Abstract
Gene expression profiling provides powerful analyses of transcriptional responses to cellular perturbation. In contrast to DNA array-based methods, reporter gene technology has been underused for this application. Here we describe a genomewide, genome-registered collection of Escherichia coli bioluminescent reporter gene fusions. DNA sequences from plasmid-borne, random fusions of E. coli chromosomal DNA to a Photorhabdus luminescens luxCDABE reporter allowed precise mapping of each fusion. The utility of this collection covering about 30% of the transcriptional units was tested by analyzing individual fusions representative of heat shock, SOS, OxyR, SoxRS, and cya/crp stress-responsive regulons. Each fusion strain responded as anticipated to environmental conditions known to activate the corresponding regulatory circuit. Thus, the collection mirrors E. coli's transcriptional wiring diagram. This genomewide collection of gene fusions provides an independent test of results from other gene expression analyses. Accordingly, a DNA microarray-based analysis of mitomycin C-treated E. coli indicated elevated expression of expected and unanticipated genes. Selected luxCDABE fusions corresponding to these up-regulated genes were used to confirm or contradict the DNA microarray results. The power of partnering gene fusion and DNA microarray technology to discover promoters and define operons was demonstrated when data from both suggested that a cluster of 20 genes encoding production of type I extracellular polysaccharide in E. coli form a single operon.
The detailed knowledge of gene expression gained from massively parallel analyses has proven to be powerful and broadly successful. DNA array technology has been widely used for analyses uncovering the consequences of individual mutations (1), gaining understanding of cellular physiology in varied growth conditions (2), and examining disease states (3). Furthermore, responses to pharmaceuticals (4) and crop protection products (5) have been analyzed. Despite the power of this methodology, there are several limitations. For example, artifacts may arise during the RNA isolation (6) or from cross hybridization (7). Moreover, development of facile, high throughput screens based on DNA array technology is unlikely due to multiple, complex steps.
An important alternative to genomewide expression profiling uses reporter gene fusions. Transcriptional fusions have been widely used (8, 9) due to straightforward means for their production (10), identification, and assay (11, 12). In the past, random collections of fusion strains were screened for a particular characteristic; the subsequent mapping of the selected fusions became rate limiting. The advent of complete genomic sequences now allows the creation and utilization of precisely defined sets of fusions. To this end, gfp fusions to a large number of genes in Saccharomyces cerevisiae were used to define transcriptional alterations due to prolonged blocks in isoprenoid biosynthesis (13). Other reporter genes allow analysis of initial responses to environmental perturbation (8). In bacterial systems, bioluminescence encoded by luxCDABE has proven to be especially useful because all components required for light production are provided by expression of this operon (14). The simplicity, sensitivity, and large dynamic range of light measurement permit facile analysis of kinetic responses and multiple samples.
Here we describe the identification and mapping of a large number of random Escherichia coli DNA fragments fused to luxCDABE, thus generating a genome-registered collection. We show that several well-characterized stress-responsive regulons were represented by one or more gene fusions in this collection. Furthermore, expected biological responses and specificity were demonstrated by monitoring the responses to chemical challenges. Among the numerous uses for a large collection of genome-registered gene fusions are operon structure elucidation and verification of results from DNA microarray analyses, examples of which are described here. These applications demonstrated the beneficial synergies of reporter gene technology with other methods of genomewide expression analysis.
Materials and Methods
Bacterial Strains and Plasmids.
The collection of random, 1.8-kbp average, E. coli genomic fragments upstream of Photorhabdus luminescens luxCDABE in a moderate copy number plasmid, pDEW201, has been described (15, 16). A lux clone identification number was assigned to each individual transformant of E. coli strain DPD1675 (ilvB2101 ara thi ΔproAB-lac tolC∷miniTn10). Strain names, where given, are found in Tables 1 and 2. One plasmid contained a chromosomal insert with the promoter region of a putative DNA damage inducible gene, yebF, oriented in the wrong direction to drive transcription of the luxCDABE reporter. After isolation of plasmid DNA, XmaI digestion to release insert DNA from the vector, religation, and transformation, a light producing transformant up-regulated by nalidixic acid was chosen. The orientation of the inserted segment in the plasmid to yield a functional fusion of yebF to the luxCDABE operon was confirmed by DNA sequence analysis.
Table 1.
Gene fusions to selected global regulatory circuits
| Strain | Regulatory circuit | Fusion to: | Basal RLU* | Inducer | Response ratio† |
|---|---|---|---|---|---|
| DPD2247 | SOS | dinF | 0.224 | Nalidixic acid | 2.2 |
| DPD2248 | SOS | dinG | 32.8 | Nalidixic acid | 2.0 |
| DPD2249 | SOS | dinP | 4.41 | Nalidixic acid | 5.0 |
| DPD2250 | SOS | recA | 97.5 | Nalidixic acid | 1.8 |
| DPD2251 | SOS | recA | 99.5 | Nalidixic acid | 2.8 |
| DPD2253 | SOS | uvrA | ND‡ | Nalidixic acid | 1.4 |
| DPD2254 | SOS | uvrD | 15.7 | Nalidixic acid | 1.2 |
| DPD2293 | SOS | ruvA | 14.3 | Nalidixic acid | 4.6 |
| DPD2242 | Heat shock | lon | 6.71 | Ethanol | 5.6 |
| DPD2246 | Heat shock | rpoD | 9.49 | Ethanol | 2.4 |
| DPD2245 | Heat shock | rpoD | 8.07 | Ethanol | 6.8 |
| DPD2243 | Heat shock | clpB | 38.5 | Ethanol | 4.7 |
| DPD2244 | Heat shock | rfaDFCL | 33.1 | Ethanol | 1.4 |
| DPD2272 | SoxR/S | zwf | 16.9 | Methyl viologen | 3.6 |
| DPD2286 | SoxR/S | ribA | 5.8 | Methyl viologen | 4.7 |
| DPD2087 | SoxR/S | inaA | 9.8 | Methyl viologen | 9.0 |
| DPD3509 | SoxR/S | poxB | 1.5 | Methyl viologen | 15.0 |
| DPD2283 | OxyR | ahpC | 92.2 | Hydrogen peroxide | 16.5§ |
| DPD2284 | OxyR | ahpC | 68.3 | Hydrogen peroxide | 15.3§ |
From 100 μl actively growing cultures of E. coli DPD1675 transformants in minimal medium (15).
For nalidixic acid the response ratio was calculated at 110 min after addition; for ethanol, at 40 min; for methyl viologen, at 120 min; and for hydrogen peroxide, at 30 min.
Not determined.
Response ratios from transformants of E. coli strain DPD2228.
Table 2.
Comparison of results from DNA microarray and lux gene fusions
| Gene | Fold induction in array experiment* | Lux fusion strain | Response ratio of lux fusion† |
|---|---|---|---|
| Known SOS genes induced by mitomycin C in array experiment | |||
| recN | 8.3 | None | |
| recA | 3.0 | DPD2250 | 4.6 |
| lexA | 3.6 | None | |
| dinI | 6.7 | None | |
| dinD | 2.2 | None | |
| uvrA | 2.3 | DPD2253 | 2.5 |
| uvrB | 2.1 | None | |
| ruvA | 2.0 | DPD2293 | 3.0 |
| sulA | 5.8 | None | |
| umuC | 2.1 | None | |
| dinP | 2.0 | DPD2249 | 10.4 |
| yebG | 5.8 | None | |
| Known SOS genes not induced by mitomycin C in array experiment | |||
| uvrD | 1.4 | DPD2254 | 2.7 |
| polB | 1.2 | None | |
| dinG | 1.4 | DPD2248 | 4.6 |
| dinF | 1.8 | DPD2247 | 4.2 |
| himA | 1.3 | None | |
| ruvB | 1.5 | None | |
| umuD | 1.2 | None | |
| Putative, novel DNA damage-inducible genes | |||
| mioC | 2.1 | None | |
| xseA | 2.0 | None | |
| insB_2 | 2.2 | None | |
| insB_1 | 2.1 | None | |
| insA_4 | 2.1 | None | |
| secG | 2.2 | None | |
| exbD | 2.2 | DPD2270 | 1.1 |
| trkH | 2.1 | DPD2274 | 1.2 |
| infA | 2.3 | None | |
| hslS | 6.7 | None | |
| hslT | 4.0 | None | |
| cspA | 2.9 | None | |
| dniR | 2.1 | DPD2273 | 1.1 |
| b0531 | 3.3 | None | |
| yebF | 3.3 | DPD3232 | 12.2 |
| b1228 | 2.5 | None | |
| b2940 | 2.3 | None | |
| ylcA | 2.2 | DPD2271 | 1.1 |
| b2559 | 2.0 | None | |
| b3199 | 2.0 | None | |
250 ng/ml treatment of E. coli MG1655 in LB medium for 40 min.
62 ng/ml treatment of E. coli DPD1675 transformants in LB medium at 100 min. For comparison with the DNA array experiment, a later time point was selected to allow for transcription and translation of the 5.8-kb luxCDABE reporter. Also, a lower dose of mitomycin C was chosen because the host strain for the luxCDABE gene fusions contains a mutation in tolC that renders it hypersensitive to many chemicals (34), including mitomycin C (35).
E. coli strain DPD2228 (F− Δlac4169 rpsL pcnB80 zad-2084∷Tn10) was derived from a previously described, plasmid-bearing E. coli strain, DPD2226 (17), by screening for sensitivity to ampicillin. E. coli strain MG1655 (rph-1) has been described (18). E. coli strain 397C contains a mutation in rpoC, whereas strain P90 is rpoC+ (19). Transformation was accomplished by using competent cells as described (20).
Isolation of Plasmid DNA and DNA Sequence Determination.
Cultures (1.2 ml) were grown to saturation in Terrific Broth (GIBCO/BRL) containing 100 μg/ml ampicillin in deep-well microplates. Plasmid DNA was extracted from the cultures by using the Qiagen (Chatsworth, CA) R.E.A.L. method with the following modification to prevent plasmid degradation by nucleases: after lysis of cells, the plates were placed in a boiling water bath for 5 min and then rapidly chilled in an ice-water bath before precipitation with buffer 3. DNA sequencing reactions were performed with approximately 1 μg of plasmid DNA under standard Applied Biosystems Prism DyeTerminator Reaction Ready conditions with previously described primers (15), yielding sequence from each end of the insert. DNA sequences were determined on ABI377-XL 96-lane upgraded Sequencers under 4× run conditions on 5% PAG LongRanger gels (FMC) and analyzed with Applied Biosystems software. DNA sequences were transferred to a unix-based utility for further analysis.
Mapping the luxCDABE Gene Fusions.
A homology search for the sequence from the beginning and end of each insert in both orientations was performed against the complete E. coli sequence (GenBank accession no. U00096) by using Pearson's fasta program (fasta3, version 3.1t13). The essential Fasta options were −nQH −m 10 −z 0. Data about each highly significant alignment (fasta score >1,000, minimum overlap length >200, and minimum identity >70%) was stored in a relational database (SYBASE SYSTEM 11, Sybase, Emeryville, CA). The location of each fragment on the E. coli genome then was based on the above computed homologies for both the beginning and end of the insert, by using the following rules: (i) both distal and proximal ends of the insert must have an unambiguously high sequence homology with E. coli; and (ii) the relative location and orientation of the matches of the sequence determined from the beginning and end of the insert implied a reasonable fragment length. Unambiguous assignments were stored in the relational database. A table of ORF annotations for E. coli was downloaded from the National Center for Biotechnology Information's entrez facility and stored in the relational database. A web-based, tabular interface to the data was created that allowed viewing of the inserts in relation to the annotated map at nucleotide resolution. A java-based graphical interface facilitated visualization of positional and directional relationships.
Bioluminescence Measurements.
For stress response tests, actively growing cultures in LB medium (21) at 37°C were used in duplicate experiments measuring light production as a function of time, as described (15, 22). The final chemical concentrations were 5 μg/ml nalidixic acid (Sigma), 3% ethanol (Quantum, Cincinnati, OH), 250 μg/ml methyl viologen (Sigma), and 0.002% hydrogen peroxide (EM Science). Response ratios were calculated by dividing the bioluminescence from the chemically treated culture by the bioluminescence of the untreated control at each time point (23). Typically, response ratios of 1.5 or greater are considered to represent induction.
Bioluminescence and turbidity, recorded with a Klett-Summerson colorimeter (24), of actively growing cultures were used for calculation of light production per 109 cells. For such experiments, transformants of P90 and 397C were grown at 30°C in LB medium containing 100 μg/ml ampicillin. The unpaired t test was used to compare quadruplicate bioluminescence measurements of strains carrying gene fusions to the control strain carrying the parental plasmid.
DNA Microarray Analyses.
Cultures of E. coli strain MG1655 grown in LB at 37°C overnight were diluted 1–250 into fresh LB and grown at 37°C with aeration. The subculture was split into two 100-ml cultures when the cell density was 1.1 × 108 colony-forming unit (cfu)/ml. Mitomycin C (Sigma), at a final concentration of 250 ng/ml, a sublethal dose, was added to one, whereas the other was a no addition control. Incubation at 37°C continued for another 40 min, then cells were collected for preparing total RNA. The mitomycin C-treated culture and its control reached 3.4 × 108 cfu/ml and 3.1 × 108 cfu/ml, respectively after 40-min incubation. E. coli strains 397C and P90 were grown as described above, but at 33°C, and collected when the cultures reached OD600 = 0.3. Total RNA purification, first-strand cDNA labeling, preparation of the E. coli whole-genome high-density microarray chips, hybridization, and data analysis were done as described (25). Both cy3-dUTP and cy5-dUTP were incorporated individually into both cDNA samples. Both combinations of reciprocally labeled samples were hybridized to microarrays. For the mitomycin C experiment, this procedure was repeated, and the average of four measurements for each gene was determined.
Results
DNA Sequence and Mapping of a Gene Fusion Collection.
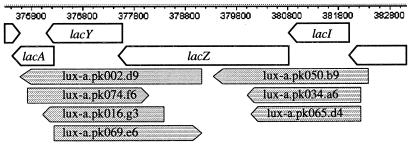
DNA sequences from both junctions of E. coli chromosomal DNA and the pDEW201 vector were obtained for each of the 8,066 individual members of the luxCDABE gene fusion collection. These sequences allowed unambiguous assignment of map position to 4,988 plasmid inserts, which were distributed randomly throughout the genome. Functional fusions, formed when an E. coli promoter region was in the proper position and orientation to drive expression of the luxCDABE reporter, to genes of interest were found with the tabular and graphical interfaces described in Materials and Methods. Fig. 1 is an example of the graphical display for the lac operon region. Three inserts mapped to this region resulted in functional fusions of the luxCDABE reporter to the lacZYA promoter, which is located between lacI and lacZ.
Figure 1.
Graphical display of mapped gene fusions near the lac operon. The open arrows indicate the chromosomal genes. The shaded arrows indicate the location and orientation of the chromosomal segments in plasmids from the collection of luxCDABE gene fusions mapped to this region. The direction of the arrowhead indicates whether the DNA of the cloned chromosomal segment is oriented such that a promoter, if present, would be driving expression of genes on the direct (arrowhead to the right) or complementary (arrowhead to the left) strand. In this example, the gene fusions lux-a.pk050.b9, lux-a.pk034.a6, and lux-a.pk065.d4 each contain functional gene fusions of the luxCDABE reporter to the lacZYA promoter.
Validation of Biological Responses and Specificity.
Having completed the sequencing and registering of the collection, strains containing luxCDABE fusions to members of several well-characterized regulatory circuits were tested with inducing environmental stimuli.
Transcription of the lac operon is negatively regulated by the lacI-encoded repressor (26) and positively activated by cAMP-CRP in response to glucose deprivation (27). Accordingly, the response of two lacZ-luxCDABE fusion strains to glucose was investigated. The bioluminescence in the presence of glucose was very low; 0.31 and 0.36 relative light units (RLU)/109 cells were produced by actively growing cultures grown at 37°C in LB medium containing 0.4% glucose. However, when these strains were grown in the same medium lacking glucose, 18.2 and 23.0 RLU/109 cells were produced. These results indicate the expected higher level of lacZYA transcription in the absence of glucose. In contrast, a strain, DPD2251, containing a recA-luxCDABE fusion, not expected to be glucose responsive, produced similar bioluminescence in both conditions, 1,090 RLU/109 cells in the presence of glucose and 1,183 RLU/109 cells in the absence. Thus, specificity of the response was shown.
Other global regulatory circuits also were examined. Fusions to genes of the SOS DNA damage response regulon (28, 29), the σ32-controlled heat shock regulon (30), the SoxR- and SoxS-regulated oxidative stress response regulon (15, 31, 32), and the OxyR-regulated peroxide stress regulon (32) were identified by using the bioinformatic tools. Each regulon was represented by at least one luxCDABE fusion (Table 1). Bioluminescence data suggested that active promoters were present within the chromosomal DNA fragment of each identified fusion (Table 1). The light production from strains carrying these fusions in the absence of stress was much greater than that from a culture carrying the parental plasmid pDEW201, which produced 0.002 RLU under the same conditions (15). Furthermore, bioluminescence was increased upon treatment with chemicals that activate each regulatory circuit. For each of the eight strains containing fusions to SOS regulon genes, nalidixic acid treatment increased bioluminescence (Table 1). Likewise, ethanol treatment yielded an increased signal for each of five strains containing fusions to heat shock regulon genes, as did methyl viologen treatment of four strains containing gene fusions to SoxR/S regulon members (Table 1). Thus, induction of the SOS response, the heat shock response, and SoxR- and SoxS-controlled oxidative stress response was accurately reflected by these fusion strains.
Specificity of the responses was shown by measuring the effect of ethanol on SOS-regulated gene fusions and of nalidixic acid on σ32-controlled fusions. For the eight SOS regulon fusions, there was little or no increased bioluminescence induced by ethanol treatment. A typical response was that of strain DPD2249 containing a dinP-luxCDABE) gene fusion. At 40 min after addition of 3% ethanol, a condition that resulted in increased light production from all of the heat shock regulon gene fusions, the response ratio for this strain was 0.80. Likewise at 110 min, there was no increase in light production by ethanol treatment. In contrast to the SOS regulon, exposure to ethanol but not nalidixic acid resulted in induction of expression of each fusion to heat shock genes. For example, strain DPD2243 containing a heat shock regulon gene fusion, clpB-luxCDABE, had a response ratio of 0.40 at 110 min after addition of 5 μg/ml nalidixic acid. Thus, appropriate specificity of the transcriptional responses reported by these luxCDABE gene fusions was observed.
The high level of light production from strains containing ahpC-luxCDABE gene fusions, representative of the OxyR-regulated oxidative stress regulon, suggested that this promoter was very strong (Table 1). When these strains were exposed to hydrogen peroxide, the bioluminescence was minimally elevated; response ratios of 1.2 to 1.6 were observed after 30 min. These responses were not far removed from that of a hydrogen peroxide nonresponsive strain containing a zwf-luxCDABE gene fusion, which had a response ratio of 1.1 under the same conditions. However, improvement in the response of the ahpC-luxCDABE gene fusions was obtained by reducing plasmid copy number with a pcnB mutation (17). An accompanying 10-fold reduction of the unstressed bioluminescence and a substantial increase in the hydrogen peroxide response ratios resulted (Table 1). Thus, the expected biological response from these highly expressed ahpC-luxCDABE fusions was obtained when the copy number of the gene fusion was reduced. Hence, predicted responses were observed in all cases, thus verifying that the collection contained representative gene fusions reflecting transcription throughout the E. coli chromosome.
Verification of DNA Microarray Results with Reporter Gene Fusions.
Independent verification of transcriptional responses discovered with DNA microarrays can be provided by reporter gene fusions. To compare these two approaches, the transcriptional responses of E. coli to mitomycin C were first analyzed by gene arrays, then followed by analyses with gene fusions. Mitomycin C, used in the initial discovery of the SOS regulon of E. coli (11), is a DNA damaging agent that intercalates into and forms a covalent attachment with double-stranded DNA. Ratios of expression in cultures treated with 250 ng/ml of mitomycin C for 40 min versus controls were calculated for all genes in the DNA array. Ratios greater than or equal to 2 were considered to identify putatively induced genes, whereas those with ratios less than 2-fold were considered uninduced. The known SOS genes (28, 29, 33) fell into both the induced and uninduced classes (Table 2). In addition, 20 genes not previously known to be induced by mitomycin C were observed to be induced in the array experiment (Table 2).
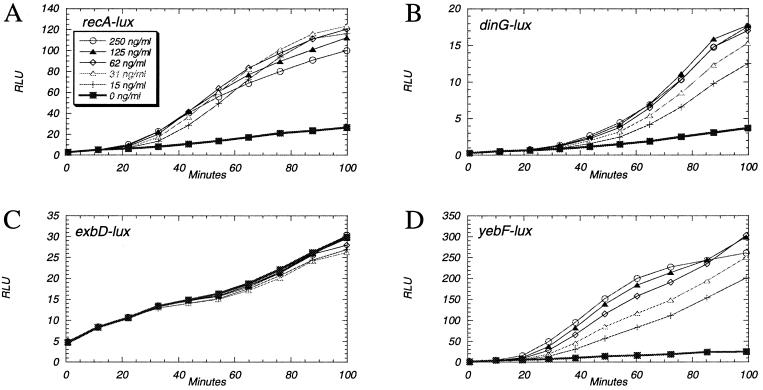
The known SOS gene fusions found in the collection were each tested for mitomycin C responses at several doses over a time course of 100 min. The expected response of a lag period followed by an induction of bioluminescence (23, 36) was observed in all four strains at several mitomycin C doses. One of these is shown in Fig. 2A. Table 2 shows the response ratios calculated at 100 min after addition of 62 ng/ml mitomycin C. Thus, up-regulation by mitomycin C of these SOS-controlled luxCDABE fusions was demonstrated.
Figure 2.
Testing DNA array results with gene fusions. (A) A fusion to a gene in the SOS regulon found to be up-regulated by mitomycin C in the DNA array experiment. (B) A fusion to a gene in the SOS regulon but not found to be up-regulated by mitomycin C in the DNA array experiment. (C and D) Fusions to genes not previously known to be DNA damage inducible found to be up-regulated by mitomycin C in the DNA array experiment. E. coli cultures grown to midexponential phase in LB medium were treated at time 0 with the indicated concentrations of mitomycin C.
In the DNA microarray experiment, the expression of several known SOS genes was elevated less than 2-fold (Table 2), placing them within a large group of 792 genes the expression of which was elevated by 20–100%. In this range, the DNA array data did not suggest authentic inductions. The collection contained three fusions to SOS genes in this class. These reporter gene fusions were tested for mitomycin C responses at several concentrations. In all three cases, increased bioluminescence resulted (Table 2 and Fig. 2B). Thus, negative results from DNA arrays, in conflict with other reports (29), were contradicted by experiments with gene fusions.
The magnitude of elevated expression after mitomycin C treatment of the 20 genes that were not previously known to be DNA damage responsive (Table 2) was in the same range as that for the known SOS genes. Of five available luxCDABE fusions corresponding to five of these 20 genes, four provided no evidence of substantially increased gene expression induced by mitomycin C (Table 2 and Fig. 2C). To eliminate strain background as the cause of this discrepancy, plasmid DNA isolated from these four gene fusion strains was used to transform E. coli strain MG1655. Similar results to mitomycin C challenge were obtained in this backcross; very slight, if any, up-regulation of the bioluminescent reporter was observed at several mitomycin C doses (data not shown), thus questioning the validity of the DNA array result. In contrast, however, the fifth luxCDABE gene fusion was strongly up-regulated by mitomycin C (Table 2 and Fig. 2D), thus verifying the up-regulation of yebF gene expression.
Functional Definition of Postulated Promoter Regions.
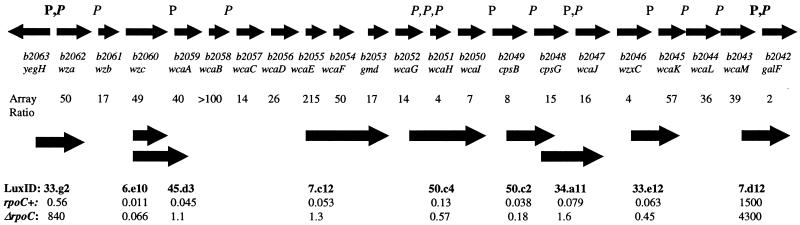
The genes encoding production of type I extracellular polysaccharide in E. coli are located in a cluster of 20 genes (37). An upstream promoter for these genes has been identified (38) and its regulation has been characterized (39, 40). Nonetheless, the transcriptional organization of this region has not been completely defined. The annotated sequence for these genes (41), which are transcribed from one strand of the genome, suggests the existence of several putative promoters and activator binding sites. Furthermore, a prediction of operon structure in this region suggests that these genes may be organized into several transcriptional units (42). Fig. 3 summarizes the predicted promoters in this region. Unexpectedly, an E. coli DNA microarray-based experiment with strains 397C, containing a truncated β′ subunit of RNA polymerase, and P90, an isogenic rpoC+ control, suggested that RNA transcripts in this region were affected by the rpoC mutation. Expression of genes b2043 through b2062 was coordinately up-regulated in the rpoC mutant (Fig. 3). The elevated expression is most readily explained if a single transcript starts before b2062 and ends between b2043 and b2042 (galF). If this is true, the region upstream of b2062 should contain a promoter. That region was fused to the luxCDABE operon in lux-a.pk033.g2. The bioluminescence produced when this gene fusion was placed into strain P90 was weak (0.56 ± 0.06 RLU/109 cfu), but yet was significantly greater (P < 0.0001) than the bioluminescence produced by the parental plasmid in the same host strain (0.028 ± 0.010 RLU/109 cfu). These data are consistent with a promoter in the region upstream of b2062 that is not very active in strain P90 growing in LB medium. The bioluminescence of this gene fusion was elevated 1,500-fold when placed in strain 397C (Fig. 3). This strong promoter activity driving luxCDABE gene expression therefore depends on the rpoC− mutation. In contrast, several other gene fusions to chromosomal DNA segments in this region, whether they contained predicted promoter regions or not, had very low levels of activity in both the wild-type and rpoC− mutant (Fig. 3). Thus, the data from both the DNA microarray experiments and from gene fusions are consistent with cotranscription of the 20 genes in this region. The end of the operon was defined by a gene fusion to the galF upstream region (in lux-a.pk07.d12) that strongly drove luxCDABE transcription. The activity of this promoter region was not dramatically effected by the rpoC mutation (Fig. 3), in agreement with the DNA array data.
Figure 3.
Promoter activity of luxCDABE gene fusions from the region encoding production of type I extracellular polysaccharide. The E. coli genes in this region are shown with the top set of arrows. Above this line are the predicted promoters regions from two sources (P, ref. 41; P, ref. 42). The two promoters supported by DNA array and gene fusion data presented here are shown in bold type. The b designation and common name for each gene is shown. The ratios of the deduced mRNA level in the rpoC mutant strain to the deduced mRNA level in the rpoC+ strain determined by the microarray method are shown on the next line. The mapped location of the chromosomal inserts of selected luxCDABE gene fusions in this region are shown with the second set of thicker arrows. Below these are the lux clone identification number and bioluminescence data (RLU/109 cfu) for each plasmid in the rpoC+ E. coli host strain and the host strain carrying an rpoC mutation. In the case of overlapping gene fusions, the shorter one is listed first. The bioluminescence of the parental plasmid in the rpoC+ host strain was 0.028 RLU/109 cfu and in the rpoC mutant strain was 0.19 RLU/109 cfu.
Discussion
Use of gene fusions has distinct advantages over other methods of expression analyses (9). Protocols are simplified because RNA isolation is not required and concerns about artifacts due to factors such as differential RNA stability are eliminated. Likewise, cross hybridization of related genes is not a concern. Among the several reporter genes available, bioluminescent reporters, such as luxCDABE in bacterial systems, provide a facile assay that readily allows dose responses and kinetic analyses. Genomic approaches to gene fusion technology are now possible in the many organisms for which genomic sequences are completed. Precise mapping of random fusions of E. coli chromosomal DNA to the luxCDABE reporter was accomplished by DNA sequencing. The resultant genomewide collection of mapped gene fusions provided an independent method for verification of results obtained by other methods.
Appropriate responses were obtained from plasmid-borne fusions to genes that respond via positive activators, cAMP-CRP, OxyR, and SoxS, genes that are negatively regulated by LexA, and genes regulated by an alternative Sigma factor, σ32. Thus, possible limitations of plasmid-borne gene fusions such as repressor titration or differing superhelicity of promoter regions on plasmids and in the chromosome did not affect detection of these responses. One limitation of elevated fusion copy number, however, was observed. Where the response from the plasmid-borne fusion to a strong promoter was minimal, reduction of copy number resulted in a more potent response. Highly expressed genes of E. coli have been cataloged (25); thus, plasmid-based fusions to these genes can be moved to a host that will reduce copy number (43) or integrated into the chromosome (44). However, an advantage of plasmid-borne fusions is the option of facile, automated transfer to other hosts. Although not comprehensive, the collection described here contains functional fusions to about 30% of the E. coli transcriptional units. This level of coverage, representative of E. coli regulatory circuitry, is useful for applications such as defining the stresses invoked by environmental manipulations or chemical treatments. Moreover, this collection contains fusions to genes of both known and unknown function, allowing discovery of novel expression patterns that have functional implications.
An example of functional implications from expression data are the up-regulation of yebF expression in response to DNA damage, suggesting a functional role for the encoded protein in DNA repair. The mitomycin C-induced up-regulation of this gene of unknown function was found by DNA array-based and gene fusion analyses. The DNA damage response reported by the yebF-luxCDABE fusion might be mediated by the SOS response because there is a LexA box upstream of yebG (33), which is just upstream of yebF and is included within the fused chromosomal fragment. The coregulation and genetic organization of yebF and yebG suggest cotranscription. However, this was not previously suspected because a promoter driving transcription of yebF is suggested by the DNA sequence of the yebF-yebG intergenic region (41).
In general, sequences used as sites of transcription initiation in E. coli cannot accurately be identified despite availability of several computational methods (45). Thus, a large collection of reporter gene fusions is useful for operon definition and promoter discovery. Active promoters not predicted by DNA sequence analysis can be defined by correlation of map position and reporter gene activity. In contrast, our data suggest that several predicted promoters within the region of the E. coli chromosome specifying production of type I extracellular polysaccharide are not functioning in rpoC or rpoC+ strains growing in LB medium. Although we cannot rule out that other conditions activate these putative internal promoters, it appears that the 20 genes form one transcriptional unit that is highly expressed in the rpoC mutant. Although the mechanism of activation is unknown, other data from the DNA array analysis suggest an indirect effect. Transcription of rcsA, encoding the limiting factor for activation of the genes in this region and its own expression (39), was up-regulated 18-fold in the mutant. This, in turn, may be due to the 6-fold decreased expression of lon, which encodes the protease responsible for RcsA instability (39).
Discovery of gene expression events diagnostic of an inhibitor's mode of action can form the basis of screens for similar antagonists. For example, yebF up-regulation is diagnostic of DNA damage; thus the strain containing the yebF-luxCDABE fusion would provide a facile means of identifying DNA damaging agents. Indeed, several luxCDABE fusion strains for detection of the SOS response have been developed (8), and one has been commercialized (46). Such an approach can be extended to other modes of action to select gene fusions useful for environmental testing or development of high throughput screens.
We have demonstrated that different gene expression profiling technologies have provided complementary insights into microbial molecular biology. It is thus to be expected that the comprehensive determination of other cellular component levels, ranging from proteins to metabolites, will result in even deeper understanding. Moreover, such biochemical measurements, as well as phenotypic ones, can be envisioned for large sets of deletion mutants, such as those now available in bakers yeast (47), and overexpression mutants. Thus an era can be foreseen where the availability of different sorts of array data, ranging from gene expression and mutants to metabolite levels and proteomics, will result in a completely integrated view of biology.
Acknowledgments
We thank Doug Wood (University of Chicago) for RNA samples from strains P90 and 397C and Dana Smulski (DuPont) for preliminary experiments with gene fusions from the type I extracellular polysaccharide biosynthesis region.
Abbreviations
- cfu
colony-forming unit
- RLU
relative light units
References
- 1.DeRisi J, van den Hazel B, Marc P, Balzi E, Brown P, Jacq C, Goffeau A. FEBS Lett. 2000;470:156–160. doi: 10.1016/s0014-5793(00)01294-1. [DOI] [PubMed] [Google Scholar]
- 2.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C F, Trent J M, Staudt L M, Hudson J J, Boguski M S, et al. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 3.DeRisi J, Penland L, Brown P, Bittner M, Meltzer P, Ray M, Chen Y, Su Y, Trent J. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 4.Marton M, DeRisi J, Bennett H, Iyer V, Meyer M, Roberts C, Stoughton R, Burchard J, Slade D, Dai H, et al. Nat Med. 1998;4:1293–1301. doi: 10.1038/3282. [DOI] [PubMed] [Google Scholar]
- 5.Jia M, LaRossa R A, Lee J-M, Rafalski A, DeRose E, Gonye G, Xue Z. Physiol Genomics. 2000;3:83–92. doi: 10.1152/physiolgenomics.2000.3.2.83. [DOI] [PubMed] [Google Scholar]
- 6.Tao H, Bausch C, Richmond C, Blattner F R, Conway T. J Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richmond C S, Glasner J D, Mau R, Jin H, Blattner F R. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LaRossa R A, Van Dyk T K. In: Bacterial Stress Responses. Storz G, Hengge-Aronis R, editors. Washington, DC: Am. Soc. Microbiol.; 2000. pp. 455–468. [Google Scholar]
- 9.Silhavy T J. J Bacteriol. 2000;182:5935–5938. doi: 10.1128/jb.182.21.5935-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casadaban M J, Cohen S N. Proc Natl Acad Sci USA. 1980;76:4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenyon C J, Walker G C. Proc Natl Acad Sci USA. 1980;77:2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wanner B L, McSharry R. J Mol Biol. 1982;158:347–363. doi: 10.1016/0022-2836(82)90202-9. [DOI] [PubMed] [Google Scholar]
- 13.Dimster-Denk D, Rine J, Phillips J, Scherer S, Cundiff P, DeBord K, Gilliland D, Hickman S, Jarvis A, Tong L, Ashby M. J Lipid Res. 1999;40:850–860. [PubMed] [Google Scholar]
- 14.Meighen E A. Microbiol Rev. 1991;55:123–142. doi: 10.1128/mr.55.1.123-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Dyk T K, Ayers B L, Morgan R W, LaRossa R A. J Bacteriol. 1998;180:785–792. doi: 10.1128/jb.180.4.785-792.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Dyk T K, Rosson R A. In: Methods in Molecular Biology: Bioluminescence Methods and Protocols. LaRossa R A, editor. Vol. 102. Towowa, NJ: Humana; 1998. pp. 85–95. [DOI] [PubMed] [Google Scholar]
- 17.Van Dyk T K, Smulski D R, Elsemore D A, LaRossa R A, Morgan R W. In: Recent Advances in Environmental Chemical Sensors and Biosensors. Mulchandani A, Sadik O A, editors. Vol. 762. Washington, DC: Am. Chem. Soc.; 2000. pp. 167–184. [Google Scholar]
- 18.Bachmann B. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2460–2488. [Google Scholar]
- 19.Christie G E, Cale S B, Isaksson L A, Jin D J, Xu M, Sauer B, Calendar R. J Bacteriol. 1996;178:6991–6993. doi: 10.1128/jb.178.23.6991-6993.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishimura A, Morita M, Nishimura Y, Sugino Y. Nucleic Acids Res. 1990;18:6169. doi: 10.1093/nar/18.20.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 22.Van Dyk T K, Majarian W R, Konstantinov K B, Young R M, Dhurjati P S, LaRossa R A. Appl Environ Microbiol. 1994;60:1414–1420. doi: 10.1128/aem.60.5.1414-1420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Dyk T K. In: Methods in Molecular Biology: Bioluminescence Methods and Protocols. LaRossa R A, editor. Vol. 102. Towowa, NJ: Humana; 1998. pp. 153–160. [Google Scholar]
- 24.Van Dyk T K, Reed T R, Vollmer A C, LaRossa R A. J Bacteriol. 1995;177:6001–6004. doi: 10.1128/jb.177.20.6001-6004.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei Y, Lee J-M, Richmond C, Blattner F, Rafalaski J A, LaRossa R A. J Bacteriol. 2001;183:545–556. doi: 10.1128/JB.183.2.545-556.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacob F, Monod J. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 27.Botsford J L, Harman J G. Microbiol Rev. 1992;56:100–122. doi: 10.1128/mr.56.1.100-122.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S-R, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H. Proc Natl Acad Sci USA. 1997;94:13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker G C. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1400–1416. [Google Scholar]
- 30.Gross C A. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1382–1399. [Google Scholar]
- 31.Koh Y S, Chung W-H, Lee J-H, Roe J-H. Mol Gen Genet. 1999;261:374–380. doi: 10.1007/s004380050978. [DOI] [PubMed] [Google Scholar]
- 32.Rosner J L, Storz G. Curr Top Cell Regul. 1997;35:163–177. doi: 10.1016/s0070-2137(97)80007-6. [DOI] [PubMed] [Google Scholar]
- 33.Lomba M R, Vasconcelos A T, Pacheco A B, de Almeida D F. FEMS Microbiol Lett. 1997;156:119–122. doi: 10.1111/j.1574-6968.1997.tb12715.x. [DOI] [PubMed] [Google Scholar]
- 34.Schnaitman C. Am Soc Microbiol News. 1991;57:612. [Google Scholar]
- 35.Davidov Y, Rozen R, Smulski D R, Van Dyk T K, Vollmer A C, Elsemore D A, LaRossa R A, Belkin S. Mutat Res. 2000;466:97–107. doi: 10.1016/s1383-5718(99)00233-8. [DOI] [PubMed] [Google Scholar]
- 36.Vollmer A C, Belkin S, Smulski D R, Van Dyk T K, LaRossa R A. Appl Environ Microbiol. 1997;63:2566–2571. doi: 10.1128/aem.63.7.2566-2571.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevenson G, Kanella A, Hobbs M, Reeves P R. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stout V. J Bacteriol. 1996;178:4273–4280. doi: 10.1128/jb.178.14.4273-4280.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gottesman S. In: Two-Component Signal Transduction. Hock J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 253–262. [Google Scholar]
- 40.Wehland M, Bernhard F. J Biol Chem. 2000;275:7013–7020. doi: 10.1074/jbc.275.10.7013. [DOI] [PubMed] [Google Scholar]
- 41.Blattner F R, Plunkett G, III, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 42.Thieffry D, Salgado H, Huerta A M, Collado-Vides J. Bioinformatics. 1998;14:391–400. doi: 10.1093/bioinformatics/14.5.391. [DOI] [PubMed] [Google Scholar]
- 43.Lopilato J, Bortner S, Beckwith J. Mol Gen Genet. 1986;205:285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- 44.Elsemore D A. In: Methods in Molecular Biology: Bioluminescencent Protocols. LaRossa R A, editor. Vol. 102. Totowa, NJ: Humana; 1998. pp. 97–104. [DOI] [PubMed] [Google Scholar]
- 45.Henaut A, Danchin A. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2047–2066. [Google Scholar]
- 46.Verschaeve L, Van Gompel J, Thilemans L, Regniers L, Vanparys P, van der Lelie D. Environ Mol Mutagen. 1999;33:240–248. [PubMed] [Google Scholar]
- 47.Winzeler E, Shoemaker D, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke J D, Bussey H, et al. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]