Summary
A single-cell assay was developed to measure the activation of phosphoinositide 3-kinase (PI3K) using microanalytical chemical separations and a fluorescently labeled lipid substrate. Phosphatidyl-inositol 4,5 bisphosphate labeled on its acyl chain with Bodipy fluorescein (Bodipy Fl PIP2) was utilized as a substrate for both in vitro and cell-based assays. Detection limits for the substrate and product of the PI3K reaction were 10 to 20 zeptomoles. In vitro assays with PI3K with and without pharmacologic inhibitors demonstrated that Bodipy Fl PIP2 was converted to phosphatidyl-inositol 3,4,5 trisphosphate (Bodipy Fl PIP3 ). Bodipy Fl PIP3 could be back converted to Bodipy Fl PIP2 by the phosphatase PTEN. When Bodipy Fl PIP2 was added to a cell lysate, 1.4 fmoles of the Bodipy Fl PIP3 were produced per ng of protein in the cytoplasmic extract in 10 min. Addition of Bodipy Fl PIP3 to a cell lysate yielded 3 fmoles of Bodipy Fl PIP2 per ng of protein in 8 min. Both Bodipy Fl PIP2 and Bodipy Fl PIP3 were measureable in single cells and the two species could be inter-converted. Under the appropriate conditions, a fluorescent diacylglycerol was also detected in single cells. When the FcεR1 receptor on the cells loaded with the fluorescent lipid was cross-linked, the amount of Bodipy Fl PIP3 generated per cell increased 4-fold over that of unstimulated cells. This production of Bodipy Fl PIP3 was blocked by wortmannin. Chemical cytometry utilizing the fluorescent lipids will be of value in understanding lipid metabolism at the single-cell level.
Introduction
Lipid signaling is now recognized to have essential roles in health and disease, particularly cancer and inflammatory diseases.1-8 The PI3K pathway is especially important as the products of numerous oncogenes provide constitutive input signals to PI3K.9, 10 PI3K itself has been shown to be mutated in a variety of cancers as has the tumor suppressor phosphatase and tensin homolog (PTEN), a 3-lipid phosphatase, which normally down regulates the PI3K pathway by dephosphorylating the PI3K product phosphatidyl-inositol 3,4,5-trisphosphate (PIP3) to generate phosphatidyl-inositol 4,5-bisphosphate (PIP2).11 Immediately downstream of PI3K lies protein kinase B (PKB, aka Akt) which functions as an important signaling node for numerous cancer-promoting activities including cell cycle entry, resistance to apoptosis, and enhanced cell migration.12, 13 Not surprisingly, the PI3K pathway is an active therapeutic target and novel PI3K inhibitors, both broad spectrum and isoform specific, have entered clinical trials.14-16
Despite its importance, direct analysis of PI3K signaling in living cells is a difficult task, particularly in small samples such as those obtained from patients.17-19 Radioactivity-based thin-layer chromatography or mass spectrometry are commonly used analytical techniques for the analysis of lipids; however, they are technically demanding, of limited sensitivity and specificity, or require sophisticated equipment.20-26 PI3K signaling has also been analyzed by fluorescence-based methods. For example, high-throughput assays are based on phosphoinositide-binding pleckstrin homology (PH) domains as detectors in measuring the production or localization of PIP3, but such assays require large numbers of cells.27, 28 GFP-tagged PH domains have been used in microscopy as an indirect assay of the enzymatic activities of PI3K and PTEN, but these molecularly engineered cell-based assays cannot be used in clinical samples.29-31 These various limitations necessitate development of new technologies for analysis of lipid signaling in primary cells and in small samples such as from needle biopsies or fine needle aspirates.
Microanalytical chemical separation applied to high-sensitivity analyses of single cells, known as chemical cytometry, has such potential.32-35 Chemical cytometry is one of the fastest growing fields within bioanalytical chemistry as it holds great promise for understanding cell metabolism and signaling.36-40 Our group has been active in this area having developed single-cell assays for protein kinases41 and sphingosine kinase.38 In these assays, cells are loaded with fluorescent substrates (reporters) followed by laser-based lysis of a single cell and loading of its contents into an overlying capillary where chemical separation is accomplished via capillary electrophoresis (CE). In CE, substrate and product forms of the reporter are readily separated, detected with high sensitivity using laser-induced-fluorescence (LIF), and identified by their characteristic migration times. The ratio of the peak areas of the substrate and product are then used as a measure of the enzyme's activation. Routine detection limits correspond to an intracellular concentration of ≤10 nM.41 The ability to detect the reporter at nanomolar concentrations means that concentrations can be used at or below those of endogenous substrates so that competition between the endogenous substrate and exogenous reporter is minimal.41 Phosphorylated products can also be dephosphorylated by phosphatases; therefore, the measurement reflects the dynamic equilibrium of the kinase and phosphatase activity in the cell.38, 41 Furthermore, the ability to analyze multiple analytes in individual cells enables the identification of cell-to-cell heterogeneity with respect to biochemical behavior.
Understanding this heterogeneity is particularly important for lipid signal transduction since lipid levels are dynamic with biosynthesis and metabolism being robust in most cells.17-19 Recently chemical reagents have become available to enable biochemical studies of these rich metabolic networks, particularly in cell-based assays.17, 42 Fluorescently labeled lipids have been shown to localize and translocate in the same manner as the endogenous substrates, and have been useful for monitoring dynamic changes in cells.42-44 Lipids tagged with fluorophores have also been developed as substrates for a variety of enzymes, often with similar kinetics to the endogenous substrate.38, 45-59 When coupled with chemical cytometry, these new chemical tools may make it possible to perform direct, quantitative measurements of the activation of PI3K and other lipid metabolizing enzymes in single cells. In the current work, a single-cell biochemical assay for PI3K was developed and validated using a tumor cell line. A fluorescently labeled PIP2 was used as a substrate of PI3K. An in vitro reaction with purified PI3K using CE-LIF to monitor the phosphorylation of the substrate demonstrated the feasibility of the assay format. The fluorescent PIP2 was then tested in a cell lysate to confirm that its phosphorylation occurred and a separation could be performed in the presence of cellular constituents. Microscopy was used to monitor and optimize the loading of the exogenous substrate into cells. Lastly, single-cell assays were undertaken to measure the phosphorylation and dephosphorylation of substrate by PI3K and PTEN, respectively. Specific inhibition of the enzymes provided additional validation for the single-cell measurements.
Results and discussion
Separation and detection limits of Bodipy Fl PIP2 and Bodipy Fl PIP3 by CE
Separation of Bodipy Fl PIP2 and Bodipy Fl PIP3 was accomplished as previously reported.60, 61 The current work sought to apply this separation scheme to single-cell analyses of PIP2 and PIP3 metabolism; therefore, it was necessary to extend the earlier work to establish that adequate sensitivity could be attained to accomplish this goal. Due to the biochemical nature of the enzyme measurements, it was desired to introduce the fluorescent substrate at the lowest possible cellular concentration; otherwise, excessive amounts of the exogenous lipid could serve to overwhelm the cellular signaling networks with a substrate that might perturb cellular physiology. In mammalian cells the estimated concentration of PIP2 (averaged across the cell volume) is 10 μM, or 10-18 mol in a typical mammalian cell of 1 pL.62
A series of separations using standard solutions of Bodipy Fl PIP2 and Bodipy Fl PIP3 were undertaken to establish the sensitivity and reproducibility in the separation of these analytes. Free fluorescein was employed as an internal standard in the electrophoretic traces. Fluorescein (1 nM), Bodipy Fl PIP2 (1 nM) and Bodipy Fl PIP3 (1nM) and were loaded simultaneously into a capillary and separated. The peak area of Bodipy Fl PIP2 and Bodipy Fl PIP3 was divided by that for the fluorescein internal standard. The migration times of Bodipy Fl PIP2 and Bodipy Fl PIP3 were also normalized by division of the migration time of the fluorescein peak.63 The relative standard deviation (RSD) for the lipids was calculated from this normalized data. The migration times for Bodipy Fl PIP2 and Bodipy Fl PIP3 were highly reproducible with respect to that of fluorescein with RSDs of 0.6% and 0.7%, respectively (n = 10). The reproducibility of the peak areas for Bodipy Fl PIP2 and Bodipy Fl PIP3 was less consistent with RSDs of 10.0% and 14.2%, respectively (n = 10). This relatively large RSD for the peak area was likely due to variability in the injected volume of sample since analyte loading into the capillary was performed manually; however, the ratio of the normalized peak areas (1.0 ± 0.1) for Bodipy Fl PIP2 and Bodipy Fl PIP3 was excellent. The method can accurately measure the ratio of these two lipid species. To determine the detection limits of the lipids in the capillary, the lipid/fluorescein solution was progressively diluted and then separated. When 10-20 mol of either lipid was loaded into the capillary, the noise height of the baseline was one third of the peak height of the lipids. Under the conditions used, the limit of detection for both species was 1× 10-20 mol, which was 100-fold less than the expected amount of endogenous PIP2 in a single cell (see above). The detection limits were approximately ten-fold higher than other small molecule analytes such as fluorescein-labeled peptides.41 This may in part be due to losses on vessel walls during dilution of the lipid prior to electrophoresis.
Phosphorylation of Bodipy Fl PIP2 by PI3K
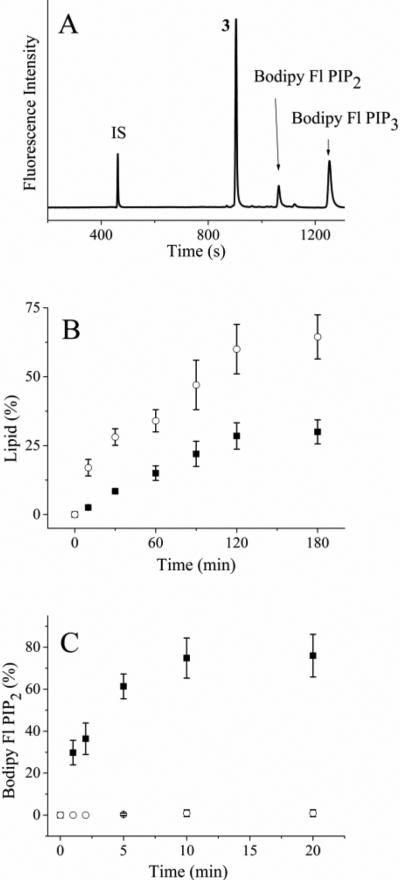
A fluorescein labeled PIP2 was previously shown to function as an in vitro substrate of PI3K.64 Thus, it was likely that the Bodipy Fl PIP2 could also serve as a substrate for PI3K. PI3K was incubated with Bodipy Fl PIP2 with or without ATP (1 mM), and the reaction mixture was analyzed by CE after varying incubation times. In the presence of ATP, the electropherogram revealed a peak possessing a longer migration time than that of Bodipy Fl PIP2 (Figure 1A). The area of this peak increased as the incubation time increased (Figure 1B). This peak co-migrated with a standard of Bodipy Fl PIP3. When the assay was performed in the absence of ATP, this additional peak was not observed. In the presence of wortmannin (100 nM), an irreversible inhibitor of PI3K65 and ATP (1 mM), the latter peak was not observed over 180 min of incubation (Figure 1B). Based on these data, this latter peak was identified as Bodipy Fl PIP3. In the presence of ATP and without wortmannin, 30 ± 4% (n = 3) of Bodipy Fl PIP2 was phosphorylated over 180 min under these conditions (Fig. 1B, Table 1). In addition to the expected lipid and internal standard peaks, an additional peak (peak 3) was observed that co-migrated with a standard of Bodipy-fluorescein labeled phosphatidyl-inositol 5-phosphate (Bodipy Fl PIP1). However, the addition of a phosphatase inhibitor cocktail containing orthovanadate, β-glycerophosphate and the PTEN inhibitor bpv(pic) was ineffective in preventing the formation of the analyte co-migrating with Bodipy Fl PIP1 (data not shown). While it was likely that peak 3 was a Bodipy Fl PIP1, its identity must be confirmed by additional means. The presence of a 3rd peak during in vitro reactions with purified PI3K has been reported previously for fluorescein-labeled PIP2.64 Similarly, PI3K assays using thin layer chromatography or HPLC have shown additional species consistent with the current findings.66, 67
Figure 1.
Incubation of PI3K and PTEN with fluorescent lipids in vitro. (A) Electropherogram of the reaction mixture of Bodipy Fl PIP2 and PI3K after 120 min. The internal standard fluorescein is marked as “IS” while the peak co-migrating with Bodipy Fl PIP1 is marked with a “3”. (B) Time course of the reaction of Bodipy Fl PIP2 with PI3K. The solid squares represent the percentage of Bodipy Fl PIP3 while the open circles reflect the percentage of the lipid in the peak co-migrating with Bodipy Fl PIP1 (peak 3 above), respectively. (C) Time course of the reaction of Bodipy Fl PIP3 with PTEN. The solid squares and open circles are the percentage of Bodipy Fl PIP3 in the absence and presence of 500 nM bpv (pic), respectively. The data in panels B and C represent the average of three experiments and the error bars depict the standard deviation of the data points.
Dephosphorylation of Bodipy Fl PIP3 by PTEN
In cells, dephosphorylation of PIP3 by the phosphatases PTEN or Src homology 2 domain-containing inositol 5' phosphatase (SHIP2) acts to convert PIP3 to PIP2. To determine if Bodipy Fl PIP3 might be dephosphorylated, this lipid was incubated with PTEN phosphatase for varying times and the reaction mixture was separated by CE. Under the conditions used, 75 ± 10% (n = 3) of Bodipy Fl PIP3 was dephosphorylated to form Bodipy Fl PIP2 over 10 min (Fig. 1C, Table 1). In contrast to the PI3K assay, only peaks co-migrating with Bodipy Fl PIP2, Bodipy Fl PIP3, or the fluorescein standard were present on the electropherogram. The addition of fresh PTEN into the reaction mixture at 10 minute intervals resulted in additional conversion of Bodipy Fl PIP3 to Bodipy Fl PIP2 suggesting that PTEN was not stable under these conditions. The assay was then performed in the presence of bpv(pic) (500 nM), a PTEN inhibitor. No peak co-migrating with a Bodipy Fl PIP2 standard was observed over 1 h even with addition of fresh PTEN at 10 min intervals. These data suggest that the Bodipy Fl PIP2 and Bodipy Fl PIP3 might serve as enzyme substrates within cells.
Separation and detection of fluorescent lipids loaded into cell lysates
A challenge that exists in the use of these fluorescent lipid probes for cell-based assays by CE is whether these probes can be successfully extracted from cellular membranes, since the lipids can be expected to partition into the lipid bilayer of the plasma membrane and organelles. Numerous intracellular proteins are also known to associate with these lipids.9 For successful CE-based analysis, these reporters must be extracted from membrane and cytosol, separated and detected. To evaluate the potential to use these compounds for performing assays in the presence of cellular constituents, Bodipy Fl PIP2 (5 μM) or Bodipy Fl PIP3 (5 μM) was incubated with a cell lysate followed by CE analysis using a protocol similar to that used for the assays with purified proteins. The RSDs for the normalized peak areas for Bodipy Fl PIP2 and Bodipy Fl PIP3 in the lysate were 9.0% and 11.5%, respectively (n =3). These results were similar to those achieved in the assay of the lipid using purified proteins. Based on the peak areas of each analyte compared with that of standards, the percentage of each analyte recovered in the sample was 73 ± 7% for Bodipy Fl PIP2 and 64 ± 5% for Bodipy Fl PIP3. Thus, recovery of each of the lipids from the cellular milieu is similar. When separated from the cell lysates, the limit of detection for both species was 2 × 10-20 mol (S/N = 3). These data support the feasibility of retrieving and accurately measuring the substrate and product lipid forms in the presence of cellular components.
Phosphorylation and dephosphorylation of fluorescent lipids in cell lysates
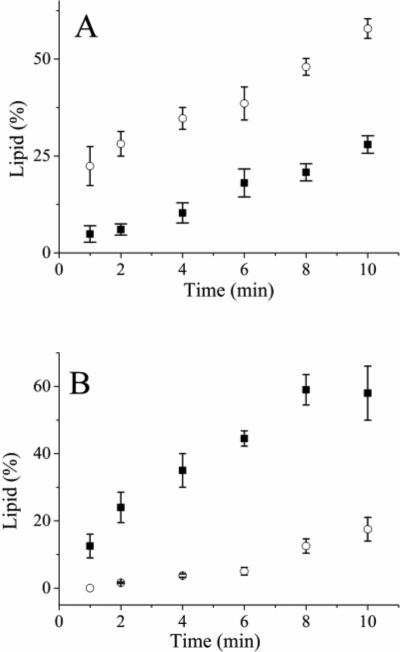
To determine whether PI3K in cell lysates might metabolize Bodipy Fl PIP2, this fluorescent lipid (5 μM) was added to cell lysates preincubated with bvp(pic) (10 μM, 30 min) to inhibit PTEN activity and the assay mix was incubated for varying times at room temperature followed by CE-based separation. A peak co-migrating with Bodipy Fl PIP3 was observed (Fig. 2A) suggesting that the conversion of Bodipy Fl PIP2 to Bodipy Fl PIP3 by PI3K in the cell lysates was occurring. Of note, a peak co-migrating with peak 3 (or Bodipy Fl PIP1) was also present on the electropherogram and the area of this peak was greater than that of Bodipy Fl PIP3. After 10 min of incubation with the cell lysate, 14 ± 3% of the Bodipy Fl PIP2 remained while 28 ± 2% had been converted to Bodipy Fl PIP3 and 58 ± 2.5% to the peak-3 analyte (Table 1). At this time point, 1.4 fmoles of the Bodipy Fl PIP3 were produced per ng of protein in the cell lysate. In the presence of the PI3K inhibitor wortmannin (100 nM) and bvp(pic), no Bodipy Fl PIP3 was present at the 10-min time point while 92 ± 5% of the fluorescent lipid was present in peak 3. In the absence of wortmannin and bpV(pic), 11 ± 4% was present in the Bodipy Fl PIP3 peak and 82 ± 5% in the peak co-migrating with Bodipy Fl PIP1 at 10 min. As with the purified PI3K, at least two enzymes metabolized the Bodipy Fl PIP2 in the cell lysate.
Figure 2.
Incubation of the fluorescent lipids with a cell lysate. (A) Time course of cell lysate incubated with Bodipy Fl PIP2. The solid squares and open circles are the percentage of Bodipy Fl PIP3 and peak 3 (identified in Fig 1A), respectively. (B) Time course of cell lysate incubated with Bodipy Fl PIP3. The solid squares and open circles are the percentage of Bodipy Fl PIP2 and peak 3, respectively. The data represent the average of three experiments and the error bars depict the standard deviation of the data points.
To determine whether Bodipy Fl PIP3 might also be metabolized in the lysate, Bodipy Fl PIP3 (5 μM) was incubated with the cell lysate and at varying times aliquots were analyzed by CE. A peak with a migration time corresponding to that of Bodipy Fl PIP2 was observed after 1 min (12 ± 3%), which suggested that Bodipy Fl PIP3 was serving as a phosphatase substrate in the lysate (Fig. 2B, Table 1). The peak area of Bodipy Fl PIP2 continued to increase with time, but after 2 min a peak corresponding to the analyte co-migrating with Bodipy Fl PIP1 peak (peak 3) was also present. After 8 min of incubation, the appearance of Bodipy Fl PIP2 reached a maximum at 59 ± 4% and then began to decline as the peak area of the peak-3 species increased proportionately. At this 8-min time point, 3 fmoles of the Bodipy Fl PIP2 were produced per ng of protein in the cell lysate. When the lysate was incubated with bpV(pic) (500 nM, 30 min) and then Bodipy Fl PIP3 added, 19 ± 2% was present as Bodipy Fl PIP2 and analyte corresponding to peak 3 was observed. When the inhibitor concentration was increased to 10 μM, no peaks co-migrating with either Bodipy Fl PIP2 or peak 3 were observed. These results suggest that the peak-3 analyte is derived from PIP2 and is very likely a form of Bodipy Fl PIP1.
Loading cells with fluorescent lipids
To detect the activation of PI3K and lipid phosphatases in intact cells, the fluorescent lipids must be translocated across the cell membrane so that the lipids are accessible to cytosolic enzymes. In the current experiments, the cells were loaded with the lipid by first forming a 1:1 complex between the lipid and histone.59, 68, 69 To examine the rate at which Bodipy Fl-PIP2 loaded into cells, the fluorescence of the cells was measured after addition of Bodipy Fl-PIP2:histone (2 μM) to the cells. Cellular fluorescence increased within 30 s of addition of the Bodipy Fl-PIP2:histone to the cell chamber suggesting that the fluorescent lipid was rapidly incorporated into the cells. The fluorescence of the cells reached a plateau within 5 min indicating that no more lipid was loaded after that time (data not shown). When cells were examined by brightfield microscopy following addition of Bodipy-Fl-PIP2:histone, the cells were noted to become more refractile with small granular objects surrounding the cell (Fig. 3A). Cells incubated under identical conditions without Bodipy-Fl-PIP2:histone did not undergo this morphologic change. Since RBL cells contain secretory granules, it was possible that the observed material surrounding the lipid-loaded cells was secreted in response to the incorporation of Bodipy Fl-PIP2 into the cells.
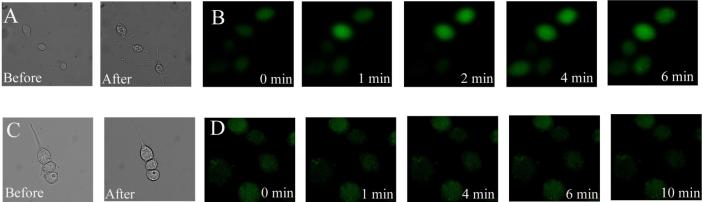
Figure 3.
Increase in [Ca2+]i in cells incubated with PIP2:histone. (A) Bright field images of RBLs before and after incubation with 2 μM PIP2:histone for 10 min. (B) Fluorescence images of Fluo-3-loaded RBL cells incubated with 2 μM PIP2:histone for varying times. (C) Bright field images of RBL cells before and after incubation with 0.5 μM PIP2:histone for 10 min. (D) Fluorescence images of Fluo-3-loaded RBL cells incubated with 0.5 μM PIP2:histone for varying times.
Prior investigators have observed increases in [Ca2+]i when cells were loaded with a high concentration of PIP2-histone complex (40 μM of a 1:1 complex).59 The elevation in [Ca2+]i was thought to be the result of conversion of the PIP2 to inositol 1,4,5-trisphosphate (IP3) by phospholipase C (PLC). This hypothesis was supported by data showing that pre-exposure of cells to the PLC inhibitor U73122 abrogated the fluorescence increase.59 Conversion of PIP2 into other active signaling species by the process of cell loading could yield substantial artifacts in a cell-based assay designed to measure the physiologic conversion of PIP2 into its metabolites.1, 70 In the current experiments, the concentration of Bodipy Fl-PIP2 incubated with the cells was 1/20 that loaded into cells by Ozaki et al and 1/5 that the averaged PIP2 cellular concentration.62 However, since Bodipy Fl-PIP2 is likely more concentrated in the membrane relative to that in the surrounding medium, the uptake into the cells may have been sufficient to increase flux through the PLC pathway, generating IP3 and increasing [Ca2+]i.
To determine whether loading cells using the PIP2:histone procedure increased [Ca2+]i, RBL cells were first loaded with the calcium indicator Fluo-3 and then incubated with nonfluorescent PIP2 in an equimolar complex with 2 μM histone. The fluorescence intensity of the cells increased within 30 s after addition of the complex and persisted beyond 10 min suggesting that incubation of cells in as little as 2 μM PIP2:histone resulted in an increase in [Ca2+]i (Figure 3B). Notably addition of histone alone (2 μM) led to a transient increase in Fluo-3 fluorescence which returned to baseline within 3 min. These data suggested that the rise in [Ca2+]i was the result of an initial perturbation of the plasma membrane by histone with a more prolonged effect due to the PIP2, perhaps as a result of its conversion to IP3.
To determine whether cells could be loaded with PIP2 without an increase in [Ca2+]i, Fluo-3-loaded cells incubated with PIP2:histone (0.5 μM) while monitoring their fluorescence intensity. No change in the fluorescence intensity of the cells was observed over 10 min (Figure 3D). Concomitant observation by brightfield demonstrated that the morphology of the cells was also not altered by the loading procedure (Figure 3C). Loading cells with a low concentration of the PIP2:histone complex did not lead to a detectable rise in [Ca2+]i and pathways downstream of PIP2 were likely not activated.
Formation of Bodipy Fl PIP3 and Bodipy Fl PIP2 in single RBL cells
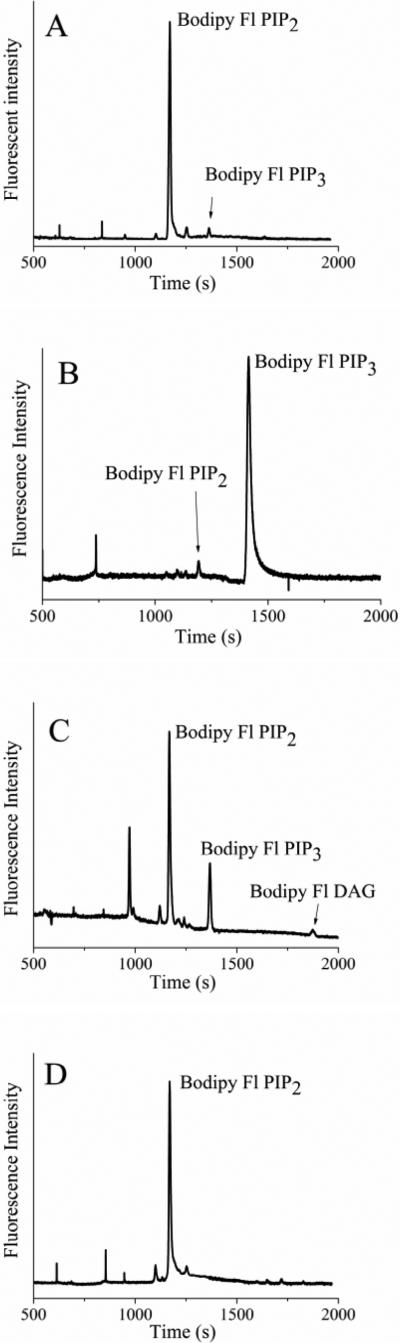
To demonstrate that fluorescent lipids from single cells could be quantified by CE, serum-starved RBL cells were loaded with Bodipy Fl PIP2:histone (0.5 μM). Single cells were then sampled and analyzed by CE. Figure 4A shows a typical electropherogram for a single cell loaded with Bodipy Fl PIP2. By 10 min of incubation, a peak co-migrating with Bodipy Fl PIP3 was present on the electropherogram. In these experiments, only 2 ± 1 % (n = 10) of Bodipy Fl PIP2 was phosphorylated in the cell (Table 1). No peak co-migrating with Bodipy Fl PIP1 was evident on the electropherogram. When cells were preincubated with wortmannin (100 nM, 30 min) and then incubated with Bodipy Fl PIP2 for 10 min, no peak corresponding to Bodipy Fl PIP3 was observed.
Figure 4.
Electropherograms obtained from single RBL cells loaded with fluorescent lipids. A) An RBL cell loaded with 0.5 μM Bodipy-Fl PIP2:histone. B) An RBL cell loaded with 0.5 μM Bodipy-Fl PIP3:histone. (C,D) RBL cells were loaded with 2 μM Bodipy-Fl PIP2. The cells were incubated with (C) 0 or (D) 100 nM wortmannin prior to loading the fluorescent lipid. The data represent the average of at least five cells and the error bars depict the standard deviation of the data points.
To investigate lipid phosphatase activity, serum-starved RBL cells were incubated with Bodipy Fl PIP3:histone (0.5 μM). Individual cells were then analyzed by CE (Figure 4B). In these experiments, 9 ± 3% (n = 5) of the Bodipy Fl PIP3 was converted to Bodipy Fl PIP2 by 10 min. No peak corresponding to Bodipy Fl PIP2 was present when cells were preincubated with the inhibitor bpV(pic) (500 nM, 30 min), and then incubated with Bodipy Fl PIP3:histone (0.5 μM). The ability of lipid phosphatases to act on Bodipy Fl PIP3 suggests that the single-cell measurements may reflect the combined actions of PI3K and phosphatase in the cells.
Formation of Bodipy Fl PIP2 metabolites in single RBL cells loaded with high concentrations of Bodipy Fl PIP2
To investigate the fate of Bodipy Fl PIP2 in activated cells, RBL cells were loaded with a high concentration Bodipy Fl PIP2:histone complex (2 μM). After incubation for 10 min, single cells were analyzed by CE (Figure 4C). A peak co-migrating with Bodipy Fl PIP3 was present on the electropherogram and comprised 15 ± 6% (n = 5) of the fluorescent lipid (Table 1). Multiple additional peaks were observed on the electropherogram which did not co-migrate with either Bodipy Fl PIP2 or peak 3. The final peak on the electropherogram co-migrated with a standard of Bodipy Fl DAG generated by reacting Bodipy Fl PIP2 with purified PLC. These data suggest that in the intact cells, Bodipy Fl PIP2 could be acted upon by both PI3K and PLC. The formation of Bodipy Fl DAG by PLC implies the formation of IP3 which is consistent with the observed rise in [Ca2+]i and degranulation of RBL cells loaded with 2 μM Bodipy Fl PIP2:histone.
To determine whether inhibition of PI3K by wortmannin could be measured in cells, RBL cells were preincubated with wortmannin (100 nM, 30 min), and then incubated with 2 μM Bodipy Fl PIP2:histone for 10 min. No peak corresponding to Bodipy Fl PIP3 or Bodipy Fl DAG was observed in the single cells (Fig. 4D). These results are consistent with wortmannin's reported inhibition of both PI3K and PLC-mediated signaling in RBL cells.71, 72
Formation of Bodipy Fl PIP3 in single cells after Fcε receptor activation
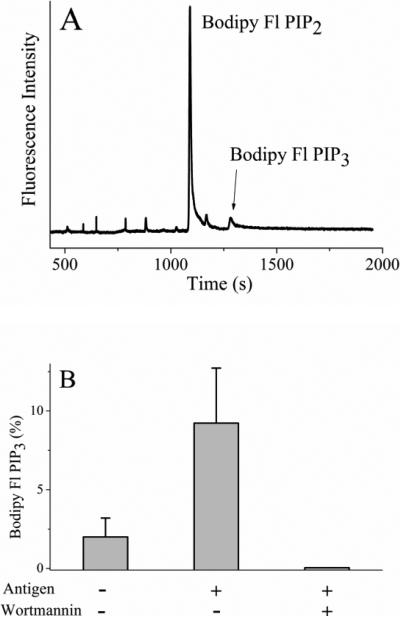
To determine if metabolism of the fluorescent lipids was increased in single cells following physiologic activation, RBL cells were loaded with Bodipy Fl PIP2:histone (0.5 μM) followed by cross-linking of the Fc receptor by antigen. Single cells were then analyzed by CE (Fig. 5A). As a percentage of the total fluorescent lipid in the cell, 4-fold more Bodipy Fl PIP3 was produced in cells stimulated by cross-linking their Fcε receptor relative to those without stimulation (Fig. 5B, Table 1). To inhibit the activity of PI3K, cells were preincubated with 100 nM wortmannin and then stimulated with antigen. No peak corresponding to Bodipy Fl PIP3 was observed suggesting that the fluorescents lipids in combination with CE can be used to assess the activation of PI3K in single cells.
Figure 5.
Physiologic activation of RBL cells loaded with Bodipy Fl PIP2. (A) Electropherogram obtained from an RBL cell loaded with 0.5 μM Bodipy Fl PIP2:histone and then stimulated with antigen. (B) The percentage of fluorescent lipid present as Bodipy Fl PIP3 in serum starved cells (control), antigen-activated cells, and antigen-activated cells in the presence of wortmannin. The data represent the average of five experiments and the error bars depict the standard deviation of the data points.
Experimental
Materials
Sodium deoxycholate (SDC), 1-propanol, adenosine triphosphate (ATP), protease inhibitor (P8340) and 4-(2-hydroxyethyl) piperazine-1-ethanesulfonic acid (HEPES) were purchased from Sigma-Aldrich Inc. (St. Louis, MO, USA). The fluorescent lipid reagents, Bodipy fluorescein phosphatidyl-inositol 4,5-bisphosphate (Bodipy Fl PIP2, C6) and Bodipy fluorescein phosphatidyl-inositol 3,4,5-trisphosphate (Bodipy Fl PIP3, C6), PIP2 and PIP3 were obtained from Echelon Biosciences Inc. (Salt Lake City, UT, USA). PI3K (p110α/p85α), and PTEN were procured from Millipore Corp. (Billerica, MA, USA). Fluo-3 AM (fluo-3 acetoxymethyl ester), fluorescein (Fl), bovine serum albumin labeled with 2,4-dinitrophenyl (DNP-BSA), anti-DNP rat IgE, DMEM media and all other tissue culture reagents were purchased from Invitrogen (Carlsbad, CA, USA). EOTrol LR (low reverse) polymer solution was acquired from Target Discovery (Palo Alto, CA, USA). Wortmannin and dipotassium bisperoxo (picolinato) oxovanadate (V) (bpV(pic)) were obtained from Enzo Life Sciences International Inc. (Plymouth Meeting, PA, USA). All other reagents were procured from Fisher Scientific (Pittsburgh, PA, USA).
PI3K assay
In vitro assays were performed at a PI3K concentration of 10 nM and a Bodipy Fl PIP2 concentration of 5 μM with a total reaction volume of 40 μL at room temperature. The assay was conducted in the presence or absence of 1 mM ATP. In some experiments, 100 nM wortmannin was added to the reaction mixture before addition of the lipid. Aliquots (5 μL) were removed from the reaction mixture at 0, 10, 30, 60, 90, 120 and 180 min time points. The reaction was stopped by diluting the aliquot in propanol (5 μL) followed by flash freezing in liquid nitrogen. Samples were thawed and diluted 200-fold in water just prior to CE analysis. The percentage of Bodipy Fl PIP3 present in the mixture was determined by dividing the peak area of the Bodipy Fl PIP3 by the area of all of the peaks on the electropherogram.
PTEN assay
The assay was performed with a PTEN concentration of 200 nM and a Bodipy Fl PIP3 concentration of 5 μM with a total reaction volume of 40 μL (5 mM HEPES, pH 8) at room temperature. The assays were performed without and with 500 nM bpV(pic), a pharmacologic inhibitor of PTEN. Aliquots (5 μL) were removed from the reaction mixture at 0, 1, 2, 5, 10, and 20 min time points. The reaction was stopped by diluting the aliquot with 5 μL propanol followed by flash freezing in liquid nitrogen. Samples were thawed and diluted 200-fold in water just prior to analysis. The percentage of Bodipy Fl PIP2 present in the mixture was determined by dividing the peak area of the Bodipy Fl PIP2 peak area by the area of all of the peaks on the electropherogram.
Preparation of Bodipy Fl diacylglycerol (DAG) standard
Bodipy Fl PIP2 (5 μM) was incubated with phospholipase C (PLC, 1 ng) in a total reaction volume of 40 μL (50 mM HEPES, 35 mM KCl, 3 mM CaCl2, 2 mM DTT, 3 mM EGTA, pH 7.2) at room temperature. The PLC protein used was provided by Professor John Sondek (Department of Biochemistry and Biophysics, UNC). An aliquot (5 μL) was taken from the reaction mixture after 120 min and the reaction was stopped by diluting the sample volume in 5 μL propanol followed by flash freezing in liquid nitrogen. Samples were thawed and diluted 200-fold in water just prior to analysis.
Cell culture
Rat basophilic leukemia 2H3 mast cells (RBL) were used since they express high levels of the p110 isoform.73 RBL cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 584 mg/L L-glutamine, 100 units/mL penicillin and 100 μg/mL streptomycin. Cells were maintained at 37 °C in a humidified 5% CO2 atmosphere. Cells for use in single-cell, CE experiments were plated the day before the experiment in custom-made cell chambers created from a #1 glass coverslip to which a silicon “O” ring was attached using poly(dimethyl siloxane) (Sylgard 184).74 For experiments using serum-starved cells, the media was exchanged with DMEM lacking FBS 8-12 h before the start of the experiment.
Preparation of RBL cell lysates
A suspension of 400,000 RBL cells in 10 mL media was centrifuged, and the pellet was washed in deionized water (1 mL) ×3 at 4 °C. The washed cell pellet was then suspended in 200 μL of deionized water and monitored under the microscope for complete lysis before use.75 Cell debris and nuclei were removed by centrifugation at 16,000 × g for 10 min at 4 °C to. The protein concentration in the lysate was determined by reaction with 3 mg/ml fluorescamine for 5 min followed by measurement of fluorescence and comparison to a standard curve. For assay of PI3K or phosphatase activity, the lysate was diluted in water to a final protein concentration of 1 mg/ml and incubated with either Bodipy Fl PIP2 or Bodipy Fl PIP3. For PI3K assays, ATP (1 mM) was added to the reaction mix.
Cell loading with fluorescent lipid reagents
Bodipy Fl PIP2 or Bodipy Fl PIP3 was loaded into RBL cells as described previously.59, 68, 69 Briefly, the lipid was mixed with histone in a 1:1 molar ratio and then incubated for 10 min at room temperature. The mixture was then diluted in a physiological extracellular buffer (ECB: 135 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 10 mM HEPES, 10 mM glucose, pH 7.4) to the desired final concentration. The lipid:histone complex was incubated with cells at room temperature for 5 min. After loading, the cells were washed five times with ECB and allowed to recover for 5 min at 37 °C.
Fluorescence microscopy and image analysis
Fluorescence imaging was performed using a 20×, 0.50 N.A., Fluor, air objective on an inverted fluorescence microscope (Nikon Ti, Melville, NY) and a standard fluorescein filter set (ex/em 488nm/535nm). Transmitted light and fluorescence images were obtained with a cooled CCD camera (Photometrix, Phoenix, AZ) using Metamorph image acquisition software (Molecular Devices, Sunnyvale, CA).
Detection of increases in intracellular free calcium ([Ca2+]i) in RBL cells
The calcium indicator dye Fluo-3 (kD = 400 nm for Ca2+) was used to measure relative increases in [Ca2+]i during the loading of lipids. Cells were cultured on a glass coverslip in DMEM without serum overnight. Prior to the experiment, the cells were incubated in ECB with 1 μM Fluo-3 AM at 22 °C for 15 min. Cells were then washed with ECB and allowed to recover in ECB with 10 mM glucose at 37 °C for 30 min. Fluorescence imaging of the cells was performed at room temperature. Images were collected every 30 s for 10 min to confirm that the fluorescence intensity was stable, then the cells were incubated with the PIPn-histone complex at various concentrations and fluorescence images were again collected every 30 s for 10 min.
Activation of RBL cells through the FcεR1 receptor
RBL cells possess the tyrosine kinase linked Fc R1 receptor that when bound to IgE and cross-linked with antigen leads to activation of PI3K.73 The cells were stimulated as previously described.76 Briefly, RBL cells were preincubated overnight with immunoglobulin E (IgE) against dinitrophenyl (DNP, 1 mg/ml). The cells were washed to remove excess IgE. At the desired time, cells were activated by the addition of polyvalent antigen (0.1 mg/ml DNP-BSA) for 10 min at 37 °C.
Capillary electrophoresis
CE analysis of lipid analytes was performed using a custom-built CE system with laser-induced fluorescence detection scheme as previously described.60 Fused-silica capillaries (40 cm length, 50 μm i.d., 360 μm o.d., Polymicro Technologies, Phoenix, AZ) were used for the analyte separations. A voltage of 15 kV was applied across the capillary during electrophoresis. For CE analysis of the in vitro reaction mixtures, sample volumes (1 nL) were loaded by hydrodynamic injection. The number of moles of analyte present in each sample was determined by comparing the peak areas on the electrophoretic trace with standards as described previously.77 Separation of Bodipy Fl PIP2 and Bodipy Fl PIP3 was performed in 100 mM Tris, 10 mM SDC, 1 mM MgCl2, 30% 1-propanol, and 5% EOTrol LR, at pH 8.5. Fluorescein (1 nM mixed with the analyte) was used as an internal standard. Prior to each run, the capillary was flushed with 1 M NaOH for 3 min, deionized H2O for 3 min, and the separation buffer for 3 min using a pressurized washing system at 20 psi.
Single-cell CE analysis
A custom-built CE-laser-induced fluorescence (LIF) system mated with a microscope was used for the single-cell analyses.77 A custom-made chamber in which cells were plated was placed on an inverted microscope (Nikon TE2000, Melville, NY), and the inlet of the capillary was positioned 50 μm above the cell to be analyzed. A pulsed Nd:YAG laser (5 ns, 532 nm, Minilase, New Wave Research, Fremont, CA) integrated with the microscope was used to lyse the cell. Simultaneously with the firing of the laser, an electric field of 25 V/cm was applied across the capillary for 10 s to load the cell contents into the capillary. The capillary was then moved to a reservoir containing the electrophoresis buffer and electrophoresis initiated (300 V/cm, inlet at ground). Peaks of fluorescent cellular analytes were detected by LIF and recorded as electropherograms. In some experiments, the sample was spiked with a standard of Bodipy Fl PIP2 or Bodipy Fl PIP3 (1 nM) immediately prior to cell lysis to confirm the identity of the peak in the electropherogram.
Conclusion
An assay has been validated for PI3K and related phosphatases, including PTEN, with a number of advantages over conventional methods, particularly for cell-based assays. This method is enabled by combining microanalytical chemical separations with fluorescently labeled lipids to provide quantitative information on the conversion of a lipid substrate to metabolic end products. The format possesses the flexibility to study purified enzymes, cell lysates, or individual cells. Importantly, the chemical separation step enables the quantitative determination of both substrate and product(s) as well as side products. The resolving power of CE permits quantitation of not only the reversible conversion of a fluorescently labeled PIP2 to the PIP3 product, but also the measurement of multiple metabolic products simultaneously in the same sample. Moreover, with the sensitivity limits of LIF detection in a capillary, analysis of these metabolites in single cells has been achieved. Such information will be critical to enhance understanding of important signal transduction and homeostatic networks at the single-cell level. With the increasing interest in biochemical measurements of the PI3K pathway for cellular signal transduction, this assay can be expected to find broad application in biological and biomedical investigations. Information rich cell-based assays of pharmaceutical compounds targeting this pathway can now be readily performed. Furthermore, the small sample size needed in this method opens up the possibility of performing drug assays directly on samples of patient cells.
Acknowledgements
This research was supported by the NIH (CA139599 and CA140173). We express our appreciation for many helpful discussions with Qisheng Zhang.
Bibliographic references
- 1.Wymann MP, Schneiter R. Nat Rev Mol Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 2.Ader I, Malavaud B, Cuvillier O. Cancer Res. 2009;69:3723–3726. doi: 10.1158/0008-5472.CAN-09-0389. [DOI] [PubMed] [Google Scholar]
- 3.Sheng H, Niu B, Sun H. Curr Med Chem. 2009;16:1561–1587. doi: 10.2174/092986709788186255. [DOI] [PubMed] [Google Scholar]
- 4.Dent P, Curiel DT, Fisher PB, Grant S. Drug Resist Updat. 2009;12:65–73. doi: 10.1016/j.drup.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlo T, Levy BD. Allergol Int. 2008;57:299–305. doi: 10.2332/allergolint.08-RAI-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duroudier NP, Tulah AS, Sayers I. Allergy. 2009;64:823–839. doi: 10.1111/j.1398-9995.2009.02015.x. [DOI] [PubMed] [Google Scholar]
- 7.Rivera J, Fierro NA, Olivera A, Suzuki R. Adv Immunol. 2008;98:85–120. doi: 10.1016/S0065-2776(08)00403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito K, Caramori G, Adcock IM. J Pharmacol Exp Ther. 2007;321:1–8. doi: 10.1124/jpet.106.111674. [DOI] [PubMed] [Google Scholar]
- 9.Engelman JA, Luo J, Cantley LC. Nature Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 10.Wymann MP, Marone R. Curr Op Cell Biol. 2005;17:141–149. doi: 10.1016/j.ceb.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 11.Castellino RC, Durden DL. Nat Clin Pract Neurol. 2007;3:682–693. doi: 10.1038/ncpneuro0661. [DOI] [PubMed] [Google Scholar]
- 12.Guertin DA, Sabatini DM. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Fritz G, Kaina B. Curr Cancer Drug Targets. 2006;6:1–4. [PubMed] [Google Scholar]
- 14.Fan QW, Weiss WA. Cell Cycle. 2006;5:2301–2305. doi: 10.4161/cc.5.20.3362. [DOI] [PubMed] [Google Scholar]
- 15.Yap TA, Garrett MD, Walton MI, Raynaud F, de Bono JS, Workman P. Curr Opin Pharmacol. 2008;8:393–412. doi: 10.1016/j.coph.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Workman P, Clarke PA, Guillard S, Raynaud FI. Nat Biotechnol. 2006;24:794–796. doi: 10.1038/nbt0706-794. [DOI] [PubMed] [Google Scholar]
- 17.Balla T, Szentpetery Z, Kim YJ. Physiology (Bethesda) 2009;24:231–244. doi: 10.1152/physiol.00014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Paolo G, De Camilli P. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 19.Rapaka RS, Piomelli D, Spiegel S, Bazan N, Dennis EA. Prostaglandins Other Lipid Mediat. 2005;77:223–234. doi: 10.1016/j.prostaglandins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Michell RH. Nature. 1997;390:187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- 21.Hokin LE, Hokin MR. J Biol Chem. 1958;233:805–810. [PubMed] [Google Scholar]
- 22.Whitman M, Downes CP, Keeler M, Keller T, Cantley L. Nature. 1988;332:644–646. doi: 10.1038/332644a0. [DOI] [PubMed] [Google Scholar]
- 23.Rusten TE, Stenmark H. Nat Methods. 2006;3:251–258. doi: 10.1038/nmeth867. [DOI] [PubMed] [Google Scholar]
- 24.Hsu FF, Turk J. J Am Soc Mass Spectrom. 2000;11:986–999. doi: 10.1016/S1044-0305(00)00172-0. [DOI] [PubMed] [Google Scholar]
- 25.Ishida Y, Nakanishi O, Hirao S, Tsuge S, Urabe J, Sekino T, Nakanishi M, Kimoto T, Ohtani H. Anal Chem. 2003;75:4514–4518. doi: 10.1021/ac030072j. [DOI] [PubMed] [Google Scholar]
- 26.Wenk MR, Lucast L, Di Paolo G, Romanelli AJ, Suchy SF, Nussbaum RL, Cline GW, Shulman GI, McMurray W, De Camilli P. Nat Biotechnol. 2003;21:813–817. doi: 10.1038/nbt837. [DOI] [PubMed] [Google Scholar]
- 27.Gray A, Olsson H, Batty IH, Priganica L, Peter Downes C. Anal Biochem. 2003;313:234–245. doi: 10.1016/s0003-2697(02)00607-3. [DOI] [PubMed] [Google Scholar]
- 28.Drees BE, Weipert A, Hudson H, Ferguson CG, Chakravarty L, Prestwich GD. Comb Chem High Throughput Screen. 2003;6:321–330. doi: 10.2174/138620703106298572. [DOI] [PubMed] [Google Scholar]
- 29.Sato M, Ueda Y, Takagi T, Umezawa Y. Nat Cell Biol. 2003;5:1016–1022. doi: 10.1038/ncb1054. [DOI] [PubMed] [Google Scholar]
- 30.Balla T, Varnai P. Sci STKE. 2002;2002:PL3. doi: 10.1126/stke.2002.125.pl3. [DOI] [PubMed] [Google Scholar]
- 31.Lemmon MA. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 32.Borland L, Kottegoda S, Phillips KS, Allbritton NL. Annu Rev Anal Chem. 2008;1:191–227. doi: 10.1146/annurev.anchem.1.031207.113100. [DOI] [PubMed] [Google Scholar]
- 33.Turner EH, Cohen D, Pugsley HR, Gonzalez Gómez D, Whitmore CD, Zhu C, Dovichi NJ. Anal Bioanal Chem. 2008;390:223–226. doi: 10.1007/s00216-007-1665-5. [DOI] [PubMed] [Google Scholar]
- 34.Kraly J, Fazal MA, Schoenherr RM, Bonn R, Harwood MM, Turner E, Jones M, Dovichi NJ. Anal Chem. 2006;78:4097–4110. doi: 10.1021/ac060704c. [DOI] [PubMed] [Google Scholar]
- 35.Dovichi NJ, Hu S. Curr Op Chem Biol. 2003;7:603–608. doi: 10.1016/j.cbpa.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 36.Whitmore CD, Hindsgaul O, Palcic MM, Schnaar RL, Dovichi NJ. Anal Chem. 2007;79:5139–5142. doi: 10.1021/ac070716d. [DOI] [PubMed] [Google Scholar]
- 37.Fuller KM, Duffy CF, Arriaga EA. Electrophoresis. 2002;23:1571–1576. doi: 10.1002/1522-2683(200206)23:11<1571::AID-ELPS1571>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 38.Lee KJ, Mwongela SM, Kottegoda S, Borland L, Nelson AR, Sims CE, Allbritton NL. Anal Chem. 2008;80:1620–1627. doi: 10.1021/ac702305q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malek A, Khaledi MG. Anal Biochem. 1999;270:50–58. doi: 10.1006/abio.1999.3096. [DOI] [PubMed] [Google Scholar]
- 40.Zhang L, Hu S, Cook L, Dovichi NJ. Electrophoresis. 2002;23:3071–3077. doi: 10.1002/1522-2683(200209)23:17<3071::AID-ELPS3071>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 41.Meredith GD, Sims CE, Soughayer JS, Allbritton NL. Nature Biotechnology. 2000;18:309–312. doi: 10.1038/73760. [DOI] [PubMed] [Google Scholar]
- 42.Prestwich GD. Chem Biol. 2004;11:619–637. doi: 10.1016/j.chembiol.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 43.Prestwich GD, Chen R, Feng L, Ozaki S, Ferguson CG, Drees BE, Neklason DA, Mostert MJ, Porter-Gill PA, Kang VH, Shope JC, Neilsen PO, Dewald DB. Adv Enzyme Regul. 2002;42:19–38. doi: 10.1016/s0065-2571(01)00039-5. [DOI] [PubMed] [Google Scholar]
- 44.Prestwich GD. Prostaglandins Other Lipid Mediat. 2005;77:168–178. doi: 10.1016/j.prostaglandins.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 45.Taylor GS, Dixon JED. Anal Biochem. 2001;295:122–126. doi: 10.1006/abio.2001.5179. [DOI] [PubMed] [Google Scholar]
- 46.Senkal CE, Ponnusamy S, Rossi MJ, Bialewski J, Sinha D, Jiang JC, Jazwinski SM, Hannun YA, Ogretmen B. Mol Cancer Ther. 2007;6:712–722. doi: 10.1158/1535-7163.MCT-06-0558. [DOI] [PubMed] [Google Scholar]
- 47.Lipsky N, Pagano R. PNAS. 1983;80:2608–2612. doi: 10.1073/pnas.80.9.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vos JP, Giudici ML, van der Bijl P, Magni P, Marchesini S, van Golde LM, Lopes-Cardozo M. FEBS Lett. 1995;368:393–396. doi: 10.1016/0014-5793(95)00695-6. [DOI] [PubMed] [Google Scholar]
- 49.Paul P, Kamisaka Y, Marks DL, Pagano RE. J Biol Chem. 1996;271:2287–2293. doi: 10.1074/jbc.271.4.2287. [DOI] [PubMed] [Google Scholar]
- 50.Hayashi Y, Horibata Y, Sakaguchi K, Okino N, Ito M. Anal Biochem. 2005;345:181–186. doi: 10.1016/j.ab.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 51.Horibata Y, Higashi H, Ito M. J Biochem. 2001;130:263–268. doi: 10.1093/oxfordjournals.jbchem.a002981. [DOI] [PubMed] [Google Scholar]
- 52.Figueiredo JM, Dias WB, Mendonça-Previato L, Previato JO, Norton Heise N. Biochem J. 2005;387:519–529. doi: 10.1042/BJ20041842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhong W, Murphy DJ, Georgopapadakou NH. FEBS Lett. 1999;463:241–244. doi: 10.1016/s0014-5793(99)01633-6. [DOI] [PubMed] [Google Scholar]
- 54.Tani M, Okino N, Mutsutake M, I. M. J Biochem. 1999;125:746–749. doi: 10.1093/oxfordjournals.jbchem.a022345. [DOI] [PubMed] [Google Scholar]
- 55.Abe A, Shayman JA, Radin NS. Lipids. 1992;27:1052–1054. doi: 10.1007/BF02535587. [DOI] [PubMed] [Google Scholar]
- 56.Mao C, Wadleigh M, Jenkins GM, Hannun YA, Obeid LM. J Biol Chem. 1997;272:28690–28694. doi: 10.1074/jbc.272.45.28690. [DOI] [PubMed] [Google Scholar]
- 57.Loidl A, Claus R, Deigner HP, Hermetter A. J Lipid Res. 2002;43:815–823. [PubMed] [Google Scholar]
- 58.Dewald DB, Ozaki S, Malaviya S, Shope JC, Manabe K, Crosby L, Neilsen P, Johnston D, Harihar S, Prestwich GD. Cell Calcium. 2005;38:59–72. doi: 10.1016/j.ceca.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 59.Ozaki S, DeWald DB, Shope JC, Chen J, Prestwich GD. PNAS. 2000;97:11286–11291. doi: 10.1073/pnas.210197897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mwongela SM, Lee K, Sims CE, Allbritton NL. Electrophoresis. 2007;28:1235–1242. doi: 10.1002/elps.200600594. [DOI] [PubMed] [Google Scholar]
- 61.Otieno AC, Quainoo EW, Mwongela SM. J Pep Sci. 2008;31:3894–3901. doi: 10.1002/jssc.200800412. [DOI] [PubMed] [Google Scholar]
- 62.McLaughlin S, Wang J, Gambhir A, Murray D. Annu Rev Biophys Biomol Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- 63.Palmer CP, Vandeginste BGM. J Chromatog A. 1995;718:153–165. [Google Scholar]
- 64.Caliper LifeSciences; Hopkinton: 2008. Anonymous. [Google Scholar]
- 65.Knight ZA, Shokat KM. Biochem. Soc. Trans. 2007;35:245–249. doi: 10.1042/BST0350245. [DOI] [PubMed] [Google Scholar]
- 66.Fry MJ. In: Lipid Signaling Protocols. Larijani B, Woscholski R, Rosser CA, editors. Human Press; Totowa: 2009. pp. 345–362. [Google Scholar]
- 67.Serunian LA, Auger KR, Cantley LC. Meth Enzymol. 1991;198:78–87. doi: 10.1016/0076-6879(91)98010-4. [DOI] [PubMed] [Google Scholar]
- 68.Wang F, Herzmark P, Weiner OD, Srinivasan S, Servant G, Bourne HR. Nat Cell Biol. 2002;4:513–518. doi: 10.1038/ncb810. [DOI] [PubMed] [Google Scholar]
- 69.Weiner OD, Neilsen PO, Prestwich GD, Kirschner MW, Cantley LC, Bourne HR. Nat Cell Biol. 2002;4:509–513. doi: 10.1038/ncb811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walker C, Thomas M, Edwards MJ. Drug Discovery Today: Disease Mechanisms. 2006;3:63–69. [Google Scholar]
- 71.Barker SA, Caldwell KK, Pfeiffer JR, Wilson BS. Mol Biol Cell. 1998;9:483–496. doi: 10.1091/mbc.9.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilde JI, Watson SP. Cell Signal. 2001;13:691–701. doi: 10.1016/s0898-6568(01)00191-7. [DOI] [PubMed] [Google Scholar]
- 73.Smith AJ, Surviladze Z, Gaudet EA, Backer JM, Mitchell CA, Wilson BS. J. Biol. Chem. 2001;276:17213–17220. doi: 10.1074/jbc.M100417200. [DOI] [PubMed] [Google Scholar]
- 74.Nelson AR, Allbritton NL, Sims CE. Meth Cell Biol. 2007;82:709–722. doi: 10.1016/S0091-679X(06)82026-1. [DOI] [PubMed] [Google Scholar]
- 75.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Short Protocols in Molecular Biology. Wiley; New York: 2002. [Google Scholar]
- 76.Allbritton NL, Oancea E, Kuhn M, Meyer T. Proc Natl Acad Sci USA. 1994;91:12458–12462. doi: 10.1073/pnas.91.26.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sims CE, Meredith GD, Krasieva TB, Berns MW, Tromberg BJ, Allbritton NL. Anal. Chem. 1998;70:4570–4577. doi: 10.1021/ac9802269. [DOI] [PubMed] [Google Scholar]