Abstract
DNA nanoparticles (DNPs) are nonviral gene transfer vectors with excellent in vivo potential. Previously, we reported that cell surface nucleolin directly binds DNPs, and functions as an important receptor for DNPs. However, the fate of the nucleolin–DNP complex following cellular uptake remains elusive. In this study, we examined the role of lipid rafts in the uptake of DNPs, and found that both nucleolin and DNPs are recovered from the low-density raft fractions of the sucrose gradient. Furthermore, nucleolin colocalizes with, and coimmunoprecipitates with a raft protein, flotillin. Disruption of lipid rafts by depleting membrane cholesterol significantly inhibited DNP transfection, while inhibition of other endocytic pathways had little effect. Following the uptake, the nuclear import of the DNPs required microtubules but not F-actin. By coimmunoprecipitation in conjunction with tandem mass spectrometry, we identified glucocorticoid receptor (GCR) as a nucleolin-associated protein, and confirmed this result by western blot. Cortisone or dexamethasone increased nucleolin's association with GCR, and transfection by DNPs. Finally, we detected the expression of nucleolin on the surface of airway epithelia in vivo. Taken together, our findings shed light on important determinants of DNP trafficking in cells and support the notion that nucleolin is a good target for nonviral gene delivery.
Introduction
Nonviral vectors for airway gene transfer have been the subject of intense investigation due to their lack of toxicity and immunogenicity.1 DNA nanoparticles (DNPs), self-assembled from polyethylene glycolated cysteine–lysine 30mer (PEG-CK30) and plasmid DNA carrying therapeutic or reporter genes, are nonviral vectors in development for human gene therapy. Preclinical studies in mice demonstrated that DNPs mediate effective airway gene transfer with minimal toxicity and immunogenicity.2,3 Similar results have been obtained in rat, rabbit, ferret, and sheep.4 They have also been tested in a phase I clinical trial to treat cystic fibrosis, an autosomal recessive, lethal disease affecting mainly the lung.5,6 Nasal delivery of DNPs carrying the normal cystic fibrosis transmembrane conductance regulator gene corrected, at least partially, the chloride transport defect in 8 out of 12 patients.7 Despite the good potential of DNPs in vivo, they are relatively inefficient compared to their viral counterparts, and thus a higher level of clinical-grade DNA is required for gene transfer. To reduce the amount of DNA needed and the associated costs, which can be prohibitive of the wide application of DNPs, researchers have focused on optimizing the composition to improve gene transfer efficiency. Another approach, which is to manipulate the cellular processes involved in the uptake and trafficking of DNPs, has not been explored.
The mechanisms involved in the transfection of target cells by nonviral vectors are poorly understood. The transfection process encompasses a range of steps, including membrane attachment, internalization, intracellular trafficking, nuclear import, dissolution of the vector and release of the transgene, and transgene expression.8 We previously reported that nucleolin expressed on the cell surface binds to DNPs directly and serves as a critical receptor for DNPs.9 Expression of cell surface nucleolin was confirmed in cultured HeLa and airway epithelial cell 16HBEo− cells. Increasing surface nucleolin by overexpression or reducing it by serum starvation or small-interfering RNA knockdown concordantly enhances or inhibits transfection by DNPs, respectively.9 As a conserved eukaryotic protein, nucleolin has essential functions in ribosomal biogenesis, mRNA metabolism, and cell cycle progression.10,11 Cell surface–expressed nucleolin was found to be a receptor for various ligands, such as human parainfluenza virus type 312 and human immunodeficiency virus type 113 and anti-human immunodeficiency virus molecules like midkine,14 lactoferrin,15 and pleiotrophin,16 antitumor agents such as endostatin,17 and a hemorrhagic strain of E. coli.18 However, little is known about its fate after ligand binding and internalization.
Following membrane attachment, cellular uptake of polyethylenimine-based vectors via clathrin-mediated endocytosis or macropinocytosis into endosomes and subsequent degradation in lysosomes has been a major barrier to efficient transfection.8 By contrast, fluorescence-labeled DNPs colocalize with nucleolin on the membrane and in the cytoplasm, but not with the endosomal marker EEA1 (early endosomal antigen-1) or the lysosomal marker cathepsin D in well-differentiated human tracheal epithelial cells, or transferrin, a marker for clathrin-dependent endocytosis, in HeLa cells,9 indicating that DNPs do not enter the clathrin-mediated endocytosis pathway. These findings prompted us to explore the possibility that DNPs enter the cell via alternative routes, such as lipid raft–mediated endocytosis,19 and to examine the role that nucleolin plays in this process.
Another barrier to efficient gene transfer in vivo is nuclear entry. Because many target cells in the body do not actively divide and vectors have to traverse the nuclear envelope, nuclear pore complex is expected to be an important determinant of nuclear entry. DNPs smaller than the nuclear pore complex can enter the nucleus and deliver transgenes in nondividing cells, albeit at a much lower efficiency than those directly injected into the nucleus.20 We previously found that nucleolin accompanies DNPs into the nucleus, where they colocalize in the nucleolus.9 Thus, in addition to the internalization and cellular trafficking of the nanoparticles, nucleolin may also play an important role in their nuclear import.
In this study, we identify lipid raft–mediated endocytosis as the major route of the cellular uptake of DNPs, and found both nucleolin and DNPs in lipid rafts. Pharmacological inhibition of this pathway blocks transfection by DNPs, while reconstitution of lipid rafts reverses this inhibition. Nucleolin associates with flotillin, a constitutive raft protein, which might explain its ability to exist in membrane rafts. Furthermore, we identify the glucocorticoid receptor (GCR) as a binding partner of nucleolin by immunoprecipitation (IP), mass spectrometry (MS), and western blot. Association of nucleolin with GCR increases rapidly with cortisone treatment, which also enhances transfection by DNPs. Finally, we confirm the expression of nucleolin in mouse airway epithelium at the apical surface. In summary, we present data on the processing of DNPs by transfected cells and our manipulation of discovered mechanisms to influence gene transfer, demonstrating the potentials for studies on the trafficking of a nonviral vector.
Results
Transfection of DNPs requires intact lipid rafts
Because we previously found little colocalization of DNPs with the endosomal marker EEA1 or the lysosomal marker cathepsin D, we suspected that endocytic pathways other than clathrin-mediated endocytosis might participate in the cellular uptake of DNPs. Therefore, we tested the effects of drugs that inhibit a range of endocytic pathways on gene transfer. Pretreatment of HeLa cells with either filipin or methyl-β-cyclodextrin significantly inhibited DNP-mediated luciferase expression (Figure 1a). This inhibition is not due to the decrease in nucleolin expression as assayed by cell surface biotinylation (Figure 1b). Chlorpromazine, which inhibits clathrin-mediated endocytosis, and ethylisopropyl amiloride and cytochalasin D, which inhibit macropinocytosis,21,22 had little effect (Figure 1a). Transfection in the presence of the drugs yielded similar results (data not shown). Repletion of cholesterol following filipin treatment reversed the inhibition, validating the specificity of the drug on cholesterol depletion and DNP transfection (Figure 1c). Therefore, the integrity of lipid rafts is critical for the uptake of DNPs.
Figure 1.
Transfection of DNA nanoparticles (DNPs) requires intact lipid rafts. (a) HeLa cells were pretreated with drugs as indicated, and transfected with DNPs carrying luciferase reporter. Luciferase activity was measured 48 hours after transfection, and compared by Student's t-test. **P < 0.01. (b) HeLa cells were treated as in a, then biotinylated on the cell surface. Surface nucleolin was pulled down using streptavidin beads and revealed by western blot. Total nucleolin and actin were also blotted as controls. (c) HeLa cells were pretreated with filipin and transfected with DNPs in the absence (−Cholesterol) or presence (+Cholesterol) of cholesterol. Luciferase activity was measured 48 hours after transfection. *P < 0.05. CPZ, chlorpromazine; CytD, cytochalasin D; DMSO, dimethyl sulfoxide; EIPA, ethylisopropyl amiloride; MβCD, methyl-β-cyclodextrin; RLU, relative light unit.
DNPs are recovered from the lipid rafts fraction from sucrose gradients
To probe more directly their trafficking route following uptake, we examined the presence of DNPs in different cellular compartments in transfected HeLa cells by sucrose-gradient separation, which allows separation of lipid rafts in the low-density fractions from other cellular components in the high-density fractions. Nucleolin, and raft and nonraft markers showed similar patterns by western blot in naked DNA- and DNP-transfected cells (Figure 2a, see also section “Nucleolin localizes to lipid rafts and associates with flotillin”). Raft and nonraft fractions (5 and 12 from the top of the gradient, respectively) were dialyzed, trypsinized to release DNA from the nanoparticles, and analyzed for plasmid DNA by semiquantitative PCR. As shown in the upper panel of Figure 2c, significantly more plasmid was recovered from both raft and cell body fractions in DNP-transfected cells (lanes 3 and 4) than in naked DNA–transfected ones (lanes 1 and 2), which was quantified by densitometry in the lower panel. By interpolating the standard curve in Figure 2b, the amounts of DNA in the DNP-transfected cells are 16.8 ± 1.5 and 54.9 ± 22.4 pg/µl (or 6.0 ± 0.6 and 19.8 ± 8.1 ng in the entire fraction) in the raft and nonraft fractions, respectively, compared to 0 and 0.9 ± 0.9 pg/µl in the naked DNA–transfected cells. No DNA was detected in the raft fractions when DNA or nanoparticles were loaded on sucrose gradients alone without cell contact (Figure 2c, middle panel) suggesting that the presence of DNPs in lipid raft fractions occurs during the transfection process. Therefore, DNPs employ lipid raft–mediated endocytosis for cellular entry.
Figure 2.
DNA nanoparticles (DNPs) enter lipid rafts. (a) HeLa cells were transfected with naked DNA or DNPs containing 25 µg plasmid DNA for 2 hours, and lysates were subject to sucrose-gradient separation. Fractions of each gradient were western blotted for nucleolin, raft marker flotillin, and nonraft marker PLCβ2. (b) Plasmid DNA of 0–10,000 pg was subjected to semiquantitative PCR. The PCR products were separated on agarose gel, quantified densitometrically, and plotted against the amounts of DNA. The inset at the right lower corner shows a representative gel image. (c) Raft and nonraft fractions (5 and 12, respectively) from gradients in a were dialyzed to removed sucrose and Na2CO3, trypsinized to release DNA, and subjected to detection of plasmid DNA by PCR (+Cell, upper panel). Naked DNA and nanoparticles without cell contact (−Cell, middle panel) were separated on a sucrose gradient, and processed and analyzed by PCR in the same fashion. The bands in the upper panel (+Cell) were quantified, and statistical analyses were performed in the lower panel. Lanes: 1, naked DNA in raft fraction; 2, naked DNA in nonraft fraction; 3, nanoparticle in raft fraction; 4, nanoparticle in nonraft fraction. PLCβ2, phospholipase C-β2.
Nucleolin localizes to lipid rafts and associates with flotillin
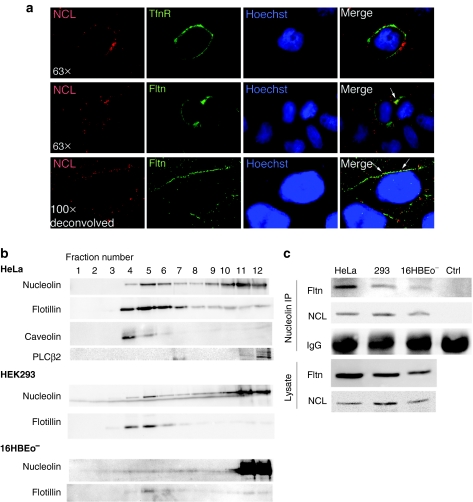
We further examined the presence of the receptor for DNPs, nucleolin, in membrane lipid rafts. In both HeLa and 16HBEo− cells, nucleolin exists in the low-density raft fractions, although the majority of nucleolin exists in the high-density fractions corresponding to the cytoplasm and nucleus (Figure 3b). Similar results were also observed in cells transfected with DNPs (Figure 2a). This distribution is consistent with our earlier findings that cell surface nucleolin constitutes a small portion of total cellular nucleolin. The colocalization of nucleolin with flotillin, as assessed by fluorescence immunostaining (Figure 3a, arrows), further confirmed the presence of nucleolin in the high-density fractions. In contrast, immunostainings for transferrin receptor and nucleolin are separated on the membrane (Figure 3a), consistent with our previous findings that DNPs do not colocalize with transferrin after cellular uptake.9
Figure 3.
Nucleolin exists in lipid rafts and associates with flotillin. (a) Colocalization of nucleolin with flotillin but not transferrin receptor. Live HeLa cells were stained for cell surface nucleolin (red), and transferrin receptor (TfnR), or flotillin (Fltn, green). Fluorescent micrographs were taken with either a ×63 or a ×100 objective, and stacks were deconvolved. Arrows indicate colocalization. (b) HeLa, HEK293 and 16HBEo− cells were separated on a sucrose gradient as described in Materials and Methods. Twelve fractions from the top were taken and immunoblotted for nucleolin, raft markers caveolin and flotillin, and nonraft marker phospholipase C (PLCβ2). (c) HeLa, HEK293, and 16HBEo− cells were immunoprecipitated with an antibody against nucleolin, and western blotted for flotillin and nucleolin in the IP fractions and lysates. Control (IP) was performed in parallel using green fluorescent protein antibody with HeLa cell lysate. Ctrl, control.
Because nucleolin has no obvious signal sequence or membrane-targeting sequence, we reasoned that its appearance in lipid rafts might be mediated by association with other raft proteins. Indeed, when endogenous nucleolin was immunoprecipitated from HeLa, HEK293, and 16HBEo− cells, flotillin could be detected in the IP fractions, but not in the control IP with a green fluorescent protein antibody (Figure 3c). Therefore, nucleolin exists in lipid rafts via association with raft protein flotillin.
Transfection of DNPs requires microtubules but not actin filaments
Numerous studies have suggested an important role of cytoskeleton in the trafficking of nonviral vectors,23,24 and nucleolin was suggested to associate with actin filaments.25 We, therefore, studied the role of cytoskeleton, including actin filaments and microtubules, in the transfection of DNPs by inhibition with cytochalasin D or phalloidin for F-actin, or taxol or nocodazole for microtubules. Both taxol and nocodazole significantly inhibit gene transfer either before or during DNP transfection, while neither cytochalasin D nor phalloidin has any effect on gene transfer (Figure 4a). As a control, neither taxol nor nocodazole significantly reduced transfection by Lipofectamine (Supplementary Figure S1), supporting that their inhibition on gene transfer by DNPs is not due to any interference with gene expression.
Figure 4.
Transfection of DNA nanoparticles (DNPs) requires intact microtubules. (a) HeLa cells were treated with cytochalasin D (CytD) and phalloidin during transfection, or taxol and nocodazole before and/or during transfection, and luciferase reporter expression from the nanoparticles were assayed in 48 hours. (b) HeLa cells were treated with vehicle DMSO or nocodazole during transfection with rhodamine-labeled DNPs, and examined under fluorescence microscope at 15 minutes to 4 hours. Arrows indicated nuclear nanoparticles. (c) HeLa and 16HBEo− cells were treated with DMSO or CytD, and transfected with rhodamine-labeled DNPs, and examined at 4 hours. Insets in each panel are composites of red and blue channels to show the fluorescence of DNPs in the nuclei. (d) HeLa and 16HBEo− cells were treated with DMSO or nocodazole, and transfected with rhodamine-labeled DNPs. At 4 hours, cells were fixed and stained for microtubules in green. Arrows indicate colocalization. DMSO, dimethyl sulfoxide; RLU, relative light unit.
We then examined the effect of nocodazole on the trafficking of DNPs by fluorescence microscopy. Rhodamine-labeled DNPs readily enter the nucleus of control HeLa cells treated with vehicle dimethyl sulfoxide, but accumulate in the cytoplasm in nocodazole-treated cells (Figure 4b). As a control, we examined dimethyl sulfoxide- and cytochalasin D–treated cells, and found no apparent difference in the nuclear entry of DNPs, even when actin filaments (stained green in Figure 4c) are disrupted by cytochalasin D treatment. Costaining of microtubules in both HeLa and 16HBEo− cells transfected with rhodamine-labeled DNPs showed colocalization of the two (Figure 4d, arrows), which was disrupted by nocodazole treatment. Therefore, DNPs require intact microtubules but not F-actin for their nuclear entry and delivery of transgene.
Nucleolin associates with GCR
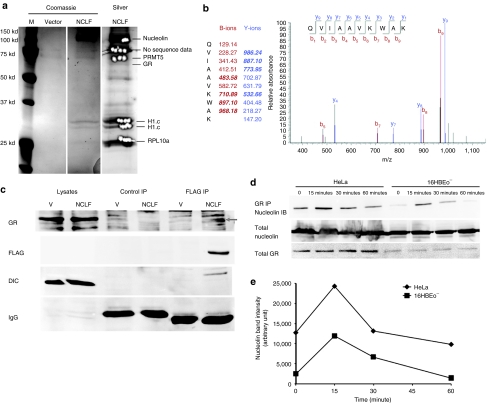
Because nuclear entry is a critical step in DNP transfection, and nucleolin plays an important role in their trafficking, we set out to determine the intracellular binding partners of nucleolin. We generated a HeLa cell line that stably expresses a C-terminal FLAG-tagged nucleolin. After lysis of the cells, total lysates were immunoprecipitated by a monoclonal antibody against FLAG. Immunoprecipitated proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and stained with Coomassie blue or silver (Figure 5a). We sequenced the in-gel digests of multiple bands by liquid chromatography (LC)/MS–MS, and identified one at ~75 kd as nuclear receptor subfamily 3 group C, or GCR (Figure 5b). Peptide sequences of other proteins identified in the MS/MS analyses are provided in Supplementary Data. The association of GCR with nucleolin was confirmed by western blot in the FLAG IP but not in the control fibrillarin IP (Figure 5c). It has been established that GCR, upon agonist binding, assembles with heat-shock proteins and adaptor proteins into a nuclear import complex, which is rapidly transported by dynein motor complex along microtubules into the nucleus.26 Interestingly, IP of nucleolin also revealed dynein intermediate chain (Figure 5c), a subunit of the dynein motor complex, associated with nucleolin and GCR, indicating that cytoplasmic/nuclear trafficking of nucleolin might involve GCR and dynein complexes. Furthermore, it was reported that nucleolin binds to the DNA-binding domain of GCR, which is inhibited by its ligand-binding domain in the absence of ligand.27 Therefore, agonists of GCR such as cortisone may increase its association with nucleolin by relieving the inhibition of the ligand-binding domain. Consistent with our hypothesis, incubation of HeLa and 16HBEo− cells with cortisone rapidly increases the association of GCR with nucleolin within 15 minutes (Figure 5d,e).
Figure 5.
Nucleolin associates with glucocorticoid receptor (GCR). (a) HeLa cells stably transfected with vector or nucleolin-FLAG (NCLF) were IPed for FLAG, and stained with Coomassie blue followed by silver stain. Bands appearing in the NCLF lane but not the vector lane were cut out, processed for LC–MS/MS analysis. MS ID of each band is labeled on the right side. (b) The spectrum of a peptide from GCR identified by MS/MS analysis. Identified b and y ions are labeled bold italic. (c) HeLa cells stably transfected with vector (V) or nucleolin-FLAG expressing vector (NCLF) were immunoprecipitated with mouse monoclonal antibodies against fibrillarin (Control IP) or FLAG (FLAG IP), and immunoblotted for GCR, FLAG, and dynein-intermediated chain (DIC). (d) HeLa and 16HBEo− cells were treated with cortisone for 0–60 minutes, immunoprecipitated for GCR, and immunoblotted for nucleolin. Total nucleolin and GCR were blotted as loading controls. (e) Nucleolin bands in the GCR IP blots in d were quantified, and plotted against time.
Cortisone increases gene transfer by DNPs
Because cortisone can increase the binding of nucleolin with GCR and nuclear entry of GCR, we tested whether it might also increase nuclear entry and gene transfer of DNPs. In HeLa cells, various concentrations of cortisone increase luciferase reporter expression by 40–100% compared to vehicle (Figure 6a). Similar trends for both cortisone and dexamethasone were observed in polarized 16HBEo− cells grown on Transwell filters (Figure 6b). This increase is unlikely to be due to interference with transcription or translation of the reporter gene, because it was only observed when cortisone was added during transfection, not after (Figure 6c). Moreover, when added before DNPs to the cells, cortisone can inhibit transfection by 50% (Figure 6d), probably due to the depletion of transport molecules, including nucleolin, GCR, and dynein, from the cytoplasm. These data indicate that the key interaction of GCR and nucleolin occurs in the cytoplasm and at rapid kinetics, and is not due to the increase of nucleolin expression in any of the compartments. Therefore, cortisone can increase gene transfer of DNPs by improving their nuclear entry.
Figure 6.
Cortisone improves transfection of DNA nanoparticles (DNPs). (a) HeLa cells were treated with different concentrations of cortisone or vehicle DMSO during nanoparticle transfection, and luciferase expression was presented as percent of vehicle DMSO-treated cells. (b) 16HBEo− cells polarized on Transwell filters 24 hours earlier were transfected with DNPs from the apical side, and treated with DMSO, 1 µmol/l cortisone or dexamethasone, and luciferase activity was assayed at 48 hours. (c) HeLa cells were treated with cortisone or dexamethasone during or after transfection with DNPs, and luciferase reporter activity assayed at 48 hours. (d) HeLa cells were treated with cortisone before or during transfection with DNPs, and luciferase activity was assayed in 48 hours. DMSO, dimethyl sulfoxide; RLU, relative light unit.
Nucleolin is expressed in mouse airway epithelium
Previously, we showed that DNPs efficiently transfect cells in the lung.2 Because nucleolin seems to have important functions during DNP transfection, and it is possible to interfere with these functions by pharmacological means, such as filipin, nocodazole, and cortisone, we sought to confirm its presence in vivo. We examined the presence of nucleolin protein in both wild-type and cystic fibrosis mice lungs by immunohistochemical staining of paraformaldehyde-fixed, paraffin-embedded sections. Nucleolin was recognized by a rabbit antinucleolin antibody (H-250), and revealed by the NovaRed (Vector Laboratories, Burlingame, CA) substrate for horseradish peroxidase, and nuclei was counterstained blue with hematoxylin. The sections were either permeabilized with 0.5% Triton X-100 (Figure 7e,f) or left untreated (Figure 7c,d) before staining. The adjacent control sections without primary antibody show no staining (Figure 7a,b). Nucleolin is widely expressed in airway epithelia and alveoli (Figure 7e,f), as well was in endothelial cells and basement membrane of the airway epithelia. Decreased staining of the airway epithelia was observed in the cytoplasm and especially in the nucleus when permeabilization was omitted (Figure 7c,d). Interestingly, the staining intensity on the airway epithelium varies among individual cells (Figure 7e,f, arrowheads for high expression and asterisks for low expression). This pattern is similar to the pattern of gene transfer by DNPs, which results in patchy expression of the transgene.2
Figure 7.
Total nucleolin expression in wild-type and cystic fibrosis (CF) mice lungs. Lung sections from (a,c,e) wild-type and (b,d,f) δF508 C57Bl6 mice were stained with rabbit antinucleolin H-250 antibody, counterstained with hematoxylin, and examined at ×200 magnification. (a,b) Triton-permeabilized sections stained without primary antibody. (c,d) Adjacent nonpermeabilized sections stained with primary antibody. (e,f) Adjacent permeabilized sections stained with primary antibody. Arrowheads in e and f represent high expression; asterisks in e and f represent low expression. BM, basement membrane; En, endothelium.
Previous and current studies suggest an important role of cell surface nucleolin in the uptake and transfection of DNPs, but nucleolin is expressed at much higher levels intracellularly than on the cell surface, and it is difficult to distinguish these two pools simply by immunohistochemical of fixed specimens. To directly pinpoint cell surface nucleolin expressed on the airway epithelium, we performed intratracheal injection of the H-250 antibody against nucleolin in live mice, and detected the antibody by immunohistochemical after harvesting the lungs. As shown in Figure 8, mice injected with phosphate-buffered saline (PBS) or an isotype control antibody show little or no staining (Figure 8a,c, ×200; d, ×400). By contrast, H-250-injected mice show a patchy staining pattern in the airway epithelium (Figure 8b,e, ×200; f, ×400) similar to the pattern of the gene expression from DNP gene transfer,2 with little staining in the endothelium or basement membrane despite their high endogenous expression (Figure 8b, endothelial cells), which confirms the specificity of the administration and staining. Therefore, because nucleolin is expressed both on the apical surface and in the cell body of the airway epithelium in vivo, it is accessible to DNPs administered intranasally and is a feasible pharmacological target.
Figure 8.
Cell surface nucleolin in mouse airway epithelium. Wild-type C57Bl6 mice were injected with (a) PBS, (c,d) GFP antibody, or (b,e,f) H-250 intratracheally, lungs were harvested 3 hours after injection, and sections stained as described. Arrows in b and f represent cells with positive surface nucleolin staining. BM, basement membrane; En, endothelium.
Discussion
DNPs are progressing toward wide clinical applications to treat cystic fibrosis and other disorders. They are nonimmunogenic and nontoxic, and no drug-related adverse event was observed in the phase I clinical trial using doses ranging from 0.8 to 8 mg in the nose.7 Despite their excellent potential for clinical use, the limited efficacy of the particles and the cost of production of sufficient amounts of clinical-grade plasmid DNA and components of the molecular conjugate for clinical application are limiting. Although we have attained considerable understanding about the extracellular stability of DNPs before cellular uptake and their ability to enter the nucleus, we lacked detailed knowledge of the fate of the vector following cellular entry, vector dissolution, and transgene expression. The rationale for this study was to better understand the molecular mechanisms governing the intracellular trafficking of the DNPs, and to identify steps that could be pharmacologic targets for improved transfection.
We first examined the role of different endocytic pathways in the transfection of DNPs by inhibiting them with drugs. Our findings were consistent with our previous reports that DNPs do not colocalize with conventional clathrin-mediated endocytosis markers EEA1, cathepsin D or transferring,9 and that chlorpromazine did not inhibit DNP-mediated transgene expression. It has been suggested that macropinocytosis might play a role in the uptake of DNPs,28 but neither the amiloride analog ethylisopropyl amiloride nor cytochalasin D inhibited transfection. In the prior study, COS-7 instead of HeLa cells were used, and in our study, amiloride was significantly toxic to HeLa cells, possibly contributing to the discrepant data. Nevertheless, macropinocytosis is unlikely to have a significant role in DNP transfection as inhibiting the remodeling of actin cytoskeleton, which is absolutely required for macropinocytosis, does not inhibit gene transfer. Finally, only lipid raft–disrupting drugs, filipin and methyl-β-cyclodextrin, significantly inhibited gene transfer, and repletion of cholesterol in filipin-treated cells reversed this inhibition, strongly suggesting that lipid raft–mediated endocytosis is the major route, by which DNPs enter the cells. Subsequent findings that both DNPs and their receptor nucleolin exist in lipid rafts, and that nucleolin associates with a lipid raft structural protein flotillin also support this theory.
Intracellularly, we discovered the importance of microtubules in nuclear import of DNPs. By affinity purification and MS analysis, we discovered the association of nucleolin with GCR, and subsequent biochemical analysis not only confirmed this association, but also extended this complex to the motor protein dynein. GCR association with nucleolin provides a potential explanation for the rapid nuclear import of DNPs, and point to possibilities of optimizing formulating of DNPs by adding binding motifs for this transport complex. Cortisone and dexamethasone enhanced transfection by DNPs in both HeLa and polarized 16HBEo− cells, a model system for airway epithelium. This indicates that similar approaches might also be effective in vivo and in humans, because for the nondividing airway epithelial cells, nuclear import is an even greater barrier to gene transfer. Glucocorticoids are suitable for clinical use for patients with cystic fibrosis, who present exaggerated lung inflammation.5 Cortisone directly affects the transport of DNPs, which happens rapidly following administration, so the timing of cortisone administration, if used in vivo, must be optimized. By this reasoning, we predict that cortisone should exert maximal effect when administered immediately after or within a few hours of administration of DNPs.
Proteomic approaches were critical to this study in identifying GCR as a potential target for therapeutic manipulation, and will undoubtedly identify more nucleolin-associated and/or DNP-associated proteins. The results of further functional proteomic studies will help answer many important questions such as how nucleolin and DNPs are internalized into the cytoplasm and how DNPs dissociate to allow transcription of the transgene. Identifying these proteins will be key to the discovery of novel targets for pharmacological manipulations.
New ligands for cell surface nucleolin are being reported, but most of these studies have been conducted only in cell culture. By directly injecting an antibody against nucleolin into the trachea, we demonstrated for the first time that nucleolin is indeed expressed on the luminal surface of the airway epithelium and may be targeted by antibodies or other ligands. The patchy expression pattern resembles the transfection by DNPs previously reported in mouse airway,2 suggesting a functional correlation between the distribution of cell surface nucleolin and gene delivery, and is consistent with our findings that nucleolin varies at least fourfold with cell cycle and can be influenced by phosphorylation (X. Chen and P. B. Davis, unpublished results). We observed wide nucleolin expression in both cytoplasm and nucleus of many cell types in the lung, but tracheal instillation of the antinucleolin antibody labeled only epithelial cells, indicating that physical targeting by compartment is feasible in the lung.
Taken together, this study not only supports our previous finding that nucleolin is a critical cell surface receptor for DNPs, but also reports novel pharmacological means to improve gene transfer. We demonstrate the importance of two cellular structures, namely membrane lipid rafts and microtubules, in the cellular trafficking of DNPs (Figure 9). We also showed the possibility to facilitate nuclear transport of DNP–nucleolin–GCR–dynein complex using cortisone. Moreover, we demonstrated the feasibility of doing so in vivo, by showing the expression of nucleolin in and on the apical surface of mouse airway epithelia. We believe that these data will aid us in the design of manipulation strategies that achieve more efficient gene transfer in vivo.
Figure 9.
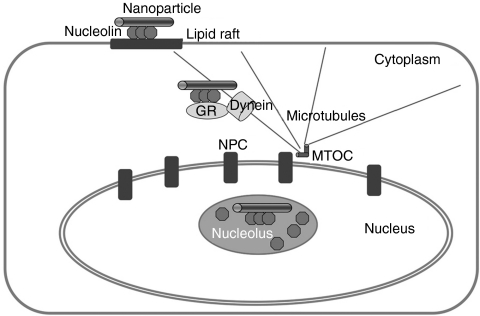
A model of the cellular uptake of DNA nanoparticles (DNPs). DNPs bind to cell surface nucleolin in lipid rafts, and are internalized via raft-mediated endocytosis. The nanoparticles traffic through the cytoplasm in association of glucocorticoid receptor (GCR) and dynein complexes along microtubules, enter the nucleus through nuclear pore complexes (NPC), and accumulate in nucleolus with nucleolin. MTOC, microtubule organizing center.
Materials and Methods
Reagents. Chemicals were purchased from Fisher Scientific (Fairlawn, NJ) or Sigma (St Louis, MO). Antibodies: MS-3 and H-250 against nucleolin, mouse antitransferrin receptor, and phospholipase C-β2 subunit were from Santa Cruz (Santa Cruz, CA); mouse anticaveolin-1 and flotillin-1 from BD Bioscience (San Jose, CA), N17 against N-terminal nucleolin, AC-40 against actin, and 2-28-33 against β-tubulin from Sigma; rabbit anti-green fluorescent protein from Invitrogen (Carlsbad, CA).
Cell culture and transfection. HeLa, HEK293, and 16HBEo− cells were maintained in Dulbecco's modified Eagle's medium or minimal essential medium (Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum and antibiotics (Mediatech). Transient transfection of HeLa cells were performed using Lipofectamine 2000 (Invitrogen) as instructed by the manufacturer. Transwell filters (Corning, Corning, NY) were coated with 1% human fibronectin (Gibco, Carlsbad, CA) and 1% collagen (Collagen Biomaterials, Palo Alto, CA) in LHC Basal Media (Gibco), and dried overnight before seeding with 0.3 million 16HBEo− cells. Cells on filters were allowed to grow for 24–48 hours before use. HeLa cell lines stably expressing nucleolin-FLAG were generated by transfecting HeLa cells with pcDNA3/nucleolin-FLAG, and selecting for zeocin (Invitrogen) resistant clones that express nucleolin-FLAG by western blot. Control cells were generated by transfecting empty pcDNA3 vector and selecting for zeocin-resistant clones.
Transfection of DNPs and reporter luciferase assay. DNPs carrying luciferase reporter gene were compacted and transfected as described before.9 In the case of 16HBEo− cells on filters, 2 µg DNPs were diluted in 350 µl full growth medium for apical transfection. Cells were harvested at 48 hours post-transfection and luciferase activity was determined by the Luciferase Assay system (Promega, Madison, WI). Protein concentrations in the lysates were determined with DC protein Assay Kit (BioRad, Hercules, CA). Final luciferase activity was presented in relative light unit per mg protein as mean ± SD, and significance was calculated by comparing drug versus vehicle treated cells using paired t-test.
IP and western blot. HeLa, HEK293, or 16HBEo− cells treated as described in the figure legends were lysed with radioimmunoprecipitation assay buffer (50 mmol/l Tris–HCl pH 8.0, 150 mmol/l NaCl, 1 mmol/l EDTA, 1% NP40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate, supplemented with protease and phosphatase inhibitors from Sigma). Lysates were incubated with M2-agarose beads against FLAG (Sigma) at 4 °C overnight. The beads were washed extensively with RIPA and Tris-buffered saline–Tween-20 buffers, and eluted with 100 µg/ml 3× FLAG peptide (Sigma) in RIPA buffer. Western membranes were developed with Super Signal West Pico kit (Pierce, Rockford, IL) and imaged with VersaDoc 3000 system (BioRad). The intensity of desired bands were quantified using Quantity One 1-D Analysis Software (BioRad).
LC/MS–MS. Bands of interest were excised from one-dimensional gels and processed for MS analysis as previously described.29 Briefly, excised bands were incubated with reducing solution (1% dithiothreitol) for 15 minutes, then incubated with alkylating solution (1% iodoacetamide). Bands were then washed in 50% ethanol three times, destained, dried under speed vacuum, and reconstituted in a 50 mmol/l sodium bicarbonate solution containing 20 µg/ml sequence grade trypsin, and incubated overnight. Tryptic peptides were extracted the next day and subjected to analysis by reverse phase liquid chromatography/tandem MS using an LCQ deca XP plus mass spectrometer (Thermo Fisher, Waltham, MA) fitted with a nanospray ion source operating at 2.4 kV and 200 °C. Ion data were collected using data-dependent sequence acquisition using the Xcalibur software. Mass spectra were assigned sequences using the Bioworks software package to search human protein database.
Isolation of lipid rafts by sucrose density gradient. Preparation of lipid rafts was performed as described before with minor modifications.30 HeLa, HEK293, or 16HBEo− cells were homogenized in 350 µl cold 0.5 mol/l Na2CO3 using a loose fitting homogenizer (20 strokes). Homogenate was placed at the bottom of ultracentrifuge tube (Beckman, Palo Alto, CA) and mixed with an equal volume of 80% sucrose (wt/vol) in 25 mmol/l 2-(N-morpholine)-ethane sulphonic acid, pH 6.5, 0.15 mol/l NaCl (2-(N-morpholine)-ethane sulphonic acid–buffered saline). The homogenate was then overlaid with 2.1 ml of 35% sucrose and 1.4 ml of 5% sucrose in 2-(N-morpholine)-ethane sulphonic acid–buffered saline containing 250 mmol/l sodium carbonate, and centrifuged at 200,000g for 20 hours in a Beckman SW41 rotor. Twelve 350-µl fractions were collected from the top of the gradient (numbered 1–12). Lipid raft–containing light-scattering band just above the 5/35% sucrose interface was mainly collected in fraction 5.
Fluorescence microscopy. Cell surface nucleolin was stained in live cells with H-250 antibody at dilution of 1:50 for 1 hour at 4 °C. The cells were then fixed in 4% paraformaldehyde for 20 minutes, blocked with 1% bovine serum albumin and 10% normal goat serum (Sigma) in PBS with 0.1% Triton X-100 and 0.1% saponin at room temperature for 30 minutes. Mouse antiflotillin or antitransferrin receptor antibody diluted at 1:100 with 1% bovine serum albumin in PBS was then added to the cells and incubated at room temperature for 1 hour. In the case of nanoparticle transfections, DNPs were labeled with rhodamine with LabelIT Tracker kit (Mirus Bio, Madison, WI). Microtubules were stained with mouse monoclonal antibody against β-tubulin from Sigma, and F-actin was stained with Alexa Fluor 488–conjugated phalloidin from Invitrogen. Appropriate secondary antibodies conjugated with Alexa Fluor 488 or 568 (Molecular Probes) were added at concentrations of 4 µg/ml at room temperature for 1 hour before the nucleus was counterstained with Hoechst 33342 (Molecular Probes). Slides were examined using a Zeiss Axiovert 200 wide field microscope with the proper filter. Image stacks were deconvolved with Hygens confocal deconvolution software and further processed with ImageJ and Photoshop software.
Animal experiments and immunohistochemistry. All animal protocols have been approved by the Institutional Animal Care and Use Committee at Case Western Reserve University. For identification of total nucleolin, 8- to 10-week-old C57Bl/6J mice carrying wild-type or homozygous deltaF508 cftr genes were euthanized with CO2 and lungs were dissected, fixed in 2% paraformaldehyde in PBS for 2 days, embedded in paraffin, and cut in 5-µm sections. Mounted tissue sections were deparaffinized, hydrated, permeabilized in 0.5% Triton X-100 in blocking buffer at room temperature for 30 minutes, and endogenous peroxidase activity was quenched in 1% H2O2 for 15 minutes. Nucleolin was probed with 2 µg/ml H-250 antibody at 4 °C overnight and stained with Vectastain Elite ABC kit following manufacturer's instructions. NovaRed was used as horseradish peroxidase substrate, and counterstained with Hematoxylin QS (Vector Laboratories). For identification of cell surface nucleolin, following intraperitoneal injection of 150 µl anesthetic cocktail (2.13 mg/ml xylazine, 0.36 mg/ml acepromazine, and 10.75 mg/ml ketamine), mice received a tracheostomy and 50 µl of PBS, green fluorescent protein antibody, or H-250 at 0.2 mg/ml through a 22-gauge catheter. Lungs were harvested and washed 3 hours following the procedure and the antibodies were stained with Vectastain Elite ABC kit (Vector Laboratories) and NovaRed substrate as described. Transmitted image analysis was performed with an Olympus Camedia C-2020 Z digital camera, Center Valley, PA attached to an inverted light microscope.
SUPPLEMENTARY MATERIAL Figure S1. Effect of microtubule disrupting drugs on the transgene expression mediated by Lipofectamine. Supplementary Data.
Acknowledgments
This work was supported by NIH grant P30DK27651, RO1HL58318, R21DK084371, the Arline & Curtis Garvin Research Endowment and a grant from the Cystic Fibrosis Foundation. We thank the CF Imaging core facility at CWRU for assistance with instruments. P.B.D. and A.G.Z. are inventors on patents that have been licensed to Copernicus Therapeutics, Inc. by Case Western Reserve University, and they hold equity in Copernicus, which is developing DNPs for clinical use in cystic fibrosis.
Supplementary Material
Effect of microtubule disrupting drugs on the transgene expression mediated by Lipofectamine.
REFERENCES
- Davis PB., and, Cooper MJ. Vectors for airway gene delivery. AAPS J. 2007;9:E11–E17. doi: 10.1208/aapsj0901002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziady AG, Gedeon CR, Miller T, Quan W, Payne JM, Hyatt SL, et al. Transfection of airway epithelium by stable PEGylated poly-l-lysine DNA nanoparticles in vivo. Mol Ther. 2003;8:936–947. doi: 10.1016/j.ymthe.2003.07.007. [DOI] [PubMed] [Google Scholar]
- Ziady AG, Gedeon CR, Muhammad O, Stillwell V, Oette SM, Fink TL, et al. Minimal toxicity of stabilized compacted DNA nanoparticles in the murine lung. Mol Ther. 2003;8:948–956. doi: 10.1016/j.ymthe.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Fink TL, Klepcyk PJ, Oette SM, Gedeon CR, Hyatt SL, Kowalczyk TH, et al. Plasmid size up to 20 kbp does not limit effective in vivo lung gene transfer using compacted DNA nanoparticles. Gene Ther. 2006;13:1048–1051. doi: 10.1038/sj.gt.3302761. [DOI] [PubMed] [Google Scholar]
- Davis PB. Cystic fibrosis since 1938. Am J Respir Crit Care Med. 2006;173:475–482. doi: 10.1164/rccm.200505-840OE. [DOI] [PubMed] [Google Scholar]
- Rowe SM, Miller S., and, Sorscher EJ. Cystic fibrosis. N Engl J Med. 2005;352:1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- Konstan MW, Davis PB, Wagener JS, Hilliard KA, Stern RC, Milgram LJ, et al. Compacted DNA nanoparticles administered to the nasal mucosa of cystic fibrosis subjects are safe and demonstrate partial to complete cystic fibrosis transmembrane regulator reconstitution. Hum Gene Ther. 2004;15:1255–1269. doi: 10.1089/hum.2004.15.1255. [DOI] [PubMed] [Google Scholar]
- Medina-Kauwe LK, Xie J., and, Hamm-Alvarez S. Intracellular trafficking of nonviral vectors. Gene Ther. 2005;12:1734–1751. doi: 10.1038/sj.gt.3302592. [DOI] [PubMed] [Google Scholar]
- Chen X, Kube DM, Cooper MJ., and, Davis PB. Cell surface nucleolin serves as receptor for DNA nanoparticles composed of pegylated polylysine and DNA. Mol Ther. 2008;16:333–342. doi: 10.1038/sj.mt.6300365. [DOI] [PubMed] [Google Scholar]
- Srivastava M., and, Pollard HB. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J. 1999;13:1911–1922. [PubMed] [Google Scholar]
- Ginisty H, Sicard H, Roger B., and, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- Bose S, Basu M., and, Banerjee AK. Role of nucleolin in human parainfluenza virus type 3 infection of human lung epithelial cells. J Virol. 2004;78:8146–8158. doi: 10.1128/JVI.78.15.8146-8158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut C, Blanco J, Benkirane N, Krust B, Jacotot E, Guichard G, et al. Identification of V3 loop-binding proteins as potential receptors implicated in the binding of HIV particles to CD4(+) cells. J Biol Chem. 1998;273:21988–21997. doi: 10.1074/jbc.273.34.21988. [DOI] [PubMed] [Google Scholar]
- Said EA, Krust B, Nisole S, Svab J, Briand JP., and, Hovanessian AG. The anti-HIV cytokine midkine binds the cell surface-expressed nucleolin as a low affinity receptor. J Biol Chem. 2002;277:37492–37502. doi: 10.1074/jbc.M201194200. [DOI] [PubMed] [Google Scholar]
- Legrand D, Vigié K, Said EA, Elass E, Masson M, Slomianny MC, et al. Surface nucleolin participates in both the binding and endocytosis of lactoferrin in target cells. Eur J Biochem. 2004;271:303–317. doi: 10.1046/j.1432-1033.2003.03929.x. [DOI] [PubMed] [Google Scholar]
- Said EA, Courty J, Svab J, Delbé J, Krust B., and, Hovanessian AG. Pleiotrophin inhibits HIV infection by binding the cell surface-expressed nucleolin. FEBS J. 2005;272:4646–4659. doi: 10.1111/j.1742-4658.2005.04870.x. [DOI] [PubMed] [Google Scholar]
- Shi H, Huang Y, Zhou H, Song X, Yuan S, Fu Y, et al. Nucleolin is a receptor that mediates antiangiogenic and antitumor activity of endostatin. Blood. 2007;110:2899–2906. doi: 10.1182/blood-2007-01-064428. [DOI] [PubMed] [Google Scholar]
- Sinclair JF., and, O'Brien AD. Cell surface-localized nucleolin is a eukaryotic receptor for the adhesin intimin-γ of enterohemorrhagic Escherichia coli O157:H7. J Biol Chem. 2002;277:2876–2885. doi: 10.1074/jbc.M110230200. [DOI] [PubMed] [Google Scholar]
- Mayor S., and, Pagano RE. Pathways of clathrin-independent endocytosis. Nat Rev Mol Cell Biol. 2007;8:603–612. doi: 10.1038/nrm2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Li D, Pasumarthy MK, Kowalczyk TH, Gedeon CR, Hyatt SL, et al. Nanoparticles of compacted DNA transfect postmitotic cells. J Biol Chem. 2003;278:32578–32586. doi: 10.1074/jbc.M305776200. [DOI] [PubMed] [Google Scholar]
- Sieczkarski SB., and, Whittaker GR. Dissecting virus entry via endocytosis. J Gen Virol. 2002;83:1535–1545. doi: 10.1099/0022-1317-83-7-1535. [DOI] [PubMed] [Google Scholar]
- Pike LJ. Lipid rafts: heterogeneity on the high seas. Biochem J. 2004;378:281–292. doi: 10.1042/BJ20031672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan EE., and, Dean DA. Intracellular trafficking of plasmids during transfection is mediated by microtubules. Mol Ther. 2006;13:422–428. doi: 10.1016/j.ymthe.2005.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan EE, Geiger RC, Miller AM, Loh-Marley PL, Suzuki T, Miyata N, et al. Microtubule acetylation through HDAC6 inhibition results in increased transfection efficiency. Mol Ther. 2008;16:1841–1847. doi: 10.1038/mt.2008.190. [DOI] [PubMed] [Google Scholar]
- Hovanessian AG, Puvion-Dutilleul F, Nisole S, Svab J, Perret E, Deng JS, et al. The cell-surface-expressed nucleolin is associated with the actin cytoskeleton. Exp Cell Res. 2000;261:312–328. doi: 10.1006/excr.2000.5071. [DOI] [PubMed] [Google Scholar]
- Heitzer MD, Wolf IM, Sanchez ER, Witchel SF., and, DeFranco DB. Glucocorticoid receptor physiology. Rev Endocr Metab Disord. 2007;8:321–330. doi: 10.1007/s11154-007-9059-8. [DOI] [PubMed] [Google Scholar]
- Schulz M, Schneider S, Lottspeich F, Renkawitz R., and, Eggert M. Identification of nucleolin as a glucocorticoid receptor interacting protein. Biochem Biophys Res Commun. 2001;280:476–480. doi: 10.1006/bbrc.2000.4141. [DOI] [PubMed] [Google Scholar]
- Walsh M, Tangney M, O'Neill MJ, Larkin JO, Soden DM, McKenna SL, et al. Evaluation of cellular uptake and gene transfer efficiency of pegylated poly-l-lysine compacted DNA: implications for cancer gene therapy. Mol Pharm. 2006;3:644–653. doi: 10.1021/mp0600034. [DOI] [PubMed] [Google Scholar]
- Ziady AG., and, Kinter M. Protein sequencing with tandem mass spectrometry. Methods Mol Biol. 2009;544:325–341. doi: 10.1007/978-1-59745-483-4_21. [DOI] [PubMed] [Google Scholar]
- Iwabuchi K, Yamamura S, Prinetti A, Handa K., and, Hakomori S. GM3-enriched microdomain involved in cell adhesion and signal transduction through carbohydrate-carbohydrate interaction in mouse melanoma B16 cells. J Biol Chem. 1998;273:9130–9138. doi: 10.1074/jbc.273.15.9130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of microtubule disrupting drugs on the transgene expression mediated by Lipofectamine.