Abstract
MAPK (mitogen-activated protein kinase) signalling pathways contribute to the regulation of diverse responses, including normal and pathological aspects of cell growth, division, differentiation and death. Their ubiquity and versatility raise the issue of how they achieve specific coupling of signal with cellular response. How do the kinases in the cascade distinguish their correct substrates from the vast excess of incorrect substrates? Furthermore, how do different signals elicit distinct responses when they are transmitted by the same components? This short review highlights several mechanisms that can promote specificity in MAPK signalling, including tethering interactions between MAPKs and their substrates and regulators mediated by docking sites, feedback loops and cross-pathway regulatory circuits, and the selective activation of scaffold proteins.
Keywords: docking site, mitogen-activated protein kinase (MAPK), scaffold protein, Ste7, yeast
Introduction
Specificity in cellular signal transduction is achieved by multiple mechanisms. First and foremost, as in all other cellular processes, the pre-eminent mechanism of biochemical specificity is molecular recognition: the ‘lock-and-key’-like complementary of interacting protein regions [1]. This paradigm can explain why X transmits a signal to Y but not to Z. But pairwise, high-fidelity protein–protein interactions are insufficient to account for some of the most puzzling aspects of signalling specificity, such as how protein kinases distinguish their correct substrates from a vast excess of incorrect substrates that contain similar target residues, or how different signals transmitted by the same components elicit distinct responses. These issues apply to most cell signalling pathways, but are particularly prominent in MAPK (mitogen-activated protein kinase) signalling. The present review highlights recent progress in understanding these issues. In keeping with the format of works in this journal, we focus on our own contributions in relation to overall progress in the field.
MAPK cascades in yeast and mammals
MAPK cascades are found in almost all eukaryotic organisms, including animals, fungi and plants. The MAPK cascade is a set of three sequentially acting protein kinases [2]. Starting from the bottom of the cascade and working back up, there is a MAPK [also termed ERK (extracellular-signal-regulated kinase)], which is phosphorylated and thereby activated by an MEK [MAPK/ERK kinase; also known as MAP2K (MAPK kinase) or MKK (MAPK kinase)]. MEK activity is regulated by phosphorylation by the topmost member of the module, an MEKK [MEK kinase; also known as MAP3K (MAPK kinase kinase)], which itself is regulated by upstream signals. Activated MEKs phosphorylate their cognate MAPKs on a TXY submotif located in the MAPK activation loop; as a result, MAPK catalytic activity is increased by over 5000-fold [3]. Activated MAPKs phosphorylate targets including transcription factors, other kinases and other enzymes. Activated MAPKs are dephosphorylated and thereby deactivated by a battery of protein phosphatases [4], some of which are dedicated MKPs (MAPK phosphatases). Scaffolds proteins often assist in MAPK activation by binding to the MAPKs and other components of the cascade.
Most species possess multiple parallel MAPK cascades. In mammalian cells, four MAPK pathways have been well characterized; these are (here named after their MEK → MAPK core) the MEK1/2 → ERK1/2 module, the MKK4/7 → JNK (c-Jun N-terminal kinase) module, the MKK3/4/6 → p38 module and the MEK5 → ERK5 module [5]. In general, ERK1/2 and ERK5 tend to regulate mitogenic and developmental responses to hormones, growth factors and morphogens, while JNK and p38 regulate stress and inflammatory responses. This is just a tendency and not a rule, however.
The yeast Saccharomyces cerevisiae contains three fully elaborated MAPK cascades: the Mkk1/2 → Slt2, Pbs2 → Hog1 and Ste7 → Fus3/Kss1 cascades [6]. Mpk1MAPK regulates the integrity of the yeast cell wall, and Hog1MAPK regulates the response to high osmolarity and various other stresses [6]. Fus3MAPK and Kss1MAPK both participate in the mating pheromone response, with Fus3 playing the major role [7,8]. In addition, Kss1 regulates aspects of the filamentous invasive growth programme [9].
MAPKs are proline-directed serine/threonine kinases; thus they phosphorylate the serine or threonine in the dipeptide motif S/T-P. Although there is some preference for leucine at −1 and proline at −2 or −3 relative to the phospho-acceptor [10], many physiological target sites do not match the ‘optimal’ consensus. Thus the minimal MAPK target site is simply S/T-P. This is problematic, as the sequence S/T-P is found in approx. 80% of all proteins, and is therefore clearly insufficient to dictate whether or not a particular protein is a MAPK substrate. Furthermore, all MAPKs appear to prefer the S/T-P target site, but not all MAPKs phosphorylate the same substrates. With regard to MEK–MAPK recognition, studies have shown that the MAPK activation loop sequence cannot be the sole determinant of specificity in MEK → MAPK transactions [11,12].
MAPK-docking interactions
What provides selectivity in kinase–substrate transactions when the target site is not enough?
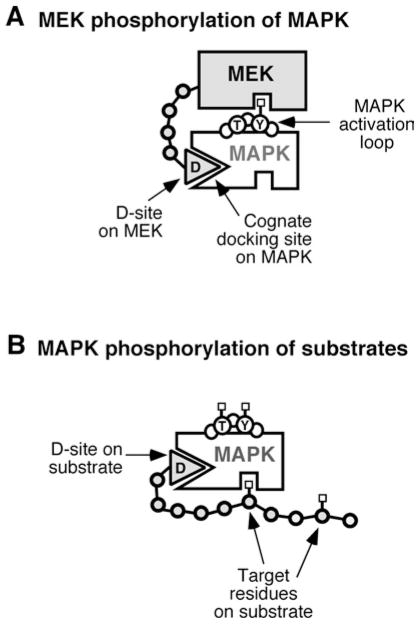
Our work has contributed to the idea that modular, adhesive protein–protein interactions, mediated by regions designated ‘docking sites’, enhance specificity in such situations by recruiting low-specificity catalytic domains to their proper substrates, and by promoting the formation of pathway-dedicated signalling complexes. MAPK-docking sites are short amino acid stretches, located on MAPK-interacting proteins, that bind to cognate binding regions on MAPKs to promote efficient enzymic transactions involving MAPKs, including MEK-mediated MAPK activation, MAPK-mediated substrate phosphorylation and MAPK dephosphorylation [13,14] (see Figure 1).
Figure 1. Tethering by MAPK-docking sites.
(A) Docking of MEK and MAPK during the process of MAPK activation.
(B) Docking of MAPK and a substrate during substrate phosphorylation.
A conserved docking sites in MEKs
The first MAPK-docking sites were discovered in the yeast MAP2K Ste7MEK and the mammalian JNK substrate c-Jun.
Our discovery of the MAPK-docking site in Ste7MEK began in 1996 with the characterization of a strikingly high-affinity (Kd = 5–100 nM), stable (half-life ~2 min) protein–protein interaction between Ste7 and its cognate MAPKs Fus3 and Kss1 [15]. This had a higher affinity and stability than would be expected for a prototypical enzyme–substrate interaction, and came at a time when many workers believed that most interactions between kinases and their substrates would be transient. Like most other MEKs, Ste7 consists of a highly conserved catalytic domain and an N-terminal extension that exhibits substantially less conservation. Unexpectedly, it was the first 20 residues of this N-terminal extension that was both necessary and sufficient for high-affinity MAPK binding [15,16]. Scrutiny of this region revealed a motif (which we now refer to as the ‘D-site’) consisting of a cluster of basic residues, a short spacer with turn-forming propensity, and then two or three hydrophobic residues spaced every other residue. Database searches revealed that this motif was present in the N-terminal extensions of MEKs from organisms representative of many different eukaryotic phyla/kingdoms [16,17] (see Table 1). Mutations in residues that were conserved between Ste7 and mammalian MEKs reduced or abolished MAPK binding [16]. We thus proposed that this conserved motif might mediate MEK–MAPK interactions in many species [17].
Table 1.
Selected examples of ‘D-site’ MAPK-docking sites
| ++++ | ϕXϕ | |
|---|---|---|
| MEKs/MKKs | ||
| MEK1 | MP KKKPTP--IQL NPAPDG | |
| MEK2 | MLA RRKPVLPALTI NPTIAE | |
| MKK3 | GKS KRKKD---LKL SCMSKP | |
| MKK6 | SKG KKRNPG--LKI PKEAFE | |
| MKK4 | MQG KRKA----LKL NFANPP | |
| MKK7-D1 | REA RRRID---LNL DISPQR | |
| MKK7-D2 | SPQ RPRPT---LQL PLANDG | |
| MKK7-D3 | PPA RPRHM---LGL PSTLFT | |
| ySte7 | TLQ RRNLKG--LNL NLHPDV | |
| Scaffolds | ||
| JIP1 | DTY RPKRPTT-LNL FPQVPR | |
| JIP3 | GRS RKERPTS-LNV FPLADG | |
| Substrates | ||
| c-Jun | SNP KILKQSMTLNL ADPVGS | |
| ATF2 | AVH KHKHE---MTL KFGPAR | |
| ELK1 | QPQ KGRKPRD-LEL PLSPSL | |
| yDig1 | KSL KRGRVPAPLNL SDSNTN | |
| yDig2 | HSL KRKRVPPALNF SDIQAS | |
| yFar1 | MMS KRGNIPKPLNL SKPISP | |
| Phosphatases | ||
| MKP1 | TIV RRRAKGA-MGL EHIVPN | |
| MKP2 | TIV RRRAKGS-VSL EQILPA | |
| MKP3 | IML RRLQKGN-LPV RALFTR | |
| PTP-SL | LQE RRGSNVS-LTL DMCTPG | |
| HePTP | LQE RRGSNVA-LML DVRSLG | |
Those proteins indicated with a ‘y’ prefix are yeast proteins and the others are human proteins. Residues matching the consensus sequence are underlined. Space limitations preclude the listing of appropriate citations. ATF2, activating transcription factor 2; HePTP, haemopoietic protein tyrosine phosphatase; PTP-SL, protein tyrosine phosphatase SL.
D-sites in human MEKs/MKKs
Consistent with the predictions made in 1996 [17], as a result of work by others and us, functional D-sites have now been identified in MEK1 [16,18], MEK2 [16], MKK3 and MKK6 [19], MKK4 [20], and MKK7 [21]. MEK5 does not contain a D-site, but contains a different type of MAPK-docking site consisting of a stretch of negatively charged residues [22]. Thus MAPK-docking sites are found in all seven MAP2Ks in the human genome. Interestingly, while MEK/MKKs 1–6 contain a single, relatively high-affinity docking site, MKK7 contains three low-affinity D-sites that interact to create a relatively high-affinity MAPK-docking platform [21].
D-sites in MEKs are targets for cleavage by the lethal factor protease of Bacillus anthracis (anthrax) [23]. Lethal factor severs the D-site from the MEK’s kinase domain, disrupting MEK–MAPK docking [24]. This impedes MAPK-transmitted signals important for the host response during early infection [25].
In vitro, MEK-derived D-site peptides are able to inhibit MEK–MAPK binding and MEK-mediated MAPK phosphorylation [16,20,26]. In cells, the ability of MEKs to efficiently phosphorylate their target MAPKs requires the integrity of the D-site [19,27]. However, when MEK2’s D-site was moved to its C-terminus, or replaced by an unrelated MAPK-binding domain from the Ets-1 (E twenty-six 1) transcription factor, ERK activation was largely retained [27]. These results indicate that a primary function of D-sites in MEKs is to tether their cognate MAPK near the MEK’s kinase domain, rather than to precisely orient the two proteins or to allosterically regulate the MAPK.
Genetics studies in yeast have also shown that docking interactions are crucial for efficient signalling [16,28,29], and suggest that a network of docking site-mediated interactions has been conserved from yeast to humans [29].
Docking sites in other proteins
Contemporaneously with our finding of the D-site in Ste7MEK, Karin and co-workers [30] delineated a short motif within the δ domain of c-Jun that promoted JNK/c-Jun binding and JNK-mediated phosphorylation of Ser63 and Ser73 in c-Jun [30]. In 1997, JIP-1, a JNK-binding protein later shown to function as a JNK-pathway scaffold, was cloned by Davis’s laboratory, and its ‘JNK-binding domain’ was identified [31]. In 1998, Sharrocks’s laboratory group identified a MAPK-docking site in the transcription factor Elk-1 (which they dubbed the ‘D-domain’) [32], and Pulido et al. [33] found a ‘kinase-interaction-motif’ in two ERK-regulating protein tyrosine phosphatases [33]. It soon became clear that these different MAPK-docking sites shared a core consensus motif, (K/R)1–3-X1–6-ϕ-X-ϕ (where ϕ is a hydrophobic residue). As stated above, we now refer to MAPK-docking sites that match this consensus as ‘D-sites’, after docking site/δ domain/D-domain.
Over the past decade, D-sites have been found in many MAPK kinases, substrates, phosphatases and scaffolds (see Table 1). Do these D-sites, found in disparate MAPK-binding proteins, compete for binding to the same region of their cognate MAPKs? Consistent this hypothesis, we found that peptide versions of the D-sites from MEKs, scaffold proteins, MKPs and transcription factor substrates all inhibited MEK–MAPK binding; in addition, these peptides also inhibited MEK-mediated MAPK phosphorylation, MKP-mediated MAPK dephosphorylation and MAPK-mediated phosphorylation of D-site-containing substrates [20,21,26]. Mutagenesis studies and crystal structures of MAPKs bound to D-site peptides also indicate that various D-sites bind to the same region of their cognate MAPKs: the basic residues of the D-site makes electrostatic contacts with two closely spaced acidic patches on the MAPK surface [28,29,34–36], while the hydrophobic residues of the D-site partially bury themselves within a nearby cluster of three hydrophobic pockets [28,35,37].
In addition to D-sites, other classes of MAPK-docking site exist [38]. Notable is the FXFP (Phe-Xaa-Phe-Pro) site identified by Kornfeld and co-workers [39], which binds to a different region of MAPKs than the D-site does [40].
Selectivity of docking interactions
Do D-sites bind selectively to their cognate MAPKs, binding poorly or not at all to non-cognate MAPKs? We have begun to explore this question for the D-sites in MEKs. In protein binding assays, JNK MAPKs do not bind to the MEK1 and MEK2 N-terminal domains or to their isolated D-sites [16,20]. However, ERK2 did bind somewhat to MKK4, and this ‘non-cognate’ interaction required the D-site in MKK4 [20]. Thus MKK4 and ERK2 are not completely specific for within-pathway docking interactions. Peptide inhibition experiments allow docking specificity to be quantified: selectivity can be quite good (20–30-fold for MKK4 versus MEK1-derived D-site peptide inhibition of JNK2) or not so good (2.3-fold for MEK1 versus MKK4-derived D-site peptide inhibition of ERK2) [20,21]. Thus the specificity of D-site-mediated interactions is quantitative rather than qualitative: it is limited in some cases.
Specificity in networks that share components
In yeast, elements of the same MAPK cascade regulate the mating and filamentous invasive growth differentiation programmes, as well as the response to osmotic stress [41] (Figure 2). Mating and filamentous invasive growth are both regulated by Ste20PAK, Ste11MEKK and Ste7MEK. In addition, Ste20PAK and Ste11MEKK are also activated during osmotic stress. Furthermore, as mentioned above, while Fus3MAPK and Hog1MAPK are activated only during mating and the stress response respectively, Kss1MAPK is activated during both mating and invasive growth. Despite this sharing of key components, the three pathways are well insulated from one another: exposure of cells to mating pheromone does not result in the hyperactivation of stress response genes, for example. Clearly there are many interesting specificity issues in this system, some of which are:
Figure 2. Yeast MAPK cascades that share components.

The yeast mating, filamentous invasive growth and osmostress response MAPK cascades share components, yet are normally well insulated from one another. See text for details.
Ste11MEKK is activated by all three pathways, yet activates Ste7MEK during mating and invasive growth, and Pbs2MEK during stress [41].
Ste7 is activated during mating and invasive growth, and activates both Fus3MAPK and Kss1MAPK during mating, but only Kss1 during invasive growth [8,42].
Although Kss1 is activated both during mating and during invasive growth, it only up-regulates invasive growth genes during the latter response [8].
What prevents Kss1MAPK from leaking out of the mating pathway? [This is issue (iii), above.] Fascinatingly, the evidence indicates that active Fus3MAPK prevents Kss1 from activating invasive growth genes, and does so by two mechanisms. First, Fus3 limits the magnitude and duration of Kss1 activation by phosphorylating an unknown upstream substrate(s) to initiate a negative-feedback circuit; in the absence of Fus3, the initial activation of Kss1 is stronger and stays on longer [8]. Secondly, in a mechanism dubbed cross-pathway inhibition, active Fus3 phosphorylates Tec1 (a transcription factor mediating Kss1 regulation of invasive growth genes), resulting Tec1’s ubiquitination and degradation [43–45]. Thus, in the absence of Fus3 activation, there is both an abundance of Tec1 complexes and an abundance of active Kss1 to up-regulate them.
What prevents invasive growth and stress signals from leaking into the mating pathway? [This is an aspect of issues (i) and (ii), above.] One popular model for pathway insulation suggests that scaffold proteins, which bind to multiple components of a given pathway, may prevent their bound components that are shared with other pathways from straying into those pathways, and protect them from intrusions from those pathways. In theory, this mechanism requires that the shared kinase be deactivated at a higher rate than the rate at which it moves on/off the scaffold [46]. However, our recent studies indicate that this might not be true for the mating pathway scaffold Ste5: Ste11 activated on Ste5 during mating can apparently either (i) dissociate from Ste5 and then activate Ste7, or (ii) activate Ste7 molecules that are not themselves scaffold-bound [47].
Furthermore, if Ste5 acts as a passive barrier to prevent leaking into the mating pathway during the stress and invasive growth responses, then such leaking should increase in the absence of Ste5. However, we found that the opposite is true: leaking into Fus3 actually decreases in ste5Δ knockout strains, and can be increased by expressing a constitutively active allele of STE5 [47]. Indeed, Fus3 activation, either by authentic or leaky signals, strictly requires Ste5 (whereas Kss1 activation does not) [47,48]. To account for these observations, we proposed a new model in which specificity is promoted by the ‘selective activation’ of the Ste5 scaffold protein. In this model, Ste5 is inactive in resting cells, and is only in a conformation and/or location capable of channelling signals to Fus3 during mating. As such, the Ste5–Fus3 interaction acts as a coincidence detector or molecular ‘AND gate’, allowing Fus3 to ignore leaking signals most of the time [47].
Acknowledgments
Work in our laboratory is supported by research grants GM60366 and GM69013 from the U.S. National Institute of General Medical Science, by an NIH (National Institutes of Health)/National Science Foundation joint initiative on Mathematical Biology through National Institute of General Medical Sciences grant GM75309, and by a grant from the National Academies Keck Futures Initiative.
Abbreviations used
- MAPK
mitogen-activated protein kinase
- ERK
extracellular-signal-regulated kinase
- JNK
c-Jun N-terminal kinase; JNK-interacting protein
- MEK
MAPK/ERK kinase
- MKP
MAPK phosphatase
- MKK
MAPK kinase
- MAP2K
MAPK kinase
- MEKK
MEK kinase
References
- 1.Pawson T. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- 2.Qi M, Elion EA. J Cell Sci. 2005;118:3569–3572. doi: 10.1242/jcs.02470. [DOI] [PubMed] [Google Scholar]
- 3.Lew J. Biochemistry. 2003;42:849–856. doi: 10.1021/bi0269761. [DOI] [PubMed] [Google Scholar]
- 4.Keyse SM. Curr Opin Cell Biol. 2000;12:186–192. doi: 10.1016/s0955-0674(99)00075-7. [DOI] [PubMed] [Google Scholar]
- 5.Johnson GL, Lapadat R. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 6.Gustin MC, Albertyn J, Alexander M, Davenport K. Microbiol Mol Biol Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farley FW, Satterberg B, Goldsmith EJ, Elion EA. Genetics. 1999;151:1425–1444. doi: 10.1093/genetics/151.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabbagh W, Jr, Flatauer LJ, Bardwell AJ, Bardwell L. Mol Cell. 2001;8:683–691. doi: 10.1016/s1097-2765(01)00322-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook JG, Bardwell L, Thorner J. Nature. 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- 10.Songyang Z, Lu KP, Kwon YT, Tsai LH, Filhol O, Cochet C, Brickey DA, Soderling TR, Bartleson C, Graves DJ, et al. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson MJ, Cheng M, Khokhlatchev A, Ebert D, Ahn N, Guan KL, Stein B, Goldsmith E, Cobb MH. J Biol Chem. 1996;271:29734–29739. doi: 10.1074/jbc.271.47.29734. [DOI] [PubMed] [Google Scholar]
- 12.Jiang Y, Li Z, Schwarz EM, Lin A, Guan K, Ulevitch RJ, Han J. J Biol Chem. 1997;272:11096–11102. doi: 10.1074/jbc.272.17.11096. [DOI] [PubMed] [Google Scholar]
- 13.Enslen H, Davis RJ. Biol Cell. 2001;93:5–14. doi: 10.1016/s0248-4900(01)01156-x. [DOI] [PubMed] [Google Scholar]
- 14.Sharrocks AD, Yang SH, Galanis A. Trends Biochem Sci. 2000;25:448–453. doi: 10.1016/s0968-0004(00)01627-3. [DOI] [PubMed] [Google Scholar]
- 15.Bardwell L, Cook JG, Chang EC, Cairns BR, Thorner J. Mol Cell Biol. 1996;16:3637–3650. doi: 10.1128/mcb.16.7.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bardwell AJ, Flatauer LJ, Matsukuma K, Thorner J, Bardwell L. J Biol Chem. 2001;276:10374–10386. doi: 10.1074/jbc.M010271200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bardwell L, Thorner J. Trends Biochem Sci. 1996;21:373–374. [PubMed] [Google Scholar]
- 18.Xu B, Stippec S, Robinson FL, Cobb MH. J Biol Chem. 2001;276:26509–26515. doi: 10.1074/jbc.M102769200. [DOI] [PubMed] [Google Scholar]
- 19.Enslen H, Brancho DM, Davis RJ. EMBO J. 2000;19:1301–1311. doi: 10.1093/emboj/19.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho DT, Bardwell AJ, Abdollahi M, Bardwell L. J Biol Chem. 2003;278:32662–32672. doi: 10.1074/jbc.M304229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho DT, Bardwell AJ, Grewal S, Iverson C, Bardwell L. J Biol Chem. 2006;281:13169–13179. doi: 10.1074/jbc.M601010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seyfried J, Wang X, Kharebava G, Tournier C. Mol Cell Biol. 2005;25:9820–9828. doi: 10.1128/MCB.25.22.9820-9828.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bodart JF, Chopra A, Liang X, Duesbery N. Cell Cycle. 2002;1:10–15. [PubMed] [Google Scholar]
- 24.Bardwell AJ, Abdollahi M, Bardwell L. Biochem J. 2004;378:569–577. doi: 10.1042/BJ20031382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agrawal A, Pulendran B. Cell Mol Life Sci. 2004;61:2859–2865. doi: 10.1007/s00018-004-4251-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bardwell AJ, Abdollahi M, Bardwell L. Biochem J. 2003;370:1077–1085. doi: 10.1042/BJ20021806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grewal S, Molina DM, Bardwell L. Cell Signalling. 2006;18:123–134. doi: 10.1016/j.cellsig.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Remenyi A, Good MC, Bhattacharyya RP, Lim WA. Mol Cell. 2005;20:951–962. doi: 10.1016/j.molcel.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 29.Kusari AB, Molina DM, Sabbagh W, Jr, Lau CS, Bardwell L. J Cell Biol. 2004;164:267–277. doi: 10.1083/jcb.200310021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kallunki T, Deng T, Hibi M, Karin M. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- 31.Dickens M, Rogers JS, Cavanagh J, Raitano A, Xia Z, Halpern JR, Greenberg ME, Sawyers CL, Davis RJ. Science. 1997;277:693–696. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- 32.Yang SH, Yates PR, Whitmarsh AJ, Davis RJ, Sharrocks AD. Mol Cell Biol. 1998;18:710–720. doi: 10.1128/mcb.18.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pulido R, Zuniga A, Ullrich A. EMBO J. 1998;17:7337–7350. doi: 10.1093/emboj/17.24.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanoue T, Adachi M, Moriguchi T, Nishida E. Nat Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 35.Heo YS, Kim SK, Seo CI, Kim YK, Sung BJ, Lee HS, Lee JI, Park SY, Kim JH, Hwang KY, et al. EMBO J. 2004;23:2185–2195. doi: 10.1038/sj.emboj.7600212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu S, Sun JP, Zhou B, Zhang ZY. Proc Natl Acad Sci USA. 2006;103:5326–5331. doi: 10.1073/pnas.0510506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang CI, Xu B, Akella R, Cobb M, Goldsmith EJ. Mol Cell. 2002;9:1241–1249. doi: 10.1016/s1097-2765(02)00525-7. [DOI] [PubMed] [Google Scholar]
- 38.Smith JA, Poteet-Smith CE, Malarkey K, Sturgill TW. J Biol Chem. 1999;274:2893–2898. doi: 10.1074/jbc.274.5.2893. [DOI] [PubMed] [Google Scholar]
- 39.Jacobs D, Glossip D, Xing H, Muslin AJ, Kornfeld K. Genes Dev. 1999;13:163–175. [PMC free article] [PubMed] [Google Scholar]
- 40.Lee T, Hoofnagle AN, Kabuyama Y, Stroud J, Min X, Goldsmith EJ, Chen L, Resing KA, Ahn NG. Mol Cell. 2004;14:43–55. doi: 10.1016/s1097-2765(04)00161-3. [DOI] [PubMed] [Google Scholar]
- 41.Schwartz MA, Madhani HD. Annu Rev Genet. 2004;38:725–748. doi: 10.1146/annurev.genet.39.073003.112634. [DOI] [PubMed] [Google Scholar]
- 42.Cullen PJ, Sabbagh W, Jr, Graham E, Irick MM, van Olden EK, Neal C, Delrow J, Bardwell L, Sprague GF., Jr Genes Dev. 2004;18:1695–1708. doi: 10.1101/gad.1178604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bao MZ, Schwartz MA, Cantin GT, Yates JR, III, Madhani HD. Cell. 2004;119:991–1000. doi: 10.1016/j.cell.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 44.Bruckner S, Kohler T, Braus GH, Heise B, Bolte M, Mosch HU. Curr Genet. 2004;46:331–342. doi: 10.1007/s00294-004-0545-1. [DOI] [PubMed] [Google Scholar]
- 45.Chou S, Huang L, Liu H. Cell. 2004;119:981–990. doi: 10.1016/j.cell.2004.11.053. [DOI] [PubMed] [Google Scholar]
- 46.Komarova NL, Zou X, Nie Q, Bardwell L. Mol Syst Biol. 2005;1:23. doi: 10.1038/msb4100031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flatauer LJ, Zadeh SF, Bardwell L. Mol Cell Biol. 2005;25:1793–1803. doi: 10.1128/MCB.25.5.1793-1803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andersson J, Simpson DM, Qi M, Wang Y, Elion EA. EMBO J. 2004;23:2564–2576. doi: 10.1038/sj.emboj.7600250. [DOI] [PMC free article] [PubMed] [Google Scholar]