Abstract
Nodamura virus (NoV; family Nodaviridae) contains a bipartite positive-strand RNA genome that replicates via negative-strand intermediates. The specific structural and sequence determinants for initiation of nodavirus RNA replication have not yet been identified. For the related nodavirus Flock House virus (FHV) undefined sequences within the 3′-terminal 50 nucleotides (nt) of FHV RNA2 are essential for its replication. We previously showed that a conserved stem-loop structure (3′SL) is predicted to form near the 3′ end of the RNA2 segments of seven nodaviruses, including NoV. We hypothesized that the 3′SL structure from NoV RNA2 is an essential cis-acting element for RNA replication. To determine whether the structure can actually form within RNA2, we analyzed the secondary structure of NoV RNA2 in vitro transcripts using nuclease mapping. The resulting nuclease maps were 86% consistent with the predicted 3′SL structure, suggesting that it can form in solution. We used a well-defined reverse genetic system for launch of NoV replication in yeast cells to test the function of the 3′SL in the viral life cycle. Deletion of the nucleotides that comprise the 3′SL from a NoV2-GFP chimeric replicon resulted in a severe defect in RNA2 replication. A minimal replicon containing the 5′-terminal 17 nt and the 3′-terminal 54 nt of RNA2 (including the predicted 3′SL) retained the ability to replicate in yeast, suggesting that this region is able to direct replication of a heterologous mRNA. These data suggest that the 3′SL plays an essential role in replication of NoV RNA2. The conservation of the predicted 3′SL suggests that this common motif may play a role in RNA replication for the other members of the Nodaviridae.
Keywords: RNA viruses, nodaviruses, Nodamura virus, RNA replication, RNA structure
1. Introduction
Members of the virus family Nodaviridae, which include Nodamura virus (NoV) and Flock House virus (FHV), are small riboviruses that contain bipartite positive strand RNA genomes (Ball and Johnson, 1998). The study of nodavirus RNA replication mechanisms has been greatly facilitated by their broad host ranges. Introduction of the isolated RNA genomes of NoV or FHV into cultured cells of mammalian, insect, plant, or yeast origin results in exponential RNA replication (Ball, 1992; Ball et al., 1992; Johnson et al., 2004; Lu et al., 2005; Price et al., 2005; Price et al., 1996; Selling et al., 1990; Tesh, 1980). The ability of cells from such a wide variety of organisms to support nodavirus RNA replication may suggest that any cellular host factors involved in this process are ubiquitous and/or highly conserved.
The viral requirements for genome replication and assembly are naturally segregated onto two separate genome segments that are co-packaged into the same virion. The larger genome segment, RNA1, encodes Protein A, the catalytic subunit of the viral RNA-dependent RNA polymerase (RdRp), while RNA2 encodes the capsid protein precursor, α. A small subgenomic RNA, RNA3, is synthesized during RNA1 replication. RNA3 encodes nonstructural proteins B1 and B2 from overlapping open reading frames (Johnson et al., 2003; Johnson et al., 2004). The function of B1 is unknown, while B2 suppresses RNA interference (RNAi) in both insect and mammalian cells (Johnson et al., 2004; Li et al., 2004; Sullivan and Ganem, 2005). Protein A catalyzes replication of both genomic RNAs and subgenomic RNA3 via negative strand replication intermediates. During RNA replication, dimeric negative-sense RNAs are synthesized for both FHV and NoV (Albariño et al., 2001; Johnson et al., 2003); these RNAs consist of covalently linked, head-to-tail monomers of RNAs 1, 2, and 3 (homodimers), and RNA2 covalently joined to RNA3 (heterodimers). When introduced into cells, synthetic FHV RNA dimeric templates of either polarity could be resolved to their monomeric counterparts by the RdRp, suggesting that the RdRp can recognize terminal cis-acting signals when they are located internally (Albariño et al., 2003; Albariño et al., 2001). For this reason, a possible role for the dimers as replication intermediates has been proposed, although it is also possible that naturally occurring dimers are dead-end products of the RdRp. Indeed, the exact mechanism of nodavirus RNA replication remains unclear. For example, the specific cis-acting signals required for initiation of negative strand synthesis have yet to be identified.
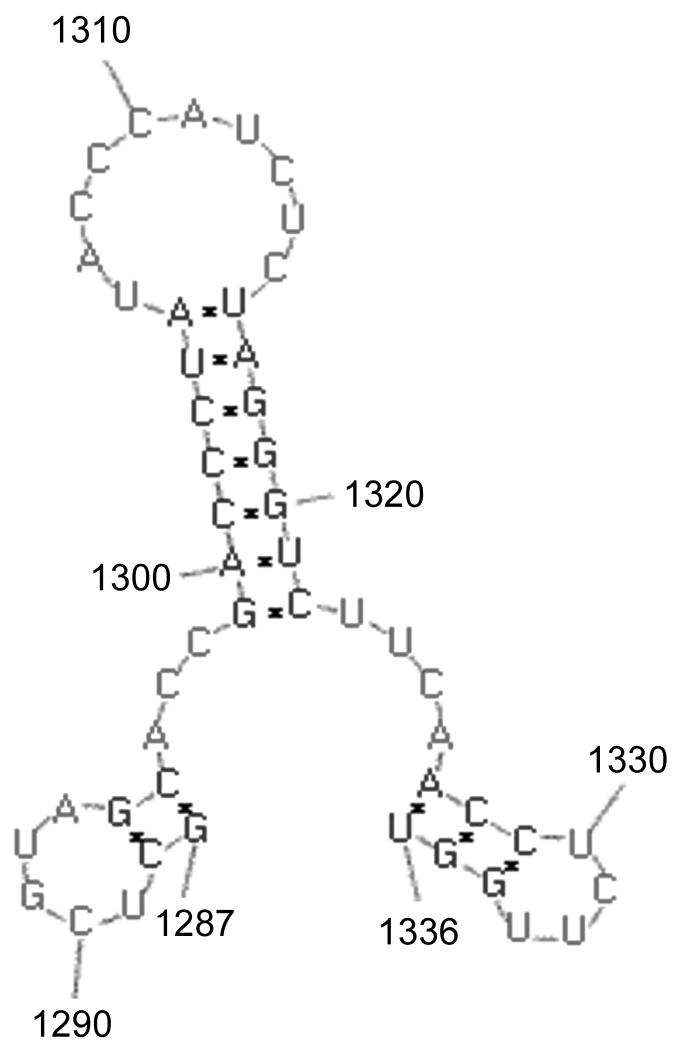
For many plant viruses with positive-strand RNA genomes, tRNA-like structures at the 3′ end of the genomic RNA play key roles in both translation and RNA replication. Indeed, the promoters for synthesis of negative strand replication intermediates of brome mosaic virus and tomato bushy stunt virus genomic RNAs have been identified within structural motifs near their 3′ termini (Chapman and Kao, 1999; Pogany et al., 2005). It was therefore reasonable to suspect that cis-acting signals (promoter elements) required for nodavirus RNA replication would lie at or near the genomic 3′ termini and would perhaps be influenced by the structures these sequences adopt. Two lines of evidence support this hypothesis. First, sequences from the 3′ terminal 50 nt of FHV RNA2 are sufficient to direct replication of a heterologous mRNA, indicating that an as-yet undefined cis-acting replication element must lie within this region (Albariño et al., 2003; Eckerle et al., 2003). Second, Kaesberg et al. (Kaesberg et al., 1990) predicted the existence of secondary structures within the 3′-noncoding regions of the RNA2 segments from BBV, BoV, FHV, and NoV. The role of these predicted structures in the viral replicative cycle was not examined, although they were hypothesized to function in RNA replication (Kaesberg et al., 1990). In order to further define the cis-acting elements required for nodavirus RNA2 replication, we previously extended these studies to include RNA2 segments from a total of seven nodaviruses: FHV, BBV, BoV, NoV, Pariacoto virus (PaV), Striped jack nervous necrosis virus (SJNNV) and Greasy grouper [Epinephelus tauvina] nervous necrosis virus (GGNNV). Since many viral RNA replication elements such as those found in plant viruses form pseudoknot structures, we used software tools that are able to predict these structures. RNA structure predictions were performed on the 3′ terminal 200 nucleotides of each segment using the RNA Virtual Laboratory (RNAVLab) platform (Taufer et al., 2008) running programs PseudoknotsRE (Rivas and Eddy, 1999), PseudoknotsRG (Reeder and Giegerich, 2004) and NuPack (Dirks and Pierce, 2003; Dirks and Pierce, 2004). RNAVLab consistently predicts a similar stem-loop structure for each virus, in each case positioned just upstream of the 3′ end of RNA2 (Taufer et al., 2008). The 3′SL predicted for NoV RNA2 is shown schematically in Figure 1; it comprises nt 1299-1322, just 14 nt upstream of the 3′ terminus located at nt 1336. Despite our use of software that allowed for the prediction of pseudoknots, our results for NoV are similar to those of the previous analysis (Kaesberg et al., 1990). In most cases, the predicted 3′SL structure formed a part of a larger pseudoknot structure, but prediction of the pseudoknot was not conserved for NoV or BBV (Taufer et al., 2008). Whether the sampled 3′ termini in NoV and BBV were too short to detect pseudoknots that might be predicted to form in a longer sequence and whether pseudoknot structures play any role in viral RNA replication remains to be determined. Nevertheless, the predicted 3′SL element is structurally conserved among all seven nodaviruses examined, although each predicted structure is comprised of different primary sequences (Taufer et al., 2008).
Figure 1. A conserved stem-loop structure (3′SL) predicted to form at the 3′ end of NoV RNA2.
RNA structure predictions of progressively shorter lengths from the 3′ end of RNA2 were performed on the RNAVLab platform (Taufer et al., 2008) using three programs: PseudoknotsRE (Rivas and Eddy, 1999), PseudoknotsRG (Reeder and Giegerich, 2004) and NuPack (Dirks and Pierce, 2003; Dirks and Pierce, 2004). Shown is the common prediction generated by all three programs for the 3′-terminal 50 nt of NoV RNA2.
In the present study, we used nuclease mapping to determine whether the predicted 3′SL structure within NoV RNA2 can form in solution and found that the resulting nuclease map was consistent with the predicted structure. Deletion of the NoV RNA2 3′SL from an RNA2-GFP subgenomic replicon resulted in a mutant that was unable to replicate in transformed yeast cells, suggesting that this structural element is essential for viral RNA replication. Finally, a minimal GFP-containing replicon that retained only the 5′-terminal 17 nt and the 3′- terminal 54 nt of NoV RNA2 (including the 3′SL structure) was still able to replicate in transformed yeast cells, suggesting that the sequences that comprise the predicted structure retain the ability to direct replication of a heterologous mRNA.
2. Materials and methods
2. 1. Cells and viruses
All recombinant plasmids were amplified in Escherichia coli strains Jm109 (Promega), NEB5α or NEB10β(New England Biolabs) grown in Luria-Bertani (LB) broth or on LB agar plates supplemented with ampicillin. RNA replication studies were performed in the S. cerevisiae synthetic deletion strain BY4733 (MATa his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0) (Brachmann et al., 1998), as described previously (Price et al., 2005; Price et al., 1996). Competent yeast cells were prepared and transformed using a Frozen-EZ Yeast Transformation II™ kit (Zymo Research Corp., Orange, CA). Plasmids containing appropriate selectable markers were co-transformed with experimental plasmids to allow growth of yeast cells in media selective for LEU2 and TRP1. For selection of leu+ trp+ colonies, yeast cells were plated on glucose-containing solid minimal medium (YNB) supplemented with histidine, methionine, and uracil and lacking leucine and tryptophan and incubated at 30°C. For induction of the GAL1 promoter, leu+ trp+ cells were inoculated into selective liquid medium containing 2% galactose and grown at 30°C for 24h prior to harvest.
2.2. Plasmids
We previously described our ability to reproduce the entire NoV replicative cycle in cells of the yeast S. cerevisiae from plasmids expressing cDNA copies of NoV RNA1 and RNA2 (plasmids pN1 and pN2, respectively) under transcriptional control of an inducible yeast GAL1 promoter (Price et al., 2005). Synthesis of replicable primary transcripts in transformed yeast cells is induced with galactose and authentic 3′ termini of the transcripts are generated by self-cleavage by the hepatitis delta virus (HDV) antigenomic ribozyme, as described previously (Price et al., 2005). Plasmids pN1 and pN2 also contain yeast TRP1 and LEU2 selectable markers, respectively, and pN2 contains the yeast ADH1 termination and polyadenylation sites immediately downstream of the HDV ribozyme (Price et al., 2005). Plasmid pNoV2(0,0), which contains the NoV RNA2 cDNA and HDV ribozyme sequences under transcriptional control of a bacteriophage T7 promoter, was described previously (Johnson et al., 2003).
2.2.1. Plasmid pT7-N2ΔMluIN2
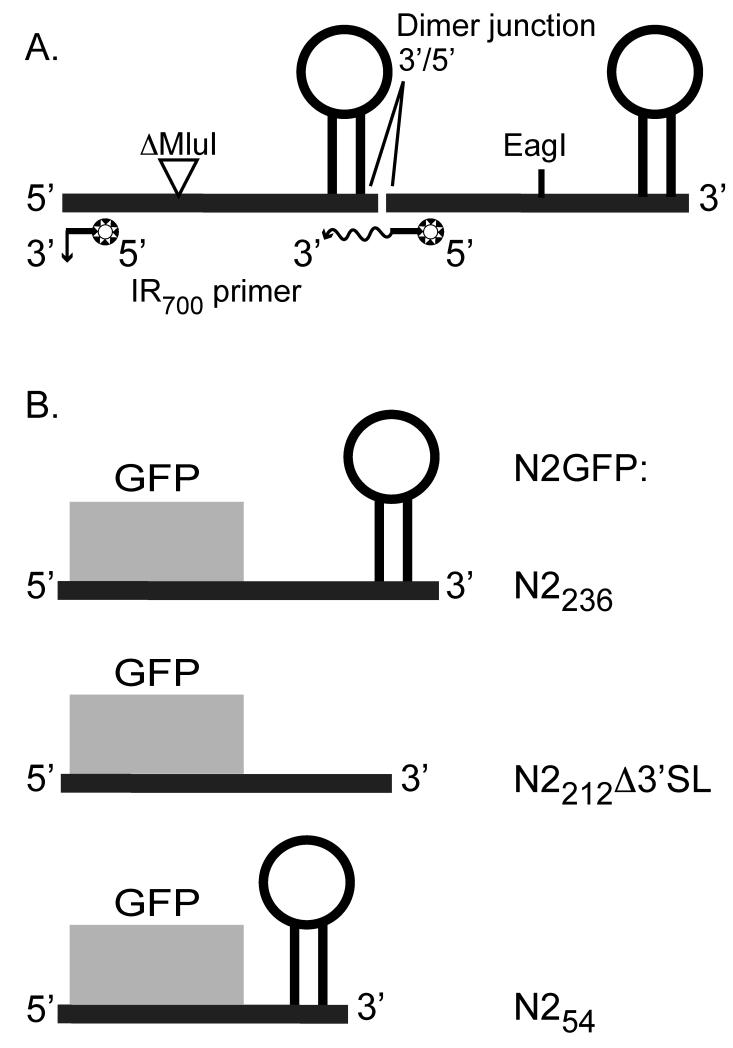
For use as a template for in vitro transcription reactions, we used standard cloning techniques to construct a plasmid, pT7-N2ΔMluIN2, which contains a head-to-tail dimer of the NoV RNA2 cDNA under control of a bacteriophage T7 promoter (Sambrook and Russell, 2001). We deleted a 319 bp MluI fragment (nt 780 – 1099) from the upstream copy of NoV2, which is followed by a complete downstream copy of NoV2, all within the plasmid backbone of pNoV2(0,0) (Johnson et al., 2003). The deletion removed an EagI site from the upstream copy, allowing us to use the single remaining EagI site in the downstream copy to linearize the plasmid. The junction between the two cDNA sequences is 5′-…CTTGGT/GTAAAC…-3′, where the 3′-terminus of the upstream copy of the NoV RNA2 cDNA is juxtaposed with the 5′ terminus of the downstream copy. A T7 transcript of the resulting dimer is shown schematically in Figure 2A.
Figure 2. Schematic of RNA transcripts used in these studies.
Panel A: A head-to-tail dimer of NoV RNA2 contains a 319 nt MluI-fragment deletion in the upstream copy and a full-length downstream copy. In vitro transcription is initiated from a T7 promoter in plasmid pT7-N2ΔMluIN2, which is linearized at the unique EagI site shown. The transcript contains two binding sites for the IR dye-labeled primer. On primer binding at the upstream site near the 5′ end, extension of the primer results in a short run-off product, while p31imer binding at the downstream site results in longer extension products that span the dimer junction. Panel B: Replicons N2GFPN2236, N2GFPN2212Δ3′SL, and N2GFPN254 contain 17 nt (nt 1 – 17) from the 5′ end of RNA2 followed by the GFP central core and 236 nt (nt 1100 – 1336), 212 nt (nt 1100 – 1298 and 1323-1336), or 54 nt (nt 1282 – 1336) from the 3′ end of RNA2. Primary transcription of each is initiated in transformed yeast cells from an inducible GAL1 promoter in plasmids pN2GFPN2236, pN2GFPN2212Δ3′SL, and pN2GFPN254, respectively.
2.2.2. Plasmid pG4-N2ΔMluIN2PvuII
We constructed an NoV2 dimer subclone in the pGEM®-4 vector (Promega), using a 1017 bp PvuII fragment from pT7-N2ΔMluIN2 that contained the primer junction, to yield plasmid pG4-N2ΔMluIN2PvuII. This subclone, which contains a single binding site for the primer used in primer extension and DNA sequencing, was used to generate the DNA sequencing ladder for the nuclease mapping analysis.
2.2.3. Plasmid pN2GFPN2236
We used PCR to construct a plasmid, pN2GFPN2236, which contains the coding region for mammalian-codon optimized green fluorescent protein (GFP), flanked by sequences from the NoV RNA2 cDNA. In this case, the GFP ORF serves as a common heterologous core region to which we can direct strand-specific probes, providing a uniform assay for testing the activity of different N2GFPN2 deletion variants, as previously described for FHV (Albariño et al., 2003; Eckerle et al., 2003). The resulting plasmid contains 17 nt (nt 1 – 17) from the 5′ end of RNA2, the GFP central core, and 236 nt (nt 1100 – 1336) from the 3′ end of RNA2. This replicon is shown schematically in Figure 2B.
2.2.4. Plasmid pN2GFPN2212Δ3′SL
We deleted the 3′SL structure from pN2GFPN2236 using PCR-based circular mutagenesis followed by Dpn1 selection, as described (Sambrook and Russell, 2001). The region of interest was confirmed by sequencing and a small DNA fragment containing the desired mutation was re-ligated into parental plasmid pN2GFPN2236 using standard techniques (Sambrook and Russell, 2001). The resulting plasmid, pN2GFPN2212Δ3′SL, contains 17 nt (nt 1 – 17) from the 5′ end of RNA2, the GFP central core, and 212 nt (nt 1100 – 1298, 1323 – 1336) from the 3′ end of RNA2, with a perfect deletion of the nt (nt 1299 – 1322) that form the predicted 3′SL; the 3′ terminal 14 nt are authentic to NoV RNA2. This replicon is shown schematically in Figure 2B.
2.2.5. Plasmid pN2GFPN254
PCR was used to remove the sequences between nt 1100 and 1281 from pN2GFPN2236, retaining the 3′ terminal sequences 1282-1336, including the nt that form the predicted structure (nt 1299-1322) as well as the authentic 3′ end. The resulting plasmid contains 17 nt (nt 1 – 17) from the 5′ end of RNA2, the GFP central core, and 54 nt (nt 1282 – 1336) from the 3′ end of RNA2. This replicon is shown schematically in Figure 2B.
2.3. Nuclease mapping and primer extension analysis
Nuclease mapping studies were performed on NoV2 in vitro transcripts as described (Hanley et al., 2002; Romero et al., 2006; Tuplin et al., 2004). Plasmid pT7-N2ΔMluIN2, which contains the head-to-tail dimer of NoV RNA2 under transcriptional control of a bacteriophage T7 promoter, was linearized with EagI and used as a template in in vitro transcription reactions using T7 RNA polymerase (Promega). The resulting transcripts were purified using an RNeasy Mini Kit (QIAGEN) and eluted in water; these transcripts are represented schematically in Figure 2A. Nuclease digestion of in vitro transcripts of the NoV2 dimer was carried out using single- or double-strand specific ribonucleases (RNases) A, I, T1, and V1 (Applied Biosystems/Ambion), as described (Romero et al., 2006; Tuplin et al., 2004) except that 5 pmol of RNA was incubated in structure buffer (10 mM Tris, pH 7, 100 mM KCl, 10 mM MgCl2) at 65°C for 2 min. and room temperature for 5 min. The RNA was treated with 0.05 U RNase A, 0.5 U RNase T1, 10 U RNase I, 0.005 U RNase V1 (Ambion), or no RNase (for no nuclease control), and incubated at room temperature for 10 minutes. Digestion was halted and samples precipitated in a solution containing 5 μg yeast tRNA, 2 μl of GlycoBlue (Ambion) and 25 μl Precipitation/Inactivation buffer (Ambion).
The digested samples were subjected to primer extension analysis using Superscript III Reverse transcriptase (Invitrogen) and an infrared dye-labeled primer (IR700; LI-COR Biosciences), which allowed us to map the sensitive sites for each nuclease. The primer contained the sequence 5′-GGGGCCGCTCTTCGGCGGCG-3′, which is complementary to nt 57-38 of NoV RNA2. A DNA sequencing ladder was generated with a SequiTherm EXCEL™ II DNA sequencing kit (Epicentre Biotechnologies) using the same IR700-labeled primer and plasmid pG4-N2ΔMluIN2PvuII as template. Samples were analyzed on a polyacrylamide sequencing gel (LI-COR Biosciences 3.7% gel matrix) using a LI-COR 4200 DNA Analyzer running e-Seq V2.0 software (LI-COR Biosciences). For ease of interpretation, the raw gel images were reversed so that the complement of the negative sense primer extension products can be read directly from the gel. Similarly, the DNA sequencing ladder was labeled with its complement, and T’s replaced with U’s, so that its correspondence to the computer prediction is readily apparent. Finally, the contrast of the entire gel image was adjusted slightly using Adobe Photoshop for optimal viewing of the bands. To calculate the percentage of consistency between the empirical map and the structure prediction, we divided the total number of enzymatic cleavages observed (22) by the number of cleavages that were consistent (19/22) or inconsistent (3/22) with the prediction and multiplied the result by 100, as described (Romero et al., 2006).
2.4. Northern blot hybridization analysis
Total RNA was isolated from plasmid-transformed yeast cells using hot phenol as described previously (Leeds et al., 1991; Newman et al., 1993). The RNA (0.5 μg for detection of positive strands and 2 μg for detection of negative strands) was separated on denaturing formaldehyde-agarose gels and stained with ethidium bromide to allow quantitation of ribosomal RNAs, as described below. The RNA was then transferred to charged nylon membranes (Nytran SuPerCharge (SPC), Whatman), and Northern blot hybridization was performed as described previously (Leeds et al., 1991; Newman et al., 1993), using strand-specific 32P-labeled RNA probes that contained positive- or negative-strand sequences of NoV RNA1 nt 2732-3204 (the entire RNA3 region) or of the entire coding region for mammalian codon-optimized GFP (nt 20-736 of plasmid pN2GFPN2236). The results were visualized with a Bio-Rad Personal Molecular Imager and quantitated using Quantity One 1-D Analysis Software (Bio-Rad Laboratories). Levels of N2GFPN2 RNA replication products were normalized to those of yeast cellular 25S ribosomal RNA (visualized by ethidium bromide staining of the gel before transfer) and are presented as a percentage of the WT N2GFPN2236 values in Figures 4 and 5. The relative RNA values from three (Figure 4) or four (Figure 5) independent experiments are presented as mean values ± standard deviations.
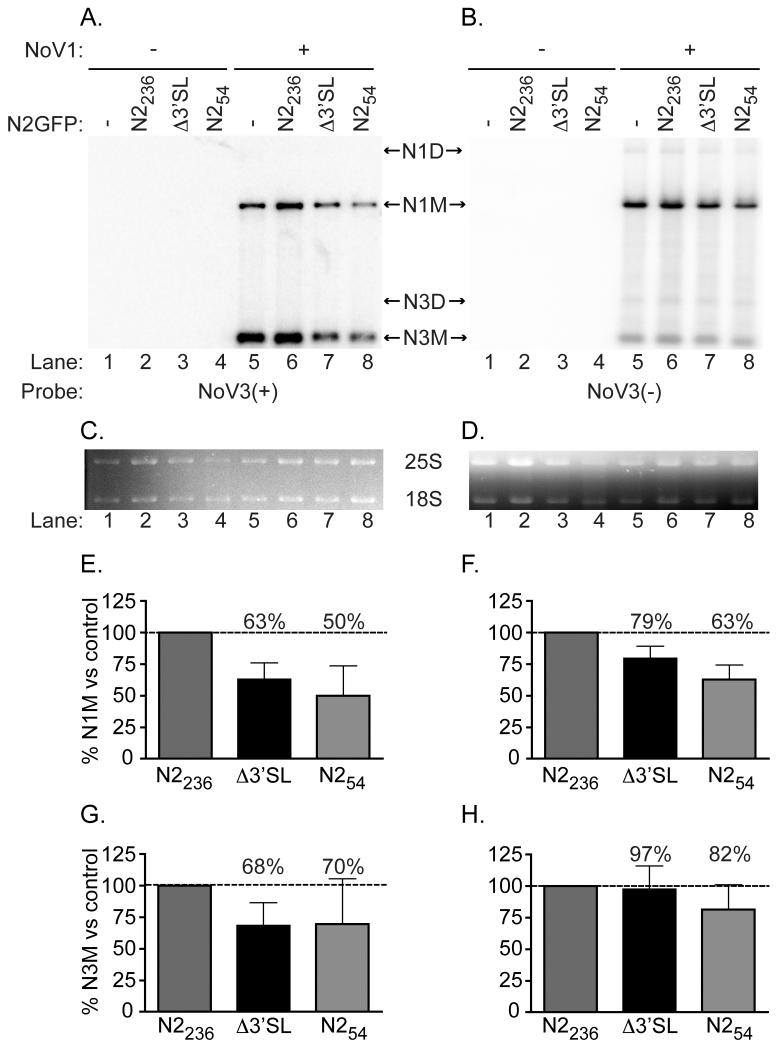
Figure 4. Δ3′SL mutant doesn’t affect RNA1 replication or RNA3 synthesis.
Total yeast RNA isolated from cells transformed with pN2GFPN2236, pN2GFPN2212Δ3′SL, or pN2GFPN254, together with pN1 as a source of RdRp, was separated on a denaturing gel and subjected to Northern blot hybridization analysis using probes specific for the positive (Panel A) or negative (Panel B) strands of RNA3 (labeled as NoV3(+) and NoV3(−), respectively). A representative blot is shown. Prior to transfer, gels were stained with ethidium bromide to allow visualization of yeast cellular 25S and 18S rRNAs (Panels C and D) for use as loading controls. Both monomeric and dimeric forms of NoV RNA1 and RNA3 were detected, indicated in the figure as N1M or N3M (monomers) and N1D or N3D (dimers). Quantitation of the positive- and negative-strand RNA1 and RNA3 species detected, compared to the levels detected for the WT N2GFP236N2 replicon in the presence of RdRp (lane 5) are shown in Panels E–H; : Panel E, RNA1(+); Panel F, RNA1(−); Panel G, RNA3(+); Panel H, RNA3(−). The relative RNA values from three independent experiments are presented as mean values ± standard deviations.
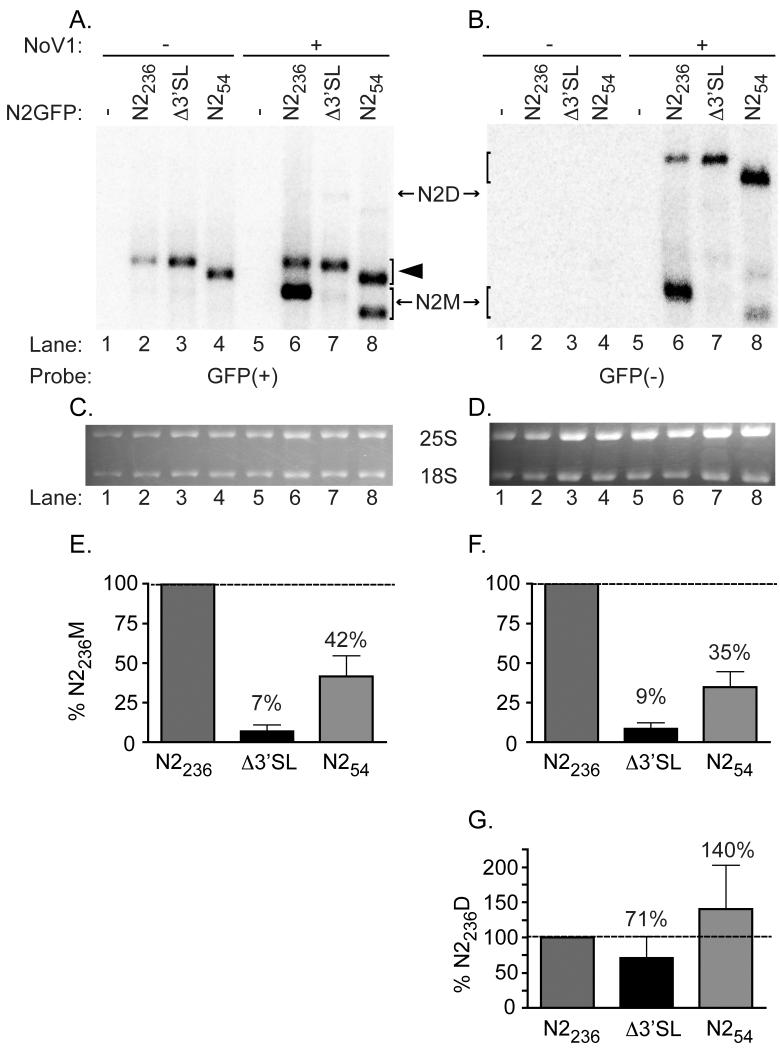
Figure 5. Stem loop mutant Δ3′SL exhibits a defect in RNA2 replication, while the 3′-terminal 54 nt of NoV RNA2 are sufficient to direct replication of a heterologous mRNA.
Total yeast RNA from cells transformed with pN2GFPN2236, pN2GFPN2212D3′SL, or pN2GFPN254, together with pN1, was subjected to Northern blot hybridization analysis as described in the legend to Figure 4, except that the probes were specific for the positive (Panel A) or negative (Panel B) strands of the GFP core region within the N2GFPN2 replicons (labeled GFP(+) and GFP(−), respectively). A representative blot is shown; ethidium bromide-stained cellular 25S rRNA was used as a loading control (Panels C and D). Both monomeric and dimeric forms of the N2GFPN2 replicons were detected, indicated in the figure as N2M (monomer) or N2D (dimer). The closed arrowhead indicates the position of the uncleaved positive-sense primary transcript. Quantitation of positive strand monomers (Panel E), negative strand monomers (Panel F), or negative strand dimers (Panel G) is shown; the relative RNA values from four independent experiments are presented as mean values ± standard deviations.
3. Results and Discussion
3.1. The predicted 3′SL structure in NoV RNA2 forms in solution
As described previously, the RNAVLab analysis predicts a conserved stem-loop structure (3′SL) involving nt 1299 – 1322 of the NoV RNA2 3′-noncoding region. 3′SL is comprised of a 7 bp stem (nt 1299 – 1305 base-pairing with nt 1316 – 1322) and a 10 nt loop (nt 1306-1315), located just 14 nt from the 3′ end (Taufer et al., 2008). This structure is shown schematically in Figure 1. Its conservation among seven nodaviruses suggests that the predicted structure may play a functional role in the nodavirus life cycle. Prior to assessing that role, we first determined whether the predicted structure could form in solution using in vitro transcripts of NoV RNA2.
As described in the Introduction, we consistently detect dimeric negative strand RNA species during NoV RNA replication. These molecules have been proposed to function as RNA replication intermediates. We hypothesized that our use of NoV RNA2 dimers as templates in the nuclease mapping studies would allow us to place a primer near the 5′ end of the downstream copy that could be extended across the dimer junction and allow mapping of the 3′SL present in the upstream copy (shown schematically in Figure 2A). PseudoknotsRE predicted formation of an internal copy of the 3′SL within the context of the RNA2 dimer (data not shown), strengthening our confidence in this approach.
Therefore, dimeric in vitro transcripts were synthesized and subjected to digestion with ribonucleases specific for single- (RNases I, A, and T1) or double-stranded RNA (RNase V1) or left untreated as controls to detect non-specific cleavage and intrinsic stops (premature transcription termination) during reverse transcription. RNase I cleaves 3′ of all four single-stranded nucleotides, RNase A cleaves 3′ of single-stranded C and U nucleotides, RNase T1 cleaves 3′ of single-stranded G nucleotides, and RNase V1 cleaves double-stranded nucleotides. The results of a representative experiment are shown in Figure 3: the nuclease mapping results are shown in Figure 3A, alongside a DNA sequencing ladder. The resulting nuclease cleavage pattern was projected onto the predicted structure in Figure 3B. It is worthwhile to note that, since RNases I, A, and T1 cleave 3′ of the reactive nucleotide, the corresponding DNA sequencing product generated by primer extension stops one base short of the reactive residue on the dideoxy sequencing ladder, while RNase V1 generates primer extension products that co-migrate with the sequencing ladder (Jacobson et al., 1993). However, the relative scarcity of G residues in the predicted structure (4 of 24 total nt) prevented us from generating a sequencing ladder by digesting denatured RNA with RNase T1.
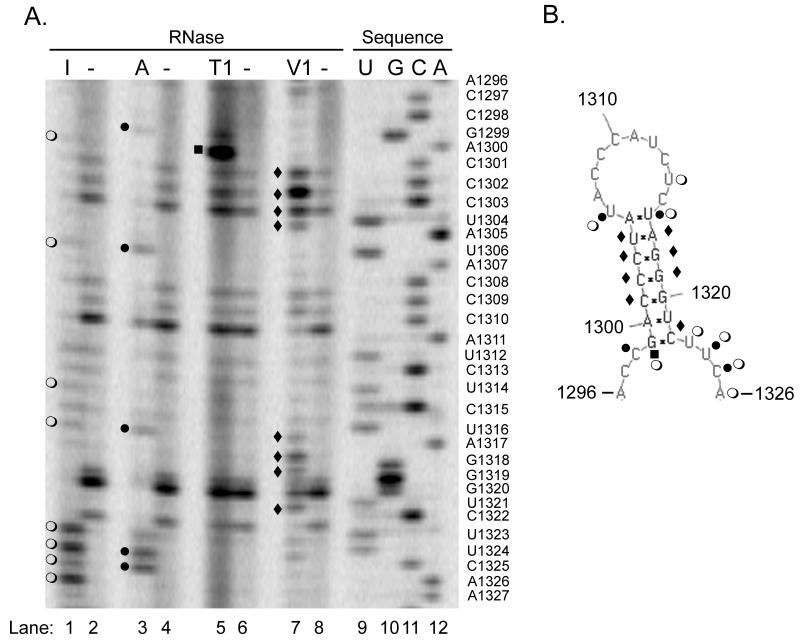
Figure 3. Nuclease mapping studies show the predicted structure can form in solution.
In vitro transcripts of the NoV RNA2 homodimer shown in Figure 2A were synthesized and subjected to digestion with ribonucleases specific for single- (RNases I, A, and T1) or double-stranded RNA (RNase V1), or left untreated. After digestion, samples were subjected to primer extension analysis using an infrared dye-labeled primer complementary to NoV RNA2 nt 57-38. A DNA sequencing ladder was generated with the same primer and the corresponding plasmid DNA template, as described in Material and Methods. Samples were analyzed on a polyacrylamide sequencing gel using a LI-COR 4200 DNA Analyzer. Panel A, nuclease map with sequencing ladder; I, RNase I (lane 1); A, RNase A (lane 3); T1, RNase T1 (lane 5); V1, RNase V1 (lane 7); minus (−), no nuclease (lanes 2, 4, 6, and 8); U, G, C, A (lanes 9 – 12), sequencing ladder. The sequencing ladder was labeled with the nucleotides complementary to the sequence obtained, and U’s were substituted for T’s, to allow direct comparison with panel B. Panel B, predicted structure annotated with the results of the nuclease mapping. Open circles (○), RNase I digestion product; closed circles (●), RNase A; closed squares (■), RNase T1; closed diamonds (◆), RNase V1.
The nuclease mapping (Figure 3) showed enzymatic cleavage of 17 out of the 24 total nucleotides in the predicted structure (71%). Of the 17 cleaved nucleotides, 5 nt were cleaved by two different nucleases for a total of 22 enzymatic cleavages. Inefficient ribonuclease cleavage may be attributable to steric radii within the nucleases that can block access to solvated nucleotides (Christiansen and Garrett, 1988). In addition, in vitro transcripts can be expected to undergo multiple, temporary conformational changes, exposing sites that may not be present in the majority structure conformation and resulting in minor cleavage events (Tuplin et al., 2004). The nuclease mapping results we obtained show that eighty-six percent of the observed enzymatic cleavages corresponded to the prediction generated by PseudoknotsRE, PseudoknotsRG, and NuPack (Figures 1 and 3). The percent consistency (86%) was calculated by dividing the total number of cleavages observed (22) with the number of cleavages that were consistent with the prediction (19), as described (Romero et al., 2006). Eight nucleotides were cleaved with RNase I, which cleaved 3′ of nt U1306, U1314, U1323, U1324, C1325, and A1326, all of which are found in regions predicted to be single-stranded (Figure 3, lane 1). These six cleavages are consistent with the predicted structure. Two additional RNase I cleavage products were detected (G1299 and U1316), resulting from cleavage of nucleotides predicted to be found in double-stranded regions; these and other inconsistent cleavages are discussed further below. We also detected five cleavages by RNase A, which cleaved 3′ of nt C1298, U1306, U1324, C1325, confirming the single-stranded nature of these residues (Figure 3, lane 3). One additional nucleotide, U1316, was cleaved with RNase A, but it was predicted to lie in a double-stranded region, as discussed further in the next paragraph. Some experiments also resulted in RNase A cleavage 3′ of nucleotides C1309, A1311, C1315, all of which lie in the predicted single-stranded loop (data not shown). However, since this additional cleavage was not seen in all experiments, these cleavages were not included in the percent consistency calculations. Digestion with RNase T1 resulted in only one unambiguous cleavage product in this region of the gel (Figure 3, lane 5); this product is considered further below. Such inefficient T1 cleavage was expected, since G residues comprised only 4 of the 24 nucleotides in the predicted structure (17%), as noted above, and all of them are predicted to base-pair with other nucleotides within the structure.
The double-strand specific ribonuclease V1 cleaved 8 residues within the predicted stem, including C1301, C1302, C1303, and U1304 from the upstream side of the stem and residues A1317, G1318, G1319, and C1322 from the downstream side (Figure 3, lane 7). In this experiment, the two additional V1 cleavages (at nt G1320 and U1321) are visible, but since there are apparent intrinsic stops at these positions in the absence of ribonuclease treatment (no nuclease; Figure 3, lanes 2, 4, 6, and 8), these potential cleavages were not included in the percent consistency calculations. Taken together, the nuclease V1 results were consistent with a double-stranded region in the area of the predicted stem.
Out of the 22 total enzymatic cleavages detected in these experiments, 3 cleavages were inconsistent with the predictions made by PseudoknotsRE, PseudoknotsRG, or NuPack (14% inconsistency; calculated by dividing the total number of cleavages observed with the number of cleavages that were inconsistent with the prediction, as described ((Romero et al., 2006). Nt U1316 was cleaved by both RNase A and RNase I, despite its position at the top of the predicted double-stranded stem (Figure 3); neither cleavage is consistent with the prediction. However, in some experiments, RNase V1 also cleaved this residue (data not shown). U1316 is predicted to form a U-A base pair with A1305 at the top of the stem, so it is conceivable that breathing at this position could result in its being sensitive to cleavage by both single- and double-strand specific ribonucleases. Alternatively, perhaps this U-A base pair at the top of the stem does not form at all and U1316 is actually single-stranded. If so, this change would have minimal impact on the overall predicted structure, resulting in a stem of six rather than seven base pairs. Also inconsistent with the model is our observation that nt G1299 was sensitive to digestion by single-strand specific RNases I and T1, since it is predicted to base pair with residue C1322 at the bottom of the stem. As noted above, C1322 was sensitive to digestion with RNase V1, suggesting that it is indeed base-paired, possibly with nt G1299 or with another nt. In the case of both U1316 and G1299, the absence of bands at these positions in the untreated controls suggest that the bands detected in the treated samples were not due to intrinsic stops (Figure 3A, lanes 2, 4, 6, and 8), but instead represented actual enzymatic cleavages. A high proportion of similar dynamic cleavages have been described during structure mapping studies of dengue and West Nile virus RNA (Romero et al., 2006; Shi et al., 1996; Tilgner et al., 2005; Tilgner and Shi, 2004). Shi et al. attributed these dynamic cleavages to higher order structures, such as pseudoknots, that could alternate between different conformations (Shi et al., 1996). For NoV RNA2, the position of both inconsistencies at the top or bottom ends of the predicted stem suggest that breathing at this position could result in its being sensitive to cleavage by both single- and double-strand specific ribonucleases. Alternatively, this structure may participate in transient, long-range interactions (pseudoknots).
Together the results of the nuclease mapping studies are 86% consistent with formation of the predicted 3′SL in solution. However, the functional significance of the structure in the viral life cycle remained unknown. To determine the functional role of the predicted structure, we used the reverse genetic system for launch of NoV RNA replication in transformed yeast cells to delete the sequences that comprise the structure from a GFP-containing RNA2 replicon and to test the effect of this deletion on RNA replication.
3.2. A 3′SL deletion mutant exhibits a severe defect in RNA2 replication
We constructed an RNA2-based replicon, pN2GFPN2236, which contains 17 nt from the 5′ end of RNA2, the GFP ORF (716 nt) as a central core region to which probes can be directed, and 236 nt from the 3′ end of RNA2, including the 3′SL. To test the role of the predicted 24 nt 3′SL in RNA replication, we deleted it from the pN2GFPN2236 replicon while leaving the 3′-terminal 14 nt unchanged, resulting in plasmid pN2GFPN2212Δ3′SL. These replicons are shown schematically in Figure 2B. Yeast cells were transformed with pN2GFPN2236 or pN2GFP212Δ3′SL, together with pN1 as a source of RdRp. Total yeast RNA was isolated and subjected to Northern blot hybridization analysis with probes specific for the positive (panel A) or negative strands (panel B) of RNA3 (which also detect RNA1 species) or of the common GFP core region within the N2GFPN2 replicons (Figures 4 and 5, respectively). Since negative strand replication intermediates are present in the cell at a much lower level than positive strands (on the order of one negative strand for each 100 positive strands), we loaded four times more RNA onto each gel, and used exposure times that were approximately ten-fold longer, for detection of negative strands than for positive strands. To allow direct comparison between the relative amounts of RNA1 and RNA3 produced by WT and mutant RNA2 replicons, we defined the levels detected in the presence of the WT replicon as 100% (Figures 4C and 4D).
As shown in Figure 4, we detected positive strands of monomeric RNA1 and RNA3 species when cells were transformed with pN1, either alone (Figure 4A, lane 5) or in the presence of WT pN2GFPN2236 (Figure 4A, lane 6). Co-transformation of yeast cells with mutant pN2GFPN2212Δ3′SL and pN1 resulted in only a modest decrease in levels of RNA1 or RNA3 (Figures 4E and 4G), suggesting that the deletion of the predicted structure in RNA2 did not primarily affect RNA1 replication or RNA3 synthesis (Figure 4A, lane 7). We also detected negative strand replication intermediates (both monomeric and dimeric) of RNA1 and RNA3 in the presence of the RdRp (Figure 4B, lanes 6 – 8). The monomeric negative strand RNAs are quantified in Figures 4F and 4H; the observed pattern was similar to that seen for the positive strands.
In Figure 5, the same RNA samples were subjected to Northern blot hybridization analysis with a probe specific for the GFP central core. When yeast cells were transformed with pN2GFPN2236 or pN2GFPN2212Δ3′SL in the absence of pN1 (Figure 5A, lanes 2 and 3), positive-strand primary transcripts were detected; these differed in size from the N2GFPN2 replication products due to the presence of uncleaved HDV ribozyme sequences (Price et al., 2005). When yeast cells were co-transformed with pN1 and pN2GFPN2236, the N2GFPN2 chimera replicated; monomeric positive strands (Figure 5A, lane 6) and both monomeric and dimeric negative strand intermediates (Figure 5B, lane 6) were detected. However, when cells were co-transformed with pN1 and the N2GFPN2212Δ3′SL mutant, only very low levels of monomer length positive (Figure 5A, lane 7, and Figure 5C) or negative strands (Figure 5B, lane 7, and Figure 5D) of the N2GFPN2 replicon were detected, indicating a severe defect in RNA2 replication in the absence of the predicted stem-loop. Quantitative analysis showed that the N2GFPN2212Δ3′SL replicon produced only 7% of the control (N2GFPN2236) level of positive strands (Figure 5E) and 9% of the control level of negative strands (Figure 5F).
Surprisingly, we also detected negative strand RNA replication products of dimer length with the N2GFPN2212Δ3′SL replicon (Figure 5B). Over the course of four independent experiments, the level of dimers averaged only 30% less than that detected for the N2GFPN2236 control (71%), with a range of values from 44 to 100% of the control (Figure 5G). For FHV, these species have been proposed to function as possible RNA replication intermediates. The exact mechanism by which dimeric nodavirus RNAs are synthesized is unclear, although it has been hypothesized that dimers form when the viral RdRp fails to release a nascent strand after synthesis and instead makes a template switch to initiate a second round of RNA replication, resulting in two nascent strands that are covalently linked (Albariño et al., 2003; Albariño et al., 2001). These authors showed that synthetic dimeric FHV RNAs could function as templates for synthesis of the corresponding monomers, suggesting that the cis-acting signals could be recognized internally within the dimeric template (Albariño et al., 2003; Albariño et al., 2001). Since the NoV RNA2 dimers were seen in the presence (Figure 5B, lanes 5-8) but not the absence of RNA1 (Figure 5B, lanes 2-4), we must conclude that their synthesis was catalyzed by the RdRp, even from a template that lacked the 3′SL (Figure 5B, lane 7). The accumulation of dimer length negative strands of NoV RNA2 in the absence of the 3′SL could mean that initiation of negative strand synthesis does not require this structural element, but rather an as-yet unidentified cis-acting signal is required instead. Alternatively, the 3′SL may indeed serve as an essential cis-acting signal for negative strand initiation but in its absence, another structural element (such as the HDV ribozyme present in an uncleaved primary transcript) may provide a substitute, leading to formation of dimer-like molecules that could contain non-NoV sequences. In either case, the 3′SL element appears to be required for resolution of the dimers, since even the WT level of dimeric negative strands synthesized by the 3′SL mutant failed to result in a WT level of monomeric positive strand synthesis. The implications of this result will be considered further at the end of section 3.3.
These results establish the necessity of the sequences that comprise the predicted 3′SL structure for efficient replication of RNA2, regardless of their mechanism of action. If this is an essential RNA replication element, we hypothesized that it should be sufficient to direct replication of an RNA2-based replicon that lacked most other RNA2 sequences. Therefore we tested whether a minimal replicon containing only the 3′SL and the authentic 3′ terminus could still replicate.
3.3. A minimal replicon containing the 3′-terminal 54 nt of NoV RNA2 (including the 3′SL) replicates in transformed yeast cells
We constructed a minimal replicon, N2GFPN254, which besides the 17 nt at the 5′ end retains only the 3′-terminal 54 nt of RNA2 downstream of GFP, a net deletion of 181 nt (RNA2 nt 1100-1281) relative to pN2GFPN2236 (shown schematically in Figure 2B). As before, the extreme 3′ terminus is authentic. The ability of this mutant to replicate was tested in yeast cells co-transformed with pN1 and pN2GFPN254. As before, total yeast RNA was subjected to Northern blot hybridization analysis with probes specific for the positive (panel A) or negative strands (panel B) of RNA3 or of the common GFP core in the N2GFPN2 replicons (Figures 4 and 5, respectively). The N2GFP54N2 replicon exhibited a modest decrease in levels of RNA1 and RNA3 detected (Figure 4A and 4B, lane 8) relative to the WT N2GFP236N2 replicon (lane 6), synthesizing 50% the WT level of monomer positive strands (Figure 4E) and 63% the WT level of monomer negative strands (Figure 4F) of RNA1; similar effects were observed for RNA3 in Figures 4G and 4H. In contrast, when we probed for the RNA2-based replicon, we detected monomeric positive strand RNA2 replication products (Figure 5A, lane 8) and both monomeric and dimeric negative strands (Figure 5B, lane 8), indicating that the minimal replicon retained the ability to replicate, albeit at a lower level than the parental replicon.
Quantitative analysis showed that the N2GFPN254 replicon produced 42% of the N2GFPN2236 level of monomeric positive strands (Figure 5E), and 35% of the N2GFPN2236 level of monomeric negative strands (Figure 5F). We also consistently detected dimer length negative strand RNA replication products (Figure 5B) at a level that averaged 1.4 times greater than that detected with the pN2GFPN2236 control, with a range of 77-200% of the control level (Figure 5G). Although in the gel shown the level of yeast 25S rRNA is slightly greater for the minimal replicon than for the parental replicon (Figure 5D, compare lanes 6 and 8), such variance was accounted for in the quantitation by normalizing all of the samples to the most intense species detected. The reason for this apparent accumulation of dimer length negative strands by the minimal replicon is unclear, but it may be related to the reduction in positive strand synthesis relative to the parental replicon (42% of the WT level). Perhaps this mutant has removed additional regulatory sequences that allow, for example, efficient use of a dimeric template for positive strand synthesis. In that case, full RNA replication may require additional sequences that are absent in the minimal replicon. The observation that this minimal replicon replicated less well than the parental N2GFPN2236 replicon, which in turn replicated less well than full length RNA2 (data not shown), lends support to this hypothesis. While the 3′SL in the context of the pN2GFPN254 replicon is sufficient to direct its replication at a basal level, full replication of RNA2 may require additional sequences. At this point, we are unable to rule out the possibility that sequences upstream of the stem-loop in full-length RNA2 contribute to the cis-acting signal for RNA replication, perhaps via long-range structural interaction. Nevertheless, these results indicate that the sequences contained in the last 54 nt of RNA2, including the predicted 24 nt 3′SL structure, were able to direct replication of RNA2.
Our results with NoV are consistent with those for FHV described by Albariño et al. (Albariño et al., 2003), who showed that a region comprising the 3′ terminal 50 nt of RNA2 was sufficient to direct replication of a heterologous RNA replicon. In the absence of FHV RNA3, replicons that contained the 3′ terminal 50 nt of FHV2 were unable to replicate, demonstrating that the cis-acting RNA2 replication signal was absolutely dependent on the presence of RNA3 (Albariño et al., 2003). Since all of the experiments described here were performed in the presence of RNA3, provided from the NoV1 plasmids used as a source of RdRp, it remains to be determined whether the 3′-terminal cis-acting element in NoV RNA2 requires RNA3 to function in RNA replication.
On the basis of the data presented here, we hypothesize two potential functions for the 3′SL structure, or the sequences that comprise it, in replication of RNA2. First, it may play a role as a key cis-acting signal for initiation of negative strand synthesis. We hypothesize that the 3′SL at the 3′ end of the RNA2 positive strand could bind the NoV RdRp and direct its synthesis of a negative strand replication intermediate. As suggested by Albariño et al. (Albariño et al., 2001), the RdRp could then either release the nascent strand (resulting in synthesis of a negative strand monomer) or retain it, leading to synthesis of a negative strand dimer. This hypothesis has yet to be fully addressed, particularly in light of our observation that the 3′SL deletion mutant appears still able to synthesize nearly WT levels of dimer length negative strands but only 9% the WT level of monomer negative strands (Figure 5). This result might be explained if the dimer length molecules produced by the Δ3′SL were the result of aberrant initiation by the RdRp on an alternate cis-acting signal. One candidate for such a cryptic initiation signal could be the pseudoknot-like structure of the HDV ribozyme that is still present on the uncleaved primary transcripts. Perhaps the RdRp binds so tightly to the putative aberrant signal that it is unable to release the nascent strand, leading to exclusive production of the dimeric rather than the monomeric form of the negative strand. Experiments are in progress to sequence the dimeric RNA species produced by the 3′SL deletion mutant and compare it to that of the dimers produced by the N2GFP replicons to determine whether there are non-NoV sequences present. This hypothesis also predicts that the 3′SL signal functions in the positive strand, which remains to be determined.
Second, we hypothesize that the complement of the 3′SL is required in the negative sense to initiate synthesis of positive strands from a dimeric template. This hypothesis predicts that the NoV RdRp could bind the complement of the 3′SL, present near the 5′ end of the negative strand and internally just 3′ of the dimer junction. Binding to the internal copy would allow synthesis of positive strand monomers. The data presented here are consistent with this hypothesis, since the 3′SL deletion mutant produced nearly WT levels of dimeric negative strand RNA2 yet was unable to produce the monomeric positive strand. This hypothesis predicts that the complement of the 3′SL functions internally and in the negative strand. Experiments are in progress to disrupt the 3′SL in a strand specific manner, to determine whether this structure is required in the positive or negative polarity, and to determine whether it can function internally within an synthetic dimeric template. The results of these experiments will help to pinpoint the mechanistic role of the 3′SL in RNA2 replication.
It remains to be determined whether it is the stem or the loop component of the 3′SL that plays a key role in NoV RNA2 replication, or if both are required. Mutations that alter the ability of the nucleotides in the stem region to base pair are currently being constructed and tested. However, preliminary results suggest that mutation of nucleotides in the loop result in decreased RNA2 replication in yeast (Rosskopf, Melendez, Betancourt, and Johnson, data not shown). If this loop region is required for RNA replication, perhaps the nucleotides in the loop participate in RNA-RNA interactions - potentially long-range interactions such as pseudoknots - or in RNA-protein interactions. perhaps with the viral RdRp. Indeed, both hypothetical functions for the 3′SL predict that the cis-acting signal might interact with the RdRp, either directly or indirectly. If so, perhaps this is a global mechanism used by NoV to synthesize not only RNA2, but also RNA1.
In that case, we hypothesize that a similar cis-acting signal exists in RNA1 to mediate its own interaction with the RdRp. Indeed, our preliminary data show that a similar stem-loop is predicted to form near the 3′ end of NoV RNA1 and that its loop region shares primary sequence identity with the 3′SL loop in RNA2 (Rosskopf, Melendez, and Johnson, unpublished observation). This apparent sequence and structural conservation lends credence to the hypothesis, but the significance of the predicted structure in RNA1 replication remains to be determined. Perhaps the 3′ terminal cis-acting signal, which is structurally conserved among seven members of the nodavirus family even if the primary sequence in this region is not (Kaesberg et al., 1990; Taufer et al., 2008), is a motif common to all nodaviruses. For example, it may form the basis for template specificity by the viral RdRp. Given the apparent preference of the NoV RdRp for its cognate template over a heterologous (FHV) template (Gallagher, 1987; Price et al., 2005), it is possible that template recognition could occur at the level of RdRp binding. Therefore, the FHV RdRp may be unable to bind to an NoV promoter sequence with which the NoV RdRp can interact. However, further work will be required to determine the exact role of the 3′SL sequence in NoV RNA2 replication.
Summary
In summary, we have shown that a sequence in the 3′noncoding region of NoV RNA2, containing the predicted 3′SL stem-loop structure, plays a key role in nodavirus RNA replication. Together, our data suggest that the sequences that comprise the 3′SL form a part of an essential cis-acting signal for replication of NoV RNA2, where they may play a role in synthesis of negative strand replication intermediates and synthesis of monomeric positive strands from a dimeric negative strand template. The conservation of the predicted structure suggests that this common motif may play a similar role in RNA2 replication for the other members of this virus family. The exact role of these sequences, and of the predicted 3′SL structure, in RNA replication remains to be determined. However, we have generated a set of experimental tools that will continue to help us to elucidate the mechanism of nodavirus RNA replication.
Acknowledgements
We thank the UTEP Border Biomedical Research Center (BBRC) DNA Analysis and Biomolecule Analysis Core Facilities and the University of Alabama at Birmingham (UAB) Center for AIDS Research DNA Sequencing Core Facility. We thank Omar Hernandez (UTEP) for DNA sequencing and assistance with the primer extension gels; Maria Salazar (UAB) for DNA sequencing; Ana Betancourt (UTEP), Alexandria Melendez (UTEP), and Abel Licon (University of Delaware) for assistance with additional RNA folding; Vincent U. Gant, Jr. and the other members of the Johnson laboratory and the RNAVLab for many helpful discussions; and Dr. Kathryn Hanley (New Mexico State University, NMSU) for assistance with the nuclease mapping. We also thank Drs. Germán Rosas-Acosta (UTEP), Siddartha Das (UTEP), and Kathryn Hanley (NMSU) and the other members of the Johnson laboratory for critical review of the manuscript.
This work was supported by NIH grants U54AI057156 and S06GM008012 to K.L.J, NSF grant DMS0800272 to M.-Y.L., M.T., and K.L.J., and Texas Higher Education Coordinating Board ARP grant 003661-0013-2007 to M.-Y.L. and K.L.J. T. R. was supported by the NMSU Minority Biomedical Research Support-Research Initiative for Scientific Enhancement (MBRS-RISE) National Institutes of Health Grant (GM61222). This work was been facilitated by the infrastructure and resources provided by NIH/NCRR grant 5G12RR008124. None of these funding sources had any role in the design of the study, the collection, analysis and interpretation of the data, the writing of the report, or the decision to submit the paper for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albariño CG, Eckerle LD, Ball LA. The cis-acting replication signal at the 3− end of Flock House virus RNA2 is RNA3-dependent. Virology. 2003;311(1):181–191. doi: 10.1016/s0042-6822(03)00190-9. [DOI] [PubMed] [Google Scholar]
- Albariño CG, Price BD, Eckerle LD, Ball LA. Characterization and template properties of RNA dimers generated during Flock House virus RNA replication. Virology. 2001;289(2):269–282. doi: 10.1006/viro.2001.1125. [DOI] [PubMed] [Google Scholar]
- Ball LA. Cellular expression of a functional nodavirus RNA replicon from vaccinia virus vectors. J. Virol. 1992;66(4):2335–2345. doi: 10.1128/jvi.66.4.2335-2345.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball LA, Amann JM, Garrett BK. Replication of Nodamura virus after transfection of viral RNA into mammalian cells in culture. J. Virol. 1992;66(4):2326–2334. doi: 10.1128/jvi.66.4.2326-2334.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball LA, Johnson KL. Nodaviruses of insects. In: Miller LK, Ball LA, editors. The Insect Viruses. Plenum Publishing Corporation; New York: 1998. pp. 225–267. [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14(2):115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Chapman MR, Kao CC. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 1999;286(3):709–720. doi: 10.1006/jmbi.1998.2503. [DOI] [PubMed] [Google Scholar]
- Christiansen J, Garrett R. Enzymatic and chemical probing of ribosomal RNA-protein interactions. Methods Enzymol. 1988;164:456–68. doi: 10.1016/s0076-6879(88)64061-4. [DOI] [PubMed] [Google Scholar]
- Dirks RM, Pierce NA. A partition function algorithm for nucleic acid secondary structure including pseudoknots. Journal of Computational Chemistry. 2003;24(13):1664–1677. doi: 10.1002/jcc.10296. [DOI] [PubMed] [Google Scholar]
- Dirks RM, Pierce NA. An algorithm for computing nucleic acid base-pairing probabilities including pseudoknots. Journal of Computational Chemistry. 2004;25(10):1295–1304. doi: 10.1002/jcc.20057. [DOI] [PubMed] [Google Scholar]
- Eckerle LD, Albariño CG, Ball LA. Flock House virus subgenomic RNA3 is replicated and its replication correlates with transactivation of RNA2. Virology. 2003;317(1):95–108. doi: 10.1016/j.virol.2003.08.029. [DOI] [PubMed] [Google Scholar]
- Gallagher TM. Ph.D. Thesis. University of Wisconsin; Madison: 1987. Synthesis and assembly of nodaviruses. [Google Scholar]
- Hanley KA, Lee JJ, Blaney JE, Jr., Murphy BR, Whitehead SS. Paired charge-to-alanine mutagenesis of dengue virus type 4 NS5 generates mutants with temperature-sensitive, host range, and mouse attenuation phenotypes. J. Virol. 2002;76(2):525–531. doi: 10.1128/JVI.76.2.525-531.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson SJ, Konings DA, Sarnow P. Biochemical and genetic evidence for a pseudoknot structure at the 3′ terminus of the poliovirus RNA genome and its role in viral RNA amplification. J. Virol. 1993;67(6):2961–2971. doi: 10.1128/jvi.67.6.2961-2971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KL, Price BD, Ball LA. Recovery of infectivity from cDNA clones of Nodamura virus and identification of small nonstructural proteins. Virology. 2003;305(2):436–451. doi: 10.1006/viro.2002.1769. [DOI] [PubMed] [Google Scholar]
- Johnson KL, Price BD, Eckerle LD, Ball LA. Nodamura virus nonstructural protein B2 can enhance viral RNA accumulation in both mammalian and insect cells. J. Virol. 2004;78(12):6698–6704. doi: 10.1128/JVI.78.12.6698-6704.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaesberg P, Dasgupta R, Sgro J-Y, Wery J-P, Selling BH, Hosur MV, Johnson JE. Structural homology among four nodaviruses as deduced by sequencing and x-ray crystallography. J. Mol. Biol. 1990;214(2):423–435. doi: 10.1016/0022-2836(90)90191-N. [DOI] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5(12A):2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Li W-X, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EWA, Johnson KL, Garcia-Sastre A, Ball LA, Palese P, Ding S-W. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. USA. 2004;101(5):1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, Li WX, Ding S-W. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436(7053):1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman TC, Ohme-Takagi M, Taylor CB, Green PJ. DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell. 1993;5(6):701–714. doi: 10.1105/tpc.5.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J, White KA, Nagy PD. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J. Virol. 2005;79(8):4859–4869. doi: 10.1128/JVI.79.8.4859-4869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price BD, Eckerle LD, Ball LA, Johnson KL. Nodamura virus RNA replication in Saccharomyces cerevisiae: heterologous gene expression allows replication-dependent colony formation. J. Virol. 2005;79(1):495–502. doi: 10.1128/JVI.79.1.495-502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price BD, Rueckert RR, Ahlquist P. Complete replication of an animal virus and maintenance of expression vectors derived from it in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1996;93(18):9465–9470. doi: 10.1073/pnas.93.18.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder J, Giegerich R. Design, implementation and evaluation of a practical pseudoknot folding algorithm based on thermodynamics. BMC Bioinformatics. 2004;5:104. doi: 10.1186/1471-2105-5-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas E, Eddy SR. A dynamic programming algorithm for RNA structure prediction including pseudoknots. J. Mol. Biol. 1999;285(5):2053–2068. doi: 10.1006/jmbi.1998.2436. [DOI] [PubMed] [Google Scholar]
- Romero TA, Tumban E, Jun J, Lott WB, Hanley KA. Secondary structure of dengue virus type 4 3′ untranslated region: impact of deletion and substitution mutations. J. Gen. Virol. 2006;87(Pt 11):3291–3296. doi: 10.1099/vir.0.82182-0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Third ed Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- Selling BH, Allison RF, Kaesberg P. Genomic RNA of an insect virus directs synthesis of infectious virions in plants. Proc. Natl. Acad. Sci. USA. 1990;87(1):434–438. doi: 10.1073/pnas.87.1.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P-Y, Brinton MA, Veal JM, Zhong YY, Wilson WD. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry. 1996;35(13):4222–4230. doi: 10.1021/bi952398v. [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Ganem D. A virus-encoded inhibitor that blocks RNA interference in mammalian cells. J. Virol. 2005;79(12):7371–7379. doi: 10.1128/JVI.79.12.7371-7379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taufer M, Leung M-Y, Solorio T, Licon A, Mireles D, Araiza R, Johnson KL. RNAVLab: A virtual laboratory for studying RNA secondary structures based on grid computing technology. Parallel Computing. 2008;34(11):661–680. doi: 10.1016/j.parco.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesh RB. Infectivity and pathogenicity of Nodamura virus for mosquitoes. J. Gen. Virol. 1980;48:177–182. [Google Scholar]
- Tilgner M, Deas TS, Shi P-Y. The flavivirus-conserved penta-nucleotide in the 3′ stem-loop of the West Nile virus genome requires a specific sequence and structure for RNA synthesis, but not for viral translation. Virology. 2005;331(2):375–386. doi: 10.1016/j.virol.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Tilgner M, Shi P-Y. Structure and function of the 3′ terminal six nucleotides of the west nile virus genome in viral replication. J. Virol. 2004;78(15):8159–8171. doi: 10.1128/JVI.78.15.8159-8171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuplin A, Evans DJ, Simmonds P. Detailed mapping of RNA secondary structures in core and NS5B-encoding region sequences of hepatitis C virus by RNase cleavage and novel bioinformatic prediction methods. J Gen Virol. 2004;85(Pt 10):3037–47. doi: 10.1099/vir.0.80141-0. [DOI] [PubMed] [Google Scholar]