Summary
Mycoplasma genitalium is one of the smallest organisms capable of self-replication and its sequence is considered a starting point for understanding the minimal genome required for life. MG289, a putative phosphonate substrate binding protein, is considered to be one of these essential genes. The crystal structure of MG289 has been solved at 1.95 Å resolution. The structurally identified thiamine binding region reveals possible mechanisms for ligand promiscuity. MG289 was determined to be an extracytoplasmic thiamine binding lipoprotein. Computational analysis, size exclusion chromatography, and small angle X-ray scattering indicates that MG289 homodimerizes in a concentration-dependant manner. Comparisons to the thiamine pyrophosphate binding homolog Cypl reveal insights into the metabolic differences between mycoplasmal species including identifying possible kinases for cofactor phosphorylation and describing the mechanism of thiamine transport into the cell. These results provide a baseline to build our understanding of the minimal metabolic requirements of a living organism.
Keywords: p37, Cypl, substrate binding protein, extracytoplasmic lipoprotein, sexually transmitted infection, X-ray crystallography, small angle X-ray scattering
Introduction
Mycoplasmas (class Mollicutes) are tiny, pleomorphic bacteria that live in association with eukaryotes either by attachment to the cell membrane of its host or intracellularly. These organisms are highly pervasive, infecting plants and animals including humans, and highly specialized to thrive within specific tissues in its host species. Mycoplasmas are believed to have evolved from low G+C content Gram-positive bacteria undergoing significant genome reduction, adapting to the resources available in its host and eliminating unnecessary genes including those responsible for oxidative phosphorylation, ATP generation via pentose phosphate pathway, and the biosynthesis of amino acids, nucleotides, lipids, and cofactors1,2. Mycoplasmas also adapted reduced substrate specificity allowing fewer proteins to accomplish the tasks of many3,4.
Mycoplasma genitalium is one of the smallest organisms capable of self-replication with a genome of 580 kb which codes for approximately 480 proteins and is considered the starting point for identifying which genes are essential for life1,2. Experimental and bioinformatic methods have identified a subset of as few as 206 to as many as 387 genes that are considered “minimal” for an organism to survive and self-replicate1,4-7. M. genitalium is also a human sexually transmitted infection that preferentially adheres to ciliated epithelial cells in the genitourinary and respiratory regions8-10. It is a cause of nongonococcal urethritis in men and several genital tract inflammatory diseases in women11. It has also been associated with infertility, increased HIV-1 transmission, and malignant transformation of prostate cells12-14. Testing for M. genitalium infections is not trivial and can be expensive, leaving most cases un- or misdiagnosed, leading to improper treatment and an increased instance of antibiotic resistance15.
MG289 is a protein from M. genitalium, located in an operon tentatively coding for a binding protein dependant substrate transporter. It is annotated as a putative phosphonate binding lipoprotein, followed by MG290, a nucleotide-binding membrane permease, and MG291, an integral membrane protein, with twelve predicted membrane spanning regions1,16,17. The gene can be disrupted non-lethally, perhaps due to the inherent redundancy of action between this and several theoretical orphaned substrate-binding lipoproteins. Because the function of this protein is critical enough for a minimal bacterium to have evolved multiple copies, it is still considered to be an “essential” gene5-7. It is homologous to the M. hyorhinis extracytoplasmic thiamine binding protein Cypl, also known as p37, though they are only 32% identical1,18. The crystal structure of Cypl was recently determined, identifying its primary ligand as the cofactor thiamine pyrophosphate (TPP), however, based on sequence alignments it was theorized that the Cypl-like proteins from other Mycoplasma species would preferentially bind to thiamine (VIB) or thiamine monophosphate due to a lack of phosphate stabilizing residues19,20.
The cloning, expression, purification and preliminary X-ray analysis of MG289 were reported previously18. Presented here is the structure solution and refinement to 1.95 Å resolution. The MG289 model is mixed α/β forming two domains with a binding cleft, similar to Cypl and the Type II periplasmic binding proteins of Gram-negative bacteria19,21,22. As predicted, the ligand found in the binding cleft was VIB. MG289 crystallizes with two molecules in the asymmetric unit. Experimental and computational analysis was undertaken to identify whether this dimerization was crystallographic or biological in nature. The discrepancy between the binding partners of Cypl and MG289 are addressed and a hypothesis regarding the phosphorylation of nucleotides and cofactors in M. genitalium discussed.
Materials and Methods
Protein expression and purification
MG289 was expressed and purified as described previously18. Briefly, a plasmid containing an N-terminally truncated coding sequence (residues 26-386) was transformed into BL21(DE3)pLysS E. coli cells. The transformation was used to inoculate 1 l LB media with 100 mg l-1 ampicillin and cultured at 37°C until the OD600nm was 0.7-1.0. Cells were induced using 1 ml of 1 M isopropyl β-D-1 thiogalactopyranoside and allowed to express for 3 h. Cells were lysed via French press in 20 mM sodium phosphate buffer, pH 7.95. Lysate was centrifuged at 40000g for 20 min at 4°C. A volume of supernatant equivalent to 125 mg of total protein was applied to a 50 ml Bio-Rad Q anion-exchange column attached to a 5 ml Bio-Rad Econo-Pac S cation-exchange column equilibrated in 20 mM sodium phosphate buffer, pH 7.95. Flow-through was collected and adjusted to pH 6.1 with acetic acid, then applied to a 5 ml Bio-Rad Econo-Pac S cation-exchange column equilibrated in 20 mM sodium acetate, pH 6.1. MG289 was isocratically eluted using 15% 1 M sodium chloride, 20 mM sodium acetate, pH 6.1. Purity was confirmed by 10% SDS-PAGE stained with Coomassie Blue. Concentrations were calculated by absorbance at 280 nm using a calculated extinction coefficient of 54620 M-1 cm-1.
Crystallization, X-ray analysis, structure solution and refinement
The purified MG289 was buffer exchanged into 50 mM Tris-HCl pH 7.5 and concentrated to ∼8 mg ml-1 using a Centriprep 10 spin column (Millipore, Bedford, Massachusetts, USA). Crystallization was performed using hanging-drop vapor diffusion23. Crystallization conditions were as follows: 5 μl protein, 5 μl precipitant solution (0.2 M ammonium sulfate, 0.1 M sodium acetate trihydrate, pH 4.6, and 15% PEG 4000) and 1μl Silver Bullet Bio condition 49 (0.16% w/v thiamine monophosphate chloride dihydrate, acetylsalicylic acid, cholic acid, 1,2,3-heptanetriol, vanillin, N-acetyl-D-mannosamine in 20 mM HEPES sodium, pH 6.8; Hampton Research, Aliso Viejo, CA, USA) equilibrated against 1 ml of precipitant solution. Useful crystals appeared in 3 weeks.
A single crystal was directly flash-cooled in liquid nitrogen without any additional cryoprotection. X-ray diffraction data was collected on beamline 5.0.2 at the Advanced Light Source (λ=1.0000 Å). Two hundred useable images were collected with data to a maximum resolution of 1.95 Å. The data was indexed and scaled using HKL-200024. Data statistics are listed in Table I.
Table I.
MG 289 Data Collection and Refinement Statistics (PDB ID 3myu).
| Data Statistics | |
|---|---|
| Space Group | P212121 |
| Cell dimensions (Å) | a = 49.3, b = 90.4, c = 175.4 |
| Resolution (Å) | 45.18-1.95 (2.02-1.95)a |
| Rsym (%)b | 7.6 (56.0) |
| I/σI | 22.0 (2.7) |
| Completeness (%) | 90.9 (71.0) |
| Redundancy | 7.2 (6.5) |
| Refinement Statistics | |
| No. Reflections | 52800 (4049) |
| Rcrystc/Rfreed/Rall data (%) | 22.0 / 25.5 / 22.2 |
| Residues in model (chain id) | 32-280, 284-367 (A) 32-280, 283-367 (B) |
| No. of Atoms | |
| Protein (dimer) | 5470 |
| VIB (2 molecules) | 36 |
| ACT (2 molecules) | 8 |
| Water | 247 |
| B-factors (Å2) | |
| Protein main-/side-chain | 37.1 / 40.8 |
| VIB / ACT | 31.2 / 46.1 |
| Water | 38.6 |
| Ramachandran Plot (%) | |
| Favored | 97.0 |
| Allowed | 3.0 |
| Disallowed | 0.0 |
| RMSDe | |
| Length (Å) | 0.006 |
| Angle (°) | 0.938 |
Values in parenthesis are for highest resolution shell.
Rsym=(Σ|I-<I>|/Σ<I>)×100.
Rwork=(Σ|Fobs|-|Fc|/Σ|Fobs|)×100.
Rfree is calculated the same as Rwork, except it uses 5% of reflection data omitted from refinement.
The root mean squared deviation from ideal values.
Molecular replacement was performed in MOLREP implemented in the CCP4i suite using the structure of M. hyorhinis Cypl (PDB 3eki) as a search model19,25,26. Refinement was carried out using alternating rounds of computation using PHENIX and manual model refinement in COOT27,28. Positive residual density in the binding cleft and the monomer interface was modeled as thiamine (VIB) and acetate (ACT) respectively. For a more detailed description of the structure solution refer to Supplemental Methods. Final refinement statistics are listed in Table II.
Table II.
List of Putative M. genitalium Thiamine Kinases
| MG locus | Gene Name | Function | UniprotKB Accession |
|---|---|---|---|
| 038 | glpK | glycerol kinase | P47284 |
| 085 | hprK | HPr kinase/phosphorylase | P47331 |
| 128 | ppnK | putative inorganic polyphosphate/ATP-NAD kinase | P47374 |
| 145 | ribF | putative riboflavin biosynthesis protein | P47391 |
| 268 | n/a | putative purine deoxynucleoside kinase | P47510 |
| 356 | n/a | putative choline/ethanolamine kinase | Q49423 |
| 382 | Udk | uridine kinase | P47622 |
Size Exclusion Chromatography
Purified MG289 and Cypl in 50 mM Tris-HCl pH 7.5 were each diluted to a concentration of 2.5 mg ml-1. 150 μl (∼400 μg of purified protein) was loaded onto a Superdex 200 10/300 GL gel filtration column (GE Healthcare) equilibrated with 150 mM sodium chloride, 50 mM sodium phosphate, pH 7.2. The flow rate was 0.4 ml min-1 and peaks were detected by absorbance at 280 nm. Molecular weight estimates were calibrated using Bio-Rad Gel Filtration Standard.
Small Angle X-ray Scattering
Purified MG289 was buffer exchanged into 50 mM sodium phosphate buffer pH 7.5 and concentrated to 0.5 mg ml-1. The SAXS data was collected at the G1 beamline at the Cornell High-Energy Synchrotron Source (CHESS). The wavelength used was 1.296 Å with a detector distance of 1210 mm. The data was collected between angular displacement values (s) of 0.001 and 0.275 Å-1. The images were processed for intensity and s values using DATASQUEEZE29. GNOM was used to compute the radii of gyration and the pairwise distribution function. DAMMIN was used to generate the 3-D ab initio model. Ten DAMMIN simulations were averaged using the DAMAVER program to generate a final model. This model was converted to a SITUS volume map for docking purposes and the MG289 X-ray dimer was docked manually into the map using the CHIMERA program30,31. Theoretical SAXS curves from the crystal structures of the monomer and the dimer were computed using the CRYSOL. GNOM, DAMMIN, DAMAVER, and CRYSOL are all located within the ATSAS suite32.
Results
Overall Structure of MG289
The structure of MG289 was solved to a resolution of 1.95 Å with an Rwork and Rfree of 22.1% and 25.5% respectively. Data and refinement statistics are listed in Table I. Solution and refinement details are described in the Experimental Methods section.
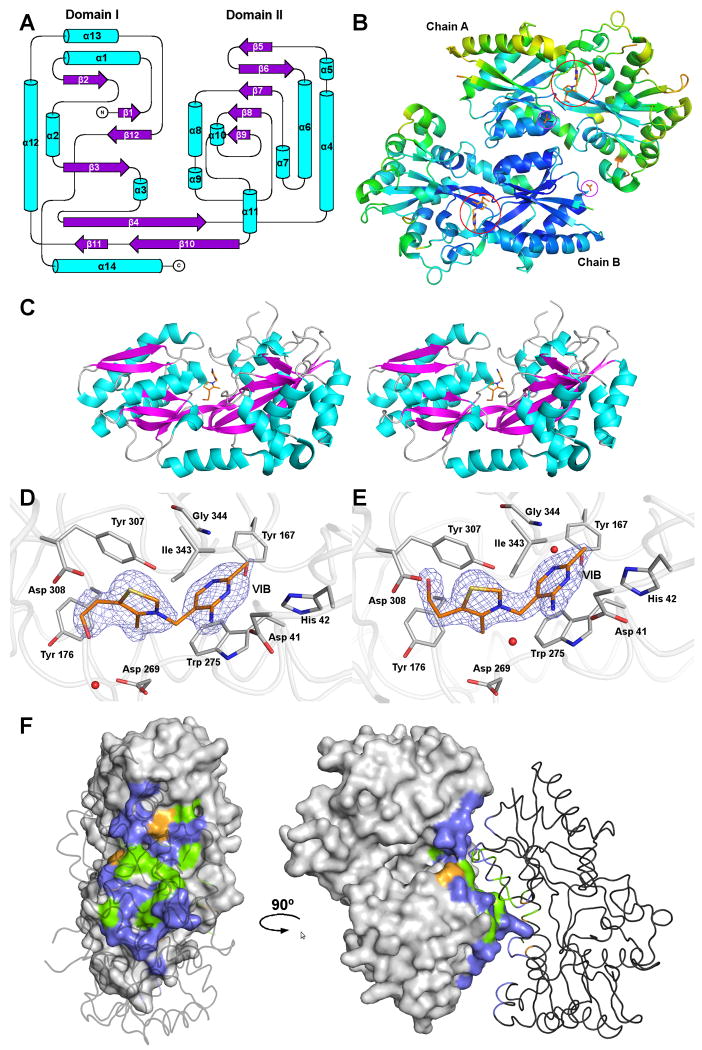
The MG289 monomer is a mixed α/β fold split into two globular domains (Domains I and II) connected by a hinge region formed by two non-sequential strands and the C-terminal helix (Figure 1A). The two domains form a deep cleft with thiamine (VIB) tightly bound between them. There are two MG289 monomers in the crystallographic asymmetric unit (ASU) (Figure 1B). The overall shape of the protein is an irregular prolate ellipsoid with the approximated dimensions of 70 × 40 × 30 Å3 (Figure 1C). Domain I is slightly smaller than Domain II. Domain I includes residues 32-115, and 308-352. It has six strands flanked by five helices with β4 crossing the hinge region into Domain II. Domain II includes residues 116-307. Residues 281-283 were not modeled due to a lack of density. Domain II contains six strands flanked by eight helices with β10 terminating at the hinge junction. The C-terminal helix, α14 (residues 353-367), lies parallel to the hinge region sitting nearly equally between Domains I and II.
Figure 1.
The structure of MG289.
(A) Secondary structural topology of the MG289 monomer. Helices are colored cyan and strands, magenta. Figure generated in Topdraw 44.
(B) A Cα ribbon trace of the MG289 dimer colored from low to high (blue to red) temperature factor. Ligands are shown as sticks with carbon colored orange; oxygen, red; nitrogen, blue; and sulfur, gold. VIB binding sites are indicated as open red circles. Open purple circles indicate ACT binding sites.
(C) Stereo image of the MG289 monomer with VIB ligand bound. Protein is colored as described in (A), and VIB as described in (B).
(D) The VIB binding site of chain A. The 2Fo-Fc average kick map (mesh) is contoured to 1.0σ 45. Protein carbons are colored grey; ligand carbons, orange; oxygen, red; nitrogen, blue; sulfur, gold. Waters are represented as red spheres.
(E) The VIB binding site of chain B. The 2Fo-Fc average kick map (mesh) is contoured to 1.8σ. Figure colored as described in (D).
(F) The dimer interface. Chain A is shown as a molecular surface, while chain B is a cartoon. Non-interface residues are colored grey; non-polar, green; polar uncharged, violet, and polar charged, orange. All figures generated in PYMOL unless stated otherwise 46.
The distribution of temperature factors (B-factors) in the structure is consistent with existing protein structures, with the core β-sheets having the lowest B-factors that increase as the residues become more solvent exposed (Figure 1C). Domain I has ∼20% lower temperature factors than Domain II partly owing to a densely packed hydrophobic core that imposes high order. This is also the result of an approximately 7% loss of helical structure with Domain II being composed of long semi-ordered loops forming 42% of the total secondary structure. An additional consideration is that there is a difference in the average temperature factor between the two chains in the ASU. Due to the high resolution of the data the refinement was carried out without non-crystallographic symmetry restraints resulting in an average B of 44.5 Å2 for Chain A and 33.6 Å2 for Chain B. This 24.5% increase in B-factor may be correlated to a 34.4% loss of crystal contact surface area in Chain A as compared to Chain B33,34.
Thiamine Binding Site
Thiamine (VIB) was located at the base of the hinge region at the junction between the two domains in both MG289 monomers in the ASU. In both chains the bulk of the residue interactions are hydrophobic (Figure 1D and E). The methyl pyrimidine ring π stacks with Trp 275, and has Van der Waals interactions with Gly 344, Ile 343, His 42, and Tyr 167. The VIB also has a direct hydrogen bond of ∼3 Å to the OD2 of Asp 41 and there is a water-mediated hydrogen bond to the OH of Tyr 167 in chain B. Also an additional water is bound to N4A at a distance of 3.2 Å in chain B. The thiazole ring engages in exclusively hydrophobic interactions binding Tyr 307, Asp 269, and Ile 343. The ethanol moiety of VIB exhibits two conformations between the two monomers of the ASU. In chain A the ethyl portion of the tail lies parallel to the phenol ring of Tyr 176 and the hydroxyl hydrogen bonds through water to Asp 269, and in chain B, C7 interacts hydrophobically with Tyr 176 and Asp 308, and has a direct hydrogen bond to the N of Asp 308 at a distance of 3.0 Å. Therefore, it appears structurally that either conformation is equally favored.
Dimer Interface
The overall accessible surface area of the MG289 monomer, as calculated by the PISA server, is 15940.0 Å2, of which 1041 Å2 (9.6%) is buried in the interface between the two molecules in the ASU34. There is an estimated gain of -4.2 kcal/mol upon complexation. The composition of the interface residues is as follows: 12 hydrophobic residues, 18 polar uncharged residues and 2 polar charged residues for chain A and 12, 17 and 3 residues respectively for chain B (Figure 1F). There are ten hydrogen bonds in the interface, mostly between Asn, Gln, and Thr residues, and no disulfides, covalent interactions, salt bridges or large continuous hydrophobic patches were detected. The overall complexation significance score (CSS) of the interface was 0.113. CSS ranges from 0 to 1 as the biological relevance of the interaction increases. This indicates that the interface may be a very weak interaction that would only play an auxiliary role in complexation.
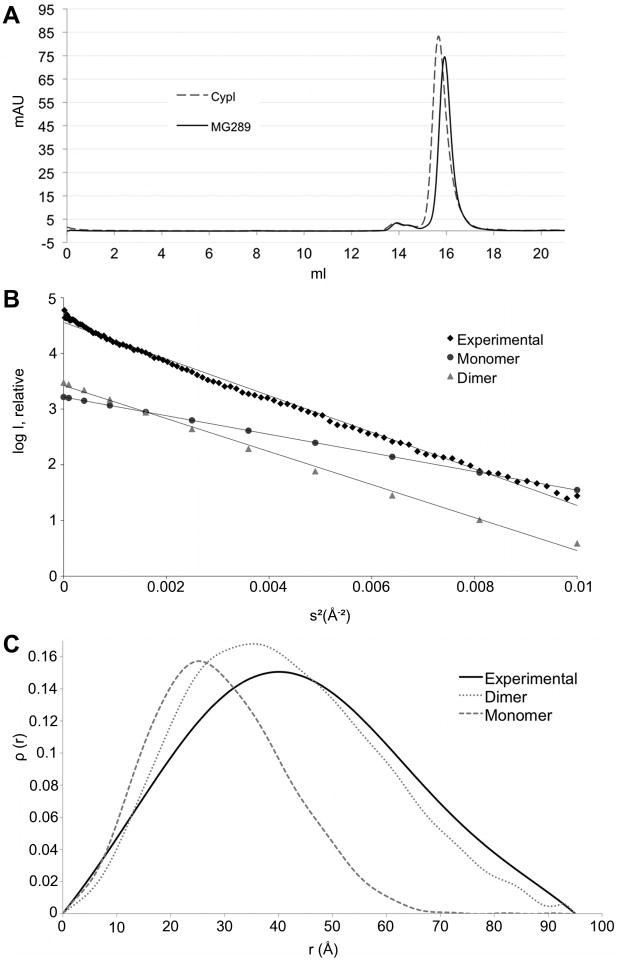
Size Exclusion Chromatography
In an effort to confirm the computational results, experimental methods were employed to identify the quaternary structure of MG289 in solution. Size exclusion chromatography (SEC) was used to derive an approximate molecular weight of the soluble species. As the shape of a protein is a factor in SEC, in addition to the globular molecular weight standards, the MG289 homolog Cypl was used for comparative purposes. Cypl is highly similar in shape to MG289 and known to be monomeric (Figure 2A)19. The MG289 has a computed molecular weight of 39.5 kDa and has an elution peak at 15.9 mL, which corresponds to approximately 45 kDa. This is compared to Cypl with a computed molecular weight of 43.4 kDa. Cypl elutes at 15.7 mL corresponding to a molecular weight of 49 kDa. There was a small peak at ∼14 mL, however SDS-PAGE of the fraction showed that it was contaminating protein (data not shown).
Figure 2.
The quaternary structure of MG289.
(A) Size exclusion chromatograph of MG289 (solid line) and Cypl (dotted line) with UV absorbance (mAU) at 280 nm plotted against elution volume (ml).
(B) Guinier plot of the squared angular displacement values (s2) versus the intensity (I). Experimental data (◆) is charted against the scaled theoretical plots of the monomer (●) and the dimer (▲).
(C) Pair distribution function (ρ(r)) versus the distance from the center of gravity (r). Experimental data (solid line) charted against the scaled theoretical plots of the monomer (dashed line) and the dimer (dotted line). where q = 4πsin(θ)/λ with λ = wavelength in Å and the 2θ = scattering angle The theoretical scale factor = [Σ (theoretical y-values)/no. points]/[Σ (experimental y-value)/no. observations].
Small Angle X-ray Scattering (SAXS)
Given the deviation of the SEC estimated molecular weight of MG289 from computed values, a second method of experimental verification was employed. The estimated radius of gyration (Rg) of the monomer and the dimer were 22.4 Å and 31.8Å respectively, indicating that SAXS would be amenable to addressing the question of monomer versus dimer. The experimental Rg of MG289 was calculated to be 33.7 Å (Figure 2B). The overall shape of the dimer is a slightly twisted disk, whereas the monomer is dumbbell shaped. The shape of the pairwise distribution curve can describe this information. The curve of the experimental data strongly resembles the dimer (Figure 2C). The experimental data was used to derive a low resolution ab initio model of MG289 in solution. The overall shape and size of this model is consistent with a dimer (Figure S1).
Comparison of MG289 to homolog Cypl
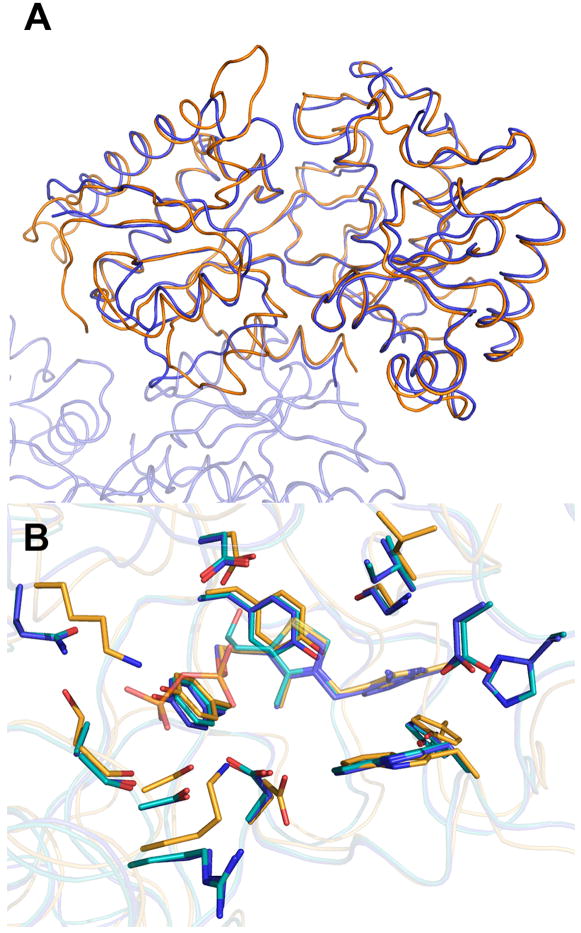
When the overall structure of MG289 is compared to the M. hyorhinis homolog Cypl, there are strong similarities between to two proteins (Figure 3A). The RMSD of the Cα is 1.36 Å for the 365 comparable atoms. As mentioned previously the two share 32% sequence identity with high conservation in the ligand binding site residues. The largest structural variation is in the truncation of two surface loops in MG289 as compared to Cypl, one along the exterior of the binding cleft and one located proximal to the dimer interface (Figure 3A). The binding cleft loop is shortened in MG289 bringing it closer to the ligand binding site, which leads to several additional residues stabilizing the methyl pyrimidine ring in VIB that do not have equivalent amino acids in Cypl (Figure 3B).
Figure 3.
MG 289 versus Cypl
A) Secondary structure matching alignment of MG289 (blue) chain A and Cypl (orange). Regions of significant difference are indicated by open red circles.
(B) Superposition of ligand binding site from MG289 and Cypl. MG289 chain A carbons are colored blue; chain B carbons, cyan; Cypl carbons, light orange; oxygen, red; nitrogen, blue; sulfur, gold; phosphate, orange. Blue text indicates MG289 residue numbering; Orange, Cypl numbering. * indicates no equivalent residues. Figure generated in Pymol 46.
Comparison of the Cypl and MG289 ligand binding sites show that, as predicted, several of the ligand binding residues are highly conserved, at least for those interacting with the methyl pyrimidine and thiazole ring (Figure 3)19. The Trp, Gly, and three Tyr residues are nearly superimposable. The conservative substitution of Ile for Leu also causes very little change in the pyrimidine ring orientation. Residues 41 and 42 of MG289, which interact with the methyl pyrimidine ring, have no equivalent amino acids in Cypl because they are located on the shortened binding cleft loop. The greatest structural differences are located in the tail region. TPP has an ethylene bridged pyrophosphate tail, whereas VIB merely has the ethanol group. In Cypl the TPP is stabilized by three Ser and two Lys residues, whereas in MG289, one of the Ser is replaced by an Ala, one Lys by a Gln, and the other Lys by an Arg.
Discussion
The overall structure of MG289 resembles a periplasmic substrate binding protein (SBP). More specifically, it resembles the Type II periplasmic binding proteins derived from domain displacement as described by Fukami-Kobayashi22. Recently a more extensive classification system for SBPs was developed35. Based on this new classification system MG289 is a class D SBP, subclass II based off of the VIB ligand. This assertion was confirmed by the secondary structural matching results obtained from the DALI server, as the first 200+ structure matches are Class D36. Since mycoplasmas lack a periplasmic space having evolved from Gram-positive bacteria and combined with the fact that they have a lipoprotein moiety, which was truncated for expression purposes, this supports the idea that MG289 is probably extracytoplasmic, being presented on the exterior of the cell membrane.
The quaternary structure of MG289 has not been fully answered. The computational and SEC data indicates a monomeric protein whereas the SAXS data indicates a dimer. Computationally the dimer lacks many of the critical features of a biological interface, including a large percentage of buried surface area, an adequate number of hydrophobic residues, and any interactions stronger than a hydrogen bond37. The SEC experiments support the computational results. Though there was a 5 kDa difference between the calculated molecular weight and the experimental one, this can be accounted for by the distinctive shape of MG289 which would affect its mobility in a size exclusion column. Its match to the elution curve of Cypl, a monomer, confirms its quaternary structure in solution19. However, the SAXS data contradicts the computation and SEC data fitting to a dimeric structure. It is likely that the interaction is concentration dependant. The SAXS data was collected at a concentration of 0.5 mg ml-1, whereas the SEC is 0.05 mg ml-1 at its most concentrated. Prokaryotic SBPs are usually monomeric when bound to an ABC transporter35. Occasionally they can purify as dimers, however in those cases the SPBs would shift towards the monomeric form upon ligand binding38,39. In the context of the native lipid-associated form the protein must interact with its transporter to transfer its ligand. The observed interface may be the result of a “sticky” integral membrane binding region of the protein surface trying to bury itself while in concentrated solution.
When compared to the M. hyorhinis homolog Cypl, there are strong similarities between the two proteins. The largest distinction is in the loss of several large loops in the binding cleft and at the binding interface (Figure 3A). This interface loop might play a role in the dimeric nature of the protein in concentrated solution. Cypl crystallizes in the P21 space group with only one molecule in the ASU. Cypl has an extended loop in what would be the dimer interface region, which may account for both the monomeric structure of Cypl in solution and the lack of a necessary crystal contact that would lead to the two molecules in the ASU and the P212121 space group of MG289 (Figure 3A)19,20.
The ligand binding residues show a high degree of conservation between MG289 and Cypl, though there are differences (Figure 3B). The residues of the tail region seem to show the greatest sequence variation. The three Ser and two Lys residues strongly stabilize the pyrophosphate in Cypl. In MG289, the residues are two Ser, an Ala, a Gln and an Arg that do not interact with the VIB. These differences would not preclude the possibility for low affinity binding of a phosphate or even pyrophosphate moiety, but they are obviously not the preferred ligand. There is also the addition of residues 41 and 42 from the shortened binding cleft loop in MG289. This does not seem to make a difference in the context of thiamine selection, however it may contribute to the affinity for other ligands. Further studies of the kinetics of MG289 and Cypl would be needed to fully characterize the significance. Both ligand binding sites lack strong hydrogen bonds in favor of less specific hydrophobic interactions that may allow for some of the predicted substrate promiscuity including the transport of unsynthesizable guanylate or phosphonates6,7,40.
When comparing MG289 and Cypl, the most obvious question that arises is why does M. hyorhinis have a preference towards binding TPP while M. genitalium binds VIB? Thiamine is a critical component of Mycoplasma minimal media41,42. This is because most Mycoplasmas are solely dependant on glycolysis, fermentation, and substrate phosphorylation to meet energetic requirements1,42. Two enzymes critical to survival are pyruvate dehydrogenase and transketolase, both of which need TPP as a cofactor4,42. Generation of TPP from thiamine requires its phosphorylation, usually by some form of nucleoside diphosphokinase (ndk). In most known genomes, including prokaryotes, there is some form of highly conserved ndk, however no such gene exists in Mollicutes. It is theorized that to compensate nonorthologous gene displacement occurred allowing other sugar kinases to perform the duties of ndk3-5,40,42. From the crystallographically observed preferential binding of Cypl for TPP rather than VIB it seems likely that M. hyorhinis lacks the kinase necessary for TPP production inside the cell; a kinase produced by M. genitalium. Several possible ndk replacements have been suggested including 6-phosphofructokinase (MG215), phosphoglycerate kinase (MG300), pyruvate kinase (MG216) and acetate kinase (MG357) that can phosphorylate nucleotide mono- and diphosphates, a dephospho-CoA kinase (MG264) and a putative deoxyribonucleoside kinase (MG268)3-5. We have recently sequenced approximately 96% of the M. hyorhinis genome, with the remainder being mostly repetitive, non-coding sequences. We searched the M. hyorhinis genome for homologs of all known or putative sugar or nucleoside kinase sequences found in M. genitalium (data not shown). Seven M. genitalium sugar or nucleoside kinases had no detectable homologs in the M. hyorhinis genome. These include MG268, mentioned above, as well as a choline/ethanolamine kinase (MG356), uridine kinase (MG382), glycerol kinase (MG038), HPr kinase/phosphorylase (MG085), an inorganic phosphate/ATP-NAD kinase (MG128), and a riboflavin biosynthetic protein (MG145). Full descriptions of the kinases and accession numbers are listed in Table II. MG289, MG382, and MG128 seem to have a higher probability of being the actual thiamine kinase given the nature of their “endogenous” substrate.
Another consideration is the mechanism of VIB transport into the cell. The remaining genes encoded on the operon include a nucleotide binding protein and a transmembrane protein, typical of prokaryotic ATP-binding cassette (ABC) transporters. It is theorized that the transporter may function as a VIB/proton symporter using proton motive force as a propellant, however this does not explain the nucleotide binding protein on the operon42. Nucleotide binding proteins or domains are equipped for substrate hydrolysis, usually ATP to power transport43. This implies that thiamine and probably other nucleoside transport is achieved via ABC transporters rather than symporters as previously theorized.
Conclusion
The structure of MG289 has revealed many interesting insights into mycoplasma metabolism as a whole. In the context of a minimal genome it discloses a possible mechanism for promiscuity in ligand binding through a dearth of specificity conferring hydrogen bonds. It behaves as a weak dimer in concentrated solution, which may reveal a biological relevant interface in the context of a plasma membrane and interactions with a transmembrane protein. The preference of the protein for VIB, the precursor rather than TPP the enzymatic cofactor, indicates that it possesses a kinase capable of processing VIB. The list of putative kinases was reduced to seven possibilities. Further studies will be necessary to identify the overall structure of the transporter, the true kinase and the extent of its binding diversity. Hopefully, this knowledge will be used to further aid our understanding about the metabolic requirements of a minimal genome and may lead to the development of better drugs for the treatment of M. genitalium infections.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Arthur H. Robbins and the presenters of the 2010 CCP4 workshop for assistance and advice during structure solution and refinement, Dr. Ranjana Sarma for providing the advice for the SAXS analysis, and Dr. Virginia Urquidi for her contribution to sequencing the M. hyorhinis genome. This work was funded by the American Cancer Society grant 58229 to CJR. The Berkeley Center for Structural Biology is supported in part by the NIH, NIGMS, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. DOE under Contract No. DE-AC02-05CH11231. The Cornell High Energy Synchrotron Source is supported by the MacCHESS grant (US NIH grant RR001646), by US DOE grant DE-FG02-97ER62443 and the US NSF grant DMR- 0225180. CCP4 workshop: From data collection to structure refinement and beyond” was funded partly by federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104); and partly by a grant from CCP4 part of the Science and Technology Facilities Council (STFC) in the UK.
References
- 1.Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM, Fritchman RD, Weidman JF, Small KV, Sandusky M, Fuhrmann J, Nguyen D, Utterback TR, Saudek DM, Phillips CA, Merrick JM, Tomb JF, Dougherty BA, Bott KF, Hu PC, Lucier TS, Peterson SN, Smith HO, Hutchison CA, 3rd, Venter JC. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270(5235):397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 2.Maniloff J. In: Phylogeny of Mycoplasmas. Maniloff J, McElheney R, Finch L, Baseman J, editors. Washington, DC, USA: American Society of Microbiology Press; 1992. [Google Scholar]
- 3.Pollack JD, Myers MA, Dandekar T, Herrmann R. Suspected utility of enzymes with multiple activities in the small genome Mycoplasma species: the replacement of the missing “household” nucleoside diphosphate kinase gene and activity by glycolytic kinases. OMICS. 2002;6(3):247–258. doi: 10.1089/15362310260256909. [DOI] [PubMed] [Google Scholar]
- 4.Mushegian AR, Koonin EV. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc Natl Acad Sci U S A. 1996;93(19):10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gil R, Silva FJ, Pereto J, Moya A. Determination of the core of a minimal bacterial gene set. Microbiol Mol Biol Rev. 2004;68(3):518–537. doi: 10.1128/MMBR.68.3.518-537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA, 3rd, Smith HO, Venter JC. Essential genes of a minimal bacterium. Proc Natl Acad Sci U S A. 2006;103(2):425–430. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hutchison CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, Smith HO, Venter JC. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286(5447):2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- 8.Baseman JB, Dallo SF, Tully JG, Rose DL. Isolation and characterization of Mycoplasma genitalium strains from the human respiratory tract. J Clin Microbiol. 1988;26(11):2266–2269. doi: 10.1128/jcm.26.11.2266-2269.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tully JG, Taylor-Robinson D, Cole RM, Rose DL. A newly discovered mycoplasma in the human urogenital tract. Lancet. 1981;1(8233):1288–1291. doi: 10.1016/s0140-6736(81)92461-2. [DOI] [PubMed] [Google Scholar]
- 10.Tully JG, Taylorrobinson D, Rose DL, Cole RM, Bove JM. Mycoplasma-Genitalium, a New Species from the Human Urogenital Tract. Int J Syst Bacteriol. 1983;33(2):387–396. [Google Scholar]
- 11.Ross JD, Jensen JS. Mycoplasma genitalium as a sexually transmitted infection: implications for screening, testing, and treatment. Sex Transm Infect. 2006;82(4):269–271. doi: 10.1136/sti.2005.017368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manhart LE, Holmes KK, Hughes JP, Houston LS, Totten PA. Mycoplasma genitalium among young adults in the United States: an emerging sexually transmitted infection. Am J Public Health. 2007;97(6):1118–1125. doi: 10.2105/AJPH.2005.074062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez G, Skurnick JH, Denny TN, Stephens R, Kennedy CA, Regivick N, Nahmias A, Lee FK, Lo SC, Wang RY, Weiss SH, Louria DB. Herpes simplex type II and Mycoplasma genitalium as risk factors for heterosexual HIV transmission: report from the heterosexual HIV transmission study. Int J Infect Dis. 1998;3(1):5–11. doi: 10.1016/s1201-9712(98)90088-1. [DOI] [PubMed] [Google Scholar]
- 14.Namiki K, Goodison S, Porvasnik S, Allan RW, Iczkowski KA, Urbanek C, Reyes L, Sakamoto N, Rosser CJ. Persistent exposure to Mycoplasma induces malignant transformation of human prostate cells. PLoS One. 2009;4(9):e6872. doi: 10.1371/journal.pone.0006872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen JS. Single-dose azithromycin treatment for Mycoplasma genitalium-positive urethritis: best but not good enough. Clin Infect Dis. 2009;48(12):1655–1656. doi: 10.1086/599034. [DOI] [PubMed] [Google Scholar]
- 16.Gilson E, Alloing G, Schmidt T, Claverys JP, Dudler R, Hofnung M. Evidence for high affinity binding-protein dependent transport systems in gram-positive bacteria and in Mycoplasma. Embo J. 1988;7(12):3971–3974. doi: 10.1002/j.1460-2075.1988.tb03284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dudler R, Schmidhauser C, Parish RW, Wettenhall RE, Schmidt T. A mycoplasma high-affinity transport system and the in vitro invasiveness of mouse sarcoma cells. Embo J. 1988;7(12):3963–3970. doi: 10.1002/j.1460-2075.1988.tb03283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sippel KH, Boehlein SK, Sakai Y, Quirit JG, Agbandje-McKenna M, Rosser CJ, McKenna R. Cloning, expression, purification, crystallization and preliminary X-ray analysis of Mycoplasma genitalium protein MG289. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009;65(Pt 9):910–912. doi: 10.1107/S1744309109030565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sippel KH, Robbins AH, Reutzel R, Boehlein SK, Namiki K, Goodison S, Agbandje-McKenna M, Rosser CJ, McKenna R. Structural insights into the extracytoplasmic thiamine-binding lipoprotein p37 of Mycoplasma hyorhinis. J Bacteriol. 2009;191(8):2585–2592. doi: 10.1128/JB.01680-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sippel KH, Robbins AH, Reutzel R, Domsic J, Boehlein SK, Govindasamy L, Agbandje-McKenna M, Rosser CJ, McKenna R. Structure determination of the cancer-associated Mycoplasma hyorhinis protein Mh-p37. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 11):1172–1178. doi: 10.1107/S0907444908030175. [DOI] [PubMed] [Google Scholar]
- 21.Quiocho FA, Ledvina PS. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol Microbiol. 1996;20(1):17–25. doi: 10.1111/j.1365-2958.1996.tb02484.x. [DOI] [PubMed] [Google Scholar]
- 22.Fukami-Kobayashi K, Tateno Y, Nishikawa K. Domain dislocation: a change of core structure in periplasmic binding proteins in their evolutionary history. J Mol Biol. 1999;286(1):279–290. doi: 10.1006/jmbi.1998.2454. [DOI] [PubMed] [Google Scholar]
- 23.McPherson A. Preparation and Analysis of Protein Crystals. New York: John Wiley and Sons; 1982. [Google Scholar]
- 24.Otwinowski Z, Minor W. In: Processing of X-ray Diffraction Data Collected in Oscillation Mode. Carter C Jr, Sweet R, editors. New Haven, CT, USA: Yale University; 1997. [DOI] [PubMed] [Google Scholar]
- 25.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 26.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heiney PA. Datasqueeze. v 2.1. Philadelphia, PA, USA: University of Pennsylvania; 2002. [Google Scholar]
- 30.Wriggers W, Milligan RA, McCammon JA. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J Struct Biol. 1999;125(2-3):185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- 31.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 32.Petoukhov MV, Konarev PV, Kikhney AG, Svergun DI. ATSAS 2.1 - towards automated and web-supported small-angle scattering data analysis. J Appl Crystallogr. 2007;40:S223–S228. [Google Scholar]
- 33.Finzel BC, Salemme FR. Anomalous Temperature Factor Behavior and Crystal Lattice Mobility in Cytochrome C′. Biophys J. 1986;49(1):73–76. doi: 10.1016/S0006-3495(86)83598-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 35.Berntsson RP, Smits SH, Schmitt L, Slotboom DJ, Poolman B. A structural classification of substrate-binding proteins. FEBS Lett. 2010 doi: 10.1016/j.febslet.2010.04.043. [DOI] [PubMed] [Google Scholar]
- 36.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24(23):2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janin J, Bahadur RP, Chakrabarti P. Protein-protein interaction and quaternary structure. Q Rev Biophys. 2008;41(2):133–180. doi: 10.1017/S0033583508004708. [DOI] [PubMed] [Google Scholar]
- 38.Richarme G. Associative properties of the Escherichia coli galactose binding protein and maltose binding protein. Biochem Biophys Res Commun. 1982;105(2):476–481. doi: 10.1016/0006-291x(82)91459-0. [DOI] [PubMed] [Google Scholar]
- 39.Richarme G. Associative properties of the Escherichia coli galactose-binding protein and maltose-binding protein. Biochim Biophys Acta. 1983;748(1):99–108. doi: 10.1016/0167-4838(83)90032-8. [DOI] [PubMed] [Google Scholar]
- 40.Pollack JD, Jones MA, Williams MV. The metabolism of AIDS-associated mycoplasmas. Clin Infect Dis. 1993;17 1:S267–271. doi: 10.1093/clinids/17.supplement_1.s267. [DOI] [PubMed] [Google Scholar]
- 41.Smith SL, Vandemark PJ, Fabricant J. Respiratory Pathways in the Mycoplasma. I. Lactate Oxidation by Mycoplasma Gallisepticum. J Bacteriol. 1963;86:893–897. doi: 10.1128/jb.86.5.893-897.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suthers PF, Dasika MS, Kumar VS, Denisov G, Glass JI, Maranas CD. A genome-scale metabolic reconstruction of Mycoplasma genitalium, iPS189. PLoS Comput Biol. 2009;5(2):e1000285. doi: 10.1371/journal.pcbi.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72(2):317–364. doi: 10.1128/MMBR.00031-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bond CS. TopDraw: a sketchpad for protein structure topology cartoons. Bioinformatics. 2003;19(2):311–312. doi: 10.1093/bioinformatics/19.2.311. [DOI] [PubMed] [Google Scholar]
- 45.Praaenikar J, Afonine PV, Guncar G, Adams PD, Turk D. Averaged kick maps: less noise, more signal… and probably less bias. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 9):921–931. doi: 10.1107/S0907444909021933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeLano WL. The Pymol Molecular Graphics System. Palo Alto, CA, USA: Delano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.