SUMMARY
Eukaryotic transcription and mRNA processing depend upon the coordinated interactions of many proteins, including Spn1 and Spt6, which are conserved across eukaryotes, are essential for viability, and associate with each other in some of their biologically important contexts. Here we report crystal structures of the Spn1 core alone and in complex with the binding determinant of Spt6. Mutating interface residues greatly diminishes binding in vitro and causes strong phenotypes in vivo, including a defect in maintaining repressive chromatin. Overexpression of Spn1 partially suppresses the defects caused by an spt6 mutation affecting the Spn1 interface, indicating that the Spn1-Spt6 interaction is important for managing chromatin. Spt6 binds nucleosomes directly in vitro, and this interaction is blocked by Spn1, providing further mechanistic insight into the function of the interaction. These data thereby reveal the structural and biochemical bases of molecular interactions that function in the maintenance of chromatin structure.
INTRODUCTION
Spn1 and Spt6 are transcription factors that interact with one another and are each essential for viability in yeast (Clark-Adams and Winston, 1987; Fischbeck et al., 2002). S. cerevisiae Spn1 is a 410 residue, 46 kDa protein with a central core domain (residues 140–300) that is flanked on both sides by regions that are predicted to be disordered (Ward et al., 2004). Spt6 is a 1451 residue, 168 kDa protein whose core (residues 300–1250) likely resembles the structure of the bacterial Tex protein (Johnson et al., 2008) with an acidic N-terminal extension that is expected to be unstructured (Ward et al., 2004) and a C-terminal domain that adopts an SH2 fold (Dengl et al., 2009; Maclennan and Shaw, 1993). Spn1 and Spt6 interact stably with one another, and they and their interaction have been implicated in several aspects of gene expression (Krogan et al., 2002; Lindstrom et al., 2003; Yoh et al., 2007; Yoh et al., 2008).
Spt6 was originally identified in a screen for factors that alter normal initiation of transcription (Clark-Adams and Winston, 1987; Denis, 1984; Neigeborn et al., 1987; Simchen et al., 1984). Subsequently, Spt6 was implicated in a variety of biological processes in organisms ranging from yeasts to humans, including embryogenesis in Zebrafish (Keegan et al., 2002; Kok et al., 2007), multiple stages of development in Drosophila (Ardehali et al., 2009), gut morphogenesis in C. elegans (Nishiwaki et al., 1993), signal transduction in mammals (Baniahmad et al., 1995; Shen et al., 2009), and HIV transcription regulation and mRNA processing in human cells (Vanti et al., 2009; Yoh et al., 2007). The role in transcription initiation has been ascribed to the ability of Spt6 to chaperone histones to promote reassembly of nucleosomes in the wake of RNA polymerase II (RNAPII), thereby reestablishing the default repressive chromatin state that prevents inappropriate initiation of transcription (Adkins and Tyler, 2006; Bortvin and Winston, 1996; Cheung et al., 2008; Kaplan et al., 2003). While of profound importance, maintaining repressive chromatin appears to be just one of Spt6’s roles. For example, Spt6 also promotes elongation by RNAPII (Hartzog et al., 1998; Kaplan et al., 2005; Kaplan et al., 2000; Lindstrom et al., 2003) on nucleosome-free DNA templates in vitro (Endoh et al., 2004; Hartzog et al., 1998; Keegan et al., 2002; Yoh et al., 2007), as well as on chromatin templates in vivo (Ardehali et al., 2009). Together, these data indicate that Spt6 plays a number of mechanistically distinct roles during transcription.
The SPN1 gene was originally identified as a key regulator of transcription from genes that are regulated post-recruitment of RNAPII (Fischbeck et al., 2002). Spn1 was also identified as a protein that interacts with Spt6 and has been reported to bind with Spt6 in some but not all of Spt6’s functional states (Lindstrom et al., 2003; Yoh et al., 2007; Zhang et al., 2008). For example, Spt6 can be coimmunopurified with three distinct Spt4/5-RNAPII complexes, whereas Spn1 is found in only two of these complexes (Lindstrom et al., 2003). The CYC1 gene of S. cerevisiae provides an example of how the Spn1-Spt6 interaction contributes to post-recruitment regulation (Zhang et al., 2008). RNAPII is constitutively bound to the CYC1 promoter, but is kept from elongating because it interacts with Spn1, which in turn inhibits the Swi/Snf nucleosome remodeling complex from promoting transcription. During activation, Spt6 binds to Spn1, and repression of Swi/Snf recruitment is relieved.
Spn1 is also needed to achieve normal recruitment of the histone methyltransferase HYPB/Setd2 (Yoh et al., 2008) and the elongation factor TFIIS (Ling et al., 2006; Zhang et al., 2008) to RNAPII complexes traversing active genes. HYPB/Setd2 methylates histone H3K36, which in turn recruits Rpd3-type histone deacetylases to restore chromatin to the repressive hypoacetylated state and block inappropriate transcription (Yoh et al., 2008). In contrast to their antagonistic relationship in activating post-recruitment initiation, Spn1 and Spt6 each contribute towards restoration of repressive chromatin. Human Spn1/IWS1 also binds the protein arginine methyltransferase PRMT5, which methylates the elongation factor Spt5 and thereby regulates its interaction with RNAPII (Liu et al., 2007). Spn1 can additionally function through interactions with pathway-specific regulatory factors, such as the Arabidopsis steroid hormone responsive transcription factor BES1, which recruits Spn1 to the promoter and transcribed regions of activated genes (Li et al.). Spn1 therefore contributes in several ways to the appropriate functioning of RNAPII.
In addition to their roles in regulating transcription, Spt6 and Spn1 also collaborate to promote mRNA processing and export. Spt6 is required for proper 3′ end formation by preventing premature 3′ processing at upstream polyadenylation signals (Bucheli and Buratowski, 2005; Kaplan et al., 2005). Further, mammalian Spt6 can bind the Ser2-phosphorylated RNAPII C-terminal domain (CTD), enhancing recruitment of RNA processing/export factors (Yoh et al., 2007; Yoh et al., 2008), and Drosophila Spt6 copurifies with the RNA processing exosome complex (Andrulis et al., 2002). Both SPN1 and SPT6 have also been implicated in mRNA splicing in S. cerevisiae (Burckin et al., 2005), and binding of human Spn1/IWS1 to the RNA export factor REF1/Aly is important for recruitment of REF1/Aly to the body of the c-Myc gene during transcription (Yoh et al., 2007).
Spt6 and Spn1 and their interaction with one another therefore play pivotal roles in defining the composition of RNAPII elongation complexes, maintaining the structure of chromatin, and modulating the production of mature mRNA transcripts. To advance mechanistic understanding of their functions, we have determined the structural basis of the Spn1-Spt6 interaction. We also demonstrate the importance of this interface in vitro and in vivo, and show that Spn1 negatively regulates binding of Spt6 to nucleosomes.
RESULTS
Mapping of the Spn1-Spt6 Interface
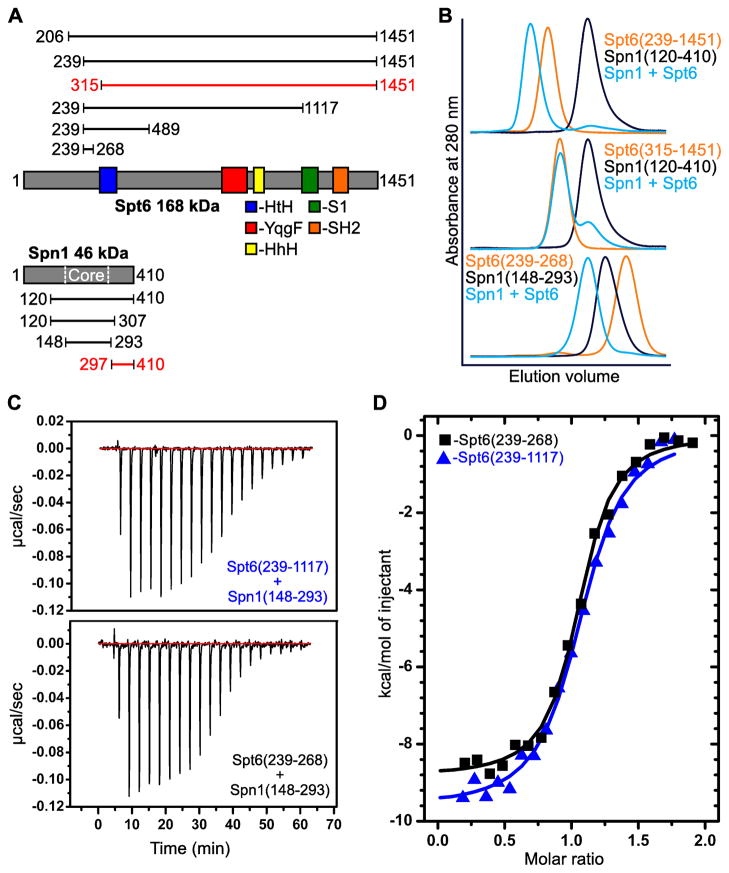
Full length S. cerevisiae Spt6 and Spn1 proteins were poorly-behaved, but deletion of much of their presumably unstructured N-terminal regions (Ward et al., 2004) allowed us to observe co-elution of a complex of recombinant Spn1(120–410) and Spt6(206–1451) by size-exclusion chromatography in sodium chloride concentrations up to 300 mM (not shown). Spt6(239–1451) also bound Spn1 whereas Spt6(315–1451) did not. Further truncations revealed that Spt6(239–268) is sufficient for Spn1 binding (Figure 1A–B). This 30 residue segment of Spt6 is predicted to be unstructured, and comes from a region that is N-terminal to the region expected to resemble the structure of the bacterial protein Tex (Johnson et al., 2008). Spn1(148–293) includes most of the Spn1 residues that are predicted to be structured and retained the ability to bind Spt6. The slightly larger Spn1(141–305) fragment was previously shown to complement a deletion of SPN1 (Fischbeck et al., 2002), indicating that this core domain provides the major function(s) of Spn1 in vivo. Isothermal titration calorimetry (ITC) was used to measure binding affinities of Spn1(148–293) for two different Spt6 constructs: Spt6(239–1117), the largest Spt6 construct that remained soluble at sufficient concentrations for these experiments, and Spt6(239–268), the smallest construct tried that retained full binding affinity. In both cases the binding displayed 1:1 stoichiometry and the mean binding constant (KD) was 170 nM (Figure 1C–D and Table S1 available online). This indicates that the 30 residue segment of Spt6, Spt6(239–268), is sufficient to recapitulate the binding energy observed for larger Spt6 constructs.
Figure 1. The Spn1 core binds 30 residues in the Spt6 N-terminal region.
(A) Spt6 and Spn1 domain organization and their interacting regions. Constructs assayed for binding are indicated above (Spt6) or below (Spn1) with their N- and C-terminal residues given. Constructs that showed binding are colored black while those that did not show binding are colored red.
(B) Overlays of size-exclusion chromatograms with Spn1 black, Spt6 orange, and binding experiments blue. Chromatograms are scaled on the y-axis to allow comparison of the elution volume profiles of each protein or binding experiment. Spn1 has a 6-fold lower molar extinction coefficient, and so at equimolar concentrations, the Spn1 peak appears approximately 6 times smaller than the Spt6 peak (blue, middle panel). The x-axis shows the 50–100 mL elution volume.
(C) Raw ITC data of the titration of Spn1(148–293) into Spt6(239–1117) (top) and the titration of Spt6(239–268) into Spn1(148–293) (bottom).
(D) Representative binding isotherms of the titration experiments from (C). See text and Table S1 for thermodynamic parameters.
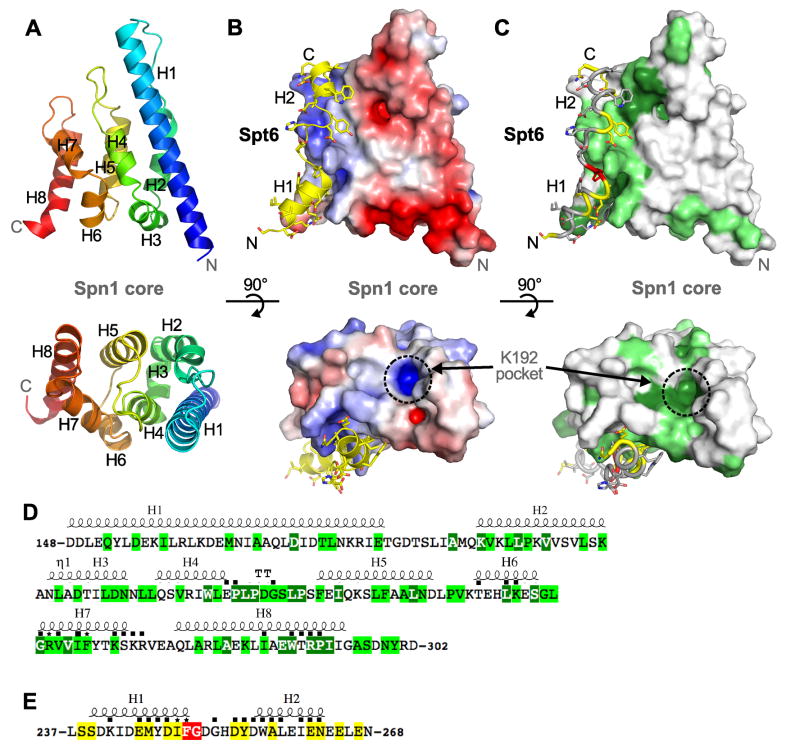
Crystal structures of the Spn1 core
The crystal structure of Spn1(148–307) was determined by the single-wavelength anomalous diffraction method using data collected to 3.0 Å resolution from a selenomethionine-substituted crystal (Figure 2A, Table 1). This unrefined model was used in molecular replacement calculations with 2.15 Å data from a native crystal that belonged to a different space group, and the native structure was refined to Rfactor/Rfree values of 18.5/22.4%. Residues 148–295 were clearly observed in the electron density as were four non-native N-terminal residues that remained after TEV digestion. The twelve C-terminal residues were disordered and are not included in the final model.
Figure 2. Structures of the Spn1 core and its complex with Spt6.
(A) The structure of the Spn1 core shown as a cartoon representation in two orthogonal views. The polypeptide chain is colored as a blue to red rainbow from N to C-terminus. Secondary structures and the N and C-termini are labeled.
(B) Structure of the Spn1-Spt6 complex. The Spn1 core is shown as a surface electrostatic (±5 kT/e) representation in the same views as panel A. Spt6 is colored yellow and the N and C-termini and the helices are labeled. Spn1 K192 has been implicated in interactions with RNAPII (Zhang et al., 2008), and lies in the basic conserved pocket indicated. Remnants of the affinity tags that remain in the crystallized Spn1 and Spt6 proteins are not shown in any of the figures and do not contribute to the Spn1 interface.
(C) Same as panel B except that the Spn1 surface and Spt6 peptide are colored according to conservation.
(D) Spn1 core amino acid sequence. Secondary structural elements are indicated above, and were defined with ESPript (Gouet et al., 2003). Residues that make direct contact across the interface are indicated with a black square or with an asterisk if they were mutated in this study. Numbering and residue identities shown here refers to the S. cerevisiae protein. Coloring represents degrees of conservation, dark green (high), light green (medium), in an alignment of proteins from Saccharomyces cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans, Drosophila melanogaster, Danio rerio, and Homo sapiens. Amino acid sequences were aligned using the T-coffee multiple sequence alignment method (Notredame et al., 2000) and slightly adjusted manually in light of the structure (Figure S2).
(E) Same as panel D but for Spt6. High conservation (red), medium conservation (yellow).
Table 1.
Data Collection and Refinement Statistics
| Spn1 Core SeMet | Spn1 Core Native | Spn1-Spt6 Complex | |
|---|---|---|---|
|
Data Collection | |||
| Beamline | SSRL 9-2 | Home source | Home source |
| Space group | P3112 | P3212 | P21212 |
| Unit cell dimensions | |||
| a, b, c (Å) | 61.9, 61.9, 240.4 | 61.3, 61.3, 116.05 | 105.9, 68.7, 73.9 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 90 |
| Resolution (Å) | 35-3.0 | 30-2.15 | 30-2.15 |
| Wavelength (Å) | 0.97923 (Peak) | 1.54178 | 1.54178 |
| I/σI | 23.2 (2.8) | 22 (4.1) | 19.7 (3.1) |
| Completeness (%) | 90.8 (92.3) | 99.8 (98.4) | 100.0 (100.0) |
| Rsym (%) | 5.2 (47.6) | 6.1 (31.9) | 6.4 (54.4) |
| Redundancy | 4.2 (4.1) | 5.8 (3.8) | 5.3 (5.0) |
|
Refinement | |||
| Resolution (Å) | 29.6-2.15 | 27.7-2.15 | |
| No. of reflections | 13,847 | 30,096 | |
| Rwork/Rfree (%) | 18.5/22.4 | 18.6/24.4 | |
| No. protein atoms | 1,216 | 2,903 | |
| No. solvent atoms | 144 | 268 | |
| RMSD Bond lengths (Å)/angles (°) | 0.007/1.001 | 0.012/1.279 | |
| φ/ψ most favored/allowed (%) | 99.3/100.0 | 99.2/100.0 | |
Values in parentheses are for the highest-resolution shell
Spn1 (148–307) forms a right-handed superhelical bundle of eight helices (named H1-8, Figure 2A). Surprisingly, this structure resembles the domains of the RNA processing factors Pcf11 (S. cerevisiae) (Meinhart and Cramer, 2004) and SCAF8 (human) (Becker et al., 2008) that bind the RNAPII CTD (Figure S1A). Spn1 is reported to associate with RNAPII (Zhang et al., 2008) and the structural similarity suggested that Spn1 might bind the RNAPII CTD. We have not, however, observed binding in a fluorescence polarization assay between the Spn1 core and synthetic peptides, either phosphorylated or unphosphoryalated, that span more than two heptad repeats of the RNAPII CTD (data not shown).
Crystal structure of an Spn1-Spt6 complex
The structure of an Spn1(148–293):Spt6(239–268) complex was determined by molecular replacement and refined to Rfactor/Rfree values of 18.6/24.4% against 2.15 Å resolution data (Figures 2B, S2, Table 1). There are two complexes in the asymmetric unit that superimpose closely with an RMSD of ~0.5 Å over all 170 native ordered Cα atoms. Residues 148–292 of Spn1 are clearly visible in the electron density as are six non-native N-terminal residues that remain after TEV digestion. Residues 239–263 of Spt6 are also clearly defined in the electron density as well as four non-native N-terminal residues. There is no clear electron density for the five C-terminal residues of Spt6 and the one C-terminal residue of Spn1, and these are not included in the final model. Spn1 retains the same globular eight helix-bundle fold in the complex, showing no notable conformational changes upon binding Spt6 (RMSD ~0.5 Å on 135/145 pairs of Cα atoms). Binding of Spt6 to Spn1 does not mimic the binding of RNAPII CTD peptides to Pcf11/SCAF8, as it involves a face of Spn1 that is structurally distinct from the binding surface of Pcf11/SCAF8 (Figure S1B).
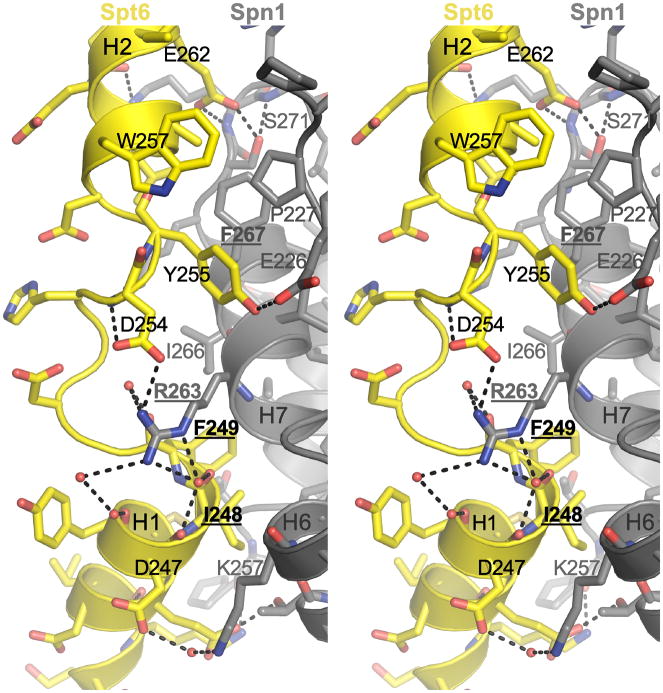
The structure of the Spn1(148–293):Spt6(239–268) complex reveals an extensive interface in which the Spt6 residues drape across the structured Spn1 core domain as two helices, H1 (residues 239–249) and H2 (256–265) that are connected by a short extended segment. There are multiple contacts along the length of the Spt6 segment that bury a total of 1,790 Å2 of accessible surface area upon complex formation. Spt6 H1 and the connecting segment, in particular, include a high fraction of conserved residues and contact a conserved patch on the Spn1 surface, thereby indicating that this interface is likely to be preserved across eukaryotes (Figures 2C–E, S1C–D)
Distinctive hydrophobic and polar interactions are made throughout the length of the Spt6 peptide (Figure 3). Starting at the Spt6 peptide N-terminus, the H1 residues M245, I248, and F249 contact a hydrophobic pocket formed by Spn1 residues L256, G262, I266, I286, and W289. The C-terminal carbonyl groups of Spt6 H1 form water-mediated hydrogen bonds with Spn1 R263 guanidinium, whose extensive network of contacts also includes direct hydrogen bonds with the carboxylate of D254 in the extended region of Spt6. The aromatic side chain of Spt6 Y255, from the extended central region, makes van der Waals contacts with Spn1 P227, V264, and F267, while its hydroxyl group forms a hydrogen bond with the carboxylate of Spn1 E226. The Spt6 H2 residues A258, L259, W257, and I261 contact two hydrophobic patches formed by Spn1 V264, F267, P227 and G231. Finally, the Spt6 E262 carboxylate hydrogen bonds with Spn1 S271 and R273, and K272 forms polar contacts with the amide of Spt6 N263. These extensive interactions are consistent with the strong specific binding observed between Spn1 and Spt6.
Figure 3. Details of the Spn1-Spt6 interface.
Stereoview of the Spn1-Spt6 interface. Residues mutated in this study are indicated in bold font and underlined. Water molecules are shown as red spheres and polar interactions are indicated by black dashes.
Mutations that disrupt Spn1-Spt6 complex formation in vitro
To validate the relevance of the interface seen in the crystal structure, Spn1(148–293) and Spt6(239–268) variants were expressed, purified, and binding affinities were measured by ITC (Figures 4, S3, Table S1). The Spt6-F249K protein bound Spn1 with a mean KD of 11 μM, a reduction in affinity compared to the WT interaction of about 60-fold. The affinity of the Spn1-R263D protein for Spt6 (mean KD of 30 μM) was reduced about 170-fold compared to WT. No binding was detected for the Spn1-F267E protein at concentrations expected to give a reliable estimate of affinity around 40 μM. These observations validate the crystallographic interface and demonstrate that single point mutations are sufficient to significantly reduce binding affinity in vitro.
Figure 4. Mutations at the Spn1-Spt6 interface disrupt binding in vitro.
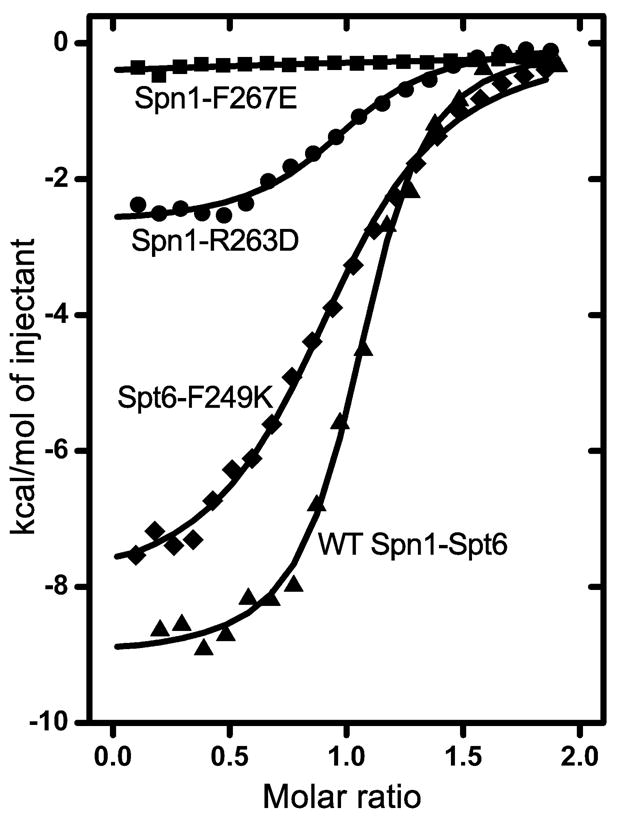
Representative ITC binding isotherms for the indicated mutant Spt6(239–268) and Spn1(148–293) proteins, Spt6-F249K (diamonds), Spn1-R263D (circles), and Spn1-F267E (squares). A WT isotherm is shown for reference (triangles). See text and Table S1 for thermodynamic parameters.
Mutating the Spn1-Spt6 interface causes profound effects in vivo
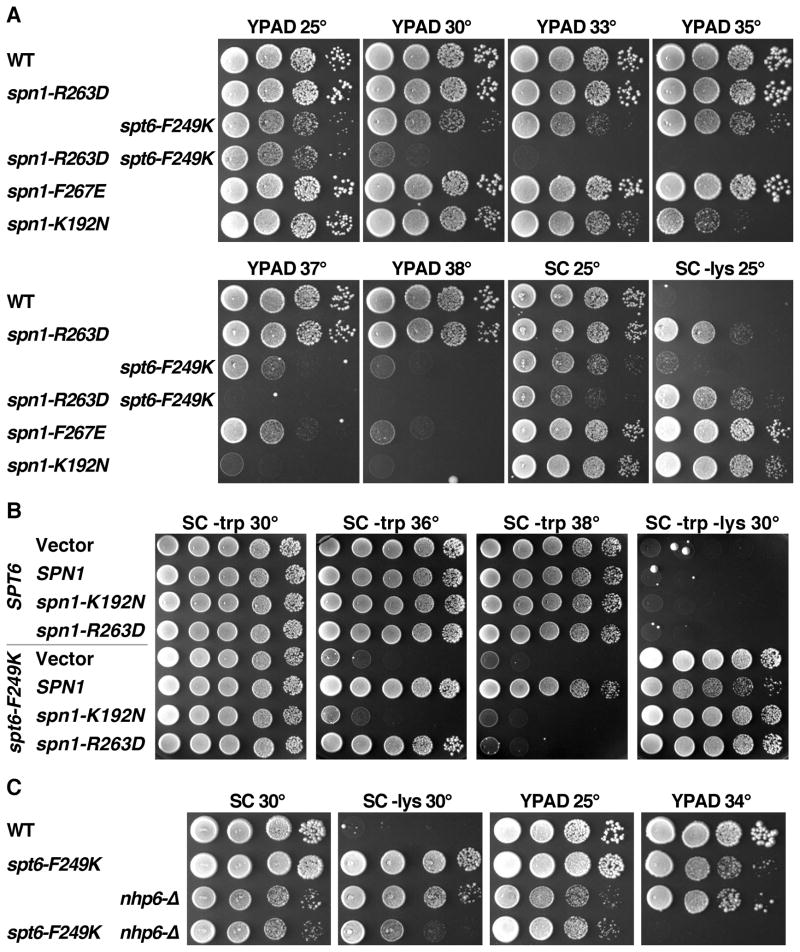
To determine the physiological importance of the Spn1-Spt6 interaction, we introduced spt6-F249K, spn1-R263D, and spn1-F267E mutations into the genomes of yeast cells such that each mutant protein was expressed from its native promoter as the sole source of the affected protein. spt6-F249K caused a moderate growth defect at low temperatures, and this was strongly enhanced at elevated temperatures (Figure 5A). Strains with the spn1-R263D mutation grew normally at all temperatures, while those with the spn1-F267E mutation were normal at low temperatures but failed to grow at high temperatures. Each of the three mutations caused a defect in transcription initiation site selection due to defective chromatin repression, as indicated by the Spt− phenotype (growth of a strain with the lys2-128∂ allele on medium lacking lysine (Simchen et al., 1984), Figure 5A–C). The strength of the defect was different for each mutation, being weakest for spt6-F249K, stronger for spn1-R263D, and strongest for spn1-F267E. This order correlates precisely with the level of perturbation of binding observed with these mutant proteins in vitro (Figure 4, Table S1), strongly supporting the importance of the Spn1-Spt6 binding interface detected in our crystal structure in maintaining a repressive chromatin state in vivo.
Figure 5. The Spn1-Spt6 interaction is significant for the essential activities of each protein in vivo.
(A) Isogenic strains from the A364a genetic background and with the relevant genotypes indicated (Supplemental Information) were grown to saturation in rich medium, then aliquots of 10-fold serial dilutions were placed on solid medium and incubated as labeled. SC is synthetic medium (complete or lacking tryptophan or lysine as noted) and YPAD is rich medium. Growth on medium lacking lysine reveals the Spt− phenotype, reporting here on aberrant transcription initiation from the lys2-128∂ allele. The strain with the spt6-F249K allele grows slowly on SC – lys at 25°, but the Spt− phenotype is more robust at 30° (panel B).
(B) As in panel A. nhp6-Δ indicates deletion of both NHP6A and NHP6B.
(C) WT and spt6-F249K strains were transformed with a high copy number vector, or the same vector carrying the version of SPN1 noted (Supplemental Information). Multiple transformants were tested to insure that the phenotypes detected are typical, then one clone of each was grown to saturation in synthetic medium lacking tryptophan to select for retention of the plasmids. Aliquots of 10-fold serial dilutions were placed on solid synthetic medium as in panel A and incubated as indicated.
Each mutation in the Spn1-Spt6 binding interface disturbed the interaction to a different extent in vitro (Figure 4, Table S1), but each individual mutation was tolerated in vivo (Figure 5). If the phenotypes caused by individual mutations result from partial disruption of binding, then combining the mutations should lead to enhanced defects. Consistent with this prediction, cells with both spt6-F249K and spn1-R263D mutations were viable but severely impaired for growth (Figure 5A), and cells with both spt6-F249K and spn1-F267E were inviable (Figure S4D). The severity of the defect caused by combining mutations therefore correlates with the level of disruption of binding by the individual mutations, suggesting that the Spn1-Spt6 interaction is essential for viability. We were unable to detect an interaction between Spt6 and Spn1-F267E in vitro (Figure 4), but the viability of the spn1-F267E strain suggests that this mutant retains some binding. Combining spt6-F249K with spn1-K192N was also lethal (Figure S4D); this mutation does not directly affect the Spn1-Spt6 interface (Figure 2B–C) but has been shown to decrease the interaction between Spn1 and RNAPII (Zhang et al., 2008), and our results with the recombinantly expressed protein suggest that Spn1-K192N protein is unstable (not shown). Synthetic growth defects therefore support the importance of the Spn1-Spt6 interaction, but defects outside this interface can also cause additive growth defects.
Another strategy for determining the importance of the Spn1-Spt6 interface is to test the effect of overexpressing one partner. If decreased affinity of Spt6-F249K protein for Spn1 is responsible for growth defects in vivo, these defects might be suppressed by increasing the level of Spn1 protein. We tested this by transforming an spt6-F249K strain with high copy plasmids containing variants of SPN1. As shown in Figure 5C, increasing the level of normal SPN1 suppressed the temperature sensitivity and partially corrected the Spt− phenotype caused by spt6-F249K. Consistent with retention of partial binding, overexpression of spn1-R263D also had an effect but suppressed less efficiently, rescuing growth at 36° but not at 38° and having no effect on the Spt− phenotype. Spn1-K192N is not active at elevated temperatures ((Zhang et al., 2008) and Figure 5A) and overexpression of this allele also did not rescue the phenotypes caused by spt6-F249K. Elevated SPN1 copy number did not suppress the temperature sensitivity or Spt− phenotypes caused by the spt6-1004 allele (data not shown), which is a deletion of the helix-hairpin-helix domain within the Tex-like core of Spt6 (Kaplan et al., 2005). The suppression of spt6 defects by increased Spn1 is therefore at least partly specific for a mutation that alters the Spn1-Spt6 interface, and supports the importance of this interaction in an essential function in vivo.
Spt6 binds nucleosomes directly and is inhibited by Spn1
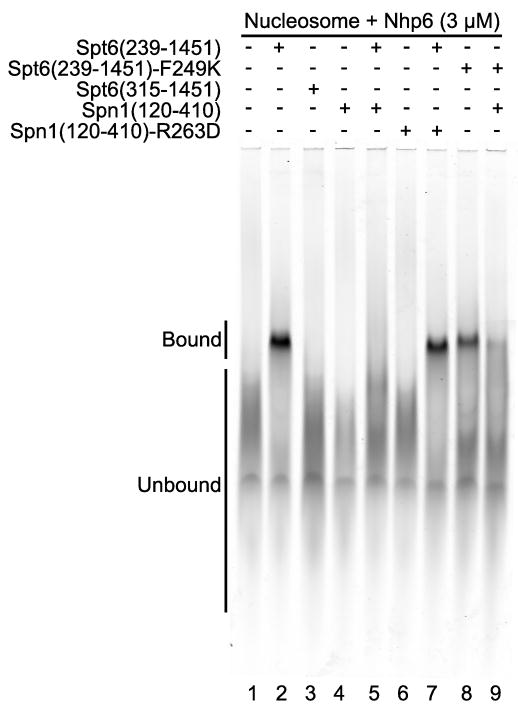
Spt6 has been shown to bind both (H3–H4)2 tetramers and H2A–H2B dimers (Bortvin and Winston, 1996), leading to models in which Spt6 acts as a histone chaperone during nucleosome eviction and redeposition. Following our earlier prediction (Johnson et al., 2008) that the N-terminal region of Spt6 binds nucleosomes, we examined purified Spt6 constructs for nucleosome-binding activity in an electrophoretic mobility shift assay (EMSA). Interestingly, we found that intact Spt6 does bind nucleosomes in this assay (Figure 6), but does so only in the presence of the small HMGB family member Nhp6 (Stillman, 2010). This is similar to the requirement for addition of Nhp6 to observe complexes between nucleosomes and a different histone chaperone, FACT (Formosa et al., 2001). As with FACT, this suggests that Spt6 may form stable complexes with nucleosomes only after the nucleosome has been partially destabilized by Nhp6. This requirement appears to be physiologically relevant, because loss of Nhp6 in vivo exacerbated the growth defects caused by any of several mutations in Spt6 (Figures 5B, S4A). Histone chaperones do not necessarily bind intact nucleosomes, in fact the Asf1-(H3–H4) interaction is incompatible with histone contacts within the nucleosome (Antczak et al., 2006; English et al., 2006; Natsume et al., 2007). Binding to both free histones and to nucleosomes therefore might indicate that Spt6 makes multiple distinct contacts with nucleosomes and their components during different steps in chromatin maintenance. Spt6(239–1451) bound nucleosomes while Spt6(315–1451) did not (Figure 6, compare lanes 2 and 3). Thus, the region of Spt6 required for nucleosome binding (residues 239–314) contains the region required for Spn1 binding (239–268), suggesting that these two interactions may be mutually exclusive.
Figure 6. Spt6 binds nucleosomes and is competed by Spn1.
Electrophoretic mobility shift assay visualizing the signal from the labeled DNA incorporated into the nucleosomes (Cy5, 633 nm). All reactions included 150 fmoles of nucleosomes, 3 μM Nhp6, and 2 μM of the indicated Spt6 or Spn1 proteins. Migration positions of bound and unbound nucleosomes are indicated. Addition of Spn1 reduced the amount of Spt6-nucleosome complex from 37% of the total signal in lane 2 to 3.2% in lane 5 (9% remaining), but Spn1-R263D only reduced it to 28% (76% remaining). Spt6-F249K formed a lower amount of stable complex (18%) but Spn1 was less effective at inhibiting this (6% bound, or 36% of the original signal remaining comparing lanes 8 and 9) than it was with WT Spt6 (9% of the original signal remaining).
Consistent with this possibility, adding Spn1(120–410) to the nucleosome-binding assay inhibited the formation of Spt6-nucleosome complexes (Figure 6, compare lanes 2 and 5). Spn1 did not form stable complexes with nucleosomes itself, so the inhibition is unlikely to be caused by competition between Spn1 and Spt6 for a common binding site on nucleosomes. Spn1 did not interact with Nhp6 genetically (Figure S4B–C), and inhibition of Spt6-nucleosome complex formation by Spn1 could not be overcome by increasing the concentration of Nhp6 (not shown), making it unlikely that the inhibition is caused by sequestering of Nhp6 by Spn1. Instead, Spn1 appears to prevent Spt6 from binding to nucleosomes directly by blocking the Spt6 binding domain. Supporting this interpretation, the Spn1-R263D variant with reduced affinity for Spt6 did not block formation of Spt6-nucleosome complexes efficiently (Figure 6, lane 7). Further, the Spt6-F249K mutation affects a region important for both nucleosome binding and Spn1 interaction (Figure 4 and Figure 6, compare lanes 2 and 8). Spt6-F249K protein was impaired for nucleosome binding but the residual binding was partially resistant to the addition of Spn1 (Figure 6, lanes 8 and 9). These results show that Spt6 residue F249 contributes to both nucleosome binding and to Spn1 binding, and that Spn1 binding can block an interaction between Spt6 and nucleosomes. The Spn1-Spt6 interaction can therefore provide a switch that controls the interaction of Spt6 with nucleosomes, and the proper functioning of this switch is important for maintaining normal chromatin structure.
DISCUSSION
We have determined a crystal structure of the ordered central domain of Spn1 and shown that it binds a 30-residue segment of Spt6. We have also determined a crystal structure of an Spn1-Spt6 complex that reveals that Spt6 binds in an extended/helical conformation that drapes the Spt6 residues along one face of Spn1. We further used site-directed mutations to disrupt this interaction and demonstrated that the interaction observed in solution depends on residues located at the interface seen in the crystal structure. Binding is not accompanied by conformational changes in Spn1. In contrast, Spt6(239–268) is almost certainly unstructured in isolation and becomes ordered upon binding Spn1. Indeed, the first 300 residues of Spt6 are likely to be unstructured in isolation (Ward et al., 2004) whereas much of the remainder of the protein appears to comprise multiple recognizable structural domains that likely fold against each other to form an elongated structure (Johnson et al., 2008). The extended and inherently flexible nature of the Spn1 binding sequence of Spt6 presumably explains why dramatic mutations in the interface substantially weaken but do not completely eliminate this interaction, as localized perturbations can be accommodated by conformational changes that do not propagate across the entire interface. Moreover, the ~35 residues separating the Spn1-binding residues from the ordered region of Spt6 likely provide a tether whose flexibility may be required to allow the Spn1-Spt6 complex to form in multiple functional contexts.
Spt6 and Spn1 were found to co-purify in high-throughput screens (Gavin et al., 2002; Krogan et al., 2002), and further studies suggested that their interaction promotes normal activation of genes regulated after recruitment of RNAPII (Fischbeck et al., 2002; Yoh et al., 2007; Zhang et al., 2008). However, Spt6 and Spn1 have also been implicated in several other distinct and overlapping roles, and each has been implicated in interactions with multiple factors. We have verified a functional interplay between Spt6 and Spn1 proteins in S. cerevisiae by demonstrating that cells cannot tolerate certain partial loss-of-function alleles of both genes simultaneously, and by showing that the defects caused by the spt6-F249K allele, which impairs binding with Spn1, can be suppressed by overexpressing SPN1. Our results therefore show that a short region of Spt6 is important for interacting with Spn1 during at least one of the essential processes mediated by these proteins.
One of the primary functions ascribed to Spt6 is as a histone chaperone that promotes the reassembly of nucleosomes following passage of RNAPII (Adkins and Tyler, 2006; Bortvin and Winston, 1996; Cheung et al., 2008; Kaplan et al., 2003). Spt6 was previously shown to bind histones and to promote nucleosome deposition in vitro (Bortvin and Winston, 1996), and we now show that it can also bind to intact nucleosomes. The dependence of this interaction upon the presence of Nhp6 is consistent with our genetic studies and with an equivalent dependence of the unrelated FACT histone chaperone for its interaction with intact nucleosomes (Formosa et al., 2001). We found that nucleosome binding requires a section of Spt6 that overlaps with the Spn1 binding site, that Spn1 antagonizes Spt6-nucleosome binding in vitro, that either SPT6 or SPN1 mutations that affect the Spn1-Spt6 interaction cause the Spt− phenotype, and that the effects of the spt6-F249K allele can be suppressed by overexpressing Spn1. These results show that the Spn1-Spt6 interaction disrupts Spt6-nucleosome binding and that this disruption has a positive role in maintaining normal chromatin. An attractive explanation is that Spn1 is actively engaged in the nucleosome reassembly process, possibly by disengaging Spt6 from nucleosomes to allow multiple rounds of reassembly (see the graphical abstract available online). Another possibility is that Spn1 binding influences the balance between Spt6’s functional roles in nucleosome reassembly and mRNA processing. Regardless of the precise mechanistic details, Spn1 appears to be important for Spt6-mediated nucleosome reassembly in vivo.
Nucleosome reassembly is just one of several processes in which Spt6 and Spn1 have been implicated. For example, Spn1, in complex with Spt6, has been reported to interact with a variety of other factors that function in mRNA processing, nucleosome modification, or transcription, including REF1/Aly, Setd2, and RNAPII (Yoh et al., 2007; Yoh et al., 2008; Zhang et al., 2008). Our structural data provide insight into how Spn1 might accommodate simultaneous interactions. Spt6 binds against a region of the Spn1 surface that is rich in conserved residues, but other regions of Spn1 display a similar level of conservation and are therefore good candidates for binding surfaces for other proteins (Figures 2C, S1C). This includes residues that are immediately adjacent to the Spt6 binding surface but extend beyond contacts with Spt6, which may indicate that other factors can bind Spn1 cooperatively or competitively with Spt6. Finally, K192, whose mutation to asparagine impairs Spn1 function (Zhang et al., 2008), and appears to function in interactions with RNAPII, is located in a conserved pocket that is formed at the ends of H1, H2, and H4 (Figure 2B–C) suggesting another potential binding surface.
In summary, we have determined the structural basis for the interaction between Spn1 and Spt6, and shown that this interaction is important in vivo and that it regulates Spt6-nucleosome binding in vitro. Overall, our data indicate that Spn1 is important for Spt6-mediated nucleosome reassembly, perhaps by regulating the process or by providing a switch that drives disengagement. This does not, however, seem to encompass all of the functional roles in which these two proteins and their interaction with each other participate (Lindstrom et al., 2003; Yoh et al., 2007; Yoh et al., 2008; Zhang et al., 2008). Sequence conservation indicates that other surfaces on the Spn1 core domain are good candidates for mediating functionally important interactions. Moreover, interactions with Setd2 and REF1/Aly have been mapped to the N- and C-terminal regions, respectively, which extend beyond the Spn1 core domain and are predicted to be unstructured (Ward et al., 2004; Yoh et al., 2007; Yoh et al., 2008). This use of inherently flexible segments, including the N-terminal region of Spt6 that binds the Spn1 core, may provide a mechanism that allows flexibility in a crowded transcriptional environment.
EXPERIMENTAL PROCEDURES
See the Supplemental Information for protein expression and purification, strains used, strain construction, plasmid construction, and tests of the effects of combining spn1 and spt6 mutations.
Crystallization and Structure Determination
All crystals were grown at 20°C by sitting drop vapor diffusion. Se-Spn1(148–307) drops comprised 2 μL of 8 mg.mL−1 protein with 2 μL of well solution (0.1 M Bis-Tris Propane pH 7.0, 1.4 M Li2SO4). Native Spn1(148–307) drops comprised two parts 7.5 mg.mL−1 protein and one part well solution (0.01 M MgCl2, 0.05 M HEPES pH 7.0, 1.6 M (NH4)2SO4. Spn1(148–293)-Spt6(239–268) drops comprised two parts 13 mg.mL−1 protein and one part well solution (0.2 M Mg(CH3CO2)2, 0.1 M MES pH 6.5, 20% PEG 8000). Crystals were cryoprotected in a solution of the reservoir made up with 30% glycerol, and cooled by plunging into liquid nitrogen.
All data were processed using HKL2000 (Otwinowski and Minor, 1997). Phases were determined for Se-Spn1(148–307) by single-wavelength anomalous diffraction. Phenix (phenix. autosol) (Adams et al., 2010) located 4 out of 6 possible selenium positions and computed a map into which a model was built. This unrefined model was used in molecular replacement using PHASER (McCoy et al., 2005) to determine the structure of native Spn1(148–307) at 2.15 Å resolution. This subsequently refined model was used to determine the Spn1(148–293):Spt6(239–268) structure by molecular replacement. In all cases, model-building, refinement and validation were performed using Coot (Emsley et al., 2010), Phenix (Adams et al., 2010), and MolProbity (Chen et al., 2010), respectively. Refinement of the Spn1(148–293)-Spt6(239–268) complex included TLS restraints (Painter and Merritt, 2006).
Electrostatic potential surfaces were calculated using APBS (Baker et al., 2001). Figures of molecular structures were generated using PyMol (DeLano, 2002).
Size-Exclusion Chromatography Binding Assay
Purified recombinant proteins were mixed at equimolar concentrations (2 μM) and incubated for 2 hours at 4°C. The protein mixture was concentrated to 15 μM and chromatographed on a 120 mL superdex 200 16/60 column (GE Healthcare) in 15 mM Tris pH 7.5, 200 mM NaCl, 5% glycerol, 0.5 mM EDTA, and 2 mM 2-mercaptoethanol.
Isothermal Titration Calorimetry
Amino-acid substitutions were made by site-directed mutagenesis and verified by DNA sequencing. Purified recombinant proteins were dialyzed overnight at 4°C against 2 L of degassed ITC buffer (20 mM Tris pH 7.5, 150 mM NaCl, 5% glycerol, 2 mM 2-mercaptoethanol, 0.5 mM EDTA). Titrations for all reactions were done at 25°C on an iTC200 (Microcal), including an initial injection of 0.4 μL (which was omitted from data analysis), and all injections were spaced 180 s apart. For Spt6(239–1117) reactions, the titrations were carried out with 18 injections of 1.8 μL 76 μM Spn1(148–293) into 8.2 μM Spt6(239–1117). For Spt6(239–268) reactions, the titrations were with 18 injections of 2 μL 74 μM Spt6(239–268) into 8 μM Spn1(148–293). For Spt6-F249K reactions, the titrations were with 18 injections of 1.8 μL 1.52 mM Spt6(239–268)-F249K into 168 μM Spn1(148–293). For Spn1-R263D reactions, the titrations were with 18 injections of 1.8 μL 3.5 mM Spt6(239–268) into 389 μM Spn1(148–293)-R263D. For Spn1-F267E reactions, the titrations were with 18 injections of 2.0 μL 5.2 mM Spt6(239–268) into 578 μM Spn1(148–293)-F267E). In all cases, three independent experiments were performed. Data were analyzed using Origin software (Microcal), and the stoichiometry (N), association constant (KA), and change in enthalpy (ΔH) were obtained by fitting the isotherm to the one-site binding model. Other thermodynamic parameters were calculated using the following relationships:
Nucleosome Preparation and Gel Mobility Shift Binding Assay
A 146 bp sea urchin 5S rDNA fragment labeled with Cy5 was generated by PCR and gel purified. Xenopus laevis histone H2A-S113C was labeled with Oregon Green488-maleimide and then assembled into nucleosome core particles as described (Xin et al., 2009). 150 fmoles of nucleosome was mixed with 2 μM Spt6 or Spn1 protein in 10 μl reactions containing 100 mM NaCl, 0.8 mg/ml HSA, 9.7% glycerol, and 3 μM S. cerevisiae Nhp6a. Following incubation at 30° for 15 min, samples were subjected to electrophoresis on native polyacrylamide gels (4.5% acrylamide (acr:bis, 37.5:1), 0.5 × TBE, 5% glycerol, 2 mM MgCl2 at 160 V for 6 hrs at 4°C). The gels were scanned using a Typhoon imager at 670 BP30/Red(633 nm) for Cy5-DNA and 520 BP40/Blue(488 nm) for Oregon Green488-H2A, and the amount of signal in the bound form quantified with ImageQuant Software (GE Health Sciences).
HIGHLIGHTS.
Spn1 core region binds directly to a segment of Spt6
Crystal structures and biochemical analysis reveal the basis for this interaction
Spn1-Spt6 binding is important in vivo for maintaining repressive chromatin
Spt6 binds nucleosomes directly and this interaction is inhibited by Spn1
Supplementary Material
Acknowledgments
Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research and by the US National Institutes of Health (NIH), National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. We thank David Stillman and Warren Voth for helpful discussions and Charisse Kettelkamp and Laura McCullough for technical assistance. This work was supported by NIH grants to (C.P.H.) and (T.F.).
Footnotes
ACCESSION NUMBERS
Protein Data Bank: Coordinates and structure factor amplitudes have been deposited for the Spn1 (3o8z) and Spn1-Spt6 (3oak) crystal structures.
Supplemental information includes one table, four figures and supplemental experimental procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkins MW, Tyler JK. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol Cell. 2006;21:405–416. doi: 10.1016/j.molcel.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- Antczak AJ, Tsubota T, Kaufman PD, Berger JM. Structure of the yeast histone H3-ASF1 interaction: implications for chaperone mechanism, species-specific interactions, and epigenetics. BMC Struct Biol. 2006;6:26. doi: 10.1186/1472-6807-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardehali MB, Yao J, Adelman K, Fuda NJ, Petesch SJ, Webb WW, Lis JT. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 2009;28:1067–1077. doi: 10.1038/emboj.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniahmad C, Nawaz Z, Baniahmad A, Gleeson MA, Tsai MJ, O’Malley BW. Enhancement of human estrogen receptor activity by SPT6: a potential coactivator. Mol Endocrinol. 1995;9:34–43. doi: 10.1210/mend.9.1.7760849. [DOI] [PubMed] [Google Scholar]
- Becker R, Loll B, Meinhart A. Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2008;283:22659–22669. doi: 10.1074/jbc.M803540200. [DOI] [PubMed] [Google Scholar]
- Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- Bucheli ME, Buratowski S. Npl3 is an antagonist of mRNA 3′ end formation by RNA polymerase II. Embo J. 2005;24:2150–2160. doi: 10.1038/sj.emboj.7600687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckin T, Nagel R, Mandel-Gutfreund Y, Shiue L, Clark TA, Chong JL, Chang TH, Squazzo S, Hartzog G, Ares M., Jr Exploring functional relationships between components of the gene expression machinery. Nat Struct Mol Biol. 2005;12:175–182. doi: 10.1038/nsmb891. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008;6:e277. doi: 10.1371/journal.pbio.0060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Adams CD, Winston F. The SPT6 gene is essential for growth and is required for delta-mediated transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:679–686. doi: 10.1128/mcb.7.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The Pymol Molecular Graphics System. Delano Scientific; Palo Alto, CA: 2002. [Google Scholar]
- Dengl S, Mayer A, Sun M, Cramer P. Structure and in vivo requirement of the yeast Spt6 SH2 domain. J Mol Biol. 2009;389:211–225. doi: 10.1016/j.jmb.2009.04.016. [DOI] [PubMed] [Google Scholar]
- Denis CL. Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics. 1984;108:833–844. doi: 10.1093/genetics/108.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh M, Zhu W, Hasegawa J, Watanabe H, Kim DK, Aida M, Inukai N, Narita T, Yamada T, Furuya A, et al. Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol Cell Biol. 2004;24:3324–3336. doi: 10.1128/MCB.24.8.3324-3336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbeck JA, Kraemer SM, Stargell LA. SPN1, a conserved gene identified by suppression of a postrecruitment-defective yeast TATA-binding protein mutant. Genetics. 2002;162:1605–1616. doi: 10.1093/genetics/162.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T, Eriksson P, Wittmeyer J, Ginn J, Yu Y, Stillman DJ. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 2001;20:3506–3517. doi: 10.1093/emboj/20.13.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SJ, Close D, Robinson H, Vallet-Gely I, Dove SL, Hill CP. Crystal structure and RNA binding of the Tex protein from Pseudomonas aeruginosa. J Mol Biol. 2008;377:1460–1473. doi: 10.1016/j.jmb.2008.01.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Holland MJ, Winston F. Interaction between transcription elongation factors and mRNA 3′-end formation at the Saccharomyces cerevisiae GAL10-GAL7 locus. J Biol Chem. 2005;280:913–922. doi: 10.1074/jbc.M411108200. [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Morris JR, Wu C, Winston F. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 2000;14:2623–2634. doi: 10.1101/gad.831900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan BR, Feldman JL, Lee DH, Koos DS, Ho RK, Stainier DY, Yelon D. The elongation factors Pandora/Spt6 and Foggy/Spt5 promote transcription in the zebrafish embryo. Development. 2002;129:1623–1632. doi: 10.1242/dev.129.7.1623. [DOI] [PubMed] [Google Scholar]
- Kok FO, Oster E, Mentzer L, Hsieh JC, Henry CA, Sirotkin HI. The role of the SPT6 chromatin remodeling factor in zebrafish embryogenesis. Dev Biol. 2007;307:214–226. doi: 10.1016/j.ydbio.2007.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ye H, Guo H, Yin Y. Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.0909198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, McCleery JA, Yates JR, 3rd, Hartzog GA. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol Cell Biol. 2003;23:1368–1378. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y, Smith AJ, Morgan GT. A sequence motif conserved in diverse nuclear proteins identifies a protein interaction domain utilised for nuclear targeting by human TFIIS. Nucleic Acids Res. 2006;34:2219–2229. doi: 10.1093/nar/gkl239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zhou Z, Chen G, Bao S. A putative transcriptional elongation factor hIws1 is essential for mammalian cell proliferation. Biochem Biophys Res Commun. 2007;353:47–53. doi: 10.1016/j.bbrc.2006.11.133. [DOI] [PubMed] [Google Scholar]
- Maclennan AJ, Shaw G. A yeast SH2 domain. Trends Biochem Sci. 1993;18:464–465. doi: 10.1016/0968-0004(93)90006-9. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- Natsume R, Eitoku M, Akai Y, Sano N, Horikoshi M, Senda T. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- Neigeborn L, Celenza JL, Carlson M. SSN20 is an essential gene with mutant alleles that suppress defects in SUC2 transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:672–678. doi: 10.1128/mcb.7.2.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki K, Sano T, Miwa J. emb-5, a gene required for the correct timing of gut precursor cell division during gastrulation in Caenorhabditis elegans, encodes a protein similar to the yeast nuclear protein SPT6. Mol Gen Genet. 1993;239:313–322. doi: 10.1007/BF00276929. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W, editors. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Academic Press; New York: 1997. [DOI] [PubMed] [Google Scholar]
- Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- Shen X, Xi G, Radhakrishnan Y, Clemmons DR. Identification of novel SHPS-1-associated proteins and their roles in regulation of insulin-like growth factor-dependent responses in vascular smooth muscle cells. Mol Cell Proteomics. 2009;8:1539–1551. doi: 10.1074/mcp.M800543-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simchen G, Winston F, Styles CA, Fink GR. Ty-mediated gene expression of the LYS2 and HIS4 genes of Saccharomyces cerevisiae is controlled by the same SPT genes. Proc Natl Acad Sci U S A. 1984;81:2431–2434. doi: 10.1073/pnas.81.8.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman DJ. Nhp6: a small but powerful effector of chromatin structure in Saccharomyces cerevisiae. Biochim Biophys Acta. 2010;1799:175–180. doi: 10.1016/j.bbagrm.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanti M, Gallastegui E, Respaldiza I, Rodriguez-Gil A, Gomez-Herreros F, Jimeno-Gonzalez S, Jordan A, Chavez S. Yeast genetic analysis reveals the involvement of chromatin reassembly factors in repressing HIV-1 basal transcription. PLoS Genet. 2009;5:e1000339. doi: 10.1371/journal.pgen.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Xin H, Takahata S, Blanksma M, McCullough L, Stillman DJ, Formosa T. yFACT induces global accessibility of nucleosomal DNA without H2A–H2B displacement. Mol Cell. 2009;35:365–376. doi: 10.1016/j.molcel.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh SM, Cho H, Pickle L, Evans RM, Jones KA. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 2007;21:160–174. doi: 10.1101/gad.1503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh SM, Lucas JS, Jones KA. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes & Development. 2008;22:3422–3434. doi: 10.1101/gad.1720008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Fletcher AG, Cheung V, Winston F, Stargell LA. Spn1 regulates the recruitment of Spt6 and the Swi/Snf complex during transcriptional activation by RNA polymerase II. Mol Cell Biol. 2008;28:1393–1403. doi: 10.1128/MCB.01733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.