Abstract
SMG-9 is part of a protein kinase complex, SMG1C, which consists of the SMG-1 kinase, SMG-8 and SMG-9. SMG1C mediated phosphorylation of Upf1 triggers nonsense-mediated mRNA decay (NMD), a eukaryotic surveillance pathway that detects and targets for degradation mRNAs harboring premature translation termination codons. Here, we have characterized SMG-9, showing that it comprises an N-terminal 180 residue intrinsically disordered region (IDR) followed by a well-folded C-terminal domain. Both domains are required for SMG-1 binding and the integrity of the SMG1C complex, whereas the C-terminus is sufficient to interact with SMG-8. In addition, we have found that SMG-9 assembles in vivo into SMG-9:SMG-9 and, most likely, SMG-8:SMG-9 complexes that are not constituents of SMG1C. SMG-9 self-association is driven by interactions between the C-terminal domains and surprisingly, some SMG-9 oligomers are completely devoid of SMG-1 and SMG-8. We propose that SMG-9 has biological functions beyond SMG1C, as part of distinct SMG-9-containing complexes. Some of these complexes may function as intermediates potentially regulating SMG1C assembly, tuning the activity of SMG-1 with the NMD machinery. The structural malleability of IDRs could facilitate the transit of SMG-9 through several macromolecular complexes.
INTRODUCTION
Eukaryotic gene expression comprises a complex set of biochemical reactions starting with the transcription of the genetic information and ending in the synthesis of proteins. Between these two events, post-transcriptional modifications and remodelling are required to assemble a mature mRNA that can be translated by the ribosome, and several surveillance mechanisms ensure the fidelity and accuracy of these processes. Nonsense-mediated mRNA decay (NMD) is a post-transcriptional surveillance mechanism that, in eukaryotes, recognizes and degrades mRNAs containing premature translation termination codons (PTCs) to prevent the accumulation of potentially harmful truncated polypeptides encoding for a truncated protein (1,2).
The NMD machinery marks a PTC-containing mRNA for degradation through a highly sophisticated sequence of protein-protein interactions involving different polypeptides (2,3). Until recently, seven conserved core factors for NMD had been identified to be present in most metazoan, SMG-1, Upf1, Upf2, Upf3, SMG-5, SMG-6 and SMG-7. Thanks to an intense current research effort, a picture of the players and the molecular mechanisms involved in NMD is starting to emerge (1,2). Yet, many aspects remain obscure and several models have been proposed to explain the molecular mechanisms by which the NMD machinery tags an mRNA for degradation (4,5). An interesting debate in current literature upholds a vision of NMD regulated by the outcome of a competition between stimulating and downregulating signals (3–6). In any case, the topic of what determines the fate of a PTC-containing mRNA is still the subject of open research (3,5,7).
One of the central players articulating the NMD response is SMG-1, a large protein of roughly 430 kDa that belongs to the phosphatidylinositol 3-kinase-related protein kinase (PIKK) family of serine-threonine kinases (8,9). SMG-1 is a component of mRNA surveillance complexes and the phosphorylation of Upf1 by SMG-1 is the single essential event in metazoans to trigger all the latter processes leading to the degradation of an mRNA (10). A complex known as SURF (SMG-1:Upf1:eRF1:eRF3) and containing SMG-1, Upf1 and the eukaryotic release factors eRF1 and eRF3 is assembled on a termination codon together with the ribosome (11–13). The ribosome:SURF complex can interact with a downstream exon–junction-complex (EJC), a protein complex deposited 20–24 nt upstream the exon–exon junction, through the Upf2 and Upf3 proteins, activating the kinase activity of SMG-1 on Upf1. Phospho-Upf1 is thought to recruit the mRNA-decapping as well as the RNA-degrading machinery that eventually degrades the mRNA containing the PTC (2,3).
SMG-1 has been shown to play other roles besides controlling NMD. Depletion of SMG-1 in human cells influences the response to DNA damage (8,14) and regulates the association of telomeric repeat-containing RNA at telomeres (15). SMG-1 is required for adequate regulation of p53 phosphorylation upon genotoxic and oxidative stress and controls cell proliferation and apoptosis (14,16–18). Although the molecular bases of all these processes are unclear, many of these functional features parallel those of other PIKKs, suggesting some cooperation among the members of this family of kinases (8,13).
Very recently, two novel components of a SMG-1 complex have been discovered and named SMG-8 and SMG-9 (12). These proteins were isolated due to their co-purification with SMG-1, with which they form a stable complex (SMG-1:SMG-8:SMG-9), named SMG1C. SMG-8 and SMG-9 are tightly associated with SMG-1 and they seem to regulate its kinase activity and the remodelling of the mRNA surveillance complex. Interestingly, additional NMD factors, Ruvbl1, Ruvbl2, RPB5 and SMG-10, have just been described, highlighting the complexity of the NMD machinery (13). SMG-8 is a 991 amino acid protein which has been proposed to regulate the correct localization of SMG-1 at the PTC-stalled ribosome to form the SURF complex (12). SMG-9 is a 520 amino acid protein comprising a central putative nucleotide-triphosphatase domain. SMG-9 seems to regulate the formation of the SMG-1:SMG-8:SMG-9 complex and probably up-regulates the kinase activity of SMG-1 since SMG-1:SMG-9 complexes show higher activity than SMG-1 (12). Both SMG-8 and SMG-9 are required for mammalian NMD and the stable association between these two proteins is an absolute requirement for the inhibition of the NMD response until a genuine PTC is recognized (12).
In order to improve our understanding of the functions and molecular structure of the SMG1C complex, here we have characterized SMG-9. We present evidence showing that SMG-9 comprises two distinct domains. The N-terminal domain is an intrinsically disordered region (IDR) essential for the maintenance of the structural integrity of the SMG-1:SMG-8:SMG-9 complex. IDRs are a hot topic of research, as they appear to provide the essential specificity and malleability for the large macromolecular machines requiring multiple and variable protein–protein interactions, as those functioning in all stages of eukaryotic gene expression (19,20). Unexpectedly, several experiments in vivo reveal that, besides is participation as a component of the SMG1C complex, SMG-9 can assembly as homodimers and, most likely, SMG-8:SMG-9 heterodimers that could represent intermediates regulating the assembly of the SMG1C complex.
MATERIALS AND METHODS
Prediction of ordered and disordered regions
We analysed the predicted ordered and disordered regions in the sequences of SMG-1, SMG-8 and SMG-9 using one of the most accepted predictors of naturally disordered regions, PONDR (http://www.pondr.com) (21). The default predictor VL-XT was used.
Cloning, expression and purification of NT-SMG-912–180
NT-SMG-9 cDNA was subcloned between the EcoRI and NcoI sites on modified N-terminal HisTag pRAT4, pRHO and pGEX-6P-2 plasmids (GE Healthcare Bio-Sciences, Buckinghamshire, UK). The initial GST fusion construct was made comprising amino acids 1–180. After the observation of spontaneous self-cleavage in the initial GST-fusion construct, the site of cleavage was identified by mass spectroscopy and the construct re-cloned comprising amino acids 12–180. All proteins were expressed in Escherichia coli strain BL21(DE3) and the expression detected in a soluble fraction after cell lysis by sonication. GST-fusion proteins were purified with GST-Trap columns (20 ml, GE Healthcare Bio-Sciences) and NT-SMG-9 was isolated from GST after digestion with the 3C protease. A second GST-trap column followed by a cation exchange chromatography in SP-sepharose HiTrap colum (5 ml, GE Healthcare Bio-Sciences) followed by a final step of gel-filtration chromatography (Sephacryl S-100, GE Healthcare Bio-Sciences) yielded a highly pure preparation as judged by SDS–PAGE after Coomassie brilliant blue staining. Protein concentration was determined by UV absorption at 280 nm. The protein solutions were concentrated with an Amicon Ultra device (Millipore, Bedford, MA, USA). Mass spectroscopy was used to assess the identity as well as the purity of final preparations.
Spectroscopic techniques
Circular dichroism (CD) spectra were recorded on a JASCO J-805 spectropolarimeter. An optical cuvette with a 1-mm path length was used. The temperature of the measuring cell was maintained at 25°C. Spectra were collected in a spectral range of 200–250 nm with a path-length of 1 nm. The NT-SMG-9 preparation was dissolved in 20 mM sodium phosphate (pH 7.2) and 50 mM NaCl at a concentration of 15 μM. Data was analysed using the software KD2.
Fluorescence spectra were acquired on an F-4500 fluorescence spectrophotometer (Hitachi, Tokyo, Japan) at 25°C. The concentration of NT-SMG-9 was 15 μM. Buffer solution contained 20 mM sodium phosphate (pH 7.2) and 50 mM NaCl. The excitation wavelength was 295 nm, and the emission spectra were recorded between 285 and 500 nm. Denaturing conditions involved the measurement with the same conditions buffer and protein concentration but in the presence of 6 M guanidinium hydrochloride (Pierce).
In the NMR spectroscopy experiments, samples for 1H monodimensional spectra were prepared in 20 mM sodium phosphate (pH 7.2) and 50 mM NaCl at a concentration of 100 µM. 15N-labelled samples were prepared in M9 medium at a concentration of 200 µM. The NMR samples contained 10% D2O.The monodimensional as well as 1H–15N heteronuclear single-quantum correlation (HSQC) spectra were acquired at 25°C on a Bruker DMX-600 spectrometer equipped with a cryoprobe.
For reagents (antibodies)
Anti-SMG-8 and -SMG-1 have been described earlier [Yamashita et al. (12)]. Anti-HA (clone 3F10) (Roche), anti-SBP (SantaCruz), anti-mTOR (Cell Signaling Technology), anti-aPKCλ (C-20) (SantaCruz) were obtained commercially.
Affinity purification, immunoprecipitation and western blot analysis
pEF_Flag-HA-SBP-SMG-9 (2–520) (for Figure 5B), pEF_Flag-HA-SBP-NT-SMG-9 (2–181), pEF_Flag-HA-SBP-CT1-SMG-9 (185–520), pEF_Flag-HA-SBP-CT2-SMG-9 (175–520), pcDNA5/NTAP(CBP-SBP)-SMG-9 (2–520) (for Figure 5C), pSR_Strep-HA-SMG-9 full (2–520), pSR-V5-SMG-9 full (2–520), pSR_V5-NT-SMG-9 (2–181) and pSR-V5-CT-SMG-9 (182–520) were constructed by cloning each cDNA fragment by standard methods. siLentGene-puro-siSMG-9UTR (siRNA sequence targeted to the 3′-UTR of the SMG-9 mRNA: GGAGAGGAATGTCATGCAC) was constructed by method described in manual.
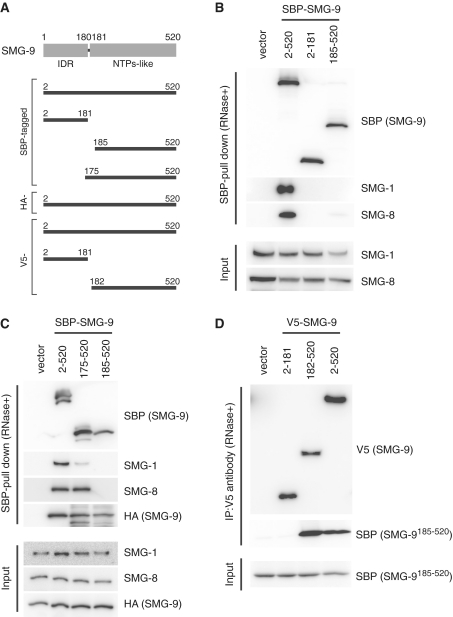
Figure 5.
Effect of NT-SMG-9 and CT-SMG-9 truncation in SMG1C assembly. (A) Schematic structures of SMG-9 construct. (B) 293T cells were transfected with the SMG-9 plasmids shown above together with plasmid expressing the siRNA targeted to 3′-UTR of SMG-9. (C) 293T cells were transfected with the SBP-tagged-SMG-9 plasmids shown above together with the HA-tagged-SMG-9 plasmid. The cells were lysed and pull downed with the streptavidn sepharose in presence of RNaseA. Pull downed products or cell lysates (input) were then probed with the antibodies shown on the right. (D) 293T cells were transfected with the V5-tagged-SMG-9 plasmids shown above together with the SBP-tagged-CT-SMG-9185–520 plasmid. The cells were lysed and immunoprecipitated with anti-V5 antibodies in presence of RNaseA. Immunoprecipitated products or cell lysates (input) were then probed with the antibodies shown on the right. ‘Vector’ indicates an empty vector.
A 293T cells were transfected using HEKfectin (Biorad), and lysed with a loose-fit Potter–Elvehjem homogenizer in T-buffer [20 mM HEPES–NaOH at pH 7.5, 50 mM NaCl, 0.05% Tween-20, 2.5 mM MgCl2, 0.5 mM DTT, protease inhibitor cocktail (Roche), phosphatase inhibitor cocktail (Roche) and 100 μg/ml RNaseA (Qiagen)]. The soluble fractions were pre-cleared with sepharose 4B (Sigma) and then incubated with streptavidin–sepharose (GE Biotech) for 2 h at 4°C with gentle rotation. Pre-cleared lysates were incubated with streptavidin–sepharose or anti-V5 antibodies for 2 h or 1 h at 4°C with gentle rotation. For antibodies, subsequently, the soluble fractions were incubated with 30 μl of protein G sepharose (GE Biotech) for an additional 1 h at 4°C with gentle rotation. After washing with RNase(−) T-lysis buffer, the affinity-purified protein complexes were eluted by incubation at 4°C for 30 min with RNase(−) lysis buffer containing 2 mM biotin (Sigma) or SDS sample buffer, respectively. All proteins in western blot experiments were detected with an ECL western blot detection kit (GE Biotech) or Lumi-Light (Roche). All experiments were performed two to three times, and typical results are shown.
Size exclusion chromatography of SMG-9 complexes
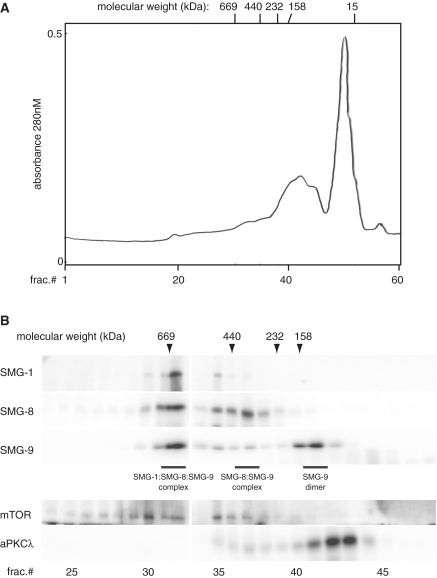
For the preparation of HeLa cell extracts for gel column fractionation, 3 × 108 HeLa cells were re-suspended in an equal volume of lysis buffer containing 10 mM Tris–HCl at pH 8.0, 150 mM NaCl, 0.4% NP40, 2 mM MgCl2, 0.2 mM DTT, 0.1 mM PMSF, 100 nM Okadaic acid and 200 μg/ml RNaseA. After incubation on ice for 10 min, cells were lysed by 15 hand-strokes of a loose-fit Potter-Elvehjem homogenizer. The cell lysate was centrifuged at 15 000g for 30 min, and the supernatant was loaded onto a 24 ml Superose 6 FPLC column (GE Biotech) equilibrated with lysis buffer. Fractions (400 μl) were collected from 5 to 24 ml elution volume. Fractions were pooled and concentrated. Proteins were detected by western blotting. A control molecular marker was obtained by running the proteins thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), and RNaseA (15 kDa) (GE Biotech) on the same column under the same conditions.
RESULTS
SMG-9 comprises two distinct structural domains
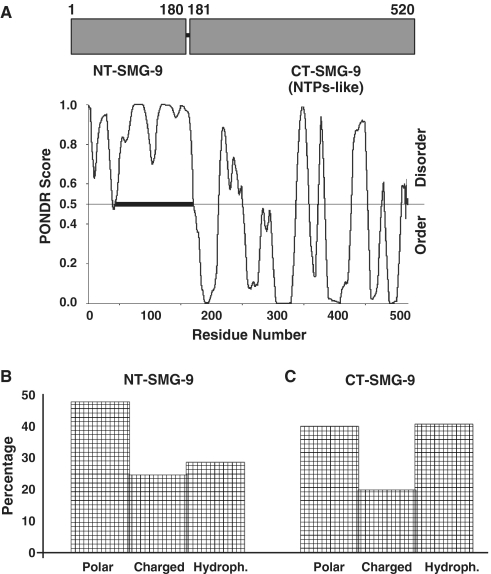
The first description of SMG-9 reported the presence of a putative nucleotide-triphosphatase domain comprising residues 181–520 (12). We now performed a more thorough analysis of the SMG-9 sequence using several bioinformatic methods, which revealed the presence of an unusual N-terminal region. The N-terminal region of SMG-9 was enriched in prolines, polar and charged residues while showing a low content in the hydrophobic residues that most frequently form the hydrophobic core of conventional protein domains (Supplementary Figure S1). These features are the signature of intrinsically disordered regions (IDRs), segments of proteins that under native conditions do not fall into a conventional fold and which participate in specific binding to targets in highly complex multi-component macromolecular machines (19). We searched for the presence of IDRs in SMG-9 based on the distinctive signature of their sequence composition and conservation of amino acids. Several in silico disordered predictors have been developed, all of them showing excellent predictive value (19,20). We used PONDR (‘Predictor Of Natural Disordered Regions’) (http://www.pondr.com/) (21) to look for potential disordered domains in SMG-9. SMG-9 exhibited two clearly distinct regions in its primary structure (Figure 1A). A C-terminal region comprising residues 181–520 was predicted to conform to a conventional well-folded domain in agreement with its description as a putative NTPase domain based on sequence homology (CT-SMG-9 from now on). In contrast, PONDR predicted a large highly disordered region encompassing the first 180 residues of SMG-9 (NT-SMG-9 from now on) (Figure 1A).
Figure 1.
Analysis of SMG-9 primary structure. (A) PONDR analysis of the SMG-9 amino acid sequence. A PONDR score >0.5 predicts those amino acids belonging to a disordered region. A stretch of disordered amino acids of more than 50 residues are usually considered a disordered domain. This analysis revealed that the sequence of SMG-9 could be divided in two regions: an N-terminal 180 residue intrinsically disordered region and a C-terminal folded domain, encoding a putative nucleotide-triphosphatase (NTPase)-like domain (12). Distribution of types of amino acid in NT-SMG-9 (B) and the C-terminal region of SMG-9 (C). NT-SMG-9 revealed a propensity towards polar and charged amino acids while the C-terminal domain was enriched in hydrophobic residues. Unstructured domains frequently exhibit a low content in hydrophobic amino acids and a bias towards charged and polar residues.
Disordered regions typically exhibit a higher ratio between the sum of polar plus charged amino acids and the hydrophobic residues than structured folded domains, since they do not form a conventional hydrophobic core. An analysis of the distribution of different types of amino acids in SMG-9 revealed a propensity of NT-SMG-9 towards polar and charged amino acids (Figure 1B), whereas the C-terminal domain of SMG-9 showed a higher proportion of hydrophobic residues (Figure 1C). Also, a comparison of the ratio between hydrophobicity versus net charge in several proteins placed the NT-SMG-9 domain well within the group of other known intrinsically unstructured proteins whereas the C-terminal domain showed the characteristic pattern of folded domains (Supplementary Figure S2).
The recombinant N-terminus of SMG-9 is a 20 kDa soluble monomeric domain
To further analyse the biophysical and structural properties of the NT-SMG-9 domain, several milligrams of highly pure protein were required, only achievable by recombinant production. Non-specific degradation of partially folded heterologous proteins expressed in Escherichia coli is a common problem, which could be potentially enhanced in the case of an IDR, due to its intrinsic disordered structure. To increase the chances of success in the production of NT-SMG-9 in E. coli, we set up three parallel strategies taking advantage of this well-established prokaryotic system. We cloned the cDNA corresponding to the first 180 residues of SMG-9 (NT-SMG-9) into three different expression plasmids containing either (i) a N-terminal hexahistidine tag (HisTag), (ii) a N-terminal OmpA peptide inducing the secretion of the expressed protein to the periplasmic space and a C-terminal HisTag and (iii) a fusion protein with a glutathione synthetase transferase (GST) and a site for the 3C protease between the GST and the target construct to remove the tag. We found no expression when using the N-terminal HisTag, whereas a small secretion of the expressed protein was observed with the periplasmic secretion-inducing vector, albeit with a high degree of non-specific degradation (not shown, see below and Figure 2A for the purification of NT-SMG-912–180). The best results were obtained with the fusion construct containing a GST tag. In standard conditions, GST-NT-SMG-9 was expressed as a soluble protein, but 20–30% of the recovered protein from the GST column was non-specifically proteolysed. We identified the non-specific cleavage site by sequencing of the N-terminus and by mass spectrometry of the spontaneous truncated product. We found that the first 11 amino acids of NT-SMG-9 were removed non-specifically during purification and we therefore recloned the fragment without these residues in the GST vector to produce a more stable product. This construct, GST-NT-SMG-912–180, missing the first 11 amino acids, expressed as a soluble protein and was entirely stable after performing the purification steps described earlier (Figure 2A). The final steps during the purification protocol involved a second GST affinity column to remove the GST-tag, an intermediate step of cationic exchange and a final gel filtration chromatography (Figure 2B). This protocol allowed the efficient production of soluble NT-SMG-912–180 with a yield of ∼3–4 mg of protein per liter of medium (Figure 2C).
Figure 2.
Expression and purification of NT-SMG-912–180 in E. coli. (A) SDS–PAGE of three different constructs assayed to produce a soluble fragment of NT-SMG-9. Left, N-terminal hexahistidine tag (pRHT vector); middle, periplasmic secretion vector (pRHO vector); and right, GST fusion protein (pGEX-6p-2 vector). The total cell extract right before induction (column T0), 5 h after induction with 1 mM IPTG (column T5) and the soluble fraction after sonication of the T5 sample (column S), are shown. For the periplasmic construct the supernatant (column SN) of the culture of T5 is shown, since the periplasmic space of E .coli is very leaky, over-expressed proteins can be easily localized in the supernatant of the centrifuged culture. Lastly, in those two constructs where some expression was detected, the elution from an in-batch incubation of the supernatant (periplasmic secretion construction) or the soluble fraction of T5 (GST-fusion construction) with HisTrap resin or GST–sepharose resin (column O), are shown. The pull-down protein in the GST-NT-SMG-9 construct is labelled. (B) SDS–PAGE of a purified NT-SMG-912–180 after the first GST-trap column before (column NC) and after a 4 h digestion (column C) with 3C protease. (C) Final preparation after applying the purification protocol described in the text.
We characterized the hydrodynamic behaviour of E. coli expressed NT-SMG-912–180 protein in solution to define its state of aggregation using analytical gel filtration and analytical ultracentrifugation (Supplementary Figure S3). NT-SMG-912–180 eluted as a single sharp peak in size exclusion chromatography, with a retention volume of 1.72 ml, compatible with a molecular mass of ∼20 kDa after calibration of the column (Supplementary Figure S3A). Less than 5% of the protein eluted as a large aggregate in the void volume, indicating that NT-SMG-912–180 behaved as expected for a single monomeric species in solution. This experiment was performed in medium-high ionic strength conditions (300 mM NaCl) that were required to avoid interaction of the protein with the column matrix. In addition, ultracentrifugation analysis at lower ionic strength conditions (50 mM NaCl) performed through sedimentation velocity experiments (Supplementary Figure S3B) unambiguously showed that NT-SMG-912–180 behaved as a single species in solution, with a Svedberg coefficient of 1.1 S corresponding to a molecular weight of 19.5 ± 0.4 kDa (Supplementary Figure S3C). These results were further confirmed by sedimentation equilibrium analysis performed at two different velocities, which notably agreed well with the previous data (Supplementary Figure S3D).
Taking all the hydrodynamic data into account, we conclude that the NT-SMG-912–180 domain did not form any major aggregate, the predominant species in solution being a monomer, at medium-high as well as low ionic strength conditions.
The N-terminal domain of SMG-9 is an intrinsically disordered region
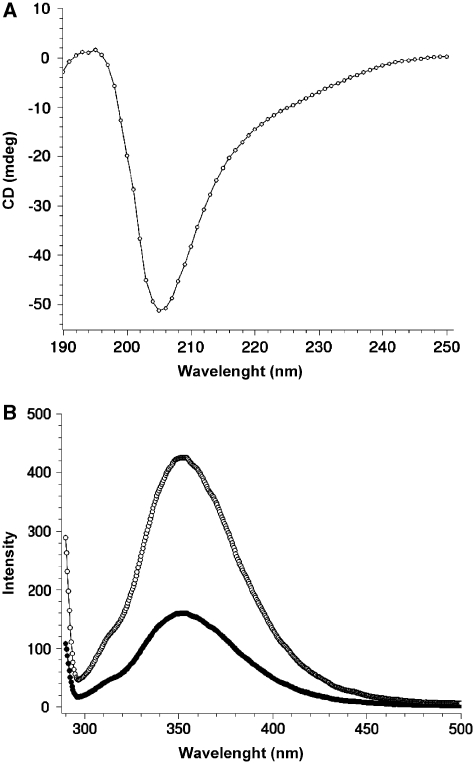
Recent studies suggest that IDRs do not show uniform structural properties, but their structure ranges from a fully unstructured protein (‘random coils’) to partially structured regions (‘pre-molten globule’) and more ‘folded’ proteins containing some elements of secondary structure (‘molten globule’) (19,20). Proteins in the first group show no secondary structure at all as well as hydrodynamic dimensions like those of coiled-coils. In contrast, pre-molten globules present a core of secondary structure, although less dense than that found in structured proteins. To gain insight into the structural properties of NT-SMG-912–180 we investigated its secondary structure content using two spectroscopic techniques, ultraviolet-circular dichroism (UV–CD) and fluorescence spectroscopy (Figure 3). Whereas completely unfolded polypeptides are characterized by a well-defined CD spectrum with a minimum in the vicinity of 200 nm and an ellipticity close to zero in the vicinity of 222 nm, the CD-spectra of NT-SMG-912–180 showed a minimum at 205 nm and negative values of ellipticity from ∼235 to 200 nm (Figure 3A). This suggested the presence of certain content in secondary structure. In addition, fluorescence spectra with an excitation wavelength of 295 nm revealed a significant increase in the fluorescence emission upon denaturation of NT-SMG-912–180 using 6 M guanidinium hydrochloride compared to native conditions (Figure 3B), strongly suggesting the presence of a certain degree of secondary structure quenching the fluorescence of the two tryptophans of NT-SMG-912–180 in the native protein.
Figure 3.
Spectroscopic analyses of NT-SMG-912–180. (A) Far UV-circular dichroism spectrum of NT-SMG-912–180. (B) Fluorescence spectra of NT-SMG-912–180 in 50 mM phosphate buffer and 50 mM NaCl (black circles) and in the same buffer but in the presence of 6 M Guanidinium hydrochloride (white circles).
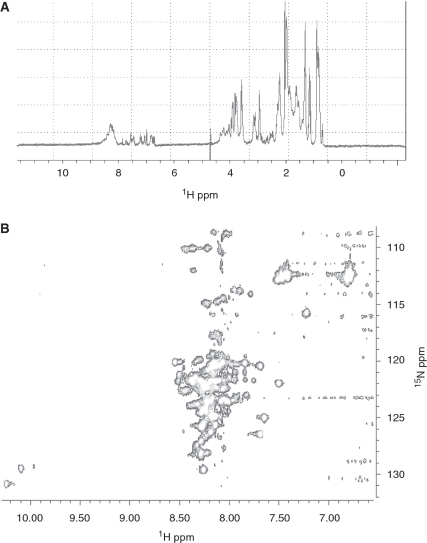
NMR spectroscopy was performed to definitively determine the presence of an IDR at the N-terminus of SMG-9. A 1H 1D spectrum of NT-SMG-912–180 showed two groups of signals, a first group of thin, well-resolved peaks (6.5–8 ppm) and a group of superposed signals accumulating between 8 and 8.5 ppm (Figure 4A). This spectrum would be compatible with an unfolded polypeptide, being the first group of sharp peaks those signals corresponding to the flexible lateral side chains and the aromatic protons whereas the N–H backbone would appear as those signals between 8 and 8.5 ppm. We also performed 1H-15N-HSQC 2D experiments after labelling NT-SMG-912–180 with 15N (Figure 4B), and we found that the majority of the signals attributable to the N-H backbone overlapped within a very narrow 1H chemical shift, ranging from 7.75 to 8.5 ppm. This is a typical spectrum for intrinsically disordered regions, where defined signals (in contrast to aggregated protein) are concentrated in a narrow range (in contrast to folded proteins). Signals of the two tryptophans present in NT-SMG-912–180 were detected at 10.25, 128.89 ppm and 10.1, 127.48 ppm, 1H, 15N chemical shifts, respectively. These chemical shifts are characteristic of solvent exposed tryptophan residues as the amino acids of a disordered protein. In addition, the HSQC spectrum showed several well-dispersed peaks typical of a folded structure, suggesting the presence of some residual structure as previously suggested by CD and fluorescence spectroscopy data.
Figure 4.
NMR spectroscopy analysis of NT-SMG-912–180. (A) 1H monodimensional spectrum of NT-SMG-912–180 showing the overlapping of signals in a narrow chemical shift in a range centered at 8.25 ppm. (B) 1H-15N HSQC spectrum of 15N-labelled NT-SMG-912–180 unambiguously identified this domain as inherently unstructured due to the absence of well dispersed cross peaks. The presence of well-resolved peaks and the absence of dispersion discarded any non-specific aggregation.
The N- and C-terminal domains of SMG-9 are required to maintain the integrity of the SMG1C complex
We examined the relevance of the two domains of SMG-9 to maintain the integrity of the SMG-1:SMG-8:SMG-9 complex in cells. For this purpose, we expressed full-length SMG-9 and fragments comprising the N-terminal (NT-SMG-92–181) and C-terminal (CT-SMG-9185–520) domains as SBP (Streptavidin Binding Peptide) tagged fusion proteins in 293T cells. To avoid the interference of endogenous SMG-9, this protein was down regulated using RNA interference targeted to 3′-untranslated region of SMG-9. The expressed proteins were bound to Streptavidin-beads in presence of RNaseA, to remove interactions mediated by RNA, eluted and the presence of SMG-1 and SMG-8 in the pull-downed material tested by western blotting (Figure 5B). Whereas each product was adequately expressed, only full-length SMG-9 co-purified with SMG-1 and SMG-8 in presence of RNaseA. These experiments indicated that both the N- and C-terminal domains of SMG-9 are necessary for the integrity of the SMG1C complex. To map the requirement of SMG-9 for these interactions more precisely, in a separate set of experiments, we simultaneously tested two C-terminal constructs comprising residues 185–520 (CT-SMG-9185–520) and 175–520 (CT-SMG-9175–520) tagged with SBP. SBP-pull-downs confirmed that CT-SMG-9185–520 was not capable of recognizing SMG-1 or SMG-8. Interestingly, CT-SMG-9175–520, where a small N-terminal segment flanking the C-terminal domain was incorporated, was sufficient to recognize SMG-8 at a similar level than full-length SMG-9 (Figure 5C), whereas the recognition of SMG-1 was heavily impaired. These results strongly suggested that CT-SMG-9 is directly responsible for the recognition of SMG-8 whereas contributions of the N- and C-terminal domains of SMG-9 are required to bind SMG-1.
The experiment described earlier was performed after simultaneous co-transfection of full-length HA-tagged SMG-9 and the SBP-tagged constructs, and surprisingly the pull-downs revealed that the full-length and two C-terminal constructs of SMG-9 tested were interacting with full-length HA-SMG-9. Unexpectedly, CT-SMG-9185–520 was forming a tight complex with full-length SMG-9, devoid of SMG-1 and SMG-8, in striking contrast with the behaviour of full length SMG-9 that reproducibly pulls downs a significant amount of SMG-1 and SMG-8 (Figure 5C). In addition, full length SBP-SMG-9 was found to interact with full length HA-SMG-9, indicating that the association between SMG-9 molecules takes place also in the context of the full protein (Figure 5C). Hence, the existence of putative SMG-9 oligomers was further investigated.
SMG-9 assembles into stable oligomers with SMG-9 (homo-oligomers) and SMG-8 (hetero-oligomers) that are not part of SMG1C
V5-tagged NT-SMG-92–181, or CT-SMG-9182–520 or full length SMG-9 were co-expressed with SBP-tagged CT-SMG-9185–520 in 293T cells (Figure 5D). Pull downs by V5 antibody revealed a significant interaction between V5-tagged CT-SMG-9182–520 and SBP-tagged CT-SMG-9185–520, indicating that the C-terminal region of SMG-9 strongly contributes to its self-association (Figure 5D). However, we failed to detect binding between NT-SMG-92–181 and CT-SMG-9185–520 (Figure 5D) and between V5-tagged NT-SMG-92–181 and SBP-tagged NT-SMG-92–181 (data not shown). These results correlate with the finding that recombinant NT-SMG-912–180 behaved as a monomer (Supplementary Figure S3).
The above experiments implied that SMG-9 could assemble other complexes besides SMG1C and we sought a further confirmation by partially resolving SMG-9 complexes by size exclusion chromatography (Figure 6). HeLa cell extracts were fractionated by gel filtration and the fractions analysed by denaturing electrophoresis and western blotting. As controls, we used markers of molecular weight (Figure 6A, top line), mTOR, a PIKK member that migrates as a monomer (∼290 kDa) and as a 0.7–0.8 MDa multi-protein complex (22), and aPKCλ (78 kDa). In addition to a well-resolved peak corresponding to SMG1C and comprising SMG-1, SMG-8 and SMG-9, SMG-9 was detected in two additional peaks. One, composed only of SMG-9 (monomer, ∼60 kDa) clearly migrated as a homo-oligomer rather than a monomer, and the apparent molecular weight correlated with the dimeric species previously detected by pull-down assays. In addition, a second peak containing SMG-9 migrated as a larger complex, which exactly co-migrated with SMG-8 as ∼400 kDa complexes, a strong indication of an SMG-8:SMG-9 complex. These results suggested that SMG-9 has biological functions beyond SMG1C, also maybe regulating SMG1C assembly by means of several SMG-9-containing sub-complexes.
Figure 6.
Several SMG-9-containing complexes can be isolated. (A and B) Size exclusion chromatography of SMG-1, SMG-8 and SMG-9 containing complexes. Fractions were run in SDS gels and the presence of either protein tested by western blot using anti-bodies specific to each component. As molecular weight markers, commercial markers (location of their elution peaks indicated in top line), mTOR (290 kDa and 0.7 MDa) and aPKCλ (78 kDa) were used.
Accordingly, we found that interfering with SMG-9 by expressing NT-SMG-9 and CT-SMG-9 truncated products, affected the normal response of cells to genotoxic stress and increased susceptibility to apoptosis (Supplementary Figure S4 and Supplementary Data), in agreement with the role described for these complexes in genome stability and apoptosis (3,13). HEK-293 cells were transfected with vectors coding for full length SMG-9, NT-SMG-9 or CT-SMG-9 fragments (see Supplementary Data for details) and tested for susceptibility to cisplatin, a DNA alkylating agent known to cause cell apoptosis. Cis-platin treatment led to a dose-dependent increase in cell apoptosis that was of higher extent in cells over-expressing the N-terminal or C-terminal domains of SMG-9 than in cells expressing the full length protein or than in mock cells. This effect was associated with reduction in pro-caspase-3 levels and with increased processing of PARP-1.
DISCUSSION
The activities of the SMG1C complex, containing SMG-1, SMG-8 and SMG-9 are essential for NMD in mammals (12). SMG-8 and SMG-9 regulate the kinase activity of SMG-1, and SMG-8 is also required to recruit SMG-1 to the mRNA surveillance complex. It has been proposed that the SMG1C complex could control NMD by inhibiting SMG-1-mediated Upf1 phosphorylation until a PTC-containing mRNA is properly recognized (12). Here, we show that SMG-9 comprises an N-terminal domain with the characteristic features of the so-called intrinsically disordered regions (IDRs) and a well-folded C-terminal domain (12). We demonstrate that both the N-terminal IDR and the C-terminal domain of SMG-9 are required for the integrity of the SMG1C complex. Both domains are implicated in SMG-1 binding, since removal of either domain disrupts, totally or partially, the interaction of SMG-9 with SMG-1. On the other hand, SMG-9 was found to interact with SMG-8 mostly through its C-terminal domain.
We have purified a protein comprising the N-terminal domain and several biophysical approaches (gel filtration chromatography, analytical ultracentrifugation, CD, and UV-spectroscopy) have confirmed unambiguously that this region behaves as a compact 20 kDa domain with the paradigmatic characteristics of unstructured domains as well as a limited presence of secondary structure. Whereas misfolded proteins usually aggregate due to the exposure of the hydrophobic residues that form the core of folded domains, an intrinsically disordered protein is soluble even in the presence of low or no secondary structure due to the unusual composition of their sequences, enriched in polar and charged residues (19,20). The results obtained by NMR spectroscopy represent the formal proof that the N-terminus of SMG-9 is an IDR. The ‘signature’ of the mono and bi-dimensional spectra of NT-SMG-9 is that typical of this group of proteins with defined signals concentrated in a narrow range (19,20). Furthermore, the combination of NMR data and the spectroscopy studies suggests that the conformation of the NT-SMG-9 domain most likely fits into the category of intrinsically unstructured proteins termed ‘pre-molten globules’, where a limited degree of secondary structure could be localized.
There is a growing interest in the functional roles of intrinsically disordered regions and intrinsically disordered proteins, since these seem to play important roles in cellular functions such as transcription regulation, genome surveillance, chromatin remodeling or mRNA processing (19,20). Algorithms designed to detect these domains in the primary structure of proteins suggests that the number and functional relevance of IDRs increase with the complexity of the organism. It has been estimated that 25% of the total number of proteins in complex eukaryotic genomes may be totally disordered and ∼50% could contain at least one disordered region. IDRs seem to be adequately suited for protein–protein interactions in large macromolecular machines involving very specific but transient interactions. These domains seem to provide high specificity sustained in a large area of contact but moderate affinities facilitating interchange of partners. A recent description of the interaction between Upf1 and the C-terminal domain of Upf2 revealed that this domain of Upf2 is intrinsically disordered and the elements of secondary structure are only co-folded upon recognition of Upf1 (23). Given the complexity of NMD and more generally of the mRNA processing machinery, intrinsically disordered regions, as the one presently described for SMG-9, could be present in other components of mRNA processing pathways.
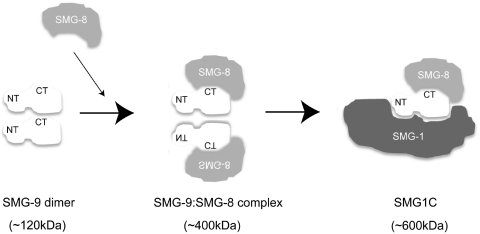
We have found that SMG-9 assembles into several complexes apart from SMG1C, SMG-9:SMG-9 complexes, and most likely also SMG-8:SMG-9 complexes. The finding that SMG-9 dimers could be isolated by gel filtration (Figure 6) and that a partially truncated dimer (CT-SMG-9185–520:SMG-9) is completely free of SMG-1 and SMG-8 (Figure 5C) suggests that SMG-9 dimers are not a component of the SMG1C complex. This finding is in striking contrast to the well-characterized behaviour of SMG-9, which pull downs SMG-1 and SMG-8 indicating that SMG-9 is a component of SMG1C (12). On the other hand, the detection of SMG-8:SMG-9 complexes indicate that an association between these two proteins may also regulate the interaction with SMG-1 and the assembly of SMG1C.
We propose that several SMG-9 containing complexes that do not contain SMG-1 could potentially have biological functions apart from SMG1C. Thus, SMG-1 has been shown to participate in the cellular stress response (14,16–18), and we find that the expression of truncated versions of SMG-9 increased the susceptibility to apoptosis (Supplementary Figure S4). In addition, we speculate that SMG-9:SMG-9 and, probably, SMG-8:SMG-9 complexes could function as intermediaries mediating the assembly of SMG1C. One possibility could be that the self-association between SMG-9 molecules through the C-terminal domains, could repress the interaction with SMG-8 and/or SMG-1, as proposed for other signalling molecules (Figure 7). For instance, ATM, a member of the PIKK family of kinases, is activated by a transition from an inhibited dimer under normal conditions converting into an active monomer after DNA damage, a transition requiring phosphorylation of Ser1981 (24). A possible regulatory event driving the transitions between the homo-oligomeric and SMG1C-assembled states of SMG-9 could be phosphorylation. Several conserved residues of SMG-9 located within the N-terminal disordered and the C-terminal domains are specifically phosphorylated by SMG-1 (12) (Akio Yamashita and Shigeo Ohno unpublished data). Under certain circumstances, SMG-9 would be activated, interacting with SMG-8 by means of its C-terminal domain. This SMG-8:SMG-9 complex could be the building block to assemble SMG1C. At this stage, we cannot completely rule out that SMG-9 could also be a dimer within SMG1C, although current data favours a model with an equimolar SMG-1:SMG-8:SMG-9 complex (12). An assembly pathway of SMG1C regulated at the level of SMG-9 and SMG-8 would allow the tuning of the biological functions of SMG1C with the rest of the NMD machinery to either restrain or promote the activation of SMG-1. The N-terminal disordered domain of SMG-9 participates in the recognition of SMG-1, and it would be the characteristic malleability of IDRs an adequate structural property to allow the transit of SMG-9 through these distinct macromolecular complexes. SMG-9 could therefore appear as an important controller of SMG-1 by regulating the formation of SMG1C and consequently the activation of its kinase activity towards Upf1(12).
Figure 7.
Model for the assembly of SMG-9-containing complexes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Science and Innovation (SAF2008-00451 to O.L., SAF2008-00479 to J.T.); ‘Red Temática de Investigación Cooperativa en Cáncer (RTICC)’ from the ‘Instituto de Salud Carlos III’ (RD06/0020/1001 to O.L. and RD06/0020/0011 to J.T.); Autonomous Region of Madrid (CAM S-BIO-0214-2006 to O.L.); Human Frontiers Science Program (RGP39/2008 to O.L.); ‘Consejería de Educación de la Comunidad de Madrid y Fondo Social Europeo’ (to E.A.P.); Japan Society for the Promotion of Science (to A.Y. and S.O.); Japan Science and Technology Corporation (to A.Y. and S.O.); Ministry of Education, Culture, Sports, Science and Technology of Japan (to S.O.); Yokohama Foundation for Advancement of Medical Science (to A.Y.). Funding for open access charge: Spanish Ministry of Science and Innovation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Prof. Jesus Jimenez-Barbero (CIB, Madrid) for his assistance and help during the NMR experiments and their analysis. The authors also thank Dr Natsuko Izumi for the plasmid construction of pcDNA5/NTAP-SMG-9 (2-520).
REFERENCES
- 1.Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci. 2009;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brogna S, Wen J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol. 2009;16:107–113. doi: 10.1038/nsmb.1550. [DOI] [PubMed] [Google Scholar]
- 5.Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr. Opin. Cell Biol. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abraham RT. The ATM-related kinase, hSMG-1, bridges genome and RNA surveillance pathways. DNA Repair. 2004;3:919–925. doi: 10.1016/j.dnarep.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Yamashita A, Kashima I, Ohno S. The role of SMG-1 in nonsense-mediated mRNA decay. Biochim. Biophys. Acta. 2005;1754:305–315. doi: 10.1016/j.bbapap.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23:1091–1105. doi: 10.1101/gad.1767209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izumi N, Yamashita A, Iwamatsu A, Kurata R, Nakamura H, Saari B, Hirano H, Anderson P, Ohno S. AAA+ proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in nonsense-mediated mRNA decay. Sci. Signal. 2010;3:ra27. doi: 10.1126/scisignal.2000468. [DOI] [PubMed] [Google Scholar]
- 14.Brumbaugh KM, Otterness DM, Geisen C, Oliveira V, Brognard J, Li X, Lejeune F, Tibbetts RS, Maquat LE, Abraham RT. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol. Cell. 2004;14:585–598. doi: 10.1016/j.molcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 16.Gehen SC, Staversky RJ, Bambara RA, Keng PC, O'Reilly MA. hSMG-1 and ATM sequentially and independently regulate the G1 checkpoint during oxidative stress. Oncogene. 2008;27:4065–4074. doi: 10.1038/onc.2008.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masse I, Molin L, Mouchiroud L, Vanhems P, Palladino F, Billaud M, Solari F. A novel role for the SMG-1 kinase in lifespan and oxidative stress resistance in Caenorhabditis elegans. PLoS One. 2008;3:e3354. doi: 10.1371/journal.pone.0003354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliveira V, Romanow WJ, Geisen C, Otterness DM, Mercurio F, Wang HG, Dalton WS, Abraham RT. A protective role for the human SMG-1 kinase against tumor necrosis factor-alpha-induced apoptosis. J. Biol. Chem. 2008;283:13174–13184. doi: 10.1074/jbc.M708008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuxreiter M, Tompa P, Simon I, Uversky VN, Hansen JC, Asturias FJ. Malleable machines take shape in eukaryotic transcriptional regulation. Nat. Chem. Biol. 2008;4:728–737. doi: 10.1038/nchembio.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tompa P, Fuxreiter M, Oldfield CJ, Simon I, Dunker AK, Uversky VN. Close encounters of the third kind: disordered domains and the interactions of proteins. Bioessays. 2009;31:328–335. doi: 10.1002/bies.200800151. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting protein disorder for N-, C-, and internal regions. Genome Inform. Ser. Workshop Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- 22.Wullschleger S, Loewith R, Oppliger W, Hall MN. Molecular organization of target of rapamycin complex 2. J. Biol. Chem. 2005;280:30697–30704. doi: 10.1074/jbc.M505553200. [DOI] [PubMed] [Google Scholar]
- 23.Clerici M, Mourao A, Gutsche I, Gehring NH, Hentze MW, Kulozik A, Kadlec J, Sattler M, Cusack S. Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J. 2009;28:2293–2306. doi: 10.1038/emboj.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.