Abstract
M-ORBIS is a Molecular Cartography approach that performs integrative high-throughput analysis of structural data to localize all types of binding sites and associated partners by homology and to characterize their properties and behaviors in a systemic way. The robustness of our binding site inferences was compared to four curated datasets corresponding to protein heterodimers and homodimers and protein–DNA/RNA assemblies. The Molecular Cartographies of structurally well-detailed proteins shows that 44% of their surfaces interact with non-solvent partners. Residue contact frequencies with water suggest that ∼86% of their surfaces are transiently solvated, whereas only 15% are specifically solvated. Our analysis also reveals the existence of two major binding site families: specific binding sites which can only bind one type of molecule (protein, DNA, RNA, etc.) and polyvalent binding sites that can bind several distinct types of molecule. Specific homodimer binding sites are for instance nearly twice as hydrophobic than previously described and more closely resemble the protein core, while polyvalent binding sites able to form homo and heterodimers more closely resemble the surfaces involved in crystal packing. Similarly, the regions able to bind DNA and to alternatively form homodimers, are more hydrophobic and less polar than previously described DNA binding sites.
INTRODUCTION
A widely used approach in Biology/Bioinformatics is to detect patterns, identify their functions and to use these patterns to gain knowledge on unknown systems. Approaches such as BLAST, or the PROSITE or Pfam databases (1–3), are now commonly used to infer and annotate molecular functions based on sequence comparisons. Similarly, the comparison of protein structures reported and summarized in databases such as CATH and SCOP (4–6) have also been widely used to classify proteins into families and subfamilies, to infer functions and to give insights into the landscape of macromolecular folds available in the cell. The detection of common patterns can also serve other purposes such as the modeling of molecular structures by homology (7) that requires structural templates to function correctly. The last decades of research in experimental and computational structural biology have been mainly devoted to the analysis, characterization and prediction of protein structures and protein assemblies. With structural genomic projects and the work realized by structural biologists, the trend is moving increasingly towards the structural characterization of proteins and nucleic acids as functional and dynamic objects by predicting protein, DNA or peptide binding sites (1–4), by studying intrinsic variability (5) or by studying local differences between unbound and bound forms (6,7). To help in the prediction of protein–protein binding sites Porollo et al. (3) for instance proposed to retrieve the homologous structural chains. Some time before, Chung et al. (8) also proposed to predict binding sites by localizing the residues which were structurally conserved in several homologous structures. More recently, a Japanese group developed a database of interaction sites also based on the inference of binding sites by homology (9).
Nevertheless, the robust and systematic retrieval of homologous structures in specific ‘molecular contexts’ (structures sharing a same set of interaction types, such as protein–protein, protein–DNA, protein–ligand, etc.) as well as the identification and mapping by homology of all the different types of binding sites onto a single protein has not yet been investigated. It allows to characterize both the molecule and its binding sites in an integrative and systemic way by extracting their properties, dynamics and functions from the different sets of structures each reflecting a ‘molecular context’. In particular, it allows to evaluate if a region of a molecule is able to bind several distinct partners of similar or different molecular types. Several major issues have to be considered carefully: the first problem is to avoid the systematic selection of non-specific binding sites due to crystal packing (10) as they can represent as much as 50% of all interactions detailed in structural databases (11). Indeed, although several protein quaternary structure databases exist (12,13) and some methods differentiate very well between biologically meaningful interfaces and crystal packing interfaces (14–17), it is still difficult to have access to a robust and automated process that tests each interface and gives full and easy access to the structures of biological assemblies. The second issue is the automatic identification of the different molecular types present in a structure file, and the distinction between each interface type, including the differentiation of true heterodimers (different molecules interacting), and false heterodimers (interaction between fragments of a same molecule). The work of scientists such as J. Janin, J.M. Thornton, S. Jones or R. Bahadur (18–23) has clearly shown the existence of distinct interface families, which suggests distinct binding site families. For instance, homodimeric binding sites are usually shown to be more hydrophobic and less planar than heterodimeric binding sites (18). As for DNA and RNA binding sites which necessarily reflect the negatively charged phosphate groups of nucleic acids, they are far more cationic than any other known binding sites (19,22). The last-but not least-issue to be considered, is that a structural homology at a global scale does not necessarily imply the same function at a local scale. For instance, even if two molecules share a global shape with a very low RMSD or a very high sequence identity, the mutation of a single residue at a binding site can still drastically change its functions (24–26).
The aims of this work are first, to propose a fully automated, yet robust and coherent approach named M-ORBIS (for Mapping of mOleculaR Binding sItes and Surfaces) to extensively describe a molecule in specific ‘molecular contexts’ from the scattered structural data; and second, to give some insights into the general properties and behaviors of molecular surfaces and interactions. The global and extensive mapping of a molecule in specific ‘molecular contexts’ (describing a precise set of interaction types), has been named ‘Molecular Cartography’, as it gives a very detailed functional and dynamic representation of molecules. This definition of ‘molecular contexts’ is used to classify the retrieved structures into groups illustrating—like snapshots—some of the dynamic events of the studied molecule. As each interaction contained in each of the structural homologues is analyzed and stored, M-ORBIS also allows to transfer the binding site locations and corresponding partners onto the studied molecule. This inference is based on several sequence and structural criteria to ensure sufficient similarities at the global and local scales. Furthermore, as M-ORBIS exploits both sequence and structure alignments, it allows to quantitatively and qualitatively analyze the change of conformations between any two molecular contexts.
The M-ORBIS approach has been validated on four curated datasets (18,19,22,27) describing different interface types and demonstrates sensitivity and specificity >90%. Next, it has been used to demonstrate the existence of binding sites specific to a particular molecular type, and polyvalent binding sites which can bind two or more different molecular types. Interestingly, polyvalent binding sites exhibit amino acid compositions that are intermediate between the specific binding sites they represent. Specific homodimer binding sites are nearly twice as hydrophobic and are two times less charged than polyvalent homodimer binding sites. Our results reveal that at least 44% of the protein surface is designed to interact with non-solvent/ion partners. The characterization of protein–water interactions at different contact frequencies (observed in homologous structures) also suggests that ∼86% of the surface can be transiently solvated, whereas only 15% appears to be specifically solvated.
MATERIALS AND METHODS
Structural databases and crystal contacts
Structural data are deposited in the Protein Data Bank (28) and are both easily accessible and retrievable. For some proteins, generally of therapeutic or cosmetic interest, there exists more than 500 structures of the same molecule (e.g. kinases), corresponding to specific environments (specific sets of interactions with different partners of different molecular types).
For the identification and mapping of non-specific binding sites due to crystal packing, another structural database, the Protein Quaternary Structure (PQS) database is used (12). PQS is based on the PDB but attempts to differentiate between specific and crystal packing interfaces based mainly on the buried ASA (Accessible Surface Area) observed at each interface and a solvation energy term. The classification error rate of PQS for the prediction of the oligomeric state of proteins is ∼16% (11,29).
Summary of the workflow
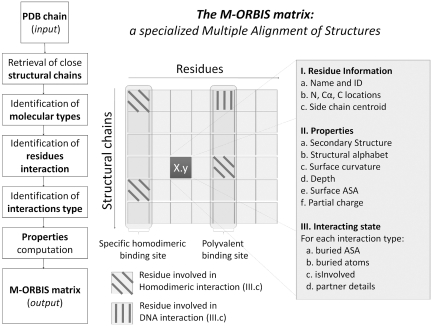
In order to compute a Molecular Cartography, the M-ORBIS process requires only the selection of a structural chain as input data. This can be done either using the M-ORBIS command line and providing a PDB file and both the molecule and chain to be treated, or using the MSVM (molecular structure visualization and mapping) graphical-user interface. Once the structural chain is selected, M-ORBIS will process the data in a 7-step workflow described in Figure 1. Finally, M-ORBIS generates an output matrix file which is a specialized multiple alignment of structures storing several physico-chemical and geometrical properties for each analyzed residue. Scripts in Java to analyze the M-ORBIS output matrices are available upon request. Each of these steps is described in more detail in the following subsections.
Figure 1.
The M-ORBIS matrix as a specialized multiple alignment of structures. The M-ORBIS output matrix is obtained via a 7-step workflow illustrated in the left part of the figure. As in traditional multiple alignments of sequences or structures, the M-ORBIS matrix gives access to every homologous chain and residue. Nevertheless, to perform the complex task of Molecular Cartography, M-ORBIS also stores information relative to the residue locations (field I.), to the physico-chemical and geometrical properties (field II.) and to the interacting state (field III.) of each analyzed residue. A cell of the M-ORBIS matrix is illustated in the right-hand table and gives insights into some of the properties that are stored. For instance, field III.c indicates that the first residue is involved in only homodimeric interaction in two of six related structural chains. A polyvalent binding site is detected when for a given residue (column), the corresponding residues of related structural chains are involved in at least two different binding site types (residue described in column 5). A specific binding site is inferred when only one binding site type is detected at that position (residue described in column 1).
Alignment and retrieval of homologous structural chains
A protein chain is used as input to a local version of the PipeAlign platform (30), in order to automatically construct a high quality hierarchical multiple alignment of sequences related to the query. To enhance the alignment quality, a maximum of 50 non-structure sequences that share <95% sequence identity (31) with the protein chain are introduced; all other sequences in the multiple alignment correspond to structure sequences derived from PQS. Once the alignment is obtained, only structures of resolution 3 Å or better are kept to ensure a high-quality analysis. Also, structures with <30% sequence identity or 75% residues aligned with the query are excluded. For each sequence alignment between the query and a retrieved structure, a structural alignment is also performed, using the CE algorithm (32) with default parameters. Together, the sequence and structure alignments combined are used in the detection of conformational moves and in the inference of functional binding sites by homology. In this study, a necessary but not sufficient criterion for two related residues to share the same interacting states is an observed distance of <1.5 Å between their two aligned Cα.
Further selection of structural chains is then achieved by M-ORBIS, by retrieving a subset of these stored chains using user criteria such as percentage of sequence identity, the presence of interaction types (depicting a molecular context), the presence of solvent molecules, etc. Figure 1 illustrates the properties stored in the M-ORBIS output matrix, for each residue of each analyzed chain, in particular their interacting status. The scripts developed in Java parse and analyze this output and allows for instance to retrieve the chains involved in protein–DNA interactions by searching for the chains that contains residues involved in protein–DNA interaction (here, first line). Similarly, it allows to retrieve the chains involved in at least homodimeric interaction (here, lines 1, 4 and 5), or exclusively homodimeric interaction (here, lines 4 and 5).
Differentiating molecular types
The MSVM research platform (http://www.bio-next.com) allows to read PDB files and to automatically differentiate between protein, peptide, small peptide, DNA, RNA, ligand, ion and solvent molecules. Each molecular type is automatically defined on the basis of written conventions defined by the PDB and the IUPAC code and is hierarchically managed via MSVM.
DNA residues are detected using both the old and new written conventions (+/D). Nevertheless, some differences in conventions are still observed in structural databases and may lead to some errors in the definition of molecular type (e.g. in the PDB 1AIS, DNA are coded with DG, DC, DT, DA, whereas in PQS, the same residues are coded with G, C, T, A). When using jointly PDB and PQS files, this problem is handled by defining the molecular types based solely on the PDB file and by further mapping this definition onto PQS chains.
Proteins, peptides and small peptides are differentiated on the basis of their size: chains composed of more than 60 amino acids are defined as protein, between 20 and 60 amino acids as peptide and as small peptides otherwise. This differentiation is important since small peptides can act as regulating keys (e.g. co-factor) of biological processes, and peptides which do not usually possess a stabilizing protein core may have several different conformations.
Ligand molecules are then defined as being the remaining unknown residues in PDB files. They are further characterized by using the database of ‘monomers’ provided by the PDB.
Residue interaction detection
The interactions between protein, peptide, small peptide, DNA and RNA residues are detected based on a change of ASA. A residue is considered to be interacting if it loses at least 1 Å2 ASA during the assembly formation (18,20,21,27). ASA values were computed with the NACCESS program (33) and default parameters. For a few of these macromolecules (like the 70S ribosome subunit: 1vp0), NACCESS was not able to compute an ASA; therefore, the detection of interacting residues was inferred by distance criteria as described below.
For ligand, solvent and ion interactions, which may involve buried residues of the protein core, residue interactions are detected on a distance criteria basis. A protein–ligand interaction is observed if at least two atoms of the ligand and two atoms of the protein are nearer than their sum of van der Waals weighted by an uncertainty factor of 1.4. For instance, a C–O contact is observed if the distance separating these two atoms is less than (1.7 + 1.5) × 1.4, i.e. 4.48 Å. Nevertheless, and as previously described (34), this cutoff may not be restrictive enough to detect specific protein–water interactions. Therefore, these particular interactions are identified if two polar atoms (N, O, S) are within 3.5 Å.
Differentiating interface types
A first level of distinction for molecular interactions is between a homodimer (assembly of identical molecules) and a heterodimer (assembly of different molecules). Protein homodimers are detected if the chains involved in the interaction share the same Uniprot ID (DBREF field of PDB file), while heterodimers are detected if the chains involved have distinct Uniprot ID. For nucleic acids and in the cases where no Uniprot ID are available, chains involved in homodimeric interactions must have >90% sequence identity and chains involved in heterodimeric interactions must have <40% sequence identity. Other cases are considered uncertain and are discarded by M-ORBIS. As PQS does not conserve the DBREF section of PDB, all chains present in a PQS file structure are assigned a chain in the PDB file structure in order to have access to this information.
The interactions can then be classified, according to the types of molecules involved, into either protein–protein, protein–peptide, protein–small peptide, protein–DNA, protein–RNA, protein–ligand, protein–ion, protein–solvent, protein–crystal packing and/or protein–fragment. As each molecule has a molecular type identified automatically, each of the detected interactions is also assigned a type automatically. True heterodimers and interactions of the same fragmented molecule are differentiated when the UniprotID information is available: if two interacting proteins have the same Uniprot ID, but are non-overlapping fragments of the full sequence protein, then they represent a protein–fragment interaction.
To take into account the diversity of interaction types considered and to further differentiate between crystal packing and biological interfaces, a few simple criteria are added: the minimum buried ASA for homodimeric and heterodimeric binding sites are set to 450 Å2 and 350 Å2, respectively, while for peptide, DNA and RNA binding sites, we mainly discard artifacts by removing interfaces of <50 Å2. Furthermore, each of the binding sites considered for homodimer, heterodimer, peptide binding sites must contain at least 10 residues, while binding sites for small peptides and ligands are simply required to have more than one interacting residue.
Phosphorylation or glycosylation sites are also mapped on the structure, using either the written conventions in PDB that indicate a phosphorylation, or using Uniprot information (35). For instance, phosphorylated serines (SEP), threonines (TPO) and phosphotyrosines (PTR) are identified from the PDB files and are used to locate the phosphorylation sites.
Molecular contexts
Each structural chain is involved in different types of interaction (protein–protein, protein–DNA, protein–RNA, protein–peptide, protein–small peptide, protein–ligand, protein–ion, protein–water) and can be assigned to a precise molecular context that can be further analyzed and compared to other molecular contexts in order to average, characterize and understand the dynamic events between two sets of contexts. More precisely, a specific molecular context is defined by selecting the structures with a requested set of interaction types and using different logical operators (Figure 1): at least one of the interaction type must be present (OR); at most one interaction type must be present (exclusive OR); all selected interaction types must be present but other are accepted (AND); only but all the selected interaction types must be present (exclusive AND); all interactions are accepted except the selected ones (NOT).
As a consequence, it is possible to examine several geometrical and physico-chemical parameters either for the structures of a same molecular context (to observe intrinsic variability) or for two sets of structures reflecting two different molecular contexts.
Contact frequencies and solvated surfaces
The residue contact frequency fcontact is a general measure that describes the fraction of interacting residues (for a specific interaction type) at a precise residue position and for a given molecular context (several related chains). Figure 1 illustrates how the fcontact is derived from the M-ORBIS matrix output file. The first residue which is seen involved in homodimeric interaction in two related structures has a fcontact of 2/6 for this particular type of interaction, whereas the fifth residue which is seen involved in one homodimeric interaction and one protein–DNA interaction in all related structures has a 1/6 fcontact for homodimer interaction and a 1/6 fcontact for DNA interaction. Thus, the fcontact can be used to indicate: (i) whether the residue is involved in a specific or polyvalent binding site and (ii) if the residue is frequently involved in a given type of interaction. In this context, the SWD10 and SWD90 parameters shown in Table 2 and computed on the SWD dataset (see below), describe the percentage of surface residues always involved (fcontact of 90%) or occasionally involved (fcontact of 10%) in a given type of interaction. As a consequence, the ratio SWD90/SWD10, indicates the fraction of interacting residues that is always seen interacting for a given type of interaction.
Table 2.
Average functional landscape of protein structures
| Dataset | Homodimer | Heterodimer | Peptide | S.Peptide | DNA | RNA | Ligand | Solvant | Int.Surface |
|---|---|---|---|---|---|---|---|---|---|
| SWD10 | 26.5 (13.5) | 28.2 (14) | 16.9 (5.9) | 10.7 (6.5) | 20.6 (8.2) | 19.8 (9.4) | 14.7 (8.6) | 86.5 (11.7) | 43.9 (16.1) |
| SWD25 | 23.4 (10.9) | 21.7 (9.1) | 16.6 (5.7) | 9.2 (5.1) | 18.7 (7.6) | 18 (8.3) | 9.9 (4.8) | 78.5 (14.5) | 37.6 (12.8) |
| SWD50 | 19.2 (8.5) | 17.4 (8) | 15.3 (5.4) | 8.1 (3.8) | 17.5 (7.6) | 16.8 (7.6) | 7.1 (4.2) | 58.4 (17.9) | 31.9 (11.1) |
| SWD75 | 16.6 (9.2) | 14.1 (6.9) | 11.9 (5.7) | 6.9 (2.8) | 15 (6.7) | 15 (7) | 5.1 (4.3) | 30 (16.9) | 26.7 (9.8) |
| SWD90 | 15.3 (8.9) | 11.4 (6.5) | 11.2 (5.8) | 5.6 (2.5) | 12.6 (5.9) | 13.6 (5.4) | 4.6 (4.4) | 15.1 (13.3) | 23.6 (9.6) |
| SWD90/SWD10 (%) | 58 | 40 | 66 | 52 | 61 | 69 | 31 | 17 | 54 |
Each line described the average fractions of protein surface involved in each interaction type for a given fcontact. These fractions have been computed on the SWD dataset for fcontact of 10, 25, 50, 75 and 90%, respectively. The contribution of a structure to each fraction of interacting surface was considered only when it presented the given interaction type. The ratio SWD90/SWD10 indicates the percentage of frequently interacting residues for a given binding site type. Numbers in parenthesis are standard deviations.
When a sufficient number of homologues is available, the fcontact measure indicates whether the residue is specific or not for the given interaction type. In this study, unless otherwise stated, the fcontact measures were computed on the SWD dataset, which is composed of proteins that are each described by at least 50 different structural chains. The solvating state of a residue is inferred from this residue contact frequency, although some more stringent criteria for the selection of related structural chains are added: (i) as the average number of water molecules observed per structure is correlated with the crystallographic resolution, only structures with a resolution between 1.5 and 2.5 Å were considered, (ii) structures containing the related chain must contain at least five water molecules to ensure they have been at least partially considered by the experimentalist. A residue is then considered as transiently solvated if it is in contact with water in at least 10% of the related structural chains. A residue that interacts with water in >90% of the related structural chains is considered to be specifically solvated.
As the contact frequency is dependent of a molecular context, it is possible to describe the variation of these contact frequencies for different sets of contexts. For solvation, this proves to be important as it allows to observe the change of residue solvation upon different assembly formation (protein–protein, protein–peptide, etc.).
Inference of the interacting state
Given a chain, it is possible to infer the interacting states of each of its residues by observing the interacting states of its aligned residues on related chains. M-ORBIS uses four main criteria to infer the interacting states of a residue from homologues: (i) the percentage of sequence identity between the studied chain and the related chain; (ii) the percentage of sequence identity for all the residues involved in the given interaction; (iii) a fcontact value for the given interaction types; (iv) the distance between the Cα of aligned residues. These four parameters are highly dependent on the molecular context as it determines the chains selected.
As for the other criteria described previously, the selection of chains also depends on resolution criteria (e.g. the selection of structures with resolutions between [R – deviation] Å and [R + deviation] Å). In some cases, as for the study of protein–water contacts, it is preferable to select only the structures with at least five water molecules and to discard the structures in which the water was not or only partially resolved. These selections are available to users via the MSVM research platform and the M-ORBIS module.
Structural datasets
Four published non-redundant datasets (18,19,22,27) representing different types of interaction as well as the protein–protein docking benchmark 3.0 (7) are used throughout this study. These datasets are composed of structures extracted from the PDB and PQS using several criteria and are further curated by checking for crystal contacts, biological units and in some cases the literature. The docking benchmark dataset differs from other datasets by describing both a bound and unbound form for each protein.
A structurally well-defined dataset (SWD) of 102 proteins has been constructed by merging the four curated datasets and keeping only the protein chains with more than 100 residues that have at least 50 known structural homologues in the PDB. The resulting dataset is composed of proteins having length varying from 107 to 796 amino acids.
RESULTS
In the following, we use the term ‘non-interacting surface’ to refer to the surface involved only in crystal packing, solvent or ion interactions, while the ‘interacting surface’ will refer to the different binding sites. Interacting residues will be referred to as IR and surface residues as SR.
Validation of the interaction analysis
The M-ORBIS Molecular Cartography of a molecule includes several annotations for each residue of each related chain analyzed (Figure 1). In particular, the interacting state of a residue, as well as its ASA values describing both its accessibility to solvent and its buried ASA are stored. The annotations present in the curated datasets (ASA and interacting state) were then compared to those provided by M-ORBIS (Supplementary Table S1). For each of the three tested datasets, the sensitivity and specificity of SR and IR detection were ∼100%. The slight differences observed in IR sensitivity and specificity could be explained either by the choice of minimal buried ASA to detect IR or by the rare but wrong assignment of a modified nucleic acid residue to another molecular type. Surface ASA and Buried ASA values for both curated analysis and M-ORBIS also have a near perfect correlation (correl1 columns). With the exception of protein–RNA assemblies, correl2 indicates that both surface ASA and buried ASA can be accurately predicted from the analysis of structural homologues. It also suggests that when a residue is involved in a particular interaction type, its buried ASA is globally conserved over its family.
Molecular context assignments
For each structural chain analyzed in M-ORBIS, the set of interacting residues and partners can be easily retrieved and it is possible to define a molecular context according to the types of interaction the chain is involved in (see ‘Materials and Methods’ section). The molecular context assignment was evaluated on the four curated datasets (18,19,22,27), and results show that with the exception of the DNA dataset, 100% of the interactions described manually were correctly characterized by M-ORBIS. In the case of the DNA dataset, six structures (1asy; 1gtr; 1zdi; 1urn; 1ttt; 1ser) were identified as participating in protein–RNA interactions rather than protein–DNA interactions but the consultation of the structures proves M-ORBIS to be right, where the nucleic acids were mainly tRNA (1asy; 1gtr; 1zdi; 1ser).
The ability to correctly define a molecular context from a structure was further tested on the docking benchmark 3.0 dataset (7) where for each protein chain, both bound and unbound forms are described. Among the 124 assemblies, 191 partners were described as single protein chains (not as a group of chains). Starting from these 191 single protein chains in bound forms, M-ORBIS was able to find (by searching for homologous chains not involved in protein–protein, protein–peptide, protein–DNA or protein–RNA interactions) 155 of the corresponding unbound chains described in the article (81% accuracy). Another 11 chains from the benchmark 3.0 were described by M-ORBIS as participating in either protein–protein or protein–peptide interactions and were therefore not considered as unbound forms. Nevertheless, these interactions were present in these 11 structures and the M-ORBIS analysis was correct according to our unbound definition, thus raising our accuracy to 87%. The remaining 25 unbound chains described in the benchmark 3.0 but not found by M-ORBIS were due to three problems: (i) a change of PDB name, (ii) a change of chain name between the PDB and PQS files (due to the adding or removal of chains needed in PQS) and (iii) the non retrieval of the PDB chain by M-ORBIS.
Binding site inferences
In a previous section, we demonstrated that M-ORBIS stores and retrieves the correct mapping for each residue of each related chain. Here, we are interested in the inference of binding sites by homology. The inference of binding sites and putative partners by M-ORBIS is influenced mainly by four parameters described in ‘Materials and Methods’ section. In the following study, the minimal percentages of sequence identity for related chains and interacting residues are set to 50%; the minimal contact frequency fcontact is set to 10%, while the accepted distance variation (in Å) between the Cα of the studied chain and the Cα of the related chains is set to 1.5 Å.
Table 1 illustrates the high sensitivity and specificity of the M-ORBIS binding site inference for each of the four interaction types considered. As the M-ORBIS annotations contains on average all the interacting residues detailed in the curated datasets (Supplementary Table S1), the Molecular Cartography of these datasets (which rely also on other homologous structures to infer binding sites) always describes a larger fraction of the surface as interacting. The fraction of surface involved in a binding site type (fcontact) was computed by considering only the structures with this type of binding site. By considering the residues described as interacting by M-ORBIS but not by the curated analysis as putative false positives, a lower bound of the binding site inference specificity is determined: 68.3% for heterodimers, and 86, 85.7% for homodimers and RNA, respectively. However, the Pearson product-moment correlations between the amino acid scales extracted from the curated and M-ORBIS analyses indicate that these new interacting residues respect the observed composition bias for each of these interaction types, suggesting they are not false positives. For instance, homodimer interacting residues are still shown to be more hydrophobic and aromatic, whereas DNA and RNA interacting residues are much more polar and cationic. To ensure the selection of only homologous chains and strengthen the inference of binding sites, a more drastic selection of related chains was performed with a minimal percentage of identity set to 90% and similar results with slightly smaller percentages of interacting surface were obtained (data not shown).
Table 1.
Comparisons of binding site annotations provided by the curated datasets and M-ORBIS
| Datasets | Sensitivity (IR) | Lower bound specificity (IR) | Pearson correlationa | Manually curated interfaceb | M-ORBIS interfacec | Interacting surfaced |
|---|---|---|---|---|---|---|
| Homodimer (18) | 99.7 | 86.4 | 0.98 | 26.8 (12.8) | 31.8 (15.4) | 33.8 (16) |
| Heterodimer (27) | 96.5 | 68.3 | 0.97 | 19.1 (9) | 32.8 (23.6) | 39.9 (18.5) |
| RNA (19) | 98.2 | 85.7 | 0.99 | 18.4 (8.3) | 21.4 (9.7) | 28.2 (16) |
| DNA (22) | – | – | 0.99 | – | 26.8 (11) | 40.3 (26.5) |
The sensitivity corresponds to the fraction of residues that are inferred as interacting by M-ORBIS, among all the interacting residues described in each curated dataset. The lower bound specificity is the precision at which a residue described as interacting by M-ORBIS was annotated as such in the curated datasets. Numbers in parenthesis are standard deviations.
aA Pearson correlation indicates the resemblance between the IR compositions calculated from the curated and M-ORBIS analysis.
bThe fractions of interacting surface described by the curated analysis.
cThe fractions of interacting surface described by M-ORBIS
dThe interacting surface corresponds to the surface involved in at least one non-solvent interaction.
The landscape of binding sites and interacting surfaces
The fractions of protein surface occupied by binding sites and solvent were first evaluated on the four manually curated datasets. With a fcontact of 10%, the interacting surface is shown to vary from 28.2% for the RNA dataset to 40.3% for the DNA dataset (Table 1). However, this first evaluation of the interacting surface suffers from the heterogeneity of the proteins studied: the M-ORBIS approach reveals that some of these proteins have less than ten homologous chains (1kq2:A, 1ser:A, etc.), thus leading to an incomplete mapping of their binding sites, whereas other proteins have more than 200 homologous chains (1us1:A, 1ajs:A, 1amk:A, etc.), leading to a more complete mapping of their binding sites and functions.
To cope with this problem and to strengthen our results, we used the SWD dataset where each protein has at least 50 structural homologous chains (see ‘Materials and Methods’ section). The results are summarized in Table 2 for different contact frequencies fcontact; interestingly, the binding site fractions of protein surface are not additive which suggests the existence of some overlap between binding sites (see specific and polyvalent binding sites). For a fcontact of 10%, the average interacting surface is 43.9% with the larger fraction of binding sites occupied by heterodimers (28.2%) and homodimers (26.5%), followed by DNA and RNA binding sites with 20.6 and 19.8%, respectively. Small peptide and ligand binding sites occupy the smallest fractions of the protein surface with 10.7 and 14.7%, respectively. We also verified that the average interacting surface described here in terms of residues (43.9%) was indeed similar to the average interacting surface in terms of accessible surface area (45%).
By increasing the minimal contact frequency to a more stringent value, the mapping can be set to emphasize the invariant interacting residues. At 50% minimal contact frequency, 31.9% of the surface is then seen as interacting, while at 90%, only 23.6% of the protein surface is described as interacting. The SWD90/SWD10 ratio indicates that 69 and 61% of RNA and DNA binding sites, respectively, are composed of residues that are seen interacting in >90% of related structures, whereas the ligand binding site is composed of only 31% of these frequently interacting residues (see ‘Discussion’ section).
The non-interacting surface landscape: the transient and specific protein–water contacts
The analysis of protein–water contact frequencies by M-ORBIS indicates that 86.5% of the surface residues are solvated in at least 10% of the structural homologues, whereas with a fcontact of 50% and 90%, the fractions of solvated residues decrease to 58.4% and 15.1%, respectively (Figure 2, Table 2). Those residues that are in contact with water molecules in at least 90% of the related structural homologues are considered to specifically bind water molecules.
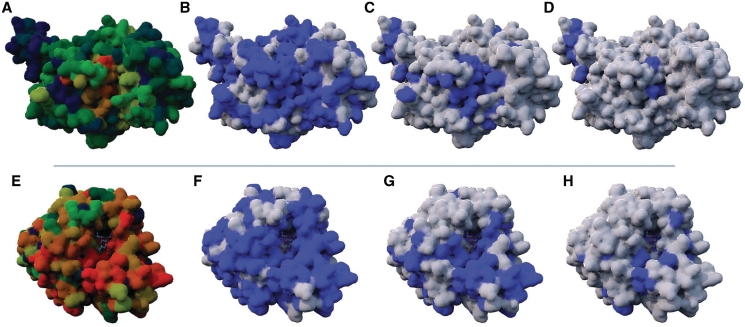
Figure 2.
Protein–water contacts at different contact frequencies. (A, B, C and D) Structures of the guanine nucleotide-binding protein G(i) (PDB: 1BOF), while (E), (F), (G) and (H) are structures of the H1N1 Neuraminidase (PDB: 3b7e). In (A) and (E), the contact frequencies fcontact with water molecules are shown, as calculated by M-ORBIS with 49 and 169 related chains, respectively; non-solvated regions are indicated in dark blue (fcontact ∼0%), while partially solvated regions are in green (fcontact ∼50%). Red surfaces correspond to the regions that specifically bind water molecules with a fcontact ∼100%. The surface in contact with water molecules is shown in light blue in (B), (C), (D), (F), (G), (H) with fcontact of 25, 50, 75, 50, 75 and 90%, respectively. For these two molecules, the central solvated regions are ligand binding sites.
The analysis of the amino acid compositions involved in protein–water contacts on the SWD dataset shows, for a fcontact of 10%, a high correlation of 0.95/0.94 with the non-interacting surfaces previously described in the literature (18,21,27). The correlation with a study dedicated to the hydratation of protein surface and interface (34) is also good, with a 0.88 Pearson correlation. Interestingly, for a fcontact of 90%, the first two correlations are decreased to 0.76/0.75, respectively, indicating some differences in the preferences of their amino acids to form contacts with water molecules. If Gly, Ala, Val, Leu, Ile, Pro and Met are considered as hydrophobic residues, and Asp, Glu, Lys and Arg as charged residues, a comparison with the previously published amino acid scales suggests that residues involved in specific water contacts are less hydrophobic (23.5% against 35.6%), and more charged (36.5% against 29.3%) than previously observed. Furthermore, the manual visualization of the SWD suggests that residues involved in specific water contacts are often co-localized with ligand binding sites. This result was partially verified in the following study concerning the polyvalent binding sites and is illustrated in Figure 2.
Specific and polyvalent binding sites
It has been observed in a previous section, that the binding site fractions of the surfaces add up to more than the global interacting surface which suggests some overlap between binding sites. Using the M-ORBIS approach, it was possible to automatically locate the residues that had the ability to alternatively participate in at least two distinct interaction types (Figure 1). More precisely, two binding sites of different types (e.g. homodimer and heterodimer) are said to be polyvalent if at least 10 of their residues overlap and participate in both interaction types. In the case of overlap with ligand or small peptide binding sites which are smaller, only two residues were required.
Two questions are investigated: first, does a specific binding site (a binding site that is seen to be involved in only one interaction type in all homologues) have the same physico-chemical properties as a polyvalent binding site (a binding site that is seen to be involved in at least two different interaction types) (Table 3); second, does a binding site observed to participate in an interaction type have some preference for participation in other interaction types (Table 4).
Table 3.
Amino acid composition/contribution of the specific versus polyvalent binding sites
| Residue | HoD (A) | HoD,HoD (B) | Log(B/A) | HoD,HeD (C) | Log(C/B) | HoD,Solvent(D) | Log(D/B) | DNA | HoD,DNA |
|---|---|---|---|---|---|---|---|---|---|
| GLY | 6.3 | 7.2 | 0.05 | 5.4 | −0.12 | 6.2 | −0.07 | 5.3 | 2.6 |
| ALA | 6.5 | 9.2 | 0.15 | 4.4 | −0.32 | 5.8 | −0.20 | 4.2 | 2.1 |
| VAL | 5.5 | 10.5 | 0.28 | 5.2 | −0.31 | 4.1 | −0.41 | 6.0 | 11.3 |
| LEU | 8.8 | 17.1 | 0.29 | 7.5 | −0.36 | 6.3 | −0.43 | 3.5 | 6.7 |
| ILE | 4.8 | 10.2 | 0.33 | 2.8 | −0.56 | 3.2 | −0.51 | 3.9 | 4.6 |
| PRO | 5.1 | 8.0 | 0.20 | 3.4 | −0.38 | 4.3 | −0.27 | 3.5 | 3.1 |
| MET | 2.6 | 4.2 | 0.22 | 2.8 | −0.18 | 2.1 | −0.31 | 4.9 | 8.7 |
| PHE | 4.4 | 6.9 | 0.20 | 8.0 | 0.07 | 3.5 | −0.29 | 0.9 | 0.0 |
| TRP | 1.6 | 1.1 | −0.16 | 1.3 | 0.07 | 1.7 | 0.18 | 0.4 | 0.0 |
| SER | 5.7 | 3.3 | −0.24 | 8.0 | 0.38 | 6.4 | 0.29 | 9.7 | 6.2 |
| THR | 5.6 | 4.0 | −0.14 | 6.7 | 0.22 | 6.1 | 0.18 | 8.8 | 8.7 |
| ASN | 4.2 | 1.5 | −0.45 | 2.8 | 0.27 | 5.0 | 0.52 | 7.2 | 6.2 |
| GLN | 4.4 | 1.9 | −0.37 | 3.4 | 0.25 | 5.2 | 0.44 | 4.6 | 5.6 |
| ASP | 5.8 | 1.9 | −0.49 | 5.2 | 0.43 | 7.0 | 0.57 | 4.6 | 2.6 |
| GLU | 6.8 | 2.3 | −0.47 | 6.7 | 0.47 | 8.1 | 0.55 | 4.1 | 5.1 |
| CYS | 1.2 | 1.5 | 0.09 | 0.5 | −0.47 | 1.1 | −0.13 | 0.9 | 0.0 |
| TYR | 4.2 | 2.6 | −0.22 | 7.7 | 0.48 | 4.7 | 0.26 | 2.8 | 1.5 |
| HIS | 3.3 | 1.5 | −0.33 | 2.3 | 0.18 | 3.8 | 0.39 | 2.7 | 3.1 |
| LYS | 6.1 | 3.1 | −0.30 | 10.6 | 0.54 | 7.1 | 0.37 | 12.7 | 11.3 |
| ARG | 6.9 | 1.9 | −0.56 | 5.4 | 0.45 | 8.4 | 0.65 | 9.2 | 10.8 |
| Hydrophob | 39.7 | 66.5 | 31.4 | 31.9 | 31.3 | 39.1 | |||
| Anionic | 12.6 | 4.2 | 11.9 | 15.1 | 8.7 | 7.7 | |||
| Cationic | 13 | 5 | 16 | 15.5 | 21.9 | 22.1 | |||
| Polar | 19.9 | 10.7 | 20.9 | 22.7 | 30.3 | 26.7 | |||
| C(HoD) | 0.97 | 0.64 | 0.61 | 0.76 | 0.45 | 0.49 | |||
| C(HeD) | 0.86 | 0.17 | 0.77 | 0.90 | 0.77 | 0.6 | |||
| C(Pcore) | 0.43 | 0.93 | 0.17 | −0.07 | −0.11 | 0.17 | |||
| C(Crystal) | 0.84 | 0.10 | 0.62 | 0.95 | 0.67 | 0.5 | |||
C(X) is a Pearson product–moment correlation between the amino acid scale described in column and the published amino acid scale X. When X is HoD (homodimer), the amino acid scale is from Bahadur et al. (18); when X is HeD (Heterodimer) or Pcore (protein core), the amino acid scales are from Albou et al. (27); when X is Crystal (crystal packing), it is from Bahadur et al. (15). HoD,X (such as HoD,HeD) indicates an homodimer/X polyvalent binding site.
Table 4.
Average log-odd ratios for two binding sites to be co-localized
| Homodimer | Heterodimer | Peptide | S.Peptide | DNA | RNA | Ligand | Solvent | |
|---|---|---|---|---|---|---|---|---|
| Homodimer | 0.12 | 0.13 | −0.35 | −0.30 | −0.02 | −0.60 | −0.09 | 0.02 |
| Heterodimer | 0.13 | 0.79 | 0.63 | 0.09 | −0.85 | 0.12 | −0.06 | |
| Peptide | −0.35 | 0.79 | −0.02 | 0.84 | −0.09 | −0.23 | ||
| S.Peptide | −0.30 | 0.63 | 0.84 | 0.30 | −0.15 | |||
| DNA | −0.02 | 0.09 | 0.34 | 0.63 | −0.12 | |||
| RNA | −0.60 | −0.85 | 2.37 | 0.43 | 0.04 | |||
| Ligand | −0.09 | 0.12 | −0.09 | 0.30 | 0.63 | 0.43 | −1.56 | 0.15 |
| Solvent | 0.02 | −0.06 | −0.23 | −0.15 | −0.12 | 0.04 | 0.15 | −0.01 |
Each cell corresponds to the log-odd ratio of the observed probability of having a binding site type A overlapping a binding site type B over the expected probability of having A and B overlapping. Positive values indicate favorable overlapping/polyvalence between the binding site types A and B, whereas negative values indicate unfavorable overlapping/polyvalence. Empty cells are unsolved values due to an insufficient number of overlaps between the two considered binding sites; however, they should indicate low log-odd values.
Differences between specific and polyvalent binding sites
The analysis of observed amino acid compositions between specific homodimer binding sites and polyvalent homodimer binding sites emphasizes several important differences. The most remarkable is the hydrophobic composition which is increased from 39.7% (for previously defined homodimer binding sites) to 66.5% for specific homodimer binding sites. Charged (Asp, Glu, Lys, Arg) and polar compositions (Ser, Thr, Asn, Gln) vary accordingly, being divided by a factor of 2. As a consequence, specific homodimer binding sites are shown to be much more correlated with the composition of the protein core than polyvalent homodimer binding sites and far less correlated with the composition of crystal packing interfaces. Indeed, homodimer interfaces had already been described as resembling the protein core (18,21). This hypothesis is further supported and detailed by our results on specific homodimer binding sites.
Concerning nucleic acids, as expected, both polyvalent homodimer/DNA and specific DNA binding sites are shown to be much more cationic than both polyvalent and specific homodimer binding sites. In addition, specific DNA binding sites tends to be less hydrophobic (31.3%) than polyvalent homodimer/DNA binding sites (39.1%) and more polar (30.3% against 26.7%).
As suspected, other homodimer polyvalent binding sites have amino acid compositions closer to what was previously described, explaining the observed differences between specific and polyvalent homodimer binding sites. For instance, polyvalent homodimer/heterodimer binding sites are more correlated to heterodimer (0.77) than specific homodimer binding sites (0.17). Furthermore, polyvalent homodimer/heterodimer and homodimer/solvent binding sites are seen to be more charged and polar which results in poor correlations with the protein core composition and higher correlations with the crystal packing binding sites.
Favorable and unfavorable polyvalent binding sites
Each binding site has different frequencies in the SWD dataset, the most frequent being the homodimer (17%), followed by ligand (15%) and heterodimer (11%) binding sites. Therefore, to perform unbiased observations of the likelihood of a binding site type A to be co-localized (polyvalent) with a binding site type B, we used the well established methodology described by Henikoff (36). A total of 525 co-localizations of binding site types were observed. Only 19 RNA binding sites were present; therefore, log-odd ratios concerning RNA should be considered as first approximations. The results are summarized in Table 4 and show that different polyvalent binding sites may be either favorable (as for the homodimer/heterodimer or heterodimer/peptide pairs), or unfavorable (homodimer/peptide or homodimer/RNA pairs). As expected, heterodimer binding sites can easily bind peptides (log odd: 0.79) and small peptides (log odd: 0.63), whereas a homodimer binding site are less able to bind peptides (−0.35) and small peptides (−0.3). Interestingly, small peptides and ligands which both serve as key regulating factors of protein activities can preferentially share a same binding site (log odds: 0.3). As for ligand binding sites, they are the most polyvalent binding sites of all, being preferentially co-localized with all other binding site types considered, with the exception of homodimer and peptide where no clear preference can be observed. Concerning water binding sites, only the ligand binding sites seem to be preferentially co-localized with a moderate log odds ratio of 0.15; nevertheless, this result agrees with the manual visualization of the SWD Molecular Cartographies (example in Figure 2).
DISCUSSION
This work was dedicated to the analysis of protein surfaces and binding sites. In addition to describing an automated approach capable of performing fast comparative and integrated analysis of structures, we have demonstrated the existence of two major families of binding sites: (i) specific binding sites that are only able to bind a specific type of molecule, (ii) polyvalent binding sites that can bind different types of molecules. Our analysis suggests that some of these specific and polyvalent binding sites can be distinguished based solely on their amino acid composition. Additionally, polyvalent binding sites often highlight an intermediate composition between the specific binding sites they represent, e.g. a polyvalent DNA/homodimer binding site exhibits properties both from specific DNA binding sites and homodimer binding sites.
The question of whether specific binding sites could demonstrate stronger or even permanent interaction remains to be answered. In the case of specific homodimer binding sites which are relatively well correlated to the core of proteins (Pearson correlation of 0.93), one should nevertheless emphasize that such localized hydrophobic regions (on average 66%) are unlikely to remain free in the cell and would indeed suggest stronger interaction. Similarly, the polyvalent binding sites which are capable of binding at least two different types of molecules are necessarily involved in weaker interactions since they need to dissociate and associate with different partners.
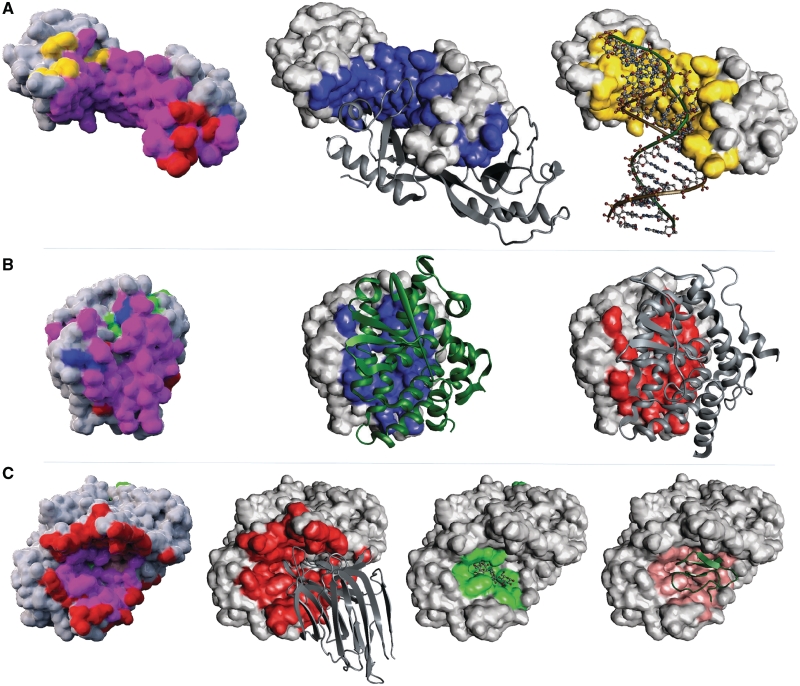
Three examples of polyvalent binding sites are reported in Figure 3. Among them, an important and well documented example is the Retinoid X Receptor (RXR) which belongs to the large and important family of nuclear receptors. Indeed, RXR differentially regulates gene expression by forming either homodimers or heterodimers with other nuclear receptors such as RAR, VDR or PPAR (25,37,38). As for the transcription factor TATA binding protein (TBP) involved in DNA melting, its homodimer binding site is also co-localized with its DNA binding site.
Figure 3.
Examples of proteins exhibiting polyvalent binding sites. Polyvalent binding sites are a general phenomena in molecular structures. (A) The TATA-binding protein (TBP) processed by the M-ORBIS Molecular Cartography approach starting from structure 1ais:A. (B) The Retinoid X Receptor-Alpha (RXR) cartography from the structure 1dkf:A. (C) The Pancreatic Alpha-Amylase (PAA) cartography from the structure 1dhk:A. Binding site types are represented in different colors: blue for homodimer, red for heterodimer, yellow for DNA, green for ligand and salmon for peptide. For TBP and RXR, the homodimer partner shown is extracted from structures 1d3u and 1dkf, respectively. For PAA, the heterodimeric and peptide partners are extracted from structures 1dhk and 1clv, respectively, whereas the ligand partner was extracted from 1g9h.
The examples above describe binary polyvalent binding sites, but the Molecular Cartography of the pancreatic α-amylase also suggests that a single binding site may accept the binding of more than three different molecular types.
Previous results based on the analysis of a single type of interaction identified on average 20% of the protein surface as interacting. Using our integrative approach, we show that by considering all types of binding sites, the average interacting surface is re-evaluated at 44% (in terms of surface residues) or 45% (in terms of accessible surface area). However, this average interacting surface should be considered as a lower approximation since it is probable that not all the biological binding sites of studied molecules were described in the PDB. Similarly, it is well known that the number of solved water molecules not only depends on the crystallographic resolution, but suffers from partial determination since their importance both in molecular stability and interaction mediation were recognized only recently (34,39,40). The averaging proposed by M-ORBIS reduces these effects and allows a mapping of protein–water contact frequencies (Figure 2). Although the specifically solvated residues can be regarded as an accurate result (criteria to detect residue–water contacts are very stringent and the contacts are seen in at least 50 structures), the transiently solvated surface should be considered as a lower approximation due to the possible omission of water molecules in structures.
Interestingly, the observation of the interacting surfaces at different minimal contact frequencies (Table 2), suggests that about half of the residues composing a binding site are not specific to the partner and always participate to a given type of interaction (ratio SWD90/SWD10). This also implies that the other half of the residues could modulate the recognition specificity of distinct partners. In particular, DNA and RNA binding sites seem to have more physico-chemical constraints since 69 and 61%, respectively, of their binding sites are composed of residues that are always seen interacting in related structures. Inversely, ligand binding sites which can bind different ligands with different affinities are shown to be composed of only 31% of those frequently interacting residues, so the remaining 69% of the residues could serve to modulate the recognition specificity of different compounds. From these observations, we would like to propose the notion of a ‘core’ binding site defined by the residues that are always seen interacting for a given type of interaction. This core binding site would meet some of the requirements required for a given type of interaction, whereas the remaining residues of the binding site would be more fuzzy, thus serving the purpose of recognition specificity. If this core binding site is indeed used to meet the requirements for a given type of interaction, it may be more evolutionarily conserved than the remainder of the binding site. This notion of core binding site may also share similarities with the existing notion of‘core’and ‘rim’residues composing a binding site which were defined according to the presence of fully buried atoms at the interface (20).
These results may have consequences for protein binding site predictions, for example to redirect attention towards the integrative prediction of the different types of binding sites (a harder computational problem) or to focus efforts towards the prediction and characterization of the core binding sites and hot spots (24). Such integrative mapping of the different types of binding sites (Molecular Cartography) of molecules will help to accelerate docking approaches by giving fast and easy access to existing structural data (Figure 5). In particular, since the docking problems now include the distinction between protein–protein, protein–nucleic acid and protein–ligand interactions, M-ORBIS should prove to be a tool of choice as it differentiates and locates all the known types of binding sites and identifies the frequently interacting residues and the frequently solvated residues.
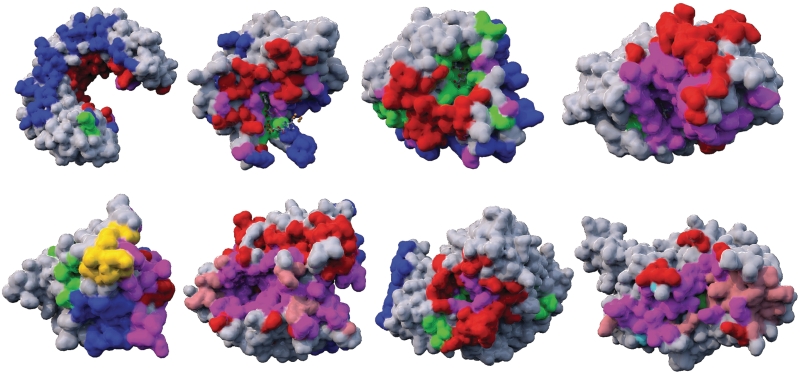
Figure 5.
Molecular Cartography of binding sites. Heterodimeric binding sites are represented in red, homodimeric and ligand binding sites in blue and green, respectively and DNA and peptide binding sites in yellow and salmon, respectively. Polyvalent binding sites are indicated in purple. From top left to bottom right, the cartography of proteins: ribonuclease inhibitor (1dfj), ferrodoxin-nadp reductase (1ewy); neuraminidase (3b7e); cAMP-dependent kinase (1ydr); p53 tumor suppressor (1tsr); neurotoxin bont/A (1xtg); acetylcholinesterase (1fss); guanine nucleotide-binding protein G(i) (1bof).
Additionally, the likelihood of polyvalent binding sites indicates that small peptides and ligands are more co-localized than peptides and ligands or proteins and ligands. The logic behind the distinction of protein, peptide and small peptide relies on the observation that proteins (defined here as a polypeptide of more than 60 residues) often have a stabilizing protein core which may reflect conformational flexibility behaviors different from those of peptides, which generally do not have a well formed protein core. Also, small peptides are separated from peptides due to their small size and the intuitive idea that they can mimic ligands (small chemical compounds). Although arbitrary, previous studies of Stanfield (41) or Petsalaki (42) also suggested differentiating small peptides from other polypeptides using a size limit near 20 residues. A clearer distinction between proteins, peptides and small peptides could be achieved either by studying the percentage of residues constituting the polypeptide core, or by studying the intrinsic conformational flexibilities, or by optimizing the likelihood of co-localization of small peptide and ligand binding sites.
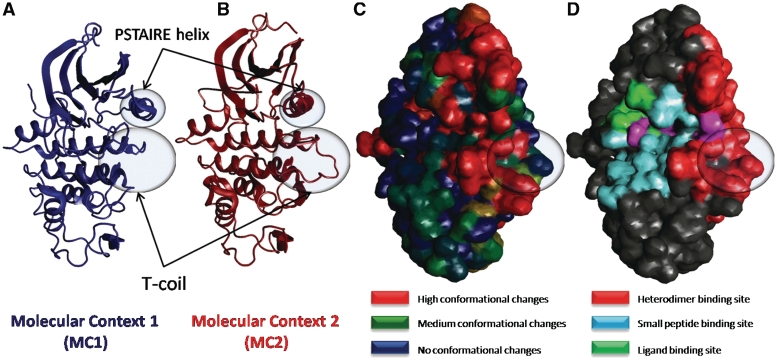
As M-ORBIS collects and classifies structures according to their molecular contexts, it is possible to analyze the conformational flexibility for a given molecular context or upon the change from a context to another. For instance, the CDK is a well studied family of enzymes which catalyses the transfer of a phosphate group from ATP onto the hydroxyl group of a serine or threonine. They play a crucial role in cell cycle regulation and are activated by their binding to different cyclins. The unbound (monomeric) form is known to be inactive due to several structural constraints (43). By defining two molecular contexts (the first corresponding to the structures of CDK in interaction only with water and ligands, the second corresponding to the structures of CDK involved only in heterodimeric, solvent and ligand interactions), the M-ORBIS cartography approach was able to automatically detect, average and map the changes of conformation between these two states (Figure 4). They correspond, with some minor differences, to what was previously observed in the literature (44), with the shifting of the T coil towards the heterodimeric partner (here the cyclin), and the displacement of the PSTAIRE helix in the opposite direction. It was also possible to define other molecular contexts, for instance representing (i) only solvent interactions, (ii) only heterodimer and solvent interactions or (iii) heterodimer, ligand, small peptide and solvent interactions. The comparison of these other molecular contexts demonstrates that the T coil displacement results only from the heterodimer formation and not from the binding of the ligand or the small peptide.
Figure 4.
Change of conformation between molecular contexts: the case of CDK. Molecular Cartography obtained from the structure of the CDK2 (1fin:A). Two molecular contexts are defined: MC1 corresponds to the CDK2 in an environment where it interacts with water and ligand only, whereas MC2 corresponds to an environment where it also interacts with another protein to form a heterodimer. For each of these molecular contexts, the corresponding sets of structures has been automatically detected by M-ORBIS and an averaged backbone is computed and shown in (A and B). Two main conformational changes are involved between MC1 and MC2: the T-Coil helix moves towards the heterodimer binding site, while the PSTAIRE-helix is pushed in the opposite direction. The amount of conformational change has been mapped onto the protein surface in (C); blue, no change; green, small change; red, important change. The molecular cartography shown in (D) allows to correlate these conformational changes with the location of each binding site type.
Most of the programs used to generate an M-ORBIS Molecular Cartography are generic and should be soon applicable to other molecules such as RNA and DNA. Furthermore, if our approach is aimed at describing the functional and dynamic behaviors of a single molecular chain, it has been observed that an assembly (group of chains) can be required to perform an interaction with another molecular partner (7,20). As a consequence, it should be possible to search not only for the structures containing a specific single chain, but also for structures containing a specific assembly. For instance, the retrieval of related structural chains is currently achieved using a sequence-based comparison engine (PipeAlign), but recent advances in structural comparisons using spherical harmonics (45) could be used to retrieve structures containing a specific assembly. Such structural comparisons will also enhance both the rapidity and sensitivity of the molecular cartographies as some homologies are easier to detect on structures than on sequences. Other comparison methods are also being investigated. In further developments, our automated analysis will benefits from other databases such as PiSA (13) to help discriminate between biological and crystal packing interfaces.
As a conclusion, starting from a molecular structure with no or little functional knowledge, the ultimate goal of Molecular Cartography is to provide an extensive description and characterization of the dynamic functions and behaviors of a molecule by integrating the analysis of related structural data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by funds from the French Fondation ‘Louis D, Institut de France’, Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale and the Université de Strasbourg. Funding for open access charge: Centre Européen de Recherche en Biologie et en Médecine (Institut de Génétique et de Biologie Moléculaire et Cellulaire).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to thank J. Janin for his teaching and passion for molecular interactions, R. Bahadur for sharing his data on molecular interactions to help in the validation of M-ORBIS, J. Thompson for her critical reading of the article and F. Gros for his ongoing encouragement and passion for molecular biology. All images were generated by the MSVM platform.
REFERENCES
- 1.Bartlett GJ, Todd AE, Thornton JM. Inferring protein function from structure. Methods Biochem. Anal. 2003;44:387–407. [PubMed] [Google Scholar]
- 2.Brylinski M, Kochanczyk M, Broniatowska E, Roterman I. Localization of ligand binding site in proteins identified in silico. J. Mol. Model. 2007;13:665–675. doi: 10.1007/s00894-007-0191-x. [DOI] [PubMed] [Google Scholar]
- 3.Porollo A, Meller J. Prediction-based fingerprints of protein-protein interactions. Proteins. 2007;66:630–645. doi: 10.1002/prot.21248. [DOI] [PubMed] [Google Scholar]
- 4.Jones S, Thornton JM. Searching for functional sites in protein structures. Curr. Opin. Chem. Biol. 2004;8:3–7. doi: 10.1016/j.cbpa.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Domingues FS, Rahnenfuhrer J, Lengauer T. Conformational analysis of alternative protein structures. Bioinformatics. 2007;23:3131–3138. doi: 10.1093/bioinformatics/btm499. [DOI] [PubMed] [Google Scholar]
- 6.Brylinski M, Skolnick J. What is the relationship between the global structures of apo and holo proteins? Proteins. 2008;70:363–377. doi: 10.1002/prot.21510. [DOI] [PubMed] [Google Scholar]
- 7.Hwang H, Pierce B, Mintseris J, Janin J, Weng Z. Protein-protein docking benchmark version 3.0. Proteins. 2008;73:705–709. doi: 10.1002/prot.22106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung JL, Wang W, Bourne PE. Exploiting sequence and structure homologs to identify protein-protein binding sites. Proteins. 2006;62:630–640. doi: 10.1002/prot.20741. [DOI] [PubMed] [Google Scholar]
- 9.Higurashi M, Ishida T, Kinoshita K. PiSite: a database of protein interaction sites using multiple binding states in the PDB. Nucleic Acids Res. 2009;37:D360–D364. doi: 10.1093/nar/gkn659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janin J. Specific versus non-specific contacts in protein crystals. Nat. Struct. Biol. 1997;4:973–974. doi: 10.1038/nsb1297-973. [DOI] [PubMed] [Google Scholar]
- 11.Janin J, Bahadur RP, Chakrabarti P. Protein-protein interaction and quaternary structure. Q. Rev. Biophys. 2008;41:133–180. doi: 10.1017/S0033583508004708. [DOI] [PubMed] [Google Scholar]
- 12.Henrick K, Thornton JM. PQS: a protein quaternary structure file server. Trends Biochem. Sci. 1998;23:358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- 13.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Ponstingl H, Henrick K, Thornton JM. Discriminating between homodimeric and monomeric proteins in the crystalline state. Proteins. 2000;41:47–57. doi: 10.1002/1097-0134(20001001)41:1<47::aid-prot80>3.3.co;2-#. [DOI] [PubMed] [Google Scholar]
- 15.Bahadur RP, Chakrabarti P, Rodier F, Janin J. A dissection of specific and non-specific protein-protein interfaces. J. Mol. Biol. 2004;336:943–955. doi: 10.1016/j.jmb.2003.12.073. [DOI] [PubMed] [Google Scholar]
- 16.Bernauer J, Bahadur RP, Rodier F, Janin J, Poupon A. DiMoVo: a Voronoi tessellation-based method for discriminating crystallographic and biological protein-protein interactions. Bioinformatics. 2008;24:652–658. doi: 10.1093/bioinformatics/btn022. [DOI] [PubMed] [Google Scholar]
- 17.Zhu H, Domingues FS, Sommer I, Lengauer T. NOXclass: prediction of protein-protein interaction types. BMC Bioinformatics. 2006;7:27. doi: 10.1186/1471-2105-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bahadur RP, Chakrabarti P, Rodier F, Janin J. Dissecting subunit interfaces in homodimeric proteins. Proteins. 2003;53:708–719. doi: 10.1002/prot.10461. [DOI] [PubMed] [Google Scholar]
- 19.Bahadur RP, Zacharias M, Janin J. Dissecting protein-RNA recognition sites. Nucleic Acids Res. 2008;36:2705–2716. doi: 10.1093/nar/gkn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakrabarti P, Janin J. Dissecting protein-protein recognition sites. Proteins. 2002;47:334–343. doi: 10.1002/prot.10085. [DOI] [PubMed] [Google Scholar]
- 21.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 22.Nadassy K, Wodak SJ, Janin J. Structural features of protein-nucleic acid recognition sites. Biochemistry. 1999;38:1999–2017. doi: 10.1021/bi982362d. [DOI] [PubMed] [Google Scholar]
- 23.Jones S, Thornton JM. Principles of protein-protein interactions. Proc. Natl Acad. Sci. USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 25.Brelivet Y, Kammerer S, Rochel N, Poch O, Moras D. Signature of the oligomeric behaviour of nuclear receptors at the sequence and structural level. EMBO Rep. 2004;5:423–429. doi: 10.1038/sj.embor.7400119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clackson T, Ultsch MH, Wells JA, de Vos AM. Structural and functional analysis of the 1:1 growth hormone:receptor complex reveals the molecular basis for receptor affinity. J. Mol. Biol. 1998;277:1111–1128. doi: 10.1006/jmbi.1998.1669. [DOI] [PubMed] [Google Scholar]
- 27.Albou LP, Schwarz B, Poch O, Wurtz JM, Moras D. Defining and characterizing protein surface using alpha shapes. Proteins. 2009;76:1–12. doi: 10.1002/prot.22301. [DOI] [PubMed] [Google Scholar]
- 28.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, et al. The Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 29.Ponstingl H, Kabir T, Thornton JM. Automatic inference of protein quaternary structure from crystals. J. Appl. Cryst. 2003;36:1116–1122. [Google Scholar]
- 30.Plewniak F, Bianchetti L, Brelivet Y, Carles A, Chalmel F, Lecompte O, Mochel T, Moulinier L, Muller A, Muller J, et al. PipeAlign: a new toolkit for protein family analysis. Nucleic Acids Res. 2003;31:3829–3832. doi: 10.1093/nar/gkg518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 32.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 33.Hubbard SJ, Thornton JM. NACCESS Computer Program. Departement of Biochemestry and Molecular Biology; 1992. [Google Scholar]
- 34.Rodier F, Bahadur RP, Chakrabarti P, Janin J. Hydration of protein-protein interfaces. Proteins. 2005;60:36–45. doi: 10.1002/prot.20478. [DOI] [PubMed] [Google Scholar]
- 35.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ijpenberg A, Tan NS, Gelman L, Kersten S, Seydoux J, Xu J, Metzger D, Canaple L, Chambon P, Wahli W, et al. In vivo activation of PPAR target genes by RXR homodimers. EMBO J. 2004;23:2083–2091. doi: 10.1038/sj.emboj.7600209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bourguet W, Vivat V, Wurtz JM, Chambon P, Gronemeyer H, Moras D. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol. Cell. 2000;5:289–298. doi: 10.1016/s1097-2765(00)80424-4. [DOI] [PubMed] [Google Scholar]
- 39.Knight JD, Hamelberg D, McCammon JA, Kothary R. The role of conserved water molecules in the catalytic domain of protein kinases. Proteins. 2009;76:527–535. doi: 10.1002/prot.22451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raschke TM. Water structure and interactions with protein surfaces. Curr. Opin. Struct. Biol. 2006;16:152–159. doi: 10.1016/j.sbi.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 41.Stanfield RL, Wilson IA. Protein-peptide interactions. Curr. Opin. Struct. Biol. 1995;5:103–113. doi: 10.1016/0959-440x(95)80015-s. [DOI] [PubMed] [Google Scholar]
- 42.Petsalaki E, Stark A, Garcia-Urdiales E, Russell RB. Accurate prediction of peptide binding sites on protein surfaces. PLoS Comput. Biol. 2009;5:e1000335. doi: 10.1371/journal.pcbi.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Bondt HL, Rosenblatt J, Jancarik J, Jones HD, Morgan DO, Kim SH. Crystal structure of cyclin-dependent kinase 2. Nature. 1993;363:595–602. doi: 10.1038/363595a0. [DOI] [PubMed] [Google Scholar]
- 44.Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich NP. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 45.Sael L, Li B, La D, Fang Y, Ramani K, Rustamov R, Kihara D. Fast protein tertiary structure retrieval based on global surface shape similarity. Proteins. 2008;72:1259–1273. doi: 10.1002/prot.22030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.