Abstract
D-sites are a class of MAPK-docking sites that have been found in many MAPK regulators and substrates. A single functional, high affinity D-site has been identified near the N terminus of each of the MAPK kinases (MKKs or MEKs) MEK1, MEK2, MKK3, MKK4, and MKK6. Here we demonstrated that MKK7 recognizes its target JNK by a novel mechanism involving a partially cooperative interaction of three low affinity D-sites in the N-terminal domain of MKK7. Mutations of the conserved residues within any one of the three docking sites (D1, D2, and D3) disrupted the ability of the N-terminal domain of MKK7β to bind JNK1 by about 50–70%. Moreover, mutation of any two of the three D-sites reduced binding by about 80–90%, and mutation of all three reduced binding by 95%. Full-length MKK7 containing combined D1/D2 mutations was compromised for binding to JNK1 and exhibited reduced JNK1 kinase activity when compared with wild-type MKK7. Peptide versions of the D-sites from MKK4 or the JIP-1 scaffold protein inhibited MKK7-JNK binding, suggesting that all three JNK regulators bind to the same region of JNK. Moreover, peptide versions of any of the three D-sites of MKK7 inhibited the ability of JNK1 and JNK2 to phosphorylate their transcription factor substrates c-Jun and ATF2, suggesting that D-site-containing substrates also compete with MKK7 for docking to JNK. Finally, MKK7-derived D-site peptides exhibited selective inhibition of JNK1 versus ERK2. We conclude that MKK7 contains three JNK-docking sites that interact to selectively bind JNK and contribute to JNK signal transmission and specificity.
Mitogen-activated protein kinases (MAPKs)4 are essential components of eukaryotic signal transduction networks that enable cells to respond appropriately to extracellular signals and stresses. In mammalian organisms, the following four major MAPK cascades have been characterized: ERK1/2, ERK5, p38, and JNK pathways (1). Each cascade contains three sequentially acting protein kinases collectively known as a MAPK module (2). This module consists of a MAPK/ERK kinase kinase (MEKK or MAP3K), which activates a downstream MAPK/ERK kinase (MEK, MKK, or MAP2K) that subsequently activates a particular set of MAPKs (2). Activated MAPKs further propagate the signal by phosphorylating downstream targets such as transcription factors and other kinases. How this ubiquitous versatile module achieves specific coupling of signal to cellular response is an important and unresolved issue that is currently the subject of intense investigation (3–7).
The c-Jun N-terminal kinase (JNK) pathway responds primarily to extracellular stress such as UV radiation and cytokines such as interleukin-1 (8–11). Dependent upon such factors as cell type and the nature of the stimulus, JNK has been shown to promote either cell survival or apoptosis (12–14). Deregulation of the JNK pathway has been implicated in the pathogenesis of many human diseases such as cancer (15), obesity and diabetes (16), muscular dystrophy (17), arthritis (18), Alzheimer disease (19), and Parkinson disease (20). Inhibition of JNK activity is being considered as a possible therapy for many of these diseases (21–24).
The JNK family of MAPKs is encoded by the three genes JNK1, JNK2, and JNK3. JNK1 and JNK2 have ubiquitous expression profiles, whereas JNK3 is primarily found in neural tissue (9). The three JNK proteins are regulated by two MAPK kinases-MKK4 (also called JNKK1 or SEK1) and MKK7/JNKK2/SEK2 (25–30). Optimal activation of JNK requires the activity of both MKK4 and MKK7; although both MEKs are capable of dual phosphorylation of JNK at the activation loop residues Thr and Tyr, MKK4 prefers the Tyr and MKK7 prefers the Thr (31, 32). MKK4 is primarily activated by environmental stresses, whereas MKK7 is primarily activated by cytokines (10, 33).
Genetic studies in mice support a critical role for MKK7 in several aspects of cell and organismal physiology. MKK7 is essential for liver formation during embryogenesis; furthermore, loss of MKK7 in fibroblasts results in premature senescence and G2/M cell cycle arrest (34). In contrast, MKK7 is a negative regulator of cell growth in multiple hematopoietic lineages (35).
MAPK-docking sites are found in the N-terminal regulatory domains of many MKKs, where they contribute to accurate and efficient enzyme substrate recognition by promoting the formation of relatively stable, high affinity MKK·MAPK complexes (2). This paradigm of MAPK recognition was first established for the yeast MEK Ste7 (36–38) and has since been extended to mammalian MEK1 (38, 39), MEK2 (38), MKK3 and MKK6 (2), MKK4 (40), and MEK5 (41). The MAPK-docking sites in many MKKs, including yeast Ste7 and human MEK1/2, MKK3/6, and MKK4, share a core consensus sequence consisting of a cluster of about three basic residues, followed by a short spacer of 1–6 residues, and finally a hydrophobic-X-hydrophobic submotif (Fig. 1B) (37, 38). Hereafter, we shall refer to this class of MAPK-docking sites as “D-sites.” D-sites have also been found in MAPK scaffolds, phosphatases, and substrates (2, 42, 43).
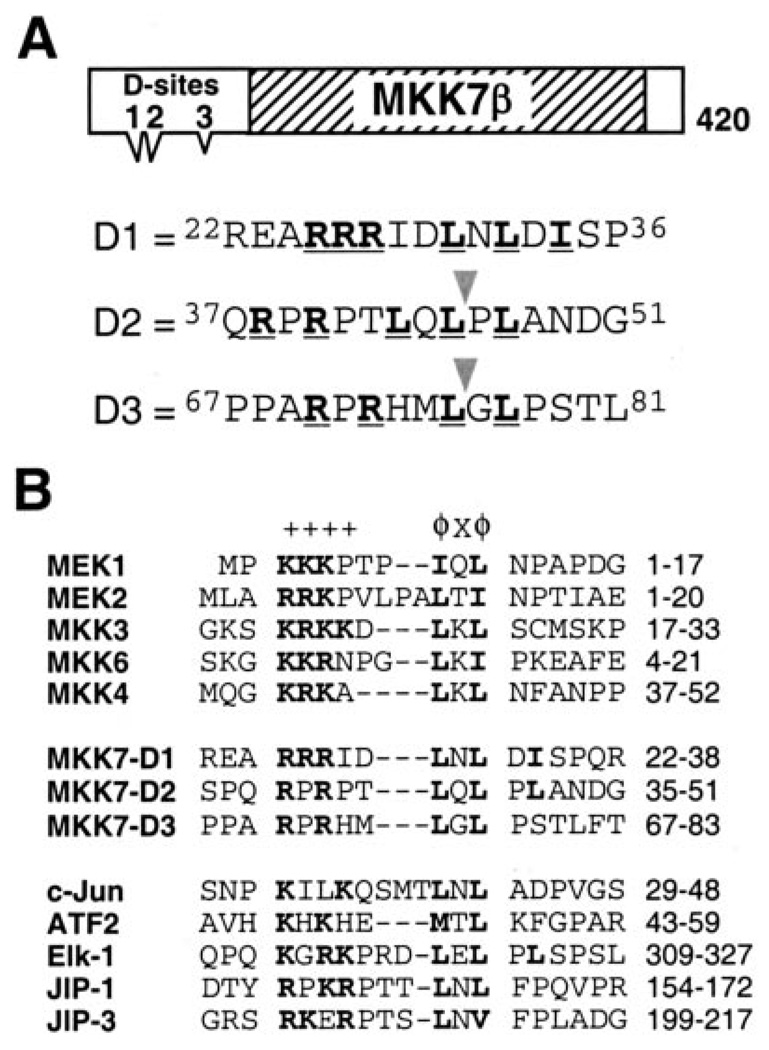
FIGURE 1. MKK7β contains three putative D-sites.
A, schematic of MKK7β1. The hatched box represents the catalytic domain; the three triangular protrusions represent the putative D-sites. Sequences of and around the putative D-sites are shown below; consensus-matching residues are boldface and underlined. The triangles indicate where anthrax lethal factor cleaves within D2 and D3 (49). B, known D-sites in JNK-binding proteins aligned with the putative D-sites in MKK7. Consensus basic (+) and hydrophobic (ϕ) residues are shown in boldface. Dashes were inserted to optimize the alignment; spaces are for visual clarity.
D-sites in MKKs are crucial for the activation of their cognate MAPKs in vivo (44, 45), where they function as portable, modular motifs that primarily serve to tether their cognate MAPKs near the kinase domain of MKKs (45). D-sites also display some selectivity in binding to MAPKs, suggesting a role in specificity (40, 46). For example, the D-sites in MEK1 and MEK2 do not bind effectively to JNK2 (40).
As noted above, functional D-sites have been characterized in MEK/MKK1–4 and MKK6. MEK5 does not contain a D-site but instead contains a novel MAPK-docking site of acidic character (41). No MAPK-docking site has heretofore been identified in MKK7; however, the N-terminal domain of the MKK7β isoform (the most prevalent isoform in humans) has been shown to be necessary and sufficient for high affinity complex formation with JNK1 (47, 48). Here we demonstrate that the N terminus of MKK7β contains three weak D-sites that interact in a partially additive, partially synergistic manner to create a high affinity JNK-docking platform.
EXPERIMENTAL PROCEDURES
Genes
The MKK7β1 clone used in this paper corresponds to Gen-Bank™ accession number NM_005043. Accession numbers of other genes used in this study have been given in an earlier work (40).
Proteins and Antisera
Fusions of glutathione S-transferase (GST) to human c-Jun-(1–89), ATF2-(19–96), and Elk-1-(307–428) were purchased from Cell Signaling Technology. Activated JNK1β1 and JNK2β2 and unactivated JNK1β1 were purchased from Upstate Cell Signaling Solutions. Activated mouse ERK2 was purchased from New England Biolabs. Anti-FLAG M2 monoclonal and anti-FLAG polyclonal antibodies were purchased from Sigma. Anti-V5 was purchased from Invitrogen.
Plasmids for in Vitro Transcription and Translation
Plasmids pGEM-MKK7β1 (48) and pGEM-MKK4 (40) have been described previously. To construct pGEM-JNK1, the JNK1α1 coding region was amplified by high fidelity PCR using Pfu DNA polymerase, primers JNK1α1up and JNK1α1down1 (see Table 1), and Genestorm clone ID RG000191 (Invitrogen) as the template. The PCR product was digested with BamHI and SalI and inserted into the corresponding sites in pGEM4Z (Promega).
TABLE 1. Oligonucleotides used in this study.
Where appropriate, restriction sites used for subcloning are underlined, and the translation initiation codon is shown in boldface.
| Name | Sequence (5′ → 3′) | Use |
|---|---|---|
| JB812B | GCGGGATCCACCATGGCGGCGTCCTCCCTG | pGEX-MKK7 (1–38, 1–60, 1–85), pGEM-MKK7 |
| JB818 | CCGCTCGACCTACCTGAAGAAGGGGCGGTG | pGEM-MKK7 |
| MKK7-(1–38)-R | GGCGTCGACTCACCGCTGGGGGCTGATATCCAG | pGEX-MKK7-(1–38) |
| MKK7-(1–60)-R | GGCGTCGACGCTCTCTGAGGATGGCGAGCGG | pGEX-MKK7-(1–60) |
| MKK7-(1–85)-R | GGCGTCGACGCGGGGTGTGAACAGGGTTG | pGEX-MKK7-(1–85) |
| M7For-(35–49) | GATCCAGCCCACAGCGGCCCAGGCCCACCCTGCAGCTCCCGCTGGCCAACG | pGEX-MKK7-D2 |
| MKK7Rev-(35–49) | TCGACGTTGGCCAGCGGGAGCTGCAGGGTGGGCCTGGGCCGCTGTGGGCTG | pGEX-MKK7-D2 |
| MKK7For-(67–80) | GATCCCTCCAGCTCGACCTCGACACATGCTGGGACTCCCTTCAACCTGAG | pGEX-MKK7-D3 |
| MKK7Rev-(67–80) | TCGACTCAGGTTGAAGGGAGTCCCAGCATCTGTCGAGGTCGAGCTGGAGGG | pGEX-MKK7-D3 |
| MKK4For-(36–49) | GATCCAGCATGCAGGGTAAACGCAAAGCACTGAAGTTGAATTTTGCAG | pGEX-MKK4-Ds |
| MKK4Rev-(36–49) | TCGACTGCAAAATTCAACTTCAGTGCTTTGCGTTTACCCTGCATGCTG | pGEX-MKK4-Ds |
| MKK7D1mutFor | GAGAACCGGGAGGCCGAGGAGGAGATCGACGCCAACGCGGATATCAGCCCGCAGCGGCCCAGG | pGEX-MKK7 D1 mutants, pcDNA-MKK7-D12-FLAG |
| MKK7D1mutRev | CCTGGGCCGCTCGGGCTGATATCCGCGTTGGCGTCGATCTCCTCCTCGGCCTCCCGGTTCTC | pGEX-MKK7 D1 mutants, pcDNA-MKK7-D12-FLAG |
| MKK7D2mutFor | CCTCAACCTGGATATCAGCCCGCAGGAGCCCGAGCCCACCGCGCAGGCCCGGCTGGCCAACGATGGG | pGEX-MKK7 D2 mutants pcDNA-MKK7-D12-FLAG |
| MKK7D2mutRev | CCCATCGTTGGCCAGCCGGGCCTGCGCGGTGGGCTCGGGCTCCTGCGGGCTGATATCCAGGTTGAGG | pGEX-MKK7 D2 mutants pcDNA-MKK7-D12-FLAG |
| MKK7D3mutFor | CCCGCAGCACCCGACGCCACCCGCCGAGCCCGAACACATGGCGGGCCTCCCGTCAACCCTGTTCACAC | pGEX-MKK7D3 mutants |
| MKK7D3mutRev | GTGTGAACAGGGTTGACGGGAGGCCCGCCATGTGTTCGGGCTCGGCGGGTGGCGTCGGGTGCTGCGGG | pGEX-MKK7D3 mutants |
| MKK7D2mutFixF | GCCAACGCGGATATCAGCCCGCAGGAGCCCGAGCCCACCGCGCAGGCC | pGEX-MKK7 D12 mutants pcDNA-MKK7-D12-FLAG |
| MKK7D2mutFixR | GGCCTGCGCGGTGGGCTCGGGCTCCTGCGGGCTGATATCCGCGTTGGC | pGEX-MKK7 D12 mutants pcDNA-MKK7-D12-FLAG |
| MKK7KDFor2 | CAGGCCACATCATTGCTGTTGCGCAAATGCGGCGCTCTGGGAAC | pcDNA-MKK7-KD-FLAG |
| MKK7KDRev2 | GTTCCCAGAGCGCCGCATTTGCGCAACAGCAATGATGTGGCCTG | pcDNA-MKK7-KD-FLAG |
Plasmids for the Production of GST Fusion Proteins
The vector used for generating the GST fusion proteins was pGEXLB (38), a derivative of pGEX-4T-1 (Amersham Biosciences). In pGEXLB, an encoded Pro residue is replaced with a Gly-Gly-Gly-Gly-Gly-Ser-Gly sequence to promote the independent functioning of the GST and fusion moieties. The cloning of pGEXLB-JNK1α1 was described previously (48). To generate GST-MKK7-(1–85), GST-MKK7-(1–60), and GST-MKK7-(1–38), PCR was used to amplify the specific fragments and introduce a BamHI at the N terminus and a SalI site at the C terminus. The primers used are shown in Table 1; a human MKK7β1 cDNA clone (48) was used as the template. The PCR products were digested with BamHI and SalI and subsequently inserted into the appropriate sites on pGEX-LB. To generate the plasmids for GST fused to the independent MKK7β-docking sites (see Fig. 3), an adaptor oligonucleotide approach was used. pGEX-LB was cut with BamHI and SalI, and the polylinker insert was removed. The excised fragment was replaced by annealed oligonucleotide pairs that encoded the independent docking sites. To encode MKK7-D2, the oligonucleotide pair used was MKK7For 35–49 and MKK7Rev 35–49; for MKK7-D3, MKK7For 67–80 and MKK7Rev 67–80; for the D-site from MKK4, MKK4For 36–49 and MKK4Rev 36–49 (Table 1).
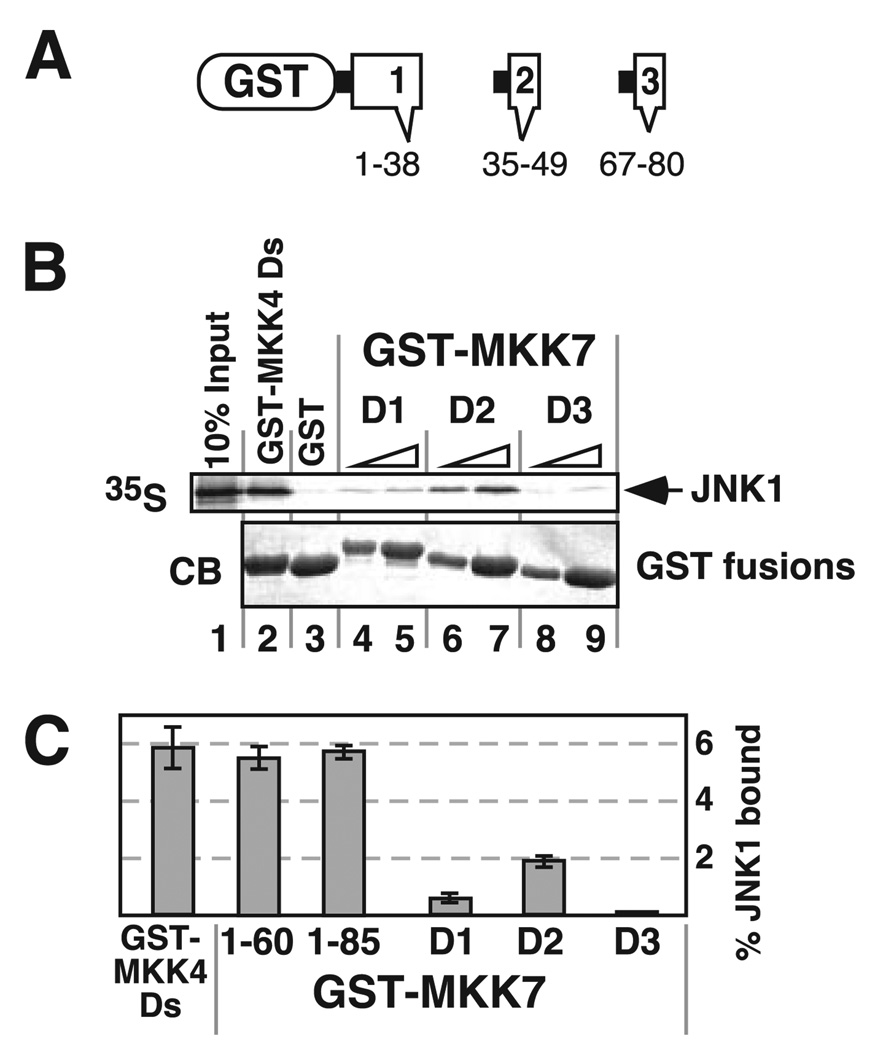
FIGURE 3. Isolated D-sites from MKK7 display weak JNK1 binding activity.
A, fusions between GST and D1, D2, or D3 were constructed and tested individually for binding to JNK1. The MKK7 residues included in each fusion are shown. B, JNK1 was tested for binding to 40 µg of GST-MKK4-(36–49) (lane 2), 40 µg of GST (lane 3), and 10 or 40 µg of GST-MKK7-D1 (lanes 4 and 5), GST-MKK7-D2 (lanes 6 and 7), or GST-MKK7-D3 (lanes 8 and 9). Other details are as in Fig. 2. C, quantification of JNK1 binding to various D-sites containing fusion proteins, using 40 µg of GST fusion per reaction. Standard error bars are shown (n = 5).
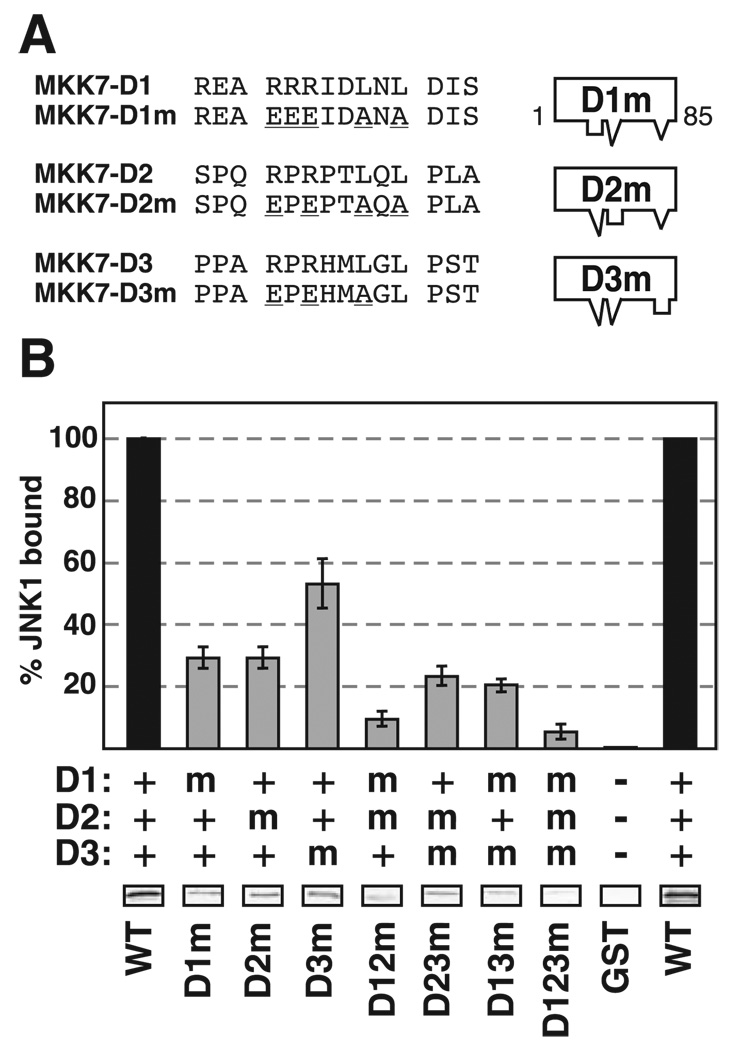
Site-directed mutagenesis (Quickchange, Stratagene) was used to generate the GST-MKK7-(1–85)-docking site mutant constructs used in Fig. 4. The template used in these mutagenesis reactions was pGEM-MKK7-(1–85). The following complement primers were used to mutate each respective docking site as follows: for D1, MKK7D1mutFor and MKK7D1mutRev; for D2, MKK7D2mutFor and MKK7D2mutRev; and for D3, MKK7D3mutFor and MKK7D3mutRev (Table 1). The double and triple D-site mutants were generated by mutating one D-site at a time. The D12 and D123 mutants required an additional primer pair to correct the return to wild-type caused by the D1 mutation primers. The accuracy of all mutant constructs was verified by sequencing.
FIGURE 4. Mutation of D-sites of MKK7.
A, D1, D2, and D3 were mutated as shown, either alone or in combination, in the context of GST-MKK7-(1–85). The mutations are underlined. B, quantification of binding of MKK7 D-site mutants (40 µg) to JNK1, normalized to the binding of wild-type GST-MKK7-(1–85). Standard error bars are shown (n = 5). The key shows whether the indicated D-sites were wild-type (WT) (+), mutated (m), or absent (−). Shown below the graph are representative data bands showing the amount of 35S-JNK1 co-sedimented with the indicated GST-MKK7 mutant in a typical experiment.
Plasmids for Tissue Culture Transfections
To generate pcDNA3.1-MKK7β1-FLAG (“MKK7-FLAG”), containing full-lengthMKK7β1 tagged at its C terminus with the FLAG epitope, the MKK7β1 coding sequence was inserted into the pcDNA3.1/FLAG vector using the HindIII and KpnI sites. MKK7-KD-FLAG, a catalytically inactive (“kinase dead”) mutant (K149A) of MKK7-FLAG, was created by site-directed mutagenesis using primers MKK7KDFor2 and MKK7KDRev2. MKK7-D12-FLAG, the D1/D2 double mutant of MKK7-FLAG, was created using the same primers and site-directed mutagenesis procedure used to make the MKK7-(1–85)-D12 mutant described above. Mutant constructs were confirmed by sequencing the full-length coding sequence. Plasmid pcDNA3.1-JNK1α1-V5-His, encoding C-terminally epitope-tagged JNK1, was obtained from the Invitrogen.
Transcription and Translation in Vitro
Proteins labeled with [35S]methionine were produced by coupled transcription and translation reactions, partially purified by ammonium sulfate precipitation, and quantified as described previously (38).
Binding Assays
GST fusion proteins were expressed in bacteria and purified by affinity chromatography using glutathione-Sepharose (Amersham Biosciences) and quantified as described elsewhere (38). Binding assays were performed, analyzed, and quantified as described previously (38). Dissociation constant (Kd) estimates are calculated from multiple replicate experiments (38, 40); an example is shown in Table 2.
TABLE 2. Binding assay data for MKK7-(1–85)-JNK1 interaction.
| Experiment a | Binding b | Kdc | |
|---|---|---|---|
| % | µ m | ||
| A-1 | 6.4 | 83 | |
| A-2 | 5.4 | 99 | |
| A-3 | 5.3 | 101 | |
| A-4 | 5.0 | 106 | |
| A-5 | 5.6 | 95 | |
| A-6 | 7.3 | 72 | |
| Mean | 93 | ||
| S.D. | 13 | ||
| S.E. | 6 |
Binding reactions (200 µl) contained ~1 pmol (~5 nm) of 35S-labeled, in vitro translated JNK1 and 40 µg (5.6 µm) of GST-MKK7-(1–85) fusion protein.
Percent of the input 35S-labeled protein that bound to the GST fusion protein.
Peptides
The soluble peptides used for the binding competition and kinase inhibition experiments were synthesized by United Biochemical Research Inc. Peptide sequences are shown in Table 3.
TABLE 3. Peptides used in this study.
Residues mutated to make the MKK4EAG and MEK2EEAA control peptides are underlined.
| Name | Sequence (NH2 → COOH) | Residues of protein |
|---|---|---|
| MKK4 | MQGKRKALKLNFANPP | 37–52 |
| MKK4EAG | MQGEAKALKGNFANPP | |
| JIP-1 | YRPKRPTTLNLF | 152–163 |
| MKK7-D1 | REARRRIDLNLDISP | 22–36 |
| MKK7-D2 | QRPRPTLQLPLANDG | 37–51 |
| MKK7-D3 | PPARPRHMLGLPSTLFT | 67–83 |
| MEK1 | MPKKKPTPIQLNPAPDG | 1–17 |
| MEK2 | MLARRKPVLPALTINPTIAE | 1–20 |
| MEK2EEAA | MLAEEKPVLPAATANPTIAE |
Protein Kinase Assays
The protein kinase assays for ERK2 phosphorylation of Elk-1 (40) and for JNK2 phosphorylation of c-Jun or ATF2 (40) have been described previously. Kinase reactions (20 µl) for JNK1 phosphorylation of c-Jun or ATF2 contained kinase assay buffer (50mm Tris-HCl (pH 7.5), 10 mm MgCl2, 1 mm EGTA, and 2 mm dithiothreitol), 1 µm (740 ng) GST-c-Jun, or 1 µm (700 ng) GST-ATF2, 0.8 milliunits (2.6 ng) of active JNK1, 50 µm ATP, 1 µCi of [γ-32P]ATP, and the indicated concentration of peptide. Reactions were for 20 min at 30 °C. Substrate phosphorylation was quantified by SDS-PAGE (12% gels), followed by analysis of relative incorporation using the PhosphorImager.
Tissue Culture and Transfections
HEK293 cells were cultured using Dulbecco’s modified Eagle’s medium enriched with 10% heat-inactivated fetal bovine serum (Invitrogen), penicillin, streptomycin, and sodium bicarbonate. The cells were seeded at a density of 5 × 105 cells per well in a 6-well dish. The culture was maintained in a humidified environment at 37 °C and 5% CO2. Transient transfections were performed with Lipofectamine (Invitrogen) following the manufacturer’s recommended procedures. Cells were harvested 48 h after transfection.
Co-immunoprecipitation Assay
HEK293 cells were transfected with 1 mg of MKK7-FLAG, MKK7-D12-FLAG, or pcDNA3.1/FLAG plasmid DNA and co-transfected with 1 µg of pcDNA3.1-JNK1α1-V5-His plasmid DNA. At 48 h post-transfection, cells from two 35-mm wells were lysed into 200 µl of lysis buffer (50 mm Hepes (pH 7.6), 150 mm NaCl, 1.5 mm MgCl2, 1 mm EDTA, 1% Triton X-100, 10% glycerol, 1 mm sodium orthovanadate, 25 mm β-glycerophosphate, 1× protease inhibitor mixture (Sigma)) and centrifuged at 14,000 × g for 15 min at 4 °C. The supernatant was then cleared with 20 µl of a 50% slurry of Protein G Plus/Protein A-agarose beads for 30 min at 4 °C. The cleared lysates were then incubated for 1 h at 4 °C with 20 µl of beads (50% slurry) that had been preincubated with 1 µl of anti-V5 antibody for 30 min at 4 °C. The beads were washed twice with 0.5 ml of wash buffer (20 mm Hepes (pH 7.6), 150 mm NaCl, 0.1% Triton X-100, 10% glycerol) and resuspended in SDS sample buffer.
Immunoprecipitation Kinase Assays
HEK293 cells were transfected with 1 µg of plasmid DNA encoding either FLAG-tagged wild-type MKK7, docking site mutated MKK7, kinase-dead MKK7, or empty vector. After 48 h, the cells were serum-starved for 30 min and then treated with anisomycin (20 µg/ml) and IL-1 (5 ng/ml) for another 30 min. Cells were lysed, and MKK7-FLAG derivatives were immunoprecipitated as above. The immunoprecipitation complexes were then washed twice with wash buffer and once with kinase buffer (50 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 1 mm EGTA, and 2 mm dithiothreitol). The activity of MKK7 was determined in a reaction at 30 °C for 30 min in 40 µl of kinase buffer containing 0.5 µg of unactivated JNK1, 2 µg of GST-c-Jun, 50 µm ATP, and 1 µCi of [γ-32P]ATP. The reactions were terminated by the addition of SDS sample buffer, resolved by SDS-PAGE, detected by immunoblot and autoradiography, and quantified on a PhosphorImager.
RESULTS
Three Putative Docking Sites in the N Terminus of MKK7β
Because all other human MKKs had been shown to contain MAPK-docking sites near their N termini, we inspected the amino acid sequence of MKK7 for potential D-sites. Three putative D-sites (hereafter D1, D2, and D3) were found in the N-terminal domain of MKK7β (Fig. 1A). All three conform to the D-site consensus, which consists of a basic submotif separated by a short spacer from a hydrophobic-X-hydrophobic submotif (Fig. 1B). Furthermore, all three have detectable similarity to identified D-sites in other MKKs and to D-sites found in JNK substrates such as the transcription factors ATF2, c-Jun, and Elk-1 and JNK scaffold proteins such as JIP-1 and JIP-3 (Fig. 1B).
The D-sites in MEK1, MEK2, MKK3, MKK4, and MKK6 have been shown to be cleaved by the anthrax lethal factor protease (48, 49). In addition, MKK4 contains a second lethal factor cleavage site that is not a functional high affinity MAPK-docking site (40, 49). MKK7 has also been shown to be cleaved by lethal factor at two sites in its N terminus (49). These correspond to putative D-sites D2 and D3 (Fig. 1A).
The N Terminus of MKKβ Binds to JNK
To confirm that the MKK7β N terminus was sufficient to bind JNK, and to evaluate whether systematic removal of the putative docking sites would compromise binding, fusions of GST to portions of the N terminus of MKK7 were tested for their ability to bind in vitro to 35S-labeled JNK1 (Fig. 2A). We used JNK1 because our previous studies showed that JNK1 exhibits substantially stronger binding to MKK7 than JNK2 (48). As shown in Fig. 2B, MKK7-(1–85), which contains all three putative D-sites, bound efficiently to JNK1 (Kd ~ 90 µm). The GST moiety by itself bound only trace amounts of JNK1, demonstrating that the observed interaction was specific to the MKK7 constituent. MKK7-(1–60), which lacks D3, exhibited only slightly decreased JNK1 binding. In contrast, MKK7-(1–38), which contains only D1, exhibited an ~7-fold decrease in JNK1 binding (Fig. 2, B and C). Taken at face value, these results imply that some sequence between residues 38 and 60 (perhaps the second putative D-site, D2) was responsible for the ability of theMKK7β N terminus to bind JNK. However, additional experiments, presented below, suggested that an unexpected, more complicated mechanism mediated JNK-MKK7 docking.
FIGURE 2. Localization of the JNK-binding region in the N-terminal domain of MKK7.
A, GST-MKK7β1 N-terminal truncations tested for JNK1 binding. GST-MKK7-(1–85) contains all three D-sites, 1–60 contains the first two, and 1–38 contains only the first. 35S-Labeled JNK1α1 was produced by in vitro translation; GST pull-down assays were used to test the binding between the truncations and JNK1α1. B, JNK1 (~1 pmol) was tested for binding to 10 and 40 µg of GST (lanes 2 and 3), GST-MKK7-(1–85) (lanes 4 and 5), GST-MKK7-(1–60) (lanes 6 and 7), or GST-MKK7-(1–38) (lanes 8 and 9). Lane 1 shows 10% of the total JNK1 input. The lower panel shows Coomassie Blue (CB) staining of the sedimented GST fusion proteins. C, quantification of JNK1-MKK7 binding, using 40 µg of GST-MKK7 truncations per reaction. Standard error bars are shown (n = 8).
None of the Three Putative D-sites Are Sufficient for High Affinity JNK Binding
To determine whether any of the D-sites were sufficient for high affinity JNK binding, we constructed GST fusions to each of them. For example, GST was fused to MKK7 residues 35–49, yielding GST-D2 (Fig. 3A). The individual fusions were then assessed for their ability to bind JNK1. When tested separately in this manner, neither D2 nor the other two putative D-sites bound to JNK1 nearly as strongly as MKK7-(1–85) or MKK7-(1–60). Of the three D-sites, D2 exhibited the strongest JNK binding followed by D1; D3 did not detectably bind JNK above background (Fig. 3, B and C). In contrast, the isolated D-site from MKK4 (MKK4-(36–49)) bound reasonably well to JNK1 when tested in the same assay (Fig. 3, B and C). MKK4-(36–49) site bound to JNK1 with about the same efficiency as did MKK7-(1–85) or MKK7-(1–60), but much more efficiently than did any of the isolated D-sites from MKK7. These results suggested that, unlike the D-site from MKK4, none of the individual D-sites from MKK7 were sufficient for high affinity JNK binding. It was intriguing, however, that MKK7-(1–60), which contains both D1 and D2, bound to JNK1 with an affinity comparable with the single MKK4 D-site; this suggested that D1 and D2 might collectively comprise a relatively high affinity JNK-binding moiety.
The Three D-sites Interact to Promote High Affinity JNK Binding
To ascertain if any of the D-sites were necessary for high affinity JNK binding, substitution mutations were introduced into D1, D2, and D3, either alone or in combination, in the context of MKK7-(1–85). The substitutions replaced conserved residues in the basic and hydrophobic submotif with nonconsensus residues (Fig. 4A). The ability of the mutant proteins to bind to JNK1 was then compared with wild-type MKK7-(1–85). As shown in Fig. 4B, mutation of either D1 or D2 resulted in an approximate 70% decrease in JNK binding, whereas mutation of D3 resulted in less than a 50% decrease in JNK1 binding. Strikingly, the double mutants displayed an even stronger binding defect than any of the single mutants; in particular, the D1/D2 mutant showed a 90% decrease in binding when compared with the wild type. Finally, the D1/D2/D3 triple mutant almost completely lost the ability to bind JNK. These results demonstrate that the three D-sites (particularly D1 and D2) jointly contribute to the binding of MKK7β’s N terminus to JNK.
The D-sites of MKK7 Are Critical for Interaction and Activation of JNK1 in Cell-based Assays
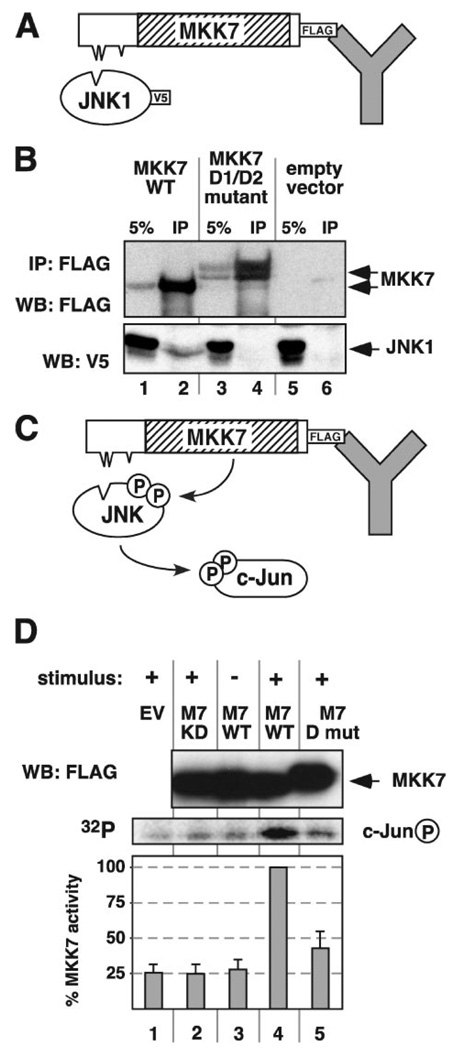
To examine the importance of D-site-mediated MKK7-JNK binding for MKK7 function in vivo, the D1/D2 double mutant used in Fig. 4 was introduced into the context of full-length of MKK7β that was tagged at its C terminus with the FLAG epitope. The resulting mutant protein was expressed in human kidney epithelial (HEK293) cells and compared with wild-type, FLAG-tagged MKK7β for its ability to bind to co-overexpressed JNK1, as assessed by co-immunoprecipitation (Fig. 5A). When wild-type MKK7β was immunoprecipitated with the anti-FLAG antibody, a detectable amount of JNK1 co-immunoprecipitated with it (Fig. 5B, lane 2). In contrast, the double D-site mutation disrupted the ability of MKK7 to bind to co-expressed JNK1 (Fig. 5B, lane 4).
FIGURE 5. A D1/D2 double mutant of MKK7 is defective for JNK1 activation.
A, D1/D2 double mutant of MKK7 is defective for JNK binding in vivo. HEK293 cells were co-transfected with plasmid DNA encoding V5-tagged JNK1 and with either empty vector, FLAG-tagged wild-type MKK7, or a FLAG-tagged D1/D2 double docking site mutant of MKK7. Cells were lysed 48 h post-transfection, and MKK7 was immunoprecipitated using an anti-FLAG antibody. B, immunoprecipitated complexes (IP) along with 5% of the input lysate (5%) were analyzed by immunoblotting using antibodies against the FLAG and V5 tags. WB, Western blot. C and D, D1/D2 double mutant of MKK7 is defective for JNK activation. HEK293 cells were transfected with either empty vector (EV, lane 1) or one of three FLAG-tagged MKK7 derivatives: catalytically inactive kinase-dead MKK7 (KD, lane 2), wild-type MKK7 (WT, lanes 3 and 4), or the MKK7 D1/D2 double mutant (D mut, lane 5). 48 h post-transfection, the cells were stimulated (+) or not (−) with both anisomycin (20 µg/ml) and interleukin-1 (10 ng/ml) and lysed, and the MKK7 derivatives were immunoprecipitated with anti-FLAG antibodies. Purified unactivated JNK1 and GST-c-Jun were added to the immunoprecipitated pellets, and the MKK7-mediated activation of JNK1 was assessed by the ability of JNK1 to subsequently phosphorylate GST-c-Jun. D, top panel, representative immunoblot of MKK7 immunoprecipitates with anti-FLAG antibodies; middle panel, representative PhosphorImager image showing c-Jun phosphorylation; bottom panel, the incorporation of radioactive phosphate into GST-c-Jun in four independent experiments was quantified and averaged; standard error bars are shown.
To assess the ability of the D1/D2 double mutant to phosphorylate and activate JNK1, wild-type or mutant MKK7 was expressed in HEK293 cells, immunoprecipitated, and mixed with purified JNK1 and c-Jun proteins in a coupled kinase assay (Fig. 5C). This assay measures the ability of MKK7 to phosphorylate and thereby activate JNK; the activated JNK then phosphorylates c-Jun, which provides the readout. As a negative control for MKK7 kinase activity, a kinase-dead mutant was generated by mutating the critical lysine residue in the ATP-binding pocket to alanine. When compared with wild-type MKK7, the kinase activity of the D-site double mutant was reduced by over 2-fold (Fig. 5D) to a level that did not greatly exceed the activity of the kinase-dead mutant. Hence, the collective functions of D-sites 1 and 2 ofMKK7 are important for the phosphorylation and activation of JNK1.
The D-sites in JIP-1 and MKK4 Compete with MKK7 for JNK Binding
The results of the previous experiment strongly suggested that the ability of MKK7-(1–85) to bind JNK can be attributed to D-site-mediated interactions. To substantiate this idea further, we used a peptide competition approach to ask if MKK7β bound to the same regions of JNK1 as the D-sites of MKK4 and JIP-1 (Fig. 6A). Full-length, radiolabeled MKK7β was produced by in vitro translation, and its ability to co-sediment with GST-JNK1 bound to glutathione-Sepharose beads was tested in the presence or absence of D-site peptides derived from MKK4, JIP-1, or MKK7 itself (see Table 3 for peptide sequences). Previously, we used this approach to show that the D-sites of JIP-1 and MKK4 compete for interaction with JNK (40).
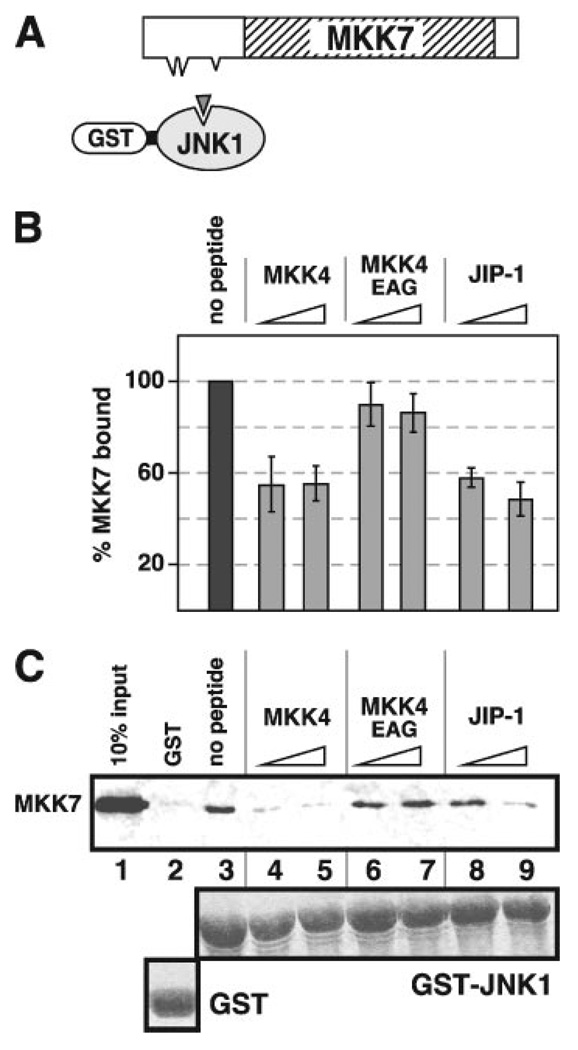
FIGURE 6. Inhibition of MKK7 binding to JNK1 by MKK4 and JIP-1 D-site peptides.
A, D-site peptides (triangle) were used to inhibit the ability of MKK7 to bind to GST-JNK1. B, quantification of peptide inhibition data. Shown is the average percent binding of MKK7 to GST-JNK1 (40 µg), normalized by setting the binding of the “no peptide” point to 100%. Standard error bars are shown (n = 4). C, 35S-labeled MKK7 was tested for binding to 40 µg of either GST or GST-JNK1. Lane 1 shows a 10% input of the MKK7 protein. Synthetic peptides (25 and 100 µm) were added to the indicated reactions to inhibit binding of MKK7 to GST-JNK1. The lower panel shows Coomassie Blue (CB) staining of the sedimented GST fusion proteins.
As shown in Fig. 6, B and C, in the absence of any added peptide, full-length MKK7β bound to GST-JNK1 (lane 3) but not to GST alone (lane 2). The addition of either the D-site peptide from MKK4 (Fig. 6C, lanes 4 and 5) or from JIP-1 (lanes 6 and 7) significantly reduced MKK7-JNK1 binding. This inhibition appeared to plateau around 50%. The lack of further inhibition may be due to relatively strong binding of the kinase domain of MKK7 to JNK, as we have observed previously that MKK7β-(77–419), which lacks all three putative D-sites, binds to GST-JNK1 at 40% of the level of full-length, wild-type MKK7β, presumably because of contacts between JNK1 and the kinase domain of MKK7 (48). In contrast to the relatively strong inhibitory potency of the MKK4 and JIP-1 D-site peptides, the MKK7-D2 peptide was a weak inhibitor, and the MKK7-D1 and -D3 peptides did not show any significant inhibition (data not shown). As expected, the negative control MKK4EAG peptide (which carries substitutions in three residues that are critical for JNK binding, see Table 3) also did not inhibit. These results indicate that MKK7 makes contacts with regions of JNK that at least partially overlap with the regions that are contacted by the MKK4 and JIP-1 D-sites.
D-site Peptides from MKK7 Inhibit JNK1/2 Phosphorylation of c-Jun and ATF2
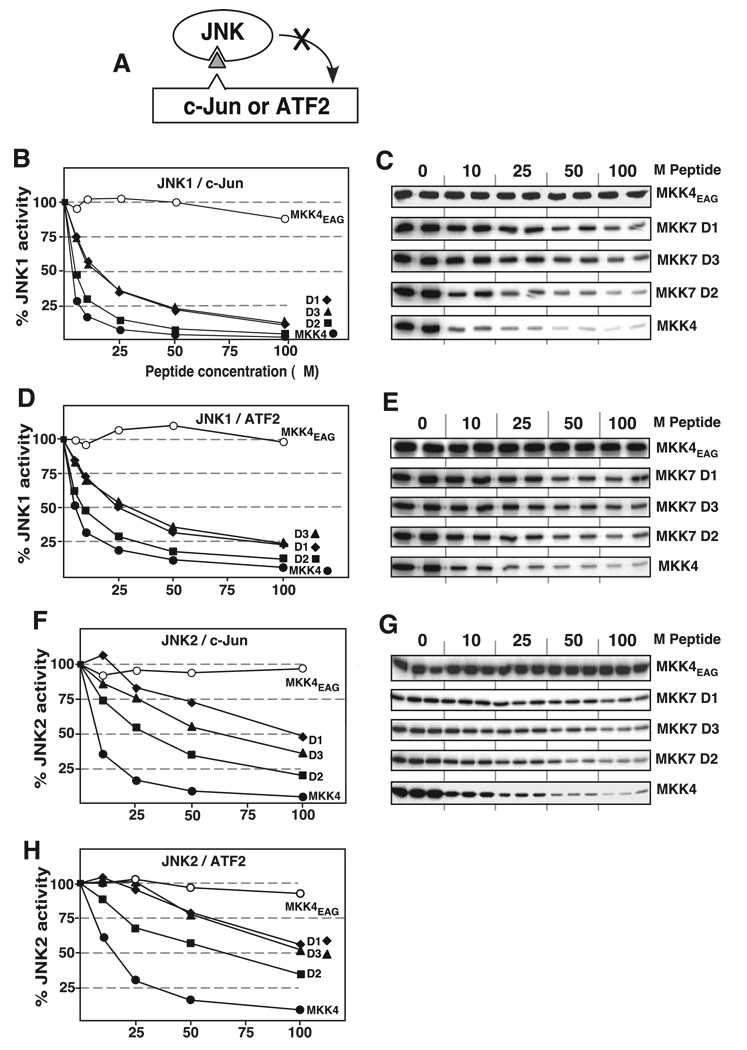
Previously, we demonstrated that the D-site peptide derived from MKK4 strongly inhibited the ability of JNK2 to phosphorylate the downstream transcription factors c-Jun and ATF2, providing evidence that the D-site of MKK4 was competing with the known c-Jun and ATF2 D-sites for binding to JNK (40). To assess the possibility that MKK7 competes with D-site-containing JNK substrates for JNK binding, and to measure the relative potency of the D-site peptides from MKK7, these peptides were evaluated for their ability to inhibit the phosphorylation of c-Jun or ATF2 by purified and active JNK1 or JNK2 (Fig. 7A).
FIGURE 7. Inhibition of JNK phosphorylation of transcription factors c-Jun and ATF2 with D-site peptides from MKK7.
A, D-site peptides (triangle) were used to inhibit JNK1 or JNK2 phosphorylation of c-Jun or ATF2. B and C, purified GST-c-Jun (1 µm) was incubated with purified active JNK1 (2.5 nm) and [γ-32P]ATP for 20 min in the absence or presence of the specific concentrations of the indicated peptides (see Table 3). B, results are plotted as percent phosphorylation relative to that observed in the absence of any added peptide. Phosphate incorporation into c-Jun was analyzed by SDS-PAGE and quantified on a PhosphorImager. Data are the average of at least two experiments, with duplicate or triplicate data points in each experiment. C, shown is an autoradiogram of a representative experiment. D and E, as for B and C, respectively, with the exception that the substrate was purified GST-ATF2 (1 µm). F and G, as for B and C, except the kinase was JNK2 (5 nm). H, as for D, except the kinase was JNK2.
As shown in Fig. 7, the D-site peptide derived from MKK4, used as a positive control, strongly inhibited JNK1 phosphorylation of c-Jun in a dose-dependent manner (IC50 ~ 5 µm), as shown previously for JNK2 (40). Also as demonstrated previously for JNK2, the MKK4EAG mutant peptide, used as a negative control, did not inhibit JNK1. All three of the D-site peptides from MKK7 inhibited the ability of JNK1 to phosphorylate c-Jun (Fig. 7, B and C). TheD2 peptide was the strongest inhibitor of the three (IC50 ~ 6 µm), whereas D1 and D3 were somewhat weaker (IC50 ~ 15 µm). Similar trends of inhibition were seen when the peptides were evaluated for the ability to inhibit JNK1 phosphorylation of ATF2 (Fig. 7, D and E). Again, MKK7-D2 was the strongest inhibitor of the three, but it was not as effective as the D-site peptide from MKK4. These results are consistent with the binding data shown in Fig. 3, where D2 also exhibited the highest binding efficiency of the three isolated D-sites from MKK7.
Compared with their potency in inhibiting JNK1, the ability of the MKK7-derived D-site peptides to inhibit JNK2 phosphorylation of c-Jun or ATF2 was about 5-fold lower (Fig. 7, F, G, and H); this is consistent with our previous data demonstrating a considerable preference (7–20-fold) of MKK7 for binding to JNK1 or JNK3 over JNK2 (48). As with JNK1, D2 was the strongest JNK2 inhibitor of the three D-site peptides from MKK7.
Specificity of the D-sites from MEK1/2 and MKK7
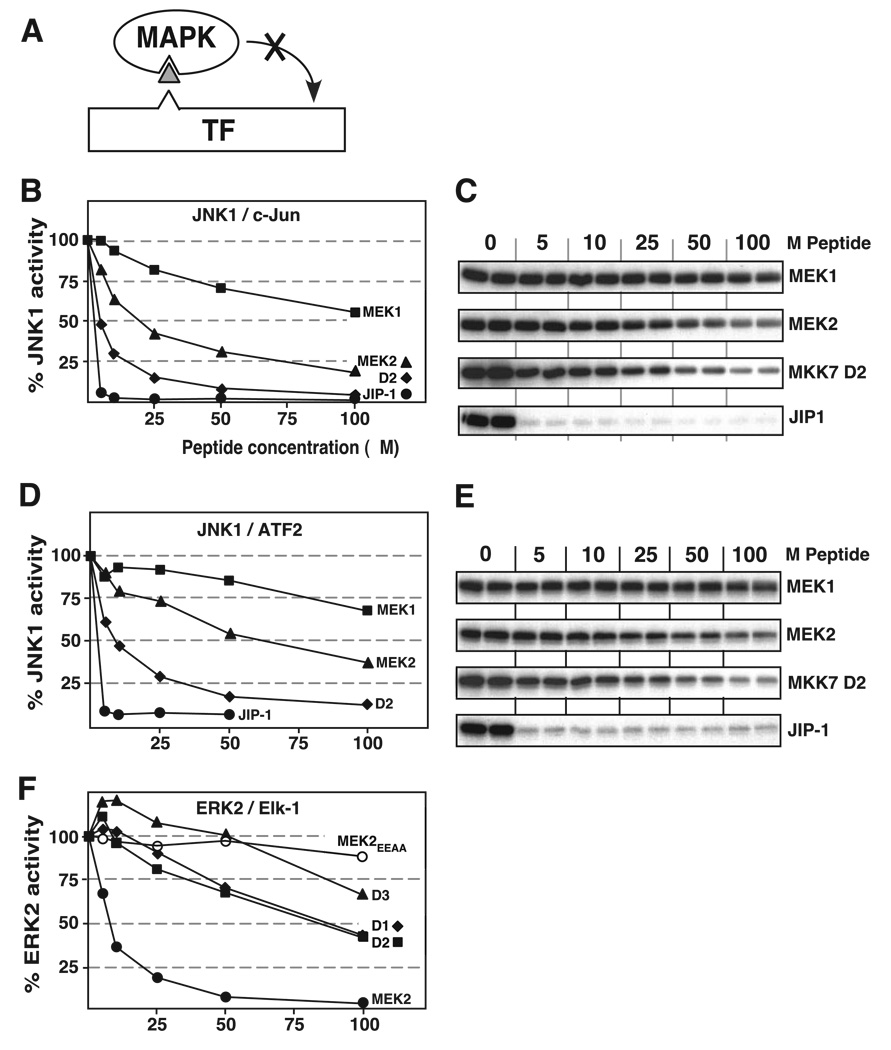
Previously we showed that D-site peptides derived from MEK1 and MEK2 were poor inhibitors of JNK2 but effective inhibitors of their cognate MAPK ERK2 (40). Conversely, the MKK4-derived D-site peptide was a better inhibitor of JNK2 than of ERK2 (40). Thus, these D-site peptides exhibited specificity, binding better to their cognate, within-the-pathway MAPKs than to noncognate MAPKs. To extend these studies to JNK1, the MEK1 and MEK2 D-site peptides were compared with the MKK7-D2 peptide for their ability to inhibit JNK1 phosphorylation of c-Jun or ATF2 (Fig. 8, A–E). The JIP-1-derived D-site peptide was used as a positive control for these experiments. Furthermore, to assess the specificity exhibited by the MKK7-derived D-site peptides for JNK (cognate) versus ERK (noncognate), their ability to inhibit ERK2 was also compared with the MEK1 and MEK2 D-site peptides (Fig. 8, A and F). ERK2 activity was assessed using Elk-1 as a substrate; Elk-1, like JNK1 and ATF2, contains a D-site necessary for efficient MAPK-mediated phosphorylation (50). A mutant MEK2 peptide, MEK2EEAA, was used as a negative control in this experiment (48).
FIGURE 8. Selectivity of MKK7 D-site interaction with JNK versus ERK.
A, D-site peptides (triangle) were used to inhibit JNK1 or ERK2 (MAPK) phosphorylation of the c-Jun, ATF2, or Elk-1 transcription factors (TF). B and C, inhibition of JNK1-dependent phosphorylation of c-Jun. The details are as described for Fig. 7, B and C, respectively. D and E, inhibition of JNK1-dependent phosphorylation of ATF2. The details are as in Fig. 7, D and E. F, inhibition of ERK2-dependent phosphorylation of Elk-1. Purified GST-Elk-1 (1 µm) was incubated with purified active ERK2 (~1 nm) and [γ-32P]ATP for 20 min in the absence or presence of the specific concentrations of the indicated peptides (see Table 3). Results are plotted as percent phosphorylation relative to that observed in the absence of any added peptide. Elk-1 phosphorylation was analyzed by SDS-PAGE and quantified on a PhosphorImager. Data are the average of at least two experiments, with duplicate data points in each experiment.
As shown in Fig. 8, B–E, MKK7-D2 was a more effective JNK inhibitor than the MEK1 or MEK2 D-site peptides. Also, as shown in Fig. 8F, the MEK2-derived D-site peptide was a more effective ERK2 inhibitor than any of the three MKK7-derived peptides. Thus, like the D-site peptides from MEK1, MEK2, and MKK4 (40), the D-site peptides from MKK7 exhibited specificity for cognate versus noncognate MAPKs. However, this specificity was not complete, because the MEK1- and (particularly) MEK2-derived D-site peptides had some activity against JNK1, and the MKK7-derived peptides had some weak inhibitory activity against ERK2.
Selectivity of the D-sites from MEK2 and MKK7
To obtain quantitative insights into D-site specificity, selectivity ratios were calculated. One type of selectivity ratio evaluates how a given kinase reaction is inhibited by a particular cognate peptide as compared with another peptide. This selectivity ratio consists of the IC50 of the comparison peptide divided by the IC50 of the cognate peptide, assessed against the same kinase. Thus, a higher ratio signifies amore selective cognate peptide, i.e. the peptide has a proportionally lower IC50 value for the chosen reaction than the comparison peptide.
The results from such calculations are shown in Table 4, in which the MKK7-D2 and MEK2 peptides are the cognate peptides. The MKK7-D2 peptide was a relatively selective inhibitor of JNK (particularly JNK1) when compared with the MEK1 D-site peptide. This is because the D2 peptide was a potent JNK1 inhibitor and the MEK1 peptide was a relatively poor JNK1 inhibitor. Similarly, the MEK1 and (particularly) MEK2 peptides were relatively selective inhibitors of ERK compared with MKK7-D2.
TABLE 4.
Selectivity ratios
| Selectivity of MAPKs for D-sites | ||
|---|---|---|
| Comparison peptide | IC50 | Selectivity of MKK7-D2a |
| µm | ||
| JNK1 phosphorylation of c-Jun | ||
| MEK1 | 105 | 17.5 |
| MEK2 | 20 | 3.3 |
| MKK7-D1 | 15 | |
| MKK7-D2 | 6 | 1.0 |
| MKK7-D3 | 15 | |
| MKK4 | <5 | |
| JNK1 phosphorylation of ATF2 | ||
| MEK1 | >150 | >18 |
| MEK2 | 60 | 7.5 |
| MKK7-D1 | 25 | |
| MKK7-D2 | 8 | 1.0 |
| MKK7-D3 | 30 | |
| MKK4 | 5 | |
| JNK2 phosphorylation of c-Jun | ||
| MEK1 | >300b | >10 |
| MEK2 | 105b | 3.5 |
| MKK7-D1 | 100 | |
| MKK7-D2 | 30 | 1.0 |
| MKK7-D3 | 65 | |
| MKK4 | 7.5 | |
| JNK2 phosphorylation of ATF2 | ||
| MEK1 | >300b | >4.6 |
| MEK2 | >150b | 2.3 |
| MKK7-D1 | >100 | |
| MKK7-D2 | 65 | 1.0 |
| MKK7-D3 | 100 | |
| MKK4 | 15 | |
| Selectivity of MAPKs for D-sites | ||
| Comparison peptide | IC50 | Selectivity vs.MEK2a |
| µm | ||
| ERK2 phosphorylation of Elk-1 | ||
| MEK1 | 18.5b | |
| MEK2 | 7.5 | 1.0 |
| MKK7-D1 | 88 | 11.7 |
| MKK7-D2 | 85 | 11.3 |
| MKK7-D3 | 125 | 16.7 |
| MKK4 | 50b | 6.6 |
Selectivity ratio = IC50 (comparison peptide)/IC50 (cognate peptide), where the cognate peptide is MKK7-D2 or MEK2.
Data were taken from Ho et al. (40).
A second type of selectivity ratio evaluates how well a particular D-site peptide inhibits the kinase reaction of a cognate MAPK versus a noncognate MAPK. This ratio consists of the IC50 value with which a given peptide inhibits a noncognate kinase divided by the IC50 value with which the same peptide inhibits a cognate kinase. Thus, a higher ratio indicates that the D-site peptide is significantly less effective inhibiting the noncognate kinase reaction versus the cognate reaction and is therefore more selective.
The results from such calculations are summarized in Table 5. The MKK7-D2 peptide stood out as a highly selective inhibitor of JNK1 (compared with its inhibition of ERK2) and was the most selective of the three MKK7 peptides in this regard. However, the MKK7 peptides were less selective inhibitors of JNK2, because of their weaker activity against this kinase. As reported previously (40), the MEK2 D-site peptide is a relatively selective inhibitor of ERK2 compared with its inhibition of JNK2. However, because the MEK2 peptide exhibits considerable activity toward JNK1, its selectivity for ERK2 versus JNK1 is lower.
TABLE 5.
Selectivity ratios
| Selectivity of D-sites for MAPKs | |||
|---|---|---|---|
| D-site peptide | Noncognate/cognate | JNK substrate | Selectivity ratioa |
| MKK7-D1 | ERK2/JNK1 | c-Jun | 5.9 |
| MKK7-D1 | ERK2/JNK1 | ATF2 | 3.5 |
| MKK7-D1 | ERK2/JNK2 | c-Jun | 0.9 |
| MKK7-D1 | ERK2/JNK2 | ATF2 | <0.9 |
| MKK7-D2 | ERK2/JNK1 | c-Jun | 14.2 |
| MKK7-D2 | ERK2/JNK1 | ATF2 | 10.6 |
| MKK7-D2 | ERK2/JNK2 | c-Jun | 2.8 |
| MKK7-D2 | ERK2/JNK2 | ATF2 | 1.3 |
| MKK7-D3 | ERK2/JNK1 | c-Jun | 8.3 |
| MKK7-D3 | ERK2/JNK1 | ATF2 | 4.2 |
| MKK7-D3 | ERK2/JNK2 | c-Jun | 1.9 |
| MKK7-D3 | ERK2/JNK2 | ATF2 | 1.3 |
| MKK4 | ERK2/JNK1 | c-Jun | >10.0 |
| MKK4 | ERK2/JNK1 | ATF2 | 10.0 |
| MKK4 | ERK2/JNK2 | c-Jun | 6.7 |
| MKK4 | ERK2/JNK2 | ATF2 | 3.3 |
| MEK2 | JNK1/ERK2 | c-Jun | 2.7 |
| MEK2 | JNK1/ERK2 | ATF2 | 8.0 |
| MEK2 | JNK2/ERK2 | c-Jun | 14.0 |
| MEK2 | JNK2/ERK2 | ATF2 | >20.0 |
Selectivity ratio = IC50 (D-site peptide with noncognate MAPK)/IC50 (D-site peptide with cognate MAPK).
DISCUSSION
Docking interactions between MKKs and their cognate MAPKs are crucial for effective and accurate signal transmission. Like other MKKs, MKK7 contains an N-terminal domain that is important for high affinity MAPK binding and efficient signaling (47). The experiments presented in this study demonstrate that, unlike other MKKs, the JNK-binding motif in the N terminus of MKK7 is not a single docking site. Rather, the N-terminal domain of MKK7 contains three functional low affinity docking sites of the class herein referred to as “D-sites” (Figs. 1–3). These D-sites were shown to be collectively necessary for the formation of stable MKK7·JNK complexes (Figs. 4 and 5), and for the efficient phosphorylation and activation of JNK1 by MKK7 (Fig. 5). In further experiments, we showed that the MKK7-JNK interaction was competed by D-site peptides from JIP-1 or MKK4 (Fig. 6), and that peptides corresponding to the three D-sites of MKK7 inhibited JNK-mediated phosphorylation of the c-Jun and ATF2 transcription factors (Fig. 7), suggesting that MKK7 competes with these JNK regulators and substrates for binding to JNK. Finally, we showed that the MKK7-derived D-sites exhibited selectivity, binding preferentially to JNK1 over ERK2 (Fig. 8). We conclude that MKK7 uses a novel MAPK-docking strategy, wherein three low affinity D-sites interact to provide a high affinity JNK docking platform, thereby facilitating signal transmission and specificity in the JNK cascade. It seems likely that some other MAPK-interacting proteins, including some substrates, will also utilize this type of docking strategy.
Three Interacting JNK-docking Sites in MKK7
Our contention that the D-sites in the N terminus of MKK7 work together to bind JNK is supported by several observations. Each of the three D-sites was genuine, because they displayed JNK binding activity when tested individually. D1 or D2 was able to bind to JNK in a pull-down assay (Fig. 3). Additionally, D1, D2, or D3 peptides were able to inhibit JNK phosphorylation of c-Jun or ATF2 (Fig. 7), indicating an ability to bind to JNK and block substrate docking. However, although they were authentic, the MKK7-derived D-sites, individually, had a relatively low affinity for JNK, compared with the isolated MKK4-derived or JIP-1-derived D-sites (Figs. 3 and 6–8 and Table 4). In contrast, full-length MKK7 and full-length MKK4 bind to JNK1 and JNK3 with comparable affinity (Kd ~ 30 µm for MKK7; Kd ~ 40 µm for MKK4 (48)). Likewise, the N-terminal domains of MKK7 and MKK4 bind to JNK1 and JNK3 with similar affinities (Kd ~90 µm for MKK7-(1–85); Kd ~ 25 µm for MKK4-(1–94) (40)). The discrepancy between the relatively high JNK-binding affinity of the N-terminal domain of MKK7 and the relatively low binding affinity of the D-sites situated therein suggested the possibility that the D-sites might interact to bind JNK. Indeed, when substitution mutations were introduced into D1, D2, or D3, the JNK-binding affinity of MKK7-(1–85) was reduced by about 50–70%. Revealingly, mutation of any two of the three D-sites reduced binding by about 80–90%, and mutation of all three reduced binding by 95% (Fig. 4). In other words, all three D-sites were required for full binding affinity, whereas any two of the three supplied partial affinity and any one of the three supplied minimal affinity.
Competition for JNK Docking
D-sites are present in many MAPK-interacting proteins, including transcription factors, MAPK phosphatases, and scaffolds (2, 43). The similar primary structure of D-sites suggests that they may bind to the same protein interaction site(s) on their cognate MAPKs. Here we showed that the D-sites in MKK4, JIP-1, and c-Jun/ATF2 compete withMKK7for docking to JNK. This suggests that two very different classes of protein, activators/regulators of JNK (MKK4, MKK7, and JIP-1) and substrates of JNK (c-Jun and ATF2), bind to the same site in the kinase.
MKK7 is found in both the nucleus and the cytoplasm of stimulated and unstimulated cells (47); therefore, competition between MKK7 and transcription factors could occur in vivo and may influence the dynamics of MAPK signaling. For example, Bruna et al. (51) recently showed that glucocorticoids induce the disassembly of JNK from MKK7 by promoting the association of JNK to a docking site in the glucocorticoid receptor.
JIP-1 is a putative scaffold protein that binds to JNK, MKK7, and certain upstream kinases (52). Perhaps the docking interaction between MKK7 and JNK is unnecessary or undesirable when both are bound to JIP-1. Competition for MAPK docking between a scaffold and an MKK is also seen in the yeast mating pathway (42, 53).
Mechanism of D-site Interaction
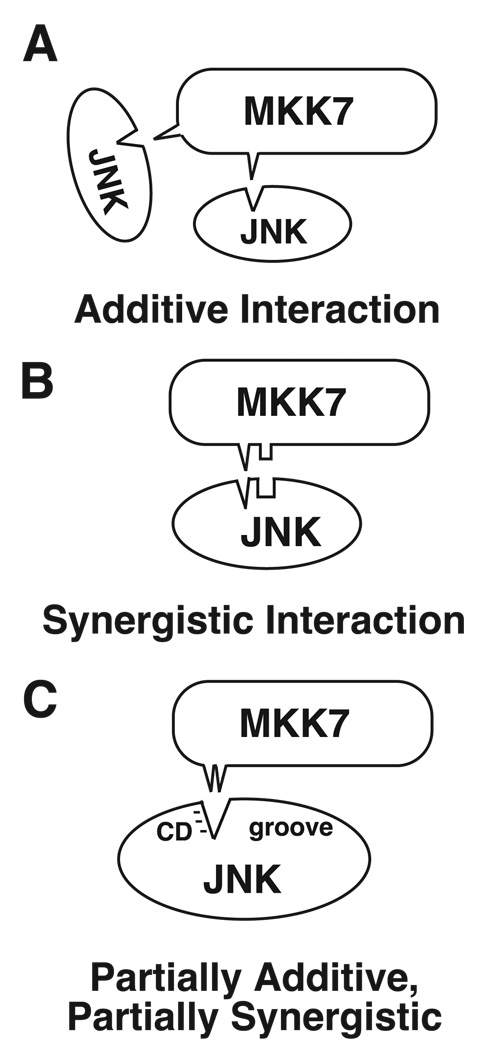
How do the D-sites in MKK7 interact to bind JNK? Two possibilities to consider are an additive mechanism versus a synergistic mechanism. In an additive mechanism, the different D-sites bind to the same region of JNK (Fig. 9A), and the effect is comparable with tripling the concentration of a single D-site. This model is consistent with the similar primary structure of the three D-sites (Fig. 1A), with data that D-sites compete for MAPK binding (40, 54), and with evidence that D-sites bind to a few conserved regions on MAPKs, the CD and ED regions and the hydrophobic docking groove (42, 55–60).
FIGURE 9. Possible mechanisms by which multiple D-sites could interact to bind JNK.
For clarity, only two D-sites are shown. A, additive interaction, in which the individual D-sites bind to the same region of JNK. B, synergistic interaction, in which the different D-sites bind weakly to distinct regions of JNK. C, an example of a partially additive, partially synergistic mechanism. In this example, the basic submotif of D1 interacts preferentially with the CD region of JNK, whereas the hydrophobic submotif of D2 interacts preferentially with the hydrophobic docking groove of JNK. If an individual D-site is absent, residues from another D-site can fill its slot, although not with the same affinity.
Alternatively, if the different D-sites bind to distinct regions of JNK, then they may act cooperatively (Fig. 9B). In a strongly cooperative mechanism, a mutation of any one single D-site should have almost the same effect as a triple D-site mutation. Based on the experiments shown in Figs. 3 and 4, the effect of mutating or deleting single D-sites is more severe than a strictly additive mechanism would predict, but less severe than a strong cooperative mechanism would entail. This suggests a partially additive, partially synergistic mechanism for JNK binding by the D-sites in MKK7 (Fig. 9C). This could occur, for instance, if the basic subdomain of D1 binds preferentially to the CD region of JNK, whereas the hydrophobic subdomain of D2 binds preferentially to the hydrophobic docking groove, and D3 makes additional weak contacts.
Ubiquity of MAPK-docking Sites in MKKs
Functional docking sites for MAPKs have now been found in the N-terminal domains of all seven human MKKs. MEK1, MEK2, MKK3, MKK4, and MKK6 have single D-sites in their N termini (38–40, 44). MEK5 does not have a D-site but has a different MAPK-docking site of acidic character (41). Finally, as shown herein, MKK7 has three low affinity D-sites.
Why should natural selection favor three low affinity D-sites in MKK7 versus one high affinity D-site in other MKKs? One speculation is that it may be advantageous for the JNK-binding affinity of MKK7 to be adjustable. Such modulation could occur, for example, if the binding of other proteins to MKK7 obstructed one or two of the D-sites. Also, the JNK-binding affinity of MKK7 could be fine-tuned by alternative splicing. Indeed, D2 is spliced out in some MKK7 isoforms (47).
Relationship of MKK D-sites to Anthrax Lethal Factor Cleavage Sites
The D-sites in MEK1, MEK2, MKK3, MKK4, and MKK6 are cleaved by the lethal factor protease of Bacillus anthracis (49), resulting in reduced MAPK binding (48). MKK7β is also cleaved by lethal factor at two sites in its N terminus that correspond to D-sites D2 and D3 (49). Thus, our results extend the correlation between bona fide D-sites in MKKs and lethal factor cleavage sites. However, our results also provide the first example of a legitimate (although weak) D-site in an MKK that is not a cleavage site for anthrax lethal factor, MKK7 D1. Lethal factor cleavage occurs within the hydrophobic submotif of the D-site. One feature of MKK7 D1 that is unusual compared with other D-sites in MKKs is that the hydrophobic submotif is flanked by acidic residues. Hence, perhaps these acidic residues are incompatible with lethal factor recognition.
Comparison and Conclusion
Recently, Mooney and Whitmarsh (60) mutated D1 and D2 in the context of full-length MKK7 and concluded, based on the modest decrease in JNK binding and phosphorylation they observed, thatMKK7must bind to JNK using distinct binding determinants. Their results are presumably attributable, at least in part, to the relatively strong JNK binding activity exhibited by the kinase domain of MKK7 (48). We found a stronger effect by examining D1/D2 double mutants in the context of MKK7-(1–85) (Fig. 4). Also, Mooney and Whitmarsh (60) used JNK2, which binds less well to MKK7, and as such may be less sensitive to docking defects. Here, we found that the D1/D2 double mutant of full-length MKK7 was defective in JNK1 binding and phosphorylation (Fig. 5).
In conclusion, the results reported herein reveal a novel mechanism of MAPK docking and highlight the versatility with which docking sites can be employed to enhance signal transmission and specificity.
Footnotes
This work was supported in part by NIGMS Research Grant GM60366 from the National Institutes of Health.
Supported in part by National Library of Medicine Training Grant LM07443.
Supported in part by a postdoctoral fellowship from the Philip Morris External Research Program and a grant from the Milheim Foundation.
The abbreviations used are: MAPK, mitogen-activated protein kinase; ERK, extracellular signal-regulated kinase; MEK, MAPK/ERK kinase; JNK, c-Jun N-terminal kinase; GST, glutathione S-transferase; MKK, MAPK kinase.
REFERENCES
- 1.Chang L, Karin M. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 2.Enslen H, Davis RJ. Biol. Cell. 2001;93:5–14. doi: 10.1016/s0248-4900(01)01156-x. [DOI] [PubMed] [Google Scholar]
- 3.Morrison DK, Davis RJ. Annu. Rev. Cell Dev. Biol. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz MA, Madhani HD. Annu. Rev. Genet. 2004;38:725–748. doi: 10.1146/annurev.genet.39.073003.112634. [DOI] [PubMed] [Google Scholar]
- 5.Ebisuya M, Kondoh K, Nishida E. J. Cell Sci. 2005;118:2997–3002. doi: 10.1242/jcs.02505. [DOI] [PubMed] [Google Scholar]
- 6.Komarova NL, Zou X, Nie Q, Bardwell L. Mol. Syst. Biol. 2005 doi: 10.1038/msb4100031. doi:10.1038/msb4100031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flatauer LJ, Zadeh SF, Bardwell L. Mol. Cell. Biol. 2005;25:1793–1803. doi: 10.1128/MCB.25.5.1793-1803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kyriakis JM, Avruch J. Physiol. Rev. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 9.Davis RJ. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 10.Weston CR, Davis RJ. Curr. Opin. Genet. Dev. 2002;12:14–21. doi: 10.1016/s0959-437x(01)00258-1. [DOI] [PubMed] [Google Scholar]
- 11.Barr RK, Bogoyevitch MA. Int. J. Biochem. Cell Biol. 2001;33:1047–1063. doi: 10.1016/s1357-2725(01)00093-0. [DOI] [PubMed] [Google Scholar]
- 12.Shaulian E, Karin M. Nat. Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 13.Lin A. BioEssays. 2002;25:17–24. doi: 10.1002/bies.10204. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Lin A. Cell Res. 2005;15:36–42. doi: 10.1038/sj.cr.7290262. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy NJ, Sluss HK, Jones SN, Bar-Sagi D, Flavell RA, Davis RJ. Genes Dev. 2003;17:629–637. doi: 10.1101/gad.1062903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirosumi J, Tuncman G, Chang L, Gorgun CZ, Uysal KT, Maeda K, Karin M, Hotamisligil GS. Nature. 2002;420:333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- 17.Kolodziejczyk SM, Walsh GS, Balazsi K, Seale P, Sandoz J, Hierlihy AM, Rudnicki MA, Chamberlain JS, Miller FD, Megeney LA. Curr. Biol. 2001;11:1278–1282. doi: 10.1016/s0960-9822(01)00397-9. [DOI] [PubMed] [Google Scholar]
- 18.Han Z, Boyle DL, Chang L, Bennett B, Karin M, Yang L, Manning AM, Firestein GS. J. Clin. Investig. 2001;108:73–81. doi: 10.1172/JCI12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida H, Hastie CJ, McLauchlan H, Cohen P, Goedert M. J. Neurochem. 2004;90:352–358. doi: 10.1111/j.1471-4159.2004.02479.x. [DOI] [PubMed] [Google Scholar]
- 20.Xia XG, Harding T, Weller M, Bieneman A, Uney JB, Schulz JB. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10433–10438. doi: 10.1073/pnas.181182298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, Shi L, Xie Y, Ma C, Li W, Su X, Huang S, Chen R, Zhu Z, Mao Z, Han Y, Li M. Neurosci. Res. 2004;48:195–202. doi: 10.1016/j.neures.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Kaneto H, Nakatani Y, Miyatsuka T, Kawamori D, Matsuoka T-A, Matsuhisa M, Kajimoto Y, Ichijo H, Yamasaki Y, Hori M. Nat. Med. 2004;10:1128–1132. doi: 10.1038/nm1111. [DOI] [PubMed] [Google Scholar]
- 23.Waetzig V, Herdegen T. Trends Pharmacol. Sci. 2005;26:455–461. doi: 10.1016/j.tips.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 24.Manning AM, Davis RJ. Nat. Rev. Drug Discov. 2003;2:554–565. doi: 10.1038/nrd1132. [DOI] [PubMed] [Google Scholar]
- 25.Holland PM, Suzanne M, Campbell JS, Noselli S, Cooper JA. J. Biol. Chem. 1997;272:24994–24998. doi: 10.1074/jbc.272.40.24994. [DOI] [PubMed] [Google Scholar]
- 26.Tournier C, Whitmarsh AJ, Cavanagh J, Barrett T, Davis RJ. Proc. Natl. Acad. Sci. U. S. A. 1997;94:7337–7342. doi: 10.1073/pnas.94.14.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moriguchi T, Toyoshima F, Masuyama N, Hanafusa H, Gotoh Y, Nishida E. EMBO J. 1997;16:7045–7053. doi: 10.1093/emboj/16.23.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao Z, Diener K, Wang XS, Zukowski M, Matsumoto G, Zhou G, Mo R, Sasaki T, Nishina H, Hui CC, Tan TH, Woodgett JP, Penninger JM. J. Biol. Chem. 1997;272:32378–32383. doi: 10.1074/jbc.272.51.32378. [DOI] [PubMed] [Google Scholar]
- 29.Wu Z, Wu J, Jacinto E, Karin M. Mol. Cell. Biol. 1997;17:7407–7416. doi: 10.1128/mcb.17.12.7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu X, Nemoto S, Lin A. J. Biol. Chem. 1997;272:24751–24754. doi: 10.1074/jbc.272.40.24751. [DOI] [PubMed] [Google Scholar]
- 31.Lawler S, Fleming Y, Goedert M, Cohen P. Curr. Biol. 1998;8:1387–1390. doi: 10.1016/s0960-9822(98)00019-0. [DOI] [PubMed] [Google Scholar]
- 32.Kishimoto H, Nakagawa K, Watanabe T, Kitagawa D, Momose H, Seo J, Nishitai G, Shimizu N, Ohata S, Tanemura S, Asaka S, Goto T, Fukushi H, Yoshida H, Suzuki A, Sasaki T, Wada T, Penninger JM, Nishina H, Katada T. J. Biol. Chem. 2003;278:16595–16601. doi: 10.1074/jbc.M213182200. [DOI] [PubMed] [Google Scholar]
- 33.Tournier C, Dong C, Turner TK, Jones SN, Flavell RA, Davis RJ. Genes Dev. 2001;15:1419–1426. doi: 10.1101/gad.888501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wada T, Joza N, Cheng HY, Sasaki T, Kozieradzki I, Bachmaier K, Katada T, Schreiber M, Wagner EF, Nishina H, Penninger JM. Nat. Cell Biol. 2004;6:215–226. doi: 10.1038/ncb1098. [DOI] [PubMed] [Google Scholar]
- 35.Sasaki T, Wada T, Kishimoto H, Irie-Sasaki J, Matsumoto G, Goto T, Yao Z, Wakeham A, Mak TW, Suzuki A, Cho SK, Zuniga-Pflucker JC, Oliveira-dos-Santos AJ, Katada T, Nishina H, Penninger JM. J. Exp. Med. 2001;194:757–768. doi: 10.1084/jem.194.6.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bardwell L, Cook JG, Chang EC, Cairns BR, Thorner J. Mol. Cell. Biol. 1996;16:3637–3650. doi: 10.1128/mcb.16.7.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bardwell L, Thorner J. Trends Biochem. Sci. 1996;21:373–374. [PubMed] [Google Scholar]
- 38.Bardwell AJ, Flatauer LJ, Matsukuma K, Thorner J, Bardwell L. J. Biol. Chem. 2001;276:10374–10386. doi: 10.1074/jbc.M010271200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu B, Wilsbacher JL, Collisson T, Cobb MH. J. Biol. Chem. 1999;274:34029–34035. doi: 10.1074/jbc.274.48.34029. [DOI] [PubMed] [Google Scholar]
- 40.Ho DT, Bardwell AJ, Abdollahi M, Bardwell L. J. Biol. Chem. 2003;278:32662–32672. doi: 10.1074/jbc.M304229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seyfried J, Wang X, Kharebava G, Tournier C. Mol. Cell. Biol. 2005;25:9820–9828. doi: 10.1128/MCB.25.22.9820-9828.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kusari AB, Molina DM, Sabbagh W, Jr, Lau CS, Bardwell L. J. Cell Biol. 2004;164:267–277. doi: 10.1083/jcb.200310021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharrocks AD, Yang SH, Galanis A. Trends Biochem. Sci. 2000;25:448–453. doi: 10.1016/s0968-0004(00)01627-3. [DOI] [PubMed] [Google Scholar]
- 44.Enslen H, Brancho DM, Davis RJ. EMBO J. 2000;19:1301–1311. doi: 10.1093/emboj/19.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grewal S, Molina DM, Bardwell L. Cell. Signal. 2006;18:123–134. doi: 10.1016/j.cellsig.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barsyte-Lovejoy D, Galanis A, Sharrocks AD. J. Biol. Chem. 2002;277:9896–9903. doi: 10.1074/jbc.M108145200. [DOI] [PubMed] [Google Scholar]
- 47.Tournier C, Whitmarsh AJ, Cavanagh J, Barrett T, Davis RJ. Mol. Cell. Biol. 1999;19:1569–1581. doi: 10.1128/mcb.19.2.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bardwell AJ, Abdollahi M, Bardwell L. Biochem. J. 2004;378:569–577. doi: 10.1042/BJ20031382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vitale G, Bernardi L, Napolitani G, Mock M, Montecucco C. Biochem. J. 2000;352:739–745. [PMC free article] [PubMed] [Google Scholar]
- 50.Yang SH, Yates PR, Whitmarsh AJ, Davis RJ, Sharrocks AD. Mol. Cell. Biol. 1998;18:710–720. doi: 10.1128/mcb.18.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bruna A, Nicolas M, Munoz A, Kyriakis JM, Caelles C. EMBO J. 2003;22:6035–6044. doi: 10.1093/emboj/cdg590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitmarsh AJ, Cavanagh J, Tournier C, Yasuda J, Davis RJ. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 53.Bhattacharyya RP, Remenyi A, Good MC, Bashor CJ, Falick AM, Lim WA. Science. 2006;311:822–826. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- 54.Bardwell AJ, Abdollahi M, Bardwell L. Biochem. J. 2003;370:1077–1085. doi: 10.1042/BJ20021806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanoue T, Adachi M, Moriguchi T, Nishida E. Nat. Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 56.Tanoue T, Maeda R, Adachi M, Nishida E. EMBO J. 2001;20:466–479. doi: 10.1093/emboj/20.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee T, Hoofnagle AN, Kabuyama Y, Stroud J, Min X, Goldsmith EJ, Chen L, Resing KA, Ahn NG. Mol. Cell. 2004;14:43–55. doi: 10.1016/s1097-2765(04)00161-3. [DOI] [PubMed] [Google Scholar]
- 58.Heo YS, Kim SK, Seo CI, Kim YK, Sung BJ, Lee HS, Lee JI, Park SY, Kim JH, Hwang KY, Hyun YL, Jeon YH, Ro S, Cho JM, Lee TG, Yang CH. EMBO J. 2004;23:2185–2195. doi: 10.1038/sj.emboj.7600212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang CI, Xu B, Akella R, Cobb M, Goldsmith EJ. Mol. Cell. 2002;9:1241–1249. doi: 10.1016/s1097-2765(02)00525-7. [DOI] [PubMed] [Google Scholar]
- 60.Mooney LM, Whitmarsh AJ. J. Biol. Chem. 2004;279:11843–11852. doi: 10.1074/jbc.M311841200. [DOI] [PubMed] [Google Scholar]