Abstract
Silica colloidal crystals formed from 330 nm nonporous silica spheres inside of 75-μm i.d. fused silica capillaries were evaluated for the efficiency of capillary electrochromatography of proteins. Three proteins, ribonuclease A, cytochrome C and lysozyme, each covalently labeled with fluorophor, were well separated over a distance of 1 cm by isocratic electromigration, using 40:60 acetonitrile:water with 0.1% formic acid. A van Deemter plot showed that the plate height for lysozyme, which was the purest of the three proteins, was diffusion-limited for electric fields ranging from 400 to 1400 V/cm. The plate height for lysozyme was below 50 nm at almost all of the migration velocities, and it approached 10 nm at the highest velocity. Eddy diffusion was negligible. Lysozyme migrated over a 12 mm separation length with more than 106 plates in 1.5 minutes. These results indicate that silica colloidal crystals are well suited for electrically driven separations of large, highly charged analytes such as proteins. The 106 plates observed for a separation length of barely more than a cm means they are potentially valuable for miniaturized separations in microchip and μTAS devices.
Keywords: protein, chromatography, silica, plate height, van Deemter, colloidal crystal
INTRODUCTION
The molecular understanding of diseases, the development of protein drugs, and the discovery of protein biomarkers for diseases all rely on protein separations. The completion of the human genome has simplified the identification of proteins by mass spectrometry,1 and mass spectrometry itself has undergone many advances in the last two decades to enable detailed characterization of proteins and peptides.2 The separation of proteins remains a vital step preceding mass spectrometric analyses, and the separation step is the bottleneck.3 Compared to protein analysis by mass spectrometry, protein separations have advanced less over the last two decades.
By improving protein separations, we mean improving resolution in a given analysis time. The resolution of two components in a separation depends on two factors: peak sharpness and selectivity, as summarized in Equation 1.
| (1) |
The term L is the separation length and H is the plate height, which is proportional to peak variance. The equation shows that the smaller the value of H, the sharper the peak. The term Δt/<t> describes the selectivity for the two components being separated, where Δt is the difference in migration time of the two components and <t> is their average migration time. Eq. 1 shows that improvements in resolution could come from increased selectivity, longer length, lower plate height, or a combination of these factors. Plate height is an attractive candidate for improving resolution because it is limited by materials science: the materials used for columns cause peak broadening due to defects or design limitations. Consequently, plate height can be improved with materials advances. Any improvements in plate height cannot be obtained at the expense of separation length because these contribute to resolution as a ratio in Eq. 1. In other words, the overall number of theoretical plates, N, which is L/H, must be increased. In addition, improvements in plate heights should not be obtained at the expense of analysis time.4–6
Recent exciting advances have improved the speed of separations by as much as four-fold, using new technology including monolithic columns,7–9 sub-2-μm particles,10–13 and fused-core particles.14 These technologies give about a two-fold improvement in minimum plate height, with the lowest reported plate heights being on the order of 6 to 8 μm for proteins.15–17 The lower column resistance afforded by monoliths potentially allows for longer column lengths to achieve more plates in a given analysis time.5 Capillary electrochromatography is an alternative way to enhance transport inside of porous particles, which also increases speed, but has given little improvement in plate height for proteins thus far.18 It is sobering to realize that the two-fold improvements in plate heights over the past decade give a very modest improvement in resolution, particularly in light of the magnitude of the need in proteomics. Technological advances are needed that improve plate height dramatically.
In this work, we explore a new material that might be able to provide a large improvement in plate height: silica colloidal crystals. This material is made of very small, nonporous silica particles, 330 nm in this case, which spontaneously crystallize to give more uniformly packed capillaries. Silica colloidal crystals in planar form have been used for several types of separations, giving rather low plate heights in each case: amino acids (H=10 μm),19 small organic molecules (H=5 μm),20 organic dye (H=4 μm),21 proteins (H=2 μm).22 A very highly ordered silica colloidal crystal of 0.4×0.4 mm2 has been made and was shown to separate DNA fragments by biased reptation.23 Packed inside capillaries, silica colloidal crystals have been demonstrated to give plate heights as low as 100 nm in the electrochromatography of a fluorescent dye, when corrected for injected width.24 The corrected plate height was contributed entirely by diffusion, within experimental error. This suggests that even smaller plate heights might be achieved for proteins, since their diffusion coefficients are an order of magnitude lower, but their charges are higher. If proteins were also to give submicrometer plate heights, such a material would potentially be valuable for analysis of protein drugs and for the study of protein isoforms for biomarker analysis. The purpose of this paper is not to develop such practical separations of proteins, but rather to characterize the contributions of the material to the plate height for proteins.
EXPERIMENTAL SECTION
The silica colloidal crystals were prepared as before.24 Briefly, 330 nm diameter silica particles (Fiber Optics Center Inc., New Bedford, MA) were calcined at 600 °C three times for 6 hours each in a box furnace, then rehydroxylated in 50% nitric acid, and finally suspended in ultrapure water and used to make a 30% (w:w) slurry. Fused silica capillaries of 75 μm i.d. with Teflon coatings (Polymicro Techniques, Phoenix, AZ) were cleaned by pumping 0.1 M NaOH for 15 min, and then rinsed with ultrapure water and ethanol for 20 min each, then cut into 12 cm sections and dried in a vacuum oven at 70° for 30 min. The slurry was wicked into the capillaries and allowed to slowly settle and dry over several days. The packed capillaries were placed in a 50% humidity chamber for 30 min to form a thin layer of water on the surface of the silica. After that, the packed capillary columns were put into a 6% n-butyltrichlorosilane and 2% methytrichlorosilane (v/v) (Gelest Inc., Morrisville, PA) in dry toluene solution under dry nitrogen atmosphere to achieve horizontally polymerization.25 The reaction was allowed to proceed overnight at room temperature. After the reaction, the capillary columns were rinsed with dry toluene by pumping using an HPLC system (Agilent 1100 series). Then the capillary columns were put into an oven at 120° for 2 h. After these steps, the capillary columns were stored in glass vials.
The mobile phase solutions consisted of different ratios of HPLC-grade acetonitrile and ultrapure water with 0.1% (v/v) formic acid, 88%. The acetonitrile and formic acid were purchased from Sigma Aldrich, St. Louis, MO. The water was supplied from an ultrapure water system (Millipore, Milford, MA).
Three proteins, ribonuclease A, cytochrome C, and lysozyme were purchased from Sigma Aldrich St. Louis, MO, and were labeled with Alexa Fluor 546 using a labeling kit (Invitrogen, Carlsbad, CA). The concentration of proteins after labeling was 10−4 M. The protein samples were diluted by 1:100 (v/v) with 1× PBS buffer (pH=7.67) to give a final solution concentration of 10−6 M.
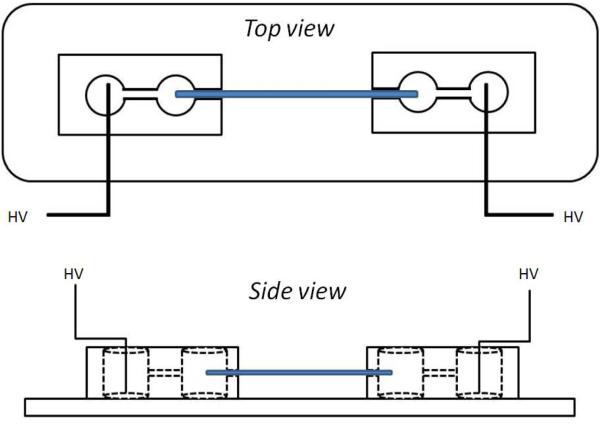
Figure 1 shows the instrument setup. The holder was fabricated by a pair of double-well reservoirs made from polydimethylsiloxane (PDMS) on glass microscope slides. The double-well reservoirs were used to prevent air bubbles from entering the capillary column. First, a polydimethylsiloxane (PDMS) sheet of 1/8” thickness was made by mixing the PDMS base and curing agent (Sylgard 184, Dow Corning, Midland, MI) at 10:1 ratio in a Petri dish. After overnight curing at 120 °C, the sheets were cut into ½” squares and the reservoir holes were cut. Then, the PDMS holders were attached to a clean glass slide after oxygen plasma oxidization (Novascan, Ames, IA) of the PDMS for 5 min. A pair of Pt electrodes connected to a high voltage power supply (Matsusada) was put in the reservoirs to control the electric field. (Caution, electrical hazard: access to the electrodes must be blocked while the power supply is operating.)
Figure 1.
Schematic of electrochromatography experiment using double reservoirs made of PDMS. The capillary (blue) spanned the two inner reservoirs, while the Pt electrodes were placed in the outer reservoirs. This design minimized bubbles entering the capillary.
Packed capillaries of 2 cm in total length were used for these studies. Before use, the capillary column was wetted by using the HPLC pump to dispense the same mobile phase as in the separation. The capillary was then conditioned with this mobile phase, under an electric field of 300 V/cm for 15 min. The sample solution was injected from the positive electrode side from PBS solution by applying a 50 V/cm electric field for 5 min. The sample solution was then removed, and after washing the reservoir with the mobile phase and refilling it with fresh mobile phase, an electric field ranging from 400 to 1400 V/cm was applied. The separation processes were monitored by using an inverted fluorescence microscope equipped with a mercury lamp and a 2× objective (Nikon Eclipse TE2000U). A Cy3 filter (Omega Optical, Brattleboro, VT) was used for the Alexa Fluor 546 conjugated protein samples. The emission was collected by a high sensitivity CCD camera (ProEm, Princeton Instruments) using an acquisition time of 0.2 s, controlled by the Winview software provided by the camera manufacturer. Data were analyzed using Origin (Microcal, Northampton, MA).
RESULTS AND DISCUSSION
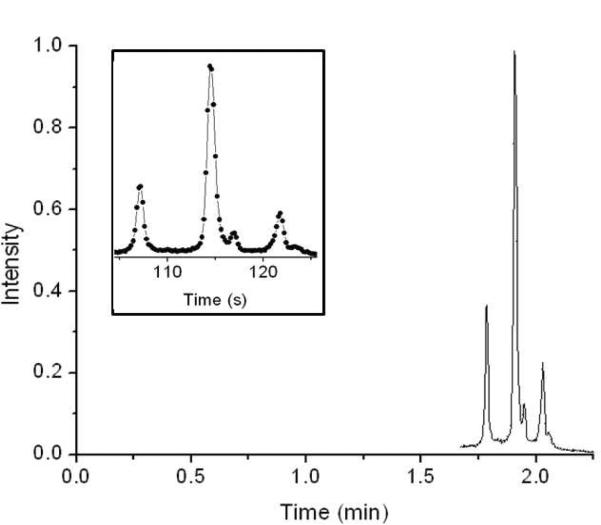
Figure 2 shows the electrochromatogram of a mixture of three labeled proteins: lysozyme (Lyz), ribonuclease A (RnaseA) and cytochrome c (CytC), for a separation length of 0.9 cm using an isocratic mobile phase composition of 40:60 ACN/water with 0.1% formic acid. The elution order was determined by injecting each protein separately. The retention order (Lyz, RnaseA, CytC) is different from what we observe in a CE separation (Lyz, CytC, RnaseA) but the same as what we observe in a reversed phase separation. The reversed phase results were obtained using the same stationary phase, but on 1 μm nonporous particles to allow for UHPLC separation showing retention factors of 0, 1.1 and 1.2 for lysozyme, ribonuclease A and cytochrome C, respectively, after accounting for the higher surface area of the smaller particles. The inset in Figure 2 shows the electrochromatogram on an expanded scale, and one can estimate that the plate heights are all 100 nm or lower. The plate height is thus lower than that for the fluorescent dye studied in our previous work for the same type of capillary,24 presumably because the injected widths and diffusion coefficients are lower. To determine how low the plate height can be for these materials, we studied lysozyme individually, rather than the mixture, because lysozyme exhibits the cleanest peak in the chromatogram of Figure 2.
Figure 2.
Electrochromatogram of a mixture of three proteins, which elute in the order of lysozyme, ribonuclease A and cytochrome C. The separation was performed isocratically using a 1,000 V/cm electric field and a mobile phase of 40:60 ACN:water with 0.1% formic acid. The separation distance was 0.91 cm. The inset shows the same electrochromatogram on an expanded time scale, with the discrete data points visible for the 0.2 s acquisition intervals.
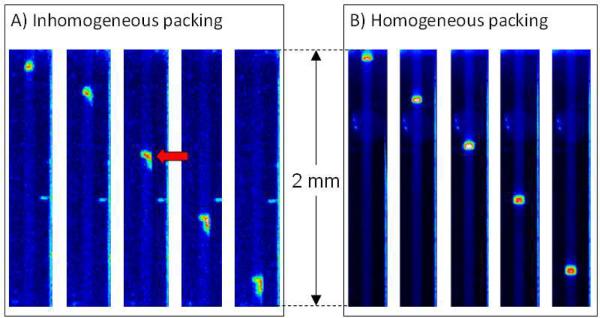
Many capillaries we prepared did not give such low plate heights because the packing was heterogeneous. Figure 3a illustrates the behavior of lysozyme electromigrating over a distance of 2 mm in a capillary in which the packing has a brief gap between the packing material and the wall, with the position of this wall-gap indicated by the position of the red arrow in the figure. This series of time-delayed images shows that the right side of the peak moves ahead of the remainder of the peak at this wall-gap, resulting in peak broadening. Such capillaries typically have multiple wall-gaps, giving significant broadening after a separation length approaching 1 cm. About 2/3 of the capillary have these wall gaps. Figure 3b illustrates the behavior of lysozyme electromigrating over the distance of 2 mm in a capillary in which the packing has no significant wall gaps. The peak retains the same shape and broadens little as it electromigrates over the 2-mm distance. A capillary that has such good homogeneity in the first 2 mm typically has good homogeneity over a distance of at least 1 cm using the conditions we used for packing. These results illustrate that homogeneous packing is essential for achieving plate heights of 100 nm and below with the silica colloidal crystals. For the remainder of these studies, we used the most uniform capillary that we were able to prepare to determine the best possible performance that one can expect to achieve from these materials.
Figure 3.
A series of fluorescence images of lysozyme electromigrating through two different capillaries. Each strip is separated by a 6 s time delay. A) Image series for lysozyme electromigrating through the first 2 mm of a capillary that has defects: a wall gap indicated by the red arrow. B) Lysozyme electromigrating from 5 to 7 mm from the entrance of a good capillary, just before it enters the reservoir. The images show that the peak remains uniform across the capillary diameter and narrow in width.
Diffusion is the ultimate limit to plate height, and the diffusion coefficient of lysozyme was determined by monitoring the broadening of the lysozyme peak electromigrated over a 1-cm distance. The images were converted to intensities and fit to Gaussians. For the lowest and highest electric fields used, 400 V/cm and 1400 V/cm, respectively, Figure 4a shows the raw images for four peaks, and Figure 4b shows the corresponding intensity profiles and their best fits to Gaussians. Figure 4c shows a linear fit of the Gaussian peak variances vs. time, the slope of which is 2γD, where D is the diffusion coefficient. The linear regression shows that γD=0.64×10−8 cm2/s. This represents the minimum contribution to the plate height.
Figure 4.
A) Images of lysozyme peaks for the indicated electric field and the elapsed time for electromigration. B) Intensity graphs for the same data as in part A (●) and best-fit Gaussians (—). C) Plot of the variances from the data of part B (●) vs. time, and the best fit to a line (—) to give the diffusion coefficient.
In determining plate height quantitatively, one must consider that the lysozyme peak is so narrow at high electric fields that the acquisition time of 0.2 s gives only a few points across the peak. There is not enough information in such a limited number of data points in the time domain to fit to a Gaussian. As an alternative, the image of the zone at a given point in time contains many data points in space, which accurately characterize peak width. These spatial data points were used to calculate the spatial standard deviation, σ, which was then used to compute the plate height from the known position of the center of the zone, L.
| (2) |
Figure 5 shows the raw images for the three replicate runs at the highest electric field, 1400 V/cm, and the position, L, of each peak. The height of each peak is 75 μm, which is the capillary diameter. In the images, the protein zones have moved from left to right, and one can see that they are uniform across the diameter of the capillary. Axially, their 4σ widths are on the order of the diameter of a human hair, which is 50 μm. The images show that the protein zones span the capillary diameter symmetrically and that they are quite symmetric along the axial direction of the capillary, as well. The excess width near the baseline is probably due to impurities. Some deviation from Gaussian shape at the peak is evident in the third zone, where the profile shows some flattening on top. This could be due to over-injection or to imperfect optical focusing of the microscope, both of which would give an overestimate of the plate height. Beneath each image is a plot of the numerical data from the image, on the same spatial scale, and one can see the high quality of the fit to a Gaussian profile.
Figure 5.
Top: Raw images of the lysozyme zones in the 75 μm i.d. capillary at their maximum migration distances, indicated in mm, for each replicate measurement at the highest electric field (1400 V/cm). Bottom: The same raw data on the same spatial scale, but converted into chromatograms (O), and showing the best fit to a Gaussian (—). The standard deviation of each Gaussian is indicated in μm.
The plate heights for the three images in Figure 5, based on the variances of the Gaussian for each, are 17, 10 and 20 nm. One other factor must be dealt with before quantitatively interpretation is the contribution from broadening during detection. At the 1400 V/cm field, the peak moves significantly during the 0.2-s time the image is acquired. This convolutes a detector response function into the peak profile. The detector contribution to plate height, σdet2/L, is readily accounted for by Eq. 3,26 where τ is the acquisition time of 0.2 s and v is the velocity of the peak.
| (3) |
A calculation using this equation shows that the detector contributes 5 nm to the plate height at the highest field. Similarly, the contributions from the detector were calculated for the peaks at the lower electric fields, and these were all subtracted to give final plate heights at all of the electric fields.
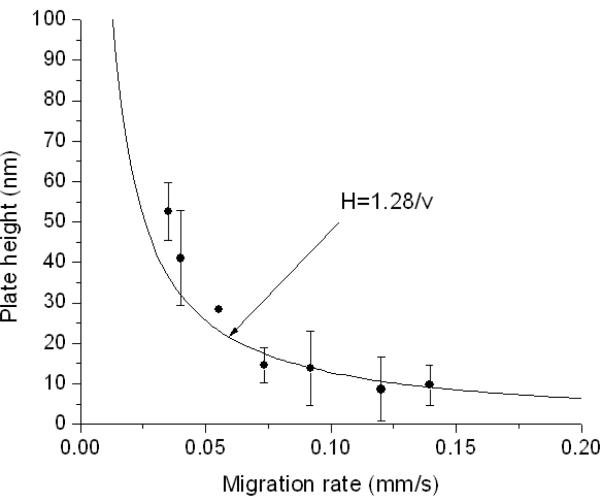
The plot of final plate height vs. analyte migration rate is shown in Figure 6. This plot is analogous to a van Deemter plot, and it has the same A, B and C terms, but has analyte migration rate, vm, plotted rather than velocity of the mobile phase, as shown in Eq. 4. The solid line in
| (4) |
Figure 6 is a plot of H=2γD/vm, where γD is the diffusion coefficient determined from Figure 4. This curve fits the van Deemter plot reasonably well. As in the case of the fluorescent dye,24 diffusion is the only significant contribution. The A term is on the nanometer scale, at most, and the C term is negligible. Since only the B term contributes, lower plate heights might be possible at higher electric fields, unless heating begins to contribute. The reason analyte migration rate is used in the plot is to account accurately for the time that a charged analyte spends in the mobile phase in CEC: tm= L/vm, where vm includes both the electrophoretic and electro-osmotic velocities. Incidentally, this type of plot could be used for retained analytes, where B=2γkD, and k is the retention factor. Figure 6 shows that the plate height reaches values as low as 10 nm at the highest electric field. This lowest plate height is 600-fold lower than that for capillary electrochromatography of proteins in polymer monoliths,16 and it is 100-fold lower than that for CZE of lysozyme.27 One caveat in this comparison with CEC is that retention can broaden peaks due to slower diffusion in and out of porous particles, nonlinearity in the adsorption isotherm, and slow desorption from the surface. While the first two factors would not contribute here, possible contribution from the others is not yet known. What Figure 6 shows is that the value of this material an eddy diffusion that is vanishing small compared to conventional separation media, and it allows for the use of high fields.
Figure 6.
Plot of plate height vs. analyte migration velocity (●) and plot of the 1.28 μm2/s (—). The error bars represents standard deviations from three replicate measurements.
There have been two earlier reports of extremely sharp peaks in CEC, and both involved focusing of the peaks as they migrated through the medium.28–29 We would not be able to conclude that the plate height for eddy diffusion if on the nanometer scale if the narrow peaks were due to a focusing phenomenon that was reducing the broadening from the packing inhomogeneities. Focusing could be accidentally obtained by the injection process used here, where water can diffuse into the column during injection, and electro-osmotic flow would then follow this with high organic content. The leading edge of the peak would be more retained, allowing the trailing edge to catch up, thereby narrowing the peak. The Fickian diffusion argues against this, but to be sure, a field-free measurement of diffusion is needed. We acquired diffusion data under conditions where the capillary never had exposure to the aqueous solution, avoiding a solvent gradient, and no electric field was used. Only diffusion was occurring, thus avoiding any possibility of focusing in this experiment. To do this, we immersed a capillary filled with mobile phase (40:60 ACN:water, and 0.1% formic acid) into a solution of lysozyme in the same mobile phase, and allowed the protein to diffuse into the capillary for 1,000 s. The results are shown in Figure 7. The graph gives fluorescence intensity along the center of the capillary axis as a function of distance from the entrance, while the inset shows the raw image, along with a 75 μm scale bar. The end of the capillary is very bright due to total internal reflection of much of the fluorescence originating inside of the capillary. This optical phenomenon is familiar to anyone who has observed the bright tips of optical fibers from the sides, and this limits a quantitative determination of the obstructed diffusion coefficient. The image demonstrates that the diffusion is indeed extremely slow in that most of the lysozyme remains at distances much less than 75 μm despite the 1,000 s time for diffusion. The ~10 μm raggedness at the cut end of the capillary limits an accurate determination of the obstructed diffusion coefficient. The data in the plot show that the obstructed diffusion coefficient, γD, for the major component is no more than 0.9×10−8 cm2/s, which is consistent with the van Deemter plot. The data also shows that there is a fluorescent impurity that diffuses four-fold faster than the lysozyme. Based on its intensity and width, and it is probably the source of the high baseline for all of the lysozyme chromatograms in Figure 4. This fluorescent impurity also interferes with determining an exact diffusion coefficient for lysozyme because it creates an unknown baseline. This is because some of this impurity near the entrance to the capillary is able to diffuse out while the capillary is being rinsed before imaging, thus its exact function over the region overlapping with the lysozyme zone is not known. Despite these limitations, the data support the conclusion that the obstructed diffusion coefficient is less than 10−8 cm2/s. Experiments on shorter time scales showed no evidence of a faster component.
Figure 7.
Image: Fluorescence micrograph of the end of the capillary after lysozyme has been allowed to diffuse for 1,000 s. A scale bar of 75 μm is shown. Plot: the intensity (O), as read from the image, for a line along the axis of the capillary, starting at the maximum intensity and moving to the right; a sum of two Gaussians (—), with the equation as given in red in the figure.
The obstructed diffusion coefficient and electromigration rate of lysozyme are about two orders of magnitude lower than in free solution. The silica colloidal crystal is expected to slow diffusion and migration, but not this much. A study of the diffusion of ions in a silica colloidal crystal explains that the diffusion coefficient is decreased by an order of magnitude relative to the free solution, where 1/10 is approximately the product of the porosity (i.e., the fractional free volume) and the tortuosity (i.e., the indirect paths that are required for displacement).30 This product gives rise to the familiar γ factor in the B term of the van Deemter equation. Lysozyme exhibits not just a one order of magnitude lower diffusion coefficient in the CEC experiment, instead, here it is two orders of magnitude. The reason for the obstructed diffusion coefficient being more than 100× slower than that in free solution is not known at this time, one can only speculate. One possibility is that there is electrostatic adsorption of the very positively charged proteins into the Debye layer, which itself could be a significant fraction of the volume in these small channels. This would give R<1 without requiring adsorption onto the hydrophobic monolayer. The UHPLC performed with 1 μm particles would not necessarily uncover this possibility because the channel volume is three-fold larger. The experiment independently supports the unexpectedly low obstructed diffusion coefficient in the chromatographic analysis, providing further evidence that focusing is not occurring in these separations.
For a diffusion limited conditions in either CZE, or in CEC with no retention, the electric field, the diffusion coefficient and the electrophoretic mobility combine to dictate the plate height.26
| (5) |
It is insightful to examine low plate heights in CZE to understand the protein CEC results here. Moore and Jorgenson showed that capillary electrophoresis attains diffusion-limited plate heights for small molecules by using very thin capillaries, 6 μm i.d., along with gated injection.26 The electrophoretic mobility of the fastest component, fluorescein-labeled arginine, is 2×10−3 cm2/V·s, which includes an unknown contribution from electro-osmotic mobility. The diffusion coefficient of this compound is D=3.5×10−5 cm2/s. For Arg-FITC, (μe+μeo)/D=57, an electric field of 1250 V/cm gave a theoretical plate height of 280 nm, which was shown to be nearly reached experimentally. At the highest field, 2500 V/cm, they observed a plate height of 75 nm, which included a 5 nm contribution from detection width. To return to the packed capillaries, one can use the ratio of 2D/(μe+μeo)E in Eq. 5 to calculate the diffusion limited plate height theoretically from values in the literature, and compare this with our experimental value. For lysozyme, μe=2.66×10−4 cm2/V·s at pH 3,31–32 and D=1.1×10−6 cm2/s,33 therefore μe/D=240 in aqueous solution. The published values of μe were obtained using coated capillaries, so electro-osmotic flow is not likely to contribute significantly. In our case, we measured electro-osmotic flow rate by the method of current monitoring,34 and the results showed that veo=0.10(±0.01) mm/s at 1400 V/cm. Since our electromigration data showed that ve+veo=0.14 mm/s at this field strength, then (μe+μeo)=(0.14/0.04)μe. Assuming the protein charge is comparable to that in aqueous solution, and using the published value of μe for lysozyme, (μe+μeo)=(2.66×10−4 cm2/V·s)(0.14/0.04)=1.3×10−3 cm2/Vs. This gives the theoretical ratio (μe+μeo)/D=1,200, therefore, substituting this into Eq. 5 and using E=1400 V/cm, the diffusion-limited plate height of lysozyme is predicted by Eq. 5 to be 15 nm at 1400 V/cm. This is in good agreement with the value of 10±5 nm for our experimentally observed plate height reported at 1400 V/cm in the van Deemter plot of Figure 6. This agreement with the predicted value further argues against the possibility of focusing to explain the low plate height. This discussion also illustrates that the ultralow diffusion coefficient is not a requirement for low plate height because it is offset by lower electrophoretic mobility; instead, it is the high ratio of μe/D for proteins that dictates the value of the diffusion-limited plate height for a given electric field. The experimental conditions attain this minimum plate height.
We emphasize that it is not the low diffusion coefficient that is giving small plate heights here. There are six factors that combine to attain the diffusion-limited plate heights of 50 nm and below for these packed capillaries. 1) The widths from injection and detection are extremely narrow. 2) There is little reversed-phase adsorption, which eliminates the C term. 3) Proteins have very high charge to give high electrophoretic mobility. 4) The electro-osmotic flow enhances the mobility four-fold. 5) The packing is sufficiently homogeneous. 6) High electric fields can be used without significant thermal broadening. These last two factors are related uniquely to the packing material, and they represent the new information that is contributed by this work. In essence, the packed capillaries are performing like the 6 μm i.d. capillaries of Moore and Jorgenson,26 giving diffusion-limited performance. The packed capillaries offer the advantages of direct injection and higher volume, and they are confirmed to work for proteins.
CONCLUSIONS
These results show that silica colloidal crystals made of 330 nm diameter silica spheres can give extremely efficient electromigration of proteins. For 75 μm i.d. capillaries, plate heights attaining the diffusion limit of 10 nm were observed even at electric fields exceeding 1,000 V/cm. Separations lengths of 12 mm were used in this study, giving more than 106 plates at the highest analyte velocity of 0.15 mm/s. The ability to compress a million plates into barely more than a cm of physical length could be valuable for miniaturized separations in lab-on-a-chip μTAS systems.
ACKNOWLEDGEMENT
This work was supported by NIH under Grant R01GM065980.
REFERENCES
- 1.Wright JC, Hubbard SJ. Comb. Chem. High Throughput Screen. 2009;12:194–202. doi: 10.2174/138620709787315508. [DOI] [PubMed] [Google Scholar]
- 2.Yates JR, Ruse CI, Nakorchevsky A. Annu. Rev. Biomed. Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 3.Bogdanov B, Smith RD. Mass Spectrom. Rev. 2005;24:168–200. doi: 10.1002/mas.20015. [DOI] [PubMed] [Google Scholar]
- 4.Carr PW, Wang XL, Stoll DR. Anal. Chem. 2009;81:5342–5353. doi: 10.1021/ac9001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desmet G, Cabooter D, Gzil P, Verelst H, Mangelings D, Vander Heyden Y, Clicq D. J. Chromatogr. A. 2006;1130:158–166. doi: 10.1016/j.chroma.2006.05.082. [DOI] [PubMed] [Google Scholar]
- 6.Poppe H. J. Chromatogr. A. 1997;778:3–21. [Google Scholar]
- 7.Cabrera K, Wieland G, Lubda D, Nakanishi K, Soga N, Minakuchi H, Unger KK. Trac. 1998;17:50–53. [Google Scholar]
- 8.Petro M, Svec F, Gitsov I, Frechet JMJ. Anal. Chem. 1996;68:315–321. doi: 10.1021/ac950726r. [DOI] [PubMed] [Google Scholar]
- 9.Svec F. J. Chromatogr. A. 2010;1217:902–924. doi: 10.1016/j.chroma.2009.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lippert JA, Xin BM, Wu NJ, Lee ML. J. Microcolumn Sep. 1999;11:631–643. [Google Scholar]
- 11.MacNair JE, Lewis KC, Jorgenson JW. Anal. Chem. 1997;69:983–989. doi: 10.1021/ac961094r. [DOI] [PubMed] [Google Scholar]
- 12.MacNair JE, Patel KD, Jorgenson JW. Anal. Chem. 1999;71:700–708. doi: 10.1021/ac9807013. [DOI] [PubMed] [Google Scholar]
- 13.Swartz ME. J. Liq. Chromatogr. Related Technol. 2005;28:1253–1263. [Google Scholar]
- 14.DeStefano JJ, Langlois TJ, Kirkland JJ. J. Chromatogr. Sci. 2008;46:254–260. doi: 10.1093/chromsci/46.3.254. [DOI] [PubMed] [Google Scholar]
- 15.Wu NJ, Liu YS, Lee ML. J. Chromatogr. A. 2006;1131:142–150. doi: 10.1016/j.chroma.2006.07.042. [DOI] [PubMed] [Google Scholar]
- 16.Zhang SH, Zhang J, Horvath C. J. Chromatogr. A. 2001;914:189–200. doi: 10.1016/s0021-9673(00)01113-4. [DOI] [PubMed] [Google Scholar]
- 17.Gritti F, Leonardis I, Shock D, Stevenson P, Shalliker A, Guiochon G. J. Chromatogr. A. 2010;1217:1589–1603. doi: 10.1016/j.chroma.2009.12.079. [DOI] [PubMed] [Google Scholar]
- 18.Zhang SH, Zhang J, Horvath C. J. Chromatogr. A. 2002;965:83–92. doi: 10.1016/s0021-9673(01)01544-8. [DOI] [PubMed] [Google Scholar]
- 19.Park J, Lee D, Kim W, Horiike S, Nishimoto T, Lee SH, Ahn CH. Anal. Chem. 2007;79:3214–3219. doi: 10.1021/ac061714g. [DOI] [PubMed] [Google Scholar]
- 20.Qu QS, Peng SW, Mangelings D, Hu XY, Yan C. Electrophoresis. 2010;31:556–562. doi: 10.1002/elps.200900375. [DOI] [PubMed] [Google Scholar]
- 21.Zheng SP, Ross E, Legg MA, Wirth MJ. J. Am. Chem. Soc. 2006;128:9016–9017. doi: 10.1021/ja062676l. [DOI] [PubMed] [Google Scholar]
- 22.Zeng Y, Harrison DJ. Anal. Chem. 2007;79:2289–2295. doi: 10.1021/ac061931h. [DOI] [PubMed] [Google Scholar]
- 23.Zeng Y, He M, Harrison DJ. Angew. Chem.-Int. Edit. 2008;47:6388–6391. doi: 10.1002/anie.200800816. [DOI] [PubMed] [Google Scholar]
- 24.Malkin DS, Wei BC, Fogiel AJ, Staats SL, Wirth MJ. Anal. Chem. 2010;82:2175–2177. doi: 10.1021/ac100062t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fairbank RWP, Xiang Y, Wirth MJ. Anal. Chem. 1995;67:3879–3885. [Google Scholar]
- 26.Moore AW, Jorgenson JW. Anal. Chem. 1993;65:3550–3560. doi: 10.1021/ac00072a004. [DOI] [PubMed] [Google Scholar]
- 27.Gilges M, Kleemiss MH, Schomburg G. Anal. Chem. 1994;66:2038–2046. [Google Scholar]
- 28.Euerby MR, Gilligan D, Johnson CM, Roulin SCP, Myers P, Bartle KD. J. Microcolumn Sep. 1997;9:373–387. [Google Scholar]
- 29.Hilder EF, Svec F, Frechet JMJ. Electrophoresis. 2002;23:3934–3953. doi: 10.1002/elps.200290011. [DOI] [PubMed] [Google Scholar]
- 30.Newton MR, Morey KA, Zhang YH, Snow RJ, Diwekar M, Shi J, White HS. Nano Lett. 2004;4:875–880. [Google Scholar]
- 31.Karger BL, Chu YH, Foret F. Annu. Rev. Biophys. Biomol. Struct. 1995;24:579–610. doi: 10.1146/annurev.bb.24.060195.003051. [DOI] [PubMed] [Google Scholar]
- 32.Ding WL, Fritz JS. HRC-J. High Resolut. Chromatogr. 1997;20:575–580. [Google Scholar]
- 33.Brune D, Kim S. Proc. Natl. Acad. Sci. U. S. A. 1993;90:3835–3839. doi: 10.1073/pnas.90.9.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang XH, Gordon MJ, Zare RN. Anal. Chem. 1988;60:1837–1838. [Google Scholar]