Abstract
TopBP1 is a scaffold protein that coordinates activation of the DNA-damage-checkpoint response by coupling binding of the 9-1-1 checkpoint clamp at sites of ssDNA, to activation of the ATR–ATRIP checkpoint kinase complex. We have now determined the crystal structure of the N-terminal region of human TopBP1, revealing an unexpected triple-BRCT domain structure. The arrangement of the BRCT domains differs significantly from previously described tandem BRCT domain structures, and presents two distinct sites for binding phosphopeptides in the second and third BRCT domains. We show that the site in the second but not third BRCT domain in the N-terminus of TopBP1, provides specific interaction with a phosphorylated motif at pSer387 in Rad9, which can be generated by CK2.
INTRODUCTION
Activation of the Chk1 arm of the DNA-damage checkpoint response is triggered by assembly of a number of multiprotein complexes at segments of single-stranded DNA resulting from replication fork stalling or from resection of a double-strand break (1). Multiple copies of replication protein A, coat the single-strand DNA, and facilitate recruitment of ATR via interaction with its constitutive binding partner ATRIP (2). The Rad9-Rad1-Hus1 DNA-damage checkpoint clamp is loaded at the junction of single- and double-stranded DNA by a specialized form of the RFC clamp loader, incorporating Rad17 in place of Rfc1 (3,4). While there may be some direct interaction, at least in yeast (5), coupling of these two major complexes into a functional system is performed by TopBP1, which binds simultaneously to the phosphorylated C-terminal tail of Rad9 (6–8), and to the ATR–ATRIP complex, upregulating its kinase activity (9,10).
TopBP1 was originally identified as a binding partner of DNA topoisomerase II in yeast 2-hybrid experiments (11), and in addition to its involvement in the DNA-damage checkpoint, it has been variously found to interact with transcription factors such as Miz-1, HPV16E2 and E2F1 (12–14), the protein kinase c-Abl (15), the DNA-damage sensor PARP-1 (16), replication protein Cdc45 (17), p53 (18) and the Nbs1 subunit of the MRN complex (19). Recently, TopBP1 has also been implicated in recruitment of DNA Pol-α, suggesting a role in the restart of stalled replication forks (20), although whether this involves a direct interaction is not clear.
Structurally, TopBP1 contains multiple copies of the BRCT domain, first identified in Schizosaccharomyces pombe Rad4 (21,22) and subsequently in the C-terminal region of the breast-cancer-associated DNA-damage mediator protein BRCA1 (23,24). Indeed S. pombe Rad4 (aka Cut5) is the orthologue of metazoan TopBP1. In Xenopus, binding of TopBP1 to the phosphorylated C-terminal tail of Rad9 has been shown to be dependent on the first-tandem pair of BRCT domains (BRCT1:2) (25). The TopBP1 orthologues Rad4 in S.pombe and Dpb11 in Saccharomyces cerevisiae also interact in a phosphorylation-dependent manner with the tail of Rad9 and Ddc1, respectively, but in these organisms it is the second-tandem pair of BRCT domains, BRCT3:4 that mediate this interaction (26–28). The first pair of BRCT domains on Rad4 is instead involved in interaction with a phosphorylated site in the N-terminus of Crb2, the S. pombe homologue of mammalian 53BP1 (29,30). Interaction of metazoan TopBP1 with ATR–ATRIP is mediated by a specific C-terminal domain of unknown structure, mapping between the sixth- and seventh-BRCT domains in the protein sequence (10), which interacts with conserved domains in ATR and ATRIP (9). A functionally comparable ATR-activating domain has also been identified in the budding yeast equivalent of TopBP1, Dpb11, at the C-terminus beyond the fourth BRCT domain (31,32).
Of the remaining BRCT domains in TopBP1, BRCT6 is implicated in binding to E2F1 (12) and PARP-1 (16), while BRCT domains 7 and 8 mediate interaction with the helicase BACH1/FANCJ (33). Although the basis of these interactions remains to be described, structural analysis of TopBP1-BRCT6 (34) suggests that it does not involve a phospho-peptide interaction of the type commonly mediated by tandem pairs of BRCT domains (35–37). A clear role for the tandem-pair BRCT4:5 have not yet emerged.
To try and gain some insight into the specific roles of the individual domains of TopBP1 in mediating specific protein interactions, we have now determined the crystal structure of an N-terminal segment of human TopBP1, from the N-terminus to beyond BRCT2 and analysed its interactions with the C-terminal tail of Rad9 in vitro.
MATERIALS AND METHODS
Cloning, expression and purification
Human TopBP1(1–290)
DNA-encoding amino acids 1–290 of human TopBP1 was amplified by PCR from IMAGE CLONE 8991925 (Geneservice, Cambridge, UK), then sub-cloned into both an in-house modified pOPIN vector (38) and pGEX-6P-1 (GE Healthcare, Chalfont St Giles, UK) encoding N-terminal, 3C-protease cleavable, octa-histidine (His8) or Glutathione S-Transferase (GST) affinity tags, respectively.
His8- or GST-tagged TopBP1(1–290) was transformed into Escherichia coli expression strain Rosetta2(DE3)pLysS (Merck, KGaA, Damstadt, Germany). A single transformed colony was used to inoculate a 250-ml flask containing 100 ml of Luria–Bertani broth (LB) supplemented with carbenicillin (100 µg/ml) and chloramphenicol (34 µg/ml). The inoculated culture was grown overnight at 37°C, at 220 rpm, in an orbital shaking incubator.
The following day, 20 ml of the overnight culture was used to inoculate a 2-l flask containing 1 l of LB, supplemented with antibiotics as before. Cultures were grown at 37°C, 220 rpm, until the optical density at 600 nm reached 0.6–0.8. They were then removed from the incubator, and rapidly cooled on ice for 30 min. Recombinant protein expression was induced by the addition of 0.4 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG), and the cultures incubated for a further 16 h, at 220 rpm, at a reduced temperature of 16°C. Cells were then harvested by centrifugation, and the resulting pellet stored at −80°C until required.
The cell pellet arising from 6 l of culture was resuspended in 100 ml of 25 mM HEPES pH 7.5, 1 M NaCl, 5% v/v glycerol, 0.1% v/v Tween 20, supplemented with protease inhibitors (Roche, Burgess Hill, UK), then lysed by sonication (18 × 5 s bursts, on ice, at 50% amplitude, Jencons Ultrasonic Processor). Cell debris and precipitated material was removed by high-speed centrifugation at 48 834g for 60 min.
The supernatant arising from this step was applied to a batch/gravity column containing 10 ml of either Talon (TaKaRa Bio, Saint-Germaine-en-Laye, France) or Glutathione Sepharose 4 Fast Flow (GE Healthcare) resin, depending on the encoded affinity tag.
The column containing the cell extract and resin was rotated/rolled at 4°C for a period of 1 h to facilitate protein binding, and then allowed to pack under gravity flow.
For the His-tagged protein, the column was washed with 250 ml of 25 mM HEPES pH 7.5, 1 M NaCl, 5% v/v glycerol, 5 mM imidazole. Bound protein was then eluted with the application of 25 mM HEPES pH 7.5, 1M NaCl, 5% v/v glycerol, 400 mM imidazole. Rhinovirus 3C-protease (PreScission, GE Healthcare) was added to the eluate, and incubated overnight at 4°C to cleave the affinity tag.
For the GST-tagged protein, the column was instead washed with 250 ml of 25 mM HEPES pH 7.5, 1 M NaCl, 5% v/v glycerol. Rhinovirus 3C-protease was then added directly to the resin/buffer slurry and incubated overnight, with rolling, at 4°C to cleave the affinity tag. The released protein was collected by repacking the column under gravity flow, and then collecting the eluate.
In both cases, the eluted TopBP1 (1–290) protein was concentrated to a final volume of 5 ml (Vivaspin 20, 10 kD MWCO, Sartorius Stedim, Epsom, UK) then applied to a HiLoad Superdex 200 16/60 size exclusion chromatography column (GE Healthcare) pre-equilibrated in 25 mM HEPES pH 7.5, 1 M NaCl, 5 mM EDTA, 10 mM DTT and 5% v/v glycerol (for crystallographic studies), or 25 mM HEPES pH 7.5, 100 mM NaCl, 5 mM EDTA, 10 mM DTT and 5% v/v glycerol (for peptide-binding studies).
Human Rad9Tail
DNA encoding the C-terminus of human Rad9 (amino acids 264–391) was amplified by PCR from a vector encoding full-length human Rad9 kindly provided by Dr Andrew Doré (ICR; The Institute of Cancer Research, UK) then sub-cloned into the expression vector pGEX-6P-1 (GE Healthcare).
Expression and purification of GST-Rad9Tail was essentially as that described for TopBP1(1–290) except that the cell pellet resulting from 4 l of cell culture was used to produce the initial cell lysate, only 5 ml of Glutathione Sepharose 4 FF resin was used in the affinity capture step, and the protein was eluted from the column using 25 mM HEPES pH 7.5, 1 M NaCl, 10 mM DTT, 5% v/v glycerol, 5 mM EDTA and 40 mM reduced glutathione and not cleaved with Rhinovirus 3C-protease.
Selenomethionine-labelled TopBP1(1–290)
The TopBP1(1–290) expression plasmid was co-transformed along with the pRARE plasmid (Merck) into the methionine auxotroph, Escherichia coli strain, B834(DE3) (Merck). Transformed colonies were selected on LB agar plates supplemented with antibiotics as before. From an overnight culture, 25 ml was used to inoculate a 2-l flask, containing 1 l of SelenoMet Medium Base plus Nutrient Mix and SelenoMethionine solution (Molecular Dimensions, Newmarket, UK) supplemented with antibiotics. Cultures were incubated at 37°C, 220 rpm, in an orbital-shaking incubator, until they reached an optical density of ∼0.7 at 600 nm, when they were removed from the incubator and rapidly cooled on ice. Protein expression was induced with IPTG as before, and cultures incubated for a further 16 h, at 220 rpm and 16°C, before the cells were harvested by centrifugation.
Incorporation of the selenomethionine label was verified using mass spectrometry (Dr WJ Mawby, Department of Biochemistry, University of Bristol).
Crystallization, data collection, phasing, model building and refinement
Selenomethionine-labelled TopBP1(1–290) was crystallized at 14°C using the hanging drop vapour diffusion method, by mixing equal volumes of the protein (10–15 mg/ml) with either 100 mM Tris–HCl pH 7.5, 400 mM MgCl2, 20–30% w/v polyethylene glycol 4000, 2–6% v/v glycerol (spacegroup P21), or 100 mM Tris–HCl pH 6.8, 500 mM KI, 18–25% w/v PEG 3350, 2–8% v/v glycerol (spacegroup P213). Crystals were visible after 24 h, but generally took a week to reach their maximum size.
Cryoprotection, for data collection, was achieved by step-wise soaking in buffers containing increasing amounts of glycerol, to a final concentration of 30% v/v.
All diffraction data were collected at 100K on station ID14.4 of the European Synchrotron Radiation Facility (ESRF), Grenoble, France. Data were processed and scaled using the software package Mosflm (39) and Scala (40).
Phases were calculated from a two-wavelength anomalous dispersion experiment with the P213 crystals, which comprised one molecule per asymmetric unit and a solvent content of 68%. Positions of eight Se atoms and five Iodide ions could be determined using SHELX (41) and refined using SHARP (42), yielding an interpretable electron density map at 2.8Å resolution.
Density modification and automated building was carried out by using the program RESOLVE (43) to produce an initial model, which was extended through a combination of manual building in Coot (44) and refinement with the PHENIX suite (45). This model was then used to solve the P21 data set by molecular replacement using PHASER (46), which contained four molecules per asymmetric unit, and a solvent content of 52%.
Peptides
Biotinylated peptides were purchased from Pepceuticals Ltd, Nottingham, UK, or from the University of Bristol Peptide Synthesis Facility, Bristol UK.
Rad9Tail: pS387, Biotin-SPVLAED[pS]EGE
H2A.1: pS129, Biotin-GSG-YSGSRTGKP-[pS]-QEL
Fluorescein-labelled peptides were purchased from Peptide Protein Research Ltd, Fareham, UK.
Rad9, pS272: fluorescein-GGSDTDSH[pS]QDLGSPE
Rad9, pT355: fluorescein-GGEPSTVPG[pT]PPPKKFR
Rad9, pS375: fluorescein-GGSILAPVR[pS]PQGPSPV
Rad9, pS380: fluorescein-GGVRSPQGP[pS]PVLAEDS
Rad9, pS387: fluorescein-GGPSPVLAED[pS]EGEG
Crb2: pT215, fluorescein-GGSGQVET[pT]PTRLAT
Where [pS] or [pT] corresponds to phospho-serine or phospho-threonine, respectively.
Co-precipitation (pull down) experiments
GST-tagged proteins
A volume of 100 µl of GST, GST-Rad9Tail WT or S387A mutant at a concentration of 31.5 μM, was mixed with 100 µl of Glutathione Sepharose 4 FF resin (GE Healthcare) pre-equilibrated in buffer A: 25 mM HEPES pH 7.5, 100 mM NaCl, 5 mM EDTA, 10 mM DTT and 5% v/v glycerol, then mixed/incubated for a period of 3 h at 4°C to facilitate protein binding.
The resin was then washed three times, with successive applications of 1 ml aliquots of buffer B: 25 mM HEPES pH 7.5, 100 mM NaCl, 5 mM EDTA, 10 mM DTT and 5% v/v glycerol supplemented with 0.1% v/v BSA (NEB, Hitchin, UK) and 0.1% v/v NP-40 (buffer B).
A volume of 100 µl of GST-CKIIα, at 31.5 µM, in buffer B supplemented with 10 mM ATP and 30 mM MgCl2 was added to the resin slurry, then mixed/incubated at 4°C for a period of 15 h. The resin was then washed with 1 ml of buffer B, and a further three times with 1 ml aliquots of buffer A.
Then, 100 µl of His-tagged TopBP1(1–290), at 94.5 µM in buffer B, was added to the beads and incubated for a further period of 3 h at 4°C. The beads were then washed four times, with 1 ml aliquots of buffer A. Samples of each co-precipitation were then analyzed by SDS–PAGE.
Dephosphorylation of immobilized GST-Rad9Tail WT, was performed after the CK2α phosphorylation step, by the addition of 40 µg of λ-phosphatase and 30 U of calf intestinal alkaline phosphatase (CIP, NEB) in 100 µl of buffer B supplemented with 30 mM MnCl2, then incubated for 15 h at 4°C.
Biotinylated peptides
A volume of 100 µl of biotinylated Rad9Tail phospho-peptide at 140 µM, or H2A.1 phospho-peptide at 100 µM, in buffer A, was added to 100 μl of NeutrAvidin agarose beads (Thermo Fisher Scientific, Loughborough, UK), then incubated/mixed at 4°C for a period of 1 h, before being washed with three successive 1 ml aliquots of buffer A. A volume of 100 µl of His-tagged TopBP1(1–260), at 42 µM in buffer B, was then added to the resin slurry and incubated for a further period of 2 h at 4°C. Samples were analysed by SDS–PAGE, after washing the beads a further four times with buffer A.
Dephoshorylation and re-phosphorylation steps were carried out using λ-phosphatase/CIP and CKIIα as before.
All samples were analysed by SDS–PAGE, on 4–12% NuPAGE Bis–Tris gels (Invitrogen, Paisley, UK) run in 1× MES buffer (Invitrogen). Western blots, used a mouse anti-his primary (TaKaRa Bio, Cat No: 631212) and an HRP-conjugated anti-mouse (GE Healthcare, Cat No: NXA931) secondary antibody.
Fluorescence polarization
Fluorescein-labelled peptides at a concentration of 10 nM, were incubated at 4°C, for a period of 30 min with increasing concentrations of WT or mutant TopBP1(1–290) in 25 mM HEPES pH 7.5, 100 mM NaCl, 5 mM EDTA, 10 mM DTT, 5% v/v glycerol, 0.01% v/v NP40. The samples (typically a volume of 50 µl) were then transferred to a black 96-well polypropylene plate (VWR, Lutterworth, UK) for measurement of fluorescence polarization in a Victor V2 Multilabel Reader (Perkin Elmer, Cambridge, UK). Two 0.1 s measurements were collected for each well with excitation/emission wavelengths of 485 and 535 nm, respectively, with either parallel or perpendicular polarizers in-line. Background fluorescence in wells containing only buffer was subtracted from all values obtained for the samples.
Polarization data were analysed using GraphPad Prism 5.0 by non-linear fitting with a one-site total binding model. The non-specific binding component was then subtracted from the data for presentation purposes. All data represent the mean of three separate experiments, and error bars represent one standard deviation.
RESULTS
Structure of TopBP1 N-terminus
An N-terminal construct of human TopBP1 (residues 1–290) was expressed in E. coli using a selenomethionine-supplemented medium, and two different crystal forms were obtained. The structure was determined by a two-wavelength anomalous diffraction experiment, and refined at a resolution of 2.6Å (‘Materials and Methods’ section and Table 1).
Table 1.
Data collection, phasing and refinement statistics
| Data collection | ||
| Space group | P213 | P1211 |
| Cell dimensions a, b, c (Å) | 115.8, 115.8, 115.8 | 39.5, 290.3, 60.6 |
| Cell angles α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 89.5, 90.0 |
| Resolution (Å) | 51.8–2.8 (2.95–2.8)* | 55.9–2.6 (2.74–2.6) |
| Rmerge | 0.091 (0.50) | 0.103 (0.38) |
| I/σI | 6.9 (1.5) | 5.0 (2.0) |
| Completeness (%) | 99.9 (99.9) | 97.2 (97.2) |
| Redundancy | 5.9 (6.1) | 2.6 (2.6) |
| Wilson B (Å2) | 75.2 | 56.2 |
| Phasing power | Acentric | Centric |
| Phasing | ||
| Se inflection anomalous | 1.313 (0.300) | |
| Se peak anomalous | 1.377 (0.310) | |
| Se peak isomorphous | 0.123 (0.065) | 0.097 (0.044) |
| FOM | 0.3972 | 0.1495 |
| Refinement | ||
| Resolution (Å) | 47.3–2.80 | 51.3–2.60 |
| Reflections (work/free) | 24687/1254 | 76051/3850 |
| Rwork/Rfree | 0.232/0.251 | 0.205/0.266 |
| Number of atoms | ||
| Protein | 2280 | 9438 |
| Ligands/ions | 17 | 66 |
| Solvent | 73 | 343 |
| B factors | ||
| Protein | 58.7 | 57.5 |
| Ligands/ions | 83.7 | 52.8 |
| Solvent | 37.1 | 43.2 |
| R.m.s.d. values | ||
| Bond lengths (Å) | 0.003 | 0.005 |
| Bond angles (°) | 0.603 | 0.806 |
| Ramachandran plot (%) | ||
| Favoured | 97.5 | 99.0 |
| Allowed | 2.5 | 1.0 |
| Outliers | 0.0 | 0.0 |
*Highest resolution shell is shown in parenthesis.
As expected from the previous amino acid sequence analysis (28), the crystal structure of TopBP1(1–290) contained the predicted BRCT domains 1 and 2, formed by residues 106–198 and 199–289, respectively. More surprising was the presence of an additional undocumented BRCT domain, N-terminal to these, consisting of residues 1–105, although the possibility of this had previously been suggested (28). To maintain consistency with published nomenclature, the second- and third-BRCT domains in this structure will be referred to as BRCT1 and 2, while the unexpected BRCT domain at the extreme N-terminus is designated BRCT0 (Figure 1A and B).
Figure 1.
Structure of the N-terminus of TopBP1. (A) Schematic representation of human TopBP1, highlighting the position of each BRCT domain. The boxed region indicates the amino acid boundaries of the TopBP1(1–290) expression construct used in this study. (B) The X-ray crystal structure of TopBP1(1–290) reveals three consecutive BRCT domains, designated as BRCT0, BRCT1 and BRCT2 (coloured grey, yellow and orange, respectively). Amino acid boundaries of each domain are indicated in parentheses. This and all other molecular graphics figures were produced with MacPyMOL unless otherwise stated. (C) Superimposition of the first BRCT domain in the consecutive TopBP1 BRCT pairs BRCT0:1 and BRCT1:2 onto the first-BRCT domain in the canonical-tandem BRCT structure (cyan) of Crb2 in a phospho-H2A.1 peptide complex (PDB: 2VXC), reveals significant differences in juxtaposition of the two domains in both TopBP1 pairs (left, middle). In contrast (right), both BRCT domains in the canonical pair from BRCA1 (pink), align with their equivalents in Crb2. The bound phospho-peptide from the Crb2–pH2A.1 complex is shown in stick representation. In order to improve the clarity of the figure, some interconnecting loops have been omitted. (D) Comparison of the TopBP1 BRCT0:1 and BRCT1:2 pairs, based on superimposition of the outlined domain from each, but with the pairs separated for clarity, reveals distinct and idiosyncratic domain juxtapositions for each, and the absence of any repeating poly BRCT superstructure.
Previous structural and bioinformatics analysis has identified two sub-classes of BRCT domains, those such as the C-terminal domain of XRCC1 (47) which occur as singletons, and those in proteins such as BRCA1, Crb2, 53BP1, Mdc1, etc., which are involved in phospho-peptide binding, and occur as tandem pairs, closely linked in sequence and in 3D structure. To our knowledge, the TopBP1(1–290) structure is the first example of a closely linked triple-BRCT, and is structurally distinct from the previously described singleton and tandem examples.
Examination of the TopBP1 BRCT0:1 or BRCT1:2 pairs, reveals a very different juxtaposition of the consecutive BRCT domains from each other, and from previously documented ‘conventional’ tandem BRCT structures (Figure 1C and D). Despite the individual BRCT domains having a standard β1-α1-β2-β3-α2-β4-α3 topology and each having its C-terminal α3 helix connected to the N-terminal β1 strand of the subsequent BRCT, the central β-sheets in the BRCT domains are perpendicular to each other rather than parallel as in canonical tandem BRCT structures. In the TopBP1(1–290) structure this spatial arrangement is dictated by the very short nature of the linkers between consecutive BRCT domains: BRCT0 α3 helix is connected to BRCT1 β1 strand through a 17-amino acid linker (91–108) and BRCT1 α3 helix is connected to BRCT2 β1 strand through a 22-amino acid linker (181–203). This contrasts with all other known tandem BRCT structures where longer inter-domain linkers permit a more extended arrangement in which the α3 helix of the upstream BRCT domain packs against the α1 helix of the downstream BRCT domain (Supplementary Figure S1).
Overall, the three consecutive BRCT domains form a continuous molecular surface of roughly cylindrical shape, with significant burial of hydrophobic surface at the interfaces. Consistent with this, the overall arrangement of the three domains seems to be rigid, and is essentially identical in the two different crystal forms (r.m.s.d of 0.91Å over 279 Cα positions), which involve distinct crystal lattice contacts.
Phospho-peptide-binding sites
Previous studies have implicated TopBP1 BRCT domains 1 and 2 in interaction with the phosphorylated tail of Rad9 (25). Structures of tandem BRCT domains bound to phosphopeptides (48–54), have identified a conserved cluster of charged and polar amino acids in the first-BRCT domain of a tandem pair, that provide generic interactions with the phosphorylated residue in a phosphopeptide ligand, while residues in the cleft formed by the junction of the two BRCT domains, furnish specificity for ligand residues C-terminal to the phosphorylated serine or threonine. Examination of the three BRCT domains in TopBP1(1–290) shows the presence of the generic phospho-peptide recognition motif in BRCT domains 1 and 2, but not in BRCT0, where the topologically equivalent residues are hydrophobic (Figure 2A). The two putative phosphopeptide-binding patches on BRCT1 and 2 are disconnected from each other, and are arrayed ∼90° apart around the overall cylindrical axis of the TopBP1(1–290) structure. The interface between BRCT1 and 2 is quite different from the more conventional tandem BRCT structures as it does not provide the peptide-binding cleft. Instead, the phosphopeptide-binding patch on BRCT1 runs into a helical channel winding around the TopBP1(1–290) structure, and connecting to the phosphopeptide-binding patch on BRCT2 (Figure 2B).
Figure 2.
Phosphopeptide-binding sites in TopBP1 N-terminus. (A) Comparison of the phosphopeptide-binding site in a canonical tandem BRCT structure, Crb2, with the topologically equivalent sites in TopBP1 BRCT0, BRCT1 and BRCT2. The pattern of residues that provide the phosphate interaction in Crb2 and in other known phosphopeptide-binding BRCT domains, are present in BRCT1 and BRCT2 but not BRCT0 where the site is hydrophobic in nature. (B) Surface representation of TopBP1(1–290) structure coloured by domain as in Figure 1. The conserved putative phosphopeptide-binding patches conserved in BRCT1 and BRCT2 are highlighted in green. (C) Difference Fourier maps from crystals of TopBP1(1–290) grown in a buffer containing MgSO4, show significant density (>2.5σ) in the putative phosphopeptide-binding sites of BRCT1 and BRCT2, corresponding to bound sulphate. This confirms that both sites are competent for interacting with the phosphate of a phosphopeptide.
Phosphate-binding sites in proteins frequently show high affinity for sulphate ions, which are iso-structural with phosphate. Crystals of TopBP1(1–290) grown in buffers containing MgSO4 instead of the original 200 mM MgCl2 displayed strong electron density for sulphate ions bound to the putative phosphopeptide-binding sites in BRCT1 and BRCT2 (Figure 2C), where the key residues are conserved, but not in BRCT0, where they are not.
Rad9 phosphorylation
The Rad9 C-terminal tail, extending beyond the structurally defined PCNA-like core (55–57), contains multiple sites of phosphorylation that are conserved to varying degrees from yeast to mammals (Supplementary Figure S2). A conserved site, Ser272 in human Rad9, occurs close to the N-terminus of the tail immediately following the PCNA-like core, and is believed to be phosphorylated by ATM/ATR on genotoxic stress (58,59). The opposite end of the Rad9 tail contains a second well-conserved site, Ser387 in humans, whose phosphorylation at least in metazoa, is constitutive and required for TopBP1 recruitment and consequent activation of ATR as part of checkpoint signalling (6,8,10,25). Multiple potential sites for phosphorylation by the cyclin-dependent kinase Cdc2 (7) can also be identified between the 272 and 387 sites, but the significance of these is not known.
The nature of the kinase responsible for providing the essential TopBP1-binding site at Rad9-pSer387, has not previously been determined, although bioinformatics analysis (60) suggested CK2, which would certainly be consistent with the constitutive nature of the phosphorylation. To determine whether CK2 could provide this essential phosphorylation, we expressed a GST-fusion with the C-terminus of Rad9 (GST-Rad9Tail; amino acids 264–391) in E. coli, incubated it with recombinant maize CK2α and Mg-ATP as described earlier (61), and tested its ability to co-precipitate TopBP1(1–290) (Figure 3A). GST alone incubated with CK2α and Mg-ATP, or GST-Rad9Tail incubated with CK2α in the absence of Mg-ATP, failed to co-precipitate detectable TopBP1(1–290). GST-Rad9Tail incubated with CK2α in the presence of Mg-ATP gave a robust co-precipitation of TopBP1(1–290), whereas a GST-Rad9Tail construct with an S387A mutation incubated in the same conditions failed to significantly co-precipitate TopBP1(1–290). To further test the specificity of the interaction, we repeated the experiment with a biotinylated-synthetic peptide consisting of the last 11 residues of human Rad9 with a phosphoserine at 387. Again we observed a robust co-precipitation of TopBP1(1–290) by neutravidin beads in the presence of the pS387 peptide or a dephosphorylated peptide that was incubated with CK2α and Mg-ATP, but not with the dephosphorylated Rad9 peptide, or with an unrelated biotinylated phosphopeptide derived from the C-terminus of S. pombe histone H2A.1 (Figure 3B). Taken together these data confirm that the C-terminal sequence of Rad9 including pSer387 is essential for interaction with TopBP1(1–290) and that the key phosphorylation is likely to be provided by CK2.
Figure 3.
Rad9-tail phosphorylation and TopBP1 binding. (A) Pull-down assay of binding of His6-TopBP1(1–290) to a GST-Rad9 tail construct. His6-TopBP1(1–290) was efficiently co-precipitated only when the GST-Rad9 tail construct was incubated with the protein kinase CK2α in the presence of Mg-ATP, but not by GST alone, or a mutant in which the target phosphorylation site, Ser387 was changed to alanine. (B) Pull-down assay using a synthetic biotin-coupled peptide incorporating a phosphoserine at the equivalent of Ser387. His6-TopBP1(1–290) was efficiently co-precipitated by the biotin-pS387 peptide, but not by a different biotinylated-phosphopeptide, nor by the biotin-pS387 peptide treated with phosphatase. Incubation of phosphatase-treated peptide with CK2α + MgATP restored binding.
Identification of the Rad9 interaction site in TopBP1
Previous studies in Xenopus, have implicated the N-terminal region of TopBP1 as necessary for interaction with the C-terminally phosphorylated Rad9 tail (25). Within that region of TopBP1 the structural data presented here reveal three BRCT domains, two of which (BRCT1 and BRCT2) possess the characteristic motifs for phosphopeptide interaction, identified from studies of conventional tandem BRCT structures (48–54,62,63). While involvement of BRCT0 in mediating part of the interaction with Rad9 cannot be totally ruled out, the strong dependence of the interaction on phosphorylation suggests that one or both of BRCT1 and BRCT2 are likely to play the dominant role.
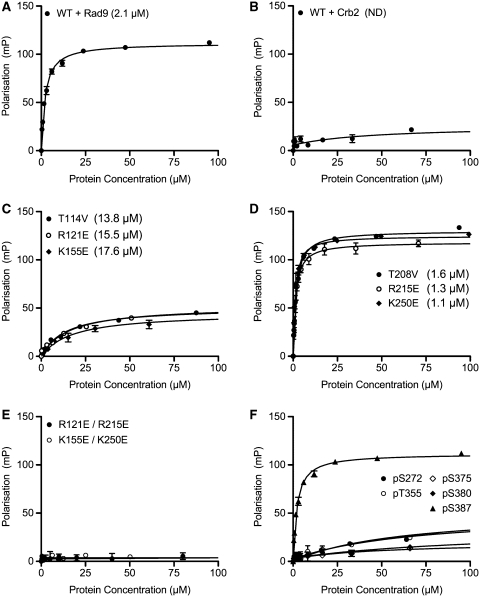
To test this, we generated a series of mutants, in which key residues in the phosphopeptide interaction motifs of BRCT1, BRCT2 or both, were changed to residues that would be likely to abrogate phosphopeptide interaction, based on analysis of other BRCT systems (49). We then determined the interaction of these with a fluorescently labelled synthetic phosphopeptide incorporating the last 13 residues of human Rad9 with the equivalent of Ser387 phosphorylated, using a fluorescence polarization (FP) assay (Figure 4A). Wild-type TopBP1(1–290) bound the Rad9 peptide with a Kd = 2.1 µM. The interaction was specific to the Rad9 sequence, and no interaction was observed with a non-cognate fluorescent phosphopeptide derived from a segment of S. pombe Crb2, implicated in binding to the TopBP1 homologue, Rad4 (Figure 4B). Mutation of Thr114, Arg121 or Lys155 in the putative phosphopeptide-binding site in BRCT1, significantly decreased the strength of the interaction, but did not totally abolish it (Figure 4C). In contrast, mutation of Thr208, Arg215 and Lys250 in BRCT2, did not weaken the interaction, but actually caused a slight increase in affinity (Figure 4D). Double mutants, in which the putative binding sites in both BRCT domains were disrupted, showed no measurable interaction with the Rad9 phosphopeptide (Figure 4E).
Figure 4.
Specificity of phospho-peptide binding to TopBP1(1–290). (A) Binding affinity of TopBP1(1–290) for a fluorescein-labeled Rad9-derived peptide, incorporating pSer387, as determined by fluorescence polarization. The calculated Kd (shown in parenthesis) indicates a specific interaction. For calculation of Kd see ‘Materials and Methods’ section. (B) As (A) but with a labelled phosphopeptide derived from the S. pombe checkpoint mediator protein Crb2. No significant binding is observed, confirming the specificity of the Rad9 interaction. C) As (A), but with TopBP1(1–290) constructs harbouring mutations in the putative phosphopeptide-binding site in BRCT1. The affinity of the interaction is substantially reduced, indicating a significant contribution to the interaction by BRCT1. (D) As (C), but with TopBP1(1–290) constructs harboring mutations in the putative phosphopeptide-binding site in BRCT2. The affinity of the interaction is comparable to that for wild-type TopBP1(1–290), indicating that there is no significant contribution to the interaction by BRCT2. (E) As (C), but with TopBP1(1–290) constructs with mutations in both putative phosphopeptide-binding sites. The weak-residual binding present in BRCT1 mutants is effectively abolished in the double mutants. (F) As (A), but with phosphopeptides corresponding to five different phosphorylation sites that have been mapped within the Rad9 tail. Only the peptide incorporating pSer387 shows significant affinity.
The substantial loss of affinity on mutation of the BRCT1 site, clearly implicates it as the primary determinant of Rad9 C-terminal phosphopeptide binding in the TopBP1(1–290) construct. The weak-residual interaction observed when the BRCT1 site, but not the BRCT2 are mutated, could suggest that BRCT2 also contributes to the interaction and that the two sites cooperate. However the observation that binding is actually tighter when the BRCT2 site is disabled, contradicts this interpretation, and is more consistent with it displaying a weak non-specific binding activity for the Rad9 phosphopeptide that competes with specific binding to BRCT1 in this assay. Together with the observation of sulphate ion-binding this does strongly support the idea that BRCT2 possesses a competent phosphopeptide-binding site, but one whose specific target is something other than the Rad9 C-terminal sequence incorporating pSer387.
Human Rad9 has a cluster of poorly conserved putative Cdc2 phosphorylation sites upstream of Ser387, at Thr355, Ser375 and Ser380 (7) (Supplementary Figure S2). The proximity of these to Ser387 in the linear sequence of the Rad9 tail, and the physical proximity of the BRCT1 and BRCT2 phosphopeptide-binding sites in the TopBP1(1–290) structure suggested the possibility that BRCT2 might provide a binding site for one of these. To test this, we synthesized fluorescently labelled phosphopeptides incorporating pThr355, pSer375 or pSer380, and used these in FP assays as above. However, unlike the pSer387 peptide, neither of these phosphopeptides showed any significant ability to bind to TopBP1(1–290) specifically (Figure 4F). Finally, we considered the possibility that BRCT2 might provide a binding site for the relatively well-conserved pSer272 site at the N-terminal end of the Rad9 tail. Again, no significant affinity was observed for a fluorescently labelled pSer272 phosphopeptide.
DISCUSSION
BRCT domain architecture of TopBP1
BRCT domains are key sites of protein–protein interaction in the regulation and assembly of numerous complexes involved in replication and repair of DNA (64). From previous biochemical and structural studies they can be roughly partitioned into two functional classes—singleton BRCTs typically involved in mediating homo or hetero-dimerization with other BRCT domains (65,66), and tandem BRCTs where pairs of closely contiguous BRCT domains cooperate to provide sequence specific binding sites for (typically) phosphorylated peptide motifs on other proteins (35). At the sequence level, TopBP1 appears to be a composite of both types of BRCT, with the tandem pairs BRCT1:2, BRCT4:5 and BRCT7:8 conforming to the consensus for phosphopeptide binding, while BRCT domains 3 and 6 resemble non-phospho-binding singletons (34). The data presented here shows the architecture of TopBP1 to be far more complicated, with an additional BRCT domain at the N-terminus of the protein, contributing to a complex triple-BRCT structure that has no precedent in previously described systems.
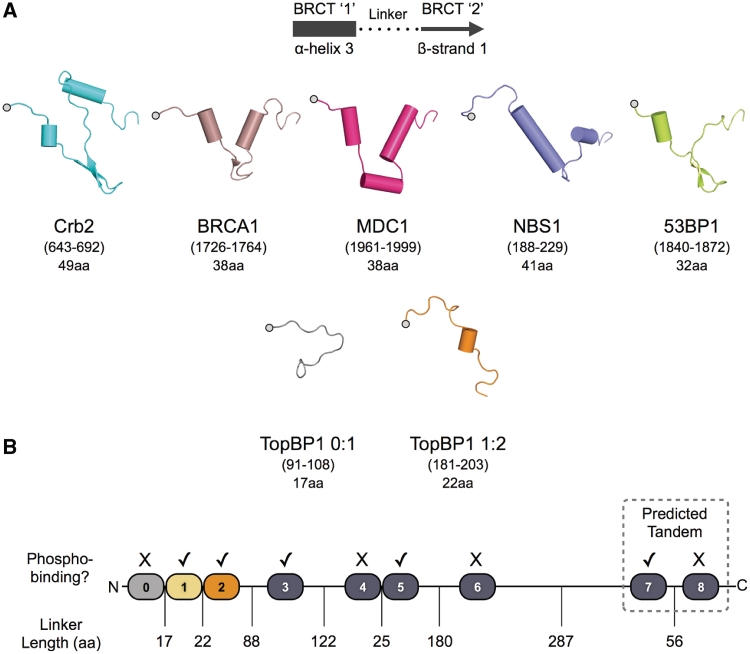
In light of this new data, we have re-examined the amino acid sequence attributes of closely contiguous tandem BRCT pairs, and find that we can distinguish between the ‘canonical’ phosphopeptide-binding tandem pairs (e.g. Crb2, BRCA1, Mdc1, etc.) and the novel structural arrangements found in TopBP1, by consideration of the size of the peptide segment that links the individual BRCT domains. Thus, the canonical tandem BRCT structures, in which the individual domains of the pair have a similar orientation and are effectively related by a translation, have a segment of ∼32–49 amino acid residues connecting the last α-helix of the first-BRCT domain to the first β-strand of the second (Figure 5A). In the novel arrangements found in TopBP1, by contrast, these linker segments are substantially shorter, with 17 residues connecting BRCT domains 0 and 1, and 22 connecting BRCT domains 1 and 2. With the shorter linker, the juxtaposition of the consecutive BRCT domains is highly constrained and results in a screw relationship, with the putative functional sites on consecutive BRCT domains presenting on opposite sides of an overall cylindrical structure.
Figure 5.
BRCT-domain classes in TopBP1 and other BRCT domain proteins. (A) Array of the linkers connecting consecutive BRCT domains in structurally characterized tandem BRCT proteins. The linker segments in the triple-BRCT structure of TopBP1(1–290) are substantially shorter, resulting in the novel non-canonical domain arrangement. (B) Assignment of BRCT domain classes in TopBP1. Domains 1, 2, 3, 5 and 7 show the characteristics of phosphopeptide binding, but only BRCT7:8 resemble the canonical tandem pair arrangement found in checkpoint mediators such as Crb2, BRCA1, etc.
Based on this, we have re-examined the BRCT domain structure of the rest of TopBP1 itself, and find that BRCT4:5, with an estimated linker length of 25 amino acids more closely resembles the novel arrangement found in BRCT0:1 and BRCT1:2, than the canonical tandem arrangement. Significantly, BRCT4 lacks the cluster of conserved residues associated with phosphopeptide-binding-tandem BRCT structures. BRCT7:8 on the other hand, conforms more to a canonical tandem pair, with a predicted linker of ∼56 amino acids, and conservation of phospho-binding residues in BRCT7 but not in BRCT8 (Figure 5B and Table 2).
Table 2.
Phospho-binding analysis of TopBP1 BRCT domains
| Phospho binding | ||||
|---|---|---|---|---|
| Crb2 (1) | Arg558 | Ser548 | Lys619 | Yes |
| BRCT0 | Phe68 | Leu14 | Val67 | No |
| BRCT1 | Arg121 | Thr114 | Lys155 | Yes |
| BRCT2 | Arg215 | Thr208 | Lys250 | Yes |
| BRCT3 | Leu374 | Cys367 | Glu407 | No |
| BRCT4 | Glu568 | Leu561 | Glu602 | No |
| BRCT5 | Lys661 | Ser654 | Lys704 | Yes |
| BRCT6 | Gln920 | Val912 | Glu957 | No |
| BRCT7 | Arg1280 | Ser1273 | Lys1317 | Yes |
| BRCT8 | Glu1408 | His1402 | Asn1452 | No |
Amino acids at the topologically equivalent positions associated with binding the phospho-group of phosphorylated peptides in a canonical phospho-binding BRCT domain (Crb-BRCT1), are conserved in TopBP1 BRCT domains 1, 2, 5 and 7.
Phosphopeptide recognition by TopBP1
The data presented here identifies Rad9 Ser387 as a probable target of CK2, that when phosphorylated, interacts specifically with the N-terminal region of TopBP1 and thereby mediates the functionally essential coupling of TopBP1 and the 9–1–1 complex (25). The presence of three distinct BRCT domains within TopBP1(1–290) considerably complicates the previous expectation that BRCT1:2 would constitute a canonical phosphopeptide-binding tandem BRCT repeat of the type previously documented in BRCA1, Mdc1, 53BP1/Crb2, Nbs1, etc. (48–54,62,63), where the first BRCT provides the phosphate recognition, while the second defines the additional sequence specificity. Instead, TopBP1 BRCT1:2 defines a new class of tandem BRCT arrangement in which the two consecutive domains independently offer binding sites for phosphorylated Ser/Thr residues.
Our mutagenesis and interaction data strongly implicates BRCT1 as the primary binding site for the Rad9 pSer387 motif. BRCT2, while conserving all the residues usually involved in phosphopeptide binding, and displaying the characteristic affinity for sulphate, only has a low affinity for the pSer387 peptide. As in other BRCT systems, we have analysed (49), while interaction with the phosphorylated residue provides a significant component of affinity for the ligand peptide, there are other overriding specificity determinants. In the canonical tandem-BRCTs, these are furnished by the second BRCT domain—the structural basis for this specificity in the non-canonical TopBP1 BRCT1:2 structure is still to be defined.
The tail of Rad9 provides a site for multiple phosphorylations in yeasts and in animals, although the location and genesis of these vary amongst different organisms. For example, the extreme C-terminal sites (Thr412/Ser423) in S. pombe Rad9 are modified by ATR (Rad3) and/or ATM (Tel1) in response to cell-cycle progression and/or DNA damage, whereas the equivalent C-terminal site (Ser387) in mammals is constitutively modified, probably by CK2 as we have shown here. In both cases however, these phosphorylations are essential for mediating the physical coupling of Rad9 to TopBP1(Rad4), that functionally connects the 9–1–1 and ATR (Rad3) complexes. Metazoan Rad9 does possess an ATR/ATM damage regulated phosphorylation site (Ser272), but at the N-terminal end of the tail segment close to the PCNA core, and several cell cycle-regulated CDK phosphorylation sites have also been mapped upstream of the TopBP1-binding pSer387. Despite the presence of multiple phosphorylation sites on Rad9, only phosphopeptides containing the pSer387 site shows the level of affinity for the TopBP1 BRCT0:1:2 segment, that is consistent with a biologically significant interaction, and while some small involvement of BRCT domains 0 and 2 in that interaction cannot be totally ruled out, the interaction with BRCT1 alone seems both necessary and sufficient. What protein–protein interactions are mediated by the other phosphorylation sites on Rad9, or by the BRCT0 and BRCT2 domains of TopBP1 remain to be defined. Experiments to address this are in progress.
During the preparation and review of this article, additional evidence supporting the role of CK2 in mediating the 9-1-1—TopBP1 interaction was published by Takeishi et al. (67); demonstrating that residues Ser341 and Ser387 are both in vitro targets of the kinase, are phosphorylated in vivo, and that HeLa cells over-expressing a phospho-deficient form of Rad9 display hypersensitivity to both UV and methyl methane sulphonate (MMS) treatment.
ACCESSION NUMBERS
2xnh and 2xnk.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cancer Research UK Programme (grant C302/A8265 to L.H.P); Infrastructure Support (grant C302/A7803 to L.H.P.). Funding for open access charge: Cancer Research UK Programme (grant C302/A8265).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to Andrew Doré and Mairi Kilkenny for help and advice during the course of the project, and to Mark Roe for assistance with data collection and processing.
REFERENCES
- 1.Smits VA, Warmerdam DO, Martin Y, Freire R. Mechanisms of ATR-mediated checkpoint signalling. Front. Biosci. 2010;15:840–853. doi: 10.2741/3649. [DOI] [PubMed] [Google Scholar]
- 2.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 3.Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, Sancar A. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc. Natl Acad. Sci. USA. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majka J, Burgers PM. The PCNA-RFC families of DNA clamps and clamp loaders. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:227–260. doi: 10.1016/S0079-6603(04)78006-X. [DOI] [PubMed] [Google Scholar]
- 5.Navadgi-Patil VM, Burgers PM. The unstructured C-terminal tail of the 9-1-1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. Mol. Cell. 2009;36:743–753. doi: 10.1016/j.molcel.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.St Onge RP, Besley BD, Pelley JL, Davey S. A role for the phosphorylation of hRad9 in checkpoint signaling. J. Biol. Chem. 2003;278:26620–26628. doi: 10.1074/jbc.M303134200. [DOI] [PubMed] [Google Scholar]
- 8.Greer DA, Besley BD, Kennedy KB, Davey S. hRad9 rapidly binds DNA containing double-strand breaks and is required for damage-dependent topoisomerase II beta binding protein 1 focus formation. Cancer Res. 2003;63:4829–4835. [PubMed] [Google Scholar]
- 9.Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 11.Yamane K, Kawabata M, Tsuruo T. A DNA-topoisomerase-II-binding protein with eight repeating regions similar to DNA-repair enzymes and to a cell-cycle regulator. Eur. J. Biochem. 1997;250:794–799. doi: 10.1111/j.1432-1033.1997.00794.x. [DOI] [PubMed] [Google Scholar]
- 12.Liu K, Paik JC, Wang B, Lin FT, Lin WC. Regulation of TopBP1 oligomerization by Akt/PKB for cell survival. EMBO J. 2006;25:4795–4807. doi: 10.1038/sj.emboj.7601355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herold S, Wanzel M, Beuger V, Frohme C, Beul D, Hillukkala T, Syvaoja J, Saluz HP, Haenel F, Eilers M. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol. Cell. 2002;10:509–521. doi: 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- 14.Boner W, Taylor ER, Tsirimonaki E, Yamane K, Campo MS, Morgan IM. A Functional interaction between the human papillomavirus 16 transcription/replication factor E2 and the DNA damage response protein TopBP1. J. Biol. Chem. 2002;277:22297–22303. doi: 10.1074/jbc.M202163200. [DOI] [PubMed] [Google Scholar]
- 15.Zeng L, Hu Y, Li B. Identification of TopBP1 as a c-Abl-interacting protein and a repressor for c-Abl expression. J. Biol. Chem. 2005;280:29374–29380. doi: 10.1074/jbc.M503016200. [DOI] [PubMed] [Google Scholar]
- 16.Wollmann Y, Schmidt U, Wieland GD, Zipfel PF, Saluz HP, Hanel F. The DNA topoisomerase IIbeta binding protein 1 (TopBP1) interacts with poly (ADP-ribose) polymerase (PARP-1) J. Cell Biochem. 2007;102:171–182. doi: 10.1002/jcb.21292. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt U, Wollmann Y, Franke C, Grosse F, Saluz HP, Hanel F. Characterization of the interaction between the human DNA topoisomerase IIbeta-binding protein 1 (TopBP1) and the cell division cycle 45 (Cdc45) protein. Biochem. J. 2008;409:169–177. doi: 10.1042/BJ20070872. [DOI] [PubMed] [Google Scholar]
- 18.Liu K, Bellam N, Lin HY, Wang B, Stockard CR, Grizzle WE, Lin WC. Regulation of p53 by TopBP1: a potential mechanism for p53 inactivation in cancer. Mol. Cell Biol. 2009;29:2673–2693. doi: 10.1128/MCB.01140-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoo HY, Kumagai A, Shevchenko A, Dunphy WG. The Mre11-Rad50-Nbs1 complex mediates activation of TopBP1 by ATM. Mol. Biol. Cell. 2009;20:2351–2360. doi: 10.1091/mbc.E08-12-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan S, Michael WM. TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. J. Cell Biol. 2009;184:793–804. doi: 10.1083/jcb.200810185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fenech M, Carr AM, Murray J, Watts FZ, Lehmann AR. Cloning and characterization of the rad4 gene of Schizosaccharomyces pombe; a gene showing short regions of sequence similarity to the human XRCC1 gene. Nucleic Acids Res. 1991;19:6737–6741. doi: 10.1093/nar/19.24.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehmann AR. Duplicated region of sequence similarity to the human XRCC1 DNA repair gene in the Schizosaccharomyces pombe rad4/cut5 gene. Nucleic Acids Res. 1993;21:5274. doi: 10.1093/nar/21.22.5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 24.Callebaut I, Mornon JP. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 1997;400:25–30. doi: 10.1016/s0014-5793(96)01312-9. [DOI] [PubMed] [Google Scholar]
- 25.Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J. Biol. Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Elledge SJ. Genetic and physical interactions between DPB11 and DDC1 in the yeast DNA damage response pathway. Genetics. 2002;160:1295–1304. doi: 10.1093/genetics/160.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furuya K, Poitelea M, Guo L, Caspari T, Carr AM. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev. 2004;18:1154–1164. doi: 10.1101/gad.291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia V, Furuya K, Carr AM. Identification and functional analysis of TopBP1 and its homologs. DNA Repair. 2005;4:1227–1239. doi: 10.1016/j.dnarep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Mochida S, Esashi F, Aono N, Tamai K, O'Connell MJ, Yanagida M. Regulation of checkpoint kinases through dynamic interaction with Crb2. EMBO J. 2004;23:418–428. doi: 10.1038/sj.emboj.7600018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 1997;11:3387–3400. doi: 10.1101/gad.11.24.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mordes DA, Nam EA, Cortez D. Dpb11 activates the Mec1-Ddc2 complex. Proc. Natl Acad. Sci. USA. 2008;105:18730–18734. doi: 10.1073/pnas.0806621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navadgi-Patil VM, Burgers PM. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J. Biol. Chem. 2008;283:35853–35859. doi: 10.1074/jbc.M807435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong Z, Kim JE, Leung CC, Glover JN, Chen J. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol. Cell. 2010;37:438–446. doi: 10.1016/j.molcel.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leung CC, Kellogg E, Kuhnert A, Hanel F, Baker D, Glover JN. Insights from the crystal structure of the sixth BRCT domain of topoisomerase IIbeta binding protein 1. Protein Sci. 2010;19:162–167. doi: 10.1002/pro.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glover JN, Williams RS, Lee MS. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem. Sci. 2004;29:579–585. doi: 10.1016/j.tibs.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 36.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 37.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 38.Berrow NS, Alderton D, Sainsbury S, Nettleship J, Assenberg R, Rahman N, Stuart DI, Owens RJ. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 2007;35:e45. doi: 10.1093/nar/gkm047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leslie AGW. MOSFLM Users Guide. Cambridge, UK: MRC Laboratory of Molecular Biology; 1995. [Google Scholar]
- 40.CCP4. Programs for protein crystallography. Acta. Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 41.Sheldrick GM. A short history of SHELX. Acta. Crystallogr. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 42.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 43.Terwilliger TC. Maximum-likelihood density modification. Acta. Crystallogr. D Biol. Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: Model-Building Tools for molecular graphics. Acta. Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Adams PD, Gopal K, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Pai RK, Read RJ, Romo TD, et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synchr. Rad. 2004;11:53–55. doi: 10.1107/s0909049503024130. [DOI] [PubMed] [Google Scholar]
- 46.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta. Crystallogr. D Biol. Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang XD, Morera S, Bates PA, Whitehead PC, Coffer AI, Hainbucher K, Nash RA, Sternberg MJE, Lindahl T, Freemont PS. Structure of an XRCC1 BRCT domain: a new protein-protein interaction module. EMBO J. 1998;17:6404–6411. doi: 10.1093/emboj/17.21.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clapperton JA, Manke IA, Lowery DM, Ho T, Haire LF, Yaffe MB, Smerdon SJ. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat. Struct. Mol. Biol. 2004;11:512–518. doi: 10.1038/nsmb775. [DOI] [PubMed] [Google Scholar]
- 49.Kilkenny ML, Dore AS, Roe SM, Nestoras K, Ho JC, Watts FZ, Pearl LH. Structural and functional analysis of the Crb2-BRCT2 domain reveals distinct roles in checkpoint signaling and DNA damage repair. Genes Dev. 2008;22:2034–2047. doi: 10.1101/gad.472808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee MS, Edwards RA, Thede GL, Glover JN. Structure of the BRCT repeat domain of MDC1 and its specificity for the free COOH-terminal end of the gamma-H2AX histone tail. J. Biol. Chem. 2005;280:32053–32056. doi: 10.1074/jbc.C500273200. [DOI] [PubMed] [Google Scholar]
- 51.Shiozaki EN, Gu L, Yan N, Shi Y. Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling. Mol. Cell. 2004;14:405–412. doi: 10.1016/s1097-2765(04)00238-2. [DOI] [PubMed] [Google Scholar]
- 52.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 53.Botuyan MV, Nomine Y, Yu X, Juranic N, Macura S, Chen J, Mer G. Structural basis of BACH1 phosphopeptide recognition by BRCA1 tandem BRCT domains. Structure. 2004;12:1137–1146. doi: 10.1016/j.str.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varma AK, Brown RS, Birrane G, Ladias JA. Structural basis for cell cycle checkpoint control by the BRCA1-CtIP complex. Biochemistry. 2005;44:10941–10946. doi: 10.1021/bi0509651. [DOI] [PubMed] [Google Scholar]
- 55.Dore AS, Kilkenny ML, Rzechorzek NJ, Pearl LH. Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex implications for clamp loading and regulation. Mol. Cell. 2009;34:735–745. doi: 10.1016/j.molcel.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 56.Sohn SY, Cho Y. Crystal structure of the human rad9-hus1-rad1 clamp. J. Mol. Biol. 2009;390:490–502. doi: 10.1016/j.jmb.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 57.Xu M, Bai L, Gong Y, Xie W, Hang H, Jiang T. Structure and functional implications of the human rad9-hus1-rad1 cell cycle checkpoint complex. J. Biol. Chem. 2009;284:20457–20461. doi: 10.1074/jbc.C109.022384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen MJ, Lin YT, Lieberman HB, Chen G, Lee EY. ATM-dependent phosphorylation of human Rad9 is required for ionizing radiation-induced checkpoint activation. J. Biol. Chem. 2001;276:16580–16586. doi: 10.1074/jbc.M008871200. [DOI] [PubMed] [Google Scholar]
- 59.St Onge RP, Besley BD, Park M, Casselman R, Davey S. DNA damage-dependent and -independent phosphorylation of the hRad9 checkpoint protein. J. Biol. Chem. 2001;276:41898–41905. doi: 10.1074/jbc.M105152200. [DOI] [PubMed] [Google Scholar]
- 60.Linding R, Jensen LJ, Pasculescu A, Olhovsky M, Colwill K, Bork P, Yaffe MB, Pawson T. NetworKIN: a resource for exploring cellular phosphorylation networks. Nucleic Acids Res. 2008;36:D695–D699. doi: 10.1093/nar/gkm902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ali AA, Jukes RM, Pearl LH, Oliver AW. Specific recognition of a multiply phosphorylated motif in the DNA repair scaffold XRCC1 by the FHA domain of human PNK. Nucleic Acids Res. 2009;37:1701–1712. doi: 10.1093/nar/gkn1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lloyd J, Chapman JR, Clapperton JA, Haire LF, Hartsuiker E, Li J, Carr AM, Jackson SP, Smerdon SJ. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell. 2009;139:100–111. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watts FZ, Brissett NC. Linking up and interacting with BRCT domains. DNA Repair. 2010;9:103–108. doi: 10.1016/j.dnarep.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 65.Dulic A, Bates PA, Zhang X, Martin SR, Freemont PS, Lindahl T, Barnes DE. BRCT domain interactions in the heterodimeric DNA repair protein XRCC1-DNA ligase III. Biochemistry. 2001;40:5906–5913. doi: 10.1021/bi002701e. [DOI] [PubMed] [Google Scholar]
- 66.Krishnan VV, Thornton KH, Thelen MP, Cosman M. Solution structure and backbone dynamics of the human DNA ligase IIIalpha BRCT domain. Biochemistry. 2001;40:13158–13166. doi: 10.1021/bi010979g. [DOI] [PubMed] [Google Scholar]
- 67.Takeishi Y, Ohashi E, Ogawa K, Masai H, Obuse C, Tsurimoto T. Casein kinase 2-dependent phosphorylation of human Rad9 mediates the interaction between human Rad9-Hus1-Rad1 complex and TopBP1. Genes Cells. 2010;15:761–771. doi: 10.1111/j.1365-2443.2010.01418.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.