Abstract
DNA double-stranded breaks (DSBs) are among the most severe forms of DNA damage and responsible for chromosomal translocations that may lead to gene fusions. The RAD51 family plays an integral role in preserving genome stability by homology directed repair of DSBs. From a proteomics screen, we recently identified SFPQ/PSF as an interacting partner with the RAD51 paralogs, RAD51D, RAD51C and XRCC2. Initially discovered as a potential RNA splicing factor, SFPQ was later shown to have homologous recombination and non-homologous end joining related activities and also to bind and modulate the function of RAD51. Here, we demonstrate that SFPQ interacts directly with RAD51D and that deficiency of both proteins confers a severe loss of cell viability, indicating a synthetic lethal relationship. Surprisingly, deficiency of SFPQ alone also leads to sister chromatid cohesion defects and chromosome instability. In addition, SFPQ was demonstrated to mediate homology directed DNA repair and DNA damage response resulting from DNA crosslinking agents, alkylating agents and camptothecin. Taken together, these data indicate that SFPQ association with the RAD51 protein complex is essential for homologous recombination repair of DNA damage and maintaining genome integrity.
INTRODUCTION
Defective DNA damage response and repair mechanisms are underlying causes for the increased genetic instability and chromosomal translocations associated with the evolution of cancer (1,2). Among the most deleterious lesions encountered are DNA double-strand breaks (DSBs) because aberrant fusions can form between breaks or with unstable telomere ends that appear as breaks. DSBs are primarily resolved by error-free pathways such as homologous recombination (HR) or error-prone pathways such as non-homologous end joining (NHEJ) (3,4). Recently, it was demonstrated that cells with HR defects are reliant upon NHEJ, which is responsible for the chromosome fusions, and absence of some HR and NHEJ components can reactivate the HR pathway (5). The mechanisms therefore appear to compete among themselves with the balance varying among different cell types as well as during different stages of the cell cycle (4).
Seven RAD51 family members form integral components of the HR machinery and interact with one another and as part of the ‘BRCA’ protein complex. Cells deficient in any of the genes encoding the RAD51 proteins have extensive levels of chromosome instability and are sensitive to complex DNA damage (6–10). The fourth member of the family, RAD51D, plays an indispensible role during both DNA repair and telomere maintenance (11,12). In addition, Rad51c-deficient mice are susceptible to specialized sebaceous gland tumors (13). Germline mutations in human RAD51C are also responsible for breast and ovarian cancer and a Fanconi anemia-like disorder (14,15).
RAD51 paralogs form at least two distinct complexes for repairing DNA damage, RAD51C–XRCC3 (CX3) and RAD51B–RAD51C–RAD51D–XRCC2 (BCDX2) (16,17). While the CX3 complex was suggested to play a role in resolution of Holliday junctions (18), the BCDX2 complex was shown to preferentially bind single-strand DNA and contribute to the strand invasion step (19). However, interacting partners involved in modulating the activities of these complexes remain to be identified, which has hindered progress towards understanding the precise mechanism of HR repair.
We recently completed a proteomic profiling study to identify candidates that specifically participate with RAD51C, RAD51D and XRCC2 protein complexes (20). Because of the suggested associations with both HR and NHEJ, one novel protein discovered to interact with all three proteins was SFPQ (splicing factor proline and glutamate-rich), which is also known as PSF (polypyrimidine tract-binding protein-associated splicing factor) (21,22). SFPQ was initially identified as a potential pre-mRNA modification protein due to its binding to the pre-mRNA splicing factor polypyrimidine tract binding protein (PTB) (23). Further evidence, however, demonstrated a clear differential nuclear matrix localization of SFPQ with the majority of the protein not co-localizing with PTB, suggesting different roles for SFPQ in addition to RNA splicing (24). In earlier studies, it was demonstrated that, together with an SFPQ paralog named NONO/p54nrb, SFPQ bound double-stranded DNA (dsDNA) and strongly bound ssDNA and RNA through RNA recognition motifs (RRMs) (25). In addition, SFPQ had DNA reannealing and strand invasion activity that formed a D-Loop, which is similar to an HR structural intermediate (21). Recent evidence for a direct role in DSB repair is based upon a direct association between the N-terminal region of SFPQ with RAD51, which stimulated RAD51 activity (26). Furthermore, SFPQ–NONO stabilized paired DNA DSB ends (22), and attenuation of NONO expression conferred a deficiency in DSB repair and increased radiation induced chromosomal aberrations (27). Even more recently, the SFPQ–NONO complex was shown to be recruited to sites of DNA damage (28). However, there still remains a gap in addressing the specific functions of these related mRNA processing factors in DNA repair.
In addition to demonstrating SFPQ directly interacts with the RAD51D protein, we report here the first direct cellular evidence for SFPQ being involved in homology-directed DNA repair. Reduced SFPQ expression conferred cellular sensitivity to DNA crosslinking and alkylating agents and reduced HR levels. SFPQ depletion also led to increased chromosomal aberrations, and quite surprisingly, SFPQ-deficient cells had substantial sister chromatid cohesion defects. To our knowledge, this is the first example of a protein involved in DNA repair that is also responsible for maintaining interchromatid cohesion between sister chromatids with normally aligned centromeres. In further support of the dual role for SFPQ during DNA repair and cohesion maintenance, combining the SFPQ depletion with a Rad51d deletion resulted in a lethal phenotype. This would be expected from disrupting HR, NHEJ, and cohesion mechanisms. These findings suggest that disruption of SFPQ activity or interaction with RAD51 proteins may provide a novel cancer therapeutic approach to simultaneously inactivate multiple DNA repair and chromosome maintenance pathways.
MATERIALS AND METHODS
Cell lines, vectors and protein purification
Mouse embryonic fibroblast (MEF) cell lines M12 (Wild-type), MEFC20 (Rad51d+/+Trp53−/−), MEF258 (Rad51d−/−Trp53−/−) and MEF172AG (Rad51d−/− Trp53−/− HARad51d) were described earlier (11). The H2B-GFP HeLa cell line was kindly provided by Dr Geoffrey Wahl (Salk Institute for Biological Studies) (29). The HeLa DRGFP cell line was derived by stable integration of the DRGFP reporter gene into the HeLa S3 cell line (30). Puromycin resistant single cell derived HeLa colonies were screened by Southern blotting for intact reporter gene integration. The pCBASce plasmid for I-SceI expression has been described earlier (31). To generate Sfpq expression constructs, full-length mSfpq cDNA was cloned in-frame from pYXASC1mSFPQ (ATCC, VA, USA), into the pET28b vector (Novagen/EMD biosciences, CA, USA) to create pET28SFPQ(His). S-tagged RAD51D(His)6, S-tagged RAD51C(His)6, XRCC2(His)6 and SFPQ(His)6 proteins were purified from respective vectors pET32Trx–RAD51D(His), pET32Trx–RAD51C(His), pET28XRCC2(His), and pET28SFPQ(His) as described earlier (20). The co-precipitation experiments were performed by mixing equimolar concentrations of each protein followed by pulldowns using the S-tag agarose matrix (Novagen, EMD Biosciences, NJ, USA) as described earlier (20).
Quantitative real-time PCR
DNA-free total RNA was isolated using RNeasy mini columns (Qiagen Inc, CA, USA) and cDNA prepared from 1 µg total RNA using Oligo(dT)20 primers (Protoscript III RT–PCR kit, New England Biolabs, MA, USA). Gene-specific primers were designed using Beacon Designer version 7.51 (Biosoft International, CA, USA). RT–PCR was performed in triplicates (iQ5 Real Time PCR detection system, Bio-Rad, CA, USA) using IQ SYBR Green Supermix (Bio-Rad). The thermo cycling protocol used was: 95°C for 3 min, followed by 40 cycles of 95°C for 10 s, and 55°C for 30 s. RNA expression levels were calculated using iQ5 optical system software using the comparative ddCt method, normalized to the expression of GAPDH, and presented as ‘normalized fold-expression’.
siRNA knockdown of SFPQ expression
Three sets of Mission siRNA oligos (Sigma Proligo, TX, USA) for the mouse Sfpq gene corresponding to nucleotides 2147–2167 (1_Sfpq_Mm and 1_Sfpq_Mm_as duplex; siRNA1), 1639–1657 (2_Sfpq_Mm and 2_Sfpq_Mm_as duplex; siRNA2) and 1509–1527 (3_Sfpq_Mm and 3_Sfpq_Mm_as duplex; siRNA3) respectively were used for transfection. Mission siRNA oligos for the mouse Gapdh housekeeping gene corresponding to nucleotides 1090–1108 (NM_008084|1 and NM_008084|1 AS duplex, Sigma Proligo) was used as a negative control. siRNAs were transfected into 6 × 104 MEF cells in 24-well plates using the nanoparticle siRNA transfection system following manufacturer’s protocol (N-TER, Sigma, MO, USA) 16–20 h after initial plating. Human Mission siRNA (Sigma Proligo) for hSFPQ (SASI_Hs01_00073163 and SASI_Hs01_00073163_AS duplex) and hGAPDH (NM_002046|1 and NM_002046|1 AS duplex) were used similarly in HeLa cells. Universal scrambled RNA duplexes that did not target human or mouse sequences were used as negative controls (DS Scrambled Neg, IDT Inc., IA, USA).
Western blot analysis
Immunoblot analysis was performed using mouse monoclonal anti-SFPQ antibody, goat polyclonal anti-RAD51B, rabbit polyclonal anti-Ku70, rabbit monoclonal anti-RPA2, mouse anti-β-actin antibody (Abcam, MA, USA), mouse anti-numatrin (Zymed laboratory, CA, USA), mouse monoclonal anti-CHK1 (Cell signaling technology, MA, USA), rabbit polyclonal anti-MSH2 (Santa Cruz, CA, USA), rabbit polyclonal anti-SMC1 (Novus Biologicals, CO, USA) for 2 h at 25°C, and goat polyclonal anti-NONO (Abcam) for 16 h at 4°C. This was followed by 2-h incubation with the respective species-specific secondary-HRP conjugate (Santa Cruz) and detection using ECL (West-Pico chemiluminescent substrate, Thermo-Scientific, IL, USA).
Cell survival assays
A total of 3 × 103 cells per well plated on a 96-well plate in quadruplicates, for each condition tested, were transfected with siRNA1 as described earlier. For MTT assays, cell viability was measured 72-h after transfection by incubation with 20 µl/well of 5 mg/ml MethylThiazolyldiphenyl-Tetrazolium bromide (MTT; Sigma), followed by measurement of A595nm (DTX880, Beckman Coulter, CA, USA). For the colony forming assays (CFUs), the cells from each well were trypsinized 24 h after transfection, plated onto two 100-mm tissue culture dishes and grown for 7 days before counting Giemsa stained colonies.
Treatment of cells with DNA damaging agents
MEF cell lines transfected with no siRNA or 30 nM of the siRNA were treated with DNA damaging agents 24 h after transfection. The cells were treated with MMS and MNNG for 1 h, while 6-TG treatment was carried out for 24 h and media replaced after the indicated times. For all other agents, treatments were performed continuously until medium was replenished. For MTT assays, the medium was replaced 72 h following treatments, and the cell viability checked 7 days after treatment. For CFUs, the cells were expanded as described above 24 h after treatment with the DNA damaging agent. The EC50 values were calculated from a semi-log plot fitted to a non-linear regression curve analysis using Graphpad Prism 4.0 software (Graphpad Software Inc., La Jolla, CA, USA). Statistical significance was determined by comparing the logEC50 for each data set using Prism 4.0, taking into account the standard error and confidence interval of the best-fit value.
Flow cytometry
A total of 5 × 105 cells were transfected with no siRNA or 30 nM of Gapdh or Sfpq siRNA in 12-well plates and treated with MMC 24 h after transfection. Floating and attached cells were collected 72 h after treatment and resuspended in plasma membrane permeabilization medium containing propidium iodide (50 µg/ml) (32). Cell-cycle analyses were performed using a Beckman Coulter FC 500 cytometer for a maximum cell count of 20 000 cells. Data were quantified using ModFit LT software version 3.1 (Verity Software House, Topsham, ME).
For recombination measurements, the no siRNA or 30 nM of respective siRNA transfected HeLa DRGFP cells in 12-well plates were co-transfected 24 h later with 1 µg pCBASce vector using Lipofectamine plus reagent (Invitrogen). Recombinant cells were detected using an FC 500 flow cytometer and quantitated using CXP analysis software (Beckman Coulter).
Chromosomal analysis
Giemsa stained metaphase spreads were prepared as described (11,33). Chromosomal aberrations were scored from a minimum of 700 chromosomes each (n ≥ 1500) from two independent experiments. Quantification of the interchromatid distance on the images was performed across each chromosome arm using Metamorph software for Olympus, Ver. 7.5.6.0 (Olympus, PA, USA). A total of 1000 chromosomes were measured from at least 20 spreads from two independent experiments. For analysis of prematurely condensed chromosomes (PCC), cells were treated with 50 nM of calyculin A (Santa Cruz) 20 min prior to harvesting. The interchromatid distances were calculated for the bivalent G2 phase chromosomes. A minimum of 500 chromosomes from at least 10 chromosome spreads from two independent experiments was scored.
Statistical analysis
Calculations of the mean, SD, and SE were performed using Microsoft Excel. Statistical analysis for comparison of each set of experimental means was performed using Graphpad Instat 3.0. Significance of variance was determined by ANOVA and post-tests performed when the variance was significant (P < 0.05). Appropriate post test comparisons for means were made for multiple groups using, Bonferroni Multiple Comparisons Test, Dunnett Multiple Comparisons Test and Tukey–Kramer Multiple Comparisons Test. For comparison of individual data pairs in cell-cycle experiments and viability data, paired t-test was performed on the raw data obtained. A value of P < 0.05 was considered statistically significant.
RESULTS
SFPQ and RAD51D directly interact and have epistatic effects on cell viability
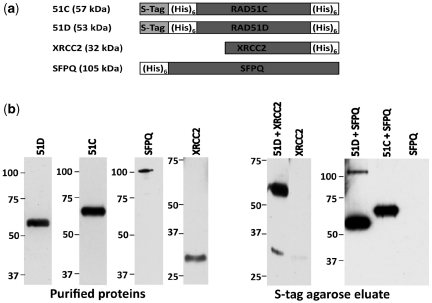
From a high-throughput co-precipitation screen to discover proteins that function with or regulate HR, the major category of candidates identified were predicted mRNA processing factors (20). Because of previous connections with DSB repair, one particularly novel candidate was the SFPQ protein, and 10 SFPQ peptides were specifically identified from the mass spectroscopy analysis of the RAD51D co-precipitation eluates [Supplementary Figure S1a; (20)]. To further validate the interaction, DNA independent binding of SFPQ with RAD51D was demonstrated (Supplementary Figure S1b), and a co-precipitation assay was performed using S-tag agarose beads with Escherichia coli purified S-tagged RAD51C(His)6, S-tagged RAD51D(His)6 and SFPQ(His)6 proteins. As a positive control, co-precipitation of S-tagged RAD51D with the known interacting partner XRCC2 was first confirmed. Similarly, the S-tag agarose pulldown of RAD51D resulted in co-precipitation of SFPQ, indicating that SFPQ directly interacts with RAD51D (Figure 1). However, SFPQ was not co-precipitated along with RAD51C, indicating SFPQ and RAD51C do not directly interact and also helped to rule out random binding with SFPQ because of any exposed dimerization domains. Taken together, these data suggest that SFPQ participates with the BCDX2 complex through direct interaction with RAD51D. Since the BCDX2 complex is known to have a strand invasion function similar to RAD51, SFPQ could modulate its strand invasion activity, in addition to RAD51 (26), during homology directed DSB repair.
Figure 1.
SFPQ interacts directly with the RAD51D protein. (a) Schematic representations of the purified tagged-proteins used for co-precipitation. Approximate sizes are indicated. (b) Purified S-TagRAD51D(His)6 (51D), S-TagRAD51C(His)6 (51C), SFPQ(His)6 (SFPQ) and XRCC2(His)6 (XRCC2) were analyzed by immunoblot using an anti-His antibody (Left panels, Purified protein). S-protein agarose pulldown experiments were performed using both 51D and XRCC2 proteins (51D + XRCC2) and XRCC2 alone, and using both 51D and SFPQ proteins (51D + SFPQ), 51C and SFPQ proteins (51C + SFPQ), and SFPQ alone. The eluted proteins, were resolved on 10% PAGE, transferred onto nitrocellulose membrane, and immunoblotted using anti-His antibody (Right panels, S-tag agarose eluate). The sizes of molecular weight markers (kDa) are indicated on the left of each blot.
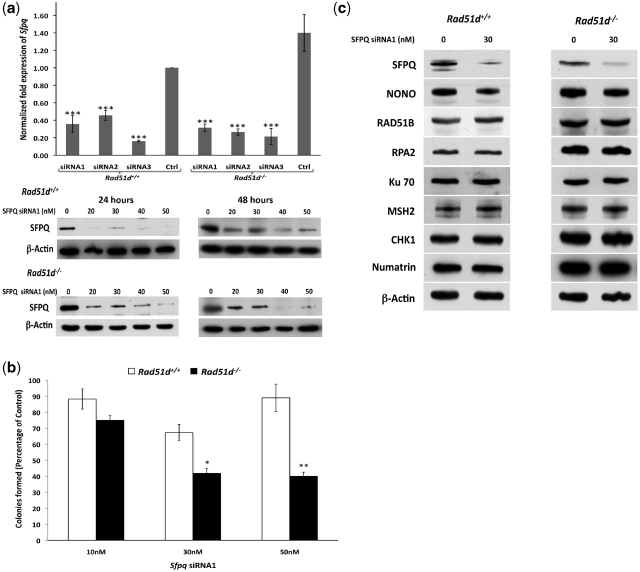
To begin testing the involvement of SFPQ during HR, knockdowns of SFPQ expression were performed in MEF cell lines. Three different siRNA duplexes decreased Sfpq transcript expression up to 70% compared to the control siRNA as determined by quantitative real-time (RT) PCR (Figure 2a). The knockdown of expression with siRNA1 was further verified at the protein level (Figure 2a and Supplementary Figure S2a). Cells deficient for RAD51 paralogs activate the p53 checkpoint response and therefore propagate only in absence of p53 (11,13). However, there was no significant effect on cell viability in Trp53-proficient nor deficient MEFs at these reduced levels of SFPQ as measured by the MTT assay (Supplementary Figure S2b). These data suggest that if SFPQ is involved in DSB repair, the form of unresolved damage fails to elicit a response that activates the p53 checkpoint.
Figure 2.
siRNA-mediated knockdown of SFPQ confers cell lethality in the absence of RAD51D. (a) Rad51d+/+ Trp53−/− (Rad51d+/+) and Rad51d−/− Trp53−/− (Rad51d−/−) MEFs were transfected with Sfpq siRNAs labeled as siRNA1, siRNA2, and siRNA3 (‘Materials and Methods’ section) and expression analyzed using quantitative real-time PCR (Top histogram) and immunoblot analysis (Bottom panels). The expression was compared to the control with no siRNA treatment (Ctrl) after the data was normalized to GAPDH expression. ***indicates P < 0.001 and error bars indicate standard deviation of values from a representative experiment performed in triplicate. Anti-SFPQ immunoblots of whole cell extracts from Rad51d+/+ and Rad51d−/− MEFs were performed 24 h and 48 h following treatment with the indicated concentrations of Sfpq siRNA1. β-actin was used as the loading control. (b) Cell viability was measured using the colony forming assay in both Rad51d+/+ Trp53−/− (Rad51d+/+) and Rad51d−/− Trp53−/− (Rad51d−/−) MEFs transfected with the indicated concentrations of Sfpq siRNA. Cell viability for each siRNA treatment was compared to and reported as percentage of control cells transfected with the same concentration of control siRNA. *P < 0.05, **P < 0.01 and error bars indicate standard error of means of values from at least two independent experiments performed in duplicate. (c) The protein levels of SFPQ, NONO, RAD51B, RPA2, Ku70, MSH2, CHK1 and Numatrin (Nucleophosmin) were measured using the respective antibodies in Rad51d-proficient (Rad51d+/+) and deficient (Rad51d−/−) cell lines 24 h after transfection with the indicated concentration of Sfpq siRNA1. β-actin was used as the loading control.
To determine whether SFPQ and RAD51D are epistatic, cell viability was measured by the clonogenic and MTT assays in MEFs proficient and deficient for RAD51D following knockdown of SFPQ expression. In the Rad51d-deficient cell line, cell viability was significantly reduced by 40–50% compared with the Rad51d-deficient cell line with no SFPQ deficiency using both assays (P < 0.05) (Figure 2b and Supplementary Figure S2b). In the colony forming assay, the SFPQ deficiency alone did confer slightly reduced cell viability, which was not significantly different from the control (P > 0.05). However, to rule out any potential siRNA off target effects or a general reduction of expression due to SFPQ depletion, protein levels of NONO and six representative DNA repair and signaling proteins were measured (Figure 2c). Overall expression levels were not substantially affected. There was a small but noticeable reduction of the NONO protein levels, but the possibility of any combinatorial influence between SFPQ and NONO was ruled out since depletion of NONO expression alone did not affect cell viability in either cell line (Data not shown). In addition, SFPQ expression was not affected by the status of either Rad51d or p53 nor by treatment of cells with the DNA crosslinking agent mitomycin C (MMC) (Supplementary Figure S2c). Such a combined synthetic lethal phenotype does resemble the results observed in PARP1 deficient BRCA1 or BRCA2 negative cells as well as mismatch repair deficient cells when combined with knockdown of PolB or PolG expression (34,35). Taken together with previous reports (21,26), these results suggest an important role for SFPQ in combination with RAD51D for repairing DSBs and maintaining cell viability. However, the synthetic lethality data do suggest that in addition to their roles in DSB repair, SFPQ or RAD51D may have independent functions.
SFPQ promotes sister chromatid cohesion and maintains chromosome integrity
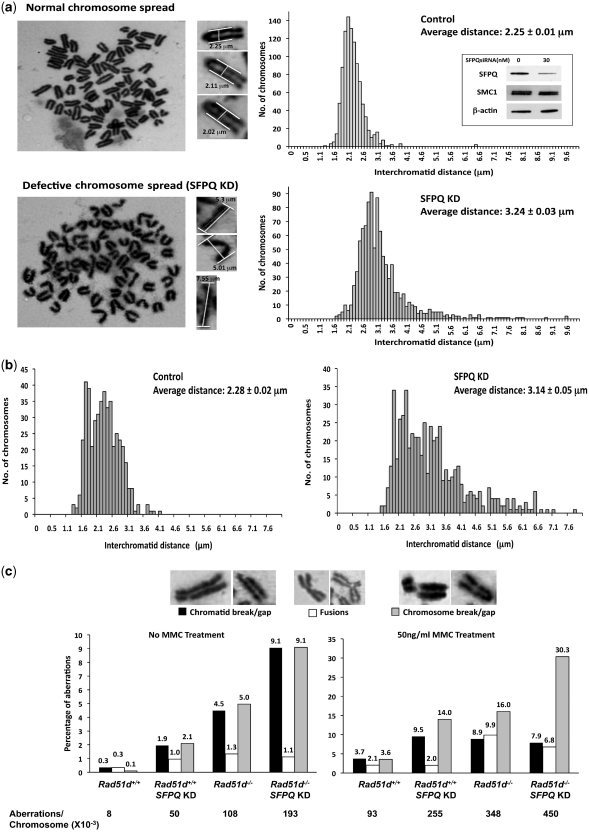
Reduced HR activity is responsible for increasing chromosomal instability (36). To study potential chromosomal defects resulting from decreased SFPQ levels, Giemsa stained metaphase spreads from Rad51d+/+ MEFs were analyzed following siRNA knockdown of SFPQ expression. Surprisingly, the most striking phenotype identified was increased interchromatid distance between sister chromatid arms from a mean of 2.25 µm for the control to 3.24 µm in SFPQ-deficient cells (P < 0.001) (Figure 3a). This chromosome phenotype was also observed in Rad51d−/− MEFs, when SFPQ expression was reduced, the average interchromatid distance increasing from 2.26 to 2.99 µm (Supplementary Figure S3). Verifying the increase is unique to SFPQ, interchromatid distance was not altered by attenuating NONO (average interchromatid distance of 2.25 µm ± 0.11 SED) nor GAPDH expression (average interchromatid distance of 2.26 µm ± 0.01 SED). Similar results were also observed in human HeLa cells with a significant increase from 2.14 µm (±0.01 SED) in controls and 2.17 µm (±0.02 SED) in NONO-deficient cells to 2.50 µm (±0.02 SED) in SFPQ-deficient cells (P < 0.001).
Figure 3.
SFPQ deficiency leads to defects in sister chromatid cohesion and chromosomal integrity. (a) Giemsa stained metaphase chromosome spreads were prepared from control or Sfpq siRNA1 treated MEFs 48 h after transfection. Representative images of a normal chromosome spread and defective chromosome spread from SFPQ deficient cells are shown. The distance between the sister chromatid arms were determined for at least 1000 chromosomes of each group from two independent experiments (Representative chromosomal measurements are indicated). Histogram for the number of chromosomes with respective interchromatid distances was plotted, with the average distance and standard error for control and SFPQKD indicated (right panel). Anti-SFPQ and anti-SMC1 (Cohesin protein) expression was verified by respective immunoblots of whole cell extracts (Inset). β-Actin was used as loading control. (b) Giemsa stained PCC spreads were prepared from control and Sfpq siRNA1 treated MEFs 48 h after transfection. A Histogram representing the number of bivalent G2 phase PCCs with respective interchromatid distances is shown. (c) Quantification of different chromosomal aberrations (Chromatid break/gap, fusions and chromosomal break/gap) observed in the Giemsa stained metaphase spreads for Rad51d+/+ and Rad51d−/− MEFs transfected with 30nM siRNA. The percentages of aberrations from untreated and MMC treated MEFs are indicated as labels above each bar, while the overall numbers of aberrations per chromosomes (×10−3) are indicated below the graphs. Note different Y-axis scales.
The effect of SFPQ deficiency on interchromatid cohesion was further validated by analysis of bivalent PCC in the G2 phase of cell cycle, generated by treating MEFs with calyculin A. Even though the interchromatid distances were more variable compared with the metaphase spreads, the average interchromatid distance observed in controls was 2.28 µm as compared with 3.14 µm in the SFPQ deficient cells (P < 0.001) (Figure 3b). In addition, SFPQ deficiency did not affect the levels of the central cohesion protein, SMC1 (Figure 3a, inset), and suggest this phenotype is not likely due to an overall decrease in cohesion protein expression. Taken together, these data suggest that SFPQ has a unique role in the assembly of sister chromatid cohesion.
Reduced expression of the SMC1 or SCC3 cohesin proteins in human cells conferred spontaneous chromatid breaks and aberrations at low levels (∼1–2%), which increased up to 8-fold when cells were irradiated (37,38). However, reduction of SFPQ expression resulted in a >5-fold increase in spontaneous chromosomal aberrations (from 8 × 10−3 to 50 × 10−3/chromosome). DNA damage deficient Rad51d−/− cells show a >10-fold increase in spontaneous aberrations (from 8 × 10−3 to 108 × 10−3/chromosome), while SFPQ deficiency in Rad51d−/− cells further increases the aberrations by about 1.8-fold to 193 × 10−3/chromosome (Figure 3c). Interestingly, the number of chromosomal and chromatid breaks increased significantly in double-deficient cells (9.1% each) as opposed to the single mutants (5.0 and 4.5% respectively), while the number of spontaneous fusions remained ∼1% (Figure 3c). Challenging each deficient cell line with 50 ng/ml of the DNA interstrand crosslinking agent MMC further increased chromosomal aberrations by 2.75-fold for SFPQ deficiency and 3.75-fold for the Rad51d−/− cell line. Double deficient MEFs exhibited elevated levels of chromosomal aberrations up to 45%, an increase of 1.7-fold and 1.3-fold compared to SFPQ or RAD51D deficient cells respectively. The elevated chromosomal breaks resulting from the deficiency of either of the two genes results in extreme chromosomal fragmentation. This extreme fragmentation phenotype results from unrepaired chromosome breaks and constituted nearly two-thirds of the aberrations identified in the double deficient cells. Taken together, these data suggest SFPQ may participate in the sister chromatid cohesion complex and enable strand invasion of RAD51 proteins during HR between sister chromatid strands.
SFPQ confers resistance to DNA-crosslinking, alkylating and DSB generating agents
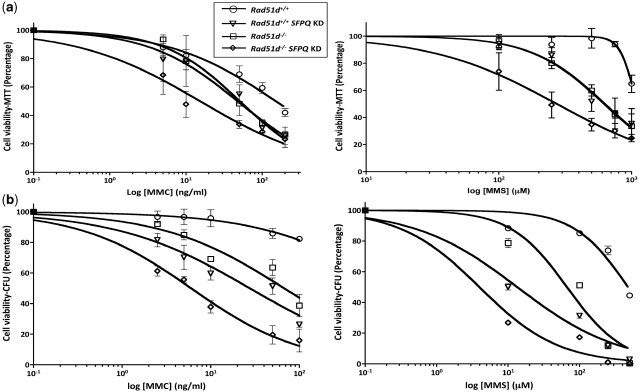
To determine whether SFPQ is necessary to repair DNA damage, Rad51d-proficient and deficient MEFs were treated with representative DNA damaging agents 24 h following siRNA knockdown and cell viability measured using the MTT assay. Reduced SFPQ expression conferred cellular sensitivity to the DNA cross-linking agents, MMC and cisplatin, and the methylating agents, MMS and MNNG when compared to control Rad51d-proficient cells (Figure 4a, Table 1). Comparison of EC50 values indicate that the sensitivity of SFPQ deficient cells is similar to Rad51d-deficient cells to DNA damage resulting from both crosslinking and alkylating agents. In the absence of both SFPQ and RAD51D, the cellular sensitivity to each of these agents was further increased by >2-fold (Table 1); however, interpretation of this data could be complicated due to the synthetic lethal phenotype exhibited in SFPQ deficient Rad51d−/− cells. In support of these findings, sensitivity to MMC and MMS induced damage was confirmed using the colony forming assay (Figure 4b). Although the EC50 values differed because of the inability of the MTT assay to measure cell proliferation, the differences in fold sensitivity in both assays were similar.
Figure 4.
Knockdown of SFPQ expression confers sensitivity to DNA damaging agents. siRNA knockdown of SFPQ expression was performed in Rad51d+/+ and Rad51d−/− cells and treated 24 h later with indicated concentrations of Mitomycin C (MMC) and Methyl methanesulfonate (MMS). Cell viability was measured 7 days after drug treatment using the (a) MTT assay or (b) Colony forming assay (CFU) and plotted as percentage of cells surviving compared to the no drug control. Error bars represent SEM from at least three independent experiments performed in quadruplicates for the MTT assay, and at least three independent experiments performed in duplicates for CFU determination.
Table 1.
Fold-sensitivity of RAD51D and SFPQ deficient MEFs to DNA damaging agents
| Agent | Rad51d−/− | SFPQ KD | Rad51d−/−SFPQ KD |
|---|---|---|---|
| MMC | 3.0 | 3.2 | 9.8 |
| Cisplatin | 1.3 | 1.8 | 4.6 |
| MMS | 1.7 | 1.7 | 4.0 |
| MNNG | 2.2 | 3.4 | 9.0 |
| Camptothecin | 4.7 | 7.5 | 12.2 |
| 6TG | 3.1 | 1.3 | 2.8 |
| FdURD | 3.2 | 1.3 | 1.3 |
| Hydroxyurea | 1.4 | 1.1 | 0.9 |
Fold sensitivities for each DNA damaging agent were calculated from the EC50 values obtained from the MTT assays. The EC50 values are from a semi-log plot fitted to a non-linear regression curve analysis (‘Materials and Methods’ section), and statistical significance was determined by comparing the logEC50 for each agent.
The topoisomerase I inhibitor camptothecin was used to determine the effect of SFPQ deficiency directly on DSBs. Camptothecin induces DSBs at replication forks by interfering with the breakage-reunion reaction of the topoisomerase I enzyme (39). Both Rad51d−/− and SFPQ deficient MEFs were 5- to 7.5-fold more sensitive to camptothecin respectively, which increased to ∼12-fold in the double-deficient MEFs compared to control Rad51d-profecient cells (Table 1). Differences in cellular sensitivity between Rad51d−/− and SFPQ deficient cells to some DNA damaging agents were identified. For example, sensitivity of MEFs to the nucleotide analogs 6-TG, which generates predominantly S6-methylguanine adducts, FdURD, a thymidylate synthase inhibitor and hydroxyurea (HU), a DNA replication-inhibitor, were not affected by reduced SFPQ expression in Rad51d-proficient nor Rad51d-deficient cell lines. Rad51d−/− cells however were 3-fold more sensitive to 6-TG and FdURD but not to HU (Table 1). Fold sensitivities were statistically significant (P < 0.01) when SFPQ-deficient cells were treated with MMC, Cisplatin, MMS, MNNG and Camptothecin. These sensitivity profiles suggest that SFPQ has a role in the HR DNA repair pathway, particularly for repairing DSBs generated directly or due to DNA crosslinks and methylation damage. However, some DNA repair intermediates that require RAD51D for processing appear to be SFPQ independent, suggesting RAD51D and SFPQ have some redundant but not identical functions.
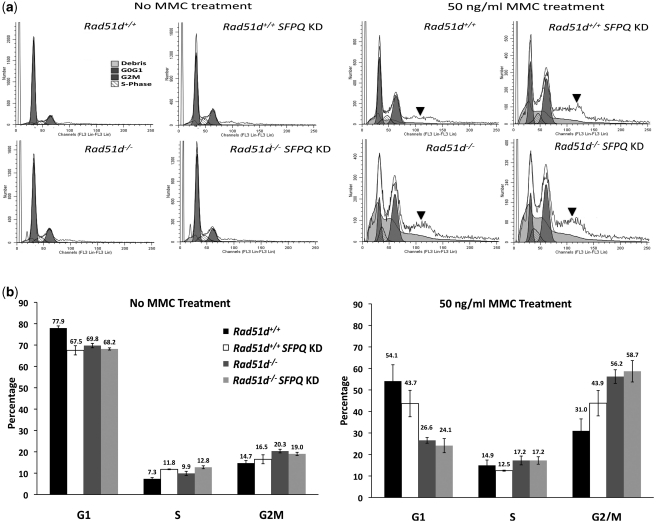
Mitomycin C induces G2M cell-cycle arrest in SFPQ-deficient cells
To ascertain the effect of SFPQ deficiency on cell-cycle progression, the cellular DNA content was analyzed by flow cytometry 72 h following knockdown of SFPQ expression (Figure 5a). Cells deficient for SFPQ show an 8–10% decrease in the percentage of cells in the G1 phase compared with controls (P < 0.05) with a small but corresponding increase of up to 5 and 6% in the S and G2M phases respectively (Figure 5b). Rad51d-deficient and SFPQ-Rad51d double deficient cells had similar cell-cycle profiles, although an increase in cell death and aneuploidy were observed in each of these experiments.
Figure 5.
MMC treatment induced G2M arrest of SFPQ-deficient cells. (a) Representative cell-cycle distribution data of Rad51d+/+ and Rad51d−/− cell lines transfected with siRNA were analyzed 72 h following MMC treatment. The proportion of cells in each cell-cycle stage (G0G1, S-phase, G2M) is indicated. The initial peak marked as debris corresponds to dead cells, while the plateau after the G2M peak (indicated by the bold arrows) corresponds to the aneuploid population of cells and were not included in the calculations. (b) The percentage of cells in the G1, S, and G2M phase 72 h following treatment. Each cell line was transfected with no siRNA or transfected with Sfpq siRNA (Rad51d+/+SFPQ KD Rad51d−/−SFPQ KD) 24 h prior to MMC treatment. The percentages of each stage of the cell cycle were calculated using ModFit software analysis (‘Materials and Methods’ section). Error bars indicate the SEM percentages from at least three independent experiments.
To determine the effect of SFPQ depletion on cell-cycle arrest following DNA damage, MEFs were treated with 50 ng/ml MMC 24 h following knockdown of SFPQ expression and cell-cycle distribution analyzed after 72 h. Treatment with MMC increased the percentage of cells in the G2M phase by 13% in SFPQ-deficient MEFs and 25% in both the Rad51d-deficient and the double-deficient cell lines when compared with the control (Figure 5b; P < 0.05). At this dose of MMC there was also a substantial increase in the number of dead and the proportion of aneuploid cells observed in MEFs lacking Rad51d or deficient in SFPQ (Figure 5a, indicated by arrows). This is consistent with the high amount of chromosomal breaks and fragmentation previously observed in cells deficient for either or both proteins. Therefore, reduced expression of SFPQ and RAD51D severely affects the ability of these cells to progress through the cell cycle following DNA damage, leading to the accumulation of cells in the G2M phase as well as chromosomal aneuploidy.
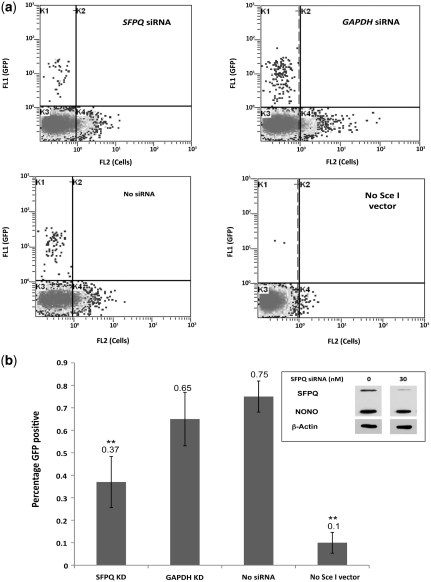
SFPQ is necessary for homologous repair of DSBs
To directly test the role of SFPQ during HR, SFPQ expression was reduced in HeLa cells containing a chromosomally integrated DRGFP recombination reporter (HeLa DRGFP). This construct consists of two tandem and differentially mutated GFP genes, SceGFP and iGFP, inactivated by a stop codon containing an I-SceI recognition sequence and by a 5′ and 3′ truncation respectively (30). Expression of an I-SceI endonuclease introduces a DSB at the SceGFP gene, and when the break is repaired by error-free HR, using iGFP as the information donor, the SceGFP allele is converted into a functional GFP gene. In the HeLa cells, siRNA directed against human SFPQ decreased SFPQ protein levels by 60–70% 24 h following siRNA transfection (Figure 6b, inset). No effect on the expression of NONO using this siRNA was observed, consistent with the siRNA not having off-target affects. HR repair was measured by the percentage of GFP positive cells, which is negligible in cells without the pCBASce vector expressing the endonuclease (Figure 6, No Sce-I vector). In the absence of siRNA, HR mediated repair of the DSB generated by the I-SceI endonuclease resulted in 0.75% GFP positive cells (Figure 6b, No siRNA) whereas siRNA knockdown of SFPQ reduced the frequency of recombinant cells by 2-fold (P < 0.05). In addition to the no-siRNA control, siRNA against GAPDH was used as a negative control, which did not affect SFPQ expression levels, and did not show significant reduction in HR repair efficiency. This assay therefore provides direct evidence that SFPQ is necessary for HR repair of DSB DNA damage.
Figure 6.
siRNA knockdown of SFPQ reduces HR mediated repair of DSBs. (a) Representative flow cytometric analyses of HeLa DRGFP cells transfected with SFPQ siRNA, GAPDH siRNA and no siRNA followed 24 h later with pSCBASce vector and no pCBASce (No Sce I vector) control. GFP positive cells from ∼20 000 counts are seen in the quadrant marked K1. (b) Effect of siRNA knockdown of SFPQ on recombination-mediated repair of Sce I induced chromosomal DSB. The GFP positive cells indicate the percentage of recombination events. The corresponding siRNA used for transfection is indicated while the HeLa DRGFP cells without the pCBASce vector (No Sce I vector) is the negative control. The error bars indicate standard errors of values from two independent experiments performed in triplicate (**P < 0.01). Anti-SFPQ and anti-NONO western blots of whole cell extracts were used to verify reduced SFPQ expression. β-Actin was used as loading control (Inset).
DISCUSSION
HR is required to efficiently repair complex DNA damage and maintain the integrity of the mammalian genome. Although great strides have been made identifying and characterizing factors involved in recognition, signaling and processing DNA breaks, the picture is far from complete (5,40). As part of an effort to discover new HR factors, we recently completed a proteomics screen for RAD51C, RAD51D and XRCC2 interacting partners (20). One intriguing protein identified from all three co-precipitations was the splicing-related factor, SFPQ. Previous findings had demonstrated that the SFPQ protein consists of an N-terminal proline glutamine rich region that also encodes the DNA-binding domain and two RRMs followed by a C-terminal region responsible for protein interaction and nuclear localization (41–43). SFPQ potentially plays important roles during different stages of mRNA processing as part of the spliceosome complex and transport (44). Additional nuclear functions include transcriptional regulation (45,46), DNA unwinding and relaxation (47), as well as DNA repair (22,27,28). The work reported here demonstrates that SFPQ is essential for HR-mediated repair of DSB DNA damage and genome maintenance, necessary to maintain sister chromatid cohesion, and directly interacts with the fourth member of the RAD51 family, RAD51D.
Direct evidence for SFPQ being involved during HR-mediated repair of DNA DSB damage was provided by reduction of the gene conversion frequency in the HeLa DRGFP cell line. The 2-fold decrease of recombinant GFP positive cells as a result of SFPQ deficiency was significant, particularly since 25–30% of SFPQ expression remained. This frequency of HR is comparable to siRNA knockdown of the RPA2 phosphorylation protein PP4 based upon a similar recombination assay (48). The role of SFPQ in DNA repair was further substantiated by the sensitivity of SFPQ deficient cells to damage caused by the DNA crosslinking and alkylating agents, as well as camptothecin. Rad51d-deficient cells were previously reported to be most sensitive to the crosslinking agents and alkylating agents that generate interstrand crosslinks (ICL) leading to DSB during replication (11,49). Similar sensitivity of the SFPQ deficient cells to these agents suggests an analogous role for SFPQ in the HR pathway. However, the lack of sensitivity of SFPQ deficient cells to DNA lesions introduced by 6-TG and FdURD differs from the Rad51d-deficient cell lines. These results suggest that the two proteins are not completely redundant. This might be important because it is becoming clear that repairing multiple forms of DNA damage requires integration of multiple cellular pathways (50–52).
Because SFPQ was predicted to be a multifunctional protein with several nuclear functions (53), we anticipated that SFPQ would be an essential gene. However, there was no significant effect conferred by the siRNA-mediated deficiency on cell viability in p53 proficient or p53 deficient mouse fibroblasts. The transient nature of siRNA knockdown as well as low levels of protein that may still be present cannot entirely rule out the possibility that SFPQ might still be indispensible. Consistent with these results, an sfpq point mutation (Whitesnake) in zebra fish embryos did not affect cell proliferation, but increased cell death by ∼2-fold (54). Surprisingly, in our experiments, there was a significant decrease in cell survival when SFPQ expression was reduced in Rad51d-deficient cells. We further demonstrate that SFPQ with RAD51D directly interact, which may lead to SFPQ modulating the strand invasion function of the BCDX2 complex, similar to that reported for RAD51 (26). An alternative possibility is the synthetic lethal phenotype occurs due to the two proteins being required for non-overlapping mutually exclusive pathways. For example, SFPQ is also known to interact as a heterodimer with NONO and was reported to protect free DSB ends that occur during NHEJ (22). Therefore, it remains a formal possibility that absence of both SFPQ and RAD51D is disrupting the HR and NHEJ repair pathways. However, this is unlikely to be fully responsible for a lethal phenotype since, to our knowledge, HR and NHEJ double deficient cells have not been reported to confer significant cell death in spite of increased DNA damage and cell proliferation defects (55). Furthermore, previous reports suggest that loss of NHEJ confers resistance to Top1 inhibitor, Camptothecin, while SFPQ deficient cells seem to be highly sensitive to Camptothecin, consistent with a primary role for SFPQ during HR (56).
Unexpectedly, our data demonstrate that SFPQ plays an important role in the maintenance of sister chromatid cohesion. A previous study showed that RAD51C defective CL-V4B hamster cells had more than double the number of cells with completely separated sister chromatids that are not attached to centromeres. This phenotype was attributed to premature sister chromatid separation at the metaphase-anaphase transition but not observed in XRCC2, XRCC3, nor BRCA1 defective cells (57). However, similar to the differences observed in cells deficient for cohesin proteins, an increase in the interchromatid distance was observed with the chromatids remaining attached to the centromeres in SFPQ deficient cells (37). These data suggest that RAD51D and SFPQ have distinct functions at different cell-cycle stages to maintain sister chromatid interaction, with SFPQ likely playing a role during cohesion formation during replication rather than cohesion maintenance. RAD51D deficiency however does not have any effect on the cohesion phenotype and a knockdown of SFPQ in these Rad51d−/− cells had a similar significant increase in interchromatid distance.
Reduced expression of SFPQ also significantly increased the number of spontaneous chromosomal aberrations. The inability to maintain sister chromatid cohesion is known to hinder the capacity to repair spontaneous and radiation induced DSBs during the G2 phase in yeast (58,59). However, the levels of spontaneous levels of chromosome breaks are low, ∼1%, in human cells deficient for cohesin proteins (37,38). Keep in mind that reduction of the recombinant GFP positive cells due to SFPQ deficiency in the HeLa DRGFP reporter system may not reflect the increased distance of homologous strands because the two GFP genes are repeated in tandem. Therefore, the increased chromosomal aberrations in SFPQ deficient cells may result from two different mechanisms being affected, DSB DNA repair (HR and NHEJ) as well as sister chromatid cohesion. A more recent report suggests early and rapid recruitment of the SFPQ–NONO complex directly to sites of DNA breaks and a delay in the repair of the DSBs in SFPQ deficient cells (28).
The importance of proteins initially implicated in spliceosome biogenesis for repairing DNA damage and maintaining genome stability is becoming increasingly clear. In a recent siRNA screen to identify genes involved in genome stabilization, an mRNA processing cluster was the most significantly enriched group of genes, with a majority having a role in RNA splicing (60). RAD51C has been attributed to maintaining genome integrity by transduction of DNA damage signals resulting from Chk2 activation as well as via the ATM/Chk1 pathway (61,62). A separate large-scale screen identified targets of ATM/ATR DNA checkpoint proteins and again discovered that a large number of RNA processing and transcription factors were ATM/ATR phosphorylation targets (63). SFPQ now adds to the growing list of splicing related factors required for HR repair that includes Pso4, Ntr1, SPF45 and Gemin2, reported to be involved in DSB and ICL repair as well as Holliday junction binding (64–67). These results are also consistent with recent evidence for transcription-associated recombination (TAR) in mammals. TAR processes are proposed to rescue stalled replication forks generated during S phase when replication encounters transcription processes or by replication forks encountering eroding telomeres (68,69). Disruptions in this pathway result in defective DNA damage response leading to more cells progressing through S and accumulating in G2M with increased mutation and chromosomal aberration frequencies.
The genome instability phenotype conferred by reduced levels of SFPQ may represent a combination of SFPQ functions to efficiently repair DNA lesions and to adequately form sister chromatid cohesion. The multiple SFPQ functions in RNA processing, transport and transcriptional regulation might also be contributing to this phenotype as well as the synthetic lethality conferred by absence of both SFPQ and RAD51D. This study therefore provides the basis to further dissect out molecular mechanisms of HR and interacting pathways. Future biochemical studies investigating SFPQ binding, protecting and resolving HR intermediates will help decipher the precise role of SFPQ during homology directed repair. Additionally, SFPQ might prove to be a useful cancer therapy target for inactivating multiple pathways in chemotherapy resistant cancer cells that may already be HR defective (70,71).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
American Cancer Society grant (RSG-030158-01-GMC to D.L.P.) and an SC Honors College Science Undergraduate Research Fellowship to D.K.B.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Marj Pena for technical help and advice with FACS analysis and Katherine Jenkins for technical assistance. The authors would also like to thank each of the anonymous reviewers for their comments and valuable suggestions.
REFERENCES
- 1.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 2.Kennedy RD, D'Andrea AD. DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes. J. Clin. Oncol. 2006;24:3799–3808. doi: 10.1200/JCO.2005.05.4171. [DOI] [PubMed] [Google Scholar]
- 3.Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair. 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 4.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 5.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 7.Shu Z, Smith S, Wang L, Rice MC, Kmiec EB. Disruption of muREC2/RAD51L1 in mice results in early embryonic lethality which can Be partially rescued in a p53(-/-) background. Mol. Cell. Biol. 1999;19:8686–8693. doi: 10.1128/mcb.19.12.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deans B, Griffin CS, Maconochie M, Thacker J. Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J. 2000;19:6675–6685. doi: 10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pittman DL, Schimenti JC. Midgestation lethality in mice deficient for the RecA-related gene, Rad51d/Rad51l3. Genesis. 2000;26:167–173. doi: 10.1002/(sici)1526-968x(200003)26:3<167::aid-gene1>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 10.Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smiraldo PG, Gruver AM, Osborn JC, Pittman DL. Extensive chromosomal instability in Rad51d-deficient mouse cells. Cancer Res. 2005;65:2089–2096. doi: 10.1158/0008-5472.CAN-04-2079. [DOI] [PubMed] [Google Scholar]
- 12.Tarsounas M, Munoz P, Claas A, Smiraldo PG, Pittman DL, Blasco MA, West SC. Telomere maintenance requires the RAD51D recombination/repair protein. Cell. 2004;117:337–347. doi: 10.1016/s0092-8674(04)00337-x. [DOI] [PubMed] [Google Scholar]
- 13.Kuznetsov SG, Haines DC, Martin BK, Sharan SK. Loss of Rad51c leads to embryonic lethality and modulation of Trp53-dependent tumorigenesis in mice. Cancer Res. 2009;69:863–872. doi: 10.1158/0008-5472.CAN-08-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, Niederacher D, Freund M, Lichtner P, Hartmann L, Schaal H, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat. Genet. 2010;42:410–414. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 15.Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, Neveling K, Endt D, Kesterton I, Autore F, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat. Genet. 2010;42:406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 16.Masson JY, Stasiak AZ, Stasiak A, Benson FE, West SC. Complex formation by the human RAD51C and XRCC3 recombination repair proteins. Proc. Natl Acad. Sci. USA. 2001;98:8440–8446. doi: 10.1073/pnas.111005698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ, Benson FE, West SC. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 2001;15:3296–3307. doi: 10.1101/gad.947001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Tarsounas M, O'regan P, West SC. Role of RAD51C and XRCC3 in genetic recombination and DNA repair. J. Biol. Chem. 2007;282:1973–1979. doi: 10.1074/jbc.M609066200. [DOI] [PubMed] [Google Scholar]
- 19.Yokoyama H, Sarai N, Kagawa W, Enomoto R, Shibata T, Kurumizaka H, Yokoyama S. Preferential binding to branched DNA strands and strand-annealing activity of the human Rad51B, Rad51C, Rad51D and Xrcc2 protein complex. Nucleic Acids Res. 2004;32:2556–2565. doi: 10.1093/nar/gkh578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajesh C, Gruver AM, Basrur V, Pittman DL. The interaction profile of homologous recombination repair proteins RAD51C, RAD51D and XRCC2 as determined by proteomic analysis. Proteomics. 2009;9:4071–4086. doi: 10.1002/pmic.200800977. [DOI] [PubMed] [Google Scholar]
- 21.Akhmedov AT, Lopez BS. Human 100-kDa homologous DNA-pairing protein is the splicing factor PSF and promotes DNA strand invasion. Nucleic Acids Res. 2000;28:3022–3030. doi: 10.1093/nar/28.16.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bladen CL, Udayakumar D, Takeda Y, Dynan WS. Identification of the polypyrimidine tract binding protein-associated splicing factor.p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J. Biol. Chem. 2005;280:5205–5210. doi: 10.1074/jbc.M412758200. [DOI] [PubMed] [Google Scholar]
- 23.Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- 24.Meissner M, Dechat T, Gerner C, Grimm R, Foisner R, Sauermann G. Differential nuclear localization and nuclear matrix association of the splicing factors PSF and PTB. J. Cell. Biochem. 2000;76:559–566. [PubMed] [Google Scholar]
- 25.Yang YS, Hanke JH, Carayannopoulos L, Craft CM, Capra JD, Tucker PW. NonO, a non-POU-domain-containing, octamer-binding protein, is the mammalian homolog of Drosophila nonAdiss. Mol. Cell. Biol. 1993;13:5593–5603. doi: 10.1128/mcb.13.9.5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morozumi Y, Takizawa Y, Takaku M, Kurumizaka H. Human PSF binds to RAD51 and modulates its homologous-pairing and strand-exchange activities. Nucleic Acids Res. 2009;37:4296–4307. doi: 10.1093/nar/gkp298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S, Kuhne WW, Kulharya A, Hudson FZ, Ha K, Cao Z, Dynan WS. Involvement of p54(nrb), a PSF partner protein, in DNA double-strand break repair and radioresistance. Nucleic Acids Res. 2009;37:6746–6753. doi: 10.1093/nar/gkp741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salton M, Lerenthal Y, Wang SY, Chen DJ, Shiloh Y. Involvement of matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle. 2010;9:1568–1576. doi: 10.4161/cc.9.8.11298. [DOI] [PubMed] [Google Scholar]
- 29.Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 30.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 32.Tate EH, Wilder ME, Cram LS, Wharton W. A method for staining 3T3 cell nuclei with propidium iodide in hypotonic solution. Cytometry. 1983;4:211–215. doi: 10.1002/cyto.990040304. [DOI] [PubMed] [Google Scholar]
- 33.Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, Heckel A, van Meel J, Rieder CL, Peters JM. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell. Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 35.Martin SA, McCabe N, Mullarkey M, Cummins R, Burgess DJ, Nakabeppu Y, Oka S, Kay E, Lord CJ, Ashworth A. DNA polymerases as potential therapeutic targets for cancers deficient in the DNA mismatch repair proteins MSH2 or MLH1. Cancer Cell. 2010;17:235–248. doi: 10.1016/j.ccr.2009.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffin CS, Thacker J. The role of homologous recombination repair in the formation of chromosome aberrations. Cytogenet. Genome Res. 2004;104:21–27. doi: 10.1159/000077462. [DOI] [PubMed] [Google Scholar]
- 37.Canudas S, Smith S. Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J. Cell. Biol. 2009;187:165–173. doi: 10.1083/jcb.200903096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bauerschmidt C, Arrichiello C, Burdak-Rothkamm S, Woodcock M, Hill MA, Stevens DL, Rothkamm K. Cohesin promotes the repair of ionizing radiation-induced DNA double-strand breaks in replicated chromatin. Nucleic Acids Res. 2010;38:477–487. doi: 10.1093/nar/gkp976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsiang YH, Liu LF. Identification of mammalian DNA topoisomerase I as an intracellular target of the anticancer drug camptothecin. Cancer Res. 1988;48:1722–1726. [PubMed] [Google Scholar]
- 40.Lisby M, Rothstein R. Choreography of recombination proteins during the DNA damage response. DNA Repair. 2009;8:1068–1076. doi: 10.1016/j.dnarep.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Urban RJ, Bodenburg Y. PTB-associated splicing factor regulates growth factor-stimulated gene expression in mammalian cells. Am. J. Physiol. Endocrinol. Metab. 2002;283:E794–E798. doi: 10.1152/ajpendo.00174.2002. [DOI] [PubMed] [Google Scholar]
- 42.Song X, Sui A, Garen A. Binding of mouse VL30 retrotransposon RNA to PSF protein induces genes repressed by PSF: effects on steroidogenesis and oncogenesis. Proc. Natl Acad. Sci. USA. 2004;101:621–626. doi: 10.1073/pnas.0307794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb. Perspect. Biol. 2010;2:a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gozani O, Patton JG, Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994;13:3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Urban RJ, Bodenburg Y, Kurosky A, Wood TG, Gasic S. Polypyrimidine tract-binding protein-associated splicing factor is a negative regulator of transcriptional activity of the porcine p450scc insulin-like growth factor response element. Mol. Endocrinol. 2000;14:774–782. doi: 10.1210/mend.14.6.0485. [DOI] [PubMed] [Google Scholar]
- 46.Mathur M, Tucker PW, Samuels HH. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol. Cell. Biol. 2001;21:2298–2311. doi: 10.1128/MCB.21.7.2298-2311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Straub T, Knudsen BR, Boege F. PSF/p54(nrb) stimulates “jumping” of DNA topoisomerase I between separate DNA helices. Biochemistry. 2000;39:7552–7558. doi: 10.1021/bi992898e. [DOI] [PubMed] [Google Scholar]
- 48.Lee DH, Pan Y, Kanner S, Sung P, Borowiec JA, Chowdhury D. A PP4 phosphatase complex dephosphorylates RPA2 to facilitate DNA repair via homologous recombination. Nat. Struct. Mol. Biol. 2010;17:365–372. doi: 10.1038/nsmb.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hinz JM, Tebbs RS, Wilson PF, Nham PB, Salazar EP, Nagasawa H, Urbin SS, Bedford JS, Thompson LH. Repression of mutagenesis by Rad51D-mediated homologous recombination. Nucleic Acids Res. 2006;34:1358–1368. doi: 10.1093/nar/gkl020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Helleday T. Pathways for mitotic homologous recombination in mammalian cells Mutat. Res. 2003;532:103–115. doi: 10.1016/j.mrfmmm.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 51.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks Chem. Res. Toxicol. 2006;19:1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rajesh P, Rajesh C, Wyatt MD, Pittman DL. RAD51D protects against MLH1-dependent cytotoxic responses to O(6)-methylguanine. DNA Repair. 2010;9:458–467. doi: 10.1016/j.dnarep.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shav-Tal Y, Zipori D. PSF and p54(nrb)/NonO–multi-functional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- 54.Lowery LA, Rubin J, Sive H. Whitesnake/sfpq is required for cell survival and neuronal development in the zebrafish. Dev. Dyn. 2007;236:1347–1357. doi: 10.1002/dvdy.21132. [DOI] [PubMed] [Google Scholar]
- 55.Couedel C, Mills KD, Barchi M, Shen L, Olshen A, Johnson RD, Nussenzweig A, Essers J, Kanaar R, Li GC, et al. Collaboration of homologous recombination and nonhomologous end-joining factors for the survival and integrity of mice and cells. Genes Dev. 2004;18:1293–1304. doi: 10.1101/gad.1209204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adachi N, So S, Koyama H. Loss of nonhomologous end joining confers camptothecin resistance in DT40 cells. Implications for the repair of topoisomerase I-mediated DNA damage. J. Biol. Chem. 2004;279:37343–37348. doi: 10.1074/jbc.M313910200. [DOI] [PubMed] [Google Scholar]
- 57.Godthelp BC, Wiegant WW, van Duijn-Goedhart A, Scharer OD, van Buul PP, Kanaar R, Zdzienicka MZ. Mammalian Rad51C contributes to DNA cross-link resistance, sister chromatid cohesion and genomic stability. Nucleic Acids Res. 2002;30:2172–2182. doi: 10.1093/nar/30.10.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sjogren C, Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr. Biol. 2001;11:991–995. doi: 10.1016/s0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 59.Strom L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjogren C. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science. 2007;317:242–245. doi: 10.1126/science.1140649. [DOI] [PubMed] [Google Scholar]
- 60.Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Badie S, Liao C, Thanasoula M, Barber P, Hill MA, Tarsounas M. RAD51C facilitates checkpoint signaling by promoting CHK2 phosphorylation. J. Cell. Biol. 2009;185:587–600. doi: 10.1083/jcb.200811079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katsura M, Tsuruga T, Date O, Yoshihara T, Ishida M, Tomoda Y, Okajima M, Takaku M, Kurumizaka H, Kinomura A, et al. The ATR-Chk1 pathway plays a role in the generation of centrosome aberrations induced by Rad51C dysfunction. Nucleic Acids Res. 2009;37:3959–3968. doi: 10.1093/nar/gkp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 64.Zeidler M, Varambally S, Cao Q, Chinnaiyan AM, Ferguson DO, Merajver SD, Kleer CG. The Polycomb group protein EZH2 impairs DNA repair in breast epithelial cells. Neoplasia. 2005;7:1011–1019. doi: 10.1593/neo.05472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Herrmann G, Kais S, Hoffbauer J, Shah-Hosseini K, Bruggenolte N, Schober H, Fasi M, Schar P. Conserved interactions of the splicing factor Ntr1/Spp382 with proteins involved in DNA double-strand break repair and telomere metabolism. Nucleic Acids Res. 2007;35:2321–2332. doi: 10.1093/nar/gkm127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Horikoshi N, Morozumi Y, Takaku M, Takizawa Y, Kurumizaka H. Holliday junction-binding activity of human SPF45. Genes Cells. 2010;15:373–383. doi: 10.1111/j.1365-2443.2010.01383.x. [DOI] [PubMed] [Google Scholar]
- 67.Takizawa Y, Qing Y, Takaku M, Ishida T, Morozumi Y, Tsujita T, Kogame T, Hirota K, Takahashi M, Shibata T, et al. GEMIN2 promotes accumulation of RAD51 at double-strand breaks in homologous recombination. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq271. doi:10.1093/nar/gkq271 [Epub ahead of print 19 April 2010] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gottipati P, Helleday T. Transcription-associated recombination in eukaryotes: link between transcription, replication and recombination. Mutagenesis. 2009;24:203–210. doi: 10.1093/mutage/gen072. [DOI] [PubMed] [Google Scholar]
- 69.Doksani Y, Bermejo R, Fiorani S, Haber JE, Foiani M. Replicon dynamics, dormant origin firing, and terminal fork integrity after double-strand break formation. Cell. 2009;137:247–258. doi: 10.1016/j.cell.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaelin WGJ. The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 71.Mukhopadhyay A, Elattar A, Cerbinskaite A, Wilkinson SJ, Drew Y, Kyle S, Los G, Hostomsky Z, Edmondson RJ, Curtin NJ. Development of a functional assay for homologous recombination status in primary cultures of epithelial ovarian tumor and correlation with sensitivity to poly(ADP-ribose) polymerase inhibitors. Clin. Cancer Res. 2010;16:2344–2351. doi: 10.1158/1078-0432.CCR-09-2758. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.