Abstract
Genome organization into transcriptionally active domains denotes one of the first levels of gene expression regulation. Although the chromatin domain concept is generally accepted, only little is known on how domain organization impacts the regulation of differential gene expression. Insulators might hold answers to address this issue as they delimit and organize chromatin domains. We have previously identified a CTCF-dependent insulator with enhancer-blocking activity embedded in the 5′ non-coding region of the chicken α-globin domain. Here, we demonstrate that this element, called the αEHS-1.4 insulator, protects a transgene against chromosomal position effects in stably transfected cell lines and transgenic mice. We found that this insulator can create a regulated chromatin environment that coincides with the onset of adult α-globin gene expression. Furthermore, such activity is in part dependent on the in vivo regulated occupancy of CTCF at the αEHS-1.4 element. Insulator function is also regulated by CTCF poly(ADP-ribosyl)ation. Our results suggest that the αEHS-1.4 insulator contributes in organizing the chromatin structure of the α-globin gene domain and prevents activation of adult α-globin gene expression at the erythroblast stage via CTCF.
INTRODUCTION
Differential gene expression in genomic domains is regulated at multiple levels. Not only does it require chromatin structure remodeling at local regulatory elements, but also the establishment of a chromatin architecture defining the domain itself. Consequently, more than proximal and distal regulatory elements are required for a gene’s coordinated transcription in order to first demarcate the genomic region in which it is located. Once the chromatin domain is delimited and organized, local regulatory actions take place to achieve expression of specific genes (1).
Insulators have been considered as regulatory elements that act as boundaries to the action of external enhancers and repressive chromatin and participate in the formation and maintenance of chromatin domains (1,2). Accumulated evidence suggests that insulators, particularly CTCF-linked insulators, do not act exclusively as boundary elements (3). For example, CTCF in coordination with cohesins, mediate the formation of chromatin loops favoring optimal spatial organization of gene domains (4,5). This vision is further supported by the presence of insulator bodies that are important for domain formation at the nuclear periphery of Drosophila cells. Insulator bodies are formed of polymorphic protein complexes including dCTCF, Su(Hw) and CP190, besides other proteins (6). Interestingly, such insulator-mediated topological organization of the genome is dynamic. For instance, CTCF and cohesin mediate chromosomal cis contacts in a regulated manner in several loci. Such loci include the Igf2/H19 imprinted locus (7,8), the cytokine locus including IL4, IL5, IL13 and IFNG (9), the mouse β-globin locus, in which the spatial organization of regulatory components to form an active chromatin hub is facilitated by CTCF during tissue-specific activation of globin genes (10), the MHC class II genes (11) and more recently, the β-globin locus, which interacts with flanking olfactory receptor genes on the human chromosome 11 (5). Notoriously, such interactions are not exclusively intrachromosomal, CTCF also mediates interchromosomal contacts between one allele of the Igf2/H19 imprinting control region on chromosome 7 and one allele of the Wsb1/Nf1 imprinted locus on chromosome 11 (12). Insulator action through CTCF enhancer blocking activity can also be regulated by other mechanisms as by thyroid hormone (13) and post-translational modifications as poly(ADP-ribosyl)ation (14) among others. Thus, CTCF is implicated in diverse regulatory and structural functions (3).
We previously described a regulatory element located in the 5′ non-coding region of the chicken α-globin domain, ∼14-kb upstream of the embryonic π gene (Figure 1A) (15). This element, named αEHS-1.4, which corresponds to a DNA fragment of 1.4 kb is an insulator with CTCF-dependent enhancer-blocking activity (15). This insulator is located within an intron of the C16orf35 gene antisense transcript (16,17), between the distal locus control region known as the α-major regulatory element (α-MRE), and the α-globin genes (Figure 1A). Since the αEHS-1.4 is located 3′ from the α-MRE it seemed unlikely that the αEHS-1.4 insulator element acts as a boundary for the 5′-side of the α-globin domain, shielding it from silent chromatin expansion or from the action of enhancers outside the domain. Consequently, we addressed the contribution of this element to the chromatin configuration of the α-globin domain and for regulated gene expression during erythropoiesis and development.
Figure 1.
Protection against position effects by the αEHS-1.4 insulator. (A) Scheme of the α-globin gene domain. The α-MRE constitutes the putative locus control region of the domain. The αEHS-1.4 insulator is located ∼14-kb upstream of the embryonic π globin gene. CpG, represents a CpG-island located ∼4-kb upstream of the embryonic π globin gene which represents the first CTCF-binding site identified in vivo, that corresponds to the promoter of the antisense transcript C16orf35 (47). Vertical arrows indicate the position of DNase I hypersensitive sites and the 3′-side enhancer (21). (B) Flow cytometry histograms (FACS) showing the GFP fluorescence level of five representative and independent stably transfected cell lines containing the control vector without insulators at days 0, 30 and 100 of continuous cell culture. (C) Fluorescence of independent cell lines incorporating the insulated plasmid with the chicken 5′cHS4 insulator of the β-globin gene locus. (D) Fluorescence of independent cell lines containing the plasmid flanked on each side of the transgene by the αEHS-1.4 insulator. (E) Chart showing the mean percentage of GFP expressing cells (±SD) at different time points during culture for the three different transgenes (pGαD3 n = 15, pGαD 5′cHS4 n = 10 and pGαD EHS-1.4 n = 10). sc = single-copy and mc = multi-copy integrants. ***P ≤ 0.001 with respect to the pGαD3 (100 days) control (ANOVA).
Here, we demonstrate that the αEHS-1.4 element shields a transgene from chromosomal position effects (CPE) and gene silencing in stably transfected cell lines and transgenic mice. In contrast to the chicken 5′ cHS4 β-globin insulator (18), the protective function of the αEHS-1.4 insulator is CTCF-dependent. Moreover, this element mediates the chromatin conformation of the 5′ non-coding region of the domain during developmentally regulated gene expression. Subsequently, we demonstrate that prior to erythroid differentiation, the αEHS-1.4 insulator is bound by CTCF and prevents adult α-globin gene expression, and, as differentiation proceeds, CTCF is deployed and the blockage is released allowing the activation of the adult α-globin genes in fully developed erythroid cells. Our results provide insight into the function of insulators as organizers of chromatin domains and regulators of differential gene expression during cell differentiation and development.
MATERIALS AND METHODS
Culture of stable cell lines
Avian arrested cell lines were maintained at 37°C, 5% CO2 with the following corresponding media: 6C2 (pre-eritroblasts) cells with α-MEM (GIBCO) supplemented with 10% (v/v) fetal bovine serum (FBS) (Multicell), 2% (v/v) chicken serum (CS), 1 mM HEPES (pH 7.2) (SIGMA) and 50 mM β-mercaptoethanol. The avian erythroblastosis virus-transformed chicken erythroblast cell line HD3 and the chicken lymphoid cell-line DT40 were cultured in DMEM (GIBCO) supplemented with 8% FBS and 2% CS (19,20). All media contain penicillin/streptomycin (500 U/ml) (GIBCO). To differentiate HD3 cells they were cultured with the same media with 2% anemic CS at 42°C for 7–15 days, as described (21,22).
Protection against chromosomal position effects assay
The protection against CPE assay was performed as previously described (18,23). Stable transfection of the different plasmids was performed with Lipofectamine (Invitrogen) following manufacturer instructions. The transfected plasmids were pGαD, containing the EGFP under the chicken αD gene promoter, pGαDHS4 (containing the β-globin 5′cHS4 insulator on each side of the αD-EGFP) and pGαD1.4 (containing the αEHS-1.4 insulator on each side of the αD-EGFP). Individual integrants were obtained by using a semisolid media (Methocel Fluka, Buchs, Switzerland) with neomycin at 0.9 mg/ml (18). After 10–14 days of selection, individual clones were isolated and their expression was determined by FACS (at Day 0). Copy number and transgene integrity was evaluated by Southern blot analysis using a DNA fragment of the EGFP as a probe. Transgene expression was evaluated twice a month until completion of at least 100 days of continuous cell culture without neomycin. The mutant αEHS1.4ΔΔ construct was obtained by nested PCR using primers containing the ΔCAG and ΔCTAG base changes at the CTCF-binding site defined previously (15) and then using primers to amplify the complete 1.4-kb DNA fragment (all primers are available upon request).
Generation of transgenic mice
The plasmid pTYR5-1.4 containing the αEHS-1.4 element was cloned in a standard 5′–3′ orientation, at the 5′ end in the unique Xba I site of the Tyr (tyrosinase)-reporter construct ptrTYR5 (24); it was digested with Ecl XI and Sal I enzymes, thereby releasing the 13-kb DNA band with the insert that was purified following described standard procedures (25). In parallel, the corresponding Ecl XI-Sal I 11.6-kb DNA band from ptrTYR5, without the insulator element, was also purified and used as the reference control for transgenesis. The resulting DNA fragments and transgenes were named as: TYR5 and TYR5–αEHS-1.4, respectively. For the generation of transgenic mice, suitable DNA solutions from each of the two transgenes were microinjected into the pronucleus of albino outbreed NMRI/Hsd fertilized mouse oocytes (Harlan Interfauna Iberica SL, Barcelona, Spain) using standard procedures (25) . Transgenic founder mice were detected at birth due to obvious pigmentation (24) and confirmed by PCR analysis, using tyrosinase minigene-specific primers, as described before (24) and also by Southern blot analysis, using the pmTyrE5 tyrosinase probe that reveals polymorphic Hind III DNA bands between the transgene (3.4 kb) and the endogenous tyrosinase locus (2.2 kb), as reported (26). Transgenic mouse lines were established and maintained, as hemizygous, in the albino outbreed NMRI/Hsd genetic background. Transgene copy-number was determined from the DNA of F1 individuals by Southern blot analysis, as reported before (26). For the TYR5–αEHS-1.4 construct, 289 fertilized oocytes were microinjected, 192 embryos were transferred to pseudopregnant females and resulted in the birth of 17 pups, out of which five independent transgenic mouse lines were established. For the TYR5 transgene, 581 fertilized oocytes were microinjected, 284 embryos were transferred and gave rise to 17 newborns, out of which three were transgenic. Subsequently, two of these mouse lines were found to carry multiple transgene insertions that could be segregated in the F1 generation, thus finally resulting in a total of six independent transgenic mouse lines with the TYR5 construct. Representative F1 individuals from each transgenic mouse line were briefly anesthetized with 1 ml/100 g of a mixture of Ketamine (10 mg/ml) and Xylazine (2 mg/ml) for obtaining the arranged pictures (Figure 2A). All the procedures that required the use of animals complied with Spanish and European legislation concerning vivisection and the use of genetically modified organisms, and the protocols were approved by the local ethics committees on animal experimentation.
Figure 2.
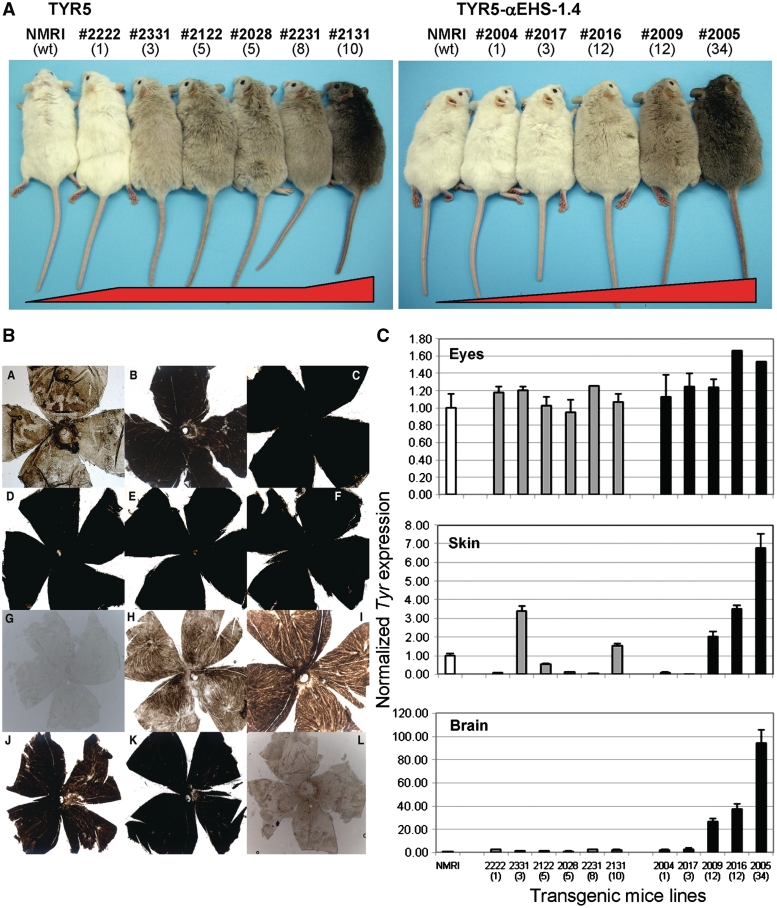
Analysis of TYR5 versusTYR5–αEHS-1.4 transgenic mice. (A) Coat color of transgenic mice. Representative F1 individuals from TYR5 (left) and TYR5–αEHS-1.4 (right) transgenic mice are shown alongside non-transgenic albino NMRI littermates (wt, for wild-type), used as the recipient mouse strain for transgenesis. The number of the independent transgenic line and number of integrated transgene copies (in brackets) are shown on the top. (B) Retinas of TYR5 and TYR5–αEHS-1.4 transgenic mice. Whole-mount retinas showing pigmentation levels due to the retinal pigment epithelium and the overlaying choroid with melanocytes, from adult TYR5 (panels A to F: A, line #2222; B, line #2332; C, line #2122; D, line #2028; E, line #2231; F, line #2131) and TYR5–αEHS1.4 (panels G to K: G, line #2004; H, line #2017; I, line #2016; J, line #2009; K, line #2005) transgenic mice. A retina from a non-transgenic albino NMRI littermate is included for reference (panel L). (C) Analysis of tyrosinase gene expression. Tyrosinase (Tyr) gene expression was measured by quantitative RT–PCR using specific TaqMan and reference (Tbp) probes in all transgenic mouse lines carrying the TYR5 (grey bars) and the TYR5–αEHS-1.4 (solid black bars) construct in total RNA isolated from three different tissues: eyes (top chart), dorsal skin (middle), and whole brain (bottom). Two F1 or F2 adult representative animals from each transgenic mouse line were used for measurements. Results are shown as averages ± SD. The number of each of the transgenic mouse lines and their corresponding transgene copy-number (in brackets) is indicated below each bar. A non-transgenic albino NMRI littermate (white bar) was used as a control for all tissues.
Analysis of transgenic mice
Total RNA was prepared from whole brain, dorsal skin and eyes from at least three adult F1 or F2 individuals from each of the transgenic mouse lines carrying the TYR5 and TYR5–αEHS-1.4 constructs, using the RNeasy Kit (Qiagen). Tyrosinase (Tyr) expression was assessed by quantitative RT–PCR using specific mouse TaqMan (Applied Biosystems) Tyr (Mm00495817_m1) and Tbp (Mm00446973_m1, included as a reference) probes, on an ABI-Prism 7000 (Applied Biosystems), using TaqMan Universal PCR Master Mix (Applied Biosystems), as described before (27). Whole-mount retinas from adult F1 or F2 individuals from each of the transgenic mouse lines carrying the TYR5 and TYR5–αEHS-1.4 constructs were prepared according to previously described methods (27,28). Quantification of the total melanin contents from eye and skin extracts was carried out from at least two adult F1 or F2 individuals from each of the transgenic mouse lines carrying the TYR5 and TYR5–αEHS-1.4 constructs, according to reported protocols (28).
Chromatin immunoprecipitation
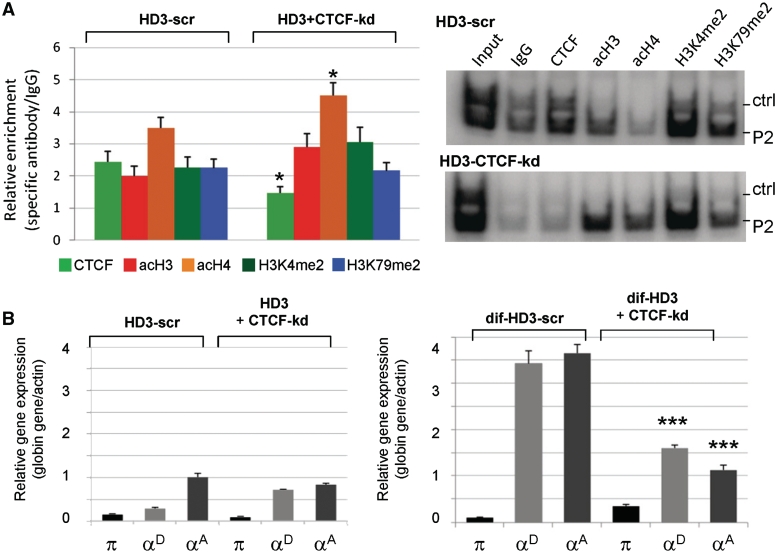
Chromatin immunoprecipitation (ChIP) analysis were performed and analyzed as described (29). Duplex PCR was used for evaluating histone marks and CTCF enrichment at different points in the αEHS-1.4 insulator region covering the CTCF-binding site (see scheme in Figure 4), the αEHS-1.4 CTCF-binding site in the transgene and a DNA fragment of EGFP. As controls we used regions in which the analyzed histone covalent modification or nuclear factor is absent; for permissive chromatin histone marks and CTCF we used two heterochromatin-associated sequences located 5′ of the chicken β-globin gene domain (30), for the repressive histone marks we used the β-globin 5′ cHS4 insulator sequence that is enriched with permissive marks in all cell types used (30). Enrichment was calculated by using the following equation: Ab test (test region/control region)/IgG (test region/control region). The antibodies employed were the following: antibodies against acetylated histone H3 and acetylated histone H4 (06-599 and 06-866) were obtained from Upstate Biotechnology (Lake Placid, NY). The antibody against H3K4me2 (ab7766) was purchased from Abcam (Cambridge, MA). The antibody against H3K79me2 was kindly provided by Fred van Leeuwen (Nederland Kanker Instituut–Antoni van Leeuwenhoek Ziekenhuis, Amsterdam, The Netherlands). Antibodies against the trimethylated versions of histone 3 on K9 and histone 4 on K20 were kindly provided by Thomas Jenuwein (Max Planck Institute of Immunology, Freiburg, Germany). The antibody against chicken CTCF [anti-cCTCF(86–233)] was previously obtained as described (15).
Figure 4.
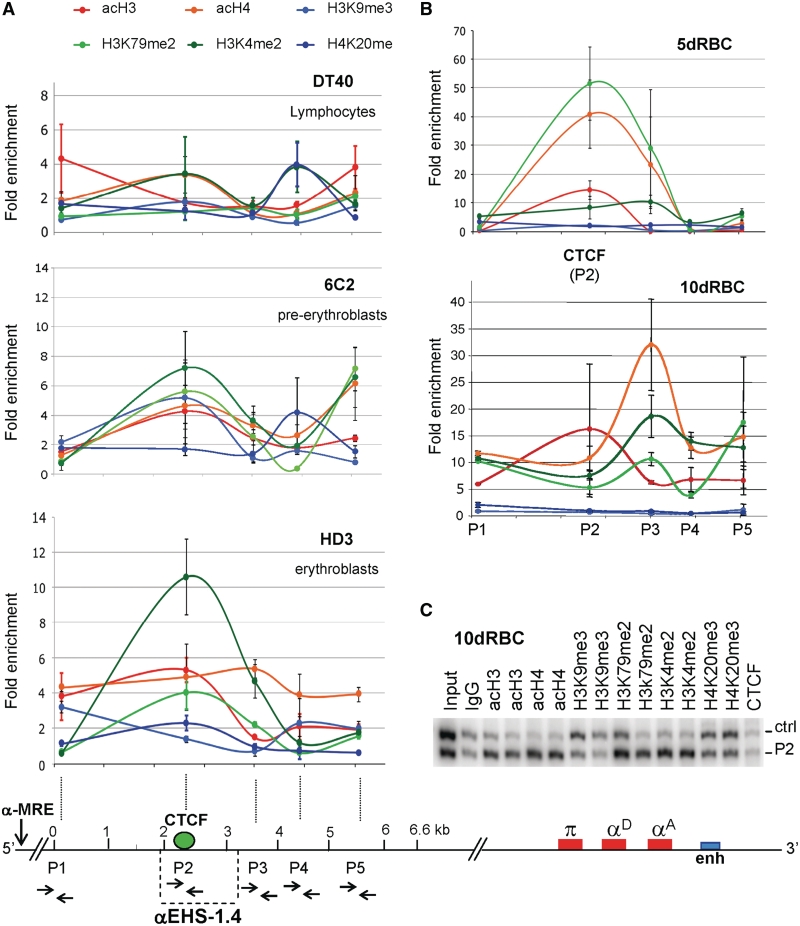
Distribution of histone post-translational modifications over the 6.6-kb genomic region containing the αEHS-1.4 insulator. (A) Histone covalent modifications were evaluated semi-quantitatively using duplex-PCR with primers P1–P5 (primers locations are shown in the bottom scheme) in DT40 (lymphoid), 6C2 (pre-erythroblasts) and HD3 (erythroblasts) cells. (B) ChIP assays as in (A) using 5d and 10dRBCs. (C) Representative gel of the duplex-PCR amplification using primers P2 located over the CTCF-binding site with its corresponding control (Ctrl). The ChIP was performed on 10-day-old embryonic erythrocytes (10dRBC). All the histone modification enrichments are statistically significant (ANOVA with P ≤ 0.001) with respect to the control IgG except for the acH3 enrichment. Results are shown as averages (±standard error).
CTCF knock down and 3-aminobenzamide treatment
For the CTCF knock down series of experiments, three different sets of small hairpin RNA interference (shRNAi) sequences against CTCF were tested with overall similar results. To generate the CTCF knock down cells we used a collection of four plasmids containing different sequences to generate shRNAi against the 5′ region of chicken CTCF mRNA. Three of them were designed from the reported sequences in (31) (pSiLefevre1, 2 and 3) and one was kindly provided by Ishihara (pCTI). Even though the pCT1 shRNAi was designed against human CTCF (32), it reduced CTCF levels in the chicken cell lines. Plasmids containing the hairpin DNAs were stably transfected as described previously, and integrants were selected with 1 µg/ml puromicin (SIGMA) for 7–15 days. CTCF reduction was assessed by western blot analysis (Supplementary Figure S4A). For 3-aminobenzamide (3-ABA) treatment, 8 mM 3-ABA was added to the culture media for 3–6 days prior to evaluation by western blotting and ChIP analysis, as described (14). The CTCF antibody used to perform the western blot (15) recognizes both the 130- and 180-kDa forms of CTCF as shown in Supplementary Figure S4A.
Statistics
Two-tailed analysis of variance (ANOVA) or multivariate analysis of variance (MANOVA) statistical analyses were performed using SPSS program (version 18) considering as significant: *P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001. Scheffe or Games-Howell post hoc tests were used when appropriate to distinguish which components analyzed are causing the significant variance between conditions.
RESULTS
The αEHS-1.4 insulator protects against chromosomal position effects in stably transfected cell lines
To test if the αEHS-1.4 insulator located 14-kb upstream of the embryonic π gene of the α-globin locus protects a trangene against chromosomal position effects, we generated stably transfected HD3 erythroblasts expressing a GFP reporter driven by the chicken αD gene promoter (αD-GFP) (Figure 1B) (29), or αD-GFP flanked on each side by the αEHS-1.4 insulator (Figure 1D). The αD-GFP flanked on each side by the 5′cHS4 core insulator of the β-globin locus was used as control (18). Transgene integrity and copy number (single- or multi-copy) was determined by Southern blot for a selected set of stable lines (data not shown). We measured GFP fluorescence by flow cytometry at different time points to assess transgene expression (18,23). As expected, the αD-GFP plasmid (without insulators) showed sensitivity to CPE, as transgene expression was extinct even before day 100 of continuous cell culture (Figure 1B). In contrast, the αEHS-1.4 insulator robustly protected against CPE even after 100 days of continuous cell culture (Figure 1D). This protective activity was comparable to that exerted by the chicken β-globin 5′cHS4 insulator (Figure 1C–E). Next, we examined the profile of histone post-translational modifications present over the GFP transgene in distinct cell lines by ChIP. Modifications corresponding to a permissive chromatin environment were present only in insulated transgenes (Supplementary Figure S1). Thus, the αEHS-1.4 insulator efficiently protects against CPE in chicken HD3 erythroblasts cells thereby favoring a transcriptionally permissive chromatin environment.
The αEHS-1.4 insulator protects against chromosomal position effects in transgenic mice
To test the functionality of the αEHS-1.4 element in vivo we addressed its role in preventing chromosomal position effects in transgenic mice. We used a reporter system in which the albino phenotype is rescued in transgenic mice expressing a functional tyrosinase gene (Tyr) (24), and which has been useful to validate the effect of the chicken 5′ cHS4 β-globin insulator in protecting transgenes from CPE (33). The αEHS-1.4 insulator was cloned as a single copy at the 5′ end of Tyr in the ptrTYR5 construct (TYR5–αEHS-1.4). We cloned the αEHS-1.4 sequence only at the 5′ end of the vector since constructs usually integrate as multi-copy tandem arrays; therefore, most transgene copies would end up shielded by αEHS-1.4 elements (25). The parental plasmid, ptrTYR5, here called TYR5, was used as a reference.
Five independent transgenic mouse lines carrying the insulated TYR5–αEHS-1.4 construct (lines #2004, #2017, #2016, #2009 and #2005), and six carrying the control TYR5 construct (lines #2222, #2331, #2122, #2028, #2231 and #2131) where generated in the albino outbreed strain NMRI/Hsd (Figure 2A). Transgene integrity and copy number was assessed by Southern blot analysis (data not shown). As anticipated, insertion of either construct rendered pigmented transgenic mice (Figure 2A). In all cases, pigmentation was uniform, without obvious signs of mosaicism [that were obvious in similar transgenic mice reported by Potts et al. (33)]. Mouse lines with higher number of TYR5 copies [i.e. #2131 (10 copies)] showed increased pigmentation, as compared to lines with fewer transgene copies [i.e. #2222 (one copy)]. However, there was little coat color variation in the remaining TYR5 transgenic lines with intermediate transgene copy-number (Figure 2A, lines #2331, #2122, #2028 and #2231). In contrast, the five transgenic mouse lines generated with the TYR5–αEHS-1.4 transgene, produced individuals with progressively darker pigmentation, mostly correlating with the corresponding transgene copy-numbers. Only one line, #2004 (one copy), showed no detectable pigmentation in skin and/or eyes (Figure 2A).
To better assess Tyr expression in transgenic mice we carried out three additional experiments. First, we prepared whole-mount retinas from representative F1 individuals from TYR5 and TYR5–αEHS-1.4 transgenic lines to directly visualize the retinal pigment epithelium cells and melanocytes present in the choroid. Pigment saturation was readily observed in retinas from TYR5 transgenic lines, without any obvious correlation with transgene copy number (Figure 2B and A–F). In contrast, progressively higher pigmentation correlated with transgene copy number in retinas from TYR5–αEHS-1.4 transgenic lines similar to coat color pigmentation (Figure 2B and G–K).
Second, we measured the Tyr gene expression by quantitative RT–PCR. Tyr expression was assessed in eyes and skin, where Tyr is normally expressed; and brain, where Tyr is not expressed (27). Tyr expression in TYR5 transgenic mouse lines was relatively uniform in the eye, erratic in the skin, and absent in the brain (Figure 2C), and did not correlate with transgene copy number. In contrast, Tyr expression in TYR5–αEHS-1.4 transgenic lines was relatively proportional to transgene copy number in the eye, while it fully correlated with transgene copy number in the skin and, surprisingly, in the brain.
Finally, we quantified the total melanin contents in the eyes and skin of all transgenic lines (Supplementary Figure S2). Eye melanin contents of TYR5 transgenic mice were relatively uniform whereas in TYR5–αEHS-1.4 mice melanin contents correlated with transgene copy number. Likewise, melanin contents in the skin of TYR5 transgenic individuals were highly variable. In contrast, melanin contents in the dorsal skin of TYR5–αEHS-1.4 transgenic mice clearly correlated with transgene copy number. In conclusion, the αEHS-1.4 insulator protects a transgene against chromosomal position effects and progressive silencing not only in stable cell lines but also in transgenic mice.
CTCF-binding site is indispensable for αEHS-1.4 insulator activity
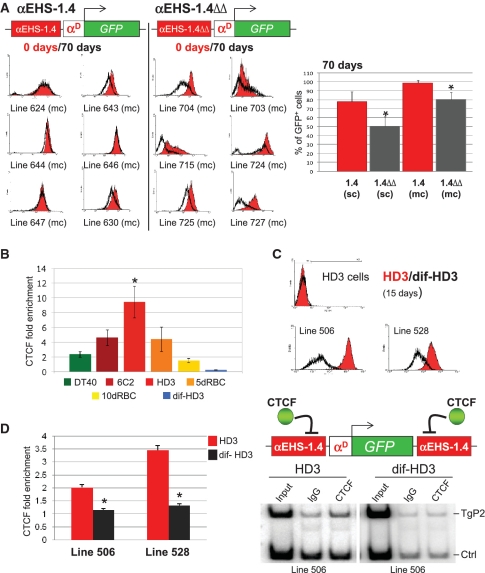
In order to address whether CTCF contributes to the protection against CPE by the αEHS-1.4 insulator, we introduced two point mutations that abolish CTCF binding to this element (15) to generate the αEHS-1.4ΔΔ insulator and assessed its protective function against CPE in stably transfected HD3 cells by flow cytometry. A very high proportion of cells from lines in which GFP was flanked on each side by the αEHS-1.4 insulator showed high levels of fluorescence even after 70 days of continuous culture. In contrast, fluorescence levels decreased in an important proportion of cells carrying the mutant insulator αEHS-1.4ΔΔ after 70 days (Figure 3A). These results are reflected in the percentages of GFP-expressing cells in selected stably transfected lines (Figure 3A). Interestingly, the protection against CPE by a single copy of the intact, or the mutant αEHS-1.4, was less efficient than by multiple copies, suggesting that optimal insulation is reached only when the αEHS-1.4 sequence flanks the transgene on each side (Figure 3A, compare single- and multi-copy cell lines). Thus, the mutant αEHS-1.4ΔΔ insulator protects against CPE less efficiently than the αEHS-1.4 insulator, suggesting that, unlike the chicken 5′ cHS4 β-globin insulator (18), CTCF binding is required for the insulator function of αEHS-1.4.
Figure 3.
CTCF is necessary for αEHS-1.4 insulator function and its in vivo occupancy varies during erythroid differentiation. (A) Transgene expression analysis by FACS at day 0 (red) and 70 (grey) in cultured cell lines with the respective transgene integrated in multi-copy (αEHS-1.4 and mutant αEHS-1.4ΔΔ insulator were placed only at the 5′-side of the transgene). Quantification of the average (±standard error) of GFP+ cells at day 0 and 70 of culture of the set of single- and multi-copy clones. αEHS-1.4 (sc) n = 16; αEHS-1.4ΔΔ (sc) n = 22; αEHS-1.4 (mc) n = 18 and αEHS-1.4ΔΔ (mc) n = 22. *P ≤ 0.05 with respect to the αEHS-1.4 (ANOVA). (B) CTCF in vivo binding at the αEHS-1.4 insulator sequence was estimated by semi-quantitative ChIP assays (29). We observed an enrichment peak of CTCF in HD3 cells (erythroblasts), and as differentiation proceeds (dif-HD3), CTCF dissociates from the insulator. *P ≤ 0.05. The number of independent ChIP assays per cell line is: DT40 n = 3, 6C2 n = 5, HD3 n = 4, 5dRBC n = 3, 10dRBC n = 3 and dif-HD3 n = 4. (C) Protection against CPE in two representative differentiated HD3 cell lines containing the plasmid insulated with the αEHS-1.4 element (the same experiment was performed using the β-globin 5′ cHS4 insulator, see Supplementary Figure S3B). (D) ChIP using an antibody against CTCF and primers recognizing the transgene. CTCF binds to the transgene in HD3 cells and is dissociated in differentiated HD3 cells mimicking what happens at the endogenous context (see B), consistent with a loss of insulator function in the differentiated clones. A representative gel of the radioactive duplex-PCR is shown. Results are shown as averages (±standard error). *P ≤ 0.05 with respect to the lines 506 or 528 (ANOVA).
The activity of the αEHS-1.4 insulator correlates with CTCF occupancy during erythroid differentiation
Since the α-MRE is located further upstream of the αEHS-1.4 in the α-globin locus, it seems unlikely that the αEHS-1.4 acts as the 5′ boundary limiting the α-globin chromatin domain (Figure 1A). Therefore, we asked if the αEHS-1.4 element regulates α-globin gene expression and if it does so via CTCF. For these purpose we first addressed the CTCF binding to the endogenous αEHS-1.4 during erythroid differentiation and chicken development by semi-quantitative ChIP. Interestingly, the occupancy of the αEHS-1.4 insulator by CTCF varied depending on the differentiation stage analyzed. CTCF abundance peaked at the pre-erythroblasts stage, represented by HD3 cells, and, it drastically diminished as differentiation proceeds into a mature erythrocyte, represented by red blood cells from 10-day-old chicken embryos (10dRBCs), or differentiated HD3 cells (Figure 3B). Western blots showed similar CTCF total protein levels in undifferentiated and differentiated HD3 cells (Supplementary Figure S4A, right panel), suggesting regulated binding of CTCF to the αEHS-1.4 element. In contrast to the αEHS-1.4 insulator, the CTCF’s highest enrichment on the chicken 5′ cHS4 β-globin insulator was detected in 10dRBCs (Supplementary Figure S3), suggesting a different mode of action of CTCF for each of these elements. Next, we analyzed the protective function of the αEHS-1.4 insulator against CPE in undifferentiated cells (HD3) and terminally differentiated erythroblasts (dif-HD3). Two HD3 cell lines carrying the αD-GFP transgene shielded by the αEHS-1.4 (lines 506 and 528) were differentiated for 15 days (21), and GFP fluorescence was measured by flow cytometry. GFP fluorescence of the differentiated cells reproducibly decreased as compared to their undifferentiated counterpart (Figure 3C). This correlates with decreased occupancy of the endogenous αEHS-1.4 insulator by CTCF in 10dRBCs (Figure 3B), and of the transgene incorporated in differentiated HD3 cells (Figure 3D). In contrast, differentiated HD3 lines carrying the 5′cHS4 β-globin insulator did not show a significant fluorescence decrease (Supplementary Figure S3B). Therefore, the decreased protective effect of the αEHS-1.4 against CPE might be due to decreased CTCF binding.
Reduction of CTCF protein levels and inhibition of PARP-1 affects insulator function
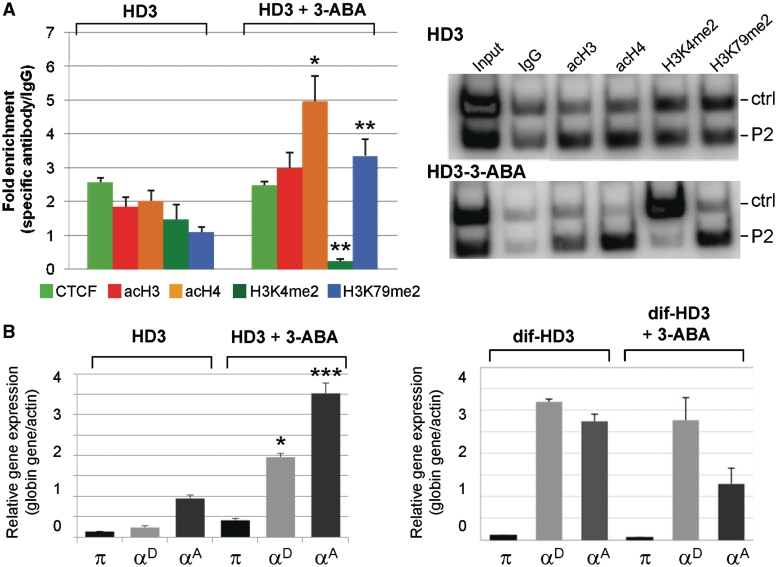
Based on the data supporting dynamic CTCF binding at the αEHS-1.4 insulator during differentiation, we decided to knock down CTCF expression and inhibit its potential poly(ADP-ribosyl)ation (Supplementary Figure S4). First, we generated a series of cell lines in which CTCF was stably knocked down by transfection of three different sets of small hairpin RNAs. Decreased CTCF protein was confirmed by western blot (Supplementary Figure S4A, left panel). The reduction of CTCF decreased the protective effect of the αEHS-1.4 insulator against CPE (Supplementary Figure S4B, left panel). ChIP assays confirmed decreased CTCF at the αEHS-1.4 insulator upon CTCF knock down (Supplementary Figure S4C). Poly(ADP-ribosyl)ation has been shown to be needed for CTCF insulator function (14,34–37), therefore we addressed whether inactivation of CTCF by inhibition of the poly(ADP) ribose polymerase-1 (PARP-1) impacts on the protection against CPE conferred by the αEHS-1.4 insulator. PARP-1 post-translationally modifies CTCF by adding ADP polymers to its N-terminal domain, resulting in two electrophoretic forms, one migrating at 130 kDa, which contains few ADP-ribose residues, and the other one migrating at 180 kDa, which contains more than 20 ADP-ribose residues (36). Treatment of the HD3 transgenic cell lines 506 and 528, which carry GFP flanked on each side by the αEHS-1.4 insulator, with 3-ABA, an inhibitor of PARP-1 (36), resulted in a diminishment of transgene protection against CPE along with a decrease of the 180 kDa form of CTCF (Supplementary Figure S4A, right panel). Interestingly, inhibition of poly(ADP-ribosyl)ation did not affect CTCF binding to the αEHS-1.4 insulator, as shown by ChIP (Figure 6A), implying that, while affecting its function on the αEHS-1.4 insulator, inhibition of CTCF PARylation does not affect its DNA-binding properties.
Figure 6.
Poly(ADP-ribose) polymerase-1 inhibitor affects histone modifications associated with the αEHS-1.4 insulator and α-globin genes expression. (A) ChIP analysis of histone covalent modifications in HD3 cells (HD3) and cells incubated with the PARP-1 inhibitor (HD3+3-ABA) using P2 primers (Figure 4). Each ChIP was performed in triplicate. Representative radioactive duplex PCR gels for HD3 cells and HD3 cells incubated with the 3-ABA inhibitor are shown. (B) Semi-quantitative duplex RT–PCR of α-globin genes expression in HD3 cells and HD3 cells treated with 8 mM of 3-ABA (left graph). The same experiment was performed in differentiated HD3 cells (dif-HD3) and differentiated HD3 cells incubated with 3-ABA (dif-HD3+3-ABA, right panel). The results are representative of nine independent PCR reactions from two independent total RNA purification and reverse transcriptase reactions. *P ≤ 0.05 (MANOVA), **P≤ 0.01 (MANOVA) and ***P ≤ 0.001 (MANOVA), with respect to the corresponding histone covalent modification or expressed gene in HD3 cells. Results are shown as averages (±standard error).
Taken together, these results further support the notion that activity of the αEHS-1.4 insulator is CTCF-dependent and that such activity can be, at least in part, modulated by post-translational modification.
The genomic region encompassing the αEHS-1.4 insulator progressively structures as transcriptionally permissive chromatin during erythroid differentiation
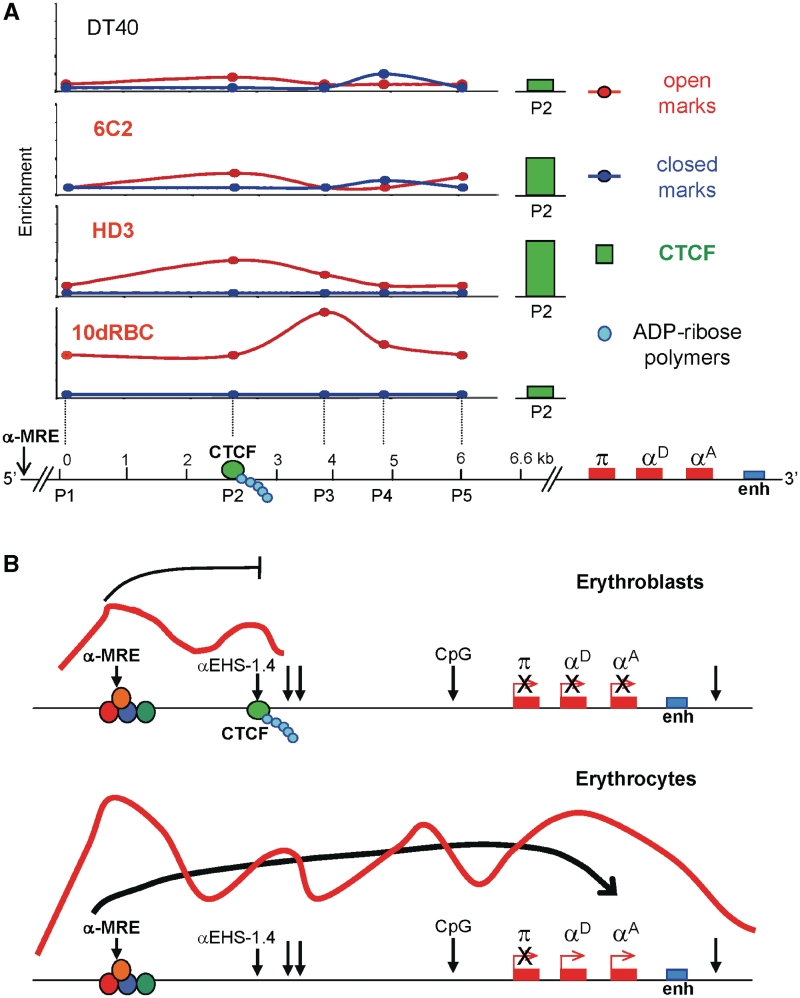
As a first approach to understand the function of αEHS-1.4 element on the chromatin organization of the α-globin domain, we characterized the chromatin structure of a 6.6-kb genomic region encompassing the αEHS-1.4 element during erythroid differentiation and development (Figure 4A). We performed ChIP assays for several histone post-translational modifications corresponding to transcriptionally permissive (acH3, acH4, H3K4me2 and H3K79me2) and repressive (H3K9me3 and H4K20me3) chromatin conformations. We found that the enrichment in histone modifications along the targeted genomic region varies during adult erythroid differentiation (Figure 4A). In 6C2 cells, which represent a pre-erythroblast differentiation stage, a modest enrichment in acetylated histones, as well as H3K4me2 and H3K79me2 was observed over the αEHS-1.4 insulator (Figure 4A). As differentiation proceeds into an erythroblast stage represented by HD3 cells, there was a notorious peak of permissive histone marks overlapping with the CTCF-binding sequence at the αEHS-1.4 element (Figure 4A). Importantly, α-globin genes are not transcribed in 6C2 or HD3 cells (21). Thus, these results suggest that the αEHS-1.4 element may contribute to the establishment of an open configuration of the domain prior to adult α-globin gene expression. Such hypothesis is further supported by notorious enrichment of transcriptionally permissive histone modifications on the same genomic region in 5dRBC [Figure 4B, CTCF(P2)], in which the embryonic π gene is transcribed (21). Then, the 6.6-kb genomic region encompassing the αEHS-1.4 insulator structures as transcriptionally permissive chromatin after globin gene expression switching, i.e. in 10dRBCs, when the adult α-globin genes (αD and αA) are robustly expressed, while the embryonic π gene is silenced (21). The DT40 lymphoid cell line, in which the α-globin gene domain is inactive, was used as a control (21,38). Thus, the progressive arrangement of the 6.6-kb genomic region containing the αEHS-1.4 insulator into transcriptionally permissive chromatin correlates with α-globin locus activation and the onset of adult α-globin gene expression. These results are in contrast to the timing of αEHS-1.4 insulator function and CTCF occupancy, which rise in erythroblasts, and decrease in mature erythrocytes (Figure 3B).
Taken together, our results support a model in which the αEHS-1.4 insulator, located between the domain’s putative α-MRE and the α-globin genes might act by blocking the transcriptional influence of the α-MRE over the α-globin adult genes at the erythroblast stage. Then, as differentiation proceeds towards mature erythrocytes, CTCF dissociates from the αEHS-1.4 insulator, releasing its blocking activity over the α-MRE and allowing the establishment of transcriptionally active chromatin and trans-activation of the adult genes.
Loss of αEHS-1.4 insulator function results in active chromatin configuration and precocious adult α-globin gene expression
To start testing the proposed model in which the αEHS-1.4 insulator blocks the α-MRE over α-globin gene expression in erythroblasts, we addressed the chromatin configuration as well as α-globin gene expression in cells with decreased or inactive CTCF. Open chromatin histone modifications at the αEHS-1.4 insulator region were increased in undifferentiated HD3 cells with knocked-down CTCF (Figure 5A). As expected, α-globin gene expression was not detected in untreated HD3 cells (Figure 5B) (21). No changes were observed in undifferentiated HD3 cells with decreased CTCF (Figure 5B, left panel). In contrast, in differentiated HD3 cells with decreased CTCF the adult αD and αA globin genes did not get fully activated (Figure 5B, right panel).
Figure 5.
Reduction of CTCF alters the profile of histone covalent modifications at the αEHS-1.4 insulator region and α-globin genes expression. (A) ChIP analysis of histone modifications in HD3 cells (HD3) and HD3 cells knocked down for CTCF (HD3+CTCF-kd). Each ChIP assay was performed in triplicate. Representative radioactive duplex PCR gels of the HD3-scramble (HD3-scr) and HD3 cells knocked down for CTCF (HD3+CTCF-kd) are shown. (B) Evaluation of α-globin gene expression in HD3 cells (HD3) and HD3 cells knocked down for CTCF in normal conditions (undifferentiated, HD3+CTCF-kd) and differentiated HD3 cells (dif-HD3). The results are representative of nine independent PCR reactions from two independent total RNA purifications and reverse transcriptase reactions. There is a premature chromatin opening of the insulator region in the CTCF knock down cells, and the adult genes are unable to be expressed in the HD3 differentiated cells. *P ≤ 0.05 (MANOVA) and ***P ≤ 0.001 (MANOVA) with respect to the corresponding histone covalent modification or expressed gene in HD3 or dif-HD3 scramble controls, respectively.
A similar set of experiments was performed in cells treated with 3-ABA inhibitor of PARP-1 enzyme. Open chromatin histone modifications at the αEHS-1.4 insulator region were generally increased in undifferentiated HD3 cells in which CTCF function was inactivated by 3-ABA, as compared to untreated cells (Figure 6A). Accordingly, HD3-treated cells showed premature activation of the adult αD and αA globin genes in contrast with CTCF knock-down cells (Figure 6B). No significant differences in histone modifications or α-globin gene expression were found in treated differentiated HD3 cells. Two main conclusions emerge from this set of experiments. First, inhibition of CTCF poly(ADP-ribosyl)ation in HD3 erythroblasts mimics, at least in part, the chromatin structure and the expression of the adult αD and αA globin genes in 10dRBCs (Figures 4B and 6). Second, knock down of CTCF perturbs more dramatically the chromatin structure of the α-globin domain, as compared to inhibition of CTCF poly(ADP-ribosyl)ation, while impeding full globin gene activation (Figure 5B, right panel). Thus, it is possible that poly(ADP-ribosyl)ation regulates the activity of CTCF on the αEHS-1.4 insulator at its natural context. Together, these results support a model in which the αEHS-1.4 promotes optimal chromatin configuration and prevents α-MRE-mediated adult α-globin genes trans-activation at the erythroblast stage.
α-MRE trans-activation potentiation by CTCF release from the αEHS-1.4 insulator during differentiation
In an attempt to validate the proposed model we generated pools of stable HD3 cell lines comparing the activity of the αD gene promoter alone, the same promoter with the α-MRE element and a construct with the αMRE, the α-EHS-1.4 and the αD gene promoter mimicking their endogenous organization (Figure 7).
Figure 7.
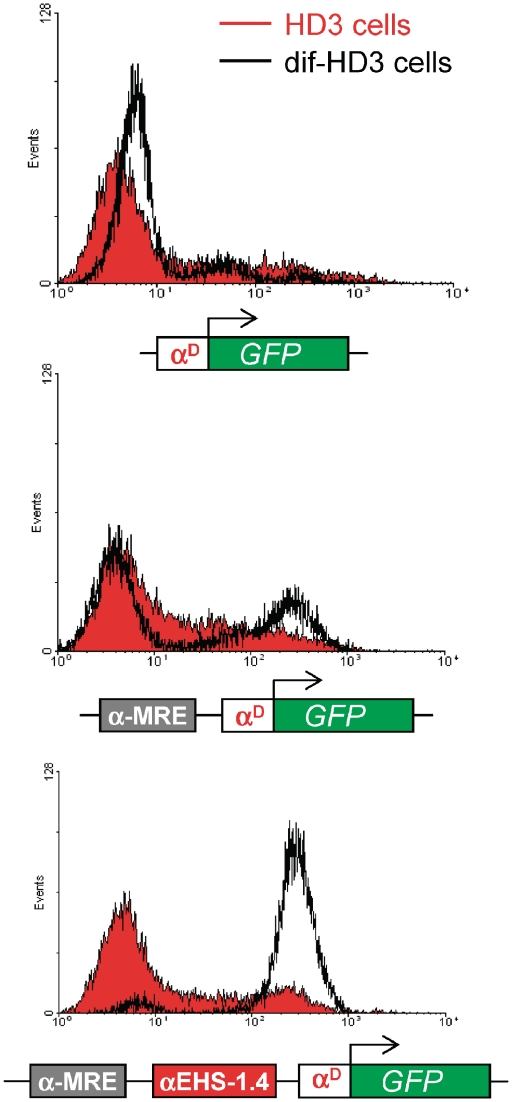
αMRE element is able to trans-activate the αD promoter upon differentiation to the mature erythrocyte stage. Flow cytometry histograms (FACS) showing the GFP fluorescence level of pools of stably transfected HD3 cells with three different constructs, αD, αMRE-αD and αMRE-αEHS-1.4-αD (showed underneath each graph). The full red graph corresponds the erythroblast stage (HD3 cells) and the black empty graph corresponds to the same pool subjected to differentiation (HD3-dif).
Based on our previous observations we propose that CTCF dissociation from the αEHS-1.4 insulator in mature erythrocytes (differentiated HD3 cells) increase α-MRE trans-activation potential. Indeed, we found that under non-differentiated conditions the majority of the reporter gene activity is not trans-activated, instead when HD3 cells are differentiated into mature erythrocytes we detected a robust trans-activation of the reporter gene (Figure 7) consistent with the dissociation of CTCF over the insulator in the transgene (Figure 3B and C).
These results further support the model in which the αEHS-1.4 insulator regulates the α-MRE trans-activating function over the adult α-globin genes during adult erythroid differentiation.
DISCUSSION
Genomic organization into chromatin domains represents an important level of gene expression regulation achieved by regulatory elements located mainly at intergenic regions (2,3). Here we studied the function of the αEHS-1.4 insulator in the chromatin organization and gene expression regulation of the α-globin gene domain. We previously demonstrated that the αEHS-1.4 insulator, embedded in the 5′ non-coding region of the α-globin domain, posses enhancer blocking activity (15). With the aim to better understand the insulator properties of the αEHS-1.4 element and its role in the chicken α-globin domain, we first demonstrate that the αEHS-1.4 element is able to shield a transgene from CPE in stably transfected cell lines and in mice in a CTCF-dependent manner. This differs from the 5′cHS4 insulator in which only enhancer blocking function is CTCF-dependent, whereas protection against CPE needs binding of USF1 and USF2 factors (18,39). Since no USF1/USF2 factors are present at the αEHS-1.4 (data not shown), a different protein complex most likely regulates αEHS-1.4 insulator activity.
Importantly, CTCF occupancy of the αEHS-1.4 insulator varies during erythroid differentiation. This raises the possibility that the αEHS-1.4 acts as a stage-specific insulator probably influencing the activity of the α-MRE on the expression of adult αD and αA genes. Indeed, the activity of αEHS-1.4 is regulated, and peaks during the switching from early to late adult erythroid differentiation, when an erythroid-specific signal might favor CTCF dissociation. We propose that the function of the αEHS-1.4 insulator is to block the activity of the α-MRE distal element during early adult erythroid differentiation. Once differentiation takes place, CTCF dissociates and open chromatin propagates 3′ towards the gene cluster (Figure 8A). Possibly the αEHS-1.4 element regulates the poised state of the RNA polymerase II and pre-initiation complex components found at the α-MRE (40). The RNA polymerase II blockage by CTCF has been described in other loci (41,42). In such a hypothetical scenario, CTCF release from the αEHS-1.4 insulator may favor the chromosomal conformation changes and the signal spreading needed for the interaction between the α-MRE and the adult αD and αA gene promoter regions and their transcriptional activation (Figure 8B). This model represents a novel regulatory role for a CTCF-dependent chromatin insulator involved in a differentiation pathway. In summary, at the endogenous context the αEHS-1.4 insulator may have enhancer blocking activity (presumably over the α-MRE) and barrier function to contribute to chromatin structure changes at the domain scale. Such dual activity is thus in part CTCF-dependent, and probably can also be driven by different stage-specific nuclear factors.
Figure 8.
Summary and model for the αEHS-1.4 insulator role at the chicken α-globin locus. (A) Summary of histone post-translational modifications and CTCF in vivo enrichment over the 6.6-kb genomic region containing the αEHS-1.4 insulator. As adult erythroid differentiation proceeds there is an increment in histone modifications corresponding to a permissive chromatin environment over the αEHS-1.4 insulator and the CTCF site (see P2 primer location) that expands and further increases over the entire region in 10dRBC when the adult genes αD and αA are switched on. CTCF is present during differentiation with a peak at the erythroblast stage (HD3 cells) and then released at the mature 10dRBC when the adult αD and αA genes are being expressed (see right side green graph bar). P1–P5 represent the primers covering the 6.6-kb genomic region used for ChIP assays and P2 are the primers located over the CTCF-binding site. (B) Model suggesting a mechanism of action for the αEHS-1.4 insulator at the level of the chromatin structure at the domain scale and adult α-globin gene expression. Vertical arrows represent DNase I hypersensitive sites. We propose that at the erythroblast stage the αEHS-1.4 insulator blocks the effects coming from the α-MRE concomitant with the fact that the adult αD and αA globin genes remain silenced. At the late erythrocyte stage CTCF dissociation from the insulator facilitates α-MRE activity allowing trans-activation of α-globin adult genes.
A possible mechanism for CTCF release regulation could come from the presence of intergenic transcripts at the α-globin domain (43 and unpublished results). It has been recently demonstrated that for lysozyme gene activation by pro-inflammatory stimuli there is a CTCF–cohesin complex eviction by the transient stimulation of a non-coding RNA (31).
Additionally, we demonstrated that poly(ADP-ribosyl)ation is modulating CTCF activity at the αEHS-1.4 insulator (Supplementary Figure S4 and Figure 6). When PARP-1 is inhibited at the erythroblast stage we are able to mimic the effect of CTCF release during differentiation, i.e. an increase in histone open marks and the activation of the adult αD and αA globin genes (Figure 6). In contrast, CTCF knock down do not cause the activation of any α-globin gene in erythroblasts (Figure 5B) and impedes α-globin gene trans-activation at the differentiated stage. From these results several interpretations emerged. One predicts that CTCF poly(ADP-ribosyl)ation does not occur in all the CTCFs bound in vivo along the domain and that the CTCF located at the αEHS-1.4 insulator is modified post-translationally by PARP-1 (36). Furthermore, it is possible that not all CTCF associated complexes located along the domain possess the same peptidic composition and activity. This is an aspect that is currently under investigation. We predict that some CTCF-associated complexes are more structural and others, like the one linked to the αEHS-1.4, should have a regulatory function. Based on such predictions, it is now attractive to look for CTCF partners at different locations in the domain, including PARP-1. For example, CHD8, cohesins, lamin or CTCF itself among others (3,32) which would further support the proposal that poly(ADP-ribosyl)ation could form a docking site for multiple protein interactions with regulatory and chromatin functions (44).
A different interpretation, that does not exclude the previous one, has to do with the spatial organization of the chicken α-globin domain. We propose that CTCF is possibly contributing to this 3D organization of the domain, and therefore when it is knocked down, there is a generalized reduction of the protein levels affecting the active chromatin hub formation and consequently the activation of the adult α-globin genes (Figure 5). This is not surprising since it has previously been demonstrated that CTCF can mediate long-distance chromatin loop formation in the mouse β-globin locus (10). We predict that CTCF is not the only factor participating as it has been shown that LCR long-distance contacts can also be dependent on EKLF and GATA-1/FOG-1 (45,46), factors that also bind different regions of the α-globin gene domain (21).
In this regard, the 3D structuring of the α-globin domain was recently suggested by a chromatin conformation capture approach, which showed the formation of an active chromatin hub by the interaction between the α-MRE, a DNase I hypersensitive site of unknown function located −9-kb upstream of the π gene (–9-kb DHS), the CpG-island corresponding to the promoter of the C16orf35 gene and the promoter region of the adult αD gene (Figure 1) (38). Interestingly, the αEHS-1.4 element, located around −14-kb upstream the embryonic π gene, is contained in the loop formed by contacts between the α-MRE and the −9-kb DHS, thus the αEHS-1.4 element appears not to contribute directly to hub formation in this context. Whether alternative CTCF sites along the α-globin domain, which we recently identified (unpublished observations), contribute to domain organization via interaction of CTCF and its co-factors, would require further investigation.
In summary, we propose that the αEHS-1.4 insulator contributes to regulate the chromatin structure and differential gene expression of the chicken α-globin gene domain throughout erythroid differentiation. We suggest that the regulated interaction of CTCF with its binding site could represent a way to control the function of the αEHS-1.4 element over α-globin genes expression, together with post-translational modifications like the poly(ADP-ribosyl)ation that modulate CTCF activity. In terms of the chromatin configuration of the locus, multiple CTCF-binding sites and their associated factors may facilitate the formation of a chromatin hub in a regulated manner. Our results suggest that the intergenic regulatory and structural components of a chromatin domain are critical to achieve regulated gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Dirección General de Asuntos del Personal Académico-Universidad Nacional Autónoma de México (IN209403 and IN214407); Consejo Nacional de Ciencia y Tecnología (CONACyT: 42653-Q and 58767); PhD fellowship from CONACyT (170087 and 207989) and Dirección General de estudios de Posgrado-Universidad Nacional Autónoma de México (DGEP) (to M.F.-M. and E.G.-B.); PhD Graduate Program of Doctorado en Ciencias Biomédicas, UNAM; Spanish Ministry of Education and Science (BFU2006-12185); Spanish Ministry of Science and Innovation (BIO2009-12697) (to L.M.). Funding for open access charge: Universidad Nacional Autónoma de México (UNAM), Instituto de Fisiologia Celular.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Paul Delgado-Olguín, Catherine Farrell and Gary Felsenfeld for invaluable suggestions on the manuscript. The authors also thank Martín Escamilla-del-Arenal, Héctor Rincón-Arano and all members of Félix Recillas-Targa laboratory for constant scientific discussions. The authors thank Fabian Flores Jasso for his help generating the shRNA used against CTCF. The authors thank L. Ongay, G. Codiz and M. Mora from the Unidad de Biología Molecular, IFC-UNAM, for the DNA sequencing and FACS facility. The CNB Transgenic Facility is acknowledged for producing the transgenic mice reported in this article. The authors are most grateful to Almudena Tello, Diego Muñoz, Zahira Corrales, and Esther Zurita for technical assistance.
REFERENCES
- 1.West AG, Fraser P. Remote control of gene transcription. Hum. Mol. Genet. 2005;14:R101–R111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- 2.Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr. Opin. Genet. Develop. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorsett D. Roles of the sister chromatin cohesion apparatus in gene expression, development and human syndromes. Chromosoma. 2007;116:1–13. doi: 10.1007/s00412-006-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc. Natl Acad. Sci. USA. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell-type-specific genomic distributions. Genes Dev. 2009;23:1338–1350. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 8.Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl Acad. Sci. USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ribeiro de Almeida C, Heath H, Krpic S, Dingjan GM, van Hamburg JP, Bergen I, van de Nobelen S, Grosveld F, Galjart N, Hendriks RW. Critical role of the transcription regulator CCCTC-binding factor in the control of the Th2 cytokine expression. J. Immunol. 2009;182:999–1010. doi: 10.4049/jimmunol.182.2.999. [DOI] [PubMed] [Google Scholar]
- 10.Splinter E, Heath H, Kooren J, Palstra R-J, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Majumder P, Gomez JA, Chadwick BP, Boss JM. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J. Exp. Med. 2008;205:785–798. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qui XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 13.Lutz M, Burke LJ, LeFevre P, Myers FA, Thorne AW, Crane-Robinson C, Bonifer C, Filippova GN, Lobanenkov V, Renkawitz R. Thyroid-hormone regulated enhancer blocking: cooperation of CTCF and thyroid hormone receptor. EMBO J. 2003;22:1579–1587. doi: 10.1093/emboj/cdg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, Farrar D, Tavoosidena G, Mukhopadhyay R, Kanduri C, et al. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat. Genet. 2004;36:1105–1110. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- 15.Valadez-Graham V, Razin SV, Recillas-Targa F. CTCF-dependent enhancer blockers at the upstream region of the chicken α-globin gene domain. Nucleic Acids Res. 2004;32:1354–1362. doi: 10.1093/nar/gkh301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vyas P, Vickers MA, Picketts DJ, Higgs DR. Conservation of position and sequence of a novel, widely expressed gene containing the major human α-globin regulatory element. Genomics. 1995;29:679–689. doi: 10.1006/geno.1995.9951. [DOI] [PubMed] [Google Scholar]
- 17.Flint J, Tufarelli C, Peden J, Clark K, Daniels RJ, Hardison R, Miller W, Philipsen S, Tan-Un KC, McMorrow T, et al. Comparative genome analysis delimits a chromosomal domain and identified key regulatory elements in the α globin cluster. Hum. Mol. Genet. 2001;10:371–382. doi: 10.1093/hmg/10.4.371. [DOI] [PubMed] [Google Scholar]
- 18.Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, Bell AC, Litt MD, West AG, Gaszner M, Felsenfeld G. Position-effect protection and enhancer blocking by the chicken β-globin insulator are separable activities. Proc. Natl Acad. Sci. USA. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beug H, von Kirchbach A, Doderlin G, Conscience JF, Graf T. Chicken haematopoietic cells transformed by seven strains of defective avian leukaemia viruses display three distinct phenotypes of differentiation. Cell. 1979;18:375–390. doi: 10.1016/0092-8674(79)90057-6. [DOI] [PubMed] [Google Scholar]
- 20.Dieken ES, Epner EM, Fiering S, Fournier RE, Groudine M. Efficient modification of human chromosomal alleles using recombination-proficient chicken/human microcell hybrids. Nat. Genet. 1996;12:174–182. doi: 10.1038/ng0296-174. [DOI] [PubMed] [Google Scholar]
- 21.Rincón-Arano H, Guerrero G, Valdes-Quezada C, Recillas-Targa F. Chicken α-globin switching depends on autonomous silencing of the embryonic π globin gene by epigenetics mechanisms. J. Cell. Biochem. 2009;108:675–687. doi: 10.1002/jcb.22304. [DOI] [PubMed] [Google Scholar]
- 22.Iarovaia O, Razin SV, Linarez-Cruz G, Sjakste N, Scherrer K. In chicken leukemia cells globin genes are fully transcribed but their RNAs are retained in the perinucleolar area. Exp. Cell. Res. 2001;270:159–165. doi: 10.1006/excr.2001.5332. [DOI] [PubMed] [Google Scholar]
- 23.Pikaart MJ, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beermann F, Ruppert S, Hummler E, Schütz G. Tyrosinase as a marker for transgenic mice. Nucleic Acids Res. 1991;19:958. doi: 10.1093/nar/19.4.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giraldo P, Martínez A, Regales L, Lavado A, García-Díaz A, Alonso A, Busturia A, Montoliu L. Functional dissection of the mouse tyrosinase locus control region identifies a new putative boundary activity. Nucleic Acids Res. 2003;31:6290–6305. doi: 10.1093/nar/gkg793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ganss R, Montoliu L, Monaghan AP, Schütz G. A cell-specific enhancer far upstream of the mouse tyrosinase gene confers high level and copy number-related expression in transgenic mice. EMBO J. 1994;13:3083–3093. doi: 10.1002/j.1460-2075.1994.tb06607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giménez E, Lavado A, Giraldo P, Montoliu L. Tyrosinase gene expression is not detected in mouse brain outside the retinal pigment epithelium cells. Eur. J. Neurosci. 2003;18:2673–2676. doi: 10.1046/j.1460-9568.2003.02992.x. [DOI] [PubMed] [Google Scholar]
- 28.Lavado A, Olivares C, García-Borrón JC, Montoliu L. Molecular basis of the extreme dilution mottled mouse mutation: a combination of coding and noncoding genomic alterations. J. Biol. Chem. 2005;280:4817–4824. doi: 10.1074/jbc.M410399200. [DOI] [PubMed] [Google Scholar]
- 29.Rincón-Arano H, Furlan-Magaril M, Recillas-Targa F. Protection against telomeric position effects by the chicken cHS4 β-globin insulator. Proc. Natl Acad. Sci. USA. 2007;104:14044–14049. doi: 10.1073/pnas.0704999104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Litt MD, Simpson M, Recillas-Targa F, Prioleau MN, Felsenfeld G. Transitions in histone acetylation reveal boundaries of three separately regulated neighbouring loci. EMBO J. 2001;20:2224–2235. doi: 10.1093/emboj/20.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lefevre P, Witham J, Lacroix CE, Cockerill PN, Bonifer C. The LPS-induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol. Cell. 2008;32:129–139. doi: 10.1016/j.molcel.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol. Cell. 2006;23:733–742. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 33.Potts W, Tucker D, Wood H, Martin C. Chicken β-globin 5'HS4 insulators function to reduce variability in transgenic founder mice. Biochem. Biophys. Res. Commun. 2000;273:1015–1018. doi: 10.1006/bbrc.2000.3013. [DOI] [PubMed] [Google Scholar]
- 34.Torrano V, Navascues J, Docquier F, Zhanf R, Burke LJ, Chernukhin I, Farrar D, León J, Berciano MT, Renkawitz R, et al. Targeting of CTCF to the nucleolus inhibits nucleolar transcription through a poly(ADP-ribosyl)ation-dependent mechanism. J. Cell. Sci. 2006;119:1746–1759. doi: 10.1242/jcs.02890. [DOI] [PubMed] [Google Scholar]
- 35.Witcher M, Emerson B. Epigenetic silencing of the p16(INK4a) tumor suppressor is associated with loss of CTCF binding and a chromatin boundary. Mol. Cell. 2009;34:271–284. doi: 10.1016/j.molcel.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farrar D, Rain S, Chernukhin I, Jagodic M, Ito Y, Yammine S, Ohlsson R, Murrel A, Klenova E. Mutational analysis of the poly(ADP-ribosyl)ation sites of the transcription factor CTCF provides an insight into the mechanism of its regulation by poly(ADP-ribosyl)ation. Mol. Cell. Biol. 2010;30:1199–1216. doi: 10.1128/MCB.00827-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soto-Reyes E, Recillas-Targa F. Epigenetic regulation of the human p53 gene promoter by the CTCF transcription factor in transformed cell lines. Oncogene. 2010;15:2217–2227. doi: 10.1038/onc.2009.509. [DOI] [PubMed] [Google Scholar]
- 38.Gavrilov AA, Razin SV. Spatial configuration of the chicken α-globin gene domain: immature and active chromatin hubs. Nucleic Acids Res. 2008;36:4629–4640. doi: 10.1093/nar/gkn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West AG, Huang S, Gaszner M, Litt MD, Felsenfeld G. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol. Cell. 2004;16:453–463. doi: 10.1016/j.molcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 40.Higgs DR, Vernimmen D, Hughes J, Gibbons R. Using genomics to study how chromatin influences gene expression. Annu. Rev. Genomics Hum. Genet. 2007;8:299–325. doi: 10.1146/annurev.genom.8.080706.092323. [DOI] [PubMed] [Google Scholar]
- 41.Zhao H, Dean A. An insulator blocks spreading of histone acetylation and interferes with RNA polymerase II transfer between an enhancer and gene. Nucleic Acids Res. 2004;32:4903–4914. doi: 10.1093/nar/gkh832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gomes NP, Espinosa JM. Gene-specific repression of the p53 target gene PUMA via intragenic CTCF-Cohesin binding. Genes Dev. 2010;24:1022–1034. doi: 10.1101/gad.1881010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borunova V, Iarovaia OV, Vassetzky YS, Razin SV. The upstream area of the chicken α-globin gene domain is transcribed in both directions in the same cells. FEBS Lett. 2005;579:4746–4750. doi: 10.1016/j.febslet.2005.07.051. [DOI] [PubMed] [Google Scholar]
- 44.Kleine H, Lüscher B. Learning how to read ADP-ribosylation. Cell. 2009;139:17–19. doi: 10.1016/j.cell.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 45.Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol. Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 47.Klochkov D, Rincón-Arano H, Ioudinkova ES, Valadez-Graham V, Gavrilov A, Recillas-Targa F, Razin SV. A CTCF-dependent silencer located in the differentially methylated area may regulate expression of a housekeeping gene overlapping a tissue-specific gene domain. Mol. Cell. Biol. 2006;26:1589–1597. doi: 10.1128/MCB.26.5.1589-1597.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.