Summary
Interspecies signaling between P. gingivalis and S. gordonii serves to constrain development of dual species communities. Contact with S. gordonii propagates a tyrosine phosphorylation dependent signal within P. gingivalis that culminates in reduced transcription of adhesin and signaling genes. Here we demonstrate the involvement of the P. gingivalis orphan LuxR family transcription factor PGN_1373, which we designate CdhR, in this control pathway. Expression of cdhR is elevated following contact with S. gordonii; however, regulation of cdhR did not occur in a mutant lacking the tyrosine phosphatase Ltp1, indicating that CdhR and Ltp1 are components of the same regulon. Contact between S. gordonii and a CdhR mutant resulted in increased transcription of mfa, encoding the subunit of the short fimbriae, along with higher levels of Mfa protein. Expression of luxS, encoding AI-2 synthase, was also increased in the cdhR mutant after contact with S. gordonii. The Mfa adhesive function and AI-2-dependent signaling participate in the formation and development of dual species communities, and consistent with this the cdhR mutant displayed elevated accumulation on a substratum of S. gordonii. Recombinant CdhR protein bound to upstream regulatory regions of both mfa and luxS, indicating that CdhR has a direct effect on gene expression. LuxS was also found to participate in a positive feedback loop that suppresses CdhR expression. Interaction of Mfa fimbriae with S. gordonii is necessary to initiate signaling through CdhR. These results reveal CdhR to be an effector molecule in a negative regulatory network that controls P. gingivalis-S. gordonii heterotypic communities.
Keywords: biofilm, fimbriae, luxS, signaling
Introduction
Bacteria on surfaces tend to accumulate into complex communities known as biofilms. Community development proceeds through distinct stages involving attachment, accretion, proliferation and dispersal (Stoodley et al., 2002, O’Toole et al., 2000). The temporal and spatial progression of communities is associated with the expression of distinct sets of genes, resulting in phenotypic differences not only between biofilm and planktonic cells, but also between cells in different stages of community development (Stanley and Lazazzera, 2004, Davey and O’Toole, 2000). On human mucosal surfaces such as in the oral cavity and lower gastrointestinal tract, bacterial biofilms comprise multiple species (Kuboniwa and Lamont, 2010, Macfarlane, 2008). Assembly of these complex communities involves interbacterial attachment and signaling, and organisms tend to accumulate in groupings that are physiologically compatible (Jenkinson and Lamont, 2005, Kolenbrander et al., 2002, Kuramitsu et al., 2007).
Dental plaque is a complex multispecies biofilm community that develops on tooth surfaces and is a direct precursor of periodontal disease, one of the most common infectious diseases in developed countries (Rosan and Lamont, 2000). While over 700 species or phylotypes of bacteria can inhabit mature dental plaque (Aas et al., 2005), a smaller subset of these are considered potential pathogens in periodontal disease. Foremost among these pathogenic biofilm constituents is the gram-negative anaerobe Porphyromonas gingivalis (Ximenez-Fyvie et al., 2000b). This organism possesses an array of virulence factors that enable colonization and persistence in dental biofilms, and mediate destruction of the periodontal tissues in susceptible hosts (Lamont and Jenkinson, 1998, Hajishengallis, 2009). P. gingivalis is a later or secondary colonizer of biofilm communities, relying on antecedent organisms to provide attachment sites and physiological support (Kuboniwa and Lamont, 2010, Socransky et al., 1999). In dental plaque, Streptococcus gordonii is one of the early colonizers that initiate biofilm formation, and P. gingivalis is capable of accumulating into a heterotypic community with S. gordonii and related oral streptococci (Demuth et al., 2001, Kuboniwa et al., 2006, Simionato et al., 2006). P. gingivalis can bind directly to S. gordonii without the need for bridging organisms (Kuboniwa and Lamont, 2010) and P. gingivalis and S. gordonii demonstrate mutualistic growth (Periasamy and Kolenbrander, 2009). Colonization of the oral cavity by P. gingivalis is facilitated by the presence of antecedent colonizers such as S. gordonii (Holt and Ebersole, 2005), and indeed introduction of P. gingivalis into the mouths of human volunteers results in almost exclusive localization in areas of streptococcal-rich supragingival plaque (Slots and Gibbons, 1978).
Community formation between P. gingivalis and S. gordonii ensues from initial co-adhesion that is mediated by at least two sets of adhesin-receptor pairs. The P. gingivalis long fimbriae, comprised of the FimA structural subunit, bind to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) present on the streptococcal surface (Maeda et al., 2004). In addition, the P. gingivalis short fimbriae, comprised of the Mfa structural subunit, engage the streptococcal SspA/B (Antigen I/II) adhesins (Park et al., 2005) through an approximately 27 aa binding epitope of SspA/B termed BAR (Demuth et al., 2001). Within the BAR domain, binding specificity is determined by amino acid motifs that resemble the consensus nuclear receptor (NR) box protein-protein interacting domain sequence of eukaryotes (Daep et al., 2008). The crystal structure of SspB shows that the BAR domain protrudes from the protein, resembling a handle that is available for attachment (Forsgren et al., 2010). Community development with P. gingivalis does not occur with streptococci that lack the BAR motif, such as S. mutans and S. intermedius (Kuboniwa and Lamont, 2010). In this manner, pioneer organisms such as the oral streptococci can influence the composition and pathogenic potential of the multispecies plaque biofilm.
Once attached to a streptococcal substratum, P. gingivalis executes a complex regulatory pathway that culminates first in the formation of heterotypic microcolonies, and ultimately in limitation of excessive microcolony development. Initially, community accumulation requires signaling mediated by AI-2 that can be derived from either P. gingivalis or S. gordonii (McNab et al., 2003). Dampening this process is a regulatory pathway involving Ltp1, a Low Molecular Weight Tyrosine Phosphatase. Levels of Ltp1 are increased following contact with S. gordonii (Maeda et al., 2008), and Ltp1 phosphatase activity downregulates transcription of luxS and of several genes involved in exopolysaccharide synthesis and transport. Ltp1 also positively regulates transcription of the hmu operon involved in hemin uptake and dephosphorylates the gingipain proteases. Thus Ltp1 may be part of a more general pathway that controls the pathogenic potential of P. gingivalis. However, the mechanisms by which Ltp1 effectuates control of gene transcription related to community development remain to be established.
The genome of P. gingivalis contains a number of transcriptional regulators, including members of the LuxR family. However, P. gingivalis does not produce acyl homoserine lactones (AHLs) and thus its LuxR regulators are considered orphans (or solos) in that they lack a cognate LuxI AHL synthase. Unpaired LuxR regulators are becoming increasingly recognized and studied, and they can control properties as diverse as type IV secretion, antibiotic production, exopolysaccharide synthesis and biofilm formation (Patankar and Gonzalez, 2009, Subramoni and Venturi, 2009). The P. gingivalis 33277 LuxR orphan PGN_1373 has been found to control transcription of the hmu operon in a cell density dependent manner (Wu et al., 2009), properties consistent with participation in the regulatory network that could also involve Ltp1. In this study we characterize the community associated role of PGN_1373, which we designate CdhR (Community Development and Hemin Regulator). Expression of cdhR was elevated by S. gordonii and this regulation requires the presence of Ltp1. Mutants lacking CdhR exhibited enhanced heterotypic community formation with S. gordonii. CdhR bound to the upstream regulatory regions of mfa and luxS resulting in reduced expression and thus contributing to restricted community development.
Results
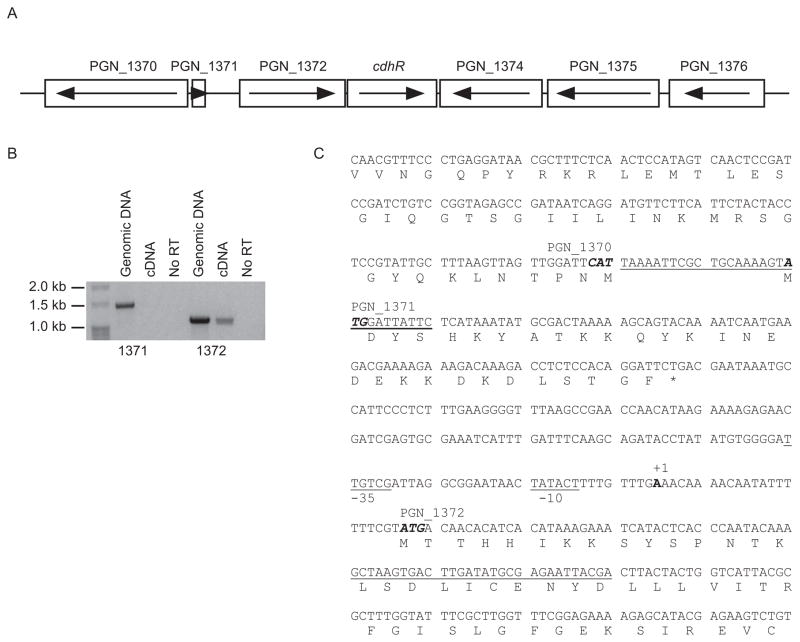
Transcriptional organization of PGN_1373 (CdhR)
The P. gingivalis orphan LuxR transcriptional regulator PGN_1373 (hereinafter called CdhR) is flanked by PGN_1372 upstream and transcribed in the same direction, and by PGN_1374 downstream and transcribed in the opposite direction (Fig. 1A). PGN_1372 is annotated as a hypothetical protein with a hemerythrin cation binding domain; and PGN_1374 is annotated as ribosomal large subunit pseudouridine synthase D. The end of the PGN_1372 coding region and the beginning of cdhR coding region overlap by 4 bases, and cotranscription of PGN_1372 and cdhR was confirmed by RT-PCR (Fig. 1B). PCR was carried out by using two different forward primers recognizing regions 250 bp upstream or 60 bp downstream of initiation codon of PGN_1372 (primers are underlined in Fig. 1C). The reverse primer was designed to bind to cdhR coding region. Genomic DNA template confirmed the fidelity of the primers which amplified the predicted 1.5 kb or 1.2 kb products (Fig. 1B). However, only the forward primer in the PGN_1372 coding region was able to amplify the 1.2 kb fragment with cDNA as a template (Fig 1B, right side). This result suggested that PGN_1372 and cdhR are in the same operon and the transcriptional start site is located within 250 bp from ATG initiation codon of PGN_1372.
Fig. 1.
Physical and transcriptional organization of the cdhR region in the P. gingivalis 33277 chromosome. (A) Genetic organization of the cdhR region. Arrows indicate the direction of transcription. (B) RT-PCR of genomic or cDNA with primers spanning cdhR and PGN_1371 (left side) or PGN_1372 (right side). The presence of an amplicon with cDNA demonstrates cotranscription of PGN_1372 and cdhR. The No RT lanes are controls without reverse transcriptase. (C) Based on 5′ RACE, the transcriptional start site for cdhR and PGN_1372 was mapped to an A (bold and labeled +1) 22 bp upstream of the ATG start codon of PGN_1372. Primers used in the RT-PCR for (B) are underlined. Putative −10 and −35 promoter regions are labeled and underlined. Initiation codons are in bold italics, and termination codon is marked with a *.
Next we focused on the upstream region of PGN_1372 as a 5′-RACE target to define the transcriptional start site (tss) for cdhR. In 6 independent RACE reactions the tss was mapped to an A residue 22 bp upstream of the ATG start codon of PGN_1372 (Fig. 1C) The tss of PGN_1372 is thus closer to the translational initiation site than has been reported for other P. gingivalis genes such as kgp (−170), rgpA (−109) and rgpB (−131) (Jackson et al., 2000). However, transcriptional initiation closer to the translational start occurs with the fimbrial genes: fimA has a tss located at −41 (Park et al., 2007), while the tss for mfa is at −44 (Park et al., 2006). Putative −10 and −35 elements for PGN_1372 were centered at −11/12 and −32/33 from the tss. The −10 sequence TATACT has a 5/6 match with the P3″ consensus element identified for a subset of P. gingivalis promoters (Jackson et al., 2000). There is also a putative P5′ element (Jackson et al., 2000) of sequence GGGATT centered at −39/40, however a P3′ element was not identified in this regulatory region.
The cdhR gene was replaced with ermF and quantitative RT-PCR (qRT-PCR) performed on total mRNA from parental and ΔcdhR strains. cdhR message was absent in ΔcdhR, and levels of PGN_1372 were similar in parental and mutant strains demonstrating that the mutation did not have an effect on the adjacent cotranscribed gene, and that cdhR is unlikely to be autoregulated (not shown).
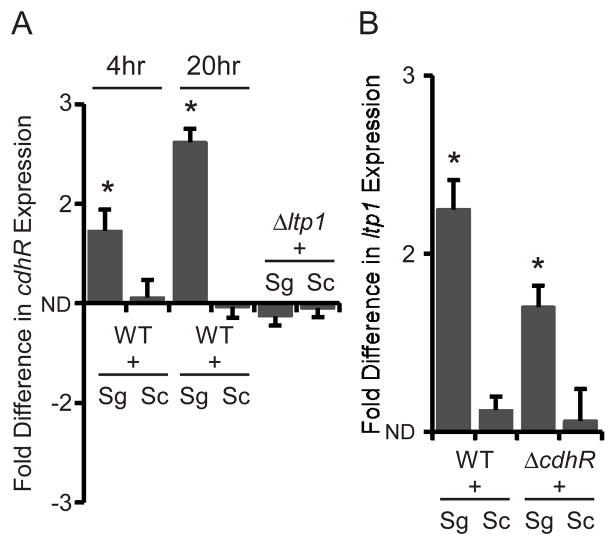
CdhR is regulated in P. gingivalis-S. gordonii heterotypic communities
On streptococcal substrata P. gingivalis cells are recruited from the fluid phase and accrete into rudimentary biofilm microcolonies. Contact and short range signaling occurs between P. gingivalis and S. gordonii, and P. gingivalis regulates its transcriptome and proteome to adapt to the community environment (Simionato et al., 2006, Kuboniwa et al., 2009). This initial increase in biomass of P. gingivalis does not require exogenous nutrients and is independent of growth and division (Kuboniwa et al., 2009, Kuboniwa et al., 2006, Maeda et al., 2008). These signaling and response interactions also occur in mixed bacterial aggregates (Park et al., 2006), and proteins that are regulated in aggregates have been found to play a role community development (Maeda et al., 2008, Kuboniwa et al., 2009). Pelleted aggregates of organisms thus provide a facile means to study regulatory circuitry (Kreth et al., 2005). Hence, to begin to assess the role of CdhR in P. gingivalis-S. gordonii community development we employed qRT-PCR of total RNA to examine expression levels in heterotypic aggregates of P. gingivalis with S. gordonii. S. cristatus which does not support P. gingivalis accumulation was employed as a control. In addition, the relationship of CdhR to Ltp1, a key component of the community constraint pathway, was investigated with an ltp1 deletion mutant.
Contact with S. gordonii, but not with S. cristatus, increased the amount of cdhR mRNA in P. gingivalis in a time dependent manner (Fig. 2A). As the tyrosine phosphatase Ltp1 is also upregulated in P. gingivalis-S. gordonii communities (Maeda et al., 2008), these results suggested that CdhR and Ltp1 may be components of the same regulatory circuit. We tested this hypothesis by measuring cdhR transcription in a Ltp1 deficient mutant. Transcriptional activity of cdhR did not increase in the ltp1 mutant when P. gingivalis cells were in contact with S. gordonii (Fig 2A), supporting the contention that CdhR is downstream of Ltp1 in a regulatory circuit that restricts community development. To further investigate the hierarchical arrangement of Ltp1 and CdhR, expression of ltp1 in the CdhR deficient mutant was tested (Fig. 2B). Both parental and CdhR mutant strains showed equivalent amounts of ltp1 mRNA when in contact with S. gordonii. Taken together, these results indicate that a community induced increase in Ltp1 subsequently leads to an increase in the amount of the transcriptional regulator CdhR.
Fig. 2.
Contact with S. gordonii induces cdhR expression when Ltp1 is present. Quantitative RT-PCR of mRNA from P. gingivalis strains in contact with S. gordonii (Sg) or S. cristatus (Sc). Fold difference is expressed relative to P. gingivalis alone. ND is no difference. (A) cdhR mRNA is increased in P. gingivalis 33277 (WT) by contact with S. gordonii but not with S. cristatus, at 4 h and 20 h. In contrast, cdhR expression in the P. gingivalis ltp1 mutant strain (Δltp1) is unaffected by S. gordonii after 20 h. (B) Expression of ltp1 is elevated by S. gordonii, but not by S. cristatus in the WT and CdhR mutant (ΔcdhR) strains at 20 h. Asterisk denotes a significant difference at P < 0.01 (t test) in levels of mRNA in P. gingivalis in contact with S. gordonii in comparison to P. gingivalis in contact with S. cristatus. Results are means with standard deviation of 3 independent experiments performed in duplicate.
CdhR constrains P. gingivalis heterotypic community development
To challenge the hypothesis that CdhR functions as a negative regulator of community development, the ability of the cdhR mutant to develop into communities on a streptococcal substratum was examined. In order to diminish the influence of possible pleiotropic effects of the mutation on the observed phenotypes, the wild-type cdhR allele was introduced into the ΔcdhR mutant using the E. coli-Bacteroides shuttle vector pT-COW, to create CΔcdhR. This complemented strain should thus display a phenotype equivalent to the parental strain. The accumulation of P. gingivalis parent and mutant strains on a substratum of S. gordonii was examined using confocal microscopy with fluorescently labeled bacteria. The CdhR deficient mutant displayed greater accumulation on S. gordonii as compared to the wild-type and complemented strains (Fig. 3A-C). Several quantitative measures, using ImageJ software, confirmed the enhanced community phenotype of the ΔcdhR mutant. Total area covered by P. gingivalis (Fig. 3D), average thickness of the P. gingivalis accumulations (Fig. 3E), and total P. gingivalis volume (Fig. 3F) were all elevated for the CdhR deficient mutant in comparison with parental or complemented strains. To ensure that different amounts of P. gingivalis were not the result of variation in the coverage of the streptococcal substratum, the ratio of P. gingivalis to S. gordonii was measured (Fig. 3G). Ratios of green (P. gingivalis) to red (S. gordonii) fluorescence confirmed the enhanced community phenotype of the CdhR mutant strain. In contrast, parental and CdhR mutant strains accumulated to the same extent in monospecies biofilms in microtiter plates (not shown). Collectively, these results demonstrate that CdhR makes an important contribution to the community restraint mechanisms operational in P. gingivalis in mixed species accumulations.
Fig. 3.
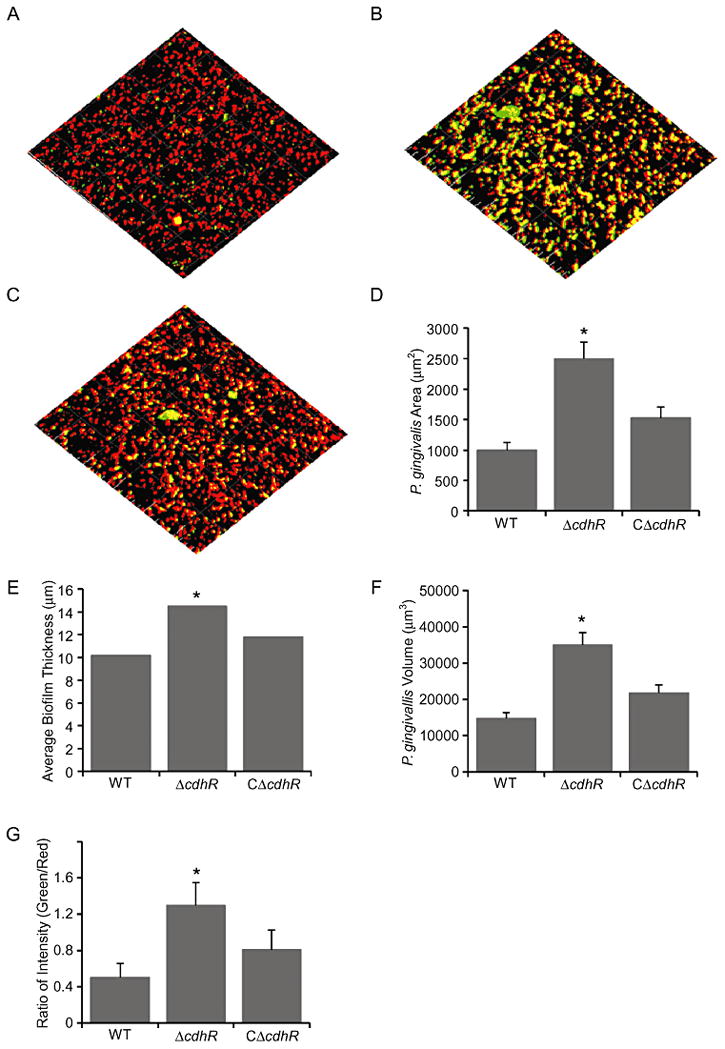
CdhR constrains heterotypic P. gingivalis-S. gordonii community development. Analysis of dual species communities by confocal laser scanning microscopy. Substrata of S. gordonii cells (red) were reacted with (A) P. gingivalis 33277 (WT), (B) mutant (ΔcdhR) or (C) complemented mutant (CΔcdhR) strains (green) for 24 h. A series of 0.2 μm deep optical fluorescent x-y sections (102 × 102 μm) were collected to create digitally reconstructed 3D images of the two species with Imaris software. Colocalized bacteria appear yellow. Images were analyzed by Image J for: (D) Total area of P. gingivalis fluorescence accumulation; (E) Average thickness of P. gingivalis accumulation; (F) P. gingivalis biovolume; and (G) Ratio of P. gingivalis to S. gordonii fluorescence. Images are representative of 3 independent experiments. Quantitative results are means with standard deviation of 3 independent experiments performed in triplicate. Asterisk denotes a significant difference at P < 0.01 compared to WT.
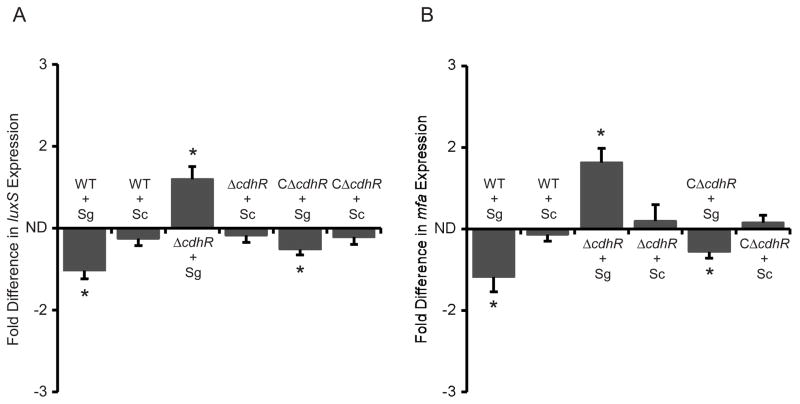
CdhR controls production of Mfa and LuxS
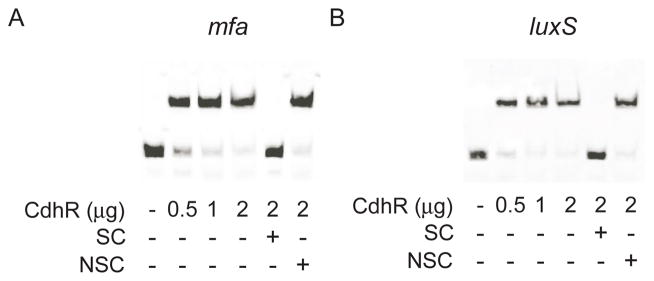
Accumulation of P. gingivalis on streptococcal substrata requires adhesins such as Mfa, and signaling molecules such as the AI-2 family of compounds produced by the enzymatic action of LuxS (McNab et al., 2003, Park et al., 2005). Mfa binds to the streptococcal Ssp receptors to initiate community formation; however, within 20 h of contact the activity of the mfa promoter is reduced, and community development is constrained (Park et al., 2006). Mfa fimbriae remain present however, and the communities do not dissociate. LuxS-dependent signaling stimulates the initial development of microcolonies (McNab et al., 2003). We reasoned, therefore, that CdhR could control community development through negative regulation of mfa and luxS. Quantitative RT-PCR was utilized to determine the transcriptional activity of mfa and luxS following 20 h of contact between P. gingivalis and S. gordonii. As shown in Fig. 4, mRNA levels for both mfa and luxS were increased in the absence of CdhR, when P. gingivalis was in contact with S. gordonii but not when in contact with S. cristatus. Moreover, the complemented CΔcdhR strain displayed a similar phenotype to the wild type strain. These findings indicate that CdhR is a negative regulator of mfa and luxS, which in turn contributes to restricted community development. To test whether CdhR regulation of mfa and luxS is direct or acts through another P. gingivalis regulator, we purified recombinant CdhR as a His6 fusion. Recombinant CdhR has been shown to be functional and bind to the promoter of the hmuY gene (Wu et al., 2009). The ability of recombinant CdhR protein to bind to the upstream regulatory regions of mfa and luxS was investigated using EMSA. The promoter region for mfa has been defined as proximal to the gene (Park et al., 2006), and the promoter that drives luxS transcription is adjacent to the upstream pfs gene (Chung et al., 2001). 200 bp fragments upstream of the initiation codons for mfa and pfs were generated for the EMSA analysis. Fig. 5 shows that purified rCdhR directly binds to the promoter regions that control both mfa and luxS. The promoter fragments were almost completely shifted with 2 μg of protein, similar to the binding affinity of CdhR for the hmuY promoter. The mobility shifts were inhibited by the addition of unlabelled target DNA, but not by the promoter fragment of fetB, indicating that the binding of CdhR to these regions is specific. Furthermore, CdhR was unable to shift a 200 bp fragment containing the promoter for the fimA gene (not shown). These data indicate that CdhR directly regulates expression of mfa and luxS.
Fig. 4.
CdhR negatively regulates expression of luxS (A) and mfa (B). mRNA levels in P. gingivalis wild type (WT), ΔcdhR and CΔcdhR strains were measured by quantitative RT-PCR. Fold difference is expressed mRNA levels after contact with S. gordonii (Sg) or S. cristatus (Sc) relative to P. gingivalis alone. ND is no difference. Data are means with standard deviation of 3 independent experiments performed in duplicate. Asterisk denotes a significant difference at P < 0.01 (t test).
Fig. 5.
CdhR protein binds to the promoter regions of mfa (A) and luxS (B). EMSA was performed with 20 fmol biotin-labeled DNA fragments containing the promoter regions and rCdhR. Non-biotinylated specific competitor oligonucleotide (SC) and non-specific competitor (fetB promoter, NCS) were used at 200-fold excess. Images are representative of three independent experiments.
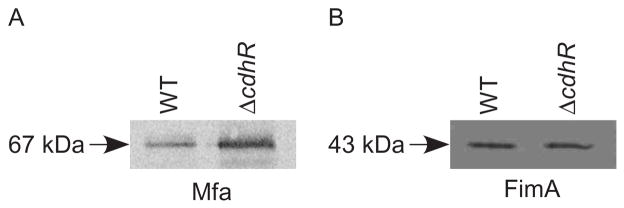
Next, we sought to determine whether the transcriptional activity controlled by CdhR is reflected at the protein level. To accomplish this we used Mfa antibodies to probe western blots of cell lysates of parental and CdhR mutant cells that were in contact with S. gordonii. As shown in Fig. 6A, the CdhR deficient mutant expressed higher levels of Mfa protein compared to parental cells, a result consistent with the mRNA data. In contrast, there was no change in the expression level of FimA (Fig. 6B). Collectively, these results demonstrate that transcriptional regulation is mediated directly by CdhR and is accompanied by changes in phenotypic properties that impact dual species community development.
Fig. 6.
CdhR regulates protein levels of Mfa. Western immunoblotting of Mfa (A) or FimA (B) expression in P. gingivalis wild type (WT) and ΔcdhR strains. Whole cell lysates were separated by SDS-PAGE and probed with antibodies to the Mfa (67 kDa) or FimA (43 kDa) subunit proteins. Images are representative of three independent experiments.
LuxS and Mfa participate in control of CdhR
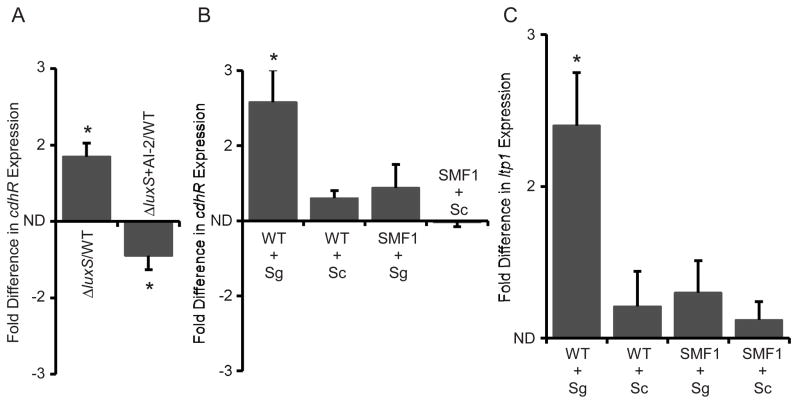
Planktonic cells of P. gingivalis express luxS, and AI-2 signaling activity negatively regulates transcription of the hmu operon (James et al., 2006). As CdhR activates hmu transcription, we investigated whether AI-2 can regulate CdhR. As shown in Fig 7A, the amount of cdhR mRNA was elevated in a luxS mutant, indicating that AI-2 negatively regulates CdhR. Chemical complementation of the luxS mutant with exogenous DPD, the precursor of AI-2, reduced expression of cdhR. Hence, the impact of AI-2 on hmu levels may be mediated by downregulation of the transcriptional activator CdhR. Furthermore, these results suggest that as the number of P. gingivalis cells increase and AI-2 levels are elevated, CdhR is down regulated and the organism expresses more Mfa fimbriae and LuxS enzyme. This positive feedback loop can be predicted to facilitate the transition to a community environment. Moreover, initial close proximity to S. gordonii and to streptococcal AI-2 may also contribute to suppression of CdhR.
Fig. 7.
(A) LuxS/AI-2 controls cdhR expression in P. gingivalis. Quantitative RT-PCR of cdhR mRNA in the ΔluxS mutant strain cultured with or without exogenous AI-2. Asterisk denotes a significant difference between WT and mutant at P < 0.01 (t test). (B) Mfa is necessary for regulation of cdhR in heterotypic communities with S. gordonii (Sg). Quantitative RT-PCR of cdhR mRNA in P. gingivalis wild type (WT) or mfa mutant (SMF1) strains in contact with S. gordonii or S. cristatus (Sc). Asterisk denotes a significant difference at P < 0.01 (t test) compared to P. gingivalis alone. (C) Mfa is necessary for regulation of ltp1 in heterotypic communities with S. gordonii. Quantitative RT-PCR of ltp1 mRNA in P. gingivalis WT or SMF1 strains in contact with S. gordonii or S. cristatus. Asterisk denotes a significant difference at P < 0.01 (t test) compared to P. gingivalis alone. Data are means with standard deviation of 3 independent experiments performed in duplicate.
In addition to chemical communication, fimbrial engagement of a receptor can be the trigger for subsequent changes in gene expression (Schwan et al., 2005, Zhang and Normark, 1996). Hence we examined the extent to which the Mfa fimbriae themselves contribute to regulation of cdhR. mRNA levels for cdhR and ltp1 were quantitated by qRT-PCR in a mfa mutant strain. Contact with S. gordonii did not induce upregulation of either ltp1 or cdhR in the absence of the Mfa fimbriae (Fig. 7B and C). These results suggest that interaction of Mfa with the streptococci is necessary for stimulation of the Ltp1-CdhR pathway.
Discussion
Communities of bacteria on solid surfaces typically contain multiple, physiologically diverse bacterial taxa. Study of interspecies signaling and response interactions in communities is an emerging field that provides insights into the behaviour of bacteria in their natural habitats. For example, quorum sensing regulated functions are important microbial competition factors in dual species communities consisting of Pseudomonas aeruginosa and Agrobacterium tumefaciens (An et al., 2006) or of Serratia plymuthica and E. coli (Moons et al., 2006). The bacterial communities that accumulate on surfaces in the oral cavity can comprise hundreds of species, and cell to cell communication shapes community composition, composite metabolic activity and pathogenic potential. Colonization of dental communities by P. gingivalis is influenced by the composition of the antecedent microbiota. While in general colonization of this anaerobe is favored by bacterial metabolism that reduces the oxygen tension, specific streptococcal species can either promote or antagonize P. gingivalis attachment. Contact with S. cristatus propagates a signal in P. gingivalis that causes downregulation of fimA expression with resultant failure of these organisms to develop into a dual species community (Xie et al., 2000, Xie et al., 2007). In contrast, S. gordonii, and related oral streptococci that possess a BAR motif in their Ag I/II family adhesins, allow P. gingivalis attachment through the shorter Mfa fimbriae, and promote two species community development (Daep et al., 2008, Demuth et al., 2001). Signaling mediated by AI-2 family molecules also stimulates dual species bacterial accumulation (McNab et al., 2003). These interspecies interactions will thus elevate the pathogenic potential of oral communities; however, realization of this potential requires that other host and bacterial factors be conducive to the onset of disease. Indeed, many individuals harbor P. gingivalis and remain healthy (Ximenez-Fyvie et al., 2000a, Mayanagi et al., 2004), indicating that mechanisms of virulence restraint are operational. We have found that the accumulation of P. gingivalis on streptococcal substrata is tightly controlled (Maeda et al., 2008, Simionato et al., 2006), which may be an adaptation to prevent reduced nutrient availability or exposure to elevated oxygen levels.
As various adhesins and signaling systems are required for P. gingivalis-S. gordonii community development, P. gingivalis requires a system to co-ordinately regulate these effectors. One component of this community regulatory pathway is the tyrosine phosphatase Ltp1. Ltp1 is upregulated following contact with S. gordonii, and tyrosine phosphatase-dependent signal transduction culminates in reduced transcription of luxS and of genes involved in exopolysaccharide production (Maeda et al., 2008). Herein we show that the LuxR family orphan transcription regulator, which we designate CdhR, is a component of the same regulatory pathway as Ltp1. CdhR is upregulated following contact with S. gordonii, but only in the presence of Ltp1, indicating that CdhR is downstream of Ltp1. In the absence of CdhR, dual species P. gingivalis-S. gordonii community formation is enhanced, also consistent with CdhR operating distal to Ltp1. CdhR was previously identified as a positive regulator of the hmu hemin uptake locus in planktonic cells of P. gingivalis (Wu et al., 2009). This activator function of CdhR also occurs in communities, as the cdhR knockout strain had less hmuR mRNA as compared to the parent (not shown). In contrast, planktonic cells of a P. gingivalis CdhR deficient strain did not exhibit differential regulation of mfa or of luxS (Wu et al., 2009). Hence the repressor functions of CdhR appear to require expression above basal levels, such as is induced following contact with S. gordonii.
P. gingivalis is one of the few organisms in which transcriptional control of luxS has been reported (James et al., 2006), although other bacteria transcriptionally control pfs (Beeston and Surette, 2002) which in P. gingivalis is part of an operon with luxS. The results from the current study show that loss of CdhR increases expression of luxS, and that CdhR can bind to the upstream regulatory region of the pfs-luxS operon. Hence, while lacking an AI-1 system, P. gingivalis appears to have utilized a LuxR homolog to control LuxS and AI-2. The GppX hybrid Two Component System (TCS) has also been implicated in regulation of luxS transcription (James et al., 2006) and we are currently investigating the relationships among GppX, Ltp1, CdhR and LuxS.
As dual species P. gingivalis-S. gordonii communities develop, the mfa gene is downregulated, one aspect of processes that regulate the level of P. gingivalis accumulation (Park et al., 2006). Control of Mfa production requires propagation of a signal from S. gordonii or related oral streptococci that are capable of supporting P. gingivalis microcolony development. The results of this study indicate that CdhR is the effector molecule of streptococcal-dependent mfa downregulation. However, transcriptional activity of mfa is also positively regulated by the FimS/FimR TCS (Lo et al., 2010, Wu et al., 2007), and Mfa expression is negatively regulated by the Clp proteolytic chaperone system components ClpXP (Capestany et al., 2008). Thus, P. gingivalis fine tunes production of Mfa according to the environmental situation.
Control of community or biofilm development through LuxR family transcriptional regulators has also been established in Vibrio cholerae. The transcriptional factor VpsT controls exopolysaccharide synthesis and biofilm formation (Yang et al., 2010). The regulatory network in which VpsT operates is complex and expression of VpsT is positively autoregulated, and also decreased through quorum-sensing dependent signaling (Yang et al., 2010). Moreover, VpsT directly detects cyclic di-guanosine monophosphate (c-di-GMP) to negatively regulate biofilm formation (Krasteva et al., 2010). A cognate kinase for VpsT has not been identified and c-di-GMP sensing results in a change in oligomerization (Krasteva et al., 2010). The mechanisms by which CdhR is regulated remain to be fully defined. Similar to VpsT, quorum sensing, in this case mediated by LuxS, decreases CdhR expression, indicating that planktonic cells become primed for community development when AI-2 reaches certain levels. CdhR also lacks a cognate kinase and is unlikely that Ltp1 directly dephosphorylates CdhR, as CdhR lacks a predicted tyrosine phosphotransferase domain and a two hybrid screen for targets of Ltp1 did not detect CdhR (unpublished). An upstream initiator of the Ltp1-CdhR pathway is the Mfa fimbriae themselves. Levels of CdhR, therefore, may be held in balance by positive and negative feedback loops involving LuxS and Mfa.
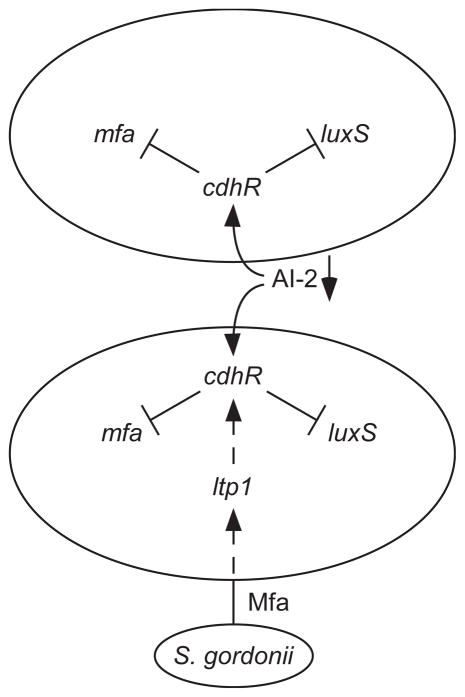
Bacterial pathogens are often adapted to colonize and persist in the absence of clinical disease in immunologically competent hosts, and bacteria that colonize host surfaces can develop mechanisms to minimize pathogenic outcomes and constrain increases in biomass and biofilm development (Galan and Zhou, 2000, Tierrez and Garcia-del Portillo, 2005, Whiteley et al., 2001, Belotserkovsky et al., 2009, Monds et al., 2007). It is becoming apparent that complex circuitry within P. gingivalis limits community development on a streptococcal substratum. Our previous results (Maeda et al., 2008, Simionato et al., 2006), along with the data in this study are consistent with the model depicted in Fig. 8. Upon encountering a streptococcal surface, a streptococcal derived signal propagating through Mfa elevates Ltp1 and which acts indirectly through a phosphorylation/dephosphorylation cascade to raise CdhR. Subsequently, CdhR reduces transcription of luxS and mfa, ultimately restricting further community development. Lower levels of LuxS, and consequently AI-2, will propagate the signaling event throughout the developing community as AI-2 is a negative regulator of CdhR. Mfa fimbriae are thought to penetrate the peptidoglycan (Murakami et al., 2004) and could thus initiate signal transduction directly. Alternatively, conformational changes in the fimbrial structure upon receptor binding may be transmitted to other outer membrane signaling initiators.
Fig. 8.
Model of circuitry governing heterotypic community formation between P. gingivalis and S. gordonii. P. gingivalis expressing Mfa and LuxS can bind and accumulate on a substratum of S. gordonii. Communication between S. gordonii and P. gingivalis requires Mfa and is transduced through Ltp1 indirectly to CdhR. Higher levels of CdhR decrease the amount of Mfa and LuxS and consequently restrain P. gingivalis accumulation. Loss of AI-2 will further increase cdhR expression which may compensate for loss of signal due to the reduction of the Mfa fimbriae.
The processes that control the assembly, function and evolution of heterotypic communities are not as fully understood as those governing homotypic community development. However, local interactions between component organisms play key roles. Contact dependent signaling has emerged as an important means by which bacteria in close proximity can modulate the growth and the behaviour of synergistic and antagonistic species (Blango and Mulvey, 2009). We have modeled an early stage in the assembly of oral biofilms, the recruitment and accretion of cells of P. gingivalis onto a streptococcal substratum prior to increases in biomass due to growth and division. This model facilitates the identification of interspecies signaling interactions that are independent of nutritional support. Components of this communication system, such as CdhR, may be of central importance in establishing the potential pathogenicity of complex oral communities.
Experimental procedures
Bacteria, plasmids and culture conditions
Bacterial strains and plasmids are listed in Table S1. P. gingivalis strains were cultured in Trypticase Soy Broth (TSB), supplemented with hemin and menadione, anaerobically at 37°C. When necessary, erythromycin (10 μg ml−1), tetracycline (1 μg ml−1), or gentamicin (100 μg ml−1) were incorporated into the medium. For AI-2 complementation, P. gingivalis ΔluxS cultures at OD600 0.2 were supplemented with10 μM DPD (OMM Scientific, Dallas, TX) and grown to OD600 0.5. S. gordonii and S. cristatus were cultured anaerobically at 37°C in TSB. Escherichia coli strains were grown aerobically at 37°C in LB media containing, when necessary, ampicillin (100μg ml−1) or kanamycin (50 μg ml−1).
Construction of complemented strain
DNA sequence containing 300 bp upstream of the PGN_1372 initiation codon and 100 bp downstream of the cdhR (PGN_1373) termination codon was amplified from P. gingivalis chromosomal DNA, using primers 1372-3f and 1372-3r (Table S2). The amplified product was cloned into shuttle vector plasmid pT-COW (Gardner et al., 1996). The resulting plasmid pT1373, was introduced into the ΔcdhR mutant by conjugation (Simionato et al., 2006) from E. coli TOP10, creating strain CΔcdhR. The conjugation reaction mixture also contained helper E. coli J53 containing R751, an IncP plasmid used to mobilize vectors from E. coli to a Bacteroides recipient (Shoemaker and Salyers, 1987). Transconjugants were selected with erythromycin and tetracycline and the presence of plasmids was confirmed by PCR and Southern hybridization. Plasmids were sequenced to confirm the construct. The conjugated strain produced similar levels of CdhR protein to the parental strain.
RT-PCR and quantitative real time RT-PCR
Primers were designed by Primer3 software. Total RNA was extracted from P. gingivalis cultures and digested with DNaseI using the Qiagen (Valencia, CA) RNeasy mini-kit. 16S and 23S rRNAs were depleted by MICROBExpress kit (Ambion, Austin, TX). The iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) was used to generate cDNA from RNA (1 μg) templates using gene specific primers (Table S2): 1372f (forward primer binding to PNG_1372); 1371f (forward primer binding to PGN_1371); and 1373r (reverse primer binding to cdhR). PCR conditions were 95°C for 6 min and 30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 3 min, followed by a final extension at 72°C for 7 min. PCR products were examined on 1.0% agarose gels. For real time quantitative RT-PCR, specific DNA standards for the genes under investigation were synthesized from chromosomal DNA using standard PCR and were quantified using an Eppendorf BioPhotometer. Real time RT-PCR was performed on a Bio-Rad iCycler using SYBR Green Supermix (Bio-Rad). Results were analyzed with the iCycler iQ Optical System software version 3.0a. The melt curve profiles were examined to verify a single peak for each sample. Transcript levels for the following genes were determined with the corresponding gene-specific primer pairs (Table S2): mfa, MFAf and MFAr; luxS, LUXf and LUXr; ltp1, LTPf and LTPr; cdhR, CDHf and CDHr; 16S, 16Sf and 16Sr. Transcript levels were normalized to 16S. In control experiments these primers did not amplify S. gordonii DNA sequences.
5′ RACE
The transcriptional start site of cdhR was determined by using the FirstChoice RNA ligase-mediated rapid amplification of cDNA ends (RACE) kit (Ambion). RNA was isolated and digested with DNaseI as described for the RT-PCR. A 45-base 5′ RNA adapter oligonucleotide was ligated to the 5′ ends of the total RNA (10 μg) using T4RNA ligase. RT was performed by using MLV reverse transcriptase with randon decamers provide by Ambion. Nested PCR was performed by first using the 5′RACE outer primer and CDHO (Table S2) a cdhR specific outer primer. Inner PCR was then conducted with the PCR product generated from the outer primers as template using the 5′ RACE inner primer that anneals to the adapter oligonucleotide, and CDHI (Table S2), a cdhR specific inner primer. The PCR product was TA cloned into the pCR4-TOPO vector and transformed into E. coli Top10. Cloning of nested PCR product was confirmed by colony PCR with M13 primers and the PCR products of six-independent clones were sequenced with an ABI capillarysequencer (Perkin-Elmer, Covina, CA).
P. gingivalis heterotypic communities
Heterotypic P. gingivalis-S. gordonii communities were generated as described previously (Kuboniwa et al., 2006). S. gordonii cells cultured for 16 h in TSB with rocking in a CultureWell coverglass system (Grace Biolabs, Bend, OR), washed three times with pre-reduced PBS and labeled with hexidium iodide (15 μg ml−1; Invitrogen, Carlsbad, CA). P. gingivalis cells were stained with 5-(and-6)-carboxyfluorescein, succinimidyl ester (4 μg ml−1; Invitrogen), and 2 × 106 cells in pre-reduced PBS reacted with the S. gordonii biofilm for 24 h anaerobically at 37°C in the dark with rocking. After washing, the resultant communities were examined on a Yokogawa spinning disc confocal scanning laser microscope system with a 60× 1.4 N.A. objective. Images were digitally reconstructed (3D image; x-y-z section), with Imaris software (Bitplane, Zurich), and quantitation of fluorescence measures was determined with ImageJ. Biofilm assays were repeated independently three times witheach strain in triplicate and analyzed with a Student’s unpaired two-tailed t test.
For gene expression studies a cell pellet assay was adopted (Kreth et al., 2005). Bacteria were cultivated to mid-exponential phase, harvested, washed and resuspended in pre-reduced PBS. P. gingivalis strains (108 cells) were mixed with an equal number of S. gordonii or S. cristatus cells in PBS. The cell mixtures were pelleted by centrifugation (10,000 g for 1 min) and incubated at 37°C anaerobically for 20 hours (except where indicated). RNA isolated from the consortia had negligible levels of streptococcal RNA as determined by RT-PCR with streptococcal specific 16S rRNA primers.
Expression of recombinant CdhR protein
The cdhR coding region was amplified using primers RCDf and RCDr (Table S2). The amplicon was confirmed by sequencing, and cloned into pCR4-TOPO (Invitrogen). For protein expression, the cdhR gene was cloned into pET30b to create pET30b-1373 which was transformed into E. coli BL21(DE3) (Novagen, Madison WI). His-tag protein was purified with a BioLogic DuoFlow chromatography system loaded with a Nickel-NTI column. Purity was greater than 97%, as determined by SDS-PAGE Coomassie staining, and the identity of the band confirmed by LC MS/MS sequencing.
EMSA
Electrophoretic mobility shift assays (EMSA) were performed using the LightShift chemiluminescent EMSA kit (Pierce, Rockford, IL) as described previously (Wu et al., 2009). Biotin-labeled DNA fragments were generated by PCR using 5′ biotin-incorporated primers: for mfa, EMFf and EMFr; for luxS ELUf and ELUr (Table S2). Binding reactions contained 20 fmol biotin-labeled DNA, 10 mM Tris (pH 7.5), 50 mM KCl, 1 mM dithiothreitol, 10 ng/μl poly(dI-dC), 2% glycerol, 0.05% NP-40, and 2 mM MgCl with varying amounts of CdhR protein and dH2O up to a total volume of 20 μl. Binding reactions were at room temperature for 30 min and samples were then loaded and run into a 5% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA buffer. The DNA and protein complexes were transferred to a positively charged nylon membrane (380 mA, 30 min) and the biotin end-labeled DNA was detected with streptavidin-horseradish peroxidase conjugate and chemiluminescent substrate. To test the specificity of binding, specific competition reactions were carried out with 200 fold excess of unlabeled target DNA (mfa or luxS promoter fragments). In addition, specificity was tested using 200 fold excess of a non specific promoter fragment of fetB generated by PCR using primers FETf and FETr (Table S2).
Western immunoblotting
P. gingivalis cells were lysed in SDS-PAGE sample buffer and proteins (5 μg) were separated by SDS-PAGE and transferred to nitrocellulose membranes. Blots were blocked with 5% BSA in TBS (PBS containing 0.1% Tween 20) for 1 h at room temperature and probed with antibodies to Mfa or FimA protein diluted 1:10,000, at 4°C overnight. Antigen–antibody binding was detected with anti-rabbit IgG peroxidase conjugate (1:5,000, Cell Signaling, Danvers, MA) and the Pierce ECL Substrate.
Supplementary Material
Acknowledgments
Supported by NIDCR DE12505 and DE14605. We thank Sarah Whitmore for assistance with the manuscript.
References
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An D, Danhorn T, Fuqua C, Parsek MR. Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc Natl Acad Sci U S A. 2006;103:3828–3833. doi: 10.1073/pnas.0511323103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeston AL, Surette MG. pfs-dependent regulation of autoinducer 2 production in Salmonella enterica serovar Typhimurium. J Bacteriol. 2002;184:3450–3456. doi: 10.1128/JB.184.13.3450-3456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovsky I, Baruch M, Peer A, Dov E, Ravins M, Mishalian I, et al. Functional analysis of the quorum-sensing streptococcal invasion locus (sil) PLoS Pathog. 2009;5:e1000651. doi: 10.1371/journal.ppat.1000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blango MG, Mulvey MA. Bacterial landlines: contact-dependent signaling in bacterial populations. Curr Opin Microbiol. 2009;12:177–181. doi: 10.1016/j.mib.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capestany CA, Tribble GD, Maeda K, Demuth DR, Lamont RJ. Role of the Clp system in stress tolerance, biofilm formation, and intracellular invasion in Porphyromonas gingivalis. J Bacteriol. 2008;190:1436–1446. doi: 10.1128/JB.01632-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WO, Park Y, Lamont RJ, McNab R, Barbieri B, Demuth DR. Signaling system in Porphyromonas gingivalis based on a LuxS protein. J Bacteriol. 2001;183:3903–3909. doi: 10.1128/JB.183.13.3903-3909.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daep CA, Lamont RJ, Demuth DR. Interaction of Porphyromonas gingivalis with oral streptococci requires a motif that resembles the eukaryotic nuclear receptor box protein-protein interaction domain. Infect Immun. 2008;76:3273–3280. doi: 10.1128/IAI.00366-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey ME, O’Toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth DR, Irvine DC, Costerton JW, Cook GS, Lamont RJ. Discrete protein determinant directs the species-specific adherence of Porphyromonas gingivalis to oral streptococci. Infect Immun. 2001;69:5736–5741. doi: 10.1128/IAI.69.9.5736-5741.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsgren N, Lamont RJ, Persson K. Two intramolecular isopeptide bonds are identified in the crystal structure of the Streptococcus gordonii SspB C-terminal domain. J Mol Biol. 2010;397:740–751. doi: 10.1016/j.jmb.2010.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE, Zhou D. Striking a balance: modulation of the actin cytoskeleton by Salmonella. Proc Natl Acad Sci U S A. 2000;97:8754–8761. doi: 10.1073/pnas.97.16.8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Russell JB, Wilson DB, Wang GR, Shoemaker NB. Use of a modified Bacteroides-Prevotella shuttle vector to transfer a reconstructed beta-1,4-D-endoglucanase gene into Bacteroides uniformis and Prevotella ruminicola B(1)4. Appl Environ Microbiol. 1996;62:196–202. doi: 10.1128/aem.62.1.196-202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. Porphyromonas gingivalis-host interactions: open war or intelligent guerilla tactics? Microbes Infect. 2009;11:637–645. doi: 10.1016/j.micinf.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- Jackson CA, Hoffmann B, Slakeski N, Cleal S, Hendtlass AJ, Reynolds EC. A consensus Porphyromonas gingivalis promoter sequence. FEMS Microbiol Lett. 2000;186:133–138. doi: 10.1111/j.1574-6968.2000.tb09094.x. [DOI] [PubMed] [Google Scholar]
- James CE, Hasegawa Y, Park Y, Yeung V, Tribble GD, Kuboniwa M, et al. LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis. Infect Immun. 2006;74:3834–3844. doi: 10.1128/IAI.01768-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13:589–595. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ., Jr Communication among oral bacteria. Microbiol Mol Biol Rev. 2002;66:486–505. doi: 10.1128/MMBR.66.3.486-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Merritt J, Shi W, Qi F. Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Mol Microbiol. 2005;57:392–404. doi: 10.1111/j.1365-2958.2005.04695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuboniwa M, Hendrickson EL, Xia Q, Wang T, Xie H, Hackett M, Lamont RJ. Proteomics of Porphyromonas gingivalis within a model oral microbial community. BMC Microbiol. 2009;9:98. doi: 10.1186/1471-2180-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuboniwa M, Lamont RJ. Subgingival biofilm formation. Periodontol 2000. 2010;52:38–52. doi: 10.1111/j.1600-0757.2009.00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuboniwa M, Tribble GD, James CE, Kilic AO, Tao L, Herzberg MC, et al. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol Microbiol. 2006;60:121–139. doi: 10.1111/j.1365-2958.2006.05099.x. [DOI] [PubMed] [Google Scholar]
- Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev. 2007;71:653–670. doi: 10.1128/MMBR.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo A, Seers C, Dashper S, Butler C, Walker G, Walsh K, et al. FimR and FimS: biofilm formation and gene expression in Porphyromonas gingivalis. J Bacteriol. 2010;192:1332–1343. doi: 10.1128/JB.01211-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane S. Microbial biofilm communities in the gastrointestinal tract. J Clin Gastroenterol. 2008;42:S142–143. doi: 10.1097/MCG.0b013e31816207df. [DOI] [PubMed] [Google Scholar]
- Maeda K, Nagata H, Yamamoto Y, Tanaka M, Tanaka J, Minamino N, Shizukuishi S. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus oralis functions as a coadhesin for Porphyromonas gingivalis major fimbriae. Infect Immun. 2004;72:1341–1348. doi: 10.1128/IAI.72.3.1341-1348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Tribble GD, Tucker CM, Anaya C, Shizukuishi S, Lewis JP, et al. A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes. Mol Microbiol. 2008;69:1153–1164. doi: 10.1111/j.1365-2958.2008.06338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayanagi G, Sato T, Shimauchi H, Takahashi N. Detection frequency of periodontitis-associated bacteria by polymerase chain reaction in subgingival and supragingival plaque of periodontitis and healthy subjects. Oral Microbiol Immunol. 2004;19:379–385. doi: 10.1111/j.1399-302x.2004.00172.x. [DOI] [PubMed] [Google Scholar]
- McNab R, Ford SK, El-Sabaeny A, Barbieri B, Cook GS, Lamont RJ. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J Bacteriol. 2003;185:274–284. doi: 10.1128/JB.185.1.274-284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monds RD, Newell PD, Gross RH, O’Toole GA. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol Microbiol. 2007;63:656–679. doi: 10.1111/j.1365-2958.2006.05539.x. [DOI] [PubMed] [Google Scholar]
- Moons P, Van Houdt R, Aertsen A, Vanoirbeek K, Engelborghs Y, Michiels CW. Role of quorum sensing and antimicrobial component production by Serratia plymuthica in formation of biofilms, including mixed biofilms with Escherichia coli. Appl Environ Microbiol. 2006;72:7294–7300. doi: 10.1128/AEM.01708-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Masuda T, Imai M, Iwami J, Nakamura H, Noguchi T, Yoshimura F. Analysis of major virulence factors in Porphyromonas gingivalis under various culture temperatures using specific antibodies. Microbiol Immunol. 2004;48:561–569. doi: 10.1111/j.1348-0421.2004.tb03552.x. [DOI] [PubMed] [Google Scholar]
- O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- Park Y, James CE, Yoshimura F, Lamont RJ. Expression of the short fimbriae of Porphyromonas gingivalis is regulated in oral bacterial consortia. FEMS Microbiol Lett. 2006;262:65–71. doi: 10.1111/j.1574-6968.2006.00357.x. [DOI] [PubMed] [Google Scholar]
- Park Y, Simionato MR, Sekiya K, Murakami Y, James D, Chen W, et al. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect Immun. 2005;73:3983–3989. doi: 10.1128/IAI.73.7.3983-3989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Xie H, Lamont RJ. Transcriptional organization of the Porphyromonas gingivalis fimA locus. FEMS Microbiol Lett. 2007;273:103–108. doi: 10.1111/j.1574-6968.2007.00782.x. [DOI] [PubMed] [Google Scholar]
- Patankar AV, Gonzalez JE. Orphan LuxR regulators of quorum sensing. FEMS Microbiol Rev. 2009;33:739–756. doi: 10.1111/j.1574-6976.2009.00163.x. [DOI] [PubMed] [Google Scholar]
- Periasamy S, Kolenbrander PE. Mutualistic biofilm communities develop with Porphyromonas gingivalis and initial, early, and late colonizers of enamel. J Bacteriol. 2009;191:6804–6811. doi: 10.1128/JB.01006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosan B, Lamont RJ. Dental plaque formation. Microbes Infect. 2000;2:1599–1607. doi: 10.1016/s1286-4579(00)01316-2. [DOI] [PubMed] [Google Scholar]
- Schwan WR, Beck MT, Hultgren SJ, Pinkner J, Woolever NL, Larson T. Down-regulation of the kps region 1 capsular assembly operon following attachment of Escherichia coli type 1 fimbriae to D-mannose receptors. Infect Immun. 2005;73:1226–1231. doi: 10.1128/IAI.73.2.1226-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker NB, Salyers AA. Facilitated transfer of IncP beta R751 derivatives from the chromosome of Bacteroides uniformis to Escherichia coli recipients by a conjugative Bacteroides tetracycline resistance element. J Bacteriol. 1987;169:3160–3167. doi: 10.1128/jb.169.7.3160-3167.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simionato MR, Tucker CM, Kuboniwa M, Lamont G, Demuth DR, Tribble GD, Lamont RJ. Porphyromonas gingivalis genes involved in community development with Streptococcus gordonii. Infect Immun. 2006;74:6419–6428. doi: 10.1128/IAI.00639-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slots J, Gibbons RJ. Attachment of Bacteroides melaninogenicus subsp. asaccharolyticus to oral surfaces and its possible role in colonization of the mouth and of periodontal pockets. Infect Immun. 1978;19:254–264. doi: 10.1128/iai.19.1.254-264.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Ximenez-Fyvie LA, Feres M, Mager D. Ecological considerations in the treatment of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis periodontal infections. Periodontol 2000. 1999;20:341–362. doi: 10.1111/j.1600-0757.1999.tb00165.x. [DOI] [PubMed] [Google Scholar]
- Stanley NR, Lazazzera BA. Environmental signals and regulatory pathways that influence biofilm formation. Mol Microbiol. 2004;52:917–924. doi: 10.1111/j.1365-2958.2004.04036.x. [DOI] [PubMed] [Google Scholar]
- Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- Subramoni S, Venturi V. LuxR-family ‘solos’: bachelor sensors/regulators of signalling molecules. Microbiol. 2009;155:1377–1385. doi: 10.1099/mic.0.026849-0. [DOI] [PubMed] [Google Scholar]
- Tierrez A, Garcia-del Portillo F. New concepts in Salmonella virulence: the importance of reducing the intracellular growth rate in the host. Cell Microbiol. 2005;7:901–909. doi: 10.1111/j.1462-5822.2005.00540.x. [DOI] [PubMed] [Google Scholar]
- Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- Wu J, Lin X, Xie H. Porphyromonas gingivalis short fimbriae are regulated by a FimS/FimR two-component system. FEMS Microbiol Lett. 2007;271:214–221. doi: 10.1111/j.1574-6968.2007.00722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Lin X, Xie H. Regulation of hemin binding proteins by a novel transcriptional activator in Porphyromonas gingivalis. J Bacteriol. 2009;191:115–122. doi: 10.1128/JB.00841-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Cook GS, Costerton JW, Bruce G, Rose TM, Lamont RJ. Intergeneric communication in dental plaque biofilms. J Bacteriol. 2000;182:7067–7069. doi: 10.1128/jb.182.24.7067-7069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Lin X, Wang BY, Wu J, Lamont RJ. Identification of a signalling molecule involved in bacterial intergeneric communication. Microbiology. 2007;153:3228–3234. doi: 10.1099/mic.0.2007/009050-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J Clin Periodontol. 2000a;27:648–657. doi: 10.1034/j.1600-051x.2000.027009648.x. [DOI] [PubMed] [Google Scholar]
- Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Microbial composition of supra- and subgingival plaque in subjects with adult periodontitis. J Clin Periodontol. 2000b;27:722–732. doi: 10.1034/j.1600-051x.2000.027010722.x. [DOI] [PubMed] [Google Scholar]
- Yang M, Frey EM, Liu Z, Bishar R, Zhu J. The virulence transcriptional activator AphA enhances biofilm formation by Vibrio cholerae by activating expression of the biofilm regulator VpsT. Infect Immun. 2010;78:697–703. doi: 10.1128/IAI.00429-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JP, Normark S. Induction of gene expression in Escherichia coli after pilus-mediated adherence. Science. 1996;273:1234–1236. doi: 10.1126/science.273.5279.1234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.