Abstract
The 1.7 Å X-ray crystal structure of the B-DNA dodecamer, [d(CGCGAATTCGCG)]2 (DDD)-bound non-covalently to a platinum(II) complex, [{Pt(NH3)3}2-µ-{trans-Pt(NH3)2(NH2(CH2)6NH2)2}](NO3)6 (1, TriplatinNC-A,) shows the trinuclear cation extended along the phosphate backbone and bridging the minor groove. The square planar tetra-am(m)ine Pt(II) units form bidentate N-O-N complexes with OP atoms, in a Phosphate Clamp motif. The geometry is conserved and the interaction prefers O2P over O1P atoms (frequency of interaction is O2P > O1P, base and sugar oxygens > N). The binding mode is very similar to that reported for the DDD and [{trans-Pt(NH3)2(NH2(CH2)6(NH3+)}2-µ-{trans-Pt(NH3)2(NH2(CH2)6NH2)2}](NO3)8 (3, TriplatinNC), which exhibits in vivo anti-tumour activity. In the present case, only three sets of Phosphate Clamps were found because one of the three Pt(II) coordination spheres was not clearly observed and was characterized as a bare Pt2+ ion. Based on the electron density, the relative occupancy of DDD and the sum of three Pt(II) atoms in the DDD-1 complex was 1:1.69, whereas the ratio for DDD-2 was 1:2.85, almost the mixing ratio in the crystallization drop. The high repetition and geometric regularity of the motif suggests that it can be developed as a modular nucleic acid binding device with general utility.
INTRODUCTION
Since the discovery of its’ anti-tumour activity the mononuclear platinum drug cis-platin {cis-[PtCl2 (NH3)2], cis-diamminedichloridoplatinum(II)}, (1,2), and subsequently the related carboplatin {[Pt(NH3)2 (CBDCA)], cis-diammine [1,1-cyclobutandicarboxylato) platinum(II)} and oxaliplatin {[Pt(R,R-dach)(ox)], 1,2-R, R-diaminocyclohexane(oxalato)platinum(II)} have contributed significantly to the anti-cancer drug armamentarium. The drugs all form predominantly short-range 1,2-intrastrand crosslinks on DNA which cause severe conformational distortion, the specific nature of which is thought to be a critical determinant of the biological consequences of platinum drug treatment (3–6). Polynuclear Pt(II) agents are structurally and biologically distinct from mononuclear Pt(II) agents, and form long-range (Pt,Pt) inter- and intra-strand crosslinks (7–14). The DNA conformational distortions induced by polynuclear agents are thus structurally distinct from those of cis-[PtX2(amine)2], and are not necessarily susceptible to the same cellular processing as cisplatin and other cis-[PtX2(amine)2] chemotypes (14). The development of cellular resistance to cisplatin (15,16) and also to carboplatin and oxaliplatin (17), through enhanced DNA repair, might thus be circumvented in polynuclear agents. The advancement to Phase II human clinical trials of the prototypical trinuclear drug BBR3464 (for all structures, see Figure 1) validates the search for structure diversity in new platinum analogues (13,18).
Figure 1.
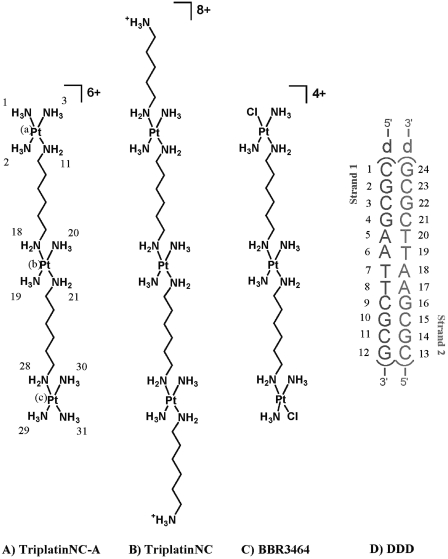
The chemical structures of the trinuclear platinum(II) compounds discussed in this article: (A) TriplatinNC-A, ([{Pt(NH3)3}2-µ-{trans-Pt(NH3)2(NH2(CH2)6NH2)2}](NO3)6, 1, 0,0,0/t,t,t,). (B) Triplatin-NC, ([{trans-Pt(NH3)2(NH2(CH2)6(NH3+)}2-µ-{trans-Pt(NH3)2 (NH2 (CH2)6 NH2)2}](NO3)8, 2). (C) BBR3464 containing substitution-labile chlorides ([{trans-PtCl(NH3)2}2-µ-{trans-Pt(NH3)2(NH2(CH2)6NH2)2}](NO3)4, 3). (D) The double-stranded self-complementary dodecamer duplex, [d(CGCGAATTCGCG)]2 (DDD), used for this crystallographic study.
Replacement of the substitution-labile chlorides of BBR3464 by ammonia (ammine) or amines gives the non-reactive polynuclear agents TriplatinNC-A (1, Figure 1A) and TriplatinNC (2, Figure 1B). Crystallographic characterization of a DNA complex with 2 (19) shows that the two mutually cis-oriented platinum-a(m)mine groups of the PtN4 units form discrete NH•••OP•••HN (amine•••phosphate•••ammine)-binding motifs. These ‘Phosphate Clamps’ represent a discrete new ligand–DNA-binding mode, extending the repertoire of binding possibilities by small ligands beyond intercalation and groove binding. Non-covalent polynuclear platinum compounds and TriplatinNC in particular, have interesting biological properties including in vitro cytotoxicity, anti-tumour activity and high cellular accumulation (20–23).
The quasi-independent and modular nature of the PtN4-based phosphate clamp as observed in the TriplatinNC–DNA complex suggests that the interaction can be used as a general DNA-binding element in combination with other nucleic acid binding motifs. The mode of association (B-form backbone tracking, A-form backbone tracking, groove spanning, intra-molecular association, etc.) can be dictated by alteration of linker chain length and rigidity, charge distribution and the number of PtN4 units. To examine the utility and generality of the phosphate clamp we have extended the study of non-covalent platinum complexes to TriplatinNC-A (Figure 1A). This compound is differentiated from TriplatinNC by the absence of dangling amines on the terminal Pt(II) atoms, resulting in a more compact molecule and an overall charge of 6+ instead of 8+. Here we describe the 3D X-ray crystal structure of 1 bound to the Dickerson–Drew Dodecamer (DDD, Figure 1D) (24), the self-complementary double-stranded DNA [d(CGCGAATTCGCG)]2. The binding modes and consequent conformational distortions of both platinum complexes are compared and the biological relevance discussed.
EXPERIMENTAL PROCEDURES
Synthesis
TriplatinNC-A, a gift of Alex Hegmans, was prepared by published procedures (20).
Crystallization
The ammonium salt of reverse-phase HPLC-purified d(CGCGAATTCGCG) (DDD, Integrated DNA Technologies, Coralville, IA, USA) was combined with TriplatinNC-A. The crystallization conditions were the same as for DDD–TriplatinNC complex (19). Crystals were grown in hanging drops by vapor diffusion from a solution initially containing 0.19 mM of the duplex, [d(CGCGAATTCGCG)]2, 0.19 mM TriplatinNC-A, 31 mM sodium cacodylate, pH 6.5, 9.6 mM magnesium chloride and 6.9% 2-methyl-2,4-pentanediol (MPD). The crystallization solution was equilibrated against a reservoir of 30% MPD at 20°C in a constant-temperature incubator. Orthorhombic (P212121) crystals appeared within a month. The crystal chosen for data collection was 0.1 × 0.1 × 0.2 mm3.
Data collection
Diffraction data were collected on beamline X26C at the National Synchrotron Light Source, Brookhaven National Laboratory. A total of 120 frames of intensity data (oscillation angle = 1.0°) were collected at 175 K at a wavelength of 1.05 Å and recorded on an ADSC-Quantum 4 CCD detector. Data were merged and reduced with HKL2000 (25).
Structure solution and refinement
The starting DNA model consisted of coordinates of [d(CGCGAATTCGCG)]2 (24), [NDB entry bdl084 (25)] excluding solvent and ions. After a rotation/translation search and rigid body refinement with CNS version 1.1 (26), the R/R-free factor were 52/55. Strong sum and difference peaks indicated positions of three Pt2+ atoms, which were added to the model, giving R/R-free factor of 38/39. The R/R-free factor dropped to 20/26 upon addition of 83 water molecules along with additional atoms of TriplatinNC-A. A partially occupied/disordered TriplatinNC-A was modeled as a bare Pt2+ ion. The initial model of TriplatinNC-A was generated with Chem3D Pro version 4.0 (CambridgeSoft).
The three Pt2+ positions were confirmed by anomalous maps, which were computed using phases of only a single Pt2+ atom, with the program CNS. During the refinement a peak of electron density surrounded by four water molecules, forming a partially disordered octahedron, was assigned as a sodium ion. The four Na+–O bond lengths are 2.03, 2.30, 2.43 and 2.52 Å, respectively for Na+–O1, Na+−O2, Na+–O3, and Na+–O4. The bond length of Na+−O1 is significantly shorter than the others and rather close to that for a Mg2+–O bond. However, replacement of Mg2+ for Na+ resulted in an increase in R/R-free factor by 0.21/0.03%. The occupancies of TriplatinNC-A were estimated using the Pt2+ atoms. Occupancies were optimized throughout the refinement by fitting statistics and by examination of ±(Fo–Fc) maps. Thirty-five cycles of refinement were carried out using the program CNS version 1.1 (27), during which isotropic thermal factor refinement was performed for all the atoms. The refinement converged to a final R/R-free factor of 19.59/25.80. Helical parameters were calculated using a program CURVES 5.3 (28).
Circular dichroism spectroscopy
Circular dichroism (CD) spectra were recorded at room temperature using a Jasco 600 CD spectrophotometer and a 10 mm quartz submicro cuvette. All samples were prepared in 10 mM phosphate buffer solution, pH 7.4 containing 50 mM sodium chloride. The dodecamer (100 µM in nucleotides) was incubated with the corresponding platinum compounds at 37°C for 1 h at variable ri values [ri = (initial number of molecules of platinum complex in solution)/(number of nucleotides of DNA in solution)]. To obtain the presented spectra six scans in the range from 200 to 350 nm were averaged. The background was subtracted electronically.
RESULTS
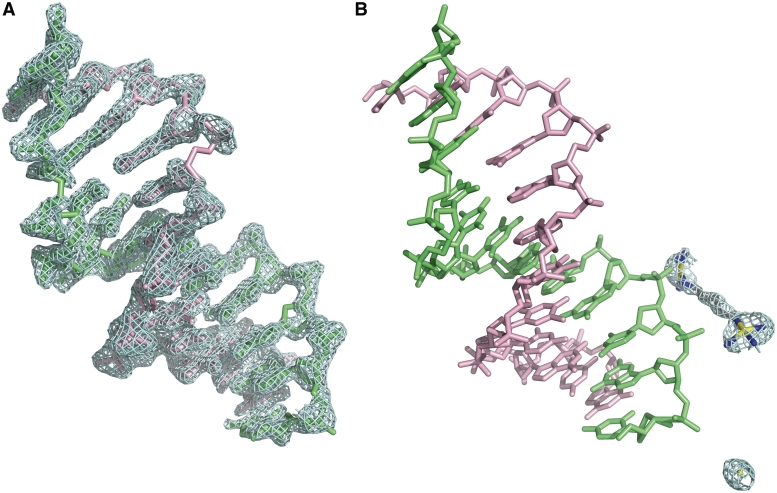
Diffraction data from crystals of the complex formed between the DDD and 1 extend to 1.70 Å. Data collection and refinement statistics are given in Table 1. The maps surrounding the DNA are clean and generally continuous (Figure 2A). The final refined model of the asymmetric unit contains (i) one DNA duplex with 12 bp, (ii) a partially ordered and partially occupied molecule of TriplatinNC-A, with two Pt coordination spheres, one hexanediamine bridge with all six carbon atoms and a total of eight nitrogen atoms from the two coordination spheres, (iii) a tetra-hydrated Na+ ion and (iv) 87 water molecules including the first shell Na+ water molecules. One Pt2+ ion lacking ligands is also observed. The occupancy of the ordered portion of 1, with two Pt2+ ions plus their coordination spheres linked by the hexanediamine bridge, is estimated to be 79%. The occupancy of the bare Pt+2 ion is estimated to be 11%.
Table 1.
Data collection and refinement statistics
| Unit cell dimensions | α = β = γ = 90° |
| a = 23.440 Å, b = 39.007 Å | |
| c = 65.922 Å | |
| DNA (asymmetric unit) | [d(CGCGAATTCGCG)]2 |
| Space group | P212121 |
| Temperature of data collection (K) | 175 |
| Number of all reflections | 10 298 |
| Number of reflections in resolution range (1.7–25 Å) | 6843 |
| Number of reflections used in refinement (F > 3σF) | 6292 |
| RMSD of bonds from ideality (Å) | 0.026 |
| RMSD of angles from ideality (°) | 1.995 |
| Max. resolution of observed reflections (Å) | 1.26 |
| Max. resolution of refinement (Å) | 1.70 |
| Number of DNA atoms | 486 |
| Number of TriplatinNC-A atoms | 15 |
| Number of water molecules, excluding sodium first shell | 83 |
| Number of sodium ions plus coordinating water molecules | 5 |
| R-factor (%, F > 3σF) | 19.64 |
| R-free (%, all) | 25.68 |
Figure 2.
Electron density maps surrounding the DDD/1 (TriplatinNC-A) complex. (A) Sum (2Fo–Fc) electron density surrounding the DNA, contoured at 1.3σ. (B) Sum electron density around 1 contoured at 1.0σ.
Platinum positions were initially established from the intense peaks in the sum (2Fo – Fc) maps, and were confirmed by the anomalous peaks (F+ – F–, Figure 2B). The anomalous electron density is consistent with the Pt2+ positions initially determined with the sum electron density map.
Phosphate clamps
The Pt-am(m)ine groups of 1 (TriplatinNC-A) form a total of 12 hydrogen bonds with DNA (Figure 3). The compound, like 2 (TriplatinNC), shows remarkable selectivity for (i) DNA oxygen atoms over nitrogen atoms, (ii) phosphate oxygen atoms over other oxygen atoms and (iii) for O2P over O1P atoms. As a result, 11 of the hydrogen bonding interactions involve oxygen atoms, 10 of which are with phosphate oxygens. The exception is a hydrogen bond to a guanine O6. Only a single hydrogen bond links an NH3 group of a PtN4 unit to a nitrogen atom (N7 of G).
Figure 3.
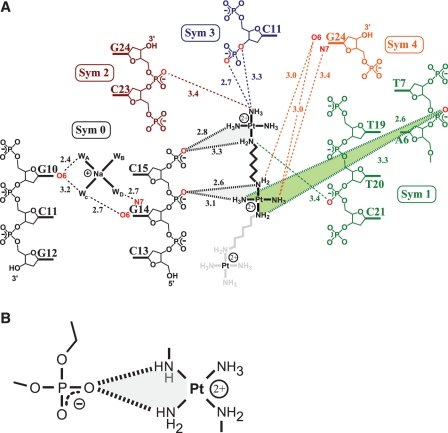
Hydrogen bonding interactions of 1 with DNA. Hydrogen bonding interactions of phosphate clamps are indicated by bold hashed lines. The phosphate clamps are highlighted by shading. Other hydrogen bonds are indicated by dashed lines. (A) 1 forms three phosphate clamps with the DNA. Oxygen and nitrogen atoms that form hydrogen bonds with 1 or the Na+ are highlighted in red. Hydrogen bonding distances are given in Ångstroms. The symmetry operators are as follows : Sym 0: x,y,z; Sym 1: –x,y–1/2,–z+3/2; Sym 2: –x–1/2,–y+1,z–1/2; Sym 3: x–1,y,z; Sym 4: –x + 1/2,–y+1,z–1/2. (B) A schematic diagram of a phosphate clamp.
The hydrogen-bonding interaction between the ligand (Pt complex) and DNA is best structurally characterized by the formation of phosphate clamps (Figure 3). In the phosphate clamp, a single OP atom accepts two hydrogen bonds, one from each of two adjacent a(m)mino groups of a PtN4 unit. Three phosphate clamps involving two PtN4 coordination spheres of TriplatinNC-A are evident in the electron density. In the case of 2, seven phosphate clamps involving four Pt2+ centers were observed (19). Similar to the previous structure, 1 forms an extensive hydrogen bonding and electrostatic network within the DDD crystal, bridging a total of five different symmetry-related duplexes (Figure 3).
In the structure discussed here and the previous one (19), the geometries of Phosphate Clamps are highly conserved. The motif again appears to require cis-orientation of the am(m)ine ligands—mutually trans ligands do not participate in phosphate clamps. Furthermore, the ligands are one Pt-NH3 (ammine) group and one Pt-NH2R group. Only two PtN4 units are observable, which collectively engage in three phosphate clamps. In this structure, one Pt atom, designated Pt(a), was assigned as a bare cation since the electron density around Pt(a) is not sufficient to determine the geometry of the coordination sphere.
Backbone tracking
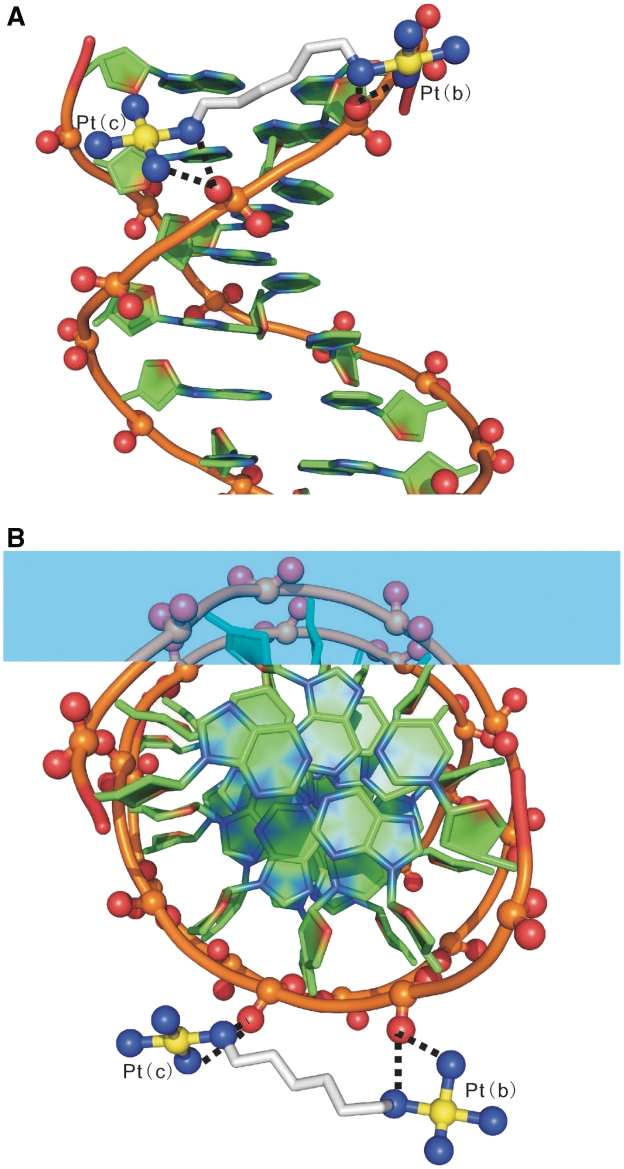
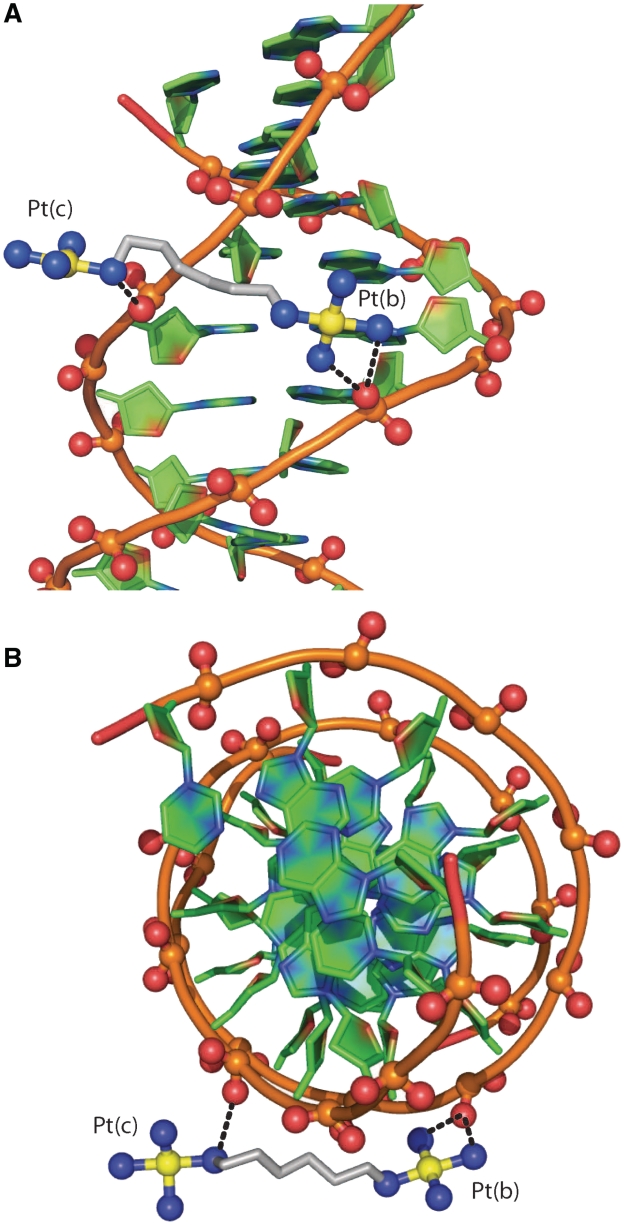
The observable portion of TriplatinNC-A is extended along the backbone of one strand of the DDD (Sym 0 in Figure 3). One Phosphate Clamp [Pt(b)] engages the O2P oxygen atom of C15 while the other [Pt(c)] engages the O2P atom of the adjacent G16 (Figure 4). In contrast, in the DDD–TriplatinNC complex the PtN4 units forms phosphate clamps with A17 and G16, but not with C15 (19). This difference in binding interactions may be due to the increased charge caused by the presence of the dangling hexanediamine, which forms hydrogen bonds with the O2P of G14 (19). This extra interaction appears to induce the trinuclear unit to bend, pushing the one PtN4 coordination sphere away from the phosphate of C15, forming a total of three phosphate clamps.
Figure 4.
Backbone tracking of the phosphate clamp. Hydrogen bonds between 1 and DNA are indicated by dashed lines. Two phosphate clamps are illustrated. (A) View perpendicular to the helical axis. (B) View along the helical axis. Atoms are colored by type with carbon (DNA), green; carbon (TriplatinNC-A) grey; nitrogen, blue; oxygen, red; platinum, yellow. The phosphodiester backbone of the DNA is represented by a tube. The P, O1P and O2P atoms are ball-and-stick. TriplatinNC-A is represented by stick, except for nitrogen and platinum atoms, which are ball-and-stick. This and some of the other figures in this manuscript were made in part with Pymol (Warren L. DeLano ‘The PyMOL Molecular Graphic System.’ DeLano Scientific LLC, San Carlos, CA, USA http://www.pymol.org).
Groove spanning
TriplatinNC-A participates in Groove Spanning by bridging between two DNA strands across the minor groove of the DNA duplex (Duplex Sym 1 in Figure 3) through clamps at the phosphate of T7 and one incomplete clamp, a PtN4-O2P hydrogen bond at C21 (Figures 3 and 5). In contrast, in the TriplatinNC–DDD complex the Groove Spanning is achieved by two intact phosphate clamps. Overall, the two compounds bind by remarkably similar motifs—the Backbone Tracking and Groove Spanning are facilitated through the Phosphate Clamp, although the specific positions and interacting parts of the DDD are not exactly the same.
Figure 5.
Groove spanning. Hydrogen bonds between 1 and DNA (Sym 1, Figure 3) are indicated by dashed lines. There are two Phosphate Clamps, with which TriplatinNC-A bridges two strands across the minor groove of a DNA duplex, at the lip of the minor groove, defined in Sines et al. (29). (A) View perpendicular to the helical axis. (B) View along the helical axis. Atoms are colored and coded as in Figure 4.
Sodium ions
In many previously-described high-resolution DDD crystal structures, a fully hydrated magnesium ion [Mg (H2O)6]2+ is located in the major-groove near one end of the duplex (25,29,30). The first shell water molecules of the Mg2+ ion form hydrogen bonds to the G(2)–C(23) and C(3)–G(22) base pairs. In the DDD-1 complex, a hydrated sodium ion [Na(H2O)4]+ is observed on the other end of the duplex in the major groove adjacent to G(10)–C(15) and C(11)–G(14). The Na+ does not form first-shell contacts with the DNA. Instead, the four first-shell water molecules that coordinate the Na+ form hydrogen bonds to the guanine O6 and N7 positions of G(14) and to the O6 position of G(10), Figure 3. These hydrogen bonds are similar to those of [Mg (H2O)6] 2+ at the other end of the DNA fragment. This Na+ ion is nearby to where 1 tracks the DNA backbone. In the DDD-2 complex a hexa-coordinate sodium ion [Na(H2O)5(O6)]+ is also observed, with the sixth ligand being the O6 from residue G(22). The first shell water ligands interact with G(2), C(21) and G(22) and the complex is located at the opposite end of DDD from where 2 forms the Backbone Tracking (19). Accordingly, the location and hydrogen bonding properties of hydrated sodium cations, although observed in both cases, were different between the two structures.
DNA conformation
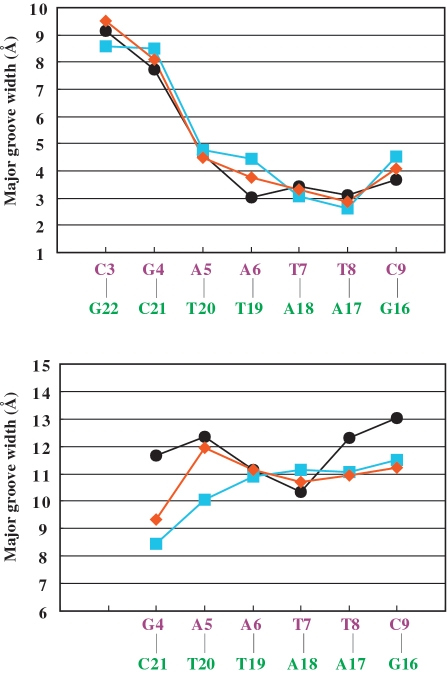
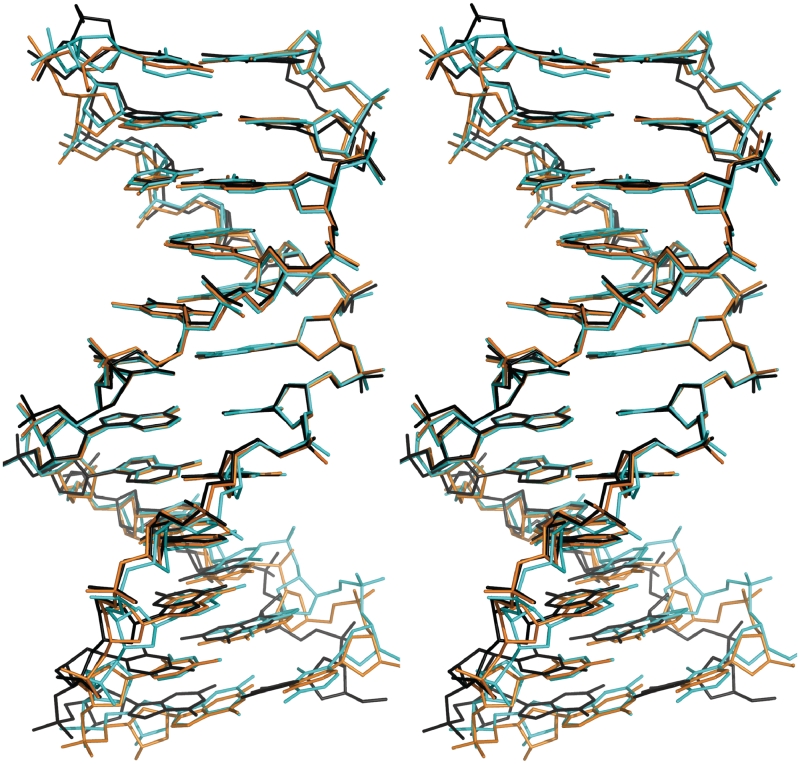
To examine the DNA conformational changes induced through binding by this class of ligand, we compare the DDD–TriplatinNC-A (1) to DDD–TriplatinNC (2) and to other high resolution DDD structures, in particular structure bdl084, which is the highest resolution, non-modified DDD structure determined thus far (25). Phosphodiester backbone torsion angles, glycosyl angles and sugar puckers are given in Table 2. The major-groove and minor-groove widths of DDD–TriplatinNC-A are intermediate between those of DDD–TriplatinNC and the control DDD, Figure 6. From these parameters, the superimposition of both polynuclear complexes onto the control DDD is shown in Figure 7. Base–base and inter-base parameters are shown in Supplementary Figures S1 and S2.
Table 2.
Backbone torsion angles (α–ζ), glycosyl angles (χ), pseudorotation phase angles and amplitudes, and sugar puckersa
| Residue | χ (°) | γ (°) | δ (°) | ε (°) | ζ (°) | α (°) | β (°) | Phase (°) | Amplitude | Pucker |
|---|---|---|---|---|---|---|---|---|---|---|
| C(1) | −121 | 71 | 128 | –179 | –124 | –38 | –177 | 133 | 40 | C1′-exo |
| G(2) | –113 | 35 | 146 | –173 | –104 | –58 | 167 | 170 | 32 | C2′-endo |
| C(3) | –129 | 55 | 112 | –164 | –105 | –49 | 176 | 111 | 34 | C1′-exo |
| G(4) | –94 | 45 | 139 | 174 | –101 | –38 | –177 | 168 | 32 | C2′-endo |
| A(5) | –100 | 33 | 138 | 179 | –105 | –53 | 174 | 169 | 31 | C2′-endo |
| A(6) | –113 | 50 | 129 | –178 | –91 | –64 | 175 | 155 | 30 | C2′-endo |
| T(7) | –125 | 53 | 110 | 178 | –102 | –53 | 166 | 108 | 39 | O1′-endo |
| T(8) | –116 | 61 | 129 | –178 | –105 | –47 | 177 | 141 | 34 | C1′-exo |
| C(9) | –107 | 46 | 134 | –165 | –92 | –68 | 167 | 154 | 33 | C2′-endo |
| G(10 | –91 | 49 | 144 | –109 | 160 | –74 | 154 | 146 | 45 | C2′-endo |
| C(11) | –106 | 48 | 137 | –163 | –88 | –61 | 170 | 157 | 30 | C2′-endo |
| G(12) | –104 | 45 | 101 | 104 | 37 | O1′-endo | ||||
| G(24) | –143 | 44 | 87 | 48 | 34 | C4′-exo | ||||
| C(23) | –125 | 52 | 117 | –171 | –95 | –57 | 168 | 116 | 31 | C1′-exo |
| G(22) | –82 | 40 | 152 | –128 | –175 | –76 | 144 | 150 | 49 | C2′-endo |
| C(21) | –109 | 42 | 123 | –169 | –88 | –59 | 173 | 129 | 35 | C1′-exo |
| T(20) | –123 | 57 | 121 | –175 | –97 | –56 | 177 | 123 | 34 | C1′-exo |
| T(19) | –127 | 59 | 115 | –179 | –91 | –62 | 175 | 117 | 36 | C1′-exo |
| A(18) | –120 | 50 | 113 | 170 | –89 | –59 | 173 | 111 | 37 | C1′-exo |
| A(17) | –103 | 51 | 155 | –173 | –108 | –60 | 169 | 181 | 38 | C3′-exo |
| G(16) | –104 | 61 | 145 | 163 | –98 | –55 | –166 | 169 | 33 | C2′-endo |
| C(15) | –133 | 54 | 100 | –176 | –89 | –65 | 179 | 70 | 30 | C4′-exo |
| G(14) | –109 | –142 | 142 | –174 | –97 | –53 | 160 | 169 | 27 | C2′-endo |
| C(13) | –175 | 172 | 155 | –108 | –77 | 73 | –152 | 207 | 35 | C3′-exo |
aBackbone torsion angles are O3′-P-α-O5′-β-C5′-γ-C4′-δ-C3′-ε-O3′-ζ-P-O5′.
Figure 6.
Minor- and major-groove widths for the DDD/TriplatinNC-A (1, orange diamonds), for the DDD–TriplatinNC complex (2, cyan squares), and for the control DDD (BDL084, black circles).
Figure 7.
Superimposition of the TriplatinNC-A (1, orange) and TriplatinNC complexes (2DYW, 2, cyan) onto the control DDD (BDL084, black). Superimposition was performed based on the set of three continuous base pairs, A(6)–T(19), A(7)–T(18) and T(8)–A(17), that give the least RMS deviation of atomic positions.
Previously we suggested that an observed increase in the minor groove width of DDD–TriplatinNC in comparison to the ‘free’ DDD is related to the groove spanning by the complex. The cationic centers of 1 interact with the same phosphates as TriplatinNC. In both structures, two Phosphate Clamps are juxtaposed across the minor groove. However, in the present case, one of the two Phosphate Clamps of the Groove Spanning mode is only partially formed (see above). The weaker Groove Spanning interactions might result in slightly lesser deformation of the duplex. For example, the major-groove width of DDD-1 at A(6)–T(19), in which the major-groove width of DDD–TriplatinNC and the control DDD show the largest deviation, was found to be the median value of the other two structures, Figure 6.
The backbone: BI versus BII
The DNA complexed with TriplatinNC-A is wholly in the BI conformation, with ε–ξ is in the range of −160 to +20. B-DNA conformation is characterized by BI, the most frequent, or BII, depending on the torsion angles ε and ξ (31,32). These torsion angles influence the positions of phosphate groups and modulate the width of the minor groove. The difference between ε and ξ is a useful indicator of BI/BII. An ε–ξ of roughly −90° indicates BI whereas a value of roughly +90° indicates BII.
The backbone: sugar puckers
Of the 24 sugar puckers of DDD-1, 11 differ from those of the corresponding residues in the control DDD. A total of 14 differ from those of DDD–TriplatinNC. Large differences in sugar pucker are found in G(12) (−103) and (C23) (+99) compared to the control DDD. Differences are also found in G(2) (+168), C(3) (+93) C23 (+99) relative to DDD–TriplatinNC.
Global conformation
The axial bending and axial shortening of the DDD-1 complex are very similar to those of DDD–TriplatinNC and significantly greater than those of the DDD. In the program Curves 5.3 (28), UU is a measure of axial bending, giving the angle formed by the axis segments of the terminal sections of the DNA duplex. PP is the angle between the vectors formed between the two axis reference points at either end of the fragment. UU and PP for the DDD-1 complex were found to be 28° and 38°, both of which are very close to the corresponding value of the DDD–TriplatinNC complex and significantly larger than those of the control DDD. This trend is also the same in axial bend and the axial path length shortening ratio (DDD-1: 28°/2.0%; DDD–TriplatinNC: 27°/2.4%; DDD: 12.9°/0.66%).
Conformation in solution
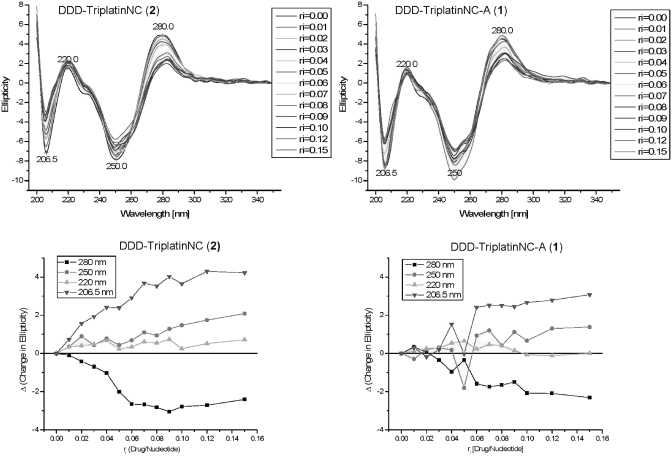
The similarity in binding mode of the two polynuclear complexes is also confirmed in solution. The CD spectra of the DDD titrated with both 1 and 2 are very similar and further confirm the essential B-nature of the conformational changes upon complex binding (Figure 8). The spectra are characterized by a negative shift (decrease) in ellipticity for the peak centered at 280 nm and a concomitant positive shift (increase) for the peak centered at 206 nm. Thus the difference in peak height is decreased.
Figure 8.
Circular Dichroism Spectra of DDD duplex titrated with aliquots of TriplatinNC-A (1) and TriplatinNC (2). Bottom panels show changes in ellipticity upon titration.
DISCUSSION
The results reported here confirm the generality and utility of the PtN4 unit for formation of Phosphate Clamps with DNA, involving two modes of binding, Backbone Tracking and Groove Spanning. Phosphate clamps form six-membered rings, with two hydrogen bonds from cis-oriented Pt-a(m)mines to a common phosphate oxygen. It is a discrete and modular DNA-binding device with high potential as a drug-design scaffold. In solution, conformational changes are equivalent for the two complexes whose structures have been determined, further emphasizing the similarity in binding mode.
Forks and clamps
The interactions of PtN4 coordination units with DNA have some analogy to the interactions of the guanidino group of arginine with polynucleotides. The limiting ‘canonical’ structures for both arginine forks and phosphate clamps are shown schematically below for comparison. It was previously suggested that arginine shows an OP clamping ability in which two adjacent NH groups of a guanidino group interact with two oxygen atoms of a common phosphate (33–35). This motif would result in an eight-membered ring (including hydrogen atoms) stabilized by two hydrogen bonds. In the ‘Arginine Fork’, one guanidino group may be engaged in two of these interactions (33). The apparent mimicry of arginine by the platinum–tetra(m)mine unit led us re-evaluate the frequency and geometry of Arginine Forks. We therefore examined ribosomal structures such as 1JJ2 (35) and 2J00/2J01 (34). The large subunit of the Haloarcula Marismortui ribosome (1JJ2), for example, contains 336 arginines and nearly 3000 phosphate groups. From analysis of such structures we conclude that the guanidino group of arginine, like PtN4, binds to nucleic acids with high selectivity for OP atoms, i.e. for non-bridging phosphate oxygens, but with phosphate oxygen selectivity: PtN4 > ARG. The guanidino group does form clamp-like structures, consisting of eight-membered rings, at some measurable frequency but again, PtN4 >> ARG. However PtN4 forms forks while the guanidino group of arginine does not; the frequency at which one guanidino group engages in two phosphate clamps is essentially zero. In sum, PtN4 appears to be unique in its combined selectivity for OP atoms, forking propensity, and highly conserved geometry of binding. Thus the PtN4 unit has potential as a general DNA/RNA-binding element and drug design scaffold.
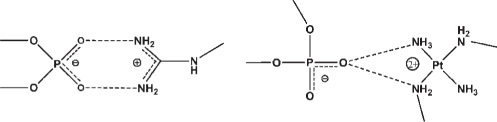
Binding modes compared to other DNA-binding ligands
Because there is some analogy between the polynuclear platinum structure (especially non-covalent compounds such as 1 and 2) and polyamines such as spermine and spermidine, it is of interest to note that polyamines generally bind preferentially in the major groove rather than to phosphates (36). Key features in differences in recognition patterns between the two classes are the distances between amino groups and the fact that the cis-[Pt(NH3(NH2R)] unit in fact is sterically rigid because of the strict adherence to square-planar geometry (∼90° bond N-Pt-N bond angle) for Pt complexes. Polyamines are conformationally flexible and optimal distances to achieve phosphate clamp binding—provided by the square planar geometry and also to a lesser extent the sterically rigid guanidine group—are not as readily available.
Non-covalent ligand–DNA complexes are stabilized by a composite of hydrogen-bonding, van der Waals and electrostatic interactions. These interactions most generally occur within the minor groove or between base pairs (37,38). More recently Arya and coworkers have developed major groove binders (39). Appropriate combinations of modular binding elements gives high affinity and specificity (37) but, comparing all cases, the discrete nature of the Phosphate Clamp is clear.
DNA distortion
We previously suggested that a change in DNA conformation may arise upon substitution of a hydrated Mg2+ in the native DDD for a partially dehydrated Na+ ion in the DDD–TriplatinNC complex (19). The Na+ ion is located directly on the floor of the major groove (interacting with O6 of a G). It was difficult to isolate the ligand, a hydrated cation or TriplatinNC, both directly interacting with DNA, as the primary contributor to DNA deformation. In the DDD-1 crystal structure, no hydrated cation is observed in the position of the hydrated Mg2+ or Na+ in the control DDD or DDD–TriplatinNC. Accordingly, all the hydrated cations found in the control DDD, DDD–TriplatinNC, and DDD-1 are different in atom species, hydrogen bonding properties, and interacting areas but deformation directions of TriplatinNC and 1 from the control DDD appear to be the same. Therefore, the ligands controlling the DNA deformation seem to be the trinuclear Pt(II) complexes, and hydrated cations are selected and fit to a preformed pocket of the DNA.
The work here expands the description of platinum–DNA-binding modes, and indeed metal complex–DNA interactions in general. The NDB database shows only a very limited number of crystal structures of duplex DNA bound with cis-platin [ntrastrand GG and the cisplatin–DNA duplex associated with the HMG protein (40), interstrand (GC)2] (41); oxaliplatin (intrastrand GG) (42) and cis-[Pt(NH3)(cyclohexylamine)Cl2] (intrastrand GG) (43) and a Z-DNA-forming sequence d(CGT NH2ACG)2 stabilized by [Pt(NH3)3]+ (44). With the exception of the latter, all of the DNA–platinum structures belong to the cis-[PtX2(amine)2] family and the work has delineated the 3D structural distortions of bifunctional binding and the factors affecting protein (HMG) recognition of the 1,2-GG intrastrand adduct. Indeed few crystal structures of non-covalently binding transition metal complexes are available—most recently the structures of a three-way DNA junction (3WJ) stabilized by metal-based supramolecular helicates have been described (45,46). The other high-resolution crystal structure entries appear limited to a sequence-specific rhodium intercalator (47,48) and the use of [Co(NH3)6]3+ and [Ru(NH3)6]3+ in stabilizing Z-DNA sequences (49,50).
Biological relevance of non-covalent Pt–DNA interactions
Both trinuclear compounds 1 and 2 bind to DNA exclusively by non-covalent interactions. Biophysical studies showed that the complexes stabilized duplex DNA and the higher the electrical charge of the complexes the greater the stabilization observed. The complexes induced both B → A and B → Z conformational changes in canonical DNA sequences (20). The binding affinity is sufficiently high such that they are not readily displaced from the helix by intercalators such as ethidium bromide, resulting in an essential irreversibility of the conformational change. The interaction with DNA of the minor-groove binding dye Hoechst 33 258 is cooperatively enhanced in the presence of the charged compound (22). Given the overall similarity of the binding modes, it is not surprising that the affinity of both molecules for DNA is also very similar—as would be expected since the overall binding differs only in charge and extra hydrogen bonding capability of the ‘dangling’ amines. Yet, the complexes differ in their biological consequences. Especially TriplatinNC (2, Figure 1) is cytotoxic at micromolar concentrations and exhibits in vivo anti-tumour activity. In contrast, 1 is ∼5–10 times less potent than TriplatinNC. The enhanced cytotoxicity has been attributed to greater cellular accumulation of the 8+ species. The differences in cytotoxicity between 1 and 2 may reflect differences in cellular accumulation—the higher positive charge produces greater accumulation which may provide a stronger driving force for access to DNA, resulting in a higher frequency of phosphate clamps. Note that TriplatinNC is also significantly more potent than other non-covalent platinum compounds reported (20–23,51,52).
CONCLUSIONS
The Phosphate Clamp is a general feature of DNA complexes of PtN4 containing non-reactive polynuclear platinum agents. The backbone selective binding is distinct from ‘classic’ minor-groove binding and intercalation or the more recently described major groove binding. The molecules described here contain numerous PtN4 groups acting as DNA-binding ligands and connected by flexible linkers—polymorphism in solution may arise from the number of PtN4 per binding ligand and flexible connections between them. Both minor-groove binders and intercalators have undergone extensive study for their cytotoxic and anti-tumor properties. The modular nature of the polynuclear platinum binding allows for significant further complex design (aliphatic/aromatic linkers, shorter/longer chain length, or cis/trans geometry) to enhance biological activity. Platinum-DNA binding triggers downstream cell cycle signals eventually leading to apoptosis and cell killing. Given that so little of cisplatin actually gets to DNA, the Phosphate Clamp motif may be a much more effective method to trigger a cellular response. The absence of covalent binding to biomolecules in general may also result in less potential side effects than currently used agents. These considerations suggest that polynuclear platinum agents capable of non-covalent interactions represent a further class of platinum-based anti-tumour agents, discrete from covalently-binding mononuclear (cis-platin) and polynuclear (BBR3464) agents. Their overall profile relative to DNA-modifying agents deserves further investigation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (RO1CA78754 to N.P.F.); the Japan Society for the Promotion of Science (to S.K., M.C. and A.O.). Funding for Open Access Charge: National Institutes of Health (grant number RO1CA78754).
Conflict of interest statement. None declared.
REFERENCES
- 1.Rosenberg B, Vancamp L, Trosko JE, Mansour VH. Platinum compounds - a new class of potent antitumour agents. Nature. 1969;222:385–386. doi: 10.1038/222385a0. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg B, Vancamp L, Krigas T. Inhibition of cell division in escherichia coli by electrolysis products from a platinum electrode. Nature. 1965;205:698–699. doi: 10.1038/205698a0. [DOI] [PubMed] [Google Scholar]
- 3.Jamieson ER, Lippard SJ. Structure, recognition, and processing of cisplatin-DNA adducts. Chemical Rev. 1999;99:2467–2498. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 4.Takahara PM, Rosenzweig AC, Frederick CA, Lippard SJ. Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin. Nature. 1995;377:649–652. doi: 10.1038/377649a0. [DOI] [PubMed] [Google Scholar]
- 5.Kelland LR, Farrell N. Platinum-based drugs in cancer therapy. In: Teicher BA, editor. Cancer Drug Discovery and Development. Humana Press; 2000. [Google Scholar]
- 6.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nature Rev. Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 7.Farrell N. Nonclassical platinum antitumor agents – perspectives for design and development of new drugs complementary to cisplatin. Cancer Invest. 1993;11:578–589. doi: 10.3109/07357909309011676. [DOI] [PubMed] [Google Scholar]
- 8.Hegmans A, Berners-Price SJ, Davies MS, Thomas DS, Humphreys AS, Farrell N. Long range 1,4 and 1,6-interstrand cross-links formed by a trinuclear platinum complex. Minor groove preassociation affects kinetics and mechanism of cross-link formation as well as adduct structure. J Am Chem Soc. 2004;126:2166–2180. doi: 10.1021/ja036105u. [DOI] [PubMed] [Google Scholar]
- 9.Farrell N, Qu Y, Feng L, Vanhouten B. Comparison of chemical-reactivity, cytotoxicity, interstrand cross-linking and DNA-sequence specificity of bis(platinum) complexes containing monodentate or bidentate coordination spheres with their monomeric analogs. Biochemistry. 1990;29:9522–9531. doi: 10.1021/bi00493a005. [DOI] [PubMed] [Google Scholar]
- 10.Zehnulova J, Kasparkova J, Farrell N, Brabec V. Conformation, recognition by HMG-domain proteins and nucleotide excision repair of DNA intrastrand cross-links of novel antitumor trinuclear platinum complex BBR3464. J. Biol. Chem. 2001;276:22191–22199. doi: 10.1074/jbc.M103118200. [DOI] [PubMed] [Google Scholar]
- 11.Cox JW, Berners-Price S, Davies MS, Qu Y, Farrell N. Kinetic analysis of the stepwise formation of a long-range DNA interstrand cross-link by a dinuclear platinum antitumor complex: evidence for aquated intermediates and formation of both kinetically and thermodynamically controlled conformers. J. Am. Chem. Soc. 2001;123:1316–1326. doi: 10.1021/ja0012772. [DOI] [PubMed] [Google Scholar]
- 12.Qu Y, Scarsdale NJ, Tran MC, Farrell NP. Cooperative effects in long-range 1,4 DNA-DNA interstrand cross-links formed by polynuclear platinum complexes: an unexpected syn orientation of adenine bases outside the binding sites. J. Biol. Inorg. Chem. 2003;8:19–28. doi: 10.1007/s00775-002-0383-x. [DOI] [PubMed] [Google Scholar]
- 13.Farrell N. Polynuclear platinum drugs. Metal Ions Biol. Sys. 2004;41:252–296. [PubMed] [Google Scholar]
- 14.Kasparkova J, Zehnulova J, Farrell NP, Brabec V. DNA interstrand crosslinks of novel antitumor trinuclear platinum complex BBR3464. Conformation, recognition by HMG-domain proteins and nucleotide excision repair. J. Biol. Chem. 2002;277:48076–48086. doi: 10.1074/jbc.M208016200. [DOI] [PubMed] [Google Scholar]
- 15.Martin LP, Hamilton TC, Schilder RJ. Platinum resistance: the role of DNA repair pathways. Clin. Cancer Res. 2008;14:1291–1295. doi: 10.1158/1078-0432.CCR-07-2238. [DOI] [PubMed] [Google Scholar]
- 16.Stewart DJ. Mechanisms of resistance to cisplatin and carboplatin. Crit. Rev. Oncol. Hematol. 2007;63:12–31. doi: 10.1016/j.critrevonc.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Reed E. ERCC1 and clinical resistance to platinum-based therapy. Clin. Cancer Res. 2005;11:6100–6105. doi: 10.1158/1078-0432.CCR-05-1083. [DOI] [PubMed] [Google Scholar]
- 18.Manzotti C, Pratesi G, Menta E, Di Domenico R, Cavalletti E, Fiebig HH, Kelland LR, Farrell N, Polizzi D, Supino R, et al. BBR 3464: a novel triplatinum complex, exhibiting a preclinical profile of antitumor efficacy different from cisplatin. Clin. Cancer Res. 2000;6:2626–2634. [PubMed] [Google Scholar]
- 19.Komeda S, Moulaei T, Woods KK, Chikuma M, Farrell NP, Williams LD. A third mode of DNA binding: phosphate clamps by a polynuclear platinum complex. J. Am. Chem. Soc. 2006;128:16092–16103. doi: 10.1021/ja062851y. [DOI] [PubMed] [Google Scholar]
- 20.Qu Y, Harris A, Hegmans A, Petz A, Kabolizadeh P, Penazova H, Farrell N. Synthesis and DNA conformational changes of non-covalent polynuclear platinum complexes. J. Inorg. Biochem. 2004;98:1591–1598. doi: 10.1016/j.jinorgbio.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 21.Harris AL, Yang X, Hegmans A, Povirk L, Ryan JJ, Kelland L, Farrell NP. Synthesis, characterization, and cytotoxicity of a novel highly charged trinuclear platinum compound. Enhancement of cellular uptake with charge. Inorg. Chem. 2005;44:9598–9600. doi: 10.1021/ic051390z. [DOI] [PubMed] [Google Scholar]
- 22.Harris A, Qu Y, Farrell NP. Unique cooperative binding interaction observed between a minor groove binding Pt anti-tumor agent and Hoeschst dye 33258. Inorg. Chem. 2005;44:1196–1198. doi: 10.1021/ic048356p. [DOI] [PubMed] [Google Scholar]
- 23.Harris AL, Ryan JJ, Farrell NP. Biological consequences of trinuclear platinum complexes: comparison of BBR 3464 to its non-covalent Congeners. Mol. Pharmacol. 2006;69:666–672. doi: 10.1124/mol.105.018762. [DOI] [PubMed] [Google Scholar]
- 24.Wing R, Drew H, Takano T, Broka C, Takana S, Itakura K, Dickerson RE. Crystal structure analysis of a complete turn of B-DNA. Nature. 1980;287:755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]
- 25.Shui X, McFail-Isom L, Hu GG, Williams LD. The B-DNA dodecamer at high resolution reveals a spine of water on sodium. Biochemistry. 1998;37:8341–8355. doi: 10.1021/bi973073c. [DOI] [PubMed] [Google Scholar]
- 26.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D-Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 27.Sheldrick GM. SHELX-97, Gottingen University, Germany. 1997. SHELX-97. [Google Scholar]
- 28.Stofer E, Lavery R. Measuring the geometry of DNA grooves. Biopolymers. 1994;34:337–346. doi: 10.1002/bip.360340305. [DOI] [PubMed] [Google Scholar]
- 29.Sines CC, McFail-Isom L, Howerton SB, VanDerveer D, Williams LD. Cations mediate B-DNA conformational heterogeneity. J. Am. Chem. Soc. 2000;122:11048–11056. [Google Scholar]
- 30.Minasov G, Tereshko V, Egli M. Atomic-resolution crystal structures of B-DNA reveal specific influences of divalent metal ions on conformation and packing. J. Mol. Biol. 1999;291:83–99. doi: 10.1006/jmbi.1999.2934. [DOI] [PubMed] [Google Scholar]
- 31.Privé GG, Heinemann U, Chandrasegaran S, Kan LS, Kopka ML, Dickerson RE. Helix geometry, hydration, and G-A mismatch in a B-DNA decamer. Science. 1987;238:498–504. doi: 10.1126/science.3310237. [DOI] [PubMed] [Google Scholar]
- 32.Hartmann B, Piazzola D, Lavery R. BI-BII transitions in B-DNA. Nucleic Acids Res. 1993;21:561–568. doi: 10.1093/nar/21.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calnan BJ, Tidor B, Biancalana S, Hudson D, Frankel A. Arginine-mediated RNA recognition: the arginine fork. Science. 1991;252:1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- 34.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 35.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woods KK, Maehigashi T, Howerton SB, Sines CC, Tannenbaum S, Williams LD. High-resolution structure of an extended A-tract: [d(CGCAAATTTGCG)]2. J. Am. Chem. Soc. 2004;126:15330–15331. doi: 10.1021/ja045207x. [DOI] [PubMed] [Google Scholar]
- 37.Strekowski L, Wilson B. Noncovalent interactions with DNA: an overview. Mutat. Res. 2007;623:3–13. doi: 10.1016/j.mrfmmm.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 38.Geierstanger BH, Wemmer DE. Complexes of the minor groove of DNA. Annu. Rev. Biophys. Biomol. Struct. 1995;24:463–493. doi: 10.1146/annurev.bb.24.060195.002335. [DOI] [PubMed] [Google Scholar]
- 39.Willis B, Arya DP. Triple recognition of B-DNA. Bioorg. Med. Chem. Lett. 2009;19:4974–4979. doi: 10.1016/j.bmcl.2009.07.079. [DOI] [PubMed] [Google Scholar]
- 40.Takahara PM, Rosenzweig AC, Frederick CA, Lippard SJ. Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin. Nature. 1995;377:649–652. doi: 10.1038/377649a0. [DOI] [PubMed] [Google Scholar]
- 41.Coste F, Malinge JM, Serre L, Shepard W, Roth M, Leng M, Zelwer C. Crystal structure of a double-stranded DNA containing a cisplatin interstrand cross-link at 1.63 Å resolution: hydration at the platinated site. Nucleic Acids Res. 1999;27:1837–1846. doi: 10.1093/nar/27.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spingler B, Whittington DA, Lippard SJ. Crystal structure of an oxaliplatin 1,2-d(GpG) intrastrand cross-link in a DNA dodecamer duplex. Inorg. Chem. 2001;40:5596–5602. doi: 10.1021/ic010790t. [DOI] [PubMed] [Google Scholar]
- 43.Silverman AP, Bu W, Cohen SM, Lippard SJ. 2.4-Å crystal structure of the asymmetric platinum complex [t(ammine)(cyclohexylamine)]2+ bound to a dodecamer DNA duplex. J. Biol. Chem. 2002;277:49743–49749. doi: 10.1074/jbc.M206979200. [DOI] [PubMed] [Google Scholar]
- 44.Parkinson GN, Arvanitis GM, Lessinger L, Ginell SL, Jones R, Gaffney B, Berman HM. Crystal and molecular structure of a new Z-DNA crystal form: d[CGT(2-NH2-A)CG] and its platinated derivative. Biochemistry. 1995;34:15487–15495. doi: 10.1021/bi00047a014. [DOI] [PubMed] [Google Scholar]
- 45.Oleksi A, Blanco AG, Boer R, Uson I, Aymami J, Rodger A, Hannon MJ, Coll M. Molecular recognition of a three-way DNA junction by a metallosupramolecular helicate. Angew Chemie, Int. Edit. 2006;45:1227–1231. doi: 10.1002/anie.200503822. [DOI] [PubMed] [Google Scholar]
- 46.Boer DR, Kerckhoffs JM, Parajo Y, Pascu M, Usón I, Lincoln P, Hannon MJ, Coll M. Self-assembly of functionalizable two-component 3D DNA arrays through the induced formation of DNA three-way-junction branch points by supramolecular cylinders. Angew. Chemie, Int. Edit. 2010;49:2336–2339. doi: 10.1002/anie.200906742. [DOI] [PubMed] [Google Scholar]
- 47.Kielkopf CL, Erkkila KE, Hudson BP, Barton JK, Rees DC. Structure of a photoactive rhodium complex intercalated into DNA. Nat. Struct. Biol. 2000;7:117–121. doi: 10.1038/72385. [DOI] [PubMed] [Google Scholar]
- 48.Pierre VC, Kaiser JT, Barton JK. Insights into finding a mismatch through the structure of a mispaired DNA bound by a rhodium intercalator. Proc. Natl Acad. Sci., USA. 2007;104:429–434. doi: 10.1073/pnas.0610170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thiyagarajan S, Rajan SS, Gautham N. Cobalt hexammine induced tautomeric shift in Z-DNA: the structure of d(CGCGCA)*d(TGCGCG) in two crystal forms. Nucleic Acids Res. 2004;32:5945–5953. doi: 10.1093/nar/gkh919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ho PS, Frederick CA, Saal D, Wang AH, Rich A. The interactions of ruthenium hexaammine with Z-DNA: crystal structure of a Ru(NH3)6+3 salt of d(CGCGCG) at 1.2 A resolution. J. Biomol. Struct. Dyn. 1987;4:521–534. doi: 10.1080/07391102.1987.10507657. [DOI] [PubMed] [Google Scholar]
- 51.Collins JG, Wheate NJ. Potential adenine and minor groove binding platinum complexes. J. Inorg. Biochem. 2004;98:1578–1584. doi: 10.1016/j.jinorgbio.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 52.Wheate NJ, Cutts SM, Phillips DR, Aldrich-Wright JR, Collins JG. The binding of [(en)Pt(μ-dpzm)2Pt(en)]4+ to G/C-rich regions of DNA. J. Inorg. Biochem. 2001;84:119–127. doi: 10.1016/s0162-0134(00)00206-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.