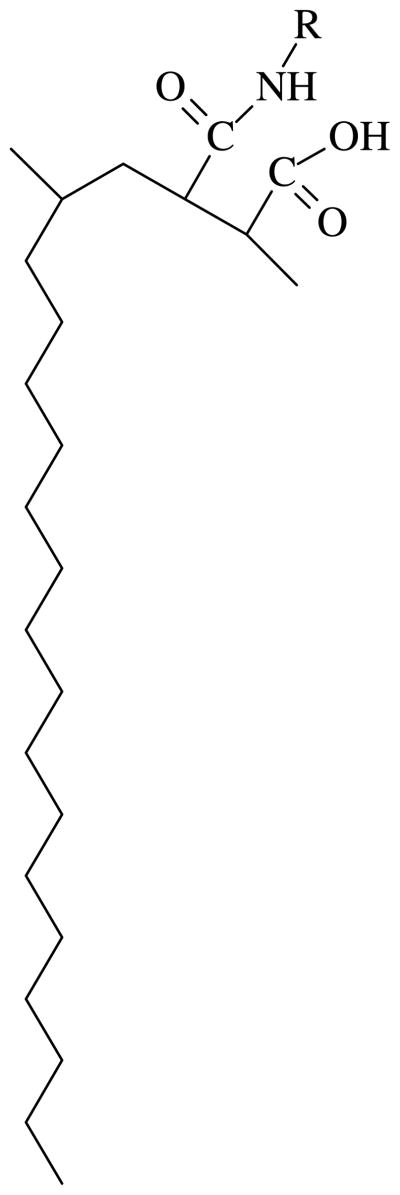
PMAOD contains an anhydride moiety in each other polymer unit, thus this moiety can easily interact with an amino group of various compounds with a formation of amide and carboxyl groups (Scheme 1). We varied the amount of grafted hydrophilic tails in the range 20–80% of all anhydride groups39,42 to determine its influence on the NP coating. In order to establish the influence of the concentration of the reagents on the degree of grafting, we used two very different concentrations: 1.67×10−5 and 6.3×10−4 mole.
Scheme 1.
A repeating unit of the modified PMAOD. R is either PEG-NH2 (MW 5,000 or 750 Da) or EG.
NMR was employed to quantitatively evaluate the proportion of grafting at different conditions. The NMR data (see the SI) revealed that at the concentration of 6.3×10−4 M, the reaction goes to completion for any degree of grafting (except for S3). At the lower concentration (1.67×10−5 M), the attachment is complete for 20% and 30% grafting while for the polymers with intended 60% or higher degree of grafting, approximately 20–30% of the PEG-NH2 molecules remained unreacted in the reaction solution. Unlike other copolymers, for S3, there is incomplete grafting even in concentrated solutions. The 13C NMR spectrum of S3 contains residual signals of NH2*CH2*CH2O at 41.7ppm and 73.3ppm, indicating the presence of unreacted PEG-NH2 (about 20%, see details in the SI).
After attaching the grafted tails, the copolymers were hydrolyzed in 20% TBE buffer to transform unreacted anhydride moieties to carboxyl groups. Because all these copolymers are amphiphilic, i.e., octadecene tail is hydrophobic, while the carboxyl groups along the PMAcOD chain and the grafted tails are hydrophilic, we expected self-assembling of these copolymers in water into micelle-like structures. These structures were characterized by DLS and ζ-potential measurements (Table 1.)
Table 1.
Hydrodynamic diameter (Dh) and ζ-potential of modified PMAcOD at the pH 7.4
| Copolymer | Dh (nm) | ζ-potential (mV) |
|---|---|---|
| G3 | 21.2 | −59.7 |
| S3 | 22.7 | −49.0 |
| P3 | 21.6 | −40.7 |
From the data presented in Table 1, the hydrodynamic diameter of the structures formed by G3, S3, and P3 does not depend on the length of the hydrophilic tail: all hydrolyzed copolymers showed approximately the same size micelles. This seems to be counterintuitive: It was believed that P3 would have the largest diameter due to the massive 5,000 Da PEG tails. Such inconsistencies of DLS data could be explained if micelles formed are not spherical. Indeed, as reported in our preceding paper41, PMAcOD forms disk-like structures in water so in the case of modified PMAOD, the analogous structures could be expected.
ζ-potential values were found to be more in accordance with our expectations. Due to the same amount of grafting, the amount of the remaining charges should be the same, but the charges can be differently exposed due the presence of the hydrophilic tails. Indeed P3 showed the smallest negative potential value at pH 7.4. It is believed that the large PEG tails are shielding the charges. The effect of shielding decreases with decreasing the molecular weight of the grafted tail.
3.2. Coating of Nanoparticles with Modified PMAcOD
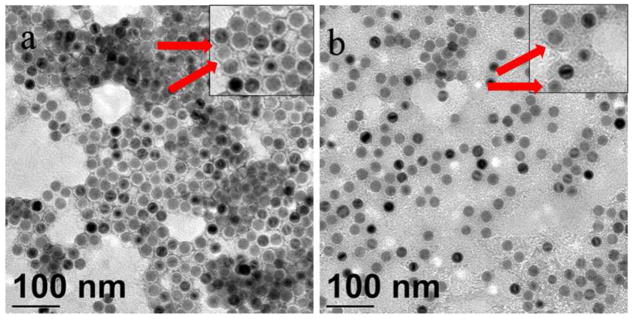
The coating procedure utilizes hydrophobic interactions of octadecene and oleic acid tails while hydrolysis of maleic anhydride moieties as well as hydrophilic PEG (EG) tails allows the NP hydrophilicity. Figure 1 presents TEM images of NPs coated with G3 and P3 copolymers. The negative staining with uranyl acetate also provides a positive staining due to interaction of carboxyl groups with uranyl cations. Thus, the PMAcOD backbone containing carboxyl group is accentuated with the stain. The thicknesses of the shells (without PEG or glycol tails which are invisible with this staining) for NP-P3 and NP-G3 are 3.4 and 3.7 nm, respectively.
Figure 1.
TEM images of NP-P3 (a) and NP-G3 (b) stained with uranyl acetate. Red arrows show the places where the positive staining can be seen despite the negative staining. Insets show higher magnification images.
3.2.1 DLS and ζ-potential data
To coat NPs with modified PMAcOD, we used a procedure described in our preceding paper41 where a solid mixture of NPs and a copolymer after evaporation of chloroform from a mixed solution was resuspended and hydrolyzed in 20% TBE buffer (see details in the Experimental part).41 This procedure allows a successful coating of various NPs with the P1, P2, P3, P4, G1, G2, and G3 copolymers, while no NPs were successfully (individually) coated with S1, S2, or S3. During the hydrolysis step only aggregates are observed. An alternative two-phase procedure as described elsewhere,39 where a chloroform solution of NPs and S1, S2 or S3 was mixed with water followed by slow removal of chloroform by rotary evaporation at room temperature, was also attempted. However, this procedure yielded a similar result: NP aggregation in aqueous solution. In the section 3.3 we discuss the causes of this phenomenon.
The DLS and ζ-potential data for the NP samples coated with modified PMAcOD are shown in Table 2.
Table 2.
Characteristics of NPs coated with modified PMAcOD
| Sample | Dha) (nm) | ζ-potential (mV) |
|---|---|---|
| NP-PMAcOD | 34.0 | −47.3 |
| NP-P1 | 39.8 | −47.0 |
| NP-P2 | 43.6 | −39.9 |
| NP-P3 | 56.0 | −30.9 |
| NP-P4 | 63.0 | −21.3 |
| NP-G1 | 58.9 | −53.2 |
| NP-G2 | 70.1 | −54.0 |
| NP-G3 | 69.3 | −33.7 |
from DLS measurements
The negative charge of NPs coated with modified PMAcOD should depend on the degree of grafting: the higher the degree of grafting, the lower the charge as a fraction of carboxylic groups decreases. Indeed, this trend is observed for both the P and G series, although for the latter it is less pronounced. The G1 and G2 and P1 and P2 samples differ only by 10% in grafting, so the ζ-potentials values can be close within an experimental error. On the other hand, for the P series, the additional effect, i.e., shielding of the charges, can take place when the grafting degree increases, thus making the difference between ζ-potentials more pronounced.
As for hydrodynamic diameters, the dependence is straightforward for the P series: the higher the degree of grafting the larger the size. This is due to increased stretching of the PEG coils when the PEG brush density increases. For the G series, one would expect the lack of the size dependence on the grafting density as EG is very short. Nevertheless, from G1 to G2 and G3 the hydrodynamic diameter increases which can be explained by increasing the fraction of the NP aggregates, when the negative charge decreases, thus weakening the electrostatic repulsion between NPs.
3.2.2 TGA data
In order to determine the amount of modified PMAcOD molecules on the NP surface, TGA measurements were carried out. First, the amount of oleic acid molecules on the as-prepared NPs has been determined by TGA (see the SI, Figure S3) using a weight loss of 14.3% upon the heating to 700 °C. One square nm of the NP surface was found to contain 7.34 OA molecules.
As is discussed earlier, coating of NPs with PMAcOD or modified PMAcOD occurs due to hydrophobic interactions of OA tails and octadecene tails,41 thus OA molecules are not removed during coating, if they are adsorbed on the NP surface. We have previously demonstrated, however, that even if NPs contain excess OA, it is perfectly removed upon coating with these copolymers.41 However, preparing the samples for TGA measurements, we made sure using FTIR that the sample contains no excessive OA (no band at 1711 cm−1), so we assumed that the amount of OA is constant in all the samples and the OA weight loss can be subtracted from the weight loss of other samples for calculation of the attached copolymer molecules. The TGA weight losses and the calculated amount of copolymer molecules per each NP are presented in Table 3 (see details in the SI, also Figure S4). It should be noted that for each data point, at least three TGA experiments have been run.
Table 3.
Weight losses and the amount of copolymer molecules per NP for the modified PMAcOD coated NPs
| Sample | Weight loss (%) | Copolymer molecules per NP |
|---|---|---|
| PMAOD | 45.5 | 417 |
| P3 | 32.3 | 21 |
| P4 | 34.3 | 19 |
| G2 | 50.0 | 398 |
| G3 | 55.4 | 465 |
The TGA data demonstrate that the PMAOD or G coated NPs carry approximately 400+ copolymer molecules on each NP, while the amount of copolymer macromolecules on the NPs coated with P copolymers is only 19–20. For the P3 and P4 samples, this can be explained by the large amount of the long PEG tails (5,000 Da) in the NP exterior creating highly hydrophilic environment with a few copolymer molecules and preventing the attachment of more copolymer molecules to the NP surface.
3.3. Small Angle X-ray Scattering
To understand the reasons for the different coating behavior of the S series copolymers compared to the copolymers of the P and G series, micellization of the modified PMAcOD samples in aqueous solutions was studied using SAXS.
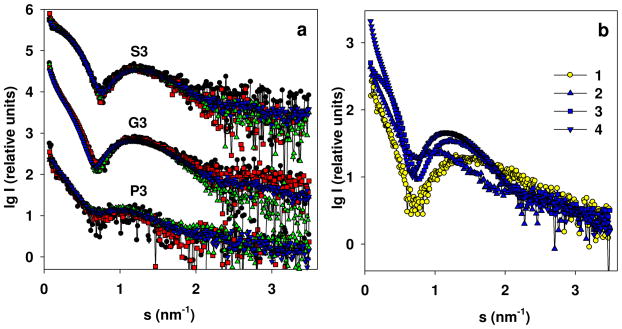
The experimental scattering profiles (Figure 2a) demonstrate that the three types of micelles have different dependence on the solute concentrations. For G3 samples, we observed a strong increase in overall size with increasing concentration; for the P3 set, the dependence was also present but weak; for the S3 samples, the scattering patterns taken at different concentrations were superimposable within the experimental noise. All the experimental profiles display a broad peak at higher angles, characteristic for micellar systems and resembling the SAXS pattern of pure PMAcOD discussed in our previous work41 (see the comparison in Figure 2b).
Figure 2.
(a) Experimental SAXS profiles from the copolymers in solution for the samples P3, G3, and S3: P3 with the solution concentrations of 1.0 (black circles), 2.0 (red squares), 4.0 (green triangles up), and 7.84 (blue triangles down); G3 with the solution concentrations of 1.0 (black circles), 2.0 (red squares), 4.0 (green triangles up), and 7.4 mg/mL (blue triangles down); S3 with the solution concentrations of 1.0 (black circles), 2.0 (red squares), 4.0 (green triangles up), and 6.4 mg/mL (blue triangles down). The patterns belonging to different copolymers are appropriately displaced along the logarithmic axis for better visualization. (b) A comparison of scattering profiles at the highest concentration for PMAcOD (1), P3 (2), G3 (3), and S3 (4).
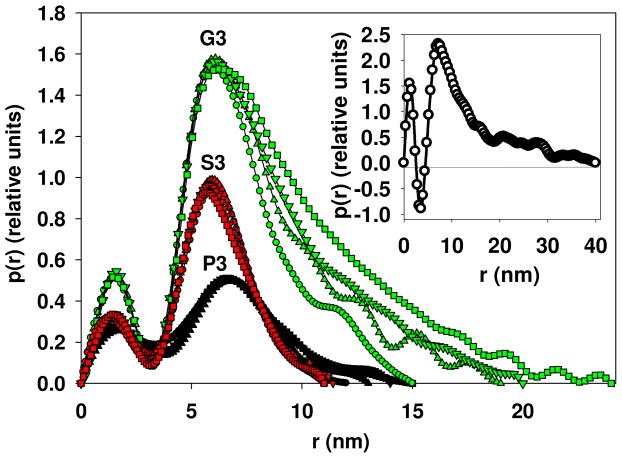
Given the concentration dependence, the distance distribution functions p(r) were calculated separately for individual concentrations (Figure 3). These functions reveal bimodal profiles characteristic for bilayer, but while p(r) for PMAcOD copolymer, displays negative values in the range of inter-atomic distances around 3–4 nm (Figure 3 inset), the distributions for the modified copolymers remain positive.
Figure 3.
Distance distribution functions for the P3, S3, and G3 samples at different concentrations: P3 with the solution concentrations of 1.0 (circles), 2.0 (triangle up), 4.0 (triangle down), and 7.84 mg/mL (square); S3 with the solution concentrations of 1.0 (circles), 2.0 (triangle up), 4.0 (triangle down), and 6.4 mg/mL (square); G3 with the solution concentrations of 1.0 (circles), 2.0 (triangle up), 4.0 (triangle down), and 7.4 mg/mL (square). In the inset: the distance distribution function for the PMAcOD copolymer in solution.41
The negative values for the pure PMAcOD copolymer are typical for lipid bilayer structures and appear due to the well-ordered hydrophobic regions in the copolymer, which have lower electron density than that of the water and thus negative contrast. The distances of 3–4 nm correspond to the bilayer width of the PMAcOD (1.6 nm for a single tail). Therefore, the observed changes in the p(r) functions for the modified PMAcOD copolymer point to alterations in the bilayer structures, which become less ordered upon modification.
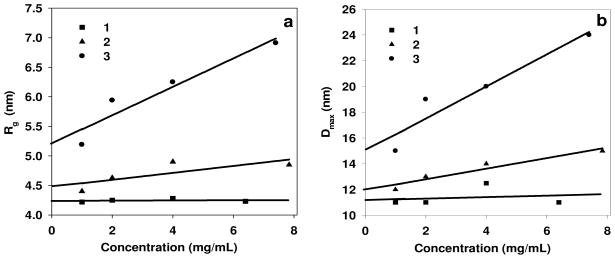
The length of the grafting tail affects the interactions between the copolymer micelles. The radii of gyration Rg and the maximum diameters Dmax depend on concentration for the P3 and G3 sets, but not for S3 (Figure 4).
Figure 4.
Dependence on the concentration (a) for radii of gyration: (1) the S3 samples (Rg0 =4.24); (2) the P3 samples (Rg0 =4.47); (3) the G3 samples (Rg0 =5.2); (b) for maximum diameters: (1) the S3 samples (D0 =11.0); (2) the P3 samples (D0 =12.0); (3) the G3 samples (D0 =15.0). Rg0 is the radius of gyration Rg extrapolated to zero concentration. D0 is the maximum size (Dmax) extrapolated to zero concentration.
As one can see from Figure 4, the micelles formed from PMAcOD modified by short glycol brushes (the G3 set) strongly interact in solution, which may facilitate interactions with NPs during coating. The regression for P3 displays a weaker dependence (less pronounced interactions) while the S3 set has no interactions at all. The latter micellar structures appear therefore as stable formations not interacting with each other, which may impair also their interactions with NPs.
To further assess the organization of the modified PMAcODs self-assemblies, models were constructed using MAcOD molecules generated in our previous work41 with an addition of appropriate glycol molecules to the head of MAcOD.
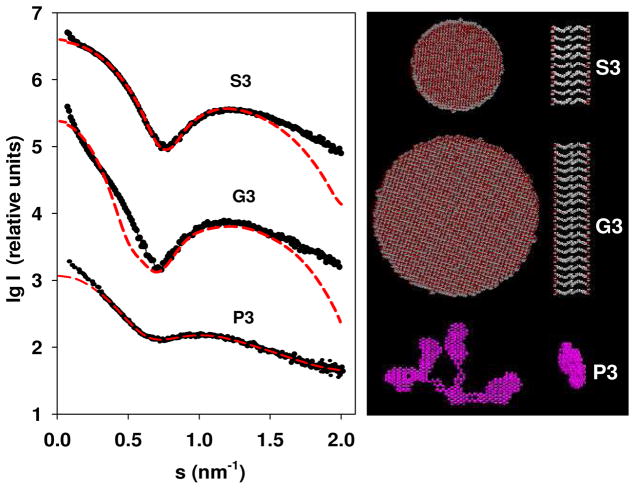
For pure PMAcOD, disc-like particles with the diameter 40 nm and the thickness of the bilayer 3.2 nm41 were found to be the only type of self-assembled structures in solution. Similar to the previous work, the modified –MA3OD- units were used to construct bilayer disks but also other geometrical structures containing bilayers. Their SAXS patterns calculated using CRYSOL48 were compared with the experimental scattering. When using disk-like models, good fits were obtained for the S3 and G3 samples (other bilayer shapes like spheres or long cylinders did not provide good fits), while for the P3 samples, no fit could be found for any geometrical shape. The experimental scattering curve for the P3 copolymer appears smeared compared to those of the G3 or S3 samples reflecting possible disruptions of the bilayer structure. This loss of regularity is probably caused by the interactions between the long and heavy PEG chains. To assess the shape of the P3 sample, ab initio dummy atom modeling was employed using the program DAMMIN.47
The best models for the G3, S3, and P3 samples along with their fits to the experimental data are presented in Figure 5. The obtained parameters of the model disk for the G3 sample were the diameter 18 nm and the width of the bilayer 4.8 nm, and for the S3 sample, the diameter was found to be 10 nm, while the width of the bilayer was estimated to be 5.0 nm. Note that the S3 pattern could be fitted very well, whereas the calculated pattern for G3 displays deviations at small angles, whereby the experimental data indicate larger overall size of the aggregates. However, further increase of the diameter of the G3 disc leads to systematic deviations at higher angles. It is conceivable that the G3 samples contain aggregates of the disc-like micelles at the concentration used in the calculations (7.4 mg/mL).
Figure 5.
Modeling and the best fits: the experimental scattering curve from the modified PMAcOD copolymer in solution (black circles) and the best fits to the experimental profile of the scattering patterns computed from models (red dotted lines). Modelling was performed at maximal concentrations to reduce the influence of the experimental noise. The right panel: disk like models for the S3 and G3 samples in two orientations, and a dummy atom model for the P3 specimen. Separately, a structure of the dense formations in the body of the P3 micelles is shown
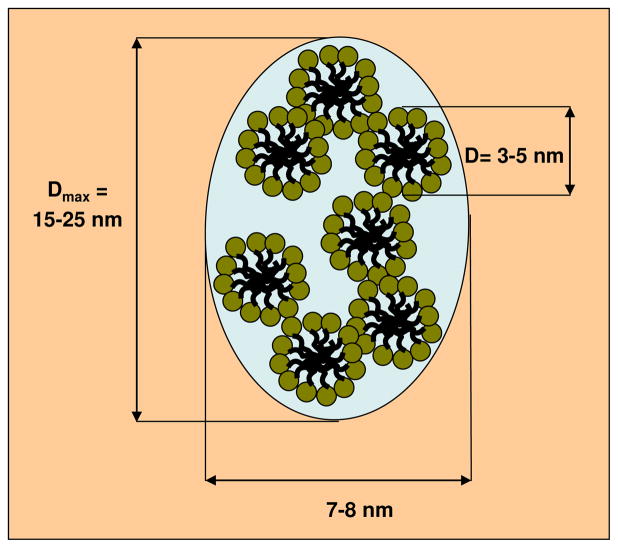
Owinig to the complexity of the modified PMAcOD units for the S3 and G3 samples, we could only use hypothetical models of the monomeric units. Nevertheless, the models obtained allow us to meaningfully analyze the possibility of creating protective shells using these structures. Both the G3 and S3 samples form ordered bilayers, but the S3 structures are much smaller, and the micelles do not interact in solution. It appears that the S3 macromolecules form quite dense, stable structures, which are energetically favorable and do not promote interactions with external hydrophobic objects like NPs. The micelles of the G3 and especially of the P3 copolymers are less stable and the NPs are able to compete with the copolymer self-assembly leading to successful coating. The ab initio model of the P3 samples appears as an agglomerate of loosely connected subunits. The shape of these subunits restored by the program DAMMIN using a portion of the experimental scattering curve at higher angles from 2.5 up to 3.5 nm−1 reveals a slightly oblate body with the diameter of about 5.5 nm (Figure 5). A tentative model of the overall structure of the P3 samples can be represented as a mesh of segregated sections of the modified copolymer, which are small micelles of the PMAcOD chains with the diameters of about 5.0 – 5.5 nm covered by layers of the PEG molecules, most probably in a bent conformation (Scheme 2). The geometrical parameters of the model indicated in Schemes 2 correspond to the dimensions calculated from the p(r) function for this sample (Figure 2).
Scheme 2.
Hypothetical model of the structure of the P3 copolymer in solution.
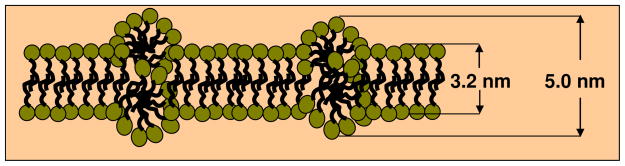
It should be noted that the bilayers of the micelles of the S3 and G3 sets are thicker than that for the pure PMAcOD evaluated previously41: 5.0 nm for the S3 sample and 4.8 nm for the G3 sample compared with 3.2 nm for PMAcOD. This can be explained by partial segregation during micellization. As a result, the total thickness of the bilayers is greater than the double thickness of the completely straightened octadecene tails of PMAcOD as depicted in Scheme 3. It is also conceivable that the PEG tails contribute to the effective thickness of the models. One should however note that the longer PEG chains are too disordered to be captured in the models together with the copolymer heads: our attempts to add longer glycol chains to the MAoCD model did not yield satisfactory fits, neither for the G3 nor for P3 molecules.
Scheme 3.
Hypothetical model of partial segregation in micellar bilayers.
3.4. Stability of NPs coated with modified PMAOD
In ref.36 encapsulation of magnetic nanoparticles by viral protein cages was reported. Protein cages of Brome Mosaic Virus (BMV) were assembled around negatively charged particles in aqueous buffers. As was shown previously, NPs coated with modified PMAOD acquire negative charge (Table 3) and display excellent stability in water, but immediately salt out in buffer solutions. We expected that grafting of hydrophilic chains to the PMAOD backbone will improve stability of NPs in buffers. The nanoparticles coated with modified PMAOD were dialyzed in a virus capsid reassembly buffer (Tris 0.05M, KCl 0.01M, MgCl2 5mM, pH 7.39). The P1, P2, G1, and G3 samples aggregated within 20 minutes, while the P3 and P4 samples remained fully soluble even after 24 hours. These samples were further dialyzed against 1M TKM buffer (Tris 0.01M, KCl 1M, and MgCl2 0.005M, pH 7.42) which is used to maintain BMV coat proteins disassembled as dimers. The nanoparticles remained stable for months in that buffer.
4. Conclusions
We established that coating of NPs with modified PMAcOD depends on the length of the grafted tails, which, in turn, determines the types of copolymer micelles in solution as revealed by SAXS. When the 5,000 Da PEG or short EG tails are attached to the PMAcOD backbone, the micelles formed (despite the difference of their shapes and ordering) exhibit intermicellar interaction in solution; the latter most likely is the crucial factor for interacting with the OA shell on the NP surface. In contrast, the copolymer formed by the attachment of the 750 Da PEG tails to the PMAcOD backbone forms small, non-interacting micelles which are presumably self-sufficient and do not exhibit interactions with a hydrophobic NP shell, thus preventing NP coating. The importance of this work is in understanding of the connection between the micelle properties and the coating ability when amphiphilic copolymers or other amphiphilic molecules are concerned, which may become a guide for future work on the NP encapsulation.
We synthesized a series of hydrophilic, negatively charged NPs using modified PMAcOD of the P and G series. The TGA data revealed that if for the G and PMAcOD coatings approximately 400 molecules need to be attached to a single NP to allow its stability in water, the P3 and P4 coatings are successful with only approximately 20 macromolecules, emphasizing the importance of the PEG tails in NP hydrophilization. Among the P coated samples, only the P3 modified NPs demonstrated both a sufficient negative charge36 and a good stability in buffers allowing further studies of virus-like particle formation with these modified cores.
Supplementary Material
Acknowledgments
This work has been supported, in part, by the NATO Science for Peace Program (grant SfP-981438), NSF award 0631982, NIH award GM081029-01, NSF award 0220560, the IU FRSP grant, and the European Union FP6 Infrastructures Program (Design Study SAXIER, RIDS 011934). B.D. acknowledges partial support from the Indiana METACyt Initiative of Indiana University, funded in part through a major grant from the Lilly Endowment, Inc. The measurements at the EMBL beamline X33 at DESY were made within the projects SAXS-06-29 and SAXS-07-29. E.S. and L.B. thank the Federal Program “Scientists and Educators of Innovative Russia” 2009-20013, contract #14.740.11.0380. We also thank resources of the IU Nanoscale Characterization Facility for access to the instrumentation.
Footnotes
Supporting Information Available: TEM image of iron oxide nanoparticles, synthesis conditions of modified PMAcOD samples, NMR studies of the degree of grafting, and TGA data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Chang SY, Zheng NY, Chen CS, Chen CD, Chen YY, Wang CRC. J Am Soc Mass Spect. 2007;18:910–918. doi: 10.1016/j.jasms.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 2.Bruls DM, Evers TH, Kahlman JAH, van Lankvelt PJW, Ovsyanko M, Pelssers EGM, Schleipen JJHB, de Theije FK, Verschuren CA, van der Wijk T, van Zon JBA, Dittmer WU, Immink AHJ, Nieuwenhuis JH, Prins MWJ. Lab on a Chip. 2009;9:3504–3510. doi: 10.1039/b913960e. [DOI] [PubMed] [Google Scholar]
- 3.Bulte JW, Kraitchman DL. NMR Biomed. 2004;17:484–499. doi: 10.1002/nbm.924. [DOI] [PubMed] [Google Scholar]
- 4.Mulder WJM, Strijkers GJ, van Tilborg GAF, Griffioen AW, Nicolay K. NMR Biomed. 2006;19:142–164. doi: 10.1002/nbm.1011. [DOI] [PubMed] [Google Scholar]
- 5.Medarova Z, Pham W, Farrar C, Petkova V, Moore A. Nature Medicine. 2006;13:372–377. doi: 10.1038/nm1486. [DOI] [PubMed] [Google Scholar]
- 6.Kumagai M, Imai Y, Nakamura T, Yamasaki Y, Sekino M, Ueno S, Hanaoka K, Kikuchi K, Nagano T, Kaneko E, Shimokado K, Kataoka K. Coll Surf B. 2007;56:174–181. doi: 10.1016/j.colsurfb.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 7.Jain TK, Foy SP, Erokwu B, Dimitrijevic S, Flask CA, Labhasetwar V. Biomaterials. 2009;30:6748–6756. doi: 10.1016/j.biomaterials.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chemla YR, Crossman HL, Poon Y, McDermott RRS, Alper MD, Clarke J. Proc Nat Acad Sci. 2000;97:14268–14272. doi: 10.1073/pnas.97.26.14268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivkov R, DeNardo SJ, Miers LA, Natarajan A, Foreman AR, Gruettner C, Adamson GN, DeNardo GL. NSTI Nanotech 2006. Vol. 2. Nano Science and Technology Institute; Boston, MA, USA: 2006. pp. 21–24. [Google Scholar]
- 10.Chen ZP, Zhang Y, Xu K, Xu RZ, Liu JW, Gu N. J Nanosci Nanotech. 2008;8:6260–6265. [PubMed] [Google Scholar]
- 11.Das M, Mishra D, Maiti TK, Basak A, Pramanik P. Nanotechnology. 2008;19:415101/1–415101/14. doi: 10.1088/0957-4484/19/41/415101. [DOI] [PubMed] [Google Scholar]
- 12.Lu AH, Salabas EL, Schueth F. Angew Chim Int Ed. 2007;46:1222–1244. doi: 10.1002/anie.200602866. [DOI] [PubMed] [Google Scholar]
- 13.Talapin DV, Shevchenko EV, Weller H. In: Nanoparticles. Schmid G, editor. Wiley-VCH; Weinheim: 2004. pp. 199–230. [Google Scholar]
- 14.Park TJ, Papaefthymiou GC, Viescas AJ, Moodenbaugh AR, Wong SS. Nano Lett. 2007;7:766–772. doi: 10.1021/nl063039w. [DOI] [PubMed] [Google Scholar]
- 15.Rong CB, Li D, Nandwana V, Poudyal N, Ding Y, Wang ZL, Zeng H, Liu JP. Adv Mater. 2006;18:2984–2988. [Google Scholar]
- 16.Hellstern D, Schulze K, Schopf B, Petri-Fink A, Steitz B, Kamau S, Hilbe M, Koch-Schneidemann S, Vaughan L, Hottiger M, Hofmann M, Hofmann H, von Rechenberg B. J Nanosci Nanotech. 2006;6:3261–3268. doi: 10.1166/jnn.2006.482. [DOI] [PubMed] [Google Scholar]
- 17.Briley-Saebo K, Bjornerud A, Grant D, Ahlstrom H, Berg T, Kindberg GM. Cell & Tissue Res. 2004;316:315–323. doi: 10.1007/s00441-004-0884-8. [DOI] [PubMed] [Google Scholar]
- 18.Sun S, Zeng H. J Am Chem Soc. 2002;124:8204–8205. doi: 10.1021/ja026501x. [DOI] [PubMed] [Google Scholar]
- 19.Li Z, Chen H, Bao H, Gao M. Chem Mater. 2004;16:1391–1393. [Google Scholar]
- 20.Redl FX, Black CT, Papaefthymiou GC, Sandstrom RL, Yin M, Zeng H, Murray CB, O’Brien SP. J Am Chem Soc. 2004;126:14583–14599. doi: 10.1021/ja046808r. [DOI] [PubMed] [Google Scholar]
- 21.Yu WW, Falkner JC, Yavuz CT, Colvin VL. Chem Comm. 2004:2306–2307. doi: 10.1039/b409601k. [DOI] [PubMed] [Google Scholar]
- 22.Park J, An K, Hwang Y, Park JG, Noh HJ, Kim JY, Park JH, Hwang NM, Hyeon T. Nature Mater. 2004;3:891–895. doi: 10.1038/nmat1251. [DOI] [PubMed] [Google Scholar]
- 23.Kwon SG, Piao Y, Park J, Angappane S, Jo Y, Hwang NM, Park JG, Hyeon T. J Am Chem Soc. 2007;129:12571–12584. doi: 10.1021/ja074633q. [DOI] [PubMed] [Google Scholar]
- 24.Jana NR, Chen Y, Peng X. Chem Mater. 2004;16:3931–3935. [Google Scholar]
- 25.Jun YW, Huh YM, Choi JS, Lee JH, Song HT, Kim S, Yoon S, Kim KS, Shin JS, Suh JS, Cheon J. J Am Chem Soc. 2005;127:5732–5733. doi: 10.1021/ja0422155. [DOI] [PubMed] [Google Scholar]
- 26.Gussin HA, Tomlinson ID, Little DM, Warnement MR, Qian H, Rosenthal SJ, Pepperberg DR. J Am Chem Soc. 2006;128:15701–15713. doi: 10.1021/ja064324k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q, Gupta S, Emrick T, Russell TP. J Am Chem Soc. 2006;128:3898–3899. doi: 10.1021/ja058615p. [DOI] [PubMed] [Google Scholar]
- 28.Han G, Ghosh P, Rotello VM. Nanomedicine. 2007;2:113–123. doi: 10.2217/17435889.2.1.113. [DOI] [PubMed] [Google Scholar]
- 29.Hostetler MJ, Templeton AC, Murray RW. Langmuir. 1999;15:3782–3789. [Google Scholar]
- 30.Wan S, Huang J, Guo M, Zhang H, Cao Y, Yan H, Liu K. J Biomed Mater Res. 2007;80A:946–954. doi: 10.1002/jbm.a.31022. [DOI] [PubMed] [Google Scholar]
- 31.Kang Y, Taton TA. Macromolecules. 2005;38:6115–6121. [Google Scholar]
- 32.Korth BD, Keng P, Shim I, Bowles SE, Tang C, Kowalewski T, Nebesny KW, Pyun J. J Am Chem Soc. 2006;128:6562–6563. doi: 10.1021/ja0609147. [DOI] [PubMed] [Google Scholar]
- 33.Nitin N, LaConte LEW, Zurkiya O, Hu X, Bao G. J Biol Inorg Chem. 2004;9:706–712. doi: 10.1007/s00775-004-0560-1. [DOI] [PubMed] [Google Scholar]
- 34.Dubertret B, Skourides P, Norris DJ, Noireaux V, Brivanlou AH, Libchaber A. Science. 2002;298:1759–1762. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- 35.Lim YT, Lee KY, Lee K, Chung BH. Biochem Biophysl Res Comm. 2006;344:926–930. doi: 10.1016/j.bbrc.2006.03.209. [DOI] [PubMed] [Google Scholar]
- 36.Huang X, Bronstein LM, Retrum JR, Dufort C, Tsvetkova I, Aniagyei S, Stein B, Stucky G, McKenna B, Remmes N, Baxter B, Kao CC, Dragnea B. Nano Lett. 2007;7:2407–2416. doi: 10.1021/nl071083l. [DOI] [PubMed] [Google Scholar]
- 37.Shtykova EV, Huang X, Remmes N, Baxter D, Stein BD, Dragnea B, Svergun DI, Bronstein LM. J Phys Chem C. 2007;111:18078–18086. [Google Scholar]
- 38.Pellegrino T, Manna L, Kudera S, Liedl T, Koktysh D, Rogach AL, Keller S, Raedler J, Natile G, Parak WJ. Nano Lett. 2004;4:703–707. [Google Scholar]
- 39.Yu WW, Chang E, Sayes CM, Drezek R, Colvin VL. Nanotechnology. 2006;17:4483–4487. [Google Scholar]
- 40.Di Corato R, Quarta A, Piacenza P, Ragusa A, Figuerola A, Buonsanti R, Cingolani R, Manna L, Pellegrino T. J Mater Chem. 2008;18:1991–1996. [Google Scholar]
- 41.Shtykova EV, Gao X, Huang X, Dyke JC, Schmucker AL, Remmes N, Baxter DV, Stein B, Dragnea B, Konarev PV, Svergun DI, Bronstein LM. J Phys Chem C. 2008;112:16809–16817. doi: 10.1021/jp8053636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu WW, Chang E, Falkner JC, Zhang J, Al-Somali AM, Sayes CM, Johns J, Drezek R, Colvin VL. J Am Chem Soc. 2007;129:2871–2879. doi: 10.1021/ja067184n. [DOI] [PubMed] [Google Scholar]
- 43.Bronstein LM, Huang X, Retrum J, Schmucker A, Pink M, Stein BD, Dragnea B. Chem Mater. 2007;19:3624–3632. doi: 10.1039/b821917f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roessle MW, Klaering R, Ristau U, Robrahn B, Jahn D, Gehrmann T, Konarev P, Round A, Fiedler S, Hermes C, Svergun D. J Appl Cryst. 2007;40:s190–s194. [Google Scholar]
- 45.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 46.Svergun DI. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- 47.Svergun DI. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Svergun DI, Barberato C, Koch MHJ. J Appl Cryst. 1995;28:768. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.