Abstract
In eukaryotes, protein-coding genes are transcribed by RNA polymerase II (pol II) together with general transcription factors (GTFs). TFIID, the largest GTF composed of TATA element-binding protein (TBP) and 14 TBP-associated factors (TAFs), plays a critical role in transcription from TATA-less promoters. In metazoans, several core promoter elements other than the TATA element are thought to be recognition sites for TFIID. However, it is unclear whether functionally homologous elements also exist in TATA-less promoters in Saccharomyces cerevisiae. Here, we identify the cis-elements required to support normal levels of transcription and accurate initiation from sites within the TATA-less and TFIID-dependent RPS5 core promoter. Systematic mutational analyses show that multiple AT-rich sequences are required for these activities and appear to function as recognition sites for TFIID. A single copy of these sequences can support accurate initiation from the endogenous promoter, indicating that they carry highly redundant functions. These results show a novel architecture of yeast TATA-less promoters and support a model in which pol II scans DNA downstream from a recruited site, while searching for appropriate initiation site(s).
INTRODUCTION
In eukaryotes, transcription of protein-coding genes is regulated by the concerted action of gene-specific transcription factors, chromatin remodeling factors, co-activators/co-repressors, mediator, general transcription factors (GTFs) and RNA polymerase II (pol II) (1–3). In principle, gene-specific transcription factors, bound to regions upstream of the transcriptional initiation site, regulate the activity of other factors, especially those of the pre-initiation complex (PIC) assembled on the core promoter surrounding the transcription initiation site. The PIC assembly is initiated via binding of TFIID, the largest GTF comprising TATA-binding protein (TBP) and 14 TBP-associated factors (TAFs) (4), to the core promoter and completed via incorporation of other GTFs, mediator and pol II in a stepwise fashion (5–7). TFIID binding is one of the most crucial rate-limiting steps for transcriptional activation (8,9) and is a target for many gene-specific transcription factors (2,10,11).
Recent studies show that most (∼80%) pol II-driven (class II) promoters do not have a TATA element, a recognition site for TBP, though TBP itself is still needed for transcription of these TATA-less promoters (12–16). Based on genome-wide expression studies, it is proposed that, in yeast cells, TBP is delivered to TATA-containing and TATA-less promoters by two distinct multi-protein complexes, SAGA (Spt-Ada-Gcn5 acetyltransferase) and TFIID, respectively (13,17). Notably, stress-responsive (inducible) and housekeeping (constitutive) genes tend to be regulated by TATA-containing or TATA-less promoters, respectively (17), suggesting two fundamentally different regulatory mechanisms for class II genes. A recent ChIP–chip study in yeast showed that in vivo turnover rates of TBP are significantly faster on SAGA-dependent and TATA-containing promoters than on TFIID-dependent and TATA-less promoters (18). Consistent with this, an in vitro study using HeLa nuclear extracts shows that PIC is assembled rapidly on the TATA-containing p21 core promoter but slowly on the TATA-less Fas/APO1 core promoter (19). Once PIC is assembled, it supports multiple rounds of transcription on the latter promoter but permits only a few rounds of transcription on the former (19). This suggests that the TATA element accelerates the turnover rate of TBP on the core promoter and may be critical for optimal regulation of a particular set of genes.
In metazoans, several core promoter elements other than the TATA element have been identified (BRE, INR/sINR, DPE, MTE, the bridge element, DCE, XCPE1/2 and CpG islands) (20–26), and are specific to a subset of class II promoters. Furthermore, each core promoter contains some of these elements in different combinations enabling a proper response to its inherent gene-specific transcription factors. In fact, some activators prefer TATA-dependent promoters, while others prefer DPE-dependent promoters (27,28). Specifically, Caudal, a key regulator of the Hox gene network, is a DPE-specific activator (29). However, even a single regulator activates different types of core promoters by different mechanisms; for example, EKLF activates the DCE-containing β-globin core promoter in a TAF9-dependent manner, but it activates the DCE-less AHSP core promoter in a TAF9-independent manner (30). Similar functional compatibility between the upstream activating sequence (UAS) and the core promoter and similar variations in factor-dependence in activation are commonly observed in yeast (31–37). Hence, core promoter architectures must play pivotal roles in the regulation of class II genes in many eukaryotes.
Most of the core promoter elements described above have not yet been identified in yeast. The TATA element has been extensively characterized (13,32,38–40) but few studies have focused on the initiator element (16,41–47). This suggests that yeast initiator elements may differ functionally from metazoan INR; the latter serves as a recognition site for the TAF1–TAF2–TBP sub-complex of TFIID (48), whereas the former represents the preferred initiation sites for pol II (20,49). In addition, there are two types of core promoter elements in metazoans, i.e. recognition sites for TFIID (e.g. TATA, DPE, DCE, MTE, the bridge element, INR) (26,50,51) and other factors (e.g. BRE, XCPE1/2) (22,23). As TBP and TAFs are evolutionally conserved in eukaryotes, yeast core promoters should contain certain as-yet unidentified recognition sites for TFIID other than the TATA element. However, such elements are largely unknown, except for a few potential candidate sequences (39,52–55).
In this study, we determine the core promoter elements within the TATA-less RPS5 promoter that are required for gene expression and that may function potentially as recognition sites for TFIID as this promoter is highly TFIID-dependent (33,55) and is bound specifically by purified TFIID (56). Since RPS5 has been extensively characterized not only in vivo (33–36,55,57) but also in vitro (56), it was chosen as a target out of many other ribosomal protein genes (RPGs). More specifically, we exploited an assay in which the core promoter activity of a given fragment can be tested by examining initiation site(s) created ectopically when the fragment is inserted at a site further upstream than the original site in the same promoter (42). This assay is more versatile than traditional mutational approaches since it has the potential to identify core promoter activity even when the target promoter contains multiple and functionally redundant cis-elements, as already observed in HIS3 (53). In addition, the RPS5 promoter was integrated at the VTC1 locus to generate the reporter. The RPS5 promoter was subjected to mutagenesis, but the original RPS5 promoter was left intact as normal expression of this gene is critical for cell growth.
As a result, we show that multiple AT-rich sequences function redundantly, not only in supporting transcription from intrinsic transcriptional initiation sites, but also in creating ectopic transcriptional initiation sites in a TFIID-dependent manner when inserted upstream of the original core promoter. Strikingly, a single copy of these sequences is sufficient to support significant transcription from the endogenous RPS5 promoter. The highly redundant and cooperative features of these sequences may make the turnover rate of TBP/TFIID slower and thereby enable TATA-less promoters to be expressed constitutively. These results are noticeably similar to those obtained previously for the HIS3 promoter containing two different types of the TATA element, TC and TR (31,32,53,54). Given that the TC element is a type of TATA-less promoter, this study demonstrates that short AT-rich stretches function redundantly as core promoter elements in TATA-less promoters either containing a typical TATA element adjacently (e.g. TR HIS3) or not at any site (e.g. RPS5). Our results also raise the possibility that similar mechanisms may operate for other genes regardless of whether they are driven by TATA-less (e.g. RPL2B) or TATA-containing (e.g. SSB1, ADH1) promoters.
MATERIALS AND METHODS
Yeast strains and media
Standard techniques were used for yeast growth and transformation (58). The yeast strains used in this study are listed in Supplementary Table S1. Oligonucleotide sequences used for the strain construction and transcription analyses are listed in Supplementary Table S2.
All strains were generated from BY4741 (EUROSCARF) or BY4742 (EUROSCARF) by a fusion PCR-based method (59,60), as described below. The endogenous promoter spanning up to 100-bp upstream of the translational initiation site (A of ATG as +1) of VTC1 of BY4741 was replaced with a PCR fragment containing a given region of RPS5 recombined with or without a certain portion of the RPL2B, SSB1, ADH1, or HIS3 promoters, and His3MX6 as a selectable marker (59). Details of PCR primers, templates and promoter regions amplified for promoter construction are summarized in Supplementary Table S3.
To examine the dependency of a subset of promoter constructs on Taf1p, TAF1 in some strains was replaced with the temperature-sensitive allele, taf1-N568Δ (61). For this replacement, three PCR fragments were amplified individually from pM1169 (62), pM1169 and pFA6a-kanMX6 (59) using the primer pairs TK3950/3955, TK176/4012 and TK4011/TK3977, respectively, and then fused together using the primer pair TK3950/3977. The resulting long DNA fragment (∼5.1 kb) was used to transform BY4742 to generate YTK6339. Similarly, YTK10001, YTK10051, YTK10053, YTK10055, YTK10057, YTK10059, YTK10061, YTK10063, YTK10065, YTK10067, YTK10069, YTK10071, YTK10073, YTK10075, YTK10077, YTK10079 and YTK10081 were transformed with a PCR fragment amplified from YTK6339 genomic DNA using the primer pair TK3950/3977 (containing taf1-N568Δ and kanMX6) to replace TAF1 in these strains with taf1-N568Δ, generating YTK10002, YTK10052, YTK10054, YTK10056, YTK10058, YTK10060, YTK10062, YTK10064, YTK10066, YTK10068, YTK10070, YTK10072, YTK10074, YTK10076, YTK10078, YTK10080 and YTK10082, respectively.
All gene or promoter replacements were confirmed by PCR, Southern blot and genomic DNA sequencing.
Northern blotting
Northern blot analysis was performed as previously described (61). To detect VTC1, a DNA fragment encoding the open reading frame (390 bp) was amplified from yeast genomic DNA using the primer pair TK2676/TK2677, purified and 32P-labeled by random priming with Klenow fragment (TOYOBO). The PCR primers used for detection of ADH1 have been previously described (61).
Primer extension analysis
Primer extension analysis was conducted as previously described (63) with some modifications. Briefly, extension reactions were conducted for 10 µg of total RNA in 25 µl of buffer A (50 mM Tris–HCl [pH 8.0], 50 mM KCl, 10 mM MgCl2, 10 mM dithiothreitol) containing four dNTPs (0.2 mM each, GE Healthcare), ribonuclease inhibitor (200 U, TaKaRa), AMV reverse transcriptase XL (25 U, TaKaRa), and [γ-32P]ATP-labeled oligonucleotide primers. The primers used were: TK8251 (+60 to +41 of VTC1), TK9044 (+120 to +101 of VTC1), TK9992 (+60 to +31 of ADH1), and TK7616 (−300 to −280 of ADH1). The cDNA products were analyzed on a 6% polyacrylamide DNA sequencing gel. Gels were exposed to imaging plates (BAS2500, Fuji Film) for visualization.
RESULTS
Mapping of the cis-elements that activate transcription and/or determine initiation sites within the RPS5 promoter
Most RPGs are activated by Rap1p and its co-activators, Fhl1p and Ifh1p, which together bind to so-called RPG-boxes (64–68). Only Rap1p recognizes RPG-boxes directly and recruits Fhl1p–Ifh1p for activation (69,70). Intriguingly, the RPS5 core promoter can be efficiently activated by its own UAS, but not by ADH1 UAS (36) or GAL1 UAS (34). This suggests that the RPS5 core promoter may contain specific cis-element(s) that are highly competent for the action of Rap1p–Fhl1p–Ifh1p.
To delineate the region(s) required for transcriptional activation and accurate initiation from the RPS5 promoter, we performed systematic deletion analyses on the fragment encompassing −631 to −1 bp (A of ATG as +1) integrated into the VTC1 locus (Figure 1). VTC1 encodes a component of the polyphosphate polymerase complex (71) and is used as a reporter gene measurable by magnetic resonance (72). In this study, however, VTC1 mRNA was measured by northern blot and/or primer extension analyses; importantly, the initiation sites of this reporter gene are the same as those of endogenous RPS5 (63). To confirm reproducibility, we conducted all experiments at least three times (or more for some constructs) by repeating the entire procedure from yeast cultivation to RNA preparation and analysis, and only representative results are shown below.
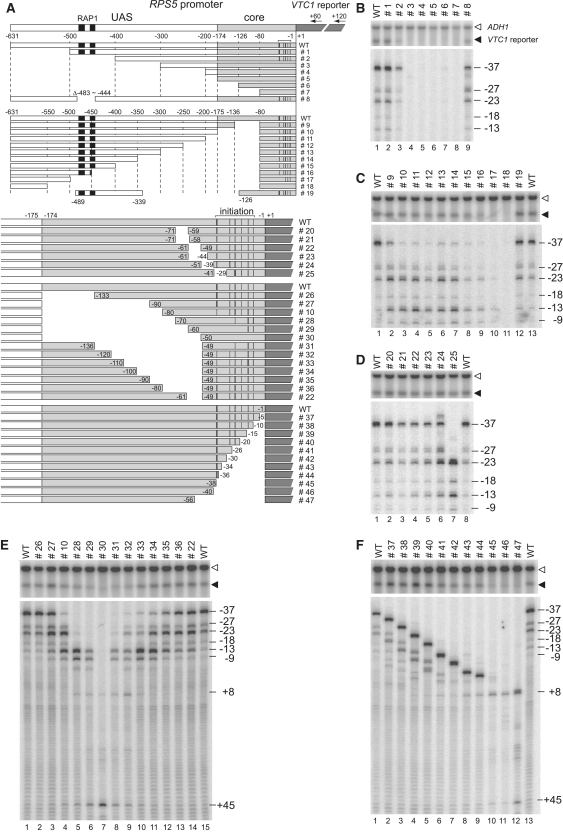
Figure 1.
Cis-element mapping of the RPS5 promoter. (A) Schematic diagram showing a series of promoter constructs with serial deletions from the 5′-end (top panel) or −81 bp (second panel) of the entire region between −631 and −1 bp (A of ATG as +1) of the RPS5 promoter and with serial deletions from the 5′-end (upper part in the fourth panel), the 3′-end (bottom panel), or internally (third panel and lower part of the fourth panel) of the core promoter region between −174 and −1 bp of the RPS5 promoter. These constructs were integrated into the VTC1 locus (dark grey rectangle). The regions corresponding to the UAS containing two potential Rap1 binding sites (black rectangles) and the core promoter (core) are white or pale grey rectangles, respectively. The number of each construct is shown on the right and the primer positions (+60 or +120) for extension analyses are shown at the top. Note that the two specifically deleted constructs (#8 and #19) are also shown (top and second panels). ‘WT’ = a wild-type construct carrying the full length RPS5 promoter. Transcriptional initiation activities from the sites at −37, −27, −23, −18 and −13 bp (marked with a horizontal bracket above the ‘WT’) were examined in (B)–(F) and summarized schematically with vertical black bars in each construct. (B) The effect of each deletion (#1 to #8) on the transcriptional activity of the RPS5 promoter. Northern blotting (upper panel) was done to determine the expression levels of VTC1 (reporter) and ADH1 (control) in the strains bearing each construct (top). Total RNA (20 µg) was isolated from each strain (cultured at 30°C in YPD media), blotted onto the membrane, and hybridized with the indicated gene-specific probes. Another sample of total RNA (20 µg) was subjected to primer extension analysis (bottom panel) using primer +60 to determine the transcriptional initiation site(s) of VTC1. The positions of major (−37) or minor (−27, −23, −18 and −13) transcriptional initiation sites are indicated (right). (C). The effect of each deletion (from #9 to #19) on the transcriptional activity of the RPS5 promoter. Northern blot and primer extension analyses were done as described in (B). (D) The effect of each deletion (from #20 to #25) on the transcriptional activity of the RPS5 promoter. Northern blot and primer extension analyses were done as described in (B). (E) The effect of each deletion (from #26 to #36, #10 and #22) on the transcriptional activity of the RPS5 promoter. Northern blot and primer extension analyses were done as described in (B) except using primer +120. (F) The effect of each deletion (#37–47) on the transcriptional activity of the RPS5 promoter. Northern blot and primer extension analyses were done as described in (E).
Serial deletions from the 5′end of this promoter showed that the −500 to −401 bp and −400 to −301 bp regions contain two distinct UAS (UAS1 and UAS2, respectively) required for activation (lanes 1–4, Figure 1B). Specifically, UAS1 corresponds to the −483 to −444 bp region containing two potential Rap1p binding sites (black rectangles, Figure 1A) (73–75) because #8, lacking this particular region, had similar activity to that of #2, lacking the entire region upstream of −401 bp (lanes 3 and 9, Figure 1B). However, UAS2 is localized to the −400 to −339 bp region because #19, in which the −489 to −339 bp region was fused to that of −126 to −1 bp, had similar activity to the full-length (wild-type; WT) promoter (lanes 1/13 and 12, Figure 1C). These results are consistent with our previous observations using a plasmid reporter system (33). In this 5′-deletion assay, it was impossible to determine the region that functions as a core promoter since the activities of #3–7 were too low to be discriminated from each other (lanes 4–8, Figure 1B).
Conversely, serial deletions from −81 bp to regions upstream (i.e. up to −550 bp; #18) of the promoter showed that the −135 to −81 bp region is required for accurate initiation because #9, lacking this region, showed weaker initiation at −37 bp but stronger initiation at the more downstream −23 and −13 bp sites (compare lanes 1 and 2, Figure 1C). Such downstream shifts are consistent with a prevailing view showing that pol II scans DNA downstream, searching for appropriate initiation site(s) (47,76–78). A further upstream deletion, from −136 to −174 bp (#10), weakened initiation from −37 bp but not from the −27, −23, −18, −13 and −9 bp sites (compare lanes 2 and 3, Figure 1C). However, several deletions between −349 and −175 bp (#11–14) did not alter the initiation profile of #10 (compare lanes 3 and 4–7, Figure 1C), suggesting that the −174 to −136 bp and −80 to −1 bp regions may support initiation from distinct sites, i.e. −37 bp and −27/−23/−18/−13/−9 bp. As expected, further upstream deletions between −499 and −350 bp (#15–17) gradually decreased the transcriptional levels (lanes 8–10, Figure 1C), probably because UAS2 (#15) and one (#16) or two (#17) potential Rap1p binding sites within UAS1, were removed sequentially from the promoter. Intriguingly, #19 showed a similar initiation profile to that of WT (lanes 12 and 13, Figure 1C), indicating that the −174 to −136 bp region is not required for initiation from −37 bp, at least when the −174 to −81 bp region is present. Therefore, these two regions (−174 to −136 and −126 to −81 bp) may function redundantly, even if their roles are not identical (compare lanes 2 and 12, Figure 1C).
Next, we sought to identify the core promoter element(s) required for normal levels of transcription and/or accurate initiation by deleting a series of small segments from the region between −174 and −1 bp (Figure 1A) containing a putative RPS5 core promoter (33,55). In metazoans, TATA-less promoters occasionally contain other core promoter elements like INR, MTE, the bridge element, DPE, DCE and XCPE1/2 that overlap with, or are located downstream of, the initiation site (22,26). However, it is unclear whether such non-TATA-type elements exist in yeast promoters, although INR-like sequences have been identified in silico (16,79).
The results showed that deleting the small segment ranging from 11 to 16 bp just upstream of the initiation site (#20–24) only slightly affected transcriptional levels and initiation profiles (compare lanes 1 and 2–6, Figure 1D). Similarly, deleting the region just downstream of the initiation site (−35 to −1 bp; #37–44) affected them only slightly (compare lanes 1 and 2–9, Figure 1F). We should note that the initiation profiles of the constructs #37–44 were the same as those of WT, although they appear rather different in Figure 1F because the distance between the initiation and primer sites was reduced due to the deletions. Remarkably, the 11-bp deletion encompassing the major initiation site (#25) abolished initiation from this site (−37 bp), but not from more downstream sites (−23/−18/−13/−9 bp; lane 7, Figure 1D). Given that initiation from the latter sites was stronger in this mutant, it is likely that pol II scans DNA from upstream and starts transcription at the nearest site that meets the requirements for productive initiation (45,49). In this model, the initiation sites function passively in yeast (20), unlike INR in metazoans that function more actively [including as a recognition element for TFIID (48)]. The observation that further upstream deletions from −37 to −55 bp (#45–47) abolished initiation from the site corresponding to the original position of −37 bp (lanes 10–12, Figure 1F) supports this model and implies that there is nothing within the region surrounding the initiation site to suppress inappropriate initiation, at least in the RPS5 promoter.
Unexpectedly, when the region between −174 and −50 bp was serially deleted from the 5′ (#26–30 and #10) or 3′end (#31–36 and #22), we found that the initiation sites shifted downstream, apparently in a manner proportional to the length of this region and independently of the particular sequences within it (Figure 1E). For instance, #10 and #34 showed similar initiation profiles (lanes 4 and 11, Figure 1E) even though they share only the region between −49 and −1 bp that, in itself, has no initiation activity (#30; lane 7, Figure 1E). Given that #19 showed similar activity to that of WT (lanes 12–13, Figure 1C), such downstream shifts should not be caused simply because the distance between the UAS and the initiation site was reduced. We prefer another possibility that there are multiple cis-elements within this region that function redundantly as binding sites for GTFs (such as TFIID) and that initiation profiles are determined by the distance between each cis-element and the initiation site.
To simplify the interpretation of the results described below, we should provisionally define two types of functional core promoter element in yeast: the Core factor binding Element (CE) and the Initiation Element (IE). CE is a recognition site for certain core factors such as free TBP, TFIID, or SAGA that bind directly to the core promoter and nucleate PIC assembly. IE is a site that allows the productive initiation of pol II that scans DNA from a site neighboring CE. This classification is consistent with a previous notion that the yeast INR is a passive contributor to promoter activity and represents a preferred initiation site for pol II (20,49). According to this definition, metazoan INR should have a dual function as CE and IE. Notably, CE and IE appear to be physically separated in the RPS5 promoter, but probably not in other yeast promoters (46).
Creation of the ectopic initiation sites by inserting authentic or putative CE into the upstream region of the RPS5 core promoter
The results described above suggested that the region between −174 and −50 bp may contain multiple CE, presumably serving as binding site(s) for TFIID (33,55). To explore this possibility, we tested whether insertion of the TATA element, known as an authentic CE, at various positions (−300, −250, −225, −213, −200, −183, −175, −163, −140, −136, −101, −81 bp) created ectopic initiation site(s) (Figure 2A, B and D). As expected (42), the results clearly showed that more upstream insertion of the TATA element creates more upstream ectopic initiation site(s) without affecting the initiation sites of the original RPS5 and endogenous ADH1 promoters (Figure 2B). Note that ectopic initiation sites were not distributed randomly but localized specifically at several preferred sites (−88 and −71 bp, Figure 2B), supporting the view that pol II scans DNA from CE to initiate transcription at specific IE. We also noticed that the distribution and shifting profiles of ectopic initiation sites could be used as a molecular ruler to assess the position of inserted CE as described below.
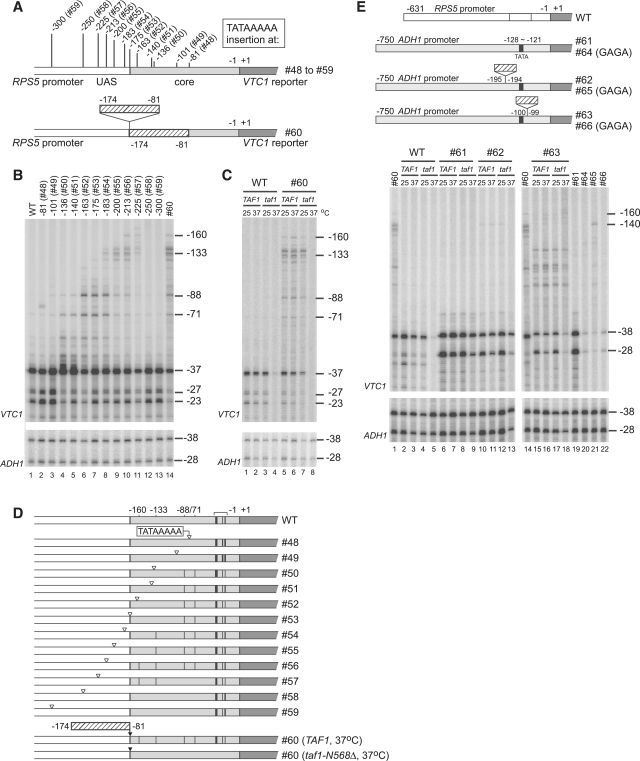
Figure 2.
Ability of the TATA element (TATAAAAA), or a portion of the RPS5 core promoter, to induce ectopic initiation sites within the RPS5 or ADH1 promoters in TAF1 and taf1-N568Δ strains. (A) Schematic diagram showing the insertion sites of the TATA element (upper panel) or a portion (−174 to −81 bp) of the RPS5 promoter (lower panel) within the RPS5 promoter. The number of constructs is shown in parenthesis (upper panel). Note that #60 contains the same promoter fragment (−174 to −81 bp) in duplicate. (B) The ability of each insertion of the TATA element (#48 − #59) or of the −174 to −81 bp fragment (#60) (top) to induce ectopic initiation site(s) in the RPS5 promoter. Primer extension analysis was done as described in Figure 1B, except that the initiation sites of both VTC1 (upper panel) and ADH1 (lower panel, control) were examined. (C) TFIID-dependency of the ectopic initiation sites induced by insertion of the −174 to −81 bp fragment (#60). Total RNA (20 µg) was isolated from the strains carrying a combination of either TAF1 or taf1-N568Δ and either WT or #60 before, or 2 h after, a temperature shift from 30 to 37°C (top). Primer extension analysis was done as described in (B). (D) Transcriptional initiation activities from the sites at −160, −133, −88, −71, −37, −27 and −23 bp (the last three endogenous sites are marked with a horizontal bracket above the ‘WT’) that were examined in (B) and (C) are summarized schematically with vertical black bars in each construct. (E) TFIID-dependency of the ectopic initiation sites induced by insertion of the −174 to −81 bp fragment of the RPS5 promoter at two distinct sites of the ADH1 promoter (−750 to −1 bp). Schematic diagram (upper panel) illustrates the constructs that contain (or do not contain) the −174 to −81 bp fragment of the RPS5 promoter as an insert at the indicated position (−195/−194 or −100/−99 bp) within the ADH1 promoter (#61–63, right) and those that contain an additional TATA to GAGA mutation (#64–66, right). Total RNA (20 µg) was isolated from the strains carrying a combination of either TAF1 or taf1-N568Δ and either WT, #61, #62 or #63 before, or 2 h after, a temperature shift from 30 to 37°C (indicated top, lanes 2–13 and 15–18). Similarly, total RNA (20 µg) was isolated from the strains carrying a combination of TAF1 and either #60, #61, #64, #65 or #66 incubated at 30°C as indicated at the top (lanes 1, 14 and 19–22). Primer extension analysis was done as described in (B).
Next, we inserted the −174 to −81 bp region at −175/−174 bp (Figure 2A) to test whether this region has TATA-like CE activity. If it did, we determined where such activity was localized within this region, and whether it was TFIID-dependent. Primer extension analyses showed that insertion of this region (#60) created ectopic initiation sites similar to those in #56 (compare lanes 10 and 14, Figure 2B). This suggests that the −174 to −81 bp region contains CE at a site corresponding to the position ‘TATAAAAA’ in #56 (39–46 bp apart from −175/−174 bp). Remarkably, such a site in #60 contained ‘CTAAAATA’ (corresponding to −126 to −119 bp of the original promoter), regarded as an atypical TATA element (underlined above) (33,55). We should note that, even if the −174 to −81 bp region contains multiple CE, only the most upstream one was detected in this assay due to the polar effect of pol II scanning.
We then tested whether CE within the −174 to −81 bp region is TFIID-dependent by comparing ectopic initiation activities from −160/−133/−88/−71 bp in TAF1 and taf1-N568Δ strains (Figure 2C and D). Importantly, these activities were specifically affected in the taf1-N568Δ strain in the same way as the intrinsic initiation sites at −37/−27/−23 bp when the strains were incubated at 37°C (lanes 4 and 8, Figure 2C) (33,55), indicating that CE within this region is TFIID-dependent. Unexpectedly, however, we also noticed that one of the two initiation sites of the ADH1 promoter was partially TFIID-dependent, since the ratio of the transcript from −28 bp to that from −38 bp was always lower in the taf1-N568Δ strain than in the TAF1 strain when the strains were incubated at 37°C (Figure 2C, E and 3A). Previous studies show that transcription from these two initiation sites occurs dependently on the same TATA element located at −128 to −121 bp (Figure 2E) (80) and on SAGA (81,82), but not on TFIID (83,84). Although the reason for the weak TFIID-dependence seen in our study is unclear, it may be due to differences in the taf1 alleles or genetic backgrounds of the yeast strains used.
Figure 3.
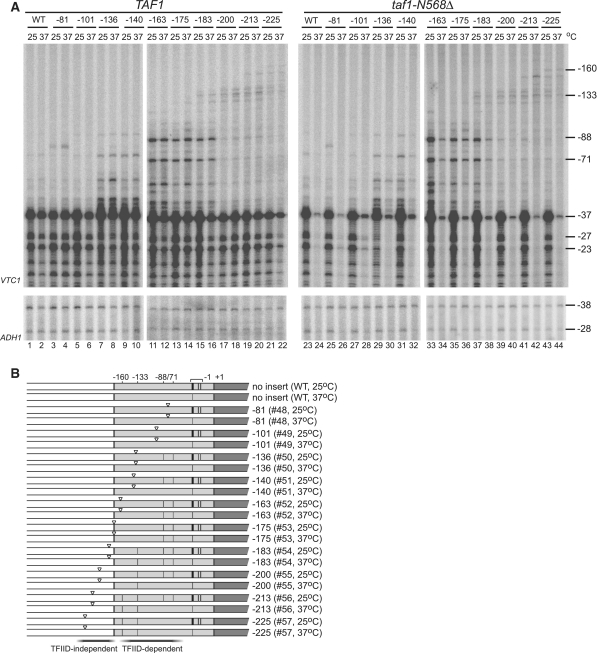
TFIID-dependency of ectopic initiation induced by the TATA insertion was altered by the position where it was inserted within the RPS5 promoter. (A). TFIID-dependency of ectopic initiation sites induced by insertion of the TATA element at various positions as depicted in Figure 2A in the RPS5 promoter. Total RNA (20 µg) was isolated from strains carrying a combination of either TAF1 (left panel) or taf1-N568Δ (right panel) and either WT, #48, #49, #50, #51, #52, #53, #54, #55, #56, or #57 before, or 2 h after, a temperature shift from 30 to 37°C as indicated at the top. Primer extension analysis was done as described in Figure 2B. (B) Transcriptional initiation activities from the sites at −160, −133, −88, −71, −37, −27 and −23 bp in a taf1-N568Δ strain (right panel in A) are summarized schematically as in Figure 2D. Note that the regions conferring TFIID-independent or TFIID-dependent ectopic initiation sites when the TATA element was inserted are indicated at the bottom.
It was shown that the RPS5 core promoter is activated very poorly by the ADH1 UAS (36). This suggests that the RPS5 CE may not function efficiently in the context of the ADH1 promoter. To confirm this, we inserted the −174 to −81 bp region of the RPS5 promoter upstream (#62) or downstream (#63) of the TATA element in the ADH1 promoter and tested whether ectopic initiation could be induced in a TFIID-dependent manner (Figure 2E). Similar to the initiation sites in the endogenous ADH1 promoter, only the initiation site located more downstream (−28 bp) of the reporter construct had weak TFIID-dependence when the insert was absent or present upstream of the TATA element (lanes 6–13). However, insertion downstream of the TATA element induced ectopic initiation in an apparently TFIID-independent manner, whereas the original initiation sites at −38/−28 bp became more TFIID-dependent (lanes 15–18). Interestingly, substitution of ‘TATA’ with ‘GAGA’ decreased transcription from both the original and ectopic initiation sites whenever the insert was absent (lanes 19–20), or present upstream (lanes 10 and 21) or downstream (lanes 15 and 22) of the TATA element. This suggests that the TATA element in #63 still allows effective transcription from both the original and ectopic initiation sites despite its position far upstream, and also that ectopic initiation is not induced by the RPS5 CE. It is unclear why initiation at −38/−28 bp was more TFIID-dependent in #63, even if the ectopic initiation sites were not TFIID-dependent. This may occur for the same reason that only the downstream site (i.e. −28 bp) of the two initiation sites in the original ADH1 promoter (−38/−28 bp) is TFIID-dependent. Therefore, we conclude that the RPS5 CE does not function efficiently within the ADH1 promoter.
The results described above show that the RPS5 CE functions efficiently in the RPS5 promoter, but not in the ADH1 promoter, although the TATA element functions well in both. Thus, we next examined whether the TATA element could induce ectopic initiation sites in the RPS5 promoter in a TFIID-dependent manner (Figure 3A and B). We constructed TAF1 or taf1-N568Δ strains carrying the same set of reporter constructs shown in Figure 2B, and compared the CE activity of the TATA element inserted at various positions in both strains. The results showed that the ectopic initiation sites at −88/−71 bp were TFIID-dependent (lanes 33 and 34, Figure 3A), similar to the endogenous sites at −37/−27/−23 bp, while the ectopic initiation sites at −160/−133 bp were TFIID-independent (lanes 41 and 42, Figure 3A). To our knowledge, this is the first evidence showing that the TFIID-dependence of CE can be altered by position, even within the same core promoter. Also, this is consistent with previous observations that the conversion of atypical TATA (−125 to −120 bp in the original promoter) to consensus TATA could not rescue RPS5 transcription in taf1 strains (33,55). Therefore, TFIID-dependency may be determined by whether the TATA element is localized within the region to which TFIID binds.
Identification of potential CE within the RPS5 promoter
It is difficult to identify CE located downstream within the inserted fragment because pol II scans DNA from upstream (Figure 2). Thus, we examined CE activity of shorter fragments (c.a. 20–50 bp) encompassing −224 to −31 bp of the RPS5 promoter by inserting them at −175/−174 bp to minimize the number of CE within each fragment (Figure 4).
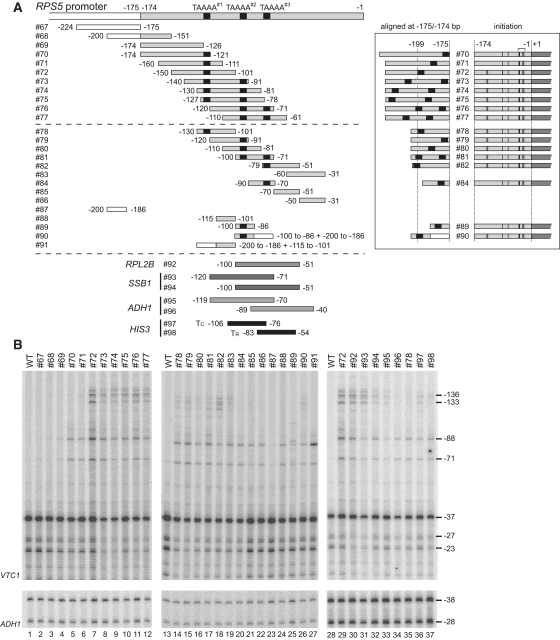
Figure 4.
CE activities of the core promoter fragments derived from RPS5 and several other genes. (A) Schematic diagram showing a series of promoter fragments from RPS5 (top and middle panels), RPL2B, SSB1, ADH1 and HIS3 (bottom panel) tested for CE activities by inserting them at −175/−174 bp of the RPS5 core promoter (as described for #60 in Figure 2B). The three ‘TAAAA’ sequences (TAAAA#1–#3) in the RPS5 promoter are marked with small black rectangles. To facilitate a comparison of the distances between ‘TAAAA’ and the −175/−174 bp insertion sites, the constructs containing these ‘TAAAA’ sequences were aligned (see inset) according to their insertion sites (i.e. −175/−174 bp). Transcriptional initiation activities from the sites at −136, −133, −88, −71, −37, −27 and −23 bp of these constructs were examined in (B) and summarized schematically as in Figure 2D (see inset). The boundaries corresponding to the 5′- or 3′-ends of each promoter fragment are indicated. The number of each construct is also indicated (left). Note that #90 and #91 are chimeric constructs between UAS (white rectangle) and core (grey rectangle). Detailed sequence information is given in Supplementary Figure 1. (B) CE activity of each insert (#67–#98 as depicted in A) to induce ectopic initiation site(s). Primer extension analysis was done as described in Figure 2B.
The results showed that the region upstream of −126 bp (#67–69) is almost inactive in this assay (lanes 1–4, Figure 4B), consistent with the result showing that #19, lacking the region between −338 and −127 bp, has similar activity to that of WT (lanes 12–13, Figure 1C). Remarkably, an extension of only 5 bp (TAAAA in #70) significantly enhanced initiation from −88/−71 bp (lanes 4 and 5), consistent with the result showing that the CE within −174 to −81 bp was assigned to the same sequence (atypical TATA element ‘CTAAAATA’, Figure 2B).
The signals for ectopic initiation sites at −136/−133 bp were stronger in #72 than in #70 and #71 (lanes 5–7), probably because the ‘TAAAA’ sequence moved upstream from −179 to −175] (#70; note that the numbering here is from A (+1) of ATG within the chimeric promoter bearing the insert) or [−189 to −185] (#71) to [−199 to −195] (#72) bp (inset, Figure 4A). Furthermore, ectopic initiation profiles for #73–77 were similar to those of #72 (lanes 7–12). In #73–77, the ‘TAAAA’ sequence was duplicated at [−209 to −205] and [−180 to −176] (#73), [−219 to −215] and [−190 to −186] (#74), [−222 to −218] and [−193 to −189] (#75), [−200 to −196] and [−182 to −178] (#76), and −210 to −206] and [−192 to −188] (#77) bp, corresponding to either pair of the three ‘TAAAA’ sequences localized at [−125 to −121] (TAAAA#1), [−96 to −92] (TAAAA#2), and [−78 to −74] (TAAAA#3) bp in the original RPS5 promoter. Importantly, the upstream ‘TAAAA’ sequence in these constructs is located further upstream than in #72, indicating that the ‘TAAAA’ sequence located at −199 to −195 bp (#72), or further upstream, (#73–77) could induce ectopic initiation at −136/−133/−88/−71 bp (inset, Figure 4A).
Next, we measured the CE activity of ∼30 bp length variants (#78–83) and found that they induced ectopic initiation from −136/−133/−88/−71 bp, but less strongly when compared with ∼50 bp length variants (#74–77) (lanes 14–19). Notably, #78, 81 and 82 contain the ‘TAAAA’ sequence upstream of −199 to −195 bp, #79 and #80 contain it further downstream, and #83 does not contain it (inset, Figure 4A). This suggests that CE other than the ‘TAAAA’ sequence must exist in #79, 80 and 83. Candidate sequences are AT-rich stretches such as ‘TATTTT’ (−204 to −199 bp [#79] corresponding to −120 to −115 bp within the original RPS5 promoter), ‘TTTTTAT’ (−204 to −198 bp [#80], −110 to −104 bp [original]), and ‘TAATTA’ (−204 to −199 bp [#83], −60 to −55 bp [original]) (Supplementary Figure S1) since they are located upstream of −199 to −195 bp in each construct. This agrees with the result showing that ∼20 bp length variants (#84–86) induced ectopic initiation at −88/−71 bp, but not at −136/−133 bp (lanes 20–22), probably because these insertions are too short to reach position −199 to −195 bp. Furthermore, they may induce ectopic initiation from −88/−71 bp because they also contain AT-rich stretches, such as ‘ATTTT’ (−192 to −188 bp [#84], −87 to −83 bp [original]), ‘TAAAA’ (−183 to −179 bp [#84], −78 to −74 bp [original]), ‘TTTTTATTTAATTA’ (−193 to −179 bp [#85], −69 to −55 bp [original]) and ‘ATAATA’ (−188 to −183 bp [#86], −44 to −39 bp [original]) (Supplementary Figure S1). To examine whether these AT-rich stretches functioned as bona fide CE, we compared the ability of three ∼15 bp length variants (#87–89) to induce ectopic initiation at −88/−71 bp (lanes 23–25). #87 is nearly inactive as expected from the results for #67 and #68, whereas #88, containing ‘TTTTTAT’ (−184 to −178 bp [#88], −110 to −104 bp [original]) (Supplementary Figure S1), showed CE activity comparable to, or stronger than, that of #89 containing ‘TAAAA’ (−185 to −181 bp [#89], −96 to −92 bp [original]) (Supplementary Figure S1). This strongly suggests that these AT-rich stretches function as CE in the RPS5 promoter. It is also notable that the ectopic initiation sites at −136/−133 bp were restored when an inactive region (−200 to −186 bp [#87]) was fused to the 3′terminus of an active region (−100 to −86 bp [#89]) (i.e. #90), but not when fused to the 5′terminus of another active region (−115 to −100 bp [#88]) (i.e. #91), again supporting the suggestion that CE must be located at, or more upstream of, −199 to −195 bp to initiate transcription at −136/−133 bp (‘TAAAA’ of #90 is located at −200 to −196 bp) (inset, Figure 4A).
Core promoter sequences derived from other genes also function as CE in the RPS5 promoter
To examine whether core promoter sequences derived from other genes function as CE in the RPS5 core promoter, we first tested CE activity of the TC or TR element of the HIS3 promoter (31,32,38,53) by inserting them at −175/−174 bp of the RPS5 promoter (#97–98, Figure 4A, B). Either element induced ectopic initiation similar to that of #54 bearing the TATA element at −190 to −183 bp (Figure 2B) or #78 bearing the ‘TAAAA’ sequence at −199 to −195 bp (lanes 14 and 35, Figure 4B). Consistently, the TATA sequence (TATATAAA) of the TR element is located at −191 to −184 bp in #98 (Supplementary Figure S1). Importantly, the TC element functioned in the RPS5 core promoter, although it is unclear whether a short AT-rich stretch (‘ATTAT’) located at −193 to −189 bp (Supplementary Figure S1) was responsible for this activity.
The insertion of −120 to −71 bp of the SSB1 promoter at the same site of the RPS5 promoter (#93) induced ectopic transcription (lane 31, Figure 4B) similar to that of #55 or #56 bearing the TATA element at −207 to −200 bp or −220 to −213 bp (lanes 9 or 10, Figure 2B). This is plausible since the TATA element (TATATAAA) of the SSB1 promoter is located at −216 to −209 bp in this construct (Supplementary Figure S1). The insertion of −100 to −51 bp of the SSB1 promoter (#94) induced ectopic transcription, albeit less strongly (lane 32), similar to #97 bearing the HIS3 TC element (lane 36). This construct lacks the TATA element but instead contains the ‘TAAAA’ sequence at −192 to −188 bp (Supplementary Figure S1), suggesting that the original SSB1 promoter has dual CE like the TC and TR elements of the HIS3 promoter. Intriguingly, the ADH1 promoter may also have multiple CE since two distinct but overlapping regions, both located downstream of the TATA element (#95–96), also showed weak CE activity (lanes 33–34). This is somewhat paradoxical, as the RPS5 CE does not appear to function in the ADH1 promoter (Figure 2E), but is consistent with the fact that the ADH1 core promoter is activated efficiently by RPS5 UAS (36). Finally, the insertion of −100 to −51 bp of the RPL2B promoter (#92) induced ectopic transcription (lane 30) similar to that of #72 (lane 29), probably because it also has the ‘TAAAA’ sequence located at −198 to −194 bp (Supplementary Figure S1). Together, this suggests that many promoters contain multiple CE that function not only in the original promoters, but also in the RPS5 promoter.
Multiple AT-rich stretches function redundantly as CE in the RPS5 promoter
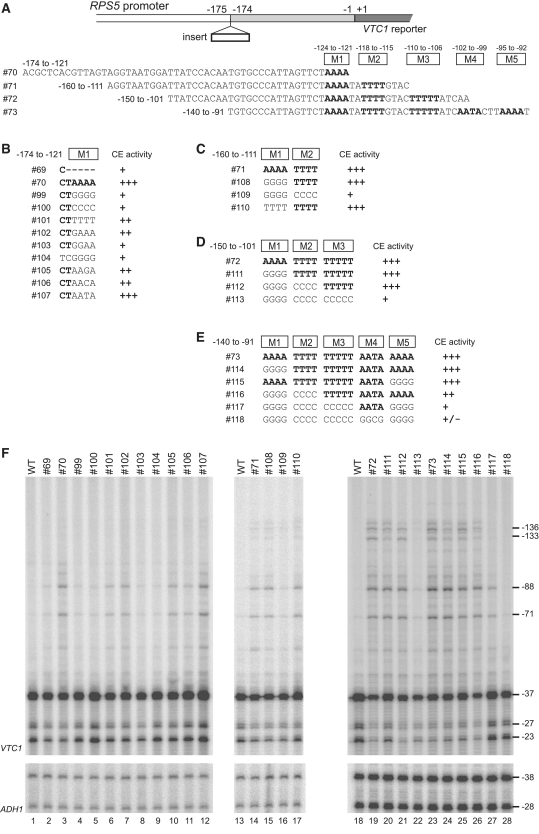
The results described above strongly suggest that several distinct AT-rich stretches function as CE in the RPS5 promoter. To test this, we conducted mutational analyses of the four overlapping promoter fragments containing different numbers of these AT-rich stretches (#70–73) and show CE activity when they are inserted at −175/−174 bp (Figure 5). The #72 and #73 showed ectopic initiation from −136/−133/−88/−71 bp, whereas #70 and #71 did only from −88/−71 bp (Figure 4B). When the ‘TAAAA’ sequence in #70 was changed to ‘TGGGG’ (#99), ‘TCCCC’ (#100), ‘TGGAA’ (#103), or ‘CGGGG’ (#104), these constructs showed minimal CE activity, similar to that of #69 (lanes 1–5, 8 and 9, Figure 5F). In contrast, the substitution mutant bearing ‘TTTTT’ (#101), ‘TGAAA’ (#102), ‘TAAGA’ (#105), or ‘TAACA’ (#106) at the same position showed stronger activity than #69 (lanes 1–3, 6–7 and 10–11), while the mutant bearing ‘TAATA’ (#107) showed similar activity to that of the wild-type #70 (lanes 3 and 12). This suggests that the number of A or T residues within the ‘TAAAA’ sequence (rather than the sequence itself) may be critical for CE function.
Figure 5.
Redundant CE function of multiple AT-rich sequences derived from the RPS5 promoter in ectopic initiation assays. (A) Schematic diagram showing the sequences of the RPS5 core promoter fragments tested for CE activity in (F) by inserting them at the −175/−174 bp position of the RPS5 promoter as indicated (top). The five mutated AT-rich regions (M1–M5 in B–E) are shown. (B) Sequences of #99–#107 generated from #70. CE activities in (F) are summarized at the right. (C) Sequences of #108–#110 generated from #71. CE activities in (F) are summarized at the right. (D) Sequences of #111–#113 generated from #72. CE activities in (F) are summarized at the right. (E) Sequences of #114–#118 generated from #73. CE activities in (F) are summarized at the right. (F) CE activity of each insert (#69–#73 and #99–#118 depicted in B–E) to induce ectopic initiation. Primer extension analysis was done as described in Figure 2B.
Substituting ‘TAAAA’ with ‘TGGGG’ greatly reduced CE activity in #70 (#99) (lanes 3–4) but not in #71 (#108) (lanes 13–15). However, when another AT-rich stretch (‘TATTTT’) located downstream was simultaneously changed to ‘TACCCC’ (#109), it almost abolished the CE activity of #71 and decreased it to the level of #69 (lanes 2, 13, 14 and 16), suggesting that the ‘TAAAA’ and ‘(TA)TTTT’ sequences function redundantly as CE in the RPS5 promoter. Analogously, the simultaneous substitution of these two sequences (#112) was not enough to abolish the CE activity of #72 (lanes 18, 19 and 21), which contains another AT-rich stretch (‘TTTTTAT’) further downstream (Figure 5A). As expected, triple substitution (including this ‘TTTTT(AT)’ sequence) (#113) decreased the CE activity to the level of #69 (lanes 2, 18, 19 and 22). Strikingly, when the −140 to −91 bp region (#73) containing two more additional AT-rich stretches (‘AATA’ and ‘TTAAAAT’, Figure 5A) was subjected to the same mutational assay, all five AT-rich sequences had to be substituted simultaneously to abolish CE activity (lanes 23–28). The ectopic initiation at −136/−133 bp was selectively weakened when the AT-rich sequences located further upstream were substituted (lanes 25–27), indicating that they function independently as CE where pol II is assembled into PIC and released to scan DNA downstream. Therefore, we conclude that multiple AT-rich sequences within the −125 to 91 bp region of the RPS5 promoter have redundant CE function.
AT-rich sequences are physiologically relevant CE for the RPS5 promoter
We showed that multiple AT-rich sequences induce ectopic initiation at −136/−133/−88/−71 bp when inserted at −175/−174 bp (Figure 5B). However, it is unclear whether these sequences play crucial roles in intrinsic initiation from the original RPS5 promoter. A search for AT-rich stretches within the RPS5 core promoter corresponding to the −174 to −51 bp region that supports intrinsic initiation from −37/−27/−23/−18/−13/−9 bp (see the result of #30 lacking this region, Figure 1E) revealed that it contains several additional AT-rich sequences downstream of the five sequences tested in Figure 5 (Figure 6A) and a region (−161 to −138 bp) with weak similarity to the TC element of the HIS3 promoter (53) (lower panel, Figure 6A). Intrinsic initiation from −13/−9 bp was observed for #31 (containing the latter TC-like region), but not for #30 (without this region) (lanes 7–8, Figure 1E). This implies that the TC-like region may have CE activity.
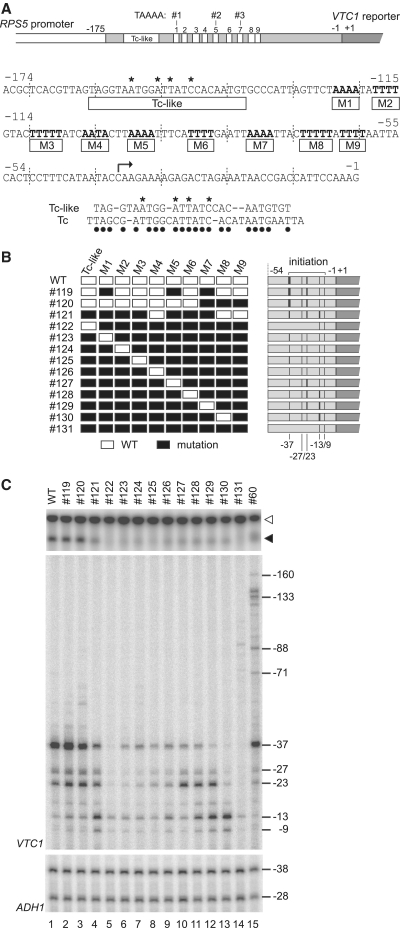
Figure 6.
Highly redundant functions of multiple AT-rich CE within the original RPS5 promoter. (A) Schematic diagram showing the sequence of the RPS5 core promoter (−174 to −1 bp) in which 10 potential CE are highlighted. The right-angled arrow indicates a major transcriptional initiation site (−37 bp) in the RPS5 promoter. Sequence comparison between the Tc-like region and the HIS3 Tc element (53) is shown below the sequence of the RPS5 core promoter. Asterisks indicate residues mutated in (B) and dots indicate identical residues in the two elements. (B) Schematic diagram showing a series of RPS5 promoter (−631 to −1 bp) constructs (#119–#131) containing different combinations of 10 intact (white rectangle) or mutated (black rectangle) potential CE. Detailed sequence information for these constructs is shown in Supplementary Figure 2. Transcriptional initiation activities from the sites at −160, −133, −88, −71, −37, −27, −23, −13 and −9 bp that were examined in (C) are summarized schematically as in Figure 2D. (C) Transcriptional activity of the RPS5 promoters constructed in (B). Northern blot analysis (top panel) to determine the expression levels of VTC1 was performed as described in Figure 1B. Primer extension analysis was performed for VTC1 (middle panel) and ADH1 (control, bottom panel) as described in Figure 2B.
To test whether these multiple AT-rich sequences and the TC-like region function as CE in the original RPS5 promoter, we mutated them in various combinations and compared CE activity in the resulting mutants (Figure 6B, C, Supplementary Figures S2 and S3). The results showed that simultaneous mutation of three elements (M1-5-7 and M7-8-9) from a total of 10 (Figure 6B) affected CE activity only slightly (lanes 1–3, Figure 6C), whereas mutation of seven elements (TC-like-M1-2-3-5-6-7) affected it more significantly (lane 4, Figure 6C). The position of non-mutated elements appears to determine the distribution profile of the intrinsic initiation sites. For instance, when M8-9 is active, but other CE located upstream are mostly inactive (#121), initiation from −37 bp becomes weaker; however, that from −13/−9 bp becomes stronger (lane 4, Figure 6C). These observations match those in Figure 5, where some of these multiple AT-rich sequences were inserted at −175/−174 bp to create ectopic initiation sites at −136/−133/−88/−71 bp. Thus, it is likely that the ectopic initiation assays explored in this study (Figures 2–4) measure CE activity in a physiologically relevant manner.
Most strikingly, we found that CE activity remained detectable even when any combination of the eight AT-rich sequences among the nine was mutated simultaneously (lanes 6–13, Figure 6C). That is, any single copy of these AT-rich sequences is enough to produce transcripts from intrinsic initiation sites, albeit less strongly than the wild-type promoter. As expected, the position of non-mutated elements correlated well with that of the intrinsic initiation site (lanes 6–13). Somewhat puzzlingly, the TC-like region appears to be inactive in this assay as there are no differences in the initiation profiles of #122 and #131 (lanes 5 and 14). Although the reason for such a discrepancy between the effects of the TC-like region on intrinsic initiation of two different mutants (#31, Figure 1E and #122, Figure 6C) is unclear, it may be due to differences in the distance between the TC-like region and the initiation site (i.e. it is too great to induce intrinsic initiation in #122). This may also explain why #69 (containing the TC-like region) was almost inactive (Figure 4B). Therefore, we conclude that multiple AT-rich sequences are physiologically-relevant CE for the RPS5 promoter and function redundantly.
DISCUSSION
This study shows that multiple AT-rich sequences function as CE within the TATA-less RPS5 core promoter in Saccharomyces cerevisiae. It is remarkable that many similarities exist between the promoter architectures of RPS5 and HIS3, the latter of which was characterized extensively in previous studies (31,32,38,52–54).
It was shown that the HIS3 promoter contains two functionally distinct CE (TC and TR), which direct transcription from the +1 and +13 sites, respectively. TR corresponds to a typical TATA element and mediates activation by Gcn4p and Gal4p, whereas TC is a relatively large (∼30 bp) TATA-less element unresponsive to these acidic activators. Notably, TC is more resistant to substitution mutations than TR. In fact, simultaneous mutation of the three short AT-rich stretches (‘ATTAT’, ‘ATAAT’ and ‘AATTA’) within TC is required for the elimination of its CE activity (53). Furthermore, short AT-rich sequences (∼7 bp) with some resemblance to a TATA element are sufficient to mediate TC function (54). These unusual properties of TC (lacks a TATA element but contains multiple, highly redundant AT-rich sequences, and a poor response to acidic activators) are also seen in the RPS5 core promoter comprising several short AT-rich CE (34) (Figure 6). Given that transcription mediated by TC and RPS5 CE are both TFIID-dependent (85) (Figure 2), these CE may be novel recognition sites for TFIID. Consistent with this, the region overlapping some of these multiple AT-rich CE in the RPS5 core promoter is bound by purified TFIID in the presence of TFIIA (56).
The reason that AT-rich CE have not been identified in endogenous yeast promoters (except TC) may be due, in part, to their highly redundant features in addition to a lack of sequence conservation. TATA elements are highly conserved (13,16) and have strict sequence requirements for CE function (38), probably because base changes affect not only TBP binding but also higher order complex formation with TFIIA and TFIIB (40). In contrast, RPS5 CE appear to have more relaxed sequence requirements. For instance, several AT-rich substitution mutants of the ‘TAAAA’ sequence (−125 to −121 bp) remained functional in the ectopic initiation assays (Figure 5). To explore this further, we randomized the ‘AATA’ sequence (M4: −102 to −99 bp) in the original RPS5 promoter lacking all potentially functional CE except M4 and compared CE activity in more than 60 mutants to determine the effect of each substitution (Supplementary Figure S4). The results showed that mutants comprising only A or T residues were mostly active (Supplementary Figure S4B), whereas those comprising only G or C residues were inactive (Supplementary Figure S4E). Although some sequence preferences may exist (Supplementary Figure S4C and D), activity tends to be correlated with the number of A or T residues included within the CE. This suggests that the reason it has been difficult to identify AT-rich CE in endogenous promoters using the usual mutational approaches is that many A/T residues need to be changed simultaneously to G/C residues after eliminating the function of neighboring CE with similar activity. In this regard, the ectopic initiation assays we have exploited here should provide an advantage in the identification of such CE, since only a portion of the original core promoter containing a minimal number of CE can be tested. In fact, several novel CE candidates were identified in both TATA-containing (ADH1 and SSB1) and TATA-less (RPL2B) promoters (Figure 4). Hence, core promoters carrying multiple CE may be more frequently utilized by yeast genes than previously anticipated.
We showed that insertion of the TATA element at −200/−199 bp (#55) induced ectopic initiation at −138/−133 bp, or further downstream (Figure 2B), in a TATA-dependent manner (Supplementary Figure S5). When we inserted randomized 10 bp sequences at the same site, six (r8, r16, r17, r37, r80 and r82) of 88 randomly selected clones showed similar ectopic initiation profiles to that of #55 (Supplementary Figure S5). However, only r80 contained a previously identified functional TATA element (TATTTA) (32). Intriguingly, the other active clones contained either TTAAA (r8, r37 and r82) or TTAAG (r16 and r17), neither of which were found in inactive clones. Remarkably, r25 contained a TAAAA sequence that was active when inserted at −175/−174 bp (Figure 5), but was not active in this assay (Supplementary Figure S5; note that the juxtaposed ‘A’ residue at −199 bp was taken into account for r37 and r25). This suggests that CE activity varies depending on the surrounding sequences, and that ‘TTAA(A/G)’ may function as CE specifically at −200/−199 bp, but not at other sites. In agreement with this, more than a dozen AT-rich sequences were previously identified as active CE by screening randomized oligonucleotides at the TR site in the HIS3 core promoter, but did not contain ‘TTAA(A/G)’ (52). However, further analysis is required to confirm the hypothesis that preferred sequences for CE are determined by surrounding sequences and/or more remote elements like UAS.
The TATA element has transcriptional directionality. Specifically, when the TATA element is reversed, the resulting core promoter does not mediate reversed transcription but simply becomes weak in the forward direction (20,42,86). As TBP binds to the TATA element in two orientations (bi-directionally), at least in solution, correct binding of TBP must be facilitated by other factors including TFIIA, TFIIB, and/or activators (20,87,88). Since RPS5 CE have more relaxed sequence requirements than the TATA element, it is likely that the former may also have weaker transcriptional directionality than the latter. To test this, the −174 to −50 bp or −134 to −50 bp regions were reversed in the RPS5 core promoter and the CE activity directing forward transcription was compared (Supplementary Figure S6). When the −174 to −50 bp region was reversed (#135), the major intrinsic initiation site at −37 bp disappeared, whereas ectopic initiation sites appeared further upstream, suggesting that the reversed Tc-like region is inactive but that some reversed CE are active, albeit weakly. When the −134 to −50 bp region was reversed (#132), intrinsic initiation at −37 bp decreased significantly but ectopic initiation was induced slightly at a region further upstream, again suggesting that the reversed CE are weakly functional. This assumption was confirmed by testing the effects of two substitution mutations within the reversed CE (#133 and #134) that abolished intrinsic and ectopic initiation. Reversed transcription was not induced by these reversed CE (unpublished observations). Therefore, we suppose that the entire set of RPS5 CE has strong transcriptional directionality similar to that of the TATA element. However, each RPS5 CE may have weaker directionality, as exemplified by the fact that randomized AT-rich sequences at the M4 region showed similar activity, even when the sequence was reversed (#r1–r13, #r2–r5, #126–r9, #r4–r7 and #r6–r12, Supplementary Figure S4A).
One of the most interesting observations was that the TFIID-dependence of the TATA element in inducing ectopic initiation varies depending on the position at which it is inserted (Figure 3A, B). Previous studies show that the conversion of atypical TATA (‘TAAAAT’ at −125 to −120 bp) to consensus TATA (‘TATAAA’) could not restore RPS5 transcription in taf1 strains (33,55). This decrease in transcription is due to core promoter-specific defects in activation by the RPS5 UAS (33). Therefore, it is likely that such activation defects cannot be restored when the TATA element is created within the region to which TFIID binds, but can be restored when it is created in regions to which it does not bind. Consistent with this, similar defects in activation by the ADH1 UAS in taf1 strains were restored when the TATA element was located at a region distant from the RPS5 core promoter (55). Therefore, we assume that the function of the TATA element may differ depending on the promoter context, since each core factor (free TBP, TFIID, SAGA etc.) regulates TATA function in its own way.
An enigmatic question is how such short AT-rich sequences could have CE function even when they exist as a single copy in the core promoter (Figure 6). It is hard to imagine that each AT-rich sequence (4–6 bp) can provide a specific recognition site for TFIID, given that it exhibits relaxed sequence requirements (Figure 5 and Supplementary Figure S4). Recent genome-wide studies show that yeast promoters contain a nucleosome free region (NFR) near the transcriptional initiation site flanked by two positionally well-defined nucleosomes (−1 and +1) (89). PIC is assembled on NFR to initiate transcription from a site within the +1 nucleosome. AT-rich CE identified here can be regarded as short segments of a poly(dA:dT) tract that plays an important role in forming NFR (90). Although each CE may be too short to exclude nucleosome formation on the core promoter, it may induce NFR in cooperation with other factors like Abf1p or Rap1p (91). If this is the case, another role of AT-rich CE may be to induce NFR that is a prerequisite for PIC assembly and subsequent transcriptional initiation. In fact, it was recently proposed that TFIID and SAGA might be recruited to the TATA-less promoters by binding to −1/+1 nucleosomes rather than to core promoter elements (89). However, the fact that the RPS5 core promoter is bound by purified TFIID (56), and that the entire set of RPS5 CE shows strong transcriptional directionality (Supplementary Figure S6), still supports a view that these CE may function as recognition sites for TFIID. Although the detailed mechanism is unclear, we prefer a model in which RPS5 CE have a dual function: one is to exclude nucleosome formation, and the other is to provide a recognition site for TFIID. The latter may depend on the entire set of CE, since a single copy showed rather weak activity in both transcriptional strength (Figure 6) and directionality (Supplementary Figure S4). Further studies are necessary to clarify the function of these AT-rich CE and elucidate the mechanism of how TATA-less promoters are transcribed in yeast.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Japan Society for the Promotion of Science; the Ministry of Education, Culture, Sports, Science and Technology of Japan; CREST program of the Japan Science and Technology Agency; Junior Research Associate Program in RIKEN (The Institute of Physical and Chemical Research), Japan. Funding for open access charge: Japan Society for the Promotion of Science.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Dr Sewon Ki for constructing yeast strain yTK6339. The authors also thank Drs Masahiro Shirakawa, Takehiko Shibata, Yoshifumi Nishimura, Hiroshi Iwasaki, Tadashi Wada and members of our laboratory for advice and comments on this work.
REFERENCES
- 1.Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 3.Venters BJ, Pugh BF. How eukaryotic genes are transcribed. Crit. Rev. Biochem. Mol. Biol. 2009;44:117–141. doi: 10.1080/10409230902858785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tora L. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 2002;16:673–675. doi: 10.1101/gad.976402. [DOI] [PubMed] [Google Scholar]
- 5.Ranish JA, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson KM, Wang J, Smallwood A, Arayata C, Carey M. TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev. 2002;16:1852–1863. doi: 10.1101/gad.995702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson KM, Carey M. Assembly of a mediator/TFIID/TFIIA complex bypasses the need for an activator. Curr. Biol. 2003;13:772–777. doi: 10.1016/s0960-9822(03)00283-5. [DOI] [PubMed] [Google Scholar]
- 8.Lieberman PM, Berk AJ. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA-promoter DNA complex formation. Genes Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- 9.Chi T, Lieberman P, Ellwood K, Carey M. A general mechanism for transcriptional synergy by eukaryotic activators. Nature. 1995;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- 10.Matangkasombut O, Auty R, Buratowski S. Structure and function of the TFIID complex. Adv. Protein Chem. 2004;67:67–92. doi: 10.1016/S0065-3233(04)67003-3. [DOI] [PubMed] [Google Scholar]
- 11.Garbett KA, Tripathi MK, Cencki B, Layer JH, Weil PA. Yeast TFIID serves as a coactivator for Rap1p by direct protein-protein interaction. Mol. Cell. Biol. 2007;27:297–311. doi: 10.1128/MCB.01558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J, Iyer VR. Global role of TATA box-binding protein recruitment to promoters in mediating gene expression profiles. Mol. Cell. Biol. 2004;24:8104–8112. doi: 10.1128/MCB.24.18.8104-8112.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 14.Gershenzon NI, Ioshikhes IP. Synergy of human Pol II core promoter elements revealed by statistical sequence analysis. Bioinformatics. 2005;21:1295–1300. doi: 10.1093/bioinformatics/bti172. [DOI] [PubMed] [Google Scholar]
- 15.Jin VX, Singer GA, Agosto-Perez FJ, Liyanarachchi S, Davuluri RV. Genome-wide analysis of core promoter elements from conserved human and mouse orthologous pairs. BMC Bioinformatics. 2006;7:114. doi: 10.1186/1471-2105-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang C, Bolotin E, Jiang T, Sladek FM, Martinez E. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene. 2007;389:52–65. doi: 10.1016/j.gene.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 18.van Werven FJ, van Teeffelen HA, Holstege FC, Timmers HT. Distinct promoter dynamics of the basal transcription factor TBP across the yeast genome. Nat. Struct. Mol. Biol. 2009;16:1043–1048. doi: 10.1038/nsmb.1674. [DOI] [PubMed] [Google Scholar]
- 19.Morachis JM, Murawsky CM, Emerson BM. Regulation of the p53 transcriptional response by structurally diverse core promoters. Genes Dev. 2010;24:135–147. doi: 10.1101/gad.1856710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 21.Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat. Rev. Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- 22.Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter - the gateway to transcription. Curr. Opin. Cell. Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anish R, Hossain MB, Jacobson RH, Takada S. Characterization of transcription from TATA-less promoters: identification of a new core promoter element XCPE2 and analysis of factor requirements. PLoS One. 2009;4:e5103. doi: 10.1371/journal.pone.0005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yarden G, Elfakess R, Gazit K, Dikstein R. Characterization of sINR, a strict version of the Initiator core promoter element. Nucleic Acids Res. 2009;37:4234–4246. doi: 10.1093/nar/gkp315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohler U, Wassarman DA. Promoting developmental transcription. Development. 2010;137:15–26. doi: 10.1242/dev.035493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theisen JW, Lim CY, Kadonaga JT. Three key subregions contribute to the function of the downstream RNA polymerase II core promoter. Mol. Cell. Biol. 2010;30:3471–3479. doi: 10.1128/MCB.00053-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butler JE, Kadonaga JT. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohtsuki S, Levine M, Cai HN. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 1998;12:547–556. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juven-Gershon T, Hsu JY, Kadonaga JT. Caudal, a key developmental regulator, is a DPE-specific transcriptional factor. Genes Dev. 2008;22:2823–2830. doi: 10.1101/gad.1698108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sengupta T, Cohet N, Morle F, Bieker JJ. Distinct modes of gene regulation by a cell-specific transcriptional activator. Proc. Natl Acad. Sci. USA. 2009;106:4213–4218. doi: 10.1073/pnas.0808347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Struhl K. Constitutive and inducible Saccharomyces cerevisiae promoters: evidence for two distinct molecular mechanisms. Mol. Cell. Biol. 1986;6:3847–3853. doi: 10.1128/mcb.6.11.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harbury PA, Struhl K. Functional distinctions between yeast TATA elements. Mol. Cell. Biol. 1989;9:5298–5304. doi: 10.1128/mcb.9.12.5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsukihashi Y, Kawaichi M, Kokubo T. Requirement for yeast TAF145 function in transcriptional activation of the RPS5 promoter that depends on both core promoter structure and upstream activating sequences. J. Biol. Chem. 2001;276:25715–25726. doi: 10.1074/jbc.M102416200. [DOI] [PubMed] [Google Scholar]
- 34.Cheng JX, Floer M, Ononaji P, Bryant G, Ptashne M. Responses of four yeast genes to changes in the transcriptional machinery are determined by their promoters. Curr. Biol. 2002;12:1828–1832. doi: 10.1016/s0960-9822(02)01257-5. [DOI] [PubMed] [Google Scholar]
- 35.Mencia M, Moqtaderi Z, Geisberg JV, Kuras L, Struhl K. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell. 2002;9:823–833. doi: 10.1016/s1097-2765(02)00490-2. [DOI] [PubMed] [Google Scholar]
- 36.Li XY, Bhaumik SR, Zhu X, Li L, Shen WC, Dixit BL, Green MR. Selective recruitment of TAFs by yeast upstream activating sequences. Implications for eukaryotic promoter structure. Curr. Biol. 2002;12:1240–1244. doi: 10.1016/s0960-9822(02)00932-6. [DOI] [PubMed] [Google Scholar]
- 37.Zhong P, Melcher K. Identification and characterization of the activation domain of Ifh1, an activator of model TATA-less genes. Biochem. Biophys. Res. Commun. 2010;392:77–82. doi: 10.1016/j.bbrc.2009.12.172. [DOI] [PubMed] [Google Scholar]
- 38.Chen W, Struhl K. Saturation mutagenesis of a yeast his3 “TATA element”: genetic evidence for a specific TATA-binding protein. Proc. Natl Acad. Sci. USA. 1988;85:2691–2695. doi: 10.1073/pnas.85.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hahn S, Buratowski S, Sharp PA, Guarente L. Yeast TATA-binding protein TFIID binds to TATA elements with both consensus and nonconsensus DNA sequences. Proc. Natl Acad. Sci. USA. 1989;86:5718–5722. doi: 10.1073/pnas.86.15.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stewart JJ, Fischbeck JA, Chen X, Stargell LA. Non-optimal TATA elements exhibit diverse mechanistic consequences. J. Biol. Chem. 2006;281:22665–22673. doi: 10.1074/jbc.M603237200. [DOI] [PubMed] [Google Scholar]
- 41.Chen W, Struhl K. Yeast mRNA initiation sites are determined primarily by specific sequences, not by the distance from the TATA element. EMBO J. 1985;4:3273–3280. doi: 10.1002/j.1460-2075.1985.tb04077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagawa F, Fink GR. The relationship between the “TATA” sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1985;82:8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hahn S, Hoar ET, Guarente L. Each of three “TATA elements” specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1985;82:8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McNeil JB, Smith M. Transcription initiation of the Saccharomyces cerevisiae iso-1-cytochrome c gene. Multiple, independent T-A-T-A sequences. J. Mol. Biol. 1986;187:363–378. doi: 10.1016/0022-2836(86)90439-0. [DOI] [PubMed] [Google Scholar]
- 45.Mosch HU, Graf R, Braus GH. Sequence-specific initiator elements focus initiation of transcription to distinct sites in the yeast TRP4 promoter. EMBO J. 1992;11:4583–4590. doi: 10.1002/j.1460-2075.1992.tb05560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohishi-Shofuda T, Suzuki Y, Yano K, Sakurai H, Fukasawa T. Transcription initiation mediated by initiator binding protein in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1999;255:157–163. doi: 10.1006/bbrc.1999.0157. [DOI] [PubMed] [Google Scholar]
- 47.Kuehner JN, Brow DA. Quantitative analysis of in vivo initiator selection by yeast RNA polymerase II supports a scanning model. J. Biol. Chem. 2006;281:14119–14128. doi: 10.1074/jbc.M601937200. [DOI] [PubMed] [Google Scholar]
- 48.Chalkley GE, Verrijzer CP. DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250- TAF(II)150 complex recognizes the initiator. EMBO J. 1999;18:4835–4845. doi: 10.1093/emboj/18.17.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Struhl K. Molecular mechanisms of transcriptional regulation in yeast. Annu. Rev. Biochem. 1989;58:1051–1077. doi: 10.1146/annurev.bi.58.070189.005155. [DOI] [PubMed] [Google Scholar]
- 50.Juven-Gershon T, Cheng S, Kadonaga JT. Rational design of a super core promoter that enhances gene expression. Nat. Methods. 2006;3:917–922. doi: 10.1038/nmeth937. [DOI] [PubMed] [Google Scholar]
- 51.Lee DH, Gershenzon N, Gupta M, Ioshikhes IP, Reinberg D, Lewis BA. Functional characterization of core promoter elements: the downstream core element is recognized by TAF1. Mol. Cell. Biol. 2005;25:9674–9686. doi: 10.1128/MCB.25.21.9674-9686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singer VL, Wobbe CR, Struhl K. A wide variety of DNA sequences can functionally replace a yeast TATA element for transcriptional activation. Genes Dev. 1990;4:636–645. doi: 10.1101/gad.4.4.636. [DOI] [PubMed] [Google Scholar]
- 53.Mahadevan S, Struhl K. Tc, an unusual promoter element required for constitutive transcription of the yeast HIS3 gene. Mol. Cell. Biol. 1990;10:4447–4455. doi: 10.1128/mcb.10.9.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iyer V, Struhl K. Mechanism of differential utilization of the his3 TR and TC TATA elements. Mol. Cell. Biol. 1995;15:7059–7066. doi: 10.1128/mcb.15.12.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen WC, Green MR. Yeast TAF(II)145 functions as a core promoter selectivity factor, not a general coactivator. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- 56.Sanders SL, Garbett KA, Weil PA. Molecular characterization of Saccharomyces cerevisiae TFIID. Mol. Cell. Biol. 2002;22:6000–6013. doi: 10.1128/MCB.22.16.6000-6013.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen WC, Bhaumik SR, Causton HC, Simon I, Zhu X, Jennings EG, Wang TH, Young RA, Green MR. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J. 2003;22:3395–3402. doi: 10.1093/emboj/cdg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amberg DC, Burke DJ, Strathern JN. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.; 2005. [Google Scholar]
- 59.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 60.Kitazono AA, Tobe BT, Kalton H, Diamant N, Kron SJ. Marker-fusion PCR for one-step mutagenesis of essential genes in yeast. Yeast. 2002;19:141–149. doi: 10.1002/yea.806. [DOI] [PubMed] [Google Scholar]
- 61.Tsukihashi Y, Miyake T, Kawaichi M, Kokubo T. Impaired core promoter recognition caused by novel yeast TAF145 mutations can be restored by creating a canonical TATA element within the promoter region of the TUB2 gene. Mol. Cell. Biol. 2000;20:2385–2399. doi: 10.1128/mcb.20.7.2385-2399.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takahata S, Ryu H, Ohtsuki K, Kasahara K, Kawaichi M, Kokubo T. Identification of a novel TATA element-binding protein binding region at the N terminus of the Saccharomyces cerevisiae TAF1 protein. J. Biol. Chem. 2003;278:45888–45902. doi: 10.1074/jbc.M306886200. [DOI] [PubMed] [Google Scholar]
- 63.Kasahara K, Ki S, Aoyama K, Takahashi H, Kokubo T. Saccharomyces cerevisiae HMO1 interacts with TFIID and participates in start site selection by RNA polymerase II. Nucleic Acids Res. 2008;36:1343–1357. doi: 10.1093/nar/gkm1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rudra D, Zhao Y, Warner JR. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J. 2005;24:533–542. doi: 10.1038/sj.emboj.7600553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wade JT, Hall DB, Struhl K. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature. 2004;432:1054–1058. doi: 10.1038/nature03175. [DOI] [PubMed] [Google Scholar]
- 66.Schawalder SB, Kabani M, Howald I, Choudhury U, Werner M, Shore D. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature. 2004;432:1058–1061. doi: 10.1038/nature03200. [DOI] [PubMed] [Google Scholar]
- 67.Martin DE, Soulard A, Hall MN. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell. 2004;119:969–979. doi: 10.1016/j.cell.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 68.Zhao Y, McIntosh KB, Rudra D, Schawalder S, Shore D, Warner JR. Fine-structure analysis of ribosomal protein gene transcription. Mol. Cell. Biol. 2006;26:4853–4862. doi: 10.1128/MCB.02367-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rudra D, Mallick J, Zhao Y, Warner JR. Potential interface between ribosomal protein production and pre-rRNA processing. Mol. Cell. Biol. 2007;27:4815–4824. doi: 10.1128/MCB.02062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gordan R, Hartemink AJ, Bulyk ML. Distinguishing direct versus indirect transcription factor-DNA interactions. Genome Res. 2009;19:2090–2100. doi: 10.1101/gr.094144.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hothorn M, Neumann H, Lenherr ED, Wehner M, Rybin V, Hassa PO, Uttenweiler A, Reinhardt M, Schmidt A, Seiler J, et al. Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science. 2009;324:513–516. doi: 10.1126/science.1168120. [DOI] [PubMed] [Google Scholar]
- 72.Ki S, Sugihara F, Kasahara K, Tochio H, Okada-Marubayashi A, Tomita S, Morita M, Ikeguchi M, Shirakawa M, Kokubo T. A novel magnetic resonance-based method to measure gene expression in living cells. Nucleic Acids Res. 2006;34:e51. doi: 10.1093/nar/gkl135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lavoie H, Hogues H, Mallick J, Sellam A, Nantel A, Whiteway M. Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 2010;8:e1000329. doi: 10.1371/journal.pbio.1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lascaris RF, Mager WH, Planta RJ. DNA-binding requirements of the yeast protein Rap1p as selected in silico from ribosomal protein gene promoter sequences. Bioinformatics. 1999;15:267–277. doi: 10.1093/bioinformatics/15.4.267. [DOI] [PubMed] [Google Scholar]
- 76.Khaperskyy DA, Ammerman ML, Majovski RC, Ponticelli AS. Functions of Saccharomyces cerevisiae TFIIF during transcription start site utilization. Mol. Cell. Biol. 2008;28:3757–3766. doi: 10.1128/MCB.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Giardina C, Lis JT. DNA melting on yeast RNA polymerase II promoters. Science. 1993;261:759–762. doi: 10.1126/science.8342041. [DOI] [PubMed] [Google Scholar]
- 78.Hampsey M. The Pol II initiation complex: finding a place to start. Nat. Struct. Mol. Biol. 2006;13:564–566. doi: 10.1038/nsmb0706-564. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Z, Dietrich FS. Mapping of transcription start sites in Saccharomyces cerevisiae using 5′ SAGE. Nucleic Acids Res. 2005;33:2838–2851. doi: 10.1093/nar/gki583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Faitar SL, Brodie SA, Ponticelli AS. Promoter-specific shifts in transcription initiation conferred by yeast TFIIB mutations are determined by the sequence in the immediate vicinity of the start sites. Mol. Cell. Biol. 2001;21:4427–4440. doi: 10.1128/MCB.21.14.4427-4440.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bhaumik SR, Green MR. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 2002;22:7365–7371. doi: 10.1128/MCB.22.21.7365-7371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shukla A, Bajwa P, Bhaumik SR. SAGA-associated Sgf73p facilitates formation of the preinitiation complex assembly at the promoters either in a HAT-dependent or independent manner in vivo. Nucleic Acids Res. 2006;34:6225–6232. doi: 10.1093/nar/gkl844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li XY, Bhaumik SR, Green MR. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science. 2000;288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- 84.Li XY, Virbasius A, Zhu X, Green MR. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;399:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- 85.Moqtaderi Z, Keaveney M, Struhl K. The histone H3-like TAF is broadly required for transcription in yeast. Mol. Cell. 1998;2:675–682. doi: 10.1016/s1097-2765(00)80165-3. [DOI] [PubMed] [Google Scholar]
- 86.Spencer JV, Arndt KM. A TATA binding protein mutant with increased affinity for DNA directs transcription from a reversed TATA sequence in vivo. Mol. Cell. Biol. 2002;22:8744–8755. doi: 10.1128/MCB.22.24.8744-8755.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cox JM, Hayward MM, Sanchez JF, Gegnas LD, van der Zee S, Dennis JH, Sigler PB, Schepartz A. Bidirectional binding of the TATA box binding protein to the TATA box. Proc. Natl Acad. Sci. USA. 1997;94:13475–13480. doi: 10.1073/pnas.94.25.13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kays AR, Schepartz A. Gal4-VP16 and Gal4-AH increase the orientational and axial specificity of TATA box recognition by TATA box binding protein. Biochemistry. 2002;41:3147–3155. doi: 10.1021/bi015817z. [DOI] [PubMed] [Google Scholar]
- 89.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Segal E, Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr. Opin. Struct. Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yarragudi A, Miyake T, Li R, Morse RH. Comparison of ABF1 and RAP1 in chromatin opening and transactivator potentiation in the budding yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:9152–9164. doi: 10.1128/MCB.24.20.9152-9164.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.