Abstract
The accuracy of the initiator tRNA (tRNAfMet) selection in the ribosomal P-site is central to the fidelity of protein synthesis. A highly conserved occurrence of three consecutive G–C base pairs in the anticodon stem of tRNAfMet contributes to its preferential selection in the P-site. In a genetic screen, using a plasmid borne copy of an inactive tRNAfMet mutant wherein the three G–C base pairs were changed, we isolated Escherichia coli strains that allow efficient initiation with the tRNAfMet mutant. Here, extensive characterization of two such strains revealed novel mutations in the metZWV promoter severely compromising tRNAfMet levels. Low cellular abundance of the chromosomally encoded tRNAfMet allows efficient initiation with the tRNAfMet mutant and an elongator tRNAGln, revealing that a high abundance of the cellular tRNAfMet is crucial for the fidelity of initiator tRNA selection on the ribosomal P-site in E. coli. We discuss possible implications of the changes in the cellular tRNAfMet abundance in proteome remodeling.
INTRODUCTION
Escherichia coli possesses four copies of the initiator tRNA genes that encode tRNAfMet. Three of these (metZ, metW and metV) are transcribed from the metZWV operon located at 63.5 min and the fourth one (metY) is transcribed as an upstream gene of the nusA-infB operon at 71.5 min in the genome (1). The three genes in the metZWV operon encode three identical tRNAs, tRNA1fMet whereas the metY encodes tRNA2fMet. In E. coli K, the tRNA1fMet and tRNA2fMet differ from each other at a single nucleotide at position 46; whereas the tRNA1fMet carries a methylated guanine (m7G), the tRNA2fMet carries an adenine (A) at this position (2,3). However, in E. coli B, all genes encode identical tRNAs corresponding to tRNA1fMet (4).
The initiator tRNAs are special in that they bind directly to the ribosomal P-site. All other tRNAs (the elongator tRNAs) first bind to the ribosomal A-site and are then, subsequent to peptide bond formation and the translocation step, located into the P-site. Selective binding of the initiator tRNAs to the P-site, crucial for the fidelity of initiation, is an outcome of the functional interaction between the unique structural features of the initiator tRNA, the elements within the ribosome, and the action of initiation factors (IFs). Among the features in tRNAfMet, a mismatch at the top of the acceptor stem (C1xA72, in E. coli) prevents its binding to the elongation factor Tu (EF-Tu); and together with the 2:71 and 3:70 base pairs, it constitutes a major determinant for formylation of the amino acid attached to tRNAfMet (5). Another major structural feature of the initiator tRNAs, highly conserved in all the three domains of life, is the presence of three consecutive G–C base pairs (G29G30G31/C39C40C41; referred to as the 3GC base pairs) in their anticodon stems. This feature is important for the preferential binding of the initiator tRNAs in the ribosomal P-site (6). Biochemical and genetic studies suggested that IF3 facilitates tRNAfMet binding to the 30S ribosomes via the 3GC base pairs (7–9). Subsequent studies showed that in the ribosome, the binding site of IF3 is located at a distance from the anticodon stem of the tRNAfMet (10,11). More recently, it has been observed that the presence of IF3 increases the rate of dissociation of all tRNAs from the 30S ribosomes. Because the presence of IF2 (along with IF1) strongly favors binding of fMet-tRNAfMet to the 30S subunit and docking of the 50S subunit, such a role of IF3, in essence allows for higher accuracy of fMet-tRNAfMet selection on the ribosome (12). Also, two of the universally conserved nucleotides G1338 and A1339 in the head region of the 16S rRNA are in juxtaposition to the minor groove of the 3GC base pairs (13,14). Mutational analysis of these residues (15) has supported their role in stabilizing tRNAfMet binding via the ‘A-minor’ interactions with the 3GC base pairs (16). In addition, C-terminals of S9 and S13 are within the contacting distance with the anticodon stem loop of the tRNAfMet (14).
To further our understanding of the biological significance of the 3GC base pairs in initiator tRNA selection on the ribosomal P-site, we developed a genetic screen (17) and isolated E. coli strains that allow initiation with tRNAs lacking the 3GC base pairs. Characterization of two such strains in this study shows that they possess novel mutations in the metZWV promoter severely compromising tRNA1fMet expression. Low cellular abundance of the tRNAfMet allows efficient initiation not only with the mutant tRNAfMet but also with tRNAGln revealing that the high abundance of the cellular tRNAfMet is crucial for the fidelity of translation in E. coli.
MATERIALS AND METHODS
Strains, plasmids and media
Strains and plasmids have been listed in Supplemetnarty Tables S1 and S2. Bacteria were grown in Luria-Bertani (LB) liquid or LB-agar plates (18) containing 1.5% bacto-agar (Difco). Unless indicated otherwise, media were supplemented with ampicillin (Amp, 100 μg ml−1), chloramphenicol (Cm, 30 μg ml−1), kanamycin (Kan, 25 μg ml−1) or tetracycline (Tet, 7.5 μg ml−1) as required.
Isolation and characterization of D4 and D27, and their genetic mapping
The isolation and preliminary characterization of the suppressors of the 3GC mutations in tRNAfMet have been described before (17). Methodologies used to characterize two of the suppressors, D4 and D27 and the genetic fine mappings of the mutations in these were as described (17).
Cloning of metZWV gene promoter regions
The promoter regions of metZWV genes were PCR amplified from genomic DNAs of E. coli KL16 (the parent strain), D4 and D27 using Pfu DNA polymerase in 50-μl reactions containing 20 pmols each of the forward (5′ ctggctggatccccagagagaa 3′) and reverse (5′ ctaccaggatcctccaccccg 3′) primers. The amplicons (∼363 bp) were digested with KpnI and BamHI, eluted from agarose gels and cloned into similarly digested promoterless pTKCAT (18).
Cloning of supE
The glnVX gene (supE) was PCR amplified from E. coli TG1 using a supEfp (5′ gccttacaagcttgccggagc 3′) and a supErp (5′ cgtagccacaagcttctgaatg 3′) primers as 0.4-kb amplicon, digested with NcoI and EcoR1 and cloned into pACDH between the same sites to generate pACDHsupE (18).
Isolation of tRNAs and northern blot analysis
Total tRNA preparations from various strains were fractionated on native 15% polyacrylamide gels (6), electroblotted onto nytran membrane and analyzed by northern blotting using a 32P 5′-end labeled DNA oligomer (5′ cttcgggttatgagcccgacgagcta 3′) essentially as described (19).
Generation of ΔmetY::kan strains
The KanR cassette from the plasmid pKD4 was amplified with metYko_fp (5′ ttcacagtatatttgaaaaaggactctaagcgaaaggtgtaggctggagctgcttcg 3′) and metYko_rp (5′ tttacccaaaacgagtagaatttgccacgtttcaggcatatgaatatcctcctta 3′) primers, electroporated into E. coli DY330 and selected on Kan plates (20). Transformants were screened for replacement of metY with the KanR cassette by PCR with the flanking primers, metYfp (5′ tgcagattttacgtcccgtc 3′) and metYrp (5′ gcactttccagaaggatttt 3′). The ΔmetY::kan locus was then mobilized into E. coli KL16, D4 and D27 strains by P1-mediated transductions to generate their ΔmetY::kan counterparts (21).
Generation of ΔmetZWV::kan strains
The KanR cassette from pKD4 was amplified with the primers, metZWVko_fp (5′ tgaaaacgcgagcggagtatagtgcgcatccacggagtgtaggctggagctgcttcg 3′) and metZWVko_rp (5′ cagaaacaaaaaaacacccgttagggtgttcaataatcatatgaatatcctcctta 3′) using Pfu DNA polymerase. The amplicon was electroporated into E. coli DY330 (20). Transformants were screened for replacement of metZWV with the KanR cassette by PCR with the flanking primers, metZWVfp and metYfp (metZWVfp 5′ tatagtgcgcatccacgga 3′ and metZWVrp 5′ agggtgttcaataat 3′) following which the ΔmetZWV::kan locus was mobilized into E. coli KL16, D4 and D27 by P1-mediated transductions to generate their ΔmetZWV::kan counterparts (21).
Preparation of cell-free extracts and chloramphenicol acetyltransferase assays
Escherichia coli cells were grown in 3-ml LB medium containing Amp to mid-log phase and processed as described (17). Total of the pixel values in the spots corresponding to 1-acetyl-, plus 3-acetyl-, chloramphenicol (Ac-Cm, P) and the left over substrate, chloramphenicol (Cm, S) were quantified using a BioImageAnalyzer (FLA5000, Fuji). The chloramphenicol acetyltransferase (CAT) assays were done for 15 min using appropriate amounts of total cell extracts as defined in the figure legends. The CAT activities were calculated as picomoles of Cm converted to Ac-Cm per microgram total protein [(by multiplying the P/(S+P) ratio with the total picomoles of Cm taken in the reaction) divided by the total protein in the cell extract]. Assays were carried out from three independent colonies and the averages (picomoles of Cm converted to Ac-Cm per microgram total protein per 15 min, along with the associated standard deviations) were used to generate histograms.
RESULTS
The assay for initiation in E. coli and characterization of D4 and D27
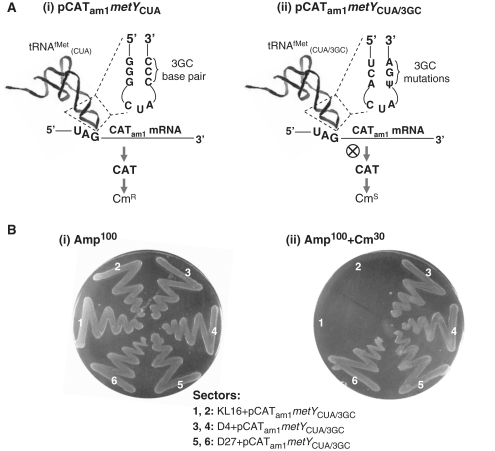
The plasmid, pCATam1metYCUA (Figure 1A) harbors a CATam1 reporter gene wherein the AUG initiation codon has been changed to UAG codon along with mutations in the second and the fifth codons to eliminate secondary sites of weak initiation (22). The presence of an initiator tRNA gene harboring a complementary mutation in the anticodon, CAU to CUA (Figure 1A, panel i) fosters efficient initiation from the UAG initiation codon of the reporter and confers resistance to chloramphenicol (CmR) to E. coli. As the metYCUA encoded tRNA is aminoacylated with Gln, initiation occurs with formyl-Gln (22). Expectedly, when the three consecutive G–C base pairs in the anticodon stem of the initiator tRNA were mutated to those found in the elongator tRNAMet (G29G30G31/C39C40C41 to U29C30A31/ψ39G40A41; hereafter referred to as ‘3GC’ mutations or the 3GC tRNA), in the plasmid pCATam1metYCUA/3GC (Figure 1A, panel ii) initiation did not occur to any detectable level and E. coli harboring the pCATam1metYCUA/3GC plasmid were sensitive to chloramphenicol (CmS) even though the encoded tRNA is fully aminoacylated and formylated [(17,23); see also Figure 1B, panel ii, sectors 1 and 2].
Figure 1.
(A) Assay for initiation in E. coli. (i) The plasmid, pCATam1metYCUA possesses pCATam1 and metYCUA genes. The pCATam1 gene produces CATam1 reporter mRNA wherein initiation occurs with the UAG codon. The metYCUA gene makes tRNAfMet wherein the CAU anticodon has been mutated to CUA. The encoded tRNA is shown as tRNAfMet(CUA) and the relevant sequence detail of the anticodon stem loop (boxed with the dotted lines) are as shown. Schematic representation shows that in the presence of the wild-type sequence (3GC base pair) in the anticodon stem of the tRNA, initiation from the reporter mRNA confers chloramphenicol resistance (CmR) to the host. (ii) The plasmid, pCATam1metYCUA/3GC is a derivative of pCATam1metYCUA wherein metYCUA gene has been replaced with metYCUA/3GC gene. The encoded tRNA is shown as tRNAfMet(CUA/3GC) and the relevant sequence detail of the anticodon stem loop (boxed with the dotted lines) are as shown. The mutated sequence in the anticodon stem (3GC mutations) of the tRNA makes it deficient in initiation and renders the host sensitive to chloramphenicol (CmS). (B) Growth of E. coli KL16 parent (KL16) and the suppressor strains (D4, D27) harboring pCATam1metYCUA/3GC (AmpR) on LB-agar plates containing, (i) Amp and, (ii) Amp plus Cm. Details of the strains (and the resident plasmids), streaked in various sectors are shown on the right. Plates were streaked with overnight cultures and incubated at 37°C for ∼15 h.
In our earlier study, we described isolation of E. coli KL16 (K strain) derivatives harboring chromosomal mutations that alleviate the initiation defect of the 3GC tRNA. Characterization of two such suppressor strains, called D4 and D27, is the focus of this study. As a part of the strain validation, introduction of pCATam1metYCUA/3GC plasmid into the plasmid-free versions of D4 and D27 conferred CmR (Figure 1B, panel ii, sectors 3 and 4 and 5 and 6). Under the same conditions, introduction of this plasmid into the parent strain (KL16) did not confer CmR (sectors 1 and 2). As a control, all transformants grew on Amp plate (Figure 1B, panel i). In yet another control, we showed that initiation from CATam1 in D4 and D27 was dependent on the presence of the 3GC tRNA (Supplementary Figure S1).
Mapping of the suppressor mutations in D4 and D27
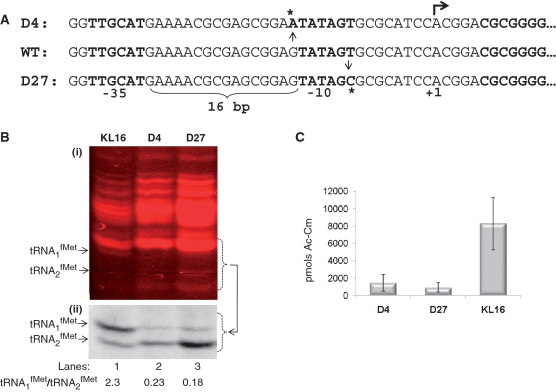
The mutation(s) responsible for the phenotypes of D4 and D27 were mapped to a site between 63.4 and 63.6 min on the E. coli genome (Supplementary Figures S2 and S3). Sequence analysis of a high-probability candidate within this region, the metZWV operon encoding for the majority of tRNAfMet in E. coli, revealed that D4 and D27 possessed novel mutations within the promoter region of metZWV (Figure 2A). We then introduced targeted mutations in the E. coli genome to ensure that these single mutations in the promoter regions of metZWV of D4 and D27 were responsible for their phenotypes (Supplementary Figure S4).
Figure 2.
Mapping of the mutations in D4 and D27 and their initial characterization. (A) Comparative analysis of the promoter regions of D4 and D27 metZWV locus with that of the parent (wild-type, WT) reveals that there is a G to A mutation in D4 just upstream to the Pribnow box (−10) and a T to C mutation in D27 within the Pribnow box (−10). The mutations have been indicated by arrows emanating from the sequence of the WT promoter and asterisks above and below the D4 and D27 sequences. The −35 region, the spacing between the −10 and the −35 sequences and the transcription start site (+1) are also indicated. (B) Northern blot analysis of tRNA1fMet and tRNA2fMet in the parent (KL16), and the D4 and D27 strains. Total tRNA preparations (∼4, 6 and 10 µg, respectively, for KL16, D4 and D27) were separated on the native polyacrylamide gel (15%) stained with ethidium bromide (panel i), transferred onto nytran membrane and analyzed by northern blotting (panel ii). Positions of tRNA1fMet and tRNA2fMet, and the ratios of the tRNA1fMet to tRNA2fMet are indicated. Lanes: 1, KL16; 2, D4; and 3, D27. (C) Promoter regions of the metZWV locus from KL16, D4 and D27 were cloned upstream of a promoterless CAT reporter in pTKCAT and introduced into E. coli KL16. Three independent colonies from each were grown to mid-log phase, and the cell-free extracts (∼90 ng total protein) used to assay for CAT activities. Average activities (±SD) are shown as picomoles Ac-Cm (1-acetyl-, and 3-acetyl-, chloramphenicol) formed per microgram total cell protein in 15 min.
The mutations in metZWV locus downregulate expression of tRNAfMet
Based on the transcriptional start site mapping of the metZWV transcripts (24), the mutations in D4 and D27 could impact the promoter quality. Hence, to analyze the influence of these mutations, we carried out northern blot analysis of the total tRNA preparations using a native polyacrylamide gel, which allows separation of tRNA1fMet and tRNA2fMet (Figure 2B). As shown in Figure 2B (panel ii), the levels of tRNA1fMet in D4 and D27 were significantly decreased [compare the relative intensities of the bands corresponding to tRNA1fMet and tRNA2fMet in KL16 (lane 1) with those in D4 and D27 (lanes 2 and 3)]. It may be noted that the decrease in the tRNA1fMet is quite severe and significantly more amounts of total tRNA were needed to detect its presence on the northern blot (Figure 2B, panel i). In fact, in the ethidium bromide stained gel, while the tRNA1fMet is detectable in the parent (KL16), the same is not seen in D4 and D27 even though the total tRNA loaded for these samples was in 1.5- and 2.5-fold excess, respectively, of the amounts used for the KL16 sample (Figure 2B, panel i). To ensure that the decreased levels of tRNA1fMet were a direct consequence of the promoter down mutation, we cloned equivalent regions of the promoters from the KL16 (wild-type), D4 and D27 strains upstream of a promoterless CAT reporter in pTKCAT vector, and assayed for the CAT activities in E. coli. As shown in Figure 2C, the mutations in D4 and D27 indeed led to drastic decreases in the promoter strength.
D4 and D27 confer a minor growth defect and cold sensitivity to E. coli
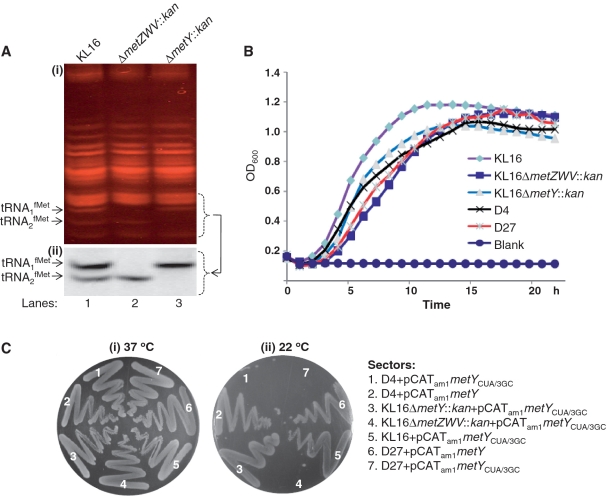
To understand the phenotypes of D4 and D27, and the mechanism of initiation by the 3GC tRNA in these strains, we generated strains of E. coli KL16 wherein either the metZWV or metY were knocked out by a KanR marker. As shown in Figure 3A, while the analysis of the KL16 total tRNA sample on the native polyacrylamide gel (panel i) revealed the presence of both the tRNA1fMet and tRNA2fMet (lane 1), the tRNA1fMet and the tRNA2fMet bands, as expected, were missing in the ΔmetZWV::kan and metY::kan strains, respectively. The observation was further confirmed by the northern blot analysis (panel ii). It may also be noted that the deletion of metZWV did not lead to an increase in the levels of tRNA2fMet. Likewise, deletion of metY did not impact the levels of tRNA1fMet. These observations suggest that the levels of the initiator tRNA in E. coli correspond to their gene copy numbers.
Figure 3.
Assay for growth and cold sensitivity. (A) Analysis of tRNA1fMet and tRNA2fMet in E. coli KL16 parent (KL16), its ΔmetZWV::kan and ΔmetY::kan derivatives. Total tRNA preparations (∼6 µg each) were separated on the native polyacrylamide (15%) gel, stained with ethidium bromide (panel i), transferred onto nytran membrane and analyzed by northern blotting (panel ii). Positions of tRNA1fMet and tRNA2fMet are indicated. (B) Growth kinetics of E. coli KL16 parent (KL16), its ΔmetZWV::kan and ΔmetY::kan; derivatives and the suppressor strains (D4, D27). Saturated cultures were diluted 1000-fold in LB broth, and growth monitored at 1-h intervals in a Bioscreen C growth reader. All strains were taken in replicates of three, and the average values were plotted. (C) Growth of E. coli KL16, its ΔmetZWV::kan and ΔmetY::kan derivatives and the suppressor strains (D4, D27) harboring pCATam1metYCUA/3GC or pCATam1metY (AmpR), as indicated, on LB-agar plates at 37°C (i) and 22°C (ii). Plates were streaked with overnight cultures and incubated at the indicated temperatures for ∼15 h.
To analyze the phenotypes of D4 and D27, we compared their growth kinetics with the metZWV and metY knockout strains. During their exponential phases, the growth of the D4 and D27 strains was slightly compromised when compared with the parent strain (KL16) harboring all four copies of the initiator tRNA genes but slightly better than its ΔmetZWV::kan derivative (Figure 3B). Further, it was earlier reported that deletion of tRNA1fMet locus resulted in cold sensitivity to E. coli (25). Consistent with these observations, deletion of metZWV locus (ΔmetZWV::kan) which encodes for the majority of the cellular initiator tRNA in E. coli, but not of metY (metY::kan) which contributes to minor amounts of the same, in the background of KL16 (harboring pCATam1metYCUA/3GC) resulted in a cold sensitive phenotype at 22°C (Figure 3C, panel ii, compare sector 4 with sectors 3 and 5). Interestingly, E. coli strains with both the D4 and D27 mutations also displayed cold sensitivity (sectors 1 and 7). However, the cold sensitivity could be rescued by the presence of a plasmid borne copy of the metY gene, which encodes for the wild-type tRNA2fMet (sectors 2 and 6). As a control, all strains grew well at 37°C (Figure 3C, panel i).
Taken together with the results shown in Figure 2, these observations suggested that a major consequence of the D4 and D27 mutations is to lower the abundance of the tRNAfMet in the cell.
Mechanism of initiation with the 3GC tRNA in D4 and D27
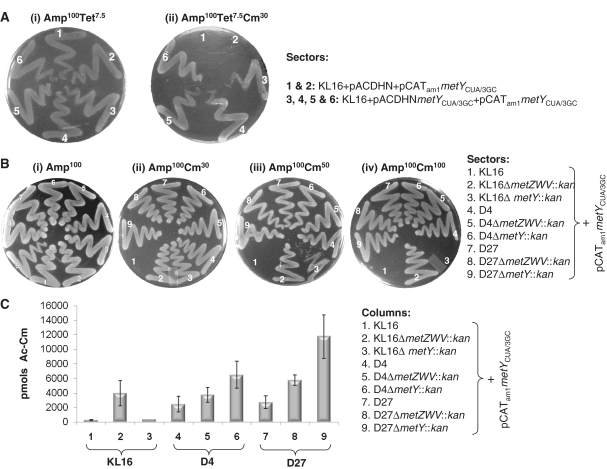
Based on the observations in Figures 2 and 3, we hypothesized that the 3GC tRNA has to compete with the chromosomally encoded tRNAfMet for its binding to the ribosomal P-site, and that the lower abundance of the chromosomally encoded tRNAfMet in D4 and D27 results in an increased probability of the P-site occupancy by the 3GC tRNA leading to an increased opportunity of initiation by it. Consistent with this hypothesis, when a compatible plasmid, pACDHNmetYCUA/3GC was introduced into the KL16 strain already harboring pCATam1metYCUA/3GC (for an elevated level of the 3GC tRNA), it conferred Cm resistance to the transformants such that they grew on a plate with 30 µg ml−1 Cm (Figure 4A, panel ii, sectors 3–6). As a control, introduction of the vector alone (pACDHN) did not confer such a resistance (sectors 1 and 2). Further, knockout of the metY gene (ΔmetY::kan), a gene which contributes to minor amounts to the cellular pools of tRNAfMet, in KL16 harboring pCATam1metYCUA/3GC resulted in gain of initiation by the 3GC tRNA such that it grows at 30–50 µg ml−1 Cm (Figure 4B, sector 3 in all panels). And, a knockout of the metZWV locus (ΔmetZWV::kan) conferred efficient initiation with the 3GC tRNA to allow good growth even at 100 µg ml−1 Cm (Figure 4B, sector 2 in all panels). As a control, in the absence of these knockouts, the KL16 strain did not foster a detectable initiation with the 3GC tRNA (Figure 4B, sector 1 in all panels).
Figure 4.
(A) Elevated expression of the 3GC tRNA allows initiation. Plasmids, pACDHN (pAC1 origin of replication, TetR) or its derivative pACDHNmetYCUA/3GC were introduced into E. coli KL16 harboring pCATam1metYCUA/3GC (a colE1 ori of replication, AmpR), and the trans-formants grown in LB containing Amp plus Tet were streaked onto Amp plus Tet, and Amp plus Tet plus Cm (as indicated) and incubated at 37°C for ∼15 h. (B) Growth of the E. coli KL16, D4 and D27 and their ΔmetZWV::kan and ΔmetY::kan derivatives harboring pCATam1metYCUA/3GC (as indicated) on LB-agar plates containing Amp and, Amp plus Cm (30–100 µg ml−1). Plates were streaked with overnight cultures and incubated at 37°C for ∼15 h. (C) CAT assays to determine initiation in E. coli KL16, D4 and D27 and their ΔmetZWV::kan and ΔmetY::kan derivatives harboring pCATam1metYCUA/3GC (as indicated). Three independent colonies of each were grown to mid-log phase, and the cell-free extracts (500-ng total protein) used to assay for CAT activities. Average activities (±SD) are shown as picomoles Ac-Cm (1-acetyl-, and 3-acetyl-, chloramphenicol) formed per microgram total cell protein in 15 min.
The knockouts of metY and metZWV were also introduced into D4 and D27 (Figure 4B, sectors 5, 6 and 8, 9). However, for a better assessment of the impact of these knockouts, we carried out CAT assays using cell-free extracts prepared from the strains (Figure 4C). The results from these assays reveal a clear correlation between increase in initiation by the 3GC tRNA and a decrease in the abundance of the chromosomally encoded tRNAfMet. For example, in the KL16 background, knockout of the metZWV gene locus (which is responsible for a major decrease in the tRNAfMet levels) leads to a greater increase in initiation by the 3GC tRNA than does the knockout of the metY gene (which is responsible for a minor decrease in the tRNAfMet levels) (Figure 4B, sectors 1–3; and Figure 4C, compare bars 1–3). On the other hand, in the D4 and D27 strains wherein the metZWV is already substantially downregulated, the metY gene knockout causes a greater increase in initiation (by the 3GC tRNA) than does the metZWV knockout (Figure 4C, compare bars 4–6 and 7–9). In fact, the knockout of metY in D27 (which sustains growth on very low levels of tRNAfMet encoded by the downregulated metZWV) has the highest level of initiation by the 3GC tRNA (bar 9). However, we should add that this strain grows slowly in the liquid medium and is unstable to continued subculturing.
Thus, data in Figure 4 support the concept that the 3GC tRNA has to compete with the wild-type tRNAfMet for the occupancy of the P-site; and the lower the cellular abundance of the wild-type tRNAfMet, the better the initiation with the 3GC mutant. Intuitively, such an inference also implies that high level of tRNAfMet in E. coli (encoded together by the four copies of the initiator tRNA genes) must contribute to the maintenance of the fidelity of initiation by competing out the elongator tRNAs from the ribosomal P-site.
Initiation with elongator tRNA (supE) and maintenance of the fidelity of initiation by the cellular levels of initiator tRNA
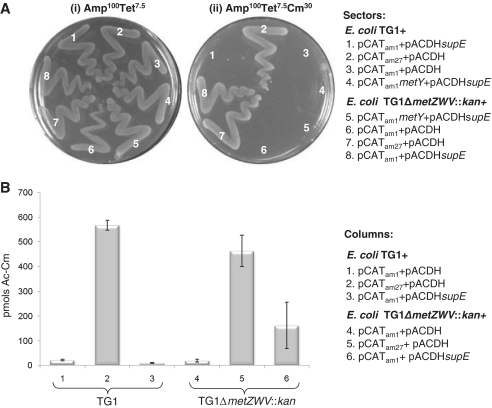
To check if the elongators could initiate in E. coli, we made use of the TG1 strain which harbors the supE tRNA (tRNAGln with CUA anticodon). To assay for the participation of supE at the step of elongation, we used pCATam27 which harbors a CAT gene possessing a UAG codon at position 27 in the reading frame (26). In addition, for these assays, we made use of a two plasmid system. The second pAC1 origin of replication-based plasmid, pACDH (TetR) is compatible for its existence with the colE1-based plasmid in E. coli, and was used to overproduce the supE tRNA. As expected, when the pCATam1 plasmid was introduced into the strain, no detectable levels of CAT protein were produced and the strain remained CmS (Figure 5A, panel ii, sector 3; and Figure 5B, bar 1). Overproduction of supE from pACDHsupE, also did not allow production of CAT to any detectable level (sector 1 and bar 3). However, as a control, when pCATam27 was introduced in TG1, it conferred CmR to the host (sector 2 and bar 2). These observations suggest that in a strain possessing full complement of the four initiator tRNA genes, the fidelity of initiation is maintained and the elongator tRNAs do not participate at the step of initiation to any detectable level.
Figure 5.
(A) Initiation with elongator tRNA. E. coli TG1 or its ΔmetZWV::kan derivative, harboring various plasmids as indicated were grown in LB containing Amp plus Tet and streaked on LB-agar plates containing Amp plus Tet, and Amp plus Tet plus Cm and incubated at 37°C for 15 h. (B) Initiation with elongator tRNA. CAT assays from the cell-free extracts of E. coli TG1 or its ΔmetZWV::kan derivative, harboring various plasmids (as indicated). CAT activities were assayed in cell-free extracts (1.2-µg total protein) prepared from three independent colonies. Average activities (±SD) are shown as picomoles Ac-Cm (1-acetyl-, and 3-acetyl-, chloramphenicol) formed per microgram total cell protein in 15 min.
We next performed these assays in the background of decreased copy number of the initiator tRNA genes (ΔmetZWV::kan). We observed that the overexpression of supE, did confer CmR to the host containing pCATam1 (Figure 5A, panel ii, sector 8; and Figure 5B, bar 6). However, in the presence of chromosomally encoded supE alone, under the assay conditions used, we did not observe CmR phenotype (sector 6) or an increase in the CAT activity above the background (compare bars 1 and 4). Although when the plates were incubated for longer periods (24–30 h), isolated colonies appeared in the transformants of pCATam1 in TG1ΔmetZWV::kan but not in TG1 (data not shown). Importantly, when we overexpressed tRNAfMet from a plasmid borne copy, it purged the initiation activity of supE (Figure 5A, panel ii, sector 5), suggesting that the initiation by supE is a specific phenomenon and that the elongator tRNAs do compete with initiator tRNAs in vivo. Furthermore, this experiment showed that higher levels of tRNAfMet in the cell are essential to maintain fidelity of initiation in that they prevent initiation by the non-initiator tRNAs. Similar observations were also made in the strains D4 and D27 (Supplementary Figure S5).
DISCUSSION
Barring a few species of mycoplasma (17,27), the presence of the three consecutive G–C base pairs in the anticodon stems of the initiator tRNAs is a highly conserved feature throughout the three domains of life, and any substitutions in the 3GC base pairs lead to a deficient utilization of the mutant tRNAfMet in initiation (6). To address the long-standing question of how the 3GC base pairs contribute to initiator tRNA selection on the ribosomal P-site and to gain insight into the mechanisms that maintain the fidelity of translation initiation in E. coli, we used a genetic screen to isolate suppressor strains of E. coli which due to mutations in their genomes, allow initiation by the 3GC mutant tRNA (17). In the present study, we have characterized two such strains, D4 and D27, and found that they both harbor single nucleotide mutations in the −10 promoter regions of the metZWV operon (a G to A mutation in D4; and a T to C mutation in D27) and possess reduced levels of tRNAfMet which allows initiation not only by the 3GC mutant but also the elongator tRNA. It is plausible that these mutations impact various aspects of promoter recognition and decrease transcription initiation by the RNA polymerase (28). Simulating conditions of reduced RNAfMet levels using knockouts of the initiator genes (Figures 4 and 5) has revealed a previously unknown connection between tRNAfMet abundance and maintenance of the fidelity of initiation by tRNAfMet by rejecting non-initiator tRNA binding in the ribosomal P-site.
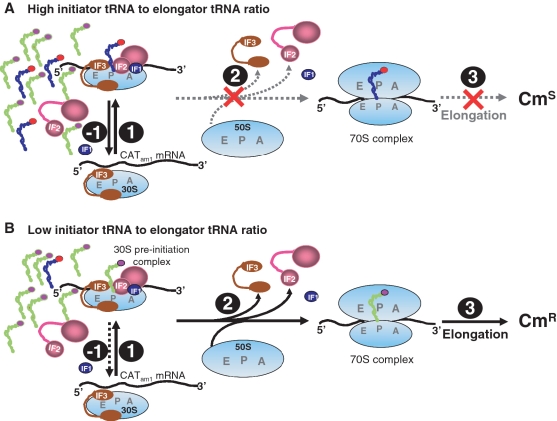
How does the cellular abundance of tRNAfMet regulate translation of CATam1 mRNA with the 3GC or the elongator tRNAs? Based on our observations, we propose the following model (Figure 6). The 3GC base pairs in the tRNAfMet are known to facilitate its binding to the ribosomal P-site. Under the normal conditions of high abundance of the tRNAfMet (i.e. under the conditions of high occupancy of the P-site with the tRNAfMet), binding of the 3GC or the supE tRNAs lacking the 3GC base pairs to the P-site is disallowed. Hence, even though the CATam1 mRNA may be engaged on the 30S subunit with the tRNAfMet containing CAU anticodon (Figure 6A, Step 1), because the mRNA lacks AUG- or AUG-related codons in the vicinity of the Shine–Dalgarno (S. D.) sequence, the codon–anticodon pairing and the conformational changes required to give rise to a 30S pre-initiation complex (29), would not take place. The initial complex would decompose (Figure 6A, Step −1), and prevent translation initiation to proceed through to Steps 2 and 3. Consequently, the CAT protein would not be synthesized and the cells remain CmS. However, when the abundance of the tRNAfMet is decreased (as is the case with D4, D27 and the KL16 strains deleted for metZWV or metY), it now allows for a finite probability of the P-site occupancy with the 3GC mutant or the supE tRNAs (possibly because of the delays in the occupancy of the P-site by the tRNAfMet due to decreased mass effect) (Figure 6B, Step 1). Even though the 3GC or supE tRNAs so bound could fall off of the 30S ribosome due to their poor affinity (Figure 6B, Step −1), the increased potential for binding of the tRNAs containing CUA anticodon (complementary to the UAG initiation codon) would lead to an increased potential for formation of productive 30S pre-initiation complexes with the CATam1 mRNA which can then proceed through steps 2 and 3. The CAT protein so synthesized would confer CmR to cells (Figure 6B).
Figure 6.
A model for the role tRNAfMet abundance in translation initiation from CATam1 mRNA. As per the established pathway, the 30S pre-initiation complexes form from 30S subunits, mRNA, initiator tRNA and the initiation factors (Step 1). Subsequently, the 50S subunit docks on to the 30S pre-initiation complex to form 70S complex (Step 2) which can then enter the elongation step (Step 3) to translate an mRNA. (A) A high occupancy of the P-site of the 30S subunit by the wild-type tRNAfMet is facilitated by its structural features (which include the 3GC base pairs in the anticodon stem) and the initiation factors. Lack of the correct pairing between the anticodon (CAU) of the P-site bound tRNA and the initiation codon, UAG of the reporter CATam1 mRNA (in the vicinity of S.D. sequence, not shown) disallows formation of a productive 30S pre-initiation complex, and the subsequent steps of initiation leading to a CmS phenotype. (B) Under the conditions of low abundance of tRNAfMet, a low occupancy of the ribosomal P-site by tRNAfMet allows for a finite probability of the P-site occupancy by tRNAs lacking the 3GC base pairs (e.g. the 3GC or the supE tRNAs), and initiation by them leading to the synthesis of CAT and a CmR phenotype (see ‘Discussion’ section for further details). Symbols for the various factors are as indicated. Letters E, P and A in gray refer to the E-, P- and A-site on the 30S and 50S subunits. The L-shaped molecules are tRNAs (blue: tRNAfMet; light green: 3GC or supE tRNAs), and the red crosses in (A) indicate failure of the 50S subunit docking and transition of the complexes into the elongation phase.
Since initiation is one of the most regulated and rate-limiting step in protein synthesis (30), a high cellular level of tRNAfMet (encoded by four genes) has thus far been solely attributed to a necessity for efficient initiation. Based on the observations that a high cellular abundance of initiator tRNA makes significant contribution to preventing initiation with the 3GC mutant, as well as with the elongator tRNAs (Figures 4, 5 and 6A), we propose that a high abundance of initiator RNA is important not only in overcoming the rate-limiting step of initiation but also crucial in maintaining the fidelity of initiation by offering an effective discrimination against selection of the elongator tRNAs in the P-site (and thus initiation from codons unrelated to AUG). This direct relationship between fidelity and levels of initiator tRNA could also be an explanation behind the multiple copies of the initiator tRNA genes in various organisms.
Fluctuations in the initiator tRNA levels have been reported during diverse physiological conditions. It has been observed that metZWV is downregulated during stringent response by ppGpp (24). Likewise, metY is downregulated by the global regulator, cAMP–CAP complex and also by ArgR, a specific regulator of arginine metabolism (31). In addition, studies have shown that there could be major decreases in the charged initiator tRNA levels in E. coli during leucine starvation (32). However, the significance of such observations has remained unknown. Although in yeast, it is known that depletion of initiator tRNA levels leads to increased translation of GCN4, and GCN4-dependent transcription of target genes (33). Also, it has been shown that the levels of initiator and elongator Met-tRNA are inversely correlated in cell proliferation versus quiescence. In quiescent cells, the initiator tRNA levels go down but the elongator tRNA levels go up (34).
Under the special circumstances of nutritional deprivation, etc., it may be advantageous for the cell to allow a ‘flexibility/relaxation’ in the fidelity of protein synthesis. Such a proposal has parallels in other biological processes. For example, fidelity of replication is paramount to the maintenance of the genetic blue print of the organism. Yet, during SOS repair/replication, error-prone incorporation of nucleotides by DNA polymerases IV and V makes an important contribution to the viability of the cell (35). Likewise, a major reprogramming of gene expression by transcriptional and translational regulation is a well-established phenomenon during conditions of stress or starvation (36). However, in these cases of proteome remodeling, the issue of possible ‘relaxation in fidelity’ for initiation has never been raised. Our preliminary computational analysis shows that in E. coli (and possibly all other eubacteria), Shine–Dalgarno (S. D.) like sequences are found in the context of a variety of non-AUG codons. This observation, together with the possibility of downregulation of the initiator tRNA abundance under stress/starvation conditions, makes it feasible that relaxation in the fidelity of initiation may offer a mechanism to contribute to regulation of the proteome remodeling/plasticity in the cell.
An accurate selection of the initiator tRNA on the ribosomal P-site is a combined outcome of several coordinated events. For example, in eubacteria, formylation of initiator tRNA which facilitates its binding to IF2, and effectively decreases its off rate upon IF3 binding (12) is perhaps one of the most crucial contributors to the specificity of the initiator tRNA selection on the ribosome. However, this feature alone is neither sufficient nor essential for the initiator tRNA selection on the ribosomal P-site. For instance, despite the fact that 3GC mutant is fully formylated in E. coli (17), it fails to initiate to any significant level in the wild-type strain background. And although they suffer a mild to severe impact on the growth rates, it is known that knockouts of the formylase gene in Pseudomonas aeruginosa and E. coli are viable (37,38). Furthermore, in Salmonella mutants, efficient initiation could occur in the absence of formylation. Interestingly, the mutants compensated for the deficiency of formylation by amplification of the initiator tRNA genes (39). Among the other mechanisms that contribute to the fidelity of initiation are the rates with which the 50S and 30S subunit interact with each other (12), and possible effects of the modified nucleosides in rRNA (both in the 30S and 50S subunits) (17,40). In this scenario, the role played by the abundance of the initiator tRNA adds another level of complexity to the fidelity maintenance.
Given the numerous factors that could influence the translation initiation fidelity, a redundancy between these is an expected possibility. Although it is difficult to determine the relative contribution of various factors, it is feasible that in organisms having a single copy of functional initiator tRNA gene (such as mycobacteria) or a single initiator tRNA gene lacking the full complement of the 3GC base pairs in their anticodon stems (such as certain species of mycoplasma), other mechanisms compensate for the decreased fidelity. Alternatively, the organism’s survival strategy in the host might itself dictate a requirement of a certain ‘relaxed’ fidelity of translation. Thus, a study of translation fidelity in these organisms (mycoplasma and mycobacteria) would be rewarding not only from the point of view of understanding basic life processes but also to gain insight into their pathogenic behavior.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants from the Department of Science and Technology (DST) and the Department of Biotechnology (DBT), New Delhi; Senior research fellowships of the Council of Scientific and Industrial Research, New Delhi (to S.K. and G.D.). Funding for open access charge: DBT, New Delhi, Government of India.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank our laboratory colleagues for their suggestions on the manuscript.
REFERENCES
- 1.Berlyn MK. Linkage map of Escherichia coli K-12, edition 10: the traditional map. Microbiol. Mol. Biol. Rev. 1998;62:814–984. doi: 10.1128/mmbr.62.3.814-984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dube SK, Marcker KA. The nucleotide sequence of N-formyl-methionyl-transfer RNA. Partial digestion with pancreatic and T-1 ribonuclease and derivation of the total primary structure. Eur. J. Biochem. 1969;8:256–262. doi: 10.1111/j.1432-1033.1969.tb00522.x. [DOI] [PubMed] [Google Scholar]
- 3.Egan BZ, Weiss JF, Kelmers AD. Separation and comparison of primary structures of three formylmethionine tRNAs from E. coli K-12 MO. Biochem. Biophys. Res. Commun. 1973;55:320–327. doi: 10.1016/0006-291x(73)91090-5. [DOI] [PubMed] [Google Scholar]
- 4.Mandal N, RajBhandary UL. Escherichia coli B lacks one of the two initiator tRNA species present in E. coli K-12. J. Bacteriol. 1992;174:7827–7830. doi: 10.1128/jb.174.23.7827-7830.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.RajBhandary UL. Initiator transfer RNAs. J. Bacteriol. 1994;176:547–552. doi: 10.1128/jb.176.3.547-552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seong BL, RajBhandary UL. Escherichia coli formylmethionine tRNA: mutations in GGG/CCC sequence conserved in anticodon stem of initiator tRNAs affect initiation of protein synthesis and conformation of anticodon loop. Proc. Natl Acad. Sci. USA. 1987;84:334–338. doi: 10.1073/pnas.84.2.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartz D, McPheeters DS, Gold L. Selection of the initiator tRNA by Escherichia coli initiation factors. Genes Dev. 1989;3:1899–1912. doi: 10.1101/gad.3.12a.1899. [DOI] [PubMed] [Google Scholar]
- 8.Hartz D, Binkley J, Hollingsworth T, Gold L. Domains of initiator-tRNA and initiation codon crucial for initiator tRNA selection by Escherichia coli IF3. Genes Dev. 1990;4:1790–1800. doi: 10.1101/gad.4.10.1790. [DOI] [PubMed] [Google Scholar]
- 9.O’Connor M, Gregory ST, RajBhandary UL, Dahlberg AE. Altered discrimination of start codons and initiator tRNAs by mutant initiation factor 3. RNA. 2001;7:969–978. doi: 10.1017/s1355838201010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCutcheon JP, Agrawal RK, Philips SM, Grassucci RA, Gerchman SE, Clemons WM, Jr, Ramakrishnan V, Frank J. Location of translational initiation factor IF3 on the small ribosomal subunit. Proc. Natl Acad. Sci. USA. 1999;96:4301–4306. doi: 10.1073/pnas.96.8.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dallas A, Noller HF. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell. 2001;8:855–864. doi: 10.1016/s1097-2765(01)00356-2. [DOI] [PubMed] [Google Scholar]
- 12.Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. How initiation factors maximize the accuracy of tRNA selection in initiation of bacterial protein synthesis. Mol. Cell. 2006;23:183–193. doi: 10.1016/j.molcel.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 13.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 14.Selmer M, Dunham CM, Murphy FV, 4th, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 15.Hui A, de Boer HA. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl Acad. Sci. USA. 1987;84:4762–4766. doi: 10.1073/pnas.84.14.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lancaster L, Noller HF. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol. Cell. 2005;20:623–632. doi: 10.1016/j.molcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Das G, Thotala TK, Kapoor S, Sheelarani K, Thakur SS, Singh NS, Varshney U. Role of 16S ribosomal RNA methylations in translation initiation in Escherichia coli. EMBO J. 2008;27:840–851. doi: 10.1038/emboj.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. NY: Cold Spring Harbor Laboratory, Cold Spring Harbor; 1989. [Google Scholar]
- 19.Thanedar S, Kumar NV, Varshney U. The fate of the initiator tRNAs is sensitive to the critical balance between interacting proteins. J. Biol. Chem. 2000;275:20361–20367. doi: 10.1074/jbc.M001238200. [DOI] [PubMed] [Google Scholar]
- 20.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller JH. In Experiments in Molecular Genetics. NY: Cold Spring Harbor Laboratory, Cold Spring Harbor; 1972. Generalized transduction: use of P1 in strain construction; pp. 201–205. [Google Scholar]
- 22.Varshney U, RajBhandary UL. Initiation of protein synthesis from a termination codon. Proc. Natl Acad. Sci. USA. 1990;87:1586–1590. doi: 10.1073/pnas.87.4.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandal N, Mangroo D, Dalluge JJ, McCloskey JA, RajBhandary UL. Role of the three consecutive G:C base pairs conserved in the anticodon stem of initiator tRNAs in initiation of protein synthesis in Escherichia coli. RNA. 1996;2:473–482. [PMC free article] [PubMed] [Google Scholar]
- 24.Nagase T, Ishii S, Imamoto F. Differential transcriptional control of the two tRNA(fMet) genes of Escherichia coli K-12. Gene. 1988;67:49–57. doi: 10.1016/0378-1119(88)90007-8. [DOI] [PubMed] [Google Scholar]
- 25.Kenri T, Kohno K, Goshima N, Imamoto F, Kano Y. Construction and characterization of an Escherichia coli mutant with a deletion of the metZ gene encoding tRNAf1Met. Gene. 1991;103:31–36. doi: 10.1016/0378-1119(91)90387-q. [DOI] [PubMed] [Google Scholar]
- 26.Capone JP, Sedivy J, Sharp PA, RajBhandary UL. Introduction of UAG, UAA and UGA nonsense mutations at a specific site in Escherichia coli chloramphenicol acetyltransferase gene: use in measurement of amber, ochre and opal suppression in mammalian cells. Mol. Cell. Biol. 1986;6:3059–3067. doi: 10.1128/mcb.6.9.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of eukarya, archaea, and bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harley CB, Reynolds RP. Analysis of E. coli promoters. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu. Rev. Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- 30.Gualerzi C, Pon CL. Initiation of mRNA translation in prokaryotes. Biochemistry. 1990;29:5881–5889. doi: 10.1021/bi00477a001. [DOI] [PubMed] [Google Scholar]
- 31.Krin E, Laurent-Winter C, Bertin PN, Danchin A, Kolb A. Transcription regulation coupling of the divergent argG and metY promoters in Escherichia coli K-12. J. Bacteriol. 2003;185:3139–3146. doi: 10.1128/JB.185.10.3139-3146.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dittmar KA, Sørensen MA, Elf J, Ehrenberg M, Pan T. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 2005;6:151–157. doi: 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conesa C, Ruotolo R, Soularue P, Simms TA, Donze D, Sentenac A, Dieci G. Modulation of yeast genome expression in response to defective RNA polymerase III-dependent transcription. Mol. Cell. Biol. 2005;25:8631–8642. doi: 10.1128/MCB.25.19.8631-8642.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanduc D. Changes of tRNA population during compensatory cell proliferation: differential expression of methionine-tRNA species. Arch. Biochem. Biophys. 1997;342:1–5. doi: 10.1006/abbi.1996.9869. [DOI] [PubMed] [Google Scholar]
- 35.Tang M, Shen X, Frank EG, O’Donnell M, Woodgate R, Goodman MF. UmuD'(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl Acad. Sci. USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishihama A. Modulation of the nucleoid, the transcription apparatus, and the translation machinery in bacteria for stationary phase survival. Genes Cells. 1999;4:135–143. doi: 10.1046/j.1365-2443.1999.00247.x. [DOI] [PubMed] [Google Scholar]
- 37.Guillon JM, Mechulam Y, Schmitter JM, Blanquet S, Fayat G. Disruption of the gene for Met-tRNA(fMet) formyltransferase severely impairs growth of Escherichia coli. J. Bacteriol. 1992;174:4294–4301. doi: 10.1128/jb.174.13.4294-4301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newton DT, Creuzenet C, Mangroo D. Formylation is not essential for initiation of protein synthesis in all eubacteria. J. Biol. Chem. 1999;274:22143–22146. doi: 10.1074/jbc.274.32.22143. [DOI] [PubMed] [Google Scholar]
- 39.Nilsson AI, Zorzet A, Kanth A, Dahlstrom S, Berg OG, Andersson DI. Reducing the fitness cost of antibiotic resistance by amplification of initiator tRNA genes. Proc. Natl Acad. Sci. USA. 2006;103:6976–6981. doi: 10.1073/pnas.0602171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seshadri A, Dubey B, Weber MH, Varshney U. Impact of rRNA methylations on ribosome recycling and fidelity of initiation in Escherichia coli. Mol. Microbiol. 2009;72:795–808. doi: 10.1111/j.1365-2958.2009.06685.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.