Abstract
p53 coordinates the expression of an intricate network of genes in response to stress signals. Sequence-specific DNA binding is essential for p53-mediated tumor suppression. We evaluated the impact of single-nucleotide polymorphisms (SNPs) in p53 response elements (p53RE) on DNA binding and gene expression in response to DNA damage. Using a bioinformatics approach based on incorporating p53 binding strength into a position weight matrix, we selected 32 SNPs in putative and validated p53REs. The microsphere assay for protein–DNA binding (MAPD) and allele-specific expression analysis was employed to assess the impact of SNPs on p53-DNA binding and gene expression, respectively. Comparing activated p53 binding in nuclear extracts from doxorubicin- or ionizing radiation (IR)-treated human cells, we observed little difference in binding profiles. Significant p53 binding was observed for most polymorphic REs and several displayed binding comparable to the p21 RE. SNP alleles predicted to lower p53 binding indeed reduced binding in 25 of the 32 sequences. Chromatin immunoprecipitation-sequencing in lymphoblastoid cells confirmed p53 binding to seven polymorphic p53 REs in response to doxorubicin. In addition, five polymorphisms were associated with altered gene expression following doxorubicin treatment. Our findings demonstrate an effective strategy to identify and evaluate SNPs that may alter p53-mediated stress responses.
INTRODUCTION
Although most of the 14.5 million validated human single-nucleotide polymorphisms (SNPs) listed in the National Center for Biotechnology Information (NCBI) dbSNP database (Build 131) are likely nonfunctional, some can alter cellular responses to a variety of perturbations such as DNA damage, and thereby increase susceptibility to diseases including cancer (1,2). Coding SNPs alter amino acid sequences and can modify structural and biological properties of encoded proteins. Alternatively, regulatory SNPs affect non-coding gene regions such as introns, enhancers, silencers and promoters. These non-coding sequence variants can modify transcription factor binding sites (3) or generate novel binding sites (4–6), which may modulate gene expression in an allele-specific manner (7–11). Nevertheless, the impact of non-coding regulatory SNPs on transcription factor binding and gene expression has been relatively unexplored.
The p53 tumor suppressor is a prominent transcription factor that regulates the expression of an intricate network of genes in response to DNA damage and other stress signals (12). This protein coordinates transcriptional programs primarily by binding to specific DNA sequences, known as p53 response elements (p53REs). The p53 consensus DNA-binding sequence is composed of two decameric half-sites, RRRCWWGYYY (where W = A or T, R = purine and Y = pyrimidine), separated by a 0–13-bp spacer (13–15). Importantly, sequence-specific DNA binding is essential for p53 to exert its tumor suppressive activity (16–18).
Although p53 plays a significant role in protecting cells against DNA damage (e.g. carcinogenesis), the biological impact of SNPs in p53 binding sites is largely unknown. Experimental studies have identified over 150 functional human p53REs (12) and genome-wide analyses have proposed many putative sites where the p53 protein may bind (19,20). It is likely that many SNPs occur within p53 binding sites, and we have previously identified several polymorphisms in putative p53REs that alter p53-DNA binding and transactivation capacity in an allele-specific manner (21,22). These findings suggest that polymorphisms in non-coding gene regulatory regions can modify transcriptional responses and may represent genetic risk factors for disease susceptibility. Using a previously described unique bioinformatics approach (22) that incorporates experimentally based p53 binding data, we conducted a comprehensive search for functional regulatory SNPs in experimentally validated (e.g. bona fide) and putative p53REs that might affect binding. This in silico strategy provides quantitative predictions of allele-specific binding and was used to identify and select candidate SNPs that were expected to alter p53 binding and gene expression in response to DNA-damaging agents.
The microsphere assay for protein–DNA binding (MAPD) (22,23) was used to evaluate allele-specific p53 binding to selected sequences. This assay utilizes fluorescent microspheres coated with oligonucleotides that contain specific binding sequences. Microsphere-oligonucleotide targets were incubated in nuclear extracts from cells that contain endogenous levels of ‘activated’ p53, and DNA-bound p53 was detected using a flow cytometer. An advantage of this assay is its ability to produce quantitative and biologically relevant protein–DNA binding measurements.
In this study, we measured the ability of p53 in nuclear extracts from human lymphoblastoid and U2OS osteosarcoma cells treated with doxorubicin (DOXO) or ionizing radiation (IR) to bind six bona fide and 26 putative p53REs containing SNPs. Most elements displayed significant binding above the negative control, and several putative elements were bound at levels similar to bona fide p53 binding sites. For most tested sequences, the alleles that were predicted to lower p53 binding indeed reduced binding, and the observed effects were independent of cell type or treatment. Chromatin immunoprecipitation-sequencing (ChIP-Seq) studies in cultured human cells confirmed that activated p53 bound to several sequences containing candidate SNPs. Furthermore, gene expression analysis demonstrated that several SNPs were associated with reduced gene transcription. These results illustrate that our bioinformatic approach combined with the MAPD binding assay and gene expression analysis is an effective strategy to identify functional SNPs that modify p53-mediated responses to stress signals and to discover novel p53REs.
MATERIALS AND METHODS
Cell culture
HapMap (http://hapmap.ncbi.nlm.nih.gov/) CEU human lymphoblastoid cells (Coriell Cell Repositories, Camden, NJ, USA) were grown in RPMI 1640 medium supplemented with 15% heat-inactivated fetal bovine serum (Invitrogen, Carlsbad, CA, USA) and 1X antibiotics/antimycotics (Gibco, Carlsbad, CA, USA). Human U2OS osteosarcoma cells (HTB-96, ATCC, Manassas, VA, USA) were grown in McCoy’s 5A medium supplemented with 10% heat-inactivated fetal bovine serum and 1X antibiotics/antimycotics. Cells were incubated at 37 °C with 5% CO2. During experiments, cells were grown in petri dishes and treated with 0.3 or 0.6 µg/ml doxorubicin (Sigma, St. Louis, MO, USA) for 18 h or exposed to 15 Gy ionizing radiation using a Shepherd 137Cesium irradiator at a dose rate of 0.85 krad/min followed by a 5-h incubation at 37°C.
Western blot
Nuclear proteins were isolated from untreated and treated cells using the Protein Extract Kit (Active Motif, Carlsbad, CA, USA). Proteins were quantified using the Bradford assay and a HTS 7000TM Bio Assay Reader (Perkin Elmer, Waltham, MA USA). Equal amounts (10 µg) of nuclear proteins were electrophoresed on a NuPAGE® 4–12% BisTris gel (Invitrogen) and transferred to a 0.45-µM nitrocellulose membrane (Invitrogen). Membranes were probed with primary anti-p53 mouse monoclonal antibodies (DO-7; BD Biosciences, San Jose, CA, USA) or anti-β-actin mouse polyclonal antibodies (C-11; Sigma). Proteins were detected using goat anti-mouse HRP-conjugated secondary antibodies (Bio-Rad, Hercules, CA, USA) and the enhanced chemiluminescence (ECL) detection system (Amersham, Piscataway, NJ, USA).
Identification of putative functional SNPs in p53REs
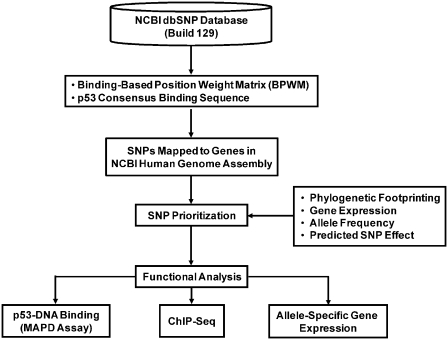
By implementing a modification of the approach of Veprintsev and Fersht (24), we created a binding-based position weight matrix (BPWM) model for quantitatively predicting p53-DNA binding (22) and incorporated this into our previous SNP identification strategy (25,26). Briefly, a delta-binding matrix was generated by subtracting experimentally measured p53 binding values for 60 variants of the ConA sequence (a perfect match to the p53 consensus binding motif) from binding to the unmodified sequence. Equations (2), (A.1) and (A.2) from Veprintsev and Fersht (24) were applied to the delta-binding matrix to generate the BPWM model.
In our current study, the BPWM model was used to identify and score SNPs within the human genome that alter p53-DNA binding. Briefly, utilizing the delta-binding matrix, each allele was scored independently, and the difference between the two scores was used to estimate the SNP impact on p53-DNA binding. We identified SNPs in putative p53REs by searching the NCBI dbSNP (Build 129) database for polymorphisms in sequences that matched the p53-DNA binding motif. We identified SNPs in validated p53REs by mapping NCBI dbSNP database entries into a list of 150 experimentally discovered p53REs.
Several SNPs in putative p53REs (rs2838769, rs3806624, rs1658728, rs3761624 and rs1465952) have previously been tested in our group using MAPD (22) and luciferase expression assays (21). These SNPs were included in this study to evaluate their effects in different cell types and under different experimental conditions.
Microsphere assay for protein–DNA binding
Allele-specific p53 binding was evaluated using the MAPD assay, as previously described (22,23). Briefly, MicroPlex™-xTAG microspheres (Luminex, Austin, TX, USA) were hybridized to double-stranded oligonucleotides (Invitrogen) containing unique p53 binding sites and a biotin moiety on the 5′-end of the reverse strand. Hybridization reactions were carried out in a polymerase chain reaction (PCR) thermocycler. Microsphere-oligonucleotide targets were pooled and washed in a MultiScreen®HTS-BV filter plate (Millipore, Bedford, MA, USA) with Assay Buffer A (Cytokine Reagent Kit, Bio-Rad).
Sequence-specific p53 binding was examined 1 h after incubating targets in reaction mixtures that contained non-competing double-stranded oligonucleotides (TransAm p53 kit, Active Motif) and nuclear extracts from doxorubicin- or IR-exposed human cells. Negative and positive control sequences were examined in each reaction. Nuclear extracts from untreated cells served as an additional negative control. Microspheres were washed and incubated for 30 min with primary anti-p53 monoclonal antibodies (DO-7) followed by a 30-min incubation with phycoerythrin-conjugated secondary antibodies. p53 DNA binding was measured by flow cytometric analysis using a Bio-Plex® 200 System (Bio-Rad).
p53 ChIP-Seq
Chromatin immunoprecipitation methods and buffers were from the Agilent Mammalian ChIP-on Chip protocol (Agilent, Version 10.0, May 2008). Briefly, 108 DOXO or untreated lymphoblastoid cells (GM06993 and GM11992) were formaldehyde crosslinked and lysed to obtain nuclear cell pellets. Pellets were resuspended, chromatin was sonicated into 200–500-bp fragments using a Misonix 3000 sonicator (30 pulses—30 s on followed by 20 s off—at 100% power) (Farmingdale, NY, USA), and the supernatant was collected after centrifugation. Using either mouse monoclonal antibody DO-7 (BD Biosciences) or a non-specific rabbit IgG (Invitrogen) conjugated to secondary Dynal magnetic beads (Invitrogen), 5 × 107 isolated nuclei were immunoprecipitated overnight (4°C), beads were washed, and protein–DNA crossslinks reversed overnight (65°C). After digestion of RNA and cellular protein, DNA was isolated using phenol chloroform extraction and a phase lock gel (Sigma), and subsequently purified using Qiaquick PCR Purification Kit (Qiagen, Germantown, MD, USA) according to the manufacturer’s instructions. Purified ChIP DNA was dried, resuspended in double-distilled H2O, and quantified using PicoGreen (Invitrogen). p53 ChIP DNA from the two cell lines were combined for each condition (DOXO-treated, untreated, and IgG ChIP).
The National Center for Genome Resources (Santa Fe, NM, USA) sequenced the immunoprecipitated samples on the Illumina Genome Analyzer II. Briefly, sequencing adapters were ligated to immunoprecipitated DNA and size-selected by gel electrophoresis (250 ± 25 bp). Fragments were PCR-amplified (18 cycles) and sequenced. Short reads were mapped to the human reference genome (Build 36.3) using Burrows-Wheeler Alignment (BWA) Tool (27). By default, BWA finds an alignment (ungapped and gapped) with maximum edit-distance 2 to the 36 bp query sequence, except for disallowing gaps close to the end of the query.
Uniquely mapped short reads from p53-immunoprecipitated DNA were used to identify regions of the genome with significant enrichment in p53-associated DNA sequences, hereafter referred to as ‘peaks’ owing to their appearance in genome-wide density plots. The peak detection was performed by QuEST 2.4 software (28) using a ‘transcription factor binding site’ setting (bandwidth of 30 bp, region size of 300 bp) and default stringency (corresponding to 50-fold ChIP to input enrichment for seeding the regions, and 3-fold ChIP enrichment for extending the regions).
Gene expression analysis
To evaluate the allele-specific effects of SNPs on gene expression, 23 HapMap CEU human lymphoblastoid cell lines were cultured in triplicate and treated with 0.6 µg/ml doxorubicin for 18 h. RNA was isolated with RNeasy columns (Qiagen) and cDNA was generated using the First-Strand Synthesis system (Invitrogen). For each biological replicate, target genes were amplified in triplicate using SYBR primers designed to span exon junctions for each target gene (see Supplementary Table S2) and Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). Fluorescence intensity was measured using a PRISM HTS 7900 System (Applied Biosystems) and the initial fluorescence (Ro value) of each amplified sample was calculated using the method described by Peirson and colleagues (29). Target values were normalized to GAPDH mRNA measurements.
RESULTS
In silico selection of candidate SNPs for functional evaluation
Using a novel bioinformatics approach (Figure 1), we identified 6538 SNPs in the NCBI dbSNP (Build 129) database that were located in putative p53 binding sites and were predicted to alter p53–DNA interactions. We also identified 33 polymorphic sequences in experimentally validated p53 binding sites. After applying additional prioritization criteria, 32 SNPs were selected for functional evaluation. Six of these variants were located within 5 kb of the transcription start site of bona fide p53-responsive genes. SNP selection was based on the requirement that: (i) each site contained all four conserved core C and G nucleotides in at least one allele; (ii) each site contained at least three of the four consensus ‘W’ (A or T) nucleotides in at least one allele; (iii) each site contained a spacer ≤1-bp separating the two half-sites; (iv) the BPWM score for the predicted strong allele was above 100 (an indicator of a moderate-to-high p53 binding sequence); and (v) the polymorphism was predicted to alter p53-DNA binding. The candidate SNPs and associated REs are described in Table 1.
Figure 1.
In silico selection of candidate p53RE SNPs. Computational analysis was used to identify human SNPs in experimentally validated and putative p53REs that are predicted to alter p53 binding.
Table 1.
Candidate p53RE SNPs
| SNP rs# | Associated gene | Sequence alignment RRRCWWGYYYRRRCWWGYYY | Strong allele (BPWM) | Weak allele (BPWM) | ΔBPWM | Cellular process |
|---|---|---|---|---|---|---|
| Validated response elements | ||||||
| rs3809024 | CASP1 | GT*CATGCATATGCATGTCT | G (123.0) | A (119.0) | 4.0 | Apoptosis |
| rs62216320 | COL18A1 | TGACA*GTGTGAGCATGTAT | T (126.1) | C (120.0) | 6.1 | Extracellular matrix |
| rs11572355 | EDN2 | CTGCAAGCCC*GGCATGCCC | G (129.4) | A (128.0) | 1.4 | Vasoconstriction |
| rs62122432 | GDF15 | CATCTTG*CCAGACTTGTCT | C (123.0) | T (117.3) | 5.7 | Cell signaling |
| rs28999675 | RRM2B | TGACATGCC*AGGCATGTCT | C (134.7) | G (132.4) | 2.3 | Nucleic acid metabolism |
| rs35926783 | TRIM22 | TGACATGTCT*GGCATGTAG | G (137.3) | A (135.0) | 2.3 | Immune response |
| Putative response elements | ||||||
| rs2838769 | ADARB1 | GGACAAGTTGAAACTT*CAC | G (113.1) | A (100.7) | 12.4 | Nucleic acid metabolism |
| rs2033654 | ADCY3 | GGACCT*GCCTAACATGTCA | G (108.3) | T (93.9) | 14.4 | Intracellular signaling |
| rs1658728 | ARHGEF7 | AAACATGTCA*CACTTGCTT | G (124.2) | T (121.8) | 2.4 | Apoptosis |
| rs3806624 | EOMES | CAACT*GCCCCAGTTTCCCC | T (117.4) | C (105.9) | 11.5 | Development |
| rs12105811 | CFLAR | GATCCTGCTTGGTC*TGTCC | A (109.5) | G (105.2) | 4.3 | Apoptosis |
| rs10917431 | FUCA1 | GGGC*TGTCCACTCAGGTCT | A (123.8) | C (112.9) | 10.9 | Carbohydrate metabolism |
| rs9282701 | HMOX1 | GAGCCAGCACGAACGAGCC* | C (103.6) | T (103.0) | 0.6 | Oxidative stress response |
| rs11855354 | IDH3A | TTTCAG*TTGTGGCCAGGCA | G (104.4) | A (90.0) | 14.4 | Carbohydrate metabolism |
| rs3755276 | IL18R1 | ATGCATGTGTTCGC*TGTGT | A (124.0) | G (119.7) | 4.3 | Immune response |
| rs2410545 | NAT1 | CCTC*TGTCAGGGCATGGGA | A (122.8) | G (111.3) | 11.5 | Xenobiotic metabolism |
| rs1941404 | NNMT | TAACA*GCACACACATGGGC | T (123.0) | C (118.7) | 4.3 | Nucleic acid metabolism |
| rs12409754 | PGD | GGGCA*GGTGTTGCATGCCT | T (118.2) | C (112.1) | 6.1 | Carbohydrate metabolism |
| rs7240884 | PMAIP1 | CAACAT*TTTGATCTTGTAG | G (112.1) | A (97.7) | 14.4 | Apoptosis |
| rs1048990 | PSMA6 | GTGCTT*TACCAACATGTCC | G (112.4) | C (97.8) | 14.6 | Protein metabolism |
| rs1003619 | RANGAP1 | CC*CAAGCCCCACCAAGCCC | G (112.1) | C (97.7) | 14.4 | Cell signaling |
| rs6017447 | RPN2 | CACCATGT*CgGCTCATGTTT | C (119.6) | G (115.8) | 3.8 | Protein metabolism |
| rs1465952 | RRM1 | GGG*ATGTGCATTCAAGTTT | C (120.6) | T (108.1) | 12.5 | Nucleic acid metabolism |
| rs27078 | SFRS12 | TGGCATGTCTTCA*ATGTAG | C (125.6) | G (111.0) | 14.6 | Nucleic acid metabolism |
| rs505802 | SLC22A12 | CCAC*TGCCCtGTACATGCTT | A (136.2) | G (132.0) | 4.2 | Ion transport |
| rs1678199 | SP100 | TGGCA*GTATTGGCATGACC | A (125.4) | C (124.8) | 0.6 | Immune response |
| rs3761624 | TLR8 | AGGCAAGATGAAACAT*TCA | G (111.9) | A (101.1) | 10.8 | Immune response |
| rs1077667 | TNFSF14 | GTGC*TGTACCCACATGTCC | A (123.5) | G (112.0) | 11.5 | Immune response |
| rs3783231 | TPP2 | CCACATG*AAAGGCATGTAT | T (126.4) | C (121.8) | 4.6 | Protein metabolism |
| rs10455025 | TSLP | GAACATGGCA*GACATGAAA | A (123.8) | C (122.6) | 1.2 | Extracellular matrix |
| rs3853154 | UBA3 | AGACATGTATCATCATGC*C | C (118.1) | A (114.6) | 3.5 | Protein metabolism |
| rs11248877 | UNKL | GGGC*TGGTGGCACATGCCT | A (119.2) | G (115.0) | 4.2 | Nucleic acid metabolism |
Thirty-two candidate p53RE SNPs were selected for functional evaluation. Six SNPs are associated with bona fide p53 regulated genes. Genes located near the SNPs are listed. Sequences and SNP position (asterisk) within the element are shown. Sequences are aligned to the p53 consensus binding sequence (lower case letters represent 1-bp spacers). The predicted strong and weak allele is shown for each site along with the corresponding predicted binding value (BPWM). ΔBPWM indicates the negative effect each SNP is predicted to have on p53 binding.
MAPD evaluation of allele-specific p53 binding to polymorphic sequences
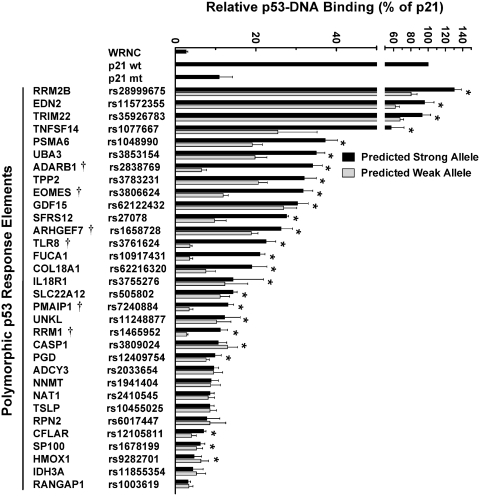
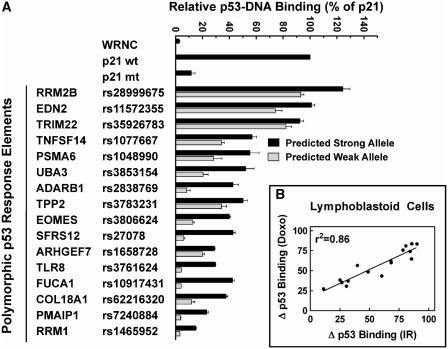
To overcome the limitations of traditional methods used to quantify protein–DNA binding, we have developed a microsphere-based DNA binding assay to measure p53-RE interactions (22). As seen in Figure 2, allele-specific binding was observed after incubating bead-oligonucleotide targets in nuclear extracts from human lymphoblastoid cells that were either untreated or treated with doxorubicin (DOXO). p53 binding levels in untreated samples were <2% of levels observed in treated samples. In addition, allele-specific differences in binding were not observed in untreated cells (data not shown). In contrast, p53 in extracts from DOXO-treated cells bound to the predicted strong allele of 29 REs at levels that were above the negative control sequence (WRNC). Moreover, the tumor suppressor bound several validated REs at levels that were comparable to or greater than those observed for the positive control (a bona fide p53RE ∼2 kb upstream of the p21 gene transcription start site) (see EDN2, RRM2B and TRIM22). Some putative REs bound p53 at levels similar to validated p53 binding sites (see ADARB1, EOMES, PSMA6, TNFSF14, TPP2 and UBA3). Compared to the predicted strong alleles, binding to the predicted weak alleles was significantly reduced for ∼80% of the elements (P < 0.05, t-test). For 10 of these variants, the SNP diminished p53 binding by ≥50%. Furthermore, the polymorphic sequences associated with FUCA1, PMAIP1, RRM1 and TLR8 nearly abolished the ability of p53 to recognize the binding sites. It should also be noted that our bioinformatic calculations were generally accurate for predicting p53 binding to REs with high scores (>100) and for estimating the SNP effect on binding.
Figure 2.
Evaluation of sequence-specific p53 DNA-binding in human cells. The MAPD assay was used to assess p53 binding to predicted strong (black bars) and weak (gray bars) alleles in nuclear extracts from DOXO-treated (0.6 µg/ml for 18 h) human lymphoblastoid cells that were supplemented with 150 pmol of non-competing oligonucleotides. p53 binding is displayed relative to a bona fide p53RE upstream of the p21 gene (set to 100%). A negative control sequence (WRNC) was included in each experiment. Polymorphic alleles that displayed significant differences in p53 binding (P < 0.05, t-test) are indicated by (asterisk). SNPs previously tested under these experimental conditions using the MAPD assay (21,22) are indicated by (†). Error bars represent the standard deviation of three independent experiments carried out in triplicate.
ChIP-Seq confirms p53 binding to sequences containing SNPs
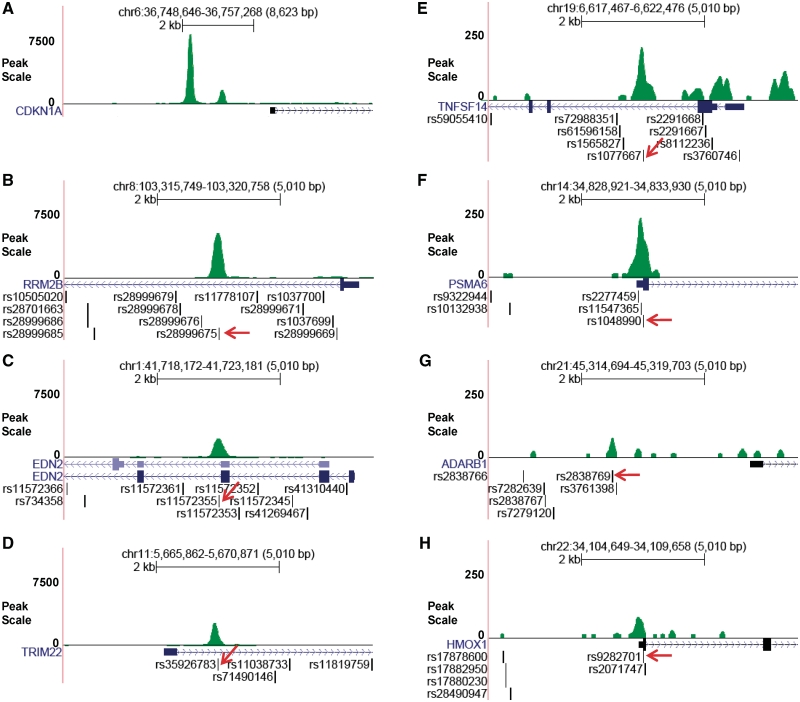
To date, only one p53 ChIP-seq study has been published, which was carried out in a colon cancer cell line treated with 5-fluorouracil (5-FU) (20). We applied ChIP-Seq to HapMap lymphoblastoid cells to determine whether DOXO induces p53 to bind to our selected sequences. Detailed information on the ChIP-Seq experiment and data analysis will be reported separately. Here, we only show limited data corresponding to the genomic regions selected for this study. After mapping sequenced p53-ChIP fragments to the human reference genome (Build 36.3), we observed that p53-bound sequences were enriched for seven of the 32 tested binding sites (Figure 3). It should be noted that four of these p53-bound sites correspond to sequences that had not been previously confirmed as p53 binding sites. Additional sequence analysis revealed that the candidate SNPs occurred within the p53-bound sites. These findings combined with our MAPD binding data suggest that a number of the bioinformatics-selected sequences are indeed bound by p53 in cultured human cells treated with DOXO and that the candidate SNPs have the potential to modify p53 binding in vivo.
Figure 3.
Evaluation of p53-DNA binding in human cells using ChIP-Seq. Human lymphoblastoid cells were treated with 0.3 µg/ml DOXO for 18 h and crosslinked with formaldehyde. Genomic DNA was sonicated, immunoprecipitated with p53 antibodies, sequenced and mapped to the human reference genome. Green peaks in (A–H) represent sequenced reads from DOXO-induced p53–DNA binding in the regulatory region of nearby genes. QuEST software was used to normalize p53 ChIP-Seq reads to control samples (IgG ChIP-Seq reads). As a positive control, panel A displays p53 ChIP-Seq peaks that contain bona fide p53REs which are located within the CDKN1A (p21) promoter region (see also Supplementary Figure S3). (B–F) illustrates that the SNPs are located within the p53 binding sites (denoted by arrows). Note: (A–D) display strong p53-DNA binding, and show relatively weak binding.
Doxorubicin induces allele-specific transcription in human cells
To determine if the selected SNPs alter gene transcription, we performed RT-qPCR on RNA isolated from untreated and DOXO-treated HapMap CEU cell lines that were homozygous for either the predicted strong or weak allele. Relative to untreated cells, nine of 11 analyzed genes were induced in response to DOXO treatment (Figure 4). Allele-specific differences in gene expression were observed for ADARB1, PSMA6, TNFSF14, TRIM22 and UBA3 where the weak allele displayed lower levels of expression (P < 0.05, t-test). Expression levels for TPP2 and SFRS12 were not induced in response to DOXO, which suggests that they are not regulated by the tumor suppressor. The other tested genes had modest allelic differences in expression. These findings suggest that SNPs in p53 binding sites can alter gene expression in response to DNA-damaging agents.
Figure 4.
Evaluation of allele-specific gene expression in DOXO-treated human cells. Allele-specific expression of genes adjacent to polymorphic REs was measured in HapMap human CEU lymphoblastoid cell lines after exposure to 0.6 µg/ml DOXO for 18 h. RT-qPCR analysis was carried out on cell lines that were homozygous for the predicted strong or weak allele. Expression was normalized to GAPDH expression levels. Values represent induction levels relative to untreated cells. The number of homozygous cell lines examined for each allele is denoted by ‘n’. Homozygous weak alleles that displayed significant differences in expression (P < 0.05, t-test) are indicated by asterisk. Error bars represent the standard deviation of triplicate RT–qPCR reactions carried out on each of three biological replicates for all cell lines.
Allele-specific p53 binding is independent of cell type-specific effects
p53 is a versatile transcription factor with distinct roles that are influenced by cell type (30) and treatment (31,32). To evaluate cell-type specific contributions to p53-DNA binding, p53 binding to a subset of REs in nuclear extracts from DOXO-treated U2OS osteosarcoma cells was compared to results observed in lymphoblastoid cells. REs that displayed either weak p53 binding (<10% relative to the positive control) or no allele-specific binding differences in DOXO-treated lymphoblastoid nuclear extracts were excluded from further analysis. As seen in Figure 5A, DOXO-activated p53 from U2OS cells bound each sequence at levels that were comparable to DOXO-activated p53 from lymphoblastoid cells (see Figure 2). A linear regression analysis of binding in lymphoblastoid versus U2OS extracts produced r2 values of 0.92 and 0.94 for strong and weak alleles, respectively (data not shown). Linear regression analysis comparing SNP-induced binding changes in both cell lines demonstrated that the SNP effect on binding were also similar (r2 = 0.87, Figure 5B).
Figure 5.
Evaluation of cell type-specific effects on p53 binding to polymorphic alleles. (A) The MAPD assay was used to assess p53 binding to predicted strong (black bars) and weak (gray bars) alleles in nuclear extracts from DOXO-treated (0.6 µg/ml for 18 h) human U2OS cells. All other experimental conditions and data analysis was performed as described in Figure 2. Each allele pair displayed significant differences in binding (P < 0.05, t-test). Error bars represent the standard deviation of three independent experiments carried out in triplicate. (B) Regression analysis was used to compare the effect of each SNP on p53 binding in nuclear extracts from DOXO-treated human lymphoblastoid versus U2OS cells.
To further evaluate cell type-specific contributions to p53-RE binding, we compared binding in nuclear extracts from IR-exposed lymphoblastoid and U2OS cells. As seen in Supplementary Figure S1A (lymphoblastoid cells) and B (U2OS cells), IR-activated p53 binding levels were similar in both extracts (r2 = 0.95 and 0.97 for strong and weak alleles, respectively; data not shown). The SNP effects on p53 binding also were similar in both cell types (r2 = 0.90, Supplementary Figure S1C). These results demonstrate that the effects of the polymorphisms on p53-DNA binding are independent of cell-type specific differences between the two cell lines.
Allele-specific p53 binding is independent of treatment-specific effects
To determine if treatment-specific effects alter p53–DNA interactions, we compared binding in nuclear extracts from IR-exposed lymphoblastoid cells to results observed in extracts from DOXO-treated cells. In Figure 6A, the pattern of IR-induced p53 binding to the predicted strong alleles was similar to the pattern induced by DOXO (see Figure 2). As seen with DOXO, reduced binding to the weak alleles was also observed in extracts from IR-exposed cells (P < 0.05 for all allele pairs, t-test). Moreover, a strong correlation was shown for SNP-induced changes on p53 binding between DOXO and IR treatment (r2 = 0.86, Figure 6B).
Figure 6.
Evaluation of treatment-specific effects on p53 binding to polymorphic alleles. (A) The MAPD assay was used to assess p53 binding to predicted strong (black bars) and weak (gray bars) RE alleles in nuclear extracts from IR-treated (15 Gy followed by a 5-h recovery) human lymphoblastoid cells. All other experimental conditions and data analysis was performed as described in Figure 2. Each allele pair displayed significant differences in p53 binding (P < 0.05, t-test). Error bars represent the standard deviation of three independent experiments carried out in triplicate. (B) Regression analysis was used to compare the effect of each SNP on p53 binding in nuclear extracts from human lymphoblastoid cells treated with DOXO versus IR.
To further determine if treatment-specific effects modify p53-RE binding, we compared p53 binding in nuclear extracts from DOXO- and IR-exposed U2OS cells. As seen in Supplementary Figure S2, the SNP effects on p53 binding were also similar in response to both treatments (r2 = 0.73). These findings indicate that the impact of sequence variation on p53-RE binding is mostly unaffected by treatment-specific effects induced by these DNA-damaging agents.
SNP sequence position influences the polymorphic effects on p53 binding
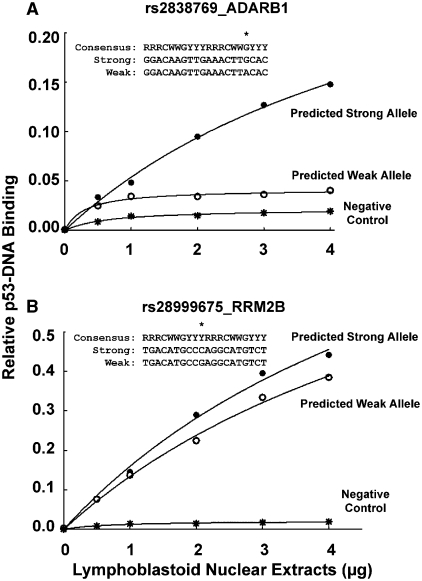
Although base changes involving the highly conserved C and G nucleotides at positions 4 and 7 in each p53RE half-site significantly reduce p53 binding, most functional REs deviate from the consensus binding sequence (22,33,34). Variations in p53RE sequences and p53 protein levels contribute greatly to gene transactivation mediated by the tumor suppressor (23,33,35). Understanding this relationship is important for determining how p53 influences cellular responses to stress. To further examine the effects of polymorphic sequences on p53-DNA binding, we evaluated ADARB1 and RRM2B RE allele pairs in the presence of increasing amounts (0–4 µg) of nuclear extracts from DOXO-treated lymphoblastoid cells. The RE associated with ADARB1 contains a G-to-A change at position 7 in the second half-site, while the RE associated with RRM2B has a C-to-G change adjacent to position 4 and 7. As seen in Figure 7A, modification of the preferred G in the ADARB1 RE dramatically reduced p53 binding. For example, at 4 µg of nuclear extracts, the predicted weak allele reduced p53 binding by 3.5-fold relative to the predicted strong allele.
Figure 7.
Evaluation of SNP sequence position on p53–DNA binding. The MAPD assay was used to assess p53 binding to predicted strong (closed circles) and weak (open circles) RE alleles that are associated with (A) ADARB1 and (B) RRM2B. Beads bearing the specific REs were incubated with increasing amounts of nuclear extracts (0–4 µg) from DOXO-treated (0.6 µg/ml for 18 h) human lymphoblastoid cells in the presence of 150 pmol of non-competing oligonucleotides. A negative control sequence (WRNC, asterisks) was included. Polymorphic sequences are aligned to the p53 consensus RE sequence. The asterisk above the sequences represents the position of the polymorphic nucleotide. Binding values represent the mean of three independent experiments carried out in triplicate.
The base change in the RRM2B RE had modest effects on p53 binding compared to the strong allele (see Figure 7B). At 4 µg of nuclear extracts, p53 binding was 0.44 and 0.39 for the predicted strong and weak allele, respectively. These results suggest that polymorphisms that alter the highly favored bases at position 4 and 7 in p53RE half-sites can have significant effects on p53-mediated responses to DNA damage, whereas other changes may be better tolerated.
DISCUSSION
Non-coding regulatory SNPs in transcription factor binding sites can have significant biological consequences (1). We carried out a comprehensive genome-wide bioinformatics search using a unique prediction model (BPWM), to identify human SNPs in validated and putative p53-targeted sequences that were predicted to alter p53 binding following exposure to DNA-damaging agents. The selected SNPs were evaluated using the MAPD binding assay and gene expression analysis to determine the functional impact of each variant. We employed ChIP-Seq analysis to confirm p53 sequence-specific binding in cultured human cells in response to doxorubicin, a DNA-damaging agent. To our knowledge, this represents the first study that examines properties of SNPs that co-locate with ChIP-seq peaks. The allele-specific differences observed for p53 target genes following exposure to doxorubicin support the hypothesis that genetic variation in non-coding regulatory sequences may impact risk in environmentally induced diseases such as cancer (1). Our findings also demonstrate that this bioinformatics strategy can identify SNPs in gene regulatory regions that alter p53–DNA binding and gene expression. In addition, we believe regulatory SNPs that affect other transcription factors or DNA binding proteins can be identified and evaluated using a similar approach.
Many of the sequences we identified have not been experimentally validated as p53REs. Our results, using MAPD, gene expression and ChIP-seq methods, demonstrate that p53 binds putative polymorphic REs near ADARB1, HMOX1, PSMA6 and TNFSF14 in cultured human cells, which suggests that these genes are novel, previously unidentified components of the p53 transcriptional network. Several polymorphic sequences that were tested are associated with genes that have tumor suppressive activity, play roles in cell cycle regulation, or function in other cellular processes associated with p53-mediated signaling pathways. ADARB1 is a member of the adenosine deaminase family, which plays important roles during developmental processes. This RNA-editing enzyme is suggested to have negative effects on the development and progression of brain cancer (36). PSMA6 is a member of the peptidase T1A family and a subunit of the proteasome 20S catalytic core. The core is involved in cell-cycle regulation and differentiation and is expressed in malignant cells (37,38). PSMA6 is involved in the development of hepatocellular carcinoma (39) and breast cancer (38). TNFSF14 is a ligand for a member of the tumor necrosis factor receptor superfamily, which stimulates the proliferation of T cells and triggers apoptosis of various tumor cells (40,41). TRIM22 is an interferon-inducible protein and is a p53 target gene. TRIM22 has antiproliferative effects in leukemic cells (42) and is suggested to play lineage-specific roles during hematopoietic differentiation (43). UBA3 is the catalytic subunit of the NEDD8-activing enzyme. NEDD8 is involved in the regulation of cell division and cell signaling. UBA3 is essential for the antiproliferative activity of an antiestrogen drug that is targeted against estrogen receptor alpha positive breast cancer cells (44).
In this work, we have used de novo approaches to identify and test polymorphic REs that alter p53–DNA binding. These SNPs may also alter cellular stress responses and increase disease susceptibility in vivo. Our results further illustrate the significance of the conserved C and G in the p53-DNA binding motif. We demonstrate that this observation is apparent in both moderate (ADARB1 and TLR8) and low (PMAIP1 and RRM1) affinity p53 binding sites, which suggests that sequence variations involving these nucleotides will have the most significant biological consequences. In contrast, SNPs that alter nucleotides outside of the conserved C and G may have little impact and produce no obvious phenotypic effects.
In this study, we identified SNPs that alter p53-RE binding and impact gene expression; however, the functional, evolutionary and risk implications of these effects are not always obvious. For example, we observe occasionally that the less common, minor allele is the predicted strong allele, displaying both stronger binding (e.g. rs1077667, TNFSF14; rs1048990, PSMA6) and stronger transactivation while the common (i.e. ancestral allele, shared with other primates) allele displays weaker binding. This suggests that the recently created, stronger allele may have some selective advantage in human lineages that have it. However, selective advantage (positive selection) or disadvantage (purifying selection) is related to reproductive success, while cancer risk is related to somatic processes that impact a post-reproductive disease outcome. Although it is unclear whether the creation of these specific strong-binding alleles in the human population affects physiology, survival or risk, transcriptional pathways can evolve in this way (45–47). Menendez et al. (6) have suggested that an SNP may create novel binding sites and connect transcriptional pathways. The impact of RE polymorphisms is further complicated by the possibility that specific p53RE ‘WW’ dinucleotide combinations can influence whether p53 activates or represses gene expression (48). Thus, an SNP that occurs in a RE from which p53 represses transcription could decrease p53 binding and thereby increase gene expression. This scenario may explain the increased expression of PMAIP1 in cell lines homozygous for the weak allele. Evaluating SNPs in p53REs that are predicted to repress transactivation would be of great interest.
The p53 protein plays key roles during the cellular stress response (12), which can be influenced by cell type (30). A number of reports have demonstrated that different stress-inducing agents trigger a variety of p53 post-translational modifications (49,50) and differential transactivation of target genes (51,52). Two recent publications disagree with regard to whether different agents produce different p53 occupancy profiles at native binding sites in cells. Shaked et al. (53), using ChIP-on-chip, observed that global patterns of p53 occupancy differed very little among gamma irradiated, DOXO, 5-FU or UV treated HCT116 and U2OS cell lines. Millau et al. (54) using a more sensitive technique, observed unique p53 occupancy patterns over time at different p21 response elements following treatment with 5-FU, Nutlin-3, UV and gamma radiation. The MAPD assay evaluates the p53 binding step and provides high resolution and discrimination of small binding differences. The present findings using DOXO and IR in lymphoblastoid and U2OS osteosarcoma cells clearly demonstrate that very similar p53 binding patterns are produced from p53 activated under all experimental conditions tested. Thus, it is likely that other features in the transcription process, such as the chromatin environment or stress-specific cofactors, may contribute to cell type- and treatment-specific p53 binding and subsequent transactivation in cells (51,52). In addition, we demonstrate for the first time that individual SNPs produced similar effects on p53–DNA binding regardless of the cell type or stimulus used for activation.
The present study suggests that sequence variation in p53 binding sites can impact binding and potentially modify p53-mediated responses to DNA-damaging agents. The MAPD binding assay is a useful way to estimate the functional impact of nucleotide changes in target binding sites and could be applied to other transcription factors. Developing quantitative binding data for many regulatory elements may allow us to elucidate the impact of polymorphic variation on transcriptional networks.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Intramural Research Program of the National Institute of Environmental Health Sciences, National Institutes of Health (ZO1-ES-100475-M-0001 and ZO1-ES065079-15). Funding for open access charge: The Intramural Research Program, National Institute of Environmental Health Sciences.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We appreciate the support of the Intramural Research Program of the National Institute of Environmental Health Sciences, National Institutes of Health. We thank Drs. Maher Noureddine, Brian Chorley, Michael Resnick, Scott Auerbach and Daniel Menendez for helpful comments on this project and review of the article. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the article.
REFERENCES
- 1.Hudson TJ. Wanted: regulatory SNPs. Nat. Genet. 2003;33:439–440. doi: 10.1038/ng0403-439. [DOI] [PubMed] [Google Scholar]
- 2.Kelada SN, Eaton DL, Wang SS, Rothman NR, Khoury MJ. The role of genetic polymorphisms in environmental health. Environ. Health Perspect. 2003;111:1055–1064. doi: 10.1289/ehp.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasiliev GV, Merkulov VM, Kobzev VF, Merkulova TI, Ponomarenko MP, Kolchanov NA. Point mutations within 663-666 bp of intron 6 of the human TDO2 gene, associated with a number of psychiatric disorders, damage the YY-1 transcription factor binding site. FEBS Lett. 1999;462:85–88. doi: 10.1016/s0014-5793(99)01513-6. [DOI] [PubMed] [Google Scholar]
- 4.Piedrafita FJ, Molander RB, Vansant G, Orlova EA, Pfahl M, Reynolds WF. An Alu element in the myeloperoxidase promoter contains a composite SP1-thyroid hormone-retinoic acid response element. J. Biol. Chem. 1996;271:14412–14420. doi: 10.1074/jbc.271.24.14412. [DOI] [PubMed] [Google Scholar]
- 5.Knight JC, Udalova I, Hill AV, Greenwood BM, Peshu N, Marsh K, Kwiatkowski D. A polymorphism that affects OCT-1 binding to the TNF promoter region is associated with severe malaria. Nat. Genet. 1999;22:145–150. doi: 10.1038/9649. [DOI] [PubMed] [Google Scholar]
- 6.Menendez D, Krysiak O, Inga A, Krysiak B, Resnick MA, Schonfelder G. A SNP in the flt-1 promoter integrates the VEGF system into the p53 transcriptional network. Proc. Natl Acad. Sci. USA. 2006;103:1406–1411. doi: 10.1073/pnas.0508103103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hohjoh H, Tokunaga K. Allele-specific binding of the ubiquitous transcription factor OCT-1 to the functional single nucleotide polymorphism (SNP) sites in the tumor necrosis factor-alpha gene (TNFA) promoter. Genes Immun. 2001;2:105–109. doi: 10.1038/sj.gene.6363721. [DOI] [PubMed] [Google Scholar]
- 8.Sugatani J, Yamakawa K, Yoshinari K, Machida T, Takagi H, Mori M, Kakizaki S, Sueyoshi T, Negishi M, Miwa M. Identification of a defect in the UGT1A1 gene promoter and its association with hyperbilirubinemia. Biochem. Biophys. Res. Commun. 2002;292:492–497. doi: 10.1006/bbrc.2002.6683. [DOI] [PubMed] [Google Scholar]
- 9.Ono S, Ezura Y, Emi M, Fujita Y, Takada D, Sato K, Ishigami T, Umemura S, Takahashi K, Kamimura K, et al. A promoter SNP (-1323T>C) in G-substrate gene (GSBS) correlates with hypercholesterolemia. J. Hum. Genet. 2003;48:447–450. doi: 10.1007/s10038-003-0055-x. [DOI] [PubMed] [Google Scholar]
- 10.Grant DJ, Hall IJ, Eastmond DA, Jones IM, Bell DA. Bilirubin UDP-glucuronosyltransferase 1A1 (UGT1A1) gene promoter polymorphisms and HPRT, glycophorin A, and micronuclei mutant frequencies in human blood. Mutat. Res. 2004;560:1–10. doi: 10.1016/j.mrgentox.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Somner J, McLellan S, Cheung J, Mak YT, Frost ML, Knapp KM, Wierzbicki AS, Wheeler M, Fogelman I, Ralston SH, et al. Polymorphisms in the P450 c17 (17-hydroxylase/17,20-Lyase) and P450 c19 (aromatase) genes: association with serum sex steroid concentrations and bone mineral density in postmenopausal women. J. Clin. Endocrinol. Metab. 2004;89:344–351. doi: 10.1210/jc.2003-030164. [DOI] [PubMed] [Google Scholar]
- 12.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 13.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat. Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 14.Funk WD, Pak DT, Karas RH, Wright WE, Shay JW. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol. Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tokino T, Thiagalingam S, el-Deiry WS, Waldman T, Kinzler KW, Vogelstein B. p53 tagged sites from human genomic DNA. Hum. Mol. Genet. 1994;3:1537–1542. doi: 10.1093/hmg/3.9.1537. [DOI] [PubMed] [Google Scholar]
- 16.Crook T, Marston NJ, Sara EA, Vousden KH. Transcriptional activation by p53 correlates with suppression of growth but not transformation. Cell. 1994;79:817–827. doi: 10.1016/0092-8674(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 17.Pietenpol JA, Tokino T, Thiagalingam S, el-Deiry WS, Kinzler KW, Vogelstein B. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc. Natl Acad. Sci. USA. 1994;91:1998–2002. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chao C, Saito S, Kang J, Anderson CW, Appella E, Xu Y. p53 transcriptional activity is essential for p53-dependent apoptosis following DNA damage. EMBO J. 2000;19:4967–4975. doi: 10.1093/emboj/19.18.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 20.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 21.Tomso DJ, Inga A, Menendez D, Pittman GS, Campbell MR, Storici F, Bell DA, Resnick MA. Functionally distinct polymorphic sequences in the human genome that are targets for p53 transactivation. Proc. Natl Acad. Sci. USA. 2005;102:6431–6436. doi: 10.1073/pnas.0501721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noureddine MA, Menendez D, Campbell MR, Bandele OJ, Horvath MM, Wang X, Pittman GS, Chorley BN, Resnick MA, Bell DA. Probing the functional impact of sequence variation on p53-DNA interactions using a novel microsphere assay for protein-DNA binding with human cell extracts. PLoS Genet. 2009;5:e1000462. doi: 10.1371/journal.pgen.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jordan JJ, Menendez D, Inga A, Noureddine M, Bell DA, Resnick MA. Noncanonical DNA motifs as transactivation targets by wild type and mutant p53. PLoS Genet. 2008;4:e1000104. doi: 10.1371/journal.pgen.1000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veprintsev DB, Fersht AR. Algorithm for prediction of tumour suppressor p53 affinity for binding sites in DNA. Nucleic Acids Res. 2008;36:1589–1598. doi: 10.1093/nar/gkm1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Tomso DJ, Liu X, Bell DA. Single nucleotide polymorphism in transcriptional regulatory regions and expression of environmentally responsive genes. Toxicol. Appl. Pharmacol. 2005;207:84–90. doi: 10.1016/j.taap.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Tomso DJ, Chorley BN, Cho HY, Cheung VG, Kleeberger SR, Bell DA. Identification of polymorphic antioxidant response elements in the human genome. Hum. Mol. Genet. 2007;16:1188–1200. doi: 10.1093/hmg/ddm066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peirson SN, Butler JN, Foster RG. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003;31:e73. doi: 10.1093/nar/gng073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fei P, Bernhard EJ, El-Deiry WS. Tissue-specific induction of p53 targets in vivo. Cancer Res. 2002;62:7316–7327. [PubMed] [Google Scholar]
- 31.Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH, Tom E, Mack DH, Levine AJ. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- 32.Krieg AJ, Hammond EM, Giaccia AJ. Functional analysis of p53 binding under differential stresses. Mol. Cell Biol. 2006;26:7030–7045. doi: 10.1128/MCB.00322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inga A, Storici F, Darden TA, Resnick MA. Differential transactivation by the p53 transcription factor is highly dependent on p53 level and promoter target sequence. Mol. Cell. Biol. 2002;22:8612–8625. doi: 10.1128/MCB.22.24.8612-8625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma B, Pan Y, Zheng J, Levine AJ, Nussinov R. Sequence analysis of p53 response-elements suggests multiple binding modes of the p53 tetramer to DNA targets. Nucleic Acids Res. 2007;35:2986–3001. doi: 10.1093/nar/gkm192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menendez D, Inga A, Jordan JJ, Resnick MA. Changing the p53 master regulatory network: ELEMENTary, my dear Mr Watson. Oncogene. 2007;26:2191–2201. doi: 10.1038/sj.onc.1210277. [DOI] [PubMed] [Google Scholar]
- 36.Paz N, Levanon EY, Amariglio N, Heimberger AB, Ram Z, Constantini S, Barbash ZS, Adamsky K, Safran M, Hirschberg A, et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17:1586–1595. doi: 10.1101/gr.6493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bureau JP, Henry L, Baz A, Scherrer K, Chateau MT. Prosomes (proteasomes) changes during differentiation are related to the type of inducer. Mol. Biol. Rep. 1997;24:57–62. doi: 10.1023/a:1006856707793. [DOI] [PubMed] [Google Scholar]
- 38.Bhui-Kaur A, Therwath A, Henry L, Chiesa J, Kurkure A, Scherrer K, Bureau JP. Increased prosomal proteins in breast cancer cells and in neighboring normal cells in Parsi and non-Parsi populations. J. Cancer Res. Clin. Oncol. 1998;124:117–126. doi: 10.1007/s004320050143. [DOI] [PubMed] [Google Scholar]
- 39.Cui F, Wang Y, Wang J, Wei K, Hu J, Liu F, Wang H, Zhao X, Zhang X, Yang X. The up-regulation of proteasome subunits and lysosomal proteases in hepatocellular carcinomas of the HBx gene knockin transgenic mice. Proteomics. 2006;6:498–504. doi: 10.1002/pmic.200500218. [DOI] [PubMed] [Google Scholar]
- 40.Mortarini R, Scarito A, Nonaka D, Zanon M, Bersani I, Montaldi E, Pennacchioli E, Patuzzo R, Santinami M, Anichini A. Constitutive expression and costimulatory function of LIGHT/TNFSF14 on human melanoma cells and melanoma-derived microvesicles. Cancer Res. 2005;65:3428–3436. doi: 10.1158/0008-5472.CAN-04-3239. [DOI] [PubMed] [Google Scholar]
- 41.Tamada K, Chen L. Renewed interest in cancer immunotherapy with the tumor necrosis factor superfamily molecules. Cancer Immunol. Immunother. 2006;55:355–362. doi: 10.1007/s00262-005-0081-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Obad S, Brunnstrom H, Vallon-Christersson J, Borg A, Drott K, Gullberg U. Staf50 is a novel p53 target gene conferring reduced clonogenic growth of leukemic U-937 cells. Oncogene. 2004;23:4050–4059. doi: 10.1038/sj.onc.1207524. [DOI] [PubMed] [Google Scholar]
- 43.Obad S, Olofsson T, Mechti N, Gullberg U, Drott K. Expression of the IFN-inducible p53-target gene TRIM22 is down-regulated during erythroid differentiation of human bone marrow. Leuk. Res. 2007;31:995–1001. doi: 10.1016/j.leukres.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 44.Fan M, Bigsby RM, Nephew KP. The NEDD8 pathway is required for proteasome-mediated degradation of human estrogen receptor (ER)-alpha and essential for the antiproliferative activity of ICI 182,780 in ERalpha-positive breast cancer cells. Mol. Endocrinol. 2003;17:356–365. doi: 10.1210/me.2002-0323. [DOI] [PubMed] [Google Scholar]
- 45.Dermitzakis ET, Clark AG. Evolution of transcription factor binding sites in Mammalian gene regulatory regions: conservation and turnover. Mol. Biol. Evol. 2002;19:1114–1121. doi: 10.1093/oxfordjournals.molbev.a004169. [DOI] [PubMed] [Google Scholar]
- 46.Contente A, Zischler H, Einspanier A, Dobbelstein M. A promoter that acquired p53 responsiveness during primate evolution. Cancer Res. 2003;63:1756–1758. [PubMed] [Google Scholar]
- 47.Horvath MM, Wang X, Resnick MA, Bell DA. Divergent evolution of human p53 binding sites: cell cycle versus apoptosis. PLoS Genet. 2007;3:e127. doi: 10.1371/journal.pgen.0030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang B, Xiao Z, Ren EC. Redefining the p53 response element. Proc. Natl Acad. Sci. USA. 2009;106:14373–14378. doi: 10.1073/pnas.0903284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chao C, Saito S, Anderson CW, Appella E, Xu Y. Phosphorylation of murine p53 at ser-18 regulates the p53 responses to DNA damage. Proc. Natl Acad. Sci. USA. 2000;97:11936–11941. doi: 10.1073/pnas.220252297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chao C, Wu Z, Mazur SJ, Borges H, Rossi M, Lin T, Wang JY, Anderson CW, Appella E, Xu Y. Acetylation of mouse p53 at lysine 317 negatively regulates p53 apoptotic activities after DNA damage. Mol. Cell Biol. 2006;26:6859–6869. doi: 10.1128/MCB.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Espinosa JM, Verdun RE, Emerson BM. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol. Cell. 2003;12:1015–1027. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- 52.Espinosa JM. Mechanisms of regulatory diversity within the p53 transcriptional network. Oncogene. 2008;27:4013–4023. doi: 10.1038/onc.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaked H, Shiff I, Kott-Gutkowski M, Siegfried Z, Haupt Y, Simon I. Chromatin immunoprecipitation-on-chip reveals stress-dependent p53 occupancy in primary normal cells but not in established cell lines. Cancer Res. 2008;68:9671–9677. doi: 10.1158/0008-5472.CAN-08-0865. [DOI] [PubMed] [Google Scholar]
- 54.Millau JF, Bastien N, Bouchard EF, Drouin R. p53 Pre- and post-binding event theories revisited: stresses reveal specific and dynamic p53-binding patterns on the p21 gene promoter. Cancer Res. 2009;69:8463–8471. doi: 10.1158/0008-5472.CAN-09-2036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.