Abstract
Purpose: To determine the diagnostic efficacy of optical coherence tomography (OCT) to identify cervical intraepithelial neoplasia (CIN) grade 2 or higher by computer-aided diagnosis (CADx).
Methods: OCT has been investigated as a screening∕diagnostic tool in the management of preinvasive and early invasive cancers of the uterine cervix. In this study, an automated algorithm was developed to extract OCT image features and identify CIN 2 or higher. First, the cervical epithelium was detected by a combined watershed and active contour method. Second, four features were calculated: The thickness of the epithelium and its standard deviation and the contrast between the epithelium and the stroma and its standard deviation. Finally, linear discriminant analysis was applied to classify images into two categories: Normal∕inflammation∕CIN 1 and CIN 2∕CIN 3. The algorithm was applied to 152 images (74 patients) obtained from an international study.
Results: The numbers of normal∕inflammatory∕CIN 1∕CIN 2∕CIN 3 images are 74, 29, 14, 24, and 11, respectively. Tenfold cross-validation predicted the algorithm achieved a sensitivity of 51% (95% CI: 36%–67%) and a specificity of 92% (95% CI: 86%–96%) with an empirical two-category prior probability estimated from the data set. Receiver operating characteristic analysis yielded an area under the curve of 0.86.
Conclusions: The diagnostic efficacy of CADx in OCT imaging to differentiate high-grade CIN from normal∕low grade CIN is demonstrated. The high specificity of OCT with CADx suggests further investigation as an effective secondary screening tool when combined with a highly sensitive primary screening tool.
Keywords: cervical intraepithelial neoplasia, optical coherence tomography, computer-aided diagnosis
BACKGROUND AND INTRODUCTION
Cervical intraepithelial neoplasia (CIN) is a precancerous condition of the uterine cervix before progression to invasive carcinoma. Treatments are recommended for high-grade CIN (CIN≥2).1 Cervical cytology screening, the Papanicolaou test (pap smear), has proved to be a successful cervical cancer prevention method and is largely responsible for the significant decrease in cervical cancer mortality in the United States.2 But the program requires a complex infrastructure that is possible to maintain in developed countries but virtually impossible in the developing world. Furthermore, cytology has a low sensitivity (53%) so that cytology programs are only successful when the patients return regularly for screening.3 Once the cytology result is abnormal, multiple visits occur, including cervical examination with or without magnification [colposcopy or visual inspection with acetic acid (VIA)] with biopsies taken for pathological diagnosis and then, eventually, treatment with excision or ablation methods. Quality management for this multivisit care is difficult and overall costs are high.4 The time required to process the cytology and biopsies also increases the risk of patient loss to follow up. Therefore, there exists a need in low resource environments for a diagnostic evaluation of the uterine cervix at the point of care to allow for a single episode of care or “see and treat” program.
Human papillomavirus (HPV) testing has been studied as an alternative screening method to cytology.3, 5, 6 It has a higher sensitivity (95%) and a lower specificity (90%) than cytology3, 4 and would benefit from a real-time secondary screening tool.
Optical coherence tomography (OCT) provides noninvasive and highly resolved images of backscattering profile from internal tissue microstructure in real-time and has the potential to be a true “optical biopsy.”7 Various research groups have demonstrated OCT images of the uterine cervix in vivo.8, 9, 10, 11, 12, 13, 14, 15, 16, 17 The visual diagnostic criteria have been proposed to differentiate stages of CIN based on OCT images.12 A series of clinical studies followed, demonstrating a specificity enhancement when using OCT as an aid to VIA and colposcopy.15, 17 The diagnostic criteria were mainly based on the observation that high-grade CIN tissue usually lacks the layered architecture or the sharp border characteristic of normal cervix and that normal and low grade tissue are highly unlikely to lack this layered appearance. These diagnostic criteria are quantifiable and can be converted into a computer-aided diagnosis (CADx) algorithm to assist and accelerate image interpretation.
CADx algorithms are image analysis and classification technologies to identify abnormalities in medical images, which provide an objective, quantitative interpretation as a “second opinion” for clinicians’ diagnosis and therapy.18, 19 These algorithms were developed for OCT images of other organs to investigate the diagnostic power.20, 21 CADx algorithms will be more important when large numbers of images need to be interpreted and as OCT imaging speed increases. Zuluaga et al.13 reported a higher average epithelial intensity in abnormal tissue than in normal tissue in premenopausal women. Turchin et al.22 proposed a scattering model based on the radiative transfer equation, which showed the potential for cervical tissue classification. However, these studies neither demonstrated a fully automated CADx algorithm which is critical in a clinical setting, nor evaluated the diagnostic power of the proposed image features. The diagnostic efficacy of CADx to identify abnormality still needs to be determined, which will justify whether CADx should be incorporated into CIN diagnosis using OCT.
In this paper, a fully automated CADx algorithm is proposed to identify OCT images of high-grade CIN (≥2). Image features were quantified and evaluated using cervical OCT images with histopathologic correlations to evaluate the diagnostic efficacy.
MATERIALS
All images were obtained from an international study approved by The Cleveland Clinic, Cleveland, OH, USA and The Hospital Maternidad Nuestra Senora de la Altagracia, Dominican Republic. Patients who were diagnosed with an abnormal Papanicolaou test or with suspicious lesions of the uterine cervix as determined by VIA, colposcopy, or both VIA and colposcopy were eligible for the study. Enrolled patients were imaged with an OCT device (Niris© Imaging System, Imalux© Corporation, Cleveland, OH) under VIA and∕or colposcopy guidance. The device operated at 980 nm center wavelength and having a lateral resolution of 25 μm and an in-depth resolution of 11 μm, respectively. Light was guided by an optical fiber within a flexible cable (2.7 mm in diameter). The imaging probe was placed perpendicularly to the tissue surface and the tissue was slightly compressed. An electromechanical actuator at the distal end steered the light for lateral scanning. A single-use probe sheath with a transparent end cap was utilized for patient protection and for facilitating the probe positioning. The frame rate was 0.67 frames per second with 200×200 pixels in each frame. Readings were taken from 2, 4, 8, and 10 o’clock locations (four quadrants) at the cervical squamous columnar junction as well as any additional abnormal areas found by VIA and∕or colposcopy. A biopsy was subsequently obtained from each OCT image site. The exclusion criteria for image∕biopsy sites are (1) multiple histopathological diagnoses present according to biopsies from the same quadrant, (2) image acquisition failure (e.g., due to loose contact and displacement between the scanner and the tissue), and (3) sites without histopathologic results available. Based on these exclusion criteria, 152 OCT images (74 patients) from 1215 images (212 patients) were selected for this study to investigate the feasibility of CADx for cervical cancer. The numbers of normal∕inflammatory∕CIN 1∕CIN 2∕CIN 3 images are 74, 29, 14, 24, and 11, respectively.
METHODS
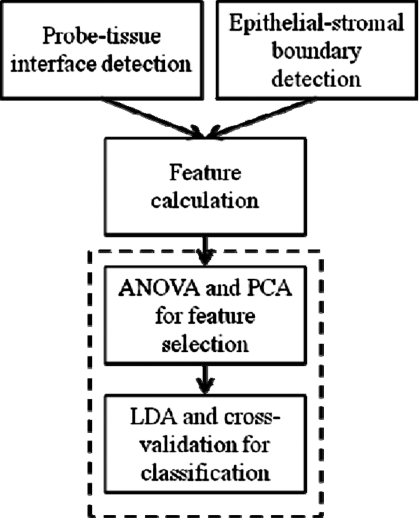
It has been observed that OCT images of healthy cervix exhibit a clear two-layered structure,12, 22 which can be seen in Fig. 1. The first layer is the epithelium. It is normally 200–400 μm thick and delineated from the second layer, the stroma, by a sharp boundary at the basement membrane. The stroma is more highly scattering than the epithelium and therefore appears brighter in the OCT image. The thickness of the epithelium and the intensity contrast between the epithelium and the stroma vary little within a single OCT image of normal cervix. As CIN progresses, the two-layered structure becomes much less distinct and regular. The intensity contrast between the first and second layers decreases. In fact, for CIN 2 and CIN 3, the two-layered structure is usually not readily observable. These changes are likely to be caused by the different scattering properties between normal and neoplastic epithelial cells. The scattering coefficient of neoplastic cells is higher than that of normal epithelial cells23, 24 and therefore closer to the scattering coefficient of the stroma. The scattering contrast between the neoplasia and stroma is low, the basement membrane is obscured, and the apparent boundary (if any is observable) in CIN≥2 may be at the boundary between neoplastic and normal epithelium, more superficial than the basement membrane. It is important to note that the OCT appearance of normal cervix is very consistent, while the OCT appearance of high-grade CIN is highly variable. The presence of the clear boundary likely indicates normal or low grade tissue, while the disruption of the boundary is very likely to indicate high-grade CIN. Therefore, the strategy adopted for this CADx algorithm was to detect the presence of a clear epithelial-stromal boundary. This is accomplished by the automated segmentation of the epithelium. The flow chart of the algorithm is shown in Fig. 2.
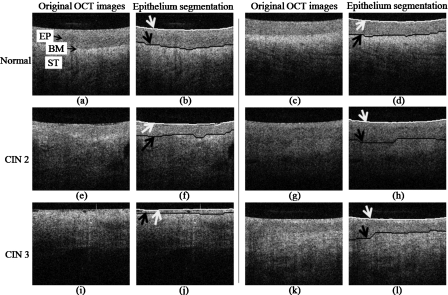
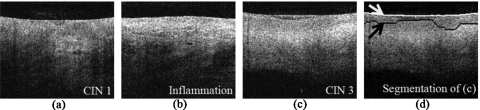
Figure 1.
Representative images of normal and high-grade CIN images with epithelium segmentation results. White curves and white arrows indicate the probe-tissue interface segmented by the CADx algorithm and black curves and black arrows indicate the segmented boundary between the epithelium and stroma. [(a)–(d)] Typical normal cervical OCT images and segmentation results. All structures are well-defined, including the epithelium (EP), the basement membrane (BM), and the stroma (ST). [(e)–(l)] CIN 2 and CIN 3 OCT images and the detections. The layered architecture becomes irregular or is not apparent. Images shown in (e) and (f) and (i) and (j) were correctly classified. Images shown in (g) and (h) and (k) and (l) were misclassified.
Figure 2.
The flow chart of the CAD algorithm. ANOVA: Analysis of variance. PCA: Principal component analysis. LDA: Linear discriminant analysis.
Epithelium segmentation
The epithelium was automatically segmented in the OCT images by detecting the upper probe-tissue interface and the lower boundary between epithelium and stroma. The basement membrane is less than 1 μm and is not resolved as an additional layer in the OCT images. The probe-tissue interface was detected by the watershed algorithm on the gradient images.25 The gradient images were preprocessed by the minimum imposition method to avoid oversegmentation, so that the watershed algorithm labeled all the tissue as one region and the background as another. The boundary between the epithelium and stroma was detected by the active contour algorithm. The active contour uses a series of anchor points to approximate a boundary by considering both the image gradient and the curvature of the estimated boundary.26 In the conventional active contour algorithm, a 2-D neighborhood around each anchor point is searched, which tends to shorten the length of the contour. For this work, the algorithm was modified to limit the searching neighborhood to one column. The anchor points therefore only moved vertically so that the contour was not shortened in the horizontal direction during iterations.
Feature calculation
Based on previously proposed diagnostic criteria and the observations, four image features were quantified from the segmented boundaries to characterize the cervical layered structure as follows: (1) The epithelial thickness. The thickness in the normal images tends to be greater. The area of epithelial tissue was first calculated. The thickness was defined as the area divided by the width of the image. (2) The standard deviation of the thickness within the image. Because the abnormal tissue tends to have a less smooth boundary, the thicknesses along the A-scans tend to have a larger variance. The epithelial thickness in each A-scan was calculated. The standard deviation of the 200 thicknesses was calculated. A smaller standard deviation represented a more consistent thickness of the epithelium. (3) The contrast between the epithelium and stroma. It is observed that the contrast in the images of normal tissue is greater. The contrast was quantified as the difference between the average pixel intensity of the lower layer (stroma) and the average pixel intensity of the upper layer (squamous epithelium) divided by the sum of those two average intensities. Here, the lower and upper layers were restricted to a region of 100 μm in depth, where the center line was the epithelial-stromal boundary. The depth of 100 μm was chosen to ensure that the field of view of the calculation included only tissue because the minimal epithelial thickness calculated was approximately 50 μm. (4) The standard deviation of the contrast within the same image. The contrasts of the individual A-scans tend to have a higher variance in abnormal tissue because of the irregular boundary. Calculating the contrast between the epithelium and the stroma along each A-scan introduced a large variance from the speckle noise. Therefore, each image was divided into 20 sections, with ten consecutive A-scans in each section. Contrast was calculated for each individual section so that the speckle variance was averaged but the structural variance was maintained. The standard deviation of the twenty values was calculated.
Classification
The images were classified into two categories: Normal∕CIN 1∕inflammation and CIN 2∕CIN 3. One-way unbalanced analysis of variance (ANOVA) was performed on each quantified feature to test if the two categories had a significant difference in mean (F-test with the null hypothesis that the means have no difference; p-value<0.01).27 Features exhibiting a significant difference in mean were chosen, normalized, and evaluated using principal component analysis (PCA) to determine the correlation among the features.28 The first several principal components representing more than 90% of the variance of the original features were then chosen as input to the classifier. Linear discriminant analysis (LDA) was then applied to classify the images into two categories.29 LDA can be understood as a linear projection of the features to the plane in feature space where the two categories are maximally separated. To provide an estimate for the overall accuracy, tenfold cross-validation was applied. Nine-tenths of the images were used to train the LDA and one-tenth of them were classified. This training-validation process was repeated ten times so that every image was used for validation. The classification results from the validation set were utilized to estimate the overall sensitivity and specificity. By varying the prior probability of the two categories, the receiver operating characteristic (ROC) curve was plotted.
All the analysis work presented here was carried out using standard functions available in the image processing and statistics toolboxes in MATLAB (The MathWorks, Inc., Natick, MA).
RESULT
Epithelium segmentation
Representative images of normal and CIN images are shown in columns 1 and 3 in Fig. 1. The typical layers of epithelium (EP), basement membrane (BM), and stroma (ST) can be clearly identified in Fig. 1a. The results of the automated segmentation method on the selected representative images are shown in columns 2 and 4. Rows 1–3 are normal, CIN 2, and CIN 3 images, respectively. The probe-tissue interface was obvious and was readily detected with few errors, as indicated by the white curves and arrows. A distinct well-defined, sharp boundary between the epithelium and stroma was consistently observed in normal images, but usually less obvious or even missing in images of high-grade CIN. The active contour algorithm accurately estimated the boundary with a curve when the boundary was clear. If the boundary was vague or did not exist, the algorithm would still provide a boundary estimate, but the curve tended to be located near the probe-tissue interface, influenced by the strong boundary of the probe-tissue interface and strongly affected by other high contrast features within the tissue. The results of the active contour algorithm were indicated by black curves and arrows in Fig. 1.
Feature calculation
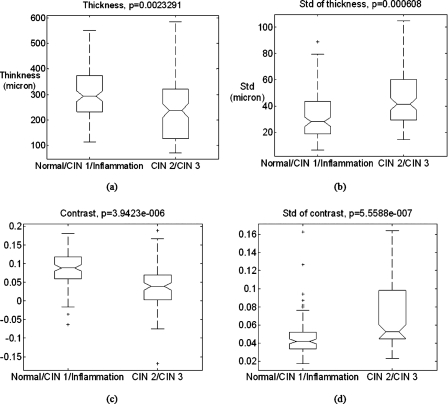
Four image features were calculated from all images and are summarized in the box plots shown in Fig. 3. On each box, the central mark is the median, the edges of the box are the 25th and 75th percentiles, the whiskers extend to the most extreme data points not considered outliers, and the outliers are marked by the plus sign. The normal∕CIN 1∕inflammation category had higher median thickness [Fig. 3a] and contrast [Fig. 3c] than the CIN 2∕CIN 3 category and the standard deviations were smaller [Figs. 3b, 3d], which is consistent with the observations.
Figure 3.
Box plots of the four quantified image features. On each box, the central mark is the median, the edges of the box are the 25th and 75th percentiles, the whiskers extend to the most extreme data points not considered outliers, and the outliers are marked by the plus sign. (a) The thickness. (b) The standard deviation (Std) of thickness within an image. (c) The contrast. (d) The standard deviation of contrast within an image. The p-values are from F-tests with the null hypothesis that the means have no difference.
Classification
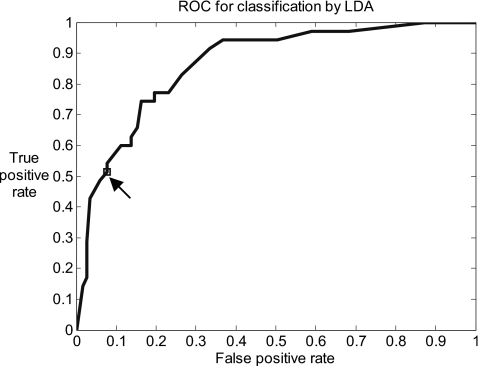
All four image features had significant differences in means between two categories (Fig. 3, all p<0.01) and therefore were considered potential features. PCA yielded four principal components which represented 38%, 32%, 18%, and 13% of the variance of four-dimensional feature space, which indicated that the four features were not highly correlated and therefore not redundant. Consequently, all four features were employed for classification by means of LDA, using tenfold cross-validation. The accuracy of the classification can be visualized using the ROC curve plotted in Fig. 4. The area under the curve is 0.86. The image data set was used to obtain an empirical prior probability that 77% images were normal∕CIN 1∕inflammation and 23% were CIN 2∕CIN 3. Using this prior probability or, in other words, the decision threshold yielding the lowest total number of misclassified images, the algorithm achieved two-category classification with a sensitivity of 51% (95% CI: 36%–67%) and a specificity of 92% (95% CI: 86%–96%) as indicated by the arrow in Fig. 4 and summarized in Table 1.
Figure 4.
The ROC curve generated by varying the prior probability in the LDA classification. The arrow indicates the true positive rate and false positive rate with the empirical prior probability estimated from the image set.
Table 1.
LDA classification results compared to histopathology.
| LDA classification | Histopathology | |
|---|---|---|
| CIN 2∕CIN 3 | Normal∕CIN 1∕inflammation | |
| CIN 2∕CIN 3 | 18 | 9 |
| Normal∕CIN 1∕inflammation | 17 | 108 |
DISCUSSION
The objective of the study was to determine the diagnostic efficacy of OCT to identify CIN grade 2 or higher by CADx. The OCT images were obtained under VIA and∕or colposcopy guidance. Therefore, this study did not validate the efficacy of OCT as a primary CIN screening tool. However, the demonstrated high specificity suggests that further investigations incorporate OCT imaging with CADx as a secondary screen after a primary screening modality such as HPV testing. An accurate and rapid CADx algorithm may assist examining clinicians by analyzing the image sequence and identifying patients with normal-appearing cervical epithelium, which therefore do not require treatment. Results from the primary screening modality could later provide the final diagnosis for the patients with positive OCT examination. For point of care use, the immediate analysis provided by CADx is necessary. In the present study, the algorithm required approximately 50 s per image for processing. However, by optimizing the algorithm and by employing parallel computing technology and the C++ programming platform, the computation time may be reduced to a fraction of a second, enabling real-time analysis.
Among the 1063 excluded images, approximately two-thirds were from quadrants with multiple histopathological conditions present according to biopsy diagnosis. These image-biopsy sites were excluded because there was no one-to-one correspondence available between OCT and histopathology. Approximately one-third of the excluded OCT images were not of sufficient quality for diagnosis. These images failed either because of motion artifact due to probe displacement during imaging or because of degradation of image signal due to loose probe-tissue contact. There were also approximately 20 cases when the biopsy results were not available due to errors in histology processing.
The proposed algorithm seeks to detect a sharp and smooth boundary within the normal range of the basement membrane in images of normal, CIN 1, and inflamed tissue. The active contour algorithm serves well the purpose of boundary detection and the extracted features further define the appearance of a normal boundary. All 74 normal images were correctly classified. Images of CIN 1 and inflammation seem to have variable characteristics between normal and high-grade CIN tissues. Most of them exhibit regular layered structure but with slightly lower contrast than normal images, while the layered structure was severely disrupted or absent in the misclassified images. Of the 14 CIN 1 images, 11 were correctly classified and three were misclassified as high-grade CIN. Of the 29 inflammation images, 23 were correctly classified and six were misclassified as high-grade because their appearance is similar to high-grade CIN. Figures 5a, 5b show one CIN 1 image and one image of inflammatory tissue, which were misclassified as high-grade CIN. Among the high-grade CIN images, the layered structure was less likely to be observed but it was present in some images, which is one reason for the moderate sensitivity. Thirteen of the total 24 CIN 2 images were misclassified as low grade. Five of the total 11 CIN 3 images were misclassified as low grade. Among the misclassified CIN 2∕CIN 3 images, eight had layered structure in the images similar to those of normal tissue [e.g., Fig. 1k]. Examples are shown in Fig. 1, where Figs. 1k, 1h were misclassified, whereas Figs. 1e, 1i were correctly classified. A limitation in the method is that the active contour algorithm requires the imaging probe to be in complete contact with the tissue. Otherwise, the gap between the probe and the tissue results in a strong boundary, which attracts the active contour, resulting in mis-segmentation. Two CIN 2 images and four CIN 3 images were misclassified by the CADx algorithm due to the irregular contact of the probe with the tissue. Figures 5c, 5d show an example of a CIN 3 image misclassified for this reason. This type of misclassification could be mitigated if the actual tissue surface is detected by a more sophisticated edge detection algorithm. In that case, the gradient at the surface will not be calculated so that the gap no longer affects the active contour.
Figure 5.
(a) CIN 1 and (b) inflammation image that was misclassified as high-grade CIN images. The layered structure in these two images was far less visible compared to normal images. (c) CIN 3 image misclassified as normal∕inflammation∕CIN 1. (d) The active contour segmentation of (c) failed due to irregular probe-tissue contact.
The image features utilized for classification in this study were selected based on experience from a series of cervical OCT imaging studies12, 14, 15, 17 and on previously published observations of the scattering properties of cervical tissue.23, 24 Numerous other image features could be quantified and analyzed for classification of cervical neoplasia. For example, texture analysis has been investigated for classifying other tissue based on OCT imaging20, 30, 31, 32 and could potentially be applied to improve the performance of CADx for CIN. However, feature mining would require a larger image data set with more high-grade cases.
Other improvements may be valuable to translate OCT with CADx technology for clinical utility. Improving image quality will always improve image analysis. An OCT probe and system capable of faster scanning would reduce motion artifacts and improve the yield of diagnostic quality images. Increasing resolution improves contrast and detectability of fine features and has been shown to detect finer structure in gastrointestinal tissue.33 An image quality assessment algorithm could identify images of low quality automatically and improve the robustness of the method. The OCT images included in this study were mostly of high quality, but image quality may be compromised by loose contact between the probe and tissue, probe displacement, or orientation of the probe with the tissue. By automated detection of images of low quality or with artifacts, the operator could be alerted in real-time to obtain adequate images and inadequate images could be rejected from analysis. More information could be incorporated into the CADx algorithm. Patient information was not available for this study, but the incorporation of information such as age and menopausal status may improve diagnosis based on OCT imaging. For example, it has been shown that the endometrium of postmenopausal women is much thinner than that of premenopausal women, which suggests that menopausal state may also affect epithelial thickness.34
CONCLUSION
In conclusion, a CADx algorithm has been described to differentiate CIN 2∕CIN 3 from normal∕CIN 1∕inflammation based on OCT images. The accurate segmentation of epithelium is the critical step for this algorithm. The classifying criteria are consistent with human interpretation. The high specificity suggests a possible role for CADx assisted OCT as a secondary screening technology to be combined with highly sensitive primary screening tests currently used in cervical cancer screening algorithms.
ACKNOWLEDGMENTS
The project described was supported by 2002 Ohio Technology Action Fund and by Grant No. RO1 CA114276 from the National Institute of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Nancy J. Tresser and Margarita Kareta are employees of Imalux Corporation, which is a medical device company and develops the optical coherence tomography system.
Nomenclature
- ANOVA
analysis of variance
- CADx
computer-aided diagnosis
- CIN
cervical intraepithelial neoplasia
- HPV
human papillomavirus
- LDA
linear discriminant analysis
- OCT
optical coherence tomography
- PCA
principle component analysis
- ROC
receiver operating characteristic
- VIA
visual inspection with acetic acid
References
- T. C.Wright, Jr., Cox J. T., Massad L. S., Carlson J., Twiggs L. B., and Wilkinson E. J., Am. J. Obstet. Gynecol. 189, 295–304 (2003). 10.1067/mob.2003.633 [DOI] [PubMed] [Google Scholar]
- Solomon D., Breen N., and McNeel T., Ca-Cancer J. Clin. 57, 105–111 (2007). 10.3322/canjclin.57.2.105 [DOI] [PubMed] [Google Scholar]
- Cuzick J., Clavel C., Petry K. U., Meijer C. J., Hoyer H., Ratnam S., Szarewski A., Birembaut P., Kulasingam S., Sasieni P., and Iftner T., Int. J. Cancer 119, 1095–1101 (2006). 10.1002/ijc.21955 [DOI] [PubMed] [Google Scholar]
- Belinson J., Qiao Y. L., Pretorius R., Zhang W. H., Elson P., Li L., Pan Q. J., Fischer C., Lorincz A., and Zahniser D., Gynecol. Oncol. 83, 439–444 (2001). 10.1006/gyno.2001.6370 [DOI] [PubMed] [Google Scholar]
- Mayrand M. H., Duarte-Franco E., Rodrigues I., Walter S. D., Hanley J., Ferenczy A., Ratnam S., Coutlee F., and Franco E. L., N. Engl. J. Med. 357, 1579–1588 (2007). 10.1056/NEJMoa071430 [DOI] [PubMed] [Google Scholar]
- Meijer C. J., Berkhof J., Castle P. E., Hesselink A. T., Franco E. L., Ronco G., Arbyn M., Bosch F. X., Cuzick J., Dillner J., Heideman D. A., and Snijders P. J., Int. J. Cancer 124, 516–520 (2009). 10.1002/ijc.24010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D., Swanson E. A., Lin C. P., Schuman J. S., Stinson W. G., Chang W., Hee H. R., Flotte T., Gregory K., Puliafito C. A., and Fujimoto J. G., Science 254, 1178–1181 (1991). 10.1126/science.1957169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeev A., Gelikonov V., Gelikonov G., Feldchtein F., Kuranov R., Gladkova N., Shakhova N., Snopova L., Shakhov A., Kuznetzova I., Denisenko A., Pochinko V., Chumakov Y., and Streltzova O., Opt. Express 1, 432–440 (1997). 10.1364/OE.1.000432 [DOI] [PubMed] [Google Scholar]
- Feldchtein F. I., Gelikonov G. V., Kuranov R. V., Sergeev A. M., Gladkova N. D., Shakhov A. V., Shakhova N. M., Snopova L. B., Terenteva A. B., Zagainova E. V., Chumakov Y. P., and Kuznetzova I. A., Opt. Express 3, 257–270 (1998). 10.1364/OE.3.000257 [DOI] [PubMed] [Google Scholar]
- Boppart S. A., Goodman A., Libus J., Pitris C., Jesser C. A., Brezinski M. E., and Fujimoto J. G., Int. J. Gynaecol. Obstet. 106, 1071–1077 (1999). [DOI] [PubMed] [Google Scholar]
- Pitris C., Goodman A., Boppart S. A., Libus J. J., Fujimoto J. G., and Brezinski M. E., Obstet. Gynecol. (N.Y., NY, U. S.) 93, 135–139 (1999). 10.1016/S0029-7844(98)00375-5 [DOI] [PubMed] [Google Scholar]
- Escobar P. F., Belinson J. L., White A., Shakhova N. M., Feldchtein F. I., Kareta M. V., and Gladkova N. D., Int. J. Gynecol. Cancer 14, 470–474 (2004). 10.1111/j.1048-891x.2004.14307.x [DOI] [PubMed] [Google Scholar]
- Zuluaga A. F., Follen M., Boiko I., Malpica A., and Richards-Kortum R., Am. J. Obstet. Gynecol. 193, 83–88 (2005). 10.1016/j.ajog.2004.11.054 [DOI] [PubMed] [Google Scholar]
- Escobar P. F., Rojas-Espaillat L., and Belinson J. L., Int. J. Gynaecol. Obstet. 89, 63–64 (2005). 10.1016/j.ijgo.2004.12.029 [DOI] [PubMed] [Google Scholar]
- Escobar P. F., Rojas-Espaillat L., Tisci S., Enerson C., Brainard J., Smith J., Tresser N. J., Feldchtein F. I., Rojas L. B., and Belinson J. L., Int. J. Gynecol. Cancer 16, 1815–1822 (2006). 10.1111/j.1525-1438.2006.00665.x [DOI] [PubMed] [Google Scholar]
- Lee S. -W., Yoo J. -Y., Kang J. -H., Kang M. -S., Jung S. -H., Chong Y., Cha D. -S., Han K. -H., and Kim B. -M., Opt. Express 16, 2709–2719 (2008). 10.1364/OE.16.002709 [DOI] [PubMed] [Google Scholar]
- Liu Z., Belinson S. E., Li J., Yang B., Wulan N., Tresser N. J., Wang C., Mohr M., Zhang L., Zhou Y., Weng L., Wu R., and Belinson J. L., Int. J. Gynecol. Cancer 20, 283–287 (2010). [DOI] [PubMed] [Google Scholar]
- Giger M. L., Karssemeijer N., and Armato S. G., IEEE Trans. Med. Imaging 20, 1205–1208 (2001). 10.1109/TMI.2001.974915 [DOI] [PubMed] [Google Scholar]
- Giger M. L., Huo Z., Kupinski M. A., and Vyborny C. J., Handbook of Medical Imaging (SPIE, Bellingham, 1999), pp. 249–272. [Google Scholar]
- Qi X., Sivak M. V., Isenberg G., Willis J. E., and Rollins A. M., J. Biomed. Opt. 11, 044010 (2006). 10.1117/1.2337314 [DOI] [PubMed] [Google Scholar]
- Qi X., Pan Y. S., Hu Z. L., Kang W., Willis J. E., Olowe K., Sivak M. V., and Rollins A. M., J. Biomed. Opt. 13, 054055 (2008). 10.1117/1.2993323 [DOI] [PubMed] [Google Scholar]
- Turchin I. V., Sergeeva E. A., Dolin L. S., Kamensky V. A., Shakhova N. M., and Richards-Kortum R., J. Biomed. Opt. 10, 064024 (2005). 10.1117/1.2137670 [DOI] [PubMed] [Google Scholar]
- Collier T., Arifler D., Malpica A., Follen M., and Richards-Kortum R., IEEE J. Sel. Top. Quantum Electron. 9, 307–313 (2003). 10.1109/JSTQE.2003.814413 [DOI] [Google Scholar]
- Arifler D., Guillaud M., Carraro A., Malpica A., Follen M., and Richards-Kortum R., J. Biomed. Opt. 8, 484–494 (2003). 10.1117/1.1578640 [DOI] [PubMed] [Google Scholar]
- Soille P., Morphological Image Analysis: Principles and Applications (Springer-Verlag, New York, 2003). [Google Scholar]
- Amini A. A., Weymouth T. E., and Jain R. C., IEEE Trans. Pattern Anal. Mach. Intell. 12, 855–867 (1990). 10.1109/34.57681 [DOI] [Google Scholar]
- Hinkelmann K. and Kempthorne O., Design and Analysis of Experiments, 2nd ed. (Wiley-Interscience, Hoboken, 2008), Vol. 1, pp. 71–136. [Google Scholar]
- Jolliffe I. T., Principal Component Analysis, 2nd ed. (Springer, New York, 2002). [Google Scholar]
- McLachlan G. J., Discriminant Analysis and Statistical Pattern Recognition (Wiley, Hoboken, 2004). [Google Scholar]
- Gossage K. W., Tkaczyk T. S., Rodriguez J. J., and Barton J. K., J. Biomed. Opt. 8, 570–575 (2003). 10.1117/1.1577575 [DOI] [PubMed] [Google Scholar]
- Gossage K. W., Smith C. M., Kanter E. M., Hariri L. P., Stone A. L., Rodriguez J. J., Williams S. K., and Barton J. K., Phys. Med. Biol. 51, 1563–1575 (2006). 10.1088/0031-9155/51/6/014 [DOI] [PubMed] [Google Scholar]
- Lingley-Papadopoulos C. A., Loew M. H., Manyak M. J., and Zara J. M., J. Biomed. Opt. 13, 024003 (2008). 10.1117/1.2904987 [DOI] [PubMed] [Google Scholar]
- Chen Y., Aguirre A. D., Hsiung P. L., Huang S. W., Mashimo H., Schmitt J. M., and Fujimoto J. G., Opt. Express 16, 2469–2485 (2008). 10.1364/OE.16.002469 [DOI] [PubMed] [Google Scholar]
- Wolman I., Amster R., Hartoov J., Gull I., Kupfermintz M., Lessing J. B., and Jaffa A. J., Gynecol. Obstet. Invest. 46, 191–194 (1998). 10.1159/000010031 [DOI] [PubMed] [Google Scholar]