Abstract
Transcription proceeding downstream of the λ phage replication origin was previously shown to support initial steps of the λ primosome assembly in vitro and to regulate frequency and directionality of λ DNA replication in vivo. In this report, the data are presented indicating that the RNA polymerase β subunit makes a direct contact with the λO protein, a replication initiator of λ phage. These results suggest that the role of RNA polymerase during the initiation of λ phage DNA replication may be more complex than solely influencing DNA topology. Results demonstrated in this study also show that gyrase supercoiling activity stimulates the formation of a complex between λO and RNA polymerase, suggesting that the introduction of negative supercoils by DNA gyrase, besides lowering the energy required for DNA strand separation, may play an additional role in modeling protein–protein interactions at early steps of DNA replication initiation.
INTRODUCTION
Bacteriophage λ DNA replication starts from binding of λO replication initiator protein to four partially symmetrical iterons present at oriλ (1). Interaction of λO with the origin sequence and formation of higher-order nucleoprotein structure induces DNA helix destabilization within the AT-rich region positioned immediately to the right from λO-binding sites (2,3). Subsequent steps, involving delivery of the host DnaB helicase to preprimosomal complex by the phage λP protein (4,5) and liberation of the helicase from inhibitory interaction with λP (6,7) by the chaperone proteins DnaK, DnaJ and GrpE, are well characterized biochemically (5,8,9). After the rearrangement of the λO–λP–DnaB complex by the heat shock proteins, replication starts in the presence of primase, DNA polymerase III, gyrase and SSB proteins (6,10).
In vivo and in a crude extract, initiation of replication of λ phage and plasmid DNA replication is dependent on transcription carried by bacterial RNA polymerase. In a system consisting of purified enzymes this dependence of λ DNA replication on ‘transcriptional activation’ is alleviated (11); however, it can be restored by the addition of bacterial histone-like protein, HU (12). HU blocks initiation of replication starting from oriλ in vitro in the absence of RNA polymerase activity (12). In the case of wild-type λ sequence, activation of oriλ is provided by transcription initiated at the pR promoter, located ∼1000 bp upstream of the origin of replication. However, in vivo results demonstrated that replication of oriλ-containing plasmids was still efficient when the promoter was situated much closer to the replication initiation point or even downstream of it (13,14). Activation of oriλ by the transcription proceeding in its vicinity plays a key role in the control of frequency of replication initiation and the switch from θ to rolling circle mode during λ phage DNA replication (15–18).
Similar dependence of the replication initiation on the transcriptional activation was found in the case of the host chromosomal DNA replication (19). DnaA, a bacterial replication initiator protein, binds to specific DnaA boxes within oriC and unwinds the DNA duplex in an AT-rich region. This reaction is stimulated by HU protein and transcription process; however, as in the case of λ DNA replication, high concentrations of HU are inhibitory to DNA unwinding step, but RNA polymerase activity overcomes this negative influence (20). Allele-specific suppression of certain dnaA (ts) mutations by the changes in the rpoB gene, coding for the β subunit of RNA polymerase, suggested direct interaction between DnaA and RNA polymerase (21), which has been recently confirmed also in vitro (22). Observation that mutations in the rpoB gene suppress, in an allele-specific manner, the inability of some λ phage P mutants to plate on certain dnaB strains, implied also the possibility of direct protein–protein interactions between RNA polymerase and the λ replication complex (23).
λO replication initiator binds as a dimer to a partially symmetric sequence and induces a strong DNA bend (24,25). Interaction of λO with four iterons within oriλ results in the formation of a large nucleoprotein complex (O-some) and condensation of a DNA fragment encompassing all iterons and their intervening spacers (2). It was also proved that λO, by binding to its recognition sequences, greatly stimulates transcription-coupled supercoiling of plasmid DNA by the gyrase. This property of λO was attributed to its capability of inducing strong DNA bends (26). The process of transcription-dependent stimulation of supercoiling was proved to rely on the twin-domain model (27), according to which counter-rotation of the transcription machinery and template, as DNA is threaded through RNA polymerase active site, generates waves of supercoiling, positive in front of elongating enzyme, and negative behind it. This concept, originally postulated by Liu and Wang (28), became an attractive mechanism for regulation of local processes involving DNA transactions, by influencing DNA topology in a sequence- and context-dependent manner (29,30). Transcription was also shown to regulate chromatin structure and, thus, selection of replication origin sites and replication initiation timing during eukaryotic chromosomes doubling (31 and references therein). Therefore, studies on coordination of replication and transcription machineries seem to be of general importance.
In this report, we demonstrate evidence that the λO protein, like the host replication initiator, DnaA, interacts directly with bacterial RNA polymerase. Moreover, the results of our studies indicate that the β subunit of RNA polymerase is a contact site for λO. In addition, we present data suggesting that gyrase activity enhances this interaction.
MATERIALS AND METHODS
Strains
Escherichia coli, strain C600 (supE44 hsdR thi-1 thr-1 leuB6 lacY1 tonA21) (32), was used for plasmid purification and strain MM294 (supE44 hsdR endA1 pro thi) (33) was used for the λO and λP protein overexpression.
Plasmids
The following plasmids were employed: pRLM4 (oriλ, cro(ts), kanR) was described previously (34); pLamberA (oriλ, colE1, ampR) was constructed by removing the 916-bp HindIII–NcoI fragment from a plasmid pLamber (35); plasmids: pGP1-2 (T7 RNA polymerase gene under the control of λ repressor CI 857(ts); kan R) (36) and pEW1 (O and P genes under the control of a T7 phage promoter) (37) were used for overproduction of the λO and λP proteins.
Proteins, antibodies and antibiotics
Both λO and λP proteins were prepared from E. coli strain MM294 bearing plasmids pGP1-2 and pEW1. Proteins were purified according to a method described previously (38). Other proteins and antibiotics were from the following sources: E. coli DNA gyrase was obtained from New England BioLabs. Bovine serum albumin (BSA) and novobiocin were from Sigma and E. coli RNA polymerase holoenzyme was provided by Eppicentre.
The following antibodies were used in this study: a polyclonal rabbit antibody against the λO protein (39) and monoclonal mouse antibodies, which were specific for α, σ, β and β′ subunits of E. coli RNA polymerase, were obtained from Neoclone; secondary HRP-conjugated anti-rabbit and anti-mouse antibodies were obtained from Sigma.
Surface plasmon resonance
To analyze a direct interaction between λO protein and E. coli RNA polymerase, surface plasmon resonance (SPR) analyses were performed using a double-stranded 141-bp linear DNA fragment containing the four iterons from oriλ. The DNA fragment was obtained by polymerase chain reaction (PCR) amplification with following program: initial denaturation at 94°C for 3 min; 36 cycles of denaturation at 94°C for 20 s; annealing at 55°C for 30 s and extension at 72°C for 30 s. The following primers were used: forward: 5′ - biotin - TCA AGC AGC AAG GCG GCA TGT TTG G-3′ and reversed: 5′ TGT CCC CCT GTT TTG AGG GAT AG-3′. Then, the DNA fragment in TNE buffer (10 mM Tris pH 7.6; 300 mM sodium chloride and 1 mM EDTA) was immobilized on a streptavidin matrix-coated Sensor Chip S.A. (Biacore) by biotin covalent linkage, following the manufacturer’s instructions. Experiments were carried out on a Biacore 2000 instrument by a 5-min injection of λO protein (50 nM) in HBS buffer (10 mM HEPES- KOH pH 7.4; 150 mM sodium chloride; 3 mM EDTA and 0.005% surfactant P20), followed with a 5-min injection of E. coli RNA polymerase (25 nM) and BSA (BSA; 25 nM) in binding buffer (10 mM HEPES pH 7.6; 10 mM MgCl2; 20 mM KCl; 0.5 mM DTT and 0.1 mM EDTA). Next, the HBS buffer was injected over the chip surface and the dissociation phase was recorded. The analyses were performed at 15 µl/min flow speed. Following the completion of each protein co-injection, the surface of the chip was regenerated by applying 0.2% sodium dodecyl sulfate (SDS), which releases all bound protein without affecting the binding capacity of the immobilized DNA. Data were evaluated using Biacore AB’s BIA evaluation software. The results are presented as the sensogram, obtained after subtraction of the background response signal and correction of the buffer effect. As a control of nonspecific interactions, an empty reference cell was used.
Detection of protein complexes by disuccinimidyl glutarate cross-linking
Reactions were performed in the buffer containing: 10 mM HEPES-KOH pH 7.6; 10 mM MgCl2; 20 mM KCl; 0.5 mM DTT, 0.1 mM EDTA and 50 µg/l poly (dI:dC). Supercoiled λ plasmid DNA (2.8 nM) (purified by ultracentrifugation in a cesium chloride/ethidium bromide gradient) was mixed with λO protein (0.5 µM) and after a 10-min incubation at 30°C reaction was applied onto a 1-ml Sepharose 4B-CL column, equilibrated with a reaction buffer. DNA–λO complexes were separated from unbound protein fraction. Next, to the reaction mixture RNA polymerase (22 nM) and NTPs (0.5 mM each) were added. After a 10-min incubation at 30°C, protein complexes were cross-linked with disuccinimidyl glutarate (DSG) reagent (a final concentration of 0.02 µM) and incubation was prolonged for an hour at 30°C. Reactions were quenched by addition of Tris–HCl pH 7.5 to 50 mM and incubation for next 10 min at 30°C. Subsequently, the cross-linked complexes were separated electrophoretically in 7.5% SDS-polyacrylamide gels, transferred onto polyvinylidene fluoride (PVDF) membrane (Millipore) and detected by immunoblot analysis using anti-λO polyclonal antibodies or monoclonal antibodies specific for various subunits of E. coli RNA polymerase. An ECL-Western Blotting Substrate Kit (Pierce) was used to detect antibody–antigen complexes.
Gel filtration
Size-exclusion chromatography was performed to investigate the influence of transcription on pre-formed O-some complexes. The Sepharose 4B-CL (Sigma) column (0.5 × 8 cm) was equilibrated with a buffer containing: 10 mM Tris–HCl pH 7.6; 10 mM MgCl2; 20 mM KCl; 0.5 mM DTT; 0.1 mM EDTA. The 120-µl reaction mixture (in the same buffer) supplemented with 10 µg of the supercoiled pRLM4 plasmid and 6 µg of λO protein was incubated for 10 min at 30°C and loaded on a Sepharose 4B-CL column at room temperature. Fractions containing O-some were collected. The presence of plasmid DNA in the O-some fractions was initially confirmed by ethidium bromide staining and visualization under ultraviolet (UV) light. During subsequent repetitions of the experiment, the presence of λO in the void volume fraction was confirmed by amide black staining; fractions containing λO were pooled and divided into two parts. Subsequently, to one of the O-some-containing samples (∼100–120 µl) 2.5 µg of RNAP polymerase was added. After 10 min at 30°C, rNTPs (2 mM each) were added to the mixture and incubation was continued for the next 10 min at 30°C. The reaction mixture containing only O-some was also incubated at 30°C. Finally, the two mixtures (separately) were loaded on a Sepharose 4B column. Two-drop fractions were collected and analyzed by SDS- polyacrylamide gel electrophoresis (PAGE), followed by silver staining. Each fraction content was quantitated densitometrically and compared to a sample containing known amount of the λO protein.
RESULTS
RNA polymerase activity facilitates λO protein–DNA interaction
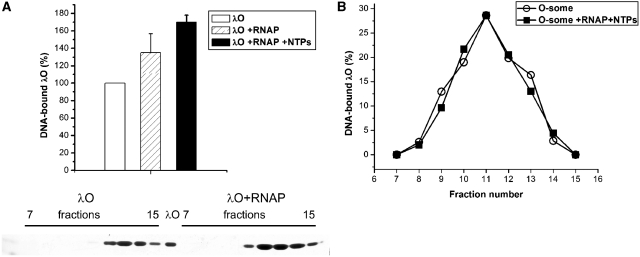
λO protein binding to its DNA recognition sequences is a primary step in the initiation of DNA replication. Nucleoprotein structure called O-some, formed due to λO oligomerization and its DNA-bending properties, serves as a landing pad for other proteins involved in the initiation of replication. As RNA polymerase activity may protect λ replication complex from disassembly during unfavorable conditions (40), we wanted to test whether RNA polymerase binding to promoter regions present in λ plasmid DNA (mainly to the strongest pR promoter) and transcription activity may influence O-some stability or the affinity of λO protein binding to the oriλ-containing plasmid DNA. To study this possibility closer, we performed reactions, using supercoiled λ plasmid DNA, and purified proteins: λO and RNA polymerase. λO–DNA complexes, formed in the presence and absence of active RNA polymerase, were resolved by gel filtration (Sepharose 4B-CL). To test whether efficient transcription occurred under the conditions we used, total RNA synthesis was tested by measuring amount of [α-32P]UTP incorporated into acid-insoluble material (data not shown). Subsequent analysis of fractions, containing λO bound to DNA sequences, demonstrated that the formation of such a nucleoprotein structure is enhanced by the transcription process. Assembly of O-some was stimulated by 70% in fractions containing RNA polymerase and nucleotides, in comparison to those where only λO and DNA were present (Figure 1A). In addition, the formation of nucleoprotein complexes by the O-protein was facilitated by RNA polymerase even in the absence of transcription process, although this effect was less pronounced than the one observed in the reactions where RNA polymerase was allowed to transcribe (Figure 1A). This result suggested that RNA polymerase could facilitate λO protein binding to the oriλ-containing plasmid. However, this result could also mean that, under the conditions we used, RNA polymerase activity enhanced stability of the formed O-some structure. To verify the latter hypothesis, we performed analogous gel filtration experiment in which nucleoprotein complexes of the λO protein were formed without RNA polymerase and separated by size-exclusion chromatography from unbound protein. Subsequently O-some containing fractions were collected and divided into equal portions. One of the samples was supplemented with RNAP and NTPs and both were subjected to a second round of gel filtration on separate columns. The amount of λO-containing nucleoprotein complexes was similar in both cases (Figure 1B), indicating that RNA polymerase may play a role in λO binding to DNA, rather than in stabilization of O-some structure during chromatography. Additionally, an experiment was performed in which pre-formed O-some complex were challenged with an excess of biotinylated DNA competitor (containing iteron sequences) in the presence and absence of RNA polymerase and transcription process. The amount of λO protein bound to competitor DNA was monitored by the separation of biotinylated DNA using magnetic beads, SDS-PAGE and blotting. The results of this experiment confirmed that RNA polymerase and transcription process do not substantially influence O-some stability (for details see Figure S1 in Supplementary Data). Stimulation of the λO protein binding to iterons by the presence of RNA polymerase and also by transcription process was further confirmed by electrophoretic mobility shift assay (EMSA) performed with supercoiled λ plasmid (see Figure S2 Supplementary Data).
Figure 1.
RNA polymerase and transcription process enhance λO binding to oriλ. (A) Formation of O-some in the presence or absence of RNA polymerase activity was analyzed by gel filtration. Nucleoprotein complexes were formed in the presence of supercoiled λ plasmid DNA (10 µg) and λO protein (1.5 µM) and RNA polymerase (2.5 µg) and nucleotides (2 mM each) (as indicated). When the influence of either RNA polymerase or transcription process on O-some formation was assessed, RNA polymerase was first incubated with plasmid DNA and nucleotides (as indicated) for 10 min at 30°C, and then the λO protein was added and incubation was prolonged for another 10 min. Samples were loaded on a Sepharose 4B column; fractions were collected and subjected to SDS–PAGE, followed by silver staining and densitometry. The panel below the graph shows an example of SDS–PAGE separation of fractions obtained after gel filtration of the λO–DNA complexes formed in the presence or absence of RNA polymerase (as indicated). (B) Stability of λO–DNA complex, formed and isolated as described above, was assessed in the presence and absence of transcription. Fractions containing O-some obtained after first round of gel filtration were pooled and divided into half. One of the samples was supplemented with RNA polymerase (2.5 µg) and NTPs (2 mM). Both samples were subsequently (separately) subjected to a second round of gel filtration. Fractions containing O-some were collected and analyzed by SDS-PAGE as described earlier. The quantity of λO protein present in the sample where RNA polymerase and nucleotides were absent was assumed as 100%, and elution profile of both samples was presented according to this value.
λO initiator protein interacts with the host RNA polymerase
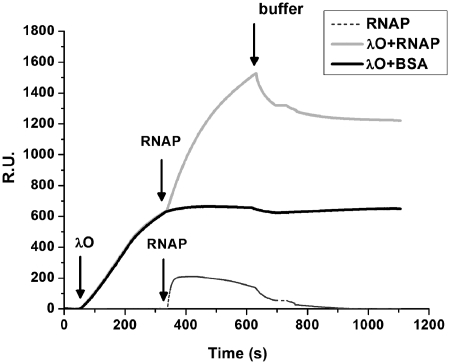
During experiments testing the influence of RNA polymerase activity on λO protein binding to oriλ, we obtained evidence suggesting that the λO initiator may interact directly with the host transcription machinery. In addition, similarity of the initial steps of replication at the E. coli and phage λ origin with respect to their dependence on transcriptional activation and recent confirmation of the direct contact between the host initiator protein DnaA and RNA polymerase (RNAP) (22), as well as genetic data suggesting that transcriptional machinery may directly associate with the components of the phage replication complex (23), encouraged us to follow that trail. Therefore, we analyzed the formation of the putative complex between RNAP and the λO protein bound at the iterons, using SPR technique. A DNA fragment encompassing all four λO binding sites of oriλ was linked to the sensor chip. λO and RNAP were injected sequentially over the sensor matrix and the signal, reflecting a change of mass on the immobilized DNA fragment, was measured. Analysis of the sensograms showed that λO bound efficiently to the sensor-attached DNA. Subsequent injection of RNAP resulted in detection of RNA polymerase binding to the pre-formed λO–DNA complex (Figure 2). The response signal, seen after injection of RNAP, was relatively high, indicating that the complex between the host and phage protein is formed with high binding affinity. As RNA polymerase displays some unspecific binding affinity to double-stranded DNA, additional control was made to assess the enzyme binding to sensor-attached DNA, containing oriλ sequence. In the absence of the λO protein, RNA polymerase bound to the DNA fragment; however, this interaction was considerably weaker than that observed with O-containing nucleoprotein complex (Figure 2).
Figure 2.
RNA polymerase forms a complex with the DNA-bound λO protein. Association of the host RNAP holoenzyme with λO nucleoprotein complex was estimated by SPR technique. Biotinylated DNA containing four iterons present at oriλ was immobilized on a sensor chip. Subsequently, a 5-min injection of λO protein (50 nM) was performed followed by a 5-min injection of RNAP (25 nM) or BSA and a change of protein mass on the sensor was monitored. In the next step, dissociation was monitored by running a buffer over the sensor chip surface (15 µl/min flow speed). Data were evaluated using Biacore AB’s BIA evaluation software. The presented sensogram reflects results obtained after subtraction of the background response signal and correction of the buffer effect.
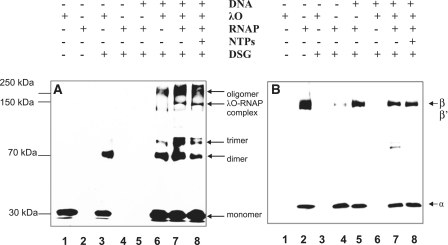
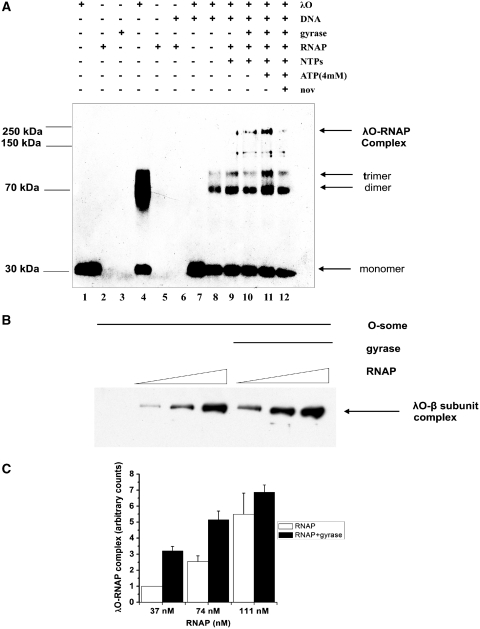
To further confirm the interaction between λO and RNA polymerase in vitro, cross-linking experiments with DSG were performed. DSG is a homobifunctional protein cross-linking reagent with amine-reactive NHS esters at both ends of the 7.7-Å long arm. The λO protein was incubated together with the supercoiled λ plasmid DNA and, subsequently, the fractions containing ‘O-some’ were separated by gel filtration from the unbound protein. The plasmid contained all λO-binding sites and λ phage promoters in their native orientation and relative position. ‘O-some’ fractions were collected and divided into individual reactions, supplemented with RNAP and nucleotides before adding DSG. The cross-linked complexes were separated in SDS-PAGE gels, blotted and probed with antibodies against λO or RNA polymerase (specific against subunits α, σ, β and β′) (Figure 3A and B). Monomeric form of the λO protein (32 kDa) was observed when no DSG was present in the reaction (Figure 3A, lane 1). In a control reaction without DNA, large fraction of λO migrated, as expected, at the dimer position in the presence of the cross-linker (Figure 3A, lane 3). DNA-bound λO protein cross-linked, forming several species of increasing molecular mass, representing λO oligomers (lane 6). When a mixture of ‘O-some’ and RNAP was subjected to cross-linking, the formation of a new complex was observed, absent from the sample containing only λO and DNA. This complex migrated at the position similar to RNAP β subunit and was also detected in a sample containing nucleotides (Figure 3A and B; lanes 7 and 8). Under these conditions, no discrete cross-linked complex of RNAP subunits was detected on a blot subjected to anti-RNAP antibodies, in the reactions where only RNAP was present, regardless of the presence or absence of plasmid DNA (Figure 3B, lanes 4 and 5). Instead, a portion of RNAP formed large cross-linked complexes that did not enter the gel. In addition, an increase in the efficiency of formation of the λO dimer and trimer was observed in the presence of DSG and RNA polymerase, in comparison to the sample where only ‘O-some’ was subjected to cross-linking (Figure 3A compare lanes 6 and 7). Moreover, much more of a large oligomeric form of the λO protein was also detected in the presence of RNA polymerase and transcription process (Figure 3A, compare lanes 6, 7 and 8). This cross-linked oligomer had an apparent molecular weight of about 250 kDa, matching the molecular weight of four λO protein dimers, present in the O-some structure. These results may suggest that some changes in the conformation of O-some structure occur in the presence of RNA polymerase activity.
Figure 3.
RNA polymerase interacts with the O-some structure. The λO protein (0.5 µM) was bound to oriλ-containing plasmid (pLamberA; 2.8 nM) and separated from unbound protein fraction by size-exclusion chromatography. Subsequently, reaction was supplemented with RNA polymerase (22 nM) and subjected to DSG cross-linking in the presence (at a final concentration of 0.05 mM) or absence of nucleotides (lanes 6–8). λO and RNA polymerase were subjected to DSG cross-linking without the presence of DNA (lanes 3 and 4, respectively), or RNA polymerase alone was cross-linked in the presence of plasmid DNA (lane 5). Protein complexes were resolved by SDS–PAGE and detected by immunoblotting, performed with polyclonal antibodies specific against λO (A) or a mixture of monoclonal antibodies against α, β and β′ subunits of RNAP (B). The presence or absence of each reaction component is indicated. λO-RNA polymerase complex was depicted by an arrow. Position of monomeric and oligomeric forms of the λO protein was marked.
Identification of the RNA polymerase β subunit as a contact site for the interaction with the λO initiator protein
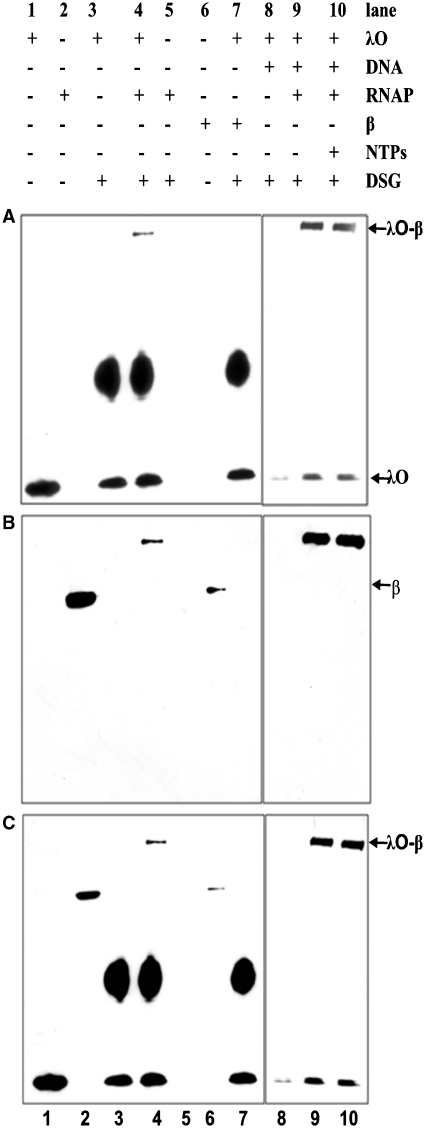
Due to complexity of the cross-linking pattern obtained with the multisubunit RNA polymerase and λO and the difficulty to distinguish individual complexes after reaction with DSG, we performed that experiment using varying concentrations of the cross-linking reagent. When the efficiency of cross-linking reaction was sufficiently high, a large portion of reacting proteins was lost from the final picture as a result of the formation of high molecular mass complexes, unable to enter the gel. However, under such conditions, we were also able to identify two products of the cross-linking reaction: λO dimer and the second one, resulting from the interaction between λO and RNA polymerase (>250 kDa), (Figure 4A–C). Subsequent analysis of cross-linked species by immunoblotting, performed with purified monoclonal antibodies specific for each of the subunits of RNA polymerase (Figure S3 Supplementary Data), revealed that the high molecular mass cross-linked product cross-reacted with antibodies directed against both λO and RNAP β subunit and most probably represents λO tetramer cross-linked to the β subunit. Moreover, we observed the formation of such a complex even in the absence of DNA, which suggest that both proteins are able to interact in solution, even in the absence of specific DNA sequences recognized by these proteins, although with a lower efficiency than in the case of DNA-bound molecules (Figure 4A–C, lane 4). RNAP β subunit alone could form no complex with the λO protein, detected after cross-linking (Figure 4A–C, lane 7). This is most probably due to altered conformation of the free β subunit in comparison to that observed when β is embedded in the RNAP core structure. These results corroborate conclusions from the SPR experiment that the λO initiator protein is able to interact directly with the host RNA polymerase in vitro and indicate that the β subunit of RNA polymerase is a target for interaction with λO.
Figure 4.
β Subunit of RNA polymerase makes a direct contact with both iteron-bound λO and a free form of the λO protein. λO (0.5 µM) was bound to oriλ-containing plasmid (2.8 nM) and separated from unbound protein fraction by gel filtration. Subsequently, reaction was supplemented with RNA polymerase (22 nM) and subjected to DSG cross-linking in the presence or absence of nucleotides (500 µM). Free λO protein was also subjected to DSG cross-linking with RNA polymerase. Subsequently, protein complexes were resolved by SDS–PAGE and detected by immunoblotting, performed with polyclonal antibodies specific against λO (A) or monoclonal antibodies against β subunit of RNAP (B) or simultaneously against both proteins (C). The presence or absence of each reaction component is indicated. λO–RNAP polymerase complex was depicted by an arrow.
DNA gyrase activity stimulates interaction between RNA polymerase and the λO protein
Results of the experiments presented to this point suggest that λ phage replication initiator protein, λO, interacts with the host RNA polymerase. This conclusion may have important implications for the role of transcription in activating replication initiated by the λ phage proteins. During transcription elongation, RNA polymerase introduces topological changes into DNA template, positive supercoils are generated ahead of the transcription machinery while negative are left in its wake (28). Transient waves of supercoiling produced by RNAP can be stabilized by the action of DNA topoisomerases. It was demonstrated that DNA gyrase rapidly converts transcription-induced positive supercoils into the negative ones (41,42). Moreover, λO protein was shown to greatly enhance transcription-coupled negative supercoiling of plasmid DNA by the gyrase in in vitro reaction (26). Therefore, we aimed to test whether gyrase activity may influence the formation of the λO–RNA polymerase complex in a reaction consisting of purified proteins and a supercoiled λ plasmid DNA. To assess this possibility, λO–plasmid DNA nucleoprotein complexes were isolated and supplemented with RNAP and/or E. coli gyrase and nucleotides. The formation of the complex between RNAP β subunit and the λO protein was analyzed by cross-linking with DSG and subsequent immunoblotting (Figure 5A). In the presence of the active RNA polymerase, a band corresponding to λO–RNAP cross-linked product was observed (Figure 5A, lane 6). Addition of DNA gyrase to the reaction, stimulated interaction between RNA polymerase and λO in a manner dependent on adenosine triphosphates (ATP) concentration (Figure 5A, lanes 7 and 8). Novobiocin, in turn, reduced this stimulatory effect (Figure 5A lane 9). This aminocoumarin antibiotic, which is a competitive inhibitor of the gyrase ATP-ase activity, also inhibits supercoiling activity of gyrase, which is exerted at the expense of ATP. We also tested λO–β subunit complex formation in the presence of transcription and gyrase activity at various ratios of RNAP to the λO protein and at higher concentration of cross-linking reagent, yielding higher molecular mass cross-linked product (Figure 5B). In this case, we also observed the enhancement of the λO–β subunit conjugation by DSG, in the reactions where gyrase was present, in comparison to the ones where transcription was allowed to proceed in the presence of the λO protein only. Stimulation of the λO–β complex formation was assessed densitometrically and summarized in the Figure 5C. These results indicate that ATP-dependent gyrase activity may facilitate the contact between the λO replication initiator and the host RNA polymerase.
Figure 5.
Gyrase activity stimulates the formation of a complex between RNA polymerase and the λO protein in an ATP-dependent manner. (A) In the presence of λO (0.5 µM) and the supercoiled λ plasmid DNA (2.8 nM) O-some structure was formed and separated from unbound protein fraction by gel filtration. Next, one of the samples was supplemented with additional ATP (final concentration of 4 mM) and novobiocin was added to the reaction mixture to a final concentration of 100 nM. Subsequently, E. coli RNA polymerase (22 nM), gyrase (9 nM) and NTPs (0.5 mM) (as indicated) were added and after a 10-min incubation at 30°C protein complexes were cross-linked with DSG. Complexes were detected by SDS–PAGE followed by immunoblotting with antibodies against λO. λO–RNAP polymerase complex was depicted by an arrow as well as oligomeric forms of the λO protein. The presence or absence of each reaction component was indicated. Lanes 1–3 and 6 contain uncross-linked λO, RNAP and gyrase and O-some, respectively. (B) Experiment was performed analogously as in point A, but higher concentration of the cross-linking reagent was added (1 µM) and various ratios of RNAP to λO were used, as indicated, in the presence of constant concentration of gyrase (8.3 nM), NTPs (500 µM) and ATP (4 mM). (C) λO–RNAP polymerase complex formation in the presence of gyrase activity was measured as described in point B in three independent experiments. Relative amount of λO–β subunit complex formed in the presence and absence of gyrase activity was measured densitometricaly and presented as a function of RNA polymerase concentration with error bars representing SD. Amount of the complex observed in the sample where the lowest concentration of RNA polymerase was present and no gyrase was added was assumed as 1 and other values reflect this value.
DISCUSSION
Transcription was shown to play a pivotal role in the regulation of bacteriophage λ replication. In vitro, transcription was necessary in the step preceding preprimosome rearrangement by the heat shock proteins and this process was also shown to be able to overcome inhibitory action of the histone-like HU protein (12,43). Both in vivo and in vitro, activity of RNA polymerase stimulated bidirectional replication starting at the λ origin (16,44). In addition, it was demonstrated that efficient transcription process was required for the λ replication complex to survive heat shock-provoked disassembly by GroELS proteins after transient DNA relaxation by gyrase (40). These results suggested that transcription stabilizes replication complex under unfavorable conditions.
Results presented in this work strongly indicate that the λO protein interacts directly with bacterial RNA polymerase (Figures 2 and 3). On RNAP part, β subunit seems to be responsible for the contact with λ phage replication initiator (Figure 4), similarly as it was suggested for the host initiator DnaA protein (21,22). This interaction seems to facilitate λO protein binding to iteron-containing DNA as we observed enhanced formation of λO nucleoprotein complex even in the absence of nucleotides (data not shown) and transcription process further supported association of λO with oriλ (Figure 1). It is unlikely that this stimulation plays a primary role in the transcriptional activation process; however, it may be helpful under conditions of limiting λO protein. Although we used under mild conditions, O-some structure is very stable and therefore we did not observe any enhancement of this nucleoprotein complex stability by transcription (Figure 1B); it cannot be excluded that direct interaction of RNA polymerase and λO may support replication complex survival under more harsh circumstances. λO–RNAP association may also help in organizing replication hyperstructure consisting of phage and host proteins necessary for phage DNA replication and progeny production, in keeping with the model of a ‘factory’ where many proteins are engaged in various DNA transactions (45).
It was demonstrated that λO protein, most probably due to its DNA-bending properties, stimulates transcription-coupled DNA supercoiling by gyrase in vitro (26,27). Here, we show that gyrase activity—in turn—stimulates formation of a complex between RNA polymerase β subunit and the λO protein. We have not determined whether this stimulatory influence of gyrase relies on, connected with supercoiling, increase in probability of juxtaposition of RNA polymerase and λO position on DNA. Supercoiling could also actively affect conformation of the λO complex in a way favoring the interaction. Topological state of the loop formed by Lac repressor bound to adjacent operator sites was also shown to influence repressor conformation (46). Similarly, DNA supercoiling was implicated in assisting in octamerization of the λ CI protein and, hence, facilitating autoregulatory repression of pRM promoter by aiding CI in binding to distant OL3 and OR3 operators and DNA looping (47,48). In addition, it was demonstrated that several promoters are sensitive to the supercoiling level (49), so the stimulatory effect of gyrase activity on the formation of RNA polymerase–λO complex may result from altered transcriptional activity of promoters located on plasmid DNA. Whatever mechanism operates in gyrase-mediated stimulation of λO and RNA polymerase interaction, this observation emphasizes the importance of DNA supercoiling, not only in lowering the energy necessary for DNA strand separation but also in modeling of protein–DNA and protein–protein interactions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
The Ministry of Science and Higher Education of Poland (grant No. N N301 4140 33). Funding for open access charge: The University of Gdańsk.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are particularly grateful to Prof. Igor Konieczny (Faculty of Biotechnology, University of Gdańsk) for his advice regarding SPR experiments.
REFERENCES
- 1.Tsurimoto T, Matsubara K. Purified bacteriophage lambda O protein binds to four repeating sequences at the lambda replication origin. Nucleic Acids Res. 1981;24:1789–1799. doi: 10.1093/nar/9.8.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dodson M, Roberts J, McMacken R, Echols H. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: complexes with lambda O protein and with lambda O, lambda P, and Escherichia coli DnaB proteins. Proc. Natl Acad. Sci. USA. 1985;82:4678–4682. doi: 10.1073/pnas.82.14.4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schnos M, Zahn K, Inman RB, Blattner FR. Initiation protein induced helix destabilization at the lambda origin: a prepriming step in DNA replication. Cell. 1988;12:385–395. doi: 10.1016/s0092-8674(88)80031-x. [DOI] [PubMed] [Google Scholar]
- 4.Alfano C, McMacken R. Ordered assembly of nucleoprotein structures at the bacteriophage lambda replication origin during the initiation of DNA replication. J. Biol. Chem. 1989;25:10699–10708. [PubMed] [Google Scholar]
- 5.Zylicz M, Ang D, Liberek K, Georgopoulos C. Initiation of lambda DNA replication with purified host- and bacteriophage-encoded proteins: the role of the dnaK, dnaJ and grpE heat shock proteins. EMBO J. 1989;8:1601–1608. doi: 10.1002/j.1460-2075.1989.tb03544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LeBowitz JH, McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J. Biol. Chem. 1986;5:4738–4748. [PubMed] [Google Scholar]
- 7.Mallory JB, Alfano C, McMacken R. Host virus interactions in the initiation of bacteriophage lambda DNA replication. Recruitment of Escherichia coli DnaB helicase by lambda P replication protein. J. Biol. Chem. 1990;5:13297–13307. [PubMed] [Google Scholar]
- 8.Liberek K, Georgopoulos C, Zylicz M. Role of the Escherichia coli DnaK and DnaJ heat shock proteins in the initiation of bacteriophage lambda DNA replication. Proc. Natl Acad. Sci. USA. 1988;85:6632–6636. doi: 10.1073/pnas.85.18.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alfano C, McMacken R. Heat shock protein-mediated disassembly of nucleoprotein structures is required for the initiation of bacteriophage lambda DNA replication. J. Biol. Chem. 1989;25:10709–10718. [PubMed] [Google Scholar]
- 10.Dodson M, Echols H, Wickner S, Alfano C, Mensa-Wilmot K, Gomes B, LeBowitz J, Roberts JD, McMacken R. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: localized unwinding of duplex DNA by a six-protein reaction. Proc. Natl Acad. Sci. USA. 1986;83:7638–7642. doi: 10.1073/pnas.83.20.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mensa-Wilmot K, Seaby R, Alfano C, Wold MC, Gomes B, McMacken R. Reconstitution of a nine-protein system that initiates bacteriophage lambda DNA replication. J. Biol. Chem. 1989;264:2853–2861. [PubMed] [Google Scholar]
- 12.Mensa-Wilmot K, Carroll K, McMacken R. Transcriptional activation of bacteriophage lambda DNA replication in vitro: regulatory role of histone-like protein HU of Escherichia coli. EMBO J. 1989;8:2393–2402. doi: 10.1002/j.1460-2075.1989.tb08369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hase T, Nakai M, Masamune Y. Transcription of a region downstream from lambda ori is required for replication of plasmids derived from coliphage lambda. Mol. Gen. Genet. 1989;216:120–125. doi: 10.1007/BF00332239. [DOI] [PubMed] [Google Scholar]
- 14.Kouhara H, Tsurimoto T, Matsubara K. Direction of transcription affects the replication mode of lambda in an in vitro system. Mol. Gen. Genet. 1987;208:428–435. doi: 10.1007/BF00328134. [DOI] [PubMed] [Google Scholar]
- 15.Szalewska-Pałasz A, Wegrzyn A, Herman A, Wegrzyn G. The mechanism of the stringent control of lambda plasmid DNA replication. EMBO J. 1994;13:5779–5785. doi: 10.1002/j.1460-2075.1994.tb06916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baranska S, Gabig M, Wegrzyn A, Konopa G, Herman-Antosiewicz A, Hernandez P, Schvartzman JB, Helinski DR, Wegrzyn G. Regulation of the switch from early to late bacteriophage lambda DNA replication. Microbiology. 2001;147:535–547. doi: 10.1099/00221287-147-3-535. [DOI] [PubMed] [Google Scholar]
- 17.Wegrzyn G, Wegrzyn A. Genetic switches during bacteriophage lambda development. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:1–48. doi: 10.1016/S0079-6603(04)79001-7. [DOI] [PubMed] [Google Scholar]
- 18.Narajczyk M, Barańska S, Wegrzyn A, Wegrzyn G. Switch from theta to sigma replication of bacteriophage lambda DNA: factors involved in the process and a model for its regulation. Mol. Genet. Genomics. 2007;278:65–74. doi: 10.1007/s00438-007-0228-y. [DOI] [PubMed] [Google Scholar]
- 19.Baker TA, Kornberg A. Transcriptional activation of initiation of replication from the E. coli chromosomal origin: an RNA-DNA hybrid near oriC. Cell. 1988;55:113–123. doi: 10.1016/0092-8674(88)90014-1. [DOI] [PubMed] [Google Scholar]
- 20.Skarstad K, Baker TA, Kornberg A. Strand separation required for initiation of replication at the chromosomal origin of E. coli is facilitated by a distant RNA-DNA hybrid. EMBO J. 1990;9:2341–2348. doi: 10.1002/j.1460-2075.1990.tb07406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atlung T. Allele-specific suppression of dnaA(Ts) mutations by rpoB mutations in Escherichia coli. Mol. Gen. Genet. 1984;197:125–128. doi: 10.1007/BF00327932. [DOI] [PubMed] [Google Scholar]
- 22.Flåtten I, Morigen, Skarstad K. DnaA protein interacts with RNA polymerase and partially protects it from the effect of rifampicin. Mol. Microbiol. 2009;71:1018–1030. doi: 10.1111/j.1365-2958.2008.06585.x. [DOI] [PubMed] [Google Scholar]
- 23.McKinney MD, Wechsler JA. RNA polymerase interaction with dnaB protein and lambda P protein during lambda replication. J. Virol. 1983;48:551–554. doi: 10.1128/jvi.48.2.551-554.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zahn K, Blattner FR. Binding and bending of the lambda replication origin by the phage O protein. EMBO J. 1985;4:3605–3616. doi: 10.1002/j.1460-2075.1985.tb04124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zahn K, Blattner FR. Direct evidence for DNA bending at the lambda replication origin. Science. 1987;236:416–422. doi: 10.1126/science.2951850. [DOI] [PubMed] [Google Scholar]
- 26.Leng F, McMacken R. Potent stimulation of transcription-coupled DNA supercoiling by sequence-specific DNA-binding proteins. Proc. Natl Acad. Sci. USA. 2002;99:9139–9144. doi: 10.1073/pnas.142002099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leng F, Amado L, McMacken R. Coupling DNA supercoiling to transcription in defined protein systems. J. Biol. Chem. 2004;279:47564–47571. doi: 10.1074/jbc.M403798200. [DOI] [PubMed] [Google Scholar]
- 28.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc. Natl Acad. Sci. USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomonaga T, Michelotti GA, Libutti D, Uy A, Sauer B, Levens D. Unrestraining genetic processes with a protein-DNA hinge. Mol. Cell. 1998;1:759–764. doi: 10.1016/s1097-2765(00)80075-1. [DOI] [PubMed] [Google Scholar]
- 30.Collins I, Weber A, Levens D. Transcriptional consequences of topoisomerase inhibition. Mol. Cell. Biol. 2001;21:8437–8451. doi: 10.1128/MCB.21.24.8437-8451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knott SR, Viggiani CJ, Aparicio OM. To promote and protect: coordinating DNA replication and transcription for genome stability. Epigenetics. 2009;4:362–365. doi: 10.4161/epi.4.6.9712. (review) [DOI] [PubMed] [Google Scholar]
- 32.Appleyard RK. Segregation of new lysogenic types during growth of a doubly lysogenic strain derived from Escherichia coli K12. Genetics. 1954;39:440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meselson M, Yuan R. DNA restriction enzyme from E. coli. Nature. 1968;217:1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- 34.Wold MS, Mallory JB, Roberts JD, LeBowitz JH, McMacken R. Initiation of bacteriophage λ DNA replication in vitro with purified λ replication proteins. Proc. Natl Acad. Sci. USA. 1982;79:6176–6180. doi: 10.1073/pnas.79.20.6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glinkowska M, Majka J, Messer W, Wegrzyn G. The mechanism of regulation of bacteriophage lambda pR promoter activity by Escherichia coli DnaA protein. J. Biol. Chem. 2003;278:22250–22256. doi: 10.1074/jbc.M212492200. [DOI] [PubMed] [Google Scholar]
- 36.Tabor S, Richardson CC. A bacteriophage T7 RNA polymerase/ promoter system for controlled exclusive expression of specific genes. Proc. Natl Acad. Sci. USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narajczyk M, Barańska S, Szambowska A, Glinkowska M, Węgrzyn A, Węgrzyn G. Modulation of λ plasmid and phage DNA replication by Escherichia coli SeqA protein. Microbiology. 2007;153:1653–1663. doi: 10.1099/mic.0.2006/005546-0. [DOI] [PubMed] [Google Scholar]
- 38.Zylicz M, Gorska I, Taylor K, Georgopoulos C. Bacteriophage lambda replication proteins: formation of a mixed oligomer and binding to the origin of lambda DNA. Mol. Gen. Genet. 1984;196:401–406. doi: 10.1007/BF00436186. [DOI] [PubMed] [Google Scholar]
- 39.Wegrzyn A, Wegrzyn G, Taylor K. Protection of coliphage λO initiator protein from proteolysis in the assembly of the replication complex in vivo. Virology. 1995;207:179–184. doi: 10.1006/viro.1995.1064. [DOI] [PubMed] [Google Scholar]
- 40.Wegrzyn A, Herman-Antosiewicz A, Taylor K, Wegrzyn G. Molecular mechanism of heat shock-provoked disassembly of the coliphage lambda replication complex. J. Bacteriol. 1998;180:2475–2483. doi: 10.1128/jb.180.9.2475-2483.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pruss GJ, Drlica K. Topoisomerase I mutants: the gene on pBR322 that encodes resistance to tetracycline affects plasmid DNA supercoiling. Proc. Natl Acad. Sci. USA. 1986;83:8952–8956. doi: 10.1073/pnas.83.23.8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu HY, Shyy SH, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto T, McIntyre J, Sell SM, Georgopoulos C, Skowyra D, Zylicz M. Enzymology of the pre-priming steps in lambda dv DNA replication in vitro. J. Biol. Chem. 1987;15:7996–7999. [PubMed] [Google Scholar]
- 44.Learn B, Karzai AW, McMacken R. Transcription stimulates the establishment of bidirectional λ DNA replication in vitro. Cold Spring Harbor Symp. Quant. Biol. 1993;58:389–402. doi: 10.1101/sqb.1993.058.01.046. [DOI] [PubMed] [Google Scholar]
- 45.Cook PR. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. (review) [DOI] [PubMed] [Google Scholar]
- 46.Normanno D, Vanzi F, Pavone FS. Single-molecule manipulation reveals supercoiling-dependent modulation of lac repressor-mediated DNA looping. Nucleic Acids Res. 2008;36:2505–2513. doi: 10.1093/nar/gkn071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dodd IB, Shearwin KE, Perkins AJ, Burr T, Hochschild A, Egan JB. Cooperativity in long-range gene regulation by the lambda CI repressor. Genes Dev. 2004;18:344–354. doi: 10.1101/gad.1167904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H, Finzi L, Lewis DE, Dunlap D. AFM studies of lambda repressor oligomers securing DNA loops. Curr. Pharm. Biotechnol. 2009;10:494–501. doi: 10.2174/138920109788922155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peter BJ, Arsuaga J, Breier AM, Khodursky AB, Brown PO, Cozzarelli NR. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 2004;5:R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.