Abstract
The pharmacological disruption of the interaction between the HIV Tat protein and its cognate transactivation response RNA (TAR) would generate novel anti-viral drugs with a low susceptibility to drug resistance, but efforts to discover ligands with sufficient potency to warrant pharmaceutical development have been unsuccessful. We have previously described a family of structurally constrained β-hairpin peptides that potently inhibits viral growth in HIV-infected cells. The nuclear magnetic resonance (NMR) structure of an inhibitory complex revealed that the peptide makes intimate contacts with the 3-nt bulge and the upper helix of the RNA hairpin, but that a single residue contacts the apical loop where recruitment of the essential cellular co-factor cyclin T1 occurs. Attempting to extend the peptide to form more interactions with the RNA loop, we examined a library of longer peptides and achieved >6-fold improvement in affinity. The structure of TAR bound to one of the extended peptides reveals that the peptide slides down the major groove of the RNA, relative to our design, in order to maintain critical interactions with TAR. These conserved contacts involve three amino acid side chains and identify critical interaction points required for potent and specific binding to TAR RNA. They constitute a template of essential interactions required for inhibition of this RNA.
INTRODUCTION
HIV-1 TAR is a stem–loop RNA found at the 5′ end of all nascent viral transcripts (Figure 1). The cooperative binding of the viral protein Tat and its cellular co-factor cyclin T1 to TAR recruits and activates the CDK9 kinase to hyperphosphorylate RNA polymerase II, leading to greatly enhanced processivity of the polymerase (1,2). If the Tat–TAR–cyclin complex does not form, the polymerase produces primarily short transcripts and viral replication is inefficient. In the past decade or two, many groups have identified small molecule and peptide mimetic ligands for TAR (3–19), but none had sufficient potency and specificity to warrant pharmaceutical development. We have previously used structure-based design methods to discover cyclic peptide structural mimics of Tat protein with nM affinity for HIV TAR (20–23). These molecules inhibit viral replication in primary human lymphocytes with activity only 10-fold lower than the non-nucleoside reverse transcriptase inhibitor nevirapine (manuscript in preparation). They are active against viral isolates from each of the HIV-1 clades and are not cytotoxic to at least 1-mM concentrations. The peptides are confined to their cyclic β-hairpin structure by a D-Pro-L-Pro hairpin-promoting template (24). Their constrained cyclic structure stabilizes them against proteolysis, and mechanistic studies show that the compounds inhibit early steps in reverse transcription as well as Tat-dependent HIV transcription (manuscript in preparation). These characteristics make the peptides exciting antiviral leads, but their activity must be improved before they can be further explored as drug candidates.
Figure 1.
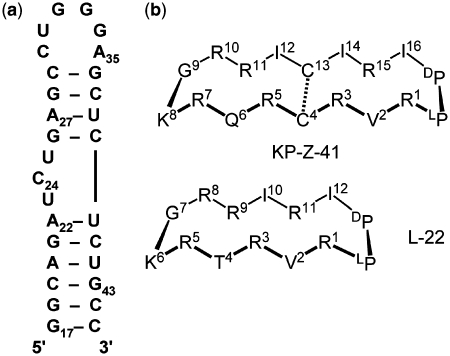
(a) Sequence and secondary structure of the HIV-1 TAR RNA construct used in these studies, numbered according to the start of transcription of the viral RNA and (b) sequences of the lead peptides L-22 and KP-Z-41 (LP = L-Pro and DP = D-Pro). The dotted line indicates a disulfide bridge.
The nuclear magnetic resonance (NMR) structure of the lead peptide L-22 (Table 1, Figure 2a) bound to TAR has been reported (23). It reveals that the peptides bind TAR in the major groove of the upper RNA helix (nucleotides 26–29 and 36–39) with the D-Pro-L-Pro template facing down towards the lower helix. Interactions important for peptide affinity and selectivity can be grouped into three categories: stacking with residue A35 of the apical loop of TAR that wraps the RNA around the solvent-exposed face of the peptide; hydrophobic contacts that drive formation of a base triple between bulge residue U23 and base pair A27-U38 in the upper helix; and the stabilization of the base triple by charged residues binding near the interhelical junction. Based on these features, we hypothesized that the peptide binding activity could be improved by lengthening the β-hairpin to establish more extensive contacts with the apical loop of the RNA.
Table 1.
Sequences of the lead peptides L-22 and KP-Z-41 and summary of their binding activities against HIV-1 TAR RNA (Kd values are in nanomolar), as determined by EMSA in the absence of competitor tRNA
| Mimetic | Residue | Kd (HIV) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L-22 | R1 | V2 | R3 | T4 | R5 | K6 | G7 | R8 | R9 | I10 | R11 | I12 | 30 | ||||
| KP-Z-41 | R1 | V2 | R3 | C4 | R5 | Q6 | R7 | K8 | G9 | R10 | R11 | I12 | C13 | I14 | R15 | I16 | 1 |
Residues are aligned in the table to represent similarity in position upon binding HIV-1 TAR.
Figure 2.
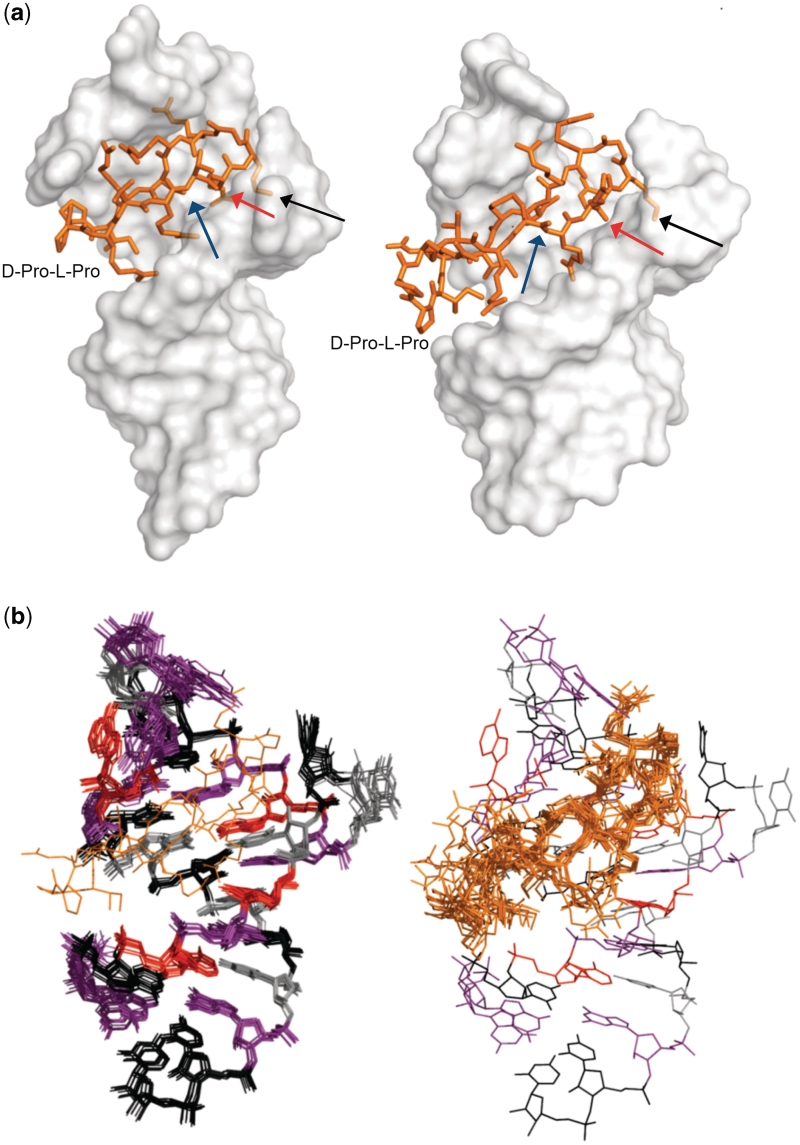
(a) Structures of the HIV-1 TAR/L-22 complex (left) and the HIV-1 TAR/KP-Z-41 complex (right). The sequences of the cyclic peptides are shown in Figure 1. The orientation of the peptide in the RNA major groove is the same in both structures. Common interaction sites are highlighted with arrows: the lysines that point toward the interhelical junction (black), the arginines that stack over the base triple (red), and the isoleucines that anchor the peptide in the major groove (blue). (b) Superposition of 10 lowest energy structures for the RNA (left) and cyclic peptide (right): guanosines are in purple, cytosines in black, adenosines in red, uracils in gray and the peptide is in orange.
In order to test this hypothesis, we synthesized a small library of 25 peptides containing 1, 2 or 4 additional amino acids and tested their binding to TAR RNA. Successful motifs were combined in a follow-up library of 12 peptides. One of these peptides, KP-Z-41, emerged as the lead molecule among these extended peptides due to its affinity for TAR and the quality of NMR data for its complex with TAR RNA. We determined the structure of this complex by NMR and observed that contacts near the interhelical junction remain almost unchanged between this structure and our previous L-22 structure, while the contacts to the loop and RNA backbone re-position themselves to accommodate the interactions at the junction. This was unexpected from our structure-based design, and this interaction positions the peptide further down the major groove, away from the apical loop, than we had predicted. Thus, while we were unsuccessful in extending the peptide toward the apical loop, the structure identifies a small set of essential interactions that are critical for recognition of HIV-1 TAR and could be exploited for the design of small molecule peptidomimetics.
METHODS
Peptide and RNA synthesis
The synthesis of the cyclic peptides was conducted as described previously (20,22). HIV-1 TAR RNA was prepared in vitro from commercially synthesized DNA templates (IDT) using in-house-purified T7 RNA polymerase and unlabeled nucleotides (Sigma), or 98% 15N, 13C labeled nucleotides (Cambridge Isotope Labs), or 98% deuterated nucleotides (Isotec). RNA products were separated by denaturing polyacrylamide gel electrophoresis. The desired RNA was excised from the gel and further purified by electroelution, desalting on a NAP-10 column (GE Healthcare), and extensive dialysis against a 10 mM potassium phosphate buffer, pH 6.6, with 0.1 mM EDTA (25,26). Prior to NMR, RNA products were exchanged into 99.99% D2O by lyophilization, and then annealed with heating to 90°C and quick cooling on ice.
Preparation of radiolabeled RNA and gel shift assays
T4 RNA ligase (Promega) and cytidine 3′,5′-bis[alpha-32P]phosphate, triethylammonium salt (pCp, GE Healthcare) were used to label purified HIV-1 TAR RNA. Free pCp remaining in solution after the ligation was largely removed by passing the RNA solution through a NAP-10 desalting column (GE Healthcare). The RNA was annealed by heating to 90°C, followed by quick cooling on ice. For each gel shift assay, peptides were incubated on ice for 30 min with labeled TAR and assay buffer (0.05 M Tris–HCl, 0.05 M KCl, 0.2 M DTT, 1% Triton X-100, and tRNA at 10 mg/ml). Samples were then fractionated by native polyacrylamide gel electrophoresis (PAGE) at 15 W and 4°C in 0.5 × TB buffer. Dried gels were exposed to a phosphor imaging plate and viewed with a Molecular Dynamics scanner. Images were analyzed with ImageQuant software.
NMR spectroscopy and spectral assignments
NMR spectra were collected on Bruker 500 and 600 MHz spectrometers equipped with cryo-cooled probes. Data were processed with NMRPipe (27) and analyzed in Sparky (28). NOESY experiments were recorded at multiple mixing times (80—300 ms) at either 25°C (D2O) or 4°C (H2O). The 2D 13C filtered NOESY and TOCSY spectra of the ‘F1fF2f’ type (29) were collected to facilitate assignments of peptide resonances. Spectra using partially perdeuterated RNA (with only H6/8, H2, H1′ and H2′ protons present) had sharper line widths that facilitated NOE assignments (SI Figure 1). Resonance assignments started from those already available for the previous L-22/HIV-1 TAR RNA complex (23) and were extended to this new complex using HCCH-COSY, HCCH-TOCSY and HSQC-NOESY experiments. Initial proton assignments for the peptide were made from F1fF2f NOESY spectra and based on the observation of the typical Hα(i) to HN(i+1) pattern of NOEs common in β-sheets, then completed with the partially perdeuterated RNA that removed much of the overlap between peptide and sugar resonances.
Structure determination
Structures of the HIV-1 TAR RNA/KP-Z-41 complex were calculated with Xplor-NIH (30) using the experimental restraints summarized in Table 2. Loose restraints on backbone dihedral angles (±40°) were introduced for base-paired regions in both helical stems based upon unambiguous spectroscopic verification of an A-form RNA conformation as determined from the pattern of cross-peaks in TOCSY and NOESY spectra (25), while backbone dihedral angle restraints for the peptide were derived with TALOS (31). The final 10 converged structures of the complex were selected from the population of structures with no NOE restraint violations >0.5 Å and had the lowest violations of all experimental restraints.
Table 3.
Sequences of selected peptides and summary of their activities against HIV-1 TAR RNA (Kd values are in nM), as determined by EMSA in the presence of competitor tRNA
| Mimetic | Position |
Kd (nM) | Error (nM) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | |||
| KP-Z-01 | R | V | R | C | R | K | L | K | G | Q | T | R | C | I | R | I | >5000 | |
| KP-Z-02 | R | V | R | C | R | K | R | DR | G | Q | T | R | C | I | R | I | 75 | ±25 |
| KP-Z-03 | R | V | R | C | R | Q | R | G | P | G | K | R | C | I | R | I | >5000 | |
| KP-Z-04 | R | V | R | C | R | K | Q | G | P | G | T | R | C | I | R | I | >5000 | |
| KP-Z-05 | R | V | R | C | R | R | K | G | P | G | Q | R | C | I | R | I | 150 | ±50 |
| KP-Z-06 | R | V | R | C | R | K | R | G | P | G | Q | R | C | I | R | I | >5000 | |
| KP-Z-07 | R | C | R | V | R | Q | R | K | G | R | A | R | K | I | C | I | 150 | ±50 |
| KP-Z-08 | R | C | R | V | R | K | Q | G | P | G | T | R | K | I | C | I | >5000 | |
| KP-Z-09 | R | C | R | V | R | K | Q | G | P | G | K | R | K | I | C | I | >5000 | |
| KP-Z-10 | R | C | R | V | R | K | R | G | P | G | Q | R | K | I | C | I | >5000 | |
| KP-Z-11 | R | C | R | V | R | R | K | G | P | G | A | R | K | I | C | I | 200 | ±50 |
| KP-Z-12 | R | V | R | C | R | R | R | G | R | C | I | R | I | >5000 | ||||
| KP-Z-13 | R | V | R | C | R | K | K | DP | K | C | I | R | I | >5000 | ||||
| KP-Z-14 | R | V | R | T | R | K | K | DP | K | R | I | R | I | 750 | ||||
| KP-Z-15 | R | V | R | V | R | K | K | G | R | T | I | R | I | >5000 | ||||
| KP-Z-16 | R | V | R | C | R | K | R | K | G | R | C | I | R | I | 500 | |||
| KP-Z-17 | R | V | R | C | R | R | K | R | G | R | C | I | R | I | 300 | |||
| KP-Z-18 | R | V | R | T | R | Q | R | K | G | R | R | I | R | I | 200 | ±50 | ||
| KP-Z-19 | R | V | R | V | R | K | R | K | G | R | Q | I | R | I | 250 | ±50 | ||
| KP-Z-20 | R | V | R | Q | R | K | R | K | G | R | V | I | R | I | 200 | ±50 | ||
| KP-Z-21 | R | V | R | C | R | K | R | K | DP | R | C | I | R | I | 300 | |||
| KP-Z-22 | R | V | R | C | R | T | R | G | K | R | R | R | C | A | R | V | 200 | ±50 |
| KP-Z-33 | R | V | R | C | R | K | R | R | G | Q | T | R | C | I | R | I | 100 | ±25 |
| KP-Z-34 | R | V | R | C | R | K | R | K | G | Q | T | R | C | I | R | I | 100 | ±25 |
| KP-Z-35 | R | V | R | T | R | K | R | DR | G | Q | T | R | A | I | K | I | 150 | ±50 |
| KP-Z-36 | R | C | R | V | R | K | R | DR | G | Q | T | R | R | I | C | I | 200 | ±50 |
| KP-Z-37 | R | C | R | V | R | R | K | K | G | R | A | R | K | I | C | I | 200 | ±50 |
| KP-Z-38 | R | C | R | V | R | Q | R | G | P | G | A | R | K | I | C | I | >1000 | |
| KP-Z-39 | R | V | R | C | R | T | R | K | G | R | A | R | C | I | K | I | 75 | ±25 |
| KP-Z-40 | R | V | R | C | R | Q | R | K | G | R | R | A | C | I | R | I | 25 | ±10 |
| KP-Z-41 | R | V | R | C | R | Q | R | K | G | R | R | I | C | I | R | I | 75 | ±25 |
All peptides are cyclized with a D-Pro-L-Pro motif not listed here to simplify the table.
Lower-case letters indicate D-amino acids.
RESULTS
Based on the interactions between the peptide and TAR observed in our previous peptide structure and the results of our mutational studies, we synthesized a small library of new peptides containing 1, 2 or 4 additional amino acids. Selected peptides from this initial set are listed in Table 3 and are named KP-Z-01 through KP-Z-22 for ease of reference. All peptides were cyclized using the D-Pro-L-Pro template, and the majority of these new peptides incorporate two central cysteine residues forming a disulfide bridge to further stabilize the hairpin fold. Peptides were tested by electrophoretic mobility shift assay (EMSA) in the presence of 10 000-fold excess tRNA to establish their affinity and selectivity for TAR RNA. Peptides containing 16 and 18 amino acids were more successful than those containing 15 amino acids, of which three out of four peptides showed no affinity for TAR even at the highest concentration tested (5 µM). This result can confidently be attributed to less stable β-hairpin formation in the cyclic peptides containing an odd number of residues. Previous studies of our initial β-hairpin peptides have shown that stability of the hairpin in solution is necessary to observe strong affinity for TAR (20,22). Even the 16-amino acid peptides bound with only high nanomolar affinity to TAR; this represented a significant loss of activity from our previous success with L-22. It was also disappointing to see that even significant changes in sequence resulted in minimal changes in affinity. For example, swapping the K-R-K motif near the hairpin turn of KP-Z-16 for R-K-R in KP-Z-17 changed the affinity of the peptide by less than two-fold. Even exchanging residues across the hairpin (compare for example KP-Z-19 and KP-Z-20) yielded almost no change in affinity. These results suggest that binding of the 16-amino acid peptides may be largely charge-mediated and is not likely to be very selective.
Table 2.
Experimental restraints and structural statistics for structure determination of the KP-Z-41/TAR complex
| Total number of restraints | 774 |
| NOE-derived restraints | 498 |
| intermolecular NOEs | 61 |
| RNA (intramolecular) | 228 |
| Peptide (intramolecular) | 209 |
| Dihedral restraints | 192 |
| Hydrogen-bonding restraints | 72 |
| Planarity restraints | 12 |
| Average rmsd values from experimental restraints | |
| Distance (Å) | 0.06 |
| Dihedral (deg) | 0.49 |
| Average rmsd values from ideal geometries | |
| bonds (Å) | 0.004 |
| Angles (deg) | 0.91 |
| Improper (deg) | 0.73 |
| Heavy atom rmsd from mean structure (Å) | |
| KP-Z-41 (all backbone atoms) | 0.29 |
| KP-Z-41 (all heavy atoms) | 1.19 |
| TAR RNA (all heavy atoms) | 0.49 |
| TAR RNA core (G18-U23, G26-C29, G36-C44) | 0.29 |
| KP-Z-41 and TAR core | 0.75 |
| Entire structure | 2.1 |
In the case of the 18-amino acid peptides, many encouragingly showed mid-nanomolar activity, even in the presence of a 10 000-fold excess of tRNA. Furthermore, small changes in sequence yielded significant changes in activity against TAR in some cases, indicating specific binding for this group of peptides. For example, the difference between KP-Z-05 and KP-Z-06 is only an exchange of residues at positions 6 and 7, but this rather small change alters binding from 150 nM to undetectable at the highest concentration tested (>5 µM). A similar change from KP-Z-10 to KP-Z-11 yields a comparable result. The sequence of KP-Z-02, one of the most successful peptides from the library, has an Arg-DArg motif at positions 7 and 8; changing these Arg to Leu-Lys in KP-Z-01 abolishes binding even at the highest concentration tested.
Encouraged by the specific binding suggested by these results, we decided to pursue the optimization of the 18-amino acid peptides by rational design. Successful motifs from these peptides and the reported L-22/TAR structure were combined in a small follow-up library (Table 3, KP-Z-33 through KP-Z-40). One of these peptides, KP-Z-40, emerged as the most potent peptide with an apparent binding constant of 25 nM, when measured in the presence of 10 000 fold excess of tRNA competitor, and Kd <1 nM (when measured in a direct binding assay in the absence of any tRNA). This result represents a >6-fold improvement in affinity over the previous lead peptide L-22.
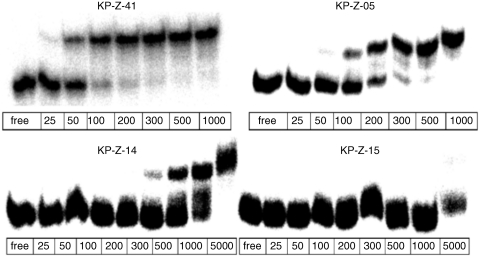
The structure of L-22 showed that the Ile methyl groups on one face of the peptide were important for locking the peptide in the major groove of TAR (23). With this in mind, we designed a new peptide KP-Z-41, based on the sequence of KP-Z-40 but with an additional Ile at position 12 in place of the Ala found in the original peptide. The affinities of KP-Z-40 and KP-Z-41, determined by EMSA (Figure 3) were similar, but the NOESY experiments for KP-Z-41 revealed a much greater number of intermolecular NOEs. For this reason, the KP-Z-41/TAR complex was selected for structure determination to better understand how increased affinity was obtained.
Figure 3.
Comparison of binding results for several peptides from the KP-Z library (all concentrations are in nM) to HIV TAR (1 nM) by electrophoretic mobility shift assay (EMSA); the buffer contains a 10 000 fold excess of tRNA to disfavor binding of peptides that form nonspecific interactions.
Hydrophobic interactions promote formation of a base triple and anchor the peptide in the major groove
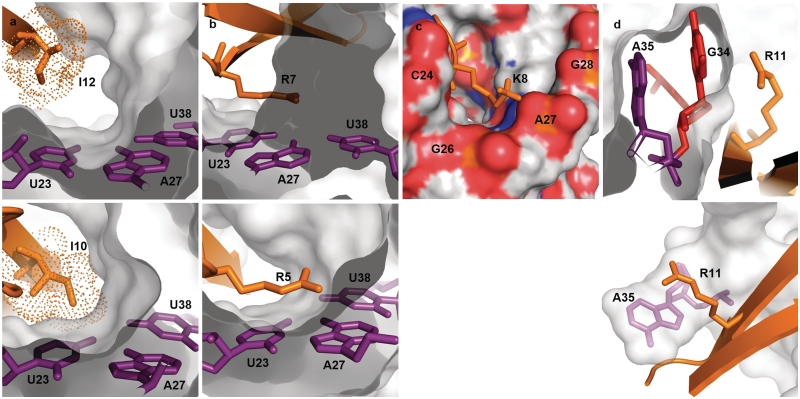
The structure contains a base triple between bulge residue U23 and the A27/U38 base pair, as also observed for L-22. The NOE data indicate a canonical pairing for U23 and A27 in the base triple; both A27 amino hydrogen atoms are protected from solvent exchange and observable in NOESY experiments. Formation of the base triple upon binding is likely driven by burial of the Ile12 methyls against U23, analogous to the interactions of Ile10 in the L22-TAR complex (Figure 4a) and Ile79 in our original model system of BIV Tat/TAR (21). Ile14 is also protected from solvent by burial in the major groove, similar to Ile12 in the L-22 peptide.
Figure 4.
(a) Comparison of the hydrophobic interactions observed in the HIV-1 TAR/KP-Z-41 complex (top) and in the HIV-1 TAR/L-22 complex (bottom) that stabilize the U23-A27-U38 base triple. The peptide L-22 penetrates deeper into the major groove compared to KP-Z-41, and thus the Ile residue in L-22 is in closer proximity to the base triple than in KP-Z-41. The RNA is represented as a gray surface with nucleotides of interest in purple, the peptide is in orange and the van der Waals surface of the Ile side chain is represented in dots. (b) An arginine guanidinium group stacks on top of the U23 base to provide a cation-π interaction that stabilizes the base triple in both the KP-Z-41/TAR complex (top) and the L-22/TAR complex (bottom). (c) The charged terminus of Lys8 is oriented toward a pocket of negative charge formed by the phosphate groups of nucleotides C24 through G28 at the interhelical junction. (d) The direct stacking between A35 and an arginine residue that is seen in the L-22/TAR complex (bottom) is replaced in this new complex by a different set of loop-peptide interactions. The base of G34 is instead flipped out from the RNA loop and sandwiched between A35 and Arg11 in the KP-Z-41/TAR complex (top).
Placement of these large hydrophobic side chains on the peptide backbone orients the correct face of the peptide toward the RNA and induces formation of the base triple; thus, location of the hydrophobic residues on the peptide is one of the most crucial elements for affinity to the TAR. The peptide KP-Z-41 contains three isoleucine residues at positions 12, 14 and 16. However, only Ile12 and Ile14 are located in the RNA major groove, forcing the D-Pro-L-Pro end of the template to extend away from the RNA into solution. We had originally hypothesized that Ile14 and Ile16 would be the hydrophobic groups docked in the major groove near the base triple, with Ile12 and the Lys8-Gly9 turn extending up the groove toward the apical loop instead. However, the structure reveals that, while burial of these hydrophobic groups is important for orienting the correct face of the peptide toward the RNA, their placement within the structure is also governed by key peptide-RNA polar interactions, as discussed below. In order to form these polar interactions at the inter-helical junction, the peptide must slide down the major groove relative to our design, leaving Ile12 and Ile 14 docked in the pocket formed in the major groove upon peptide binding.
Interactions between polar side chains and the bulge region of TAR position the peptide relative to the RNA
Two important interactions between polar side chains and the RNA can be seen near the TAR bulge. The guanidinium group of Arg7 lies over U23 and stabilizes the base triple (Figure 4b). At the same time, the charged terminus of Lys8 is favorably oriented by the positioning of Arg7 and anchor residue Ile12 (as discussed above) toward a pocket of negative charge formed by the phosphate groups of nucleotides C24 through G28 at the interhelical junction (Figure 4c). The roles of Arg7 and Lys8 in KP-Z-41 are nearly identical to those of Arg5 and Lys6 in L-22. This Arg-Lys combination, along with the hydrophobic residue buried beside the base triple, appear to be the best maintained features between the two structures. Other interactions adjust themselves to keep these contacts intact. These two polar interactions alter the way the peptide contacts the apical loop, as discussed in the following section, and force the D-Pro-L-Pro motif and adjacent residues to slide down the major groove and extend away from the RNA, contrary to what we had expected in the rational design of the longer peptide library.
The cyclic peptide contacts the apical loop of TAR in a novel manner
A promising feature of these β-hairpin peptides for RNA specificity is their ability to simultaneously contact multiple unique secondary structure elements of the RNA. In the L-22/TAR complex, the Arg11 guanidinium group stacks on the base of loop residue A35, and this interaction pulls the apical loop to partially close around the RNA. An interaction with A35 is present for KP-Z-41 as well, but the contact is different, because the space occupied by Arg11 in the L-22 peptide is replaced by a Cys in KP-Z-41. Nevertheless, the contact to the loop is of such importance that the RNA adjusts itself to maintain the interaction. Instead of direct stacking between A35 and an arginine residue, the base of G34 is flipped out from the rest of the RNA and sandwiched between A35 and Arg11 of KP-Z-41 (Figure 4d). In this new arrangement, G34 completes the Arg–A35 interaction previously seen in L-22.
Comparison of NOESY data for complexes of lead peptides KP-Z-40 and KP-Z-41 with HIV-1 TAR RNA
As mentioned previously, cyclic peptide KP-Z-40 has the strongest affinity for the TAR RNA based on EMSA results, but the NOE data for this peptide-RNA complex were of lower quality than for KP-Z-41, so that the latter peptide was selected for structure determination. Comparison of the intermolecular NOEs for both complexes reveals strong similarities for cross-peaks defining the peptide contact to the apical loop. Specifically, NOEs between Val2 and A35, Arg10 and G34 and Arg11 and G34 are present for both complexes. These pairings represent several of the most intense intermolecular NOEs for the KP-Z-40/HIV-1 TAR complex. From this NOE comparison, we conclude that the peptides bind to the RNA in a similar manner. However, without a structure for both peptides, it is difficult to conclusively identify the structural or dynamics basis for the difference in activity between the two peptides.
DISCUSSION
The Tat–TAR interaction has long been considered a target for the development of novel antivirals but the difficulty of inhibiting RNA–protein interfaces with small molecules has prevented the successful pre-clinical or clinical development of Tat–TAR inhibitors (3–19,32–34). To overcome these difficulties, our group has focused on cyclic β-hairpin peptide mimics of Tat protein, developed through a structural rationale (20–23,35). We reasoned that the rigidity of these pre-structured peptides would increase their selectivity, compared to other flexible RNA-binding peptide mimics (3,4), and their larger molecular weight would allow more potent and specific interactions compared to small organic structures. Based on this rationale, we succeeded in developing cyclic peptide structural mimics of Tat protein with nM affinity for HIV TAR and with an in vivo activity that makes them attractive antiviral leads (22,23). In order to further improve their potency, we extended them by 1, 2 or 4 amino acids, reasoning that a longer hairpin would make more extensive contacts with the single-stranded nucleotides in the apical loop than we had previously observed in the structure of a lead peptide-RNA complex (23). Some of the new peptides (Table 3) were found to bind tightly to HIV-1 TAR, but only when the hairpin was extended by 4 amino acids. For this group of molecules, even small variations in the peptide sequence led to relatively large changes in activity, suggesting a specific interaction. The specificity of the peptides for HIV TAR RNA is also demonstrated by their ability to retain strong affinity for the target even in the presence of very large excess of tRNA. Furthermore, when we compared binding for other members of this family of cyclic peptides with various mutated versions of HIV TAR, including mutation and deletion of residues U23 and A35, we demonstrated that the absence of a single critical nucleotide abolishes peptide activity, lending additional evidence for the specificity of these peptides (23). A second set of peptides were designed based on the features of the more active library members, and a molecule, KP-Z-41, emerged with sub-nanomolar affinity for HIV-1 TAR. An improvement >6-fold was thus successfully obtained in binding activity.
We determined the structure of the KP-Z-41/TAR complex in order to verify whether our rationale of extending the peptide to interact with the loop was correct and to guide our next steps of peptide development. We were surprised to discover that the additional residues did not bring the peptide closer to the loop; instead, the D-Pro-L-Pro end of the peptide hairpin extended away from the RNA into solution. The structural reasons for this effect were nonetheless very revealing. Interactions between the peptide and the RNA interhelical junction are nearly identical for KP-Z-41 and the previous lead peptide L-22, at residues Arg7, Lys8 and Ile12. In order to form these interactions, however, the peptide slides down the major groove away from the apical loop and pushes residues near the D-Pro-L-Pro template into solution. Not only are the peptide-RNA interactions at Ile12, Arg7 and Lys8 preferably formed over other possible interactions near the D-Pro-L-Pro end of the peptide, but they are also apparently favorable enough to drive the increased affinity of the peptide for the RNA despite the entropic costs of leaving several hydrophobic residues (an isoleucine and both prolines) exposed to solvent. The placement of these three critical residues (Ile12, Arg7 and Lys8) should be regarded as the minimum requirement for peptide mimics of Tat targeting this HIV TAR conformation. Their requirement to organize the RNA–peptide complex will be taken into account in our future peptide designs and should be considered when designing small molecule ligands for TAR as well.
Because of these unexpected changes in the peptide–RNA interactions, Arg10 and Arg11 are the only residues in the KP-Z-41/TAR structure positioned near the loop, and only Arg11 forms a stable enough interaction to be detected by NOESY experiments, stacking over G34 and A35. The placement of the disulfide bridge used for reinforcing hairpin formation in KP-Z-41 prohibits the direct arginine stacking with RNA residue A35 that was seen in the L-22/TAR structure. Thus, interactions of the amino acids at the C-terminal side of the KG turn are fairly weak and should be optimized further to improve peptide binding. Our future work on peptide design will focus on development of new contacts at the apical loop based on re-organization of the peptide, while maintaining features that are shown in this study to be crucial for specific HIV TAR binding. By fully understanding the essential hydrophobic and polar interactions that lead to strong binding activity, we should now be able to design intermolecular contacts at other locations in our peptide. By moving the crucial Ile–Arg–Lys combination to new locations within the peptide, we may be able to induce the peptide to slide upwards along the major groove and reach the apical loop of the RNA to form more extensive contacts.
ACCESSION NUMBER
2kx5.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
National Institutes of Health-National Institute of Allergy and Infectious Diseases (to G.V.); Swiss National Science Foundation (to J.R.). Funding for open access charge: NIH-AI 070090.
Conflict of interest statement. None declared.
REFERENCES
- 1.Karn J. Tackling Tat. J. Mol. Biol. 1999;293:235–254. doi: 10.1006/jmbi.1999.3060. [DOI] [PubMed] [Google Scholar]
- 2.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 3.Hamy F, Felder ER, Heizmann G, Lazdins J, Aboul-ela F, Varani G, Karn J, Klimkait T. An inhibitor of the Tat/TAR RNA interaction that effectively suppresses HIV-1 Replication. Proc. Natl Acad. Sci. USA. 1997;94:3548–3553. doi: 10.1073/pnas.94.8.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huq I, Wang X, Rana TM. Specific recognition of HIV-1 TAR RNA by a D-Tat Peptide. Nat. Struct. Biol. 1997;4:881–882. doi: 10.1038/nsb1197-881. [DOI] [PubMed] [Google Scholar]
- 5.Wang S, Huber PW, Cui M, Czarnik AW, Mei H-Y. Binding of neomycin to TAR element of HIV-1 RNA induces dissociation of Tat protein by an allosteric mechanism. Biochemistry. 1998;37:5549–5557. doi: 10.1021/bi972808a. [DOI] [PubMed] [Google Scholar]
- 6.Tamilarasu N, Huq I, Rana TM. Targeting RNA with peptidomimetic oligomers in human cells. Bioorg. Med. Chem. Lett. 2001;11:505–507. doi: 10.1016/s0960-894x(00)00709-5. [DOI] [PubMed] [Google Scholar]
- 7.Gelman MA, Richter S, Cao H, Umezawa N, Gellman SH, Rana TM. Selective binding of TAR RNA by a tat-derived beta-peptide. Org. Lett. 2003;5:3563–3565. doi: 10.1021/ol034977v. [DOI] [PubMed] [Google Scholar]
- 8.Davis B, Afshar M, Varani G, Murchie AIH, Karn J, Lentzen G, Drysdale M, Bower J, Potter AJ, Starkey ID, et al. Rational design of inhibitors of HIV-1 TAR RNA through the stabilisation of electrostatic “hot spots”. J. Mol. Biol. 2004;336:343–356. doi: 10.1016/j.jmb.2003.12.046. [DOI] [PubMed] [Google Scholar]
- 9.Aboul-ela F, Varani G. Recognition of HIV-1 TAR RNA by Tat protein and Tat-derived peptides. J. Mol. Struct. 1998;423:29–39. [Google Scholar]
- 10.Afshar M, Prescott CD, Varani G. Structure-based and combinatorial search for new RNA-binding drugs. Curr. Opin. Biotechnol. 1999;10:59–63. doi: 10.1016/s0958-1669(99)80011-1. [DOI] [PubMed] [Google Scholar]
- 11.Burns VA, Bobay BG, Basso A, Cavanagh J, Melander C. Targeting RNA with cysteine-constrained peptides. Bioorg. Med. Chem. Lett. 2008;18:565–567. doi: 10.1016/j.bmcl.2007.11.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darfeuille F, Hansen JB, Orum H, Primo CD, Toulme J-J. LNA/DNA chimeric oligomers mimic RNA aptamers targeted to the TAR RNA element of HIV-1. Nucleic Acids Res. 2004;32:3101–3107. doi: 10.1093/nar/gkh636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mei H-Y, Galan AA, Halim NS, Mack DP, Moreland DW, Sanders KB, Truong HN, Czarnik AW. Inhibition of an HIV-1 Tat-derived peptide binding to TAR RNA by aminoglycoside antibiotics. Bioorg. Med. Chem. Lett. 1995;22:2755–2760. [Google Scholar]
- 14.Murchie AIH, Davis B, Isel C, Afshar M, Drysdale MJ, Bower J, Potter AJ, Starkey ID, Swarbrick TM, Mirza S, et al. Structure-based drug design targeting an inactive RNA conformation: exploiting the flexibility of HIV-1 TAR RNA. J. Mol. Biol. 2004;336:625–638. doi: 10.1016/j.jmb.2003.12.028. [DOI] [PubMed] [Google Scholar]
- 15.Raghunathan D, Sanchez-Pedregal VM, Junker J, Schwiegk C, Kalesse M, Kirschning A, Carlomagno T. TAR-RNA recognition by a novel cyclic aminoglycoside analogue. Nucleic Acids Res. 2006;34:3599–3608. doi: 10.1093/nar/gkl494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turner JJ, Ivanova GD, Verbeure B, Williams D, Arzumanov AA, Abes S, Lebleu B, Gait MJ. Cell-penetrating peptide conjugates of peptide nucleic acids (PNA) as inhibitors of HIV-1 Tat-dependent trans-activation in cells. Nucleic Acids Res. 2005;33:6837–6849. doi: 10.1093/nar/gki991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan D, He M, Pang R, Lin S-s, Li Z, Yang M. The design, synthesis, and biological evaluation of novel substituted purines as HIV-1 Tat-TAR inhibitors. Bioorg. Med. Chem. 2007;15:265–272. doi: 10.1016/j.bmc.2006.09.062. [DOI] [PubMed] [Google Scholar]
- 18.Wang D, Iera J, Baker H, Hogan P, Ptak R, Yang L, Hartman T, Buckheit RW, Jr, Desjardins A, Yang A, et al. Multivalent binding oligomers inhibit HIV Tat-TAR interaction critical for viral replication. Bioorg. Med. Chem. Lett. 2009;19:6893–6897. doi: 10.1016/j.bmcl.2009.10.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayer M, Lang PT, Gerber S, Madrid PB, Pinto IG, Guy RK, James TL. Synthesis and testing of a focused phenothiazine library for binding to HIV-1 TAR RNA. 2006;13:993–1000. doi: 10.1016/j.chembiol.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 20.Athanassiou Z, Dias RLA, Moehle K, Dobson N, Varani G, Robinson JA. Structural mimicry of retroviral Tat proteins by constrained β-hairpin peptidomimetics - new ligands with high affinity and selectivity for viral TAR RNA regulatory elements. J. Am. Chem. Soc. 2004;126:6906–6913. doi: 10.1021/ja0497680. [DOI] [PubMed] [Google Scholar]
- 21.Leeper TC, Athanassiou Z, Dias RLA, Robinson JA, Varani G. TAR RNA recognition by a cyclic peptidomimetic of Tat protein. Biochemistry. 2005;44:12362–12372. doi: 10.1021/bi0510532. [DOI] [PubMed] [Google Scholar]
- 22.Athanassiou Z, Patora K, Dias RLA, Moehle K, Robinson JA, Varani G. Structure-guided peptidomimetic design leads to nanomolar inhibitors of the Tat-TAR interaction of bovine immunodeficiency virus. Biochemistry. 2007;46:741–751. doi: 10.1021/bi0619371. [DOI] [PubMed] [Google Scholar]
- 23.Davidson A, Leeper TC, Athanassiou Z, Patora-Komisarska K, Karn J, Robinson JA, Varani G. Simultaneous recognition of HIV-1 TAR RNA bulge and loop sequences by cyclic peptide mimics of Tat protein Proc. Natl Acad. Sci. USA. 2009;106:11931–11936. doi: 10.1073/pnas.0900629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson JA. The design, synthesis and conformation of some new β-hairpin mimetics: novel reagents for drug and vaccine discovery. SynLett. 1999;4:429–441. [Google Scholar]
- 25.Varani G, Aboul-ela F, Allain FH-T. NMR investigations of RNA structure. Progr. NMR Spectr. 1996;29:51–127. [Google Scholar]
- 26.Price SR, Oubridge C, Varani G, Nagai K. In: RNA-Protein Interaction: Practical Approach. Smith C, editor. New York: Oxford University Press; 1998. pp. 37–74. [Google Scholar]
- 27.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 28.Goddard TD, Kneller DG. Sparky 3. San Francisco: University of California; [Google Scholar]
- 29.Peterson RD, Theimer CA, Wu H, Feigon J. New applications of 2D filtered/edited NOESY for assignment and structure elucidation of RNA and RNA-protein complexes. J. Biomol. NMR. 2004;28:59–67. doi: 10.1023/B:JNMR.0000012861.95939.05. [DOI] [PubMed] [Google Scholar]
- 30.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn. Res. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 31.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 32.Hamy F, Gelus N, Zeller M, Lazdins JL, Bailly C, Klimkait T. Blocking HIV replication by targeting Tat protein. Chem. Biol. 2000;7:669–676. doi: 10.1016/s1074-5521(00)00012-0. [DOI] [PubMed] [Google Scholar]
- 33.Gallego J, Varani G. Targeting RNA with small molecule drugs: therapeutic promises and chemical challenges. Acc. Chem. Res. 2001;34:836–843. doi: 10.1021/ar000118k. [DOI] [PubMed] [Google Scholar]
- 34.Baba M. Recent status of HIV-1 gene expression inhibitors. Antiviral Res. 2006;71:301–306. doi: 10.1016/j.antiviral.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 35.Moehle K, Athanassiou Z, Patora K, Davidson A, Varani G, Robinson JA. Design of β-hairpin peptidomimetics that inhibit binding of α-helical HIV-1 Rev protein to the Rev response element RNA. Angew. Chemie. 2007;46:1–5. doi: 10.1002/anie.200702801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.