Abstract
Mitochondrial-DNA diseases have no effective treatments. Allotopic expression—synthesis of a wild-type version of the mutated protein in the nuclear-cytosolic compartment and its importation into mitochondria—has been proposed as a gene-therapy approach. Allotopic expression has been successfully demonstrated in yeast, but in mammalian mitochondria results are contradictory. The evidence available is based on partial phenotype rescue, not on the incorporation of a functional protein into mitochondria. Here, we show that reliance on partial rescue alone can lead to a false conclusion of successful allotopic expression. We recoded mitochondrial mt-Nd6 to the universal genetic code, and added the N-terminal mitochondrial-targeting sequence of cytochrome c oxidase VIII (C8) and the HA epitope (C8Nd6HA). The protein apparently co-localized with mitochondria, but a significant part of it seemed to be located outside mitochondria. Complex I activity and assembly was restored, suggesting successful allotopic expression. However, careful examination of transfected cells showed that the allotopically-expressed protein was not internalized in mitochondria and that the selected clones were in fact revertants for the mt-Nd6 mutation. These findings demonstrate the need for extreme caution in the interpretation of functional rescue experiments and for clear-cut controls to demonstrate true rescue of mitochondrial function by allotopic expression.
INTRODUCTION
Mitochondria originated ∼1.5 billion years ago, as a result of a symbiotic relationship between a primitive anaerobic eukaryotic cell and an aerobic bacterial cell (α-proteobacteria). During evolution, most of the original bacterial genetic material was transferred to the nucleus. Some of these DNA fragments underwent codon mutations and acquired mitochondrial targeting sequences that enable their correct translation on cytosolic ribosomes and subsequent targeting to mitochondria. More than 600 recognizable mitochondrial DNA (mtDNA)-derived fragments have been detected in the nuclear genome of Homo sapiens, mostly DNA fragments that have migrated from the mitochondria (1). In eukaryotes that have lost their mitochondria, such as Archaeozoa, this transfer has been completed. However, in the vast majority of eukaryotes a mitochondrial genome has been maintained, consisting of a small set of the original bacterial genes (2).
The mitochondrial protein synthesis system is similar to that of prokaryotes, including specific ribosomes and factors. In mammals, all the proteins of this system are encoded in the nuclear genome. The significant energy needed to maintain a second translation apparatus for the synthesis of just 13 proteins suggests that there is strong selection pressure to retain the genes for these proteins in mtDNA. Various hypotheses have been proposed to explain why this should be the case (3). It may be that genetic code divergences preclude translation of the mtDNA polypeptide genes by the nuclear-cytosolic machinery (4,5). Other authors suggest that intra-mitochondrial translation of the retained proteins is needed to limit assembly of the oxidative phosphorylation (OXPHOS) complexes to the inner mitochondrial membrane, thus preventing their misallocation in other membranes where they could contribute to cell damage and dysfunction through ROS generation (6,7). Finally, the proteins encoded in mtDNA might be so hydrophobic that they could not be imported into the organelle from the cytosol to be assembled in the mitochondrial inner membrane; maintenance of their synthesis inside mitochondria would circumvent this problem (5,8).
There are currently no effective treatments for mitochondrial-DNA associated disorders. Gene-therapy approaches have been proposed but so far no practical method has been developed to transfect mammalian mitochondria with exogenous nucleic acids, even though successful procedures have been developed for Saccharomyces cerevisiae and Chlamydomonas reinhardtii (9). A proposed alternative approach is allotopic expression, in which the mtDNA-encoded gene is transferred to the nucleus and the protein is synthesized in the cytosol and subsequently imported into mitochondria. However, although allotopic expression has been successfully demonstrated in yeast (10), results obtained in human and mouse cells are unclear. In 2002, Schon and co-workers reported the successful allotopic expression of human and C. reinhardtii ATPase6 subunit in human cells (11,12). However, using the same model, Holt and co-workers found that C. reinhardtii ATPase6 protein was not integrated into mature complex V (13). Similar findings were obtained with ND4, with Guy and colleagues reporting positive results (14), followed one year later by negative results from Moraes and colleagues (15). An as yet unexplored cause of these contradictory findings is the possibility that recipient mitochondria were not devoid of functional endogenous protein in all cases.
We have explored the application of allotopic expression in a mouse cell line with a homoplasmic KO mutation in the mt-Nd6 gene. We show that partial rescue experiments can lead to a false conclusion of successful allotopic expression, when restored OXPHOS function is in fact due to the artefactual selection of revertants for the mutated gene. These findings demonstrate the need for extreme caution in the interpretation of functional rescue experiments and for clear-cut controls to demonstrate true allotopic rescue of mitochondrial function.
MATERIALS AND METHODS
Cell lines and strategy for allotopic expression of recoded mitochondrial genes
The ND6dKO cell line used in this study was generated by random mutagenesis in the NIH3T3 mtDNA background, as described elsewhere (16). The ND6dKO line has a deletion of cytosine 13887 in the mt-Nd6 gene, which occurs in a run of 6 cytosines in the wild-type gene. The mutation results in the expression of a 72 amino acid-long truncated polypeptide instead of the 169 aa wild-type protein (17).
All cells were grown in DMEM (GibcoBRL) supplemented with 5% fetal bovine serum (FBS, Gibco BRL).
C8ND6, C8ND6HA and NDI1HA constructs
mtNd6 and yeast mtNdi1 were recoded using Backtranslation-Tool v2 (Entelechon http://www.entelechon.com/index.php?id=tools/backtranslation&lang=eng) to generate mouse codon-usage optimized nuclear encoded sequences. The recoded genes were ordered from GenScript Corporation, cloned in pUC57. The haemagglutinin epitope (HA tag) (YPYDVPDYA) was added to the C-termiminus by PCR. C8Nd6 was subcloned using KpnI/BamHI sites in the pcDNA3.1 hygro. C8Nd6HA and NDI1HA genes were subcloned in the lentiviral vector p156RRLsinPPThCMVMCSpre (from Tronolab), using XbaI/BamHI and XbaI/MluI sites, respectively.
Lentiviral vector production and cell infection
The 2.5 × 106 human 293T cells were plated 24 h before co-transfection with 10 μg transfer vector (C8ND6HA-p156RRLsinPPThCMVMCSpre or NDI1HA-p156RRL sinPPThCMVMCSpre), 7.5 μg of the second generation packaging plasmid pCMVdR8.74 and 3 μg envelope plasmid (pMD2.VSVG). Transfections were carried out with FuGENE® 6 Transfection Reagent (Roche). Infectious particles were collected 24 and 48 h after transfection (18). Lentiviral particles were used to transduce ND6dKO cells (80% confluent). The pool of cells expressing the gene of interest was isolated by selection in galactose-containing medium (DMEM, 5% FBS, 4.5 g/l galactose and 110 µg/ml pyruvate).
Inmunological techniques
For immunocytochemistry, cells were incubated with 200 nM mitochondrial dye Mitotracker red (Invitrogen) for 30 min and anti-HA primary antibody (Roche). The secondary antibody was Alexa Fluor 488 IgG anti-rat (Invitrogen).
For western blot, cell proteins were extracted in RIPA buffer (Pierce). Twenty micrograms of total protein were separated by 12.5% SDS polyacrylamide gel electrophoresis (SDS–PAGE), electroblotted onto PVDF membrane, and sequentially probed with specific antibodies. Antibodies used were anti-ND6 (which in fact recognizes NDUFB8; Molecular Probes), anti-HA (Roche), anti-COI (Molecular Probes), anti-Hsp60 (SIGMA), anti-TOM20 (Santa Cruz), anti-IκB-α (Santa Cruz) and anti-β actin (SIGMA).
Pulse-chase experiments: mtDNA-encoded subunit labelling
mtDNA-encoded proteins were labelled with [35S]-methionine/cysteine (EXPRE35S35S Protein Labeling Mix; Perkin Elmer Life Sciences) in intact cells as described elsewhere (19,20).
Blue native electrophoresis
Mitochondria were isolated from cells according to Schägger (1995), with some modifications (21). Blue Native gradient gels (5–13%) were cast as described earlier (22) and run with 100 μg of DDM solubilized mitochondrial protein (1.6 g DDM/1g mitochondrial protein). For 2D electrophoresis, bands from the blue native (BN) gel corresponding to assembled mitochondrial complexes were excised and separated by SDS–PAGE (23).
After electrophoresis, the gels were electroblotted onto ‘Hybond-P’ PVDF membranes (GE Healthcare Life Sciences) and probed with antibodies against complex I (anti-ND6 from Molecular Probes, which in fact recognizes NDUFB8), and complex IV (anti-COI, Molecular Probes). The secondary antibody was peroxidase-conjugated anti-mouse (Invitrogen), and signal was revealed with ECL® Plus (GE Healthcare Life Sciences).
In vitro reticulocyte protein expression
Nuclear NDUFB8 was amplified from mouse cDNA using the following primers: forward, ctcgagGGAGAAGGTGAAGATGGCTG (XhoI site indicated in lower case) and reverse tctgagTTAAGCCTCTAGGAACGAGG (XbaI site indicated in lower case). The PCR product was cloned into the pCR®2.1 vector (Invitrogen) and sequenced. The NDUFB8 CDS was released from pCR®2.1 by digestion with XhoI and XbaI, and this fragment was subsequently ligated into pTNTTM (Promega). In the same way, nuclear recoded Nd6 was amplified from C8ND6-pUC57 with the following primers: forward, acgcgtATGAACAACTACATCTTCGTG (MluI site in lower case) and reverse gtcgacTTAATCCCTTGTGATCTCG (SalI site in lower case). The sequence was cloned into pTNTTM (Promega). The C8Nd6, Nd6 and Ndufb8 genes were expressed with the TNT® T7/SP6 Coupled Reticulocyte Lysate System (Promega) in the presence of [35S]-methionine. The radiolabelled proteins were run in a 12% SDS–PAGE gel and blotted onto a ‘Hybond–P’ PVDF membrane (Amersham).
Oxygen consumption measurement
O2 consumption determinations in digitonin-permeabilized cells were carried out with an oxytherm Clark-type electrode (Hansatech) as described earlier (24) with small modifications (25).
RESULTS
Analysis of allotopic expression in cells containing mutated mitochondrial DNA
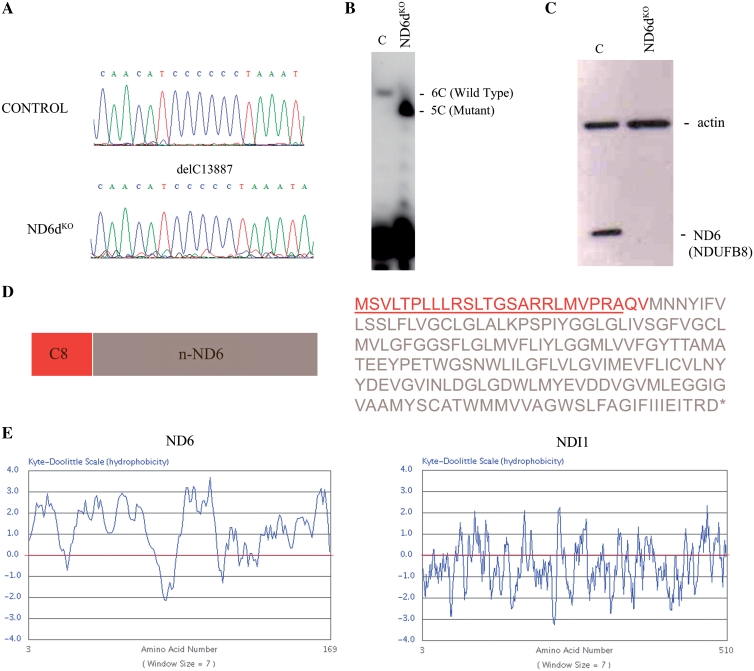
To explore the potential of allotopic expression, we recoded mt-Nd6 and expressed it in ND6dKO cells (Figure 1). Recoded mt-Nd6 was tagged with an N-terminal sequence encoding the entire 25-amino acid mitochondrial targeting signal (MTS) of cytochrome c oxidase subunit VIII (C8) plus the first two amino acids of the mature COX VIII polypeptide (C8ND6) (Figure 1D, Supplementary Figure S2). The major impediment to allotopic expression is the highly hydrophobic nature of the mitochondrial-endoced polypeptides. As a positive control, we therefore also transformed ND6dKO cells with a recoded version of the relatively hydrophilic yeast mitochondrial-encoded protein NDI1. Kyte and Doolittle hydrophobicity profiles of ND6 and NDI1 are shown in Figure 1E. Transformed cells were isolated by antibiotic selection, first and were then transferred to medium containing galactose instead of glucose for metabolic selection. Growth in galactose medium severely limits generation of ATP via glycolysis, forcing cells to rely on oxidative phosphorylation; OXPHOS mutant cells are therefore unable to grow in this medium. We reasoned that only clones that correctly import C8ND6 protein into mitochondria would form a functional complex I and survive under this metabolic restriction. Nine cell clones were isolated from galactose cultures, of which four (1.0, 1.1, 1.6 and 1.7) were selected for further analysis.
Figure 1.
Allotopic expression strategy. (A) Chromatograms showing the homoplasmic mutations found in the mt-Nd6 gene. (B) Allele-specific termination of primer extension assay to confirm the homoplasmy of the deletion of one C in ND6dKO cells. (C) Western blot of total protein from control and mutant cells with the anti-ND6 antibody marketed by Molecular Probes (which in fact recognizes NDUFB8; see text and Figure 3). Actin expression was probed as a loading control. (D) Amino acid sequence and map of C8ND6 (the mt-Nd6 gene sequence recoded to the universal genetic code using Backtranslation software). Red letters correspond to the C8 MTS sequence plus the first two amino acids of mature COX8 (QV). (E) Kyte and Doolittle plots illustrating the highly hydrophobic character of the mitochondrial ND6 protein, compared with the yeast protein NDI1.
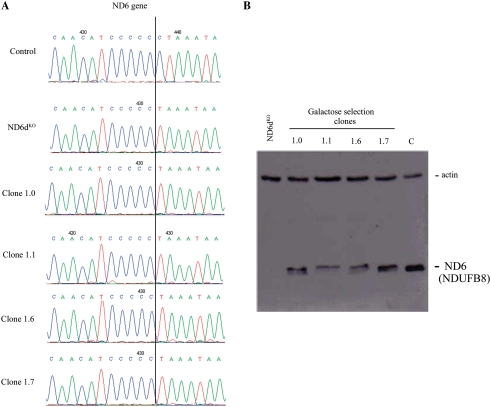
Sequencing analysis confirmed that the four ND6dKOC8ND6 cell clones still contained the mutation in the mt-Nd6 gene in apparently homoplasmic form, with no traces of the wild-type gene (Figure 2A). Moreover, western blot analysis detected ND6 protein in all four galactose-resistant clones, strongly suggesting that allotopic expression had been successfully achieved (Figure 2B).
Figure 2.
Analysis of galactose-resistant clones. (A) Sequence chromatograms showing conservation of the 13887 delC mutation in the mt-Nd6 gene in four clones (1.0, 1.1, 1.6 and 1.7) obtained after galactose selection. (B) Western blot of total protein from wild-type (C), ND6dKO cells and galactose-resistant clones allotopically expressing C8ND6 protein. The blot was probed with the anti-ND6 from Molecular Probes, which in fact recognizes NDUFB8 (see text and Figure 3) and with anti-actin as a loading control.
Four clones expressing yeast NDI1 were analysed after galactose selection (Supplementary Figure S1A). To assay the activity of the yeast protein in mouse cells, the respiration properties of the ND6dKONDI1 cells were investigated in detail (Supplementary Figure S1C). Polarographic measurements of ND6dKONDI1 cells showed that glutamate/malate driven respiration, which usually reflects the rate-limiting activity of complex I, was insensitive to the complex I inhibitor rotenone but sensitive to the NDI1 inhibitor flavone, indicating that this respiration was due to the function of the yeast enzyme. Allele-specific termination of primer extension was performed to confirm the homoplasmy of the mt-Nd6 gene in the clones analysed (Supplementary Figure S1B).
Is allotopically expressed ND6 protein inside the mitochondria?
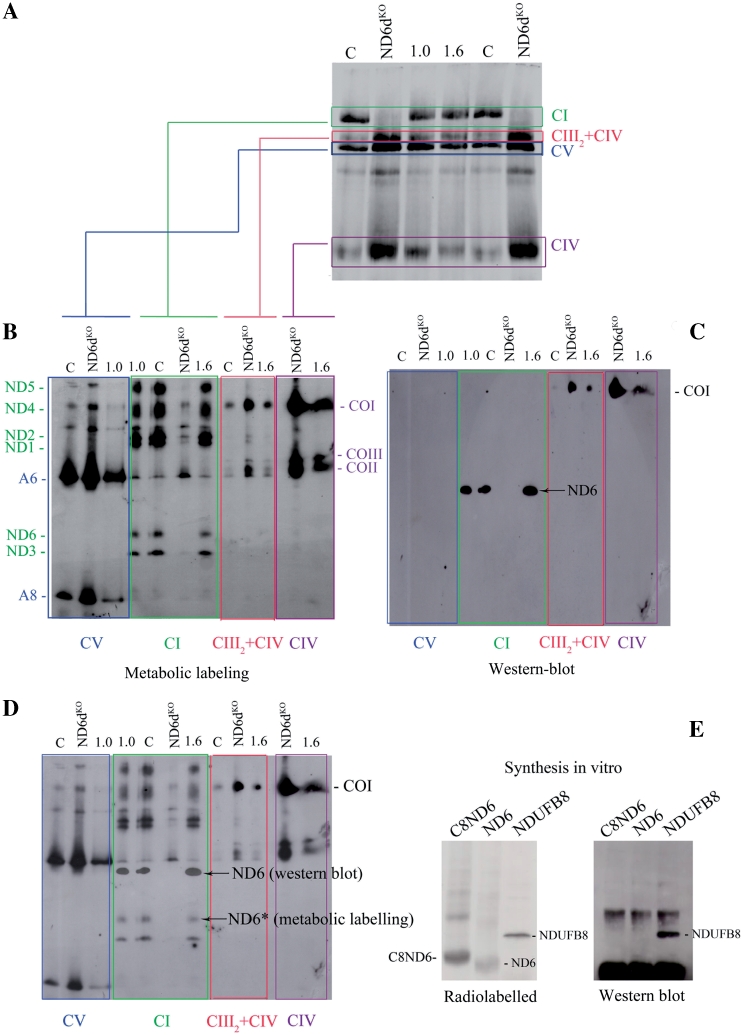
The experiments shown above strongly suggested that allotopically expressed ND6 can be properly imported and assembled to restore complex I activity. To confirm this we performed a routine set of control experiments. We first selectively labelled mtDNA-encoded OXPHOS subunits by pulsing cells with [35S] Met-Cys for 2 h in the presence of cycloheximide, which blocks cytosolic protein synthesis. Cycloheximide was then removed and the cells were cultured for a further 12 h to allow importation of allotopically-expressed C8ND6 and full assembly of OXPHOS complexes. Dodecyl-maltoside (DDM) solubilized mitochondria were then isolated and analysed by BN–PAGE. These experiments demonstrated that complex I assembly was impaired in ND6dKO cells but restored in the two ND6dKOC8ND6 clones analysed (1.0 and 1.6) (Figure 3A).
Figure 3.
Galactose-resistant ND6dKOC8ND6 clones express wild-type mt-Nd6. (A) Metabolic labelling of assembled OXPHOS complexes. Fluorogram after Blue-Native gel electrophoresis of the mitochondrial translation products of wild-type (C), mutant (ND6dKO) and two galactose-resistant ND6dKOC8ND6 clones (1.0 and 1.6). Before protein extraction, cells were pulse-labelled with [35S]-methionine for 1 h in the presence of cycloheximide and chased for 12 h. (B). Fluorogram of mitochondrial translation products separated by 2D electrophoresis. Isolated OXPHOS complexes obtained in the first dimension were resolved on denaturing gels and electro-transferred to PVDF membrane as indicated. (C) Western blot of the second dimension gel with the Molecular Probes anti-ND6 antibody (complex I) and anti-CO1 (complex IV). (D). Superposition of the fluorogram and western blot from B and C, showing that the anti-ND6 antibody does not recognize ND6. (E) Identification of the specificity of the anti-ND6 antibody. C8ND6, ND6 and NDUFB8, which participates in the assembly of mitochondrial complex I, were synthesized in vitro in the presence of [35S] methionine. The left panel shows an autoradiogram of a PVDF membrane blotted from a 12% SDS–polyacrylamide gel. The right panel shows a western blot of the membrane with the Molecular Probes anti-ND6 antibody.
Since this procedure labels only the mtDNA-encoded proteins (13 in wild-type cells and 12 in the mutant), we reasoned that complex I in wild-type cells should contain all seven mtDNA-encoded complex I proteins (ND1 to 6 and ND4L), whereas the rescued ND6dKOC8ND6 cells should contain only six (not ND6). To confirm this, we cut the bands corresponding to isolated complex I (and complexes V, IV and supercomplex III2+IV as controls) from the BN gels and resolved them by 2D SDS–PAGE to identify the individual proteins contained (Figure 3B). As expected, all seven ND subunits observed in wild-type cells, and all except ND6 were detected in ND6dKO cells. However, to our surprise all seven ND subunits, including ND6, were labelled in the two galactose-resistant ND6dKOC8ND6 cell clones. Thus the ND6 protein in these clones is of mitochondrial origin, bringing into question our initial interpretation of successful importation of allotopically-expressed ND6 and its functional assembly into complex I.
Moreover, immunodetection of ND6 with an antibody from Molecular Probes revealed that the protein signal recognized by the antibody is not the same as the one corresponding to metabolically labelled ND6 (Figure 3C and D). In fact, no labelled protein co-migrated with the protein detected by the anti-ND6 antibody. After contacting the antibody distributor, we suspect that the protein targeted by the antibody might be NDUFB8, which participates in the assembly of mitochondrial complex I. To test this, we synthesized C8ND6, native ND6 (without the MTS or C8) and NDUFB8 proteins in the presence of [35S] methionine, to analyse which protein is recognized by the antibody. The radiolabelled proteins were run in an SDS–PAGE gel and immunodetected with the Molecular Probes antibody, demonstrating that the antibody does in fact recognize NDUFB8 and not ND6 (Figure 3E).
Allotopically expressed ND6 protein is not imported into the mitochondrial inner membrane
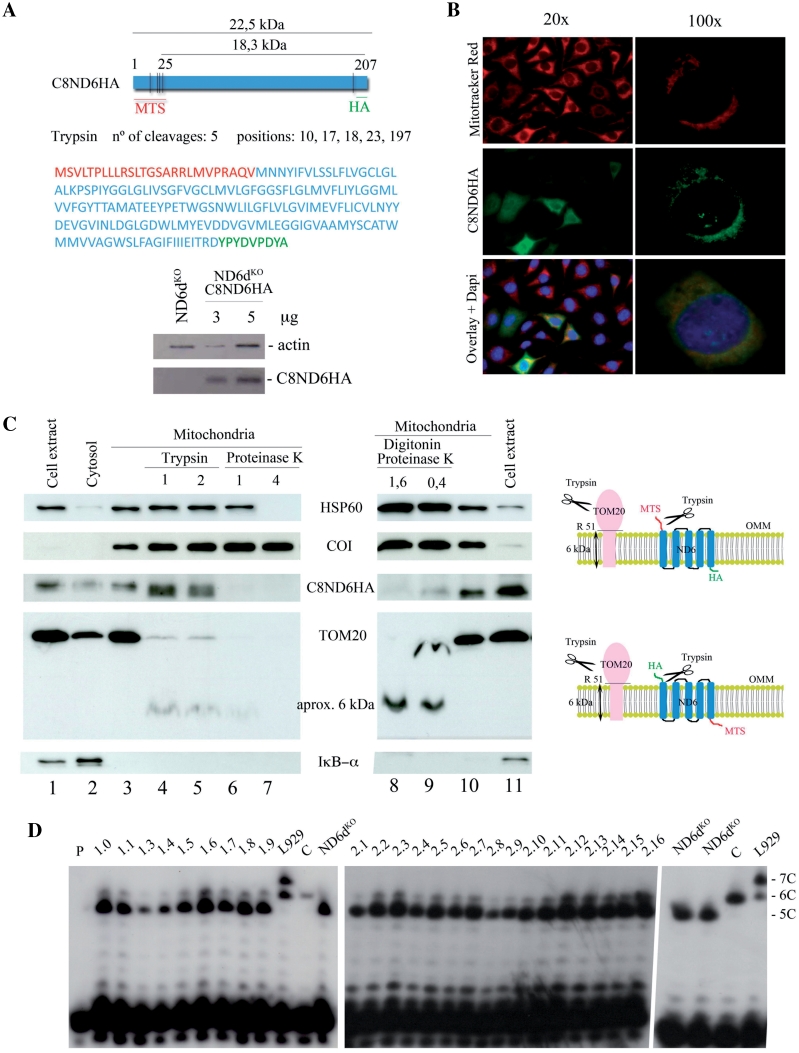
In the absence of an antibody to detect the expression of ND6 protein, we added the HA epitope to the carboxy terminus of the recoded ND6 gene (C8ND6) to generate C8ND6HA (Figure 4A). To increase transfection efficiency we used lentiviral vectors to transform ND6dKO cells with C8ND6HA. The allotopically expressed C8ND6HA protein was expressed and apparently colocalized with mitochondria, but a significant amount seemed to be located outside mitochondria (Figure 4B). After metabolic selection in galactose containing medium, we isolated 16 cell galactose-resistant C8ND6HA clones.
Figure 4.
Allotopically expressed ND6 is not incorporated into the inner mitochondrial membrane. (A) Upper panel: map and amino acid sequence of C8ND6HA. The COX8 MTS is shown in red, the recoded mt-Nd6 sequence in blue and the HA epitope in green. Lower panel: western blot showing expression of C8ND6HA in lentivirally transformed ND6dKO cells. The blot was probed with anti-HA (Roche) and with anti-actin as a loading control. (B) Subcellular localization of C8ND6HA in transformed cells. ND6kdKOC8ND6HA cells were visualized by indirect inmunofluorescence using anti-HA antibody and Mitotracker® Red (Invitrogen). The precursor polypeptide was expressed and the protein apparently co-localized with mitochondria. (C) Left panel: western blot of isolated mitochondria from ND6dKOC8ND6HA cells, showing that allotopically expressed C8ND6HA is not internalized. Lanes (from left to right): 1, cell extract; 2, cytosol; 3, untreated isolated mitochondria; 4 and 5, mitochondria treated with 1 or 2 µg trypsin/100 µg mitochondrial protein; 6 and 7, mitochondria treated with 1 or 4 µg proteinase K/100 µg mitochondria; 8 and 9, mitochondria treated with 1.6 or 0.4 µg proteinase K/100 µg mitochondria plus 0.15 mg/ml digitonin; 10, isolated mitochondria and 11, cell extract. The protein loading was 20 µg for lanes 1, 2 and 11; 10 µg for lanes 3–7 and 10 and 50 µg for lanes 8 and 9. Right panel: diagram showing the likely actions of trypsin on TOM20 and allotopically expressed C8ND6HA proteins. (D) Allele-specific primer extension termination analysis. All the clones obtained after galactose selection were revertants for the mt-Nd6 mutation, in both experiments (C8ND6 and C8ND6HA expression). P: primer; 1.0–1.9, galactose-resistant clones obtained after allotopic expression of C8ND6; L929, mouse cell line heteroplasmic for the 13887iC mutation in mt-ND6 (25); 2.1-2.16, galactose-resistant clones obtained after allotopic expression of C8ND6HA; C, wild-type cells.
Analysis of mitochondria isolated from ND6dKO C8ND6HA cells showed that the organelles were enriched in C8ND6HA protein as well as the outer membrane protein TOM20, the inner membrane protein COI, and the matrix protein HSP60, but were devoid of IkB-α (a cytosolic protein) (Figure 4C). Treatment of intact mitochondria with trypsin did not remove COI or HSP60, since the enzyme cannot access these proteins (Figure 4C, lines 4 and 5). In contrast, trypsin partially digested TOM20 (the 6 kDa hydrophobic inner-membrane domain of the protein is protected) (Figure 4C, lines 4 and 5). Trypsin also cut the C8ND6HA polypeptide, changing its electrophoretic mobility from 22.5 kDa to 18.7 kDa (Figure 4C). The distribution of K and R residues in C8ND6HA is very asymmetric (Figure 4A). Four R residues are located within the MTS sequence (N-terminus) and a fifth R residue is located immediately before the HA tag (C-terminus). Since C8ND6HA protein is immunodetected with anti-HA, and the HA tag is at the protein C-terminus, the size of the tryspin digested C8ND6HA protein is only compatible with exposure of the N-terminus on the cytosolic side of the outer mitochondrial membrane. The continued detection of the HA tagged protein after trypsin digestion indicates that the protein is partially protected, probably by location of the C-terminal portion in the inter-membrane space.
More aggressive protease digestion with proteinase K is able to digest outer-membrane proteins at low concentration and even matrix proteins at higher concentration, suggesting that it can generate holes across both membranes. This treatment completely removed the C8ND6HA HA signal (Figure 4C, lines 6 and 7). Indeed, permeabilization of the outer membrane with low doses of digitonin in the presence of low doses of proteinase K abolished the protection of the HA epitope without affecting the protection of the 6 kDa Tom 20 fragment or the stability of matrix located proteins (Figure 4C, lines 8 and 9). These results indicate that although the MTS directs allotopically expressed C8ND6HA to mitochondria, the protein cannot properly internalize to insert into the inner mitochondrial membrane (Figure 4B and C).
Since sequencing analysis is insufficiently sensitive to determine low levels of heteroplasmy, we used allele-specific termination of primer extension (Figure 4D). Using this approach we found that all the C8ND6 and C8ND6HA clones analysed were heteroplasmic for the mt-Nd6 mutation: all of them harboured a small proportion of the wild-type gene. Thus, our results show that a low gene dose of wild-type mt-Nd6 (<10%; undetectable by sequencing) is sufficient to drive assembly of high levels of Complex I, as assessed by metabolic labelling (Figure 3A). ND6 was unequivocally present in the assembled Complex I (Figure 3B), demonstrating that, under selective pressure, low doses of wild-type mt-Nd6 gene allow the assembly of near normal levels of Complex I.
DISCUSSION
In 1988, Nagley and coworkers were able to show that the respiratory defect in yeast carrying mutations in the mitochondrial mt-Atp8 gene could be rescued by engineering a normal copy of the gene tagged with the mitochondrial targeting sequence of nuclear-encoded Neurospora crassa ATPase9, and introducing this gene into the nucleus, resulting in cytosolic translation of the ATPase8 gene product (10).
Manfredi and colleagues showed that allotopic expression of ATPase6 can improve ATP synthesis in human cells with a pathogenic mt-Atp6 mutation. The mitochondrial targeting sequence of C8 (the same used in the present study) allowed ATPase6 to be targeted to mitochondria and be correctly processed by the mitochondrial proteases (11). The same group also had success with the nuclear-encoded Atp6 gene from C. reinhardtii, which they expressed in human control cells to rescue the ATP synthesis defect in human cells harbouring an mt-Atp6 mutation (12). Recently, Bokori-Brown et al. (13) carried out similar allotopic experiments to rescue a cell line carrying an identical mutation in mt-Atp6 (8993T > G), but with very different conclusions. The authors optimized the import of nuclear-encoded ATPase6 into the mitochondria of human cell lines, but after careful analysis of assembled complexes were unable to show integration of the imported protein into mature functional ATP-synthase. Thus, restoration of complex V activity in the original experiments was suggested to have been due to random clonal variations in ATP synthesis as a consequence of aneuploidy in the transfected cell population.
Recently, Qi and colleagues showed that allotopic expression of a nuclear version of the mutant human mt-Nd4 gene in mouse eyes led to optic nerve degeneration, prompting them to propose the use of these mice to test the effectiveness of treatments for Leber's hereditary optic neuropathy (LHON) disease (26). However, this approach is limited by the inefficient mitochondrial import of the hydrophobic ND4 protein, as reported by Oca-Cossio et al. in 2003 (15). Therefore retinal toxicity in this model might reflect a collapse of the mitochondrial import machinery rather than the possible effects produced by a mutant ND4 subunit. More recently, the same group has evaluated the potential of allotopic expression of a normal human mt-Nd4 gene in the mouse visual system (27). In their opinion, this approach appears safe, and these authors have proposed allotopic expression of ND4 as an effective gene therapy in patients with LHON disease (G11778A mtDNA mutation). However, our findings indicate that the evidence for functional allotopic expression is insufficient to initiate clinical trials in humans. There are two broad reasons for our caution. First, successful allotopic expression is not definitively demonstrated, and additional analyses are needed to show unequivocally that the imported proteins are indeed integrated into fully assembled OXPHOS complexes. These experiments are easy to perform and will provide a clear-cut conclusion. Second, incompatibilities between nuclear DNA and mtDNA from different species have been repeatedly reported to have a significant effect on complex I activity (28–30). For example, rat mtDNA is unable to build a functional respiratory chain when introduced in mouse cells (31,32). Therefore, the use of human mtDNA sequences in a mouse model is questionable.
Corral-Debrinski and colleagues have attempted to optimize the allotopic expression of highly hydrophobic mitochondrial proteins by targeting the transcript to the outer mitochondrial membrane, with the aim of facilitating co-translational translocation of the gene product and thus preventing the accumulation of cytosolic aggregates (33–36). Our results suggest that the import process can begin, but it is aborted because the allotopically expressed C8ND6 protein is wrongly placed in the outer mitochondrial membrane. In addition, very likely the C-terminus of the C8ND6HA protein faces the intermembrane space while the N-terminus the cytosolic side of the outer membrane (Figure 4). This suggests that despite having the C8 targeting sequence, the mitochondrial import apparatus interprets the C8ND6HA protein as an outer membrane protein. Thus, even targeting the allotopically encoded mRNA to the mitochondrial surface by a 3′-UTR sequence would not overcome the inability of the translated protein to translocate toward the inner mitochondrial membrane if it is processed as an outer membrane protein (34).
Like Guy and colleagues, Corral-Debrinski and co-workers propose their optimized allotopic expression as a decisive and promising treatment for patients with LHON disease. However, their reports did not show analysis demonstrating the assembly of the allotopically-expressed protein into the holoenzyme.
Many groups have failed in the attempt to achieve allotopic expression in mammalian cells because none of them was able to show unequivocally that the allotopic protein was responsible for the improved function in the mutant cells tested. Several studies suggest that allotopic expression of most mitochondrial proteins is not feasible because of their high mesohydrophobicity, a parameter that can be used to predict the importability of hydrophobic peptides (37). With few exceptions, the 13 proteins encoded in the mitochondrial genome are conserved among all species. Although the reason of this conservation has been the subject of speculation, the more widely held view is that these proteins are so hydrophobic that they are unable to be imported from the cytosol. Interestingly, in the few cases where these genes have transferred to the nucleus naturally, this was accompanied by a significant reduction in hydrophobicity. This is the case, for example, in C. reinhardtii, which expresses several genes in the nuclear-cytosolic compartment that are generally encoded in the mitochondrial genome in other species (COII, COIII and ATPase6). The fact that the mitochondrial and nuclear genetic codes in C. reinhardtii are identical might have facilitated this transfer of genes from mitochondria to cell nucleus (38). However, even in this case some genes apparently cannot be transferred: cytochrome c oxidase subunit I and apocytochrome b are the two exceptions that have remained in the mtDNA in all species, probably due to their high mesohydrophobicity. Potential strategies to overcome these difficulties include substituting amino acids that reduce hydrophobicity without affecting biological activity (39), or dividing the mitochondrial gene into two pieces that are subsequently transferred to the nucleus, as happened in the case of mt-Co2 (40).
The findings presented here show that the behaviour of mitochondria in cultured cells can give a false impression that allotopic expression of mtDNA encoded proteins is successful. Workers in this field should be aware that the only definitive evidence for successful allotopic expression is the demonstration that the protein expressed in the cytosol is assembled in the full OXPHOS complex, restoring its activity. Fortunately, this can be easily confirmed or discounted by electrophoresis and determination of complex activity. This is particularly important when proposing a clinical trial to test the potential of allotopic expression of mtDNA-encoded proteins for the treatment of mtDNA-linked diseases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: The Spanish Ministry of Education (SAF2009-08007 and CSD2007-00020); EUmitocombat (EUMITOCOMBAT-LSHM-CT-2004-503116); Group of Excellence grant DGA (B55); Spanish Ministry of Education (pre-doctoral fellowship to E.P.-C.); Spanish Ministry of Science and Innovation and the Pro-CNIC Foundation (to CNIC).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are very grateful to Doug Turnbull and Simon Bartlett for critical reading of the article. We would like to thank Nieves Movilla and Santiago Morales for their technical assistance.
REFERENCES
- 1.Woischnik M, Moraes CT. Pattern of organization of human mitochondrial pseudogenes in the nuclear genome. Genome Res. 2002;12:885–893. doi: 10.1101/gr.227202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saccone C, Gissi C, Reyes A, Larizza A, Sbisa E, Pesole G. Mitochondrial DNA in metazoa: degree of freedom in a frozen event. Gene. 2002;286:3–12. doi: 10.1016/s0378-1119(01)00807-1. [DOI] [PubMed] [Google Scholar]
- 3.Wallace DC. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu. Rev. Biochem. 2007;76:781–821. doi: 10.1146/annurev.biochem.76.081205.150955. [DOI] [PubMed] [Google Scholar]
- 4.Wallace DC. Structure and evolution of organelle genomes. Microbiol. Rev. 1982;46:208–240. doi: 10.1128/mr.46.2.208-240.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Heijne G. Why mitochondria need a genome. FEBS Lett. 1986;198:1–4. doi: 10.1016/0014-5793(86)81172-3. [DOI] [PubMed] [Google Scholar]
- 6.Race HL, Herrmann RG, Martin W. Why have organelles retained genomes? Trends Genet. 1999;15:364–370. doi: 10.1016/s0168-9525(99)01766-7. [DOI] [PubMed] [Google Scholar]
- 7.Allen JF. Control of gene expression by redox potential and the requirement for chloroplast and mitochondrial genomes. J. Theor. Biol. 1993;165:609–631. doi: 10.1006/jtbi.1993.1210. [DOI] [PubMed] [Google Scholar]
- 8.Claros MG, Perea J, Shu Y, Samatey FA, Popot JL, Jacq C. Limitations to in vivo import of hydrophobic proteins into yeast mitochondria. The case of a cytoplasmically synthesized apocytochrome b. Eur. J. Biochem. 1995;228:762–771. [PubMed] [Google Scholar]
- 9.Bonnefoy N, Remacle C, Fox TD. Genetic transformation of Saccharomyces cerevisiae and Chlamydomonas reinhardtii mitochondria. Methods Cell Biol. 2007;80:525–548. doi: 10.1016/S0091-679X(06)80026-9. [DOI] [PubMed] [Google Scholar]
- 10.Nagley P, Farrell LB, Gearing DP, Nero D, Meltzer S, Devenish RJ. Assembly of functional proton-translocating ATPase complex in yeast mitochondria with cytoplasmically synthesized subunit 8, a polypeptide normally encoded within the organelle. Proc. Natl Acad. Sci. USA. 1988;85:2091–2095. doi: 10.1073/pnas.85.7.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manfredi G, Fu J, Ojaimi J, Sadlock JE, Kwong JQ, Guy J, Schon EA. Rescue of a deficiency in ATP synthesis by transfer of MTATP6, a mitochondrial DNA-encoded gene, to the nucleus. Nat. Genet. 2002;30:394–399. doi: 10.1038/ng851. [DOI] [PubMed] [Google Scholar]
- 12.Ojaimi J, Pan J, Santra S, Snell WJ, Schon EA. An algal nucleus-encoded subunit of mitochondrial ATP synthase rescues a defect in the analogous human mitochondrial-encoded subunit. Mol. Biol. Cell. 2002;13:3836–3844. doi: 10.1091/mbc.E02-05-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bokori-Brown M, Holt IJ. Expression of algal nuclear ATP synthase subunit 6 in human cells results in protein targeting to mitochondria but no assembly into ATP synthase. Rejuvenation Res. 2006;9:455–469. doi: 10.1089/rej.2006.9.455. [DOI] [PubMed] [Google Scholar]
- 14.Guy J, Qi X, Pallotti F, Schon EA, Manfredi G, Carelli V, Martinuzzi A, Hauswirth WW, Lewin AS. Rescue of a mitochondrial deficiency causing Leber Hereditary Optic Neuropathy. Ann. Neurol. 2002;52:534–542. doi: 10.1002/ana.10354. [DOI] [PubMed] [Google Scholar]
- 15.Oca-Cossio J, Kenyon L, Hao H, Moraes CT. Limitations of allotopic expression of mitochondrial genes in mammalian cells. Genetics. 2003;165:707–720. doi: 10.1093/genetics/165.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bayona-Bafaluy MP, Movilla N, Perez-Martos A, Fernandez-Silva P, Enriquez JA. Functional genetic analysis of the mammalian mitochondrial DNA encoded peptides: a mutagenesis approach. Methods Mol. Biol. 2008;457:379–390. doi: 10.1007/978-1-59745-261-8_28. [DOI] [PubMed] [Google Scholar]
- 17.Perales-Clemente E, Fernandez-Vizarra E, Acin-Perez R, Movilla N, Bayona-Bafaluy MP, Moreno-Loshuertos R, Perez-Martos A, Fernandez-Silva P, Enriquez JA. Five entry points of the mitochondrially encoded subunits in Mammalian complex I assembly. Mol. Cell Biol. 30:3038–3047. doi: 10.1128/MCB.00025-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naldini L, Blomer U, Gage FH, Trono D, Verma IM. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc. Natl Acad. Sci. USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chomyn A. In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol. 1996;264:197–211. doi: 10.1016/s0076-6879(96)64020-8. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez-Silva P, Acin-Perez R, Fernandez-Vizarra E, Perez-Martos A, Enriquez JA. In vivo and in organello analyses of mitochondrial translation. Methods Cell Biol. 2007;80:571–588. doi: 10.1016/S0091-679X(06)80028-2. [DOI] [PubMed] [Google Scholar]
- 21.Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA. Respiratory active mitochondrial supercomplexes. Mol. Cell. 2008;32:529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 22.Schagger H. Native electrophoresis for isolation of mitochondrial oxidative phosphorylation protein complexes. Methods Enzymol. 1995;260:190–202. doi: 10.1016/0076-6879(95)60137-6. [DOI] [PubMed] [Google Scholar]
- 23.Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat. Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 24.Hofhaus G, Shakeley RM, Attardi G. Use of polarography to detect respiration defects in cell cultures. Methods Enzymol. 1996;264:476–483. doi: 10.1016/s0076-6879(96)64043-9. [DOI] [PubMed] [Google Scholar]
- 25.Bayona-Bafaluy MP, Acin-Perez R, Mullikin JC, Park JS, Moreno-Loshuertos R, Hu P, Perez-Martos A, Fernandez-Silva P, Bai Y, Enriquez JA. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 2003;31:5349–5355. doi: 10.1093/nar/gkg739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi X, Sun L, Lewin AS, Hauswirth WW, Guy J. The mutant human ND4 subunit of complex I induces optic neuropathy in the mouse. Invest. Ophthalmol. Vis. Sci. 2007;48:1–10. doi: 10.1167/iovs.06-0789. [DOI] [PubMed] [Google Scholar]
- 27.Guy J, Qi X, Koilkonda RD, Arguello T, Chou TH, Ruggeri M, Porciatti V, Lewin AS, Hauswirth WW. Efficiency and safety of AAV-mediated gene delivery of the human ND4 complex I subunit in the mouse visual system. Invest. Ophthalmol. Vis. Sci. 2009;50:4205–4214. doi: 10.1167/iovs.08-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dey R, Barrientos A, Moraes CT. Functional constraints of nuclear-mitochondrial DNA interactions in xenomitochondrial rodent cell lines. J. Biol. Chem. 2000;275:31520–31527. doi: 10.1074/jbc.M004053200. [DOI] [PubMed] [Google Scholar]
- 29.Barrientos A, Kenyon L, Moraes CT. Human xenomitochondrial cybrids. Cellular models of mitochondrial complex I deficiency. J. Biol. Chem. 1998;273:14210–14217. doi: 10.1074/jbc.273.23.14210. [DOI] [PubMed] [Google Scholar]
- 30.Kenyon L, Moraes CT. Expanding the functional human mitochondrial DNA database by the establishment of primate xenomitochondrial cybrids. Proc. Natl Acad. Sci. USA. 1997;94:9131–9135. doi: 10.1073/pnas.94.17.9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKenzie M, Chiotis M, Pinkert CA, Trounce IA. Functional respiratory chain analyses in murid xenomitochondrial cybrids expose coevolutionary constraints of cytochrome b and nuclear subunits of complex III. Mol. Biol. Evol. 2003;20:1117–1124. doi: 10.1093/molbev/msg132. [DOI] [PubMed] [Google Scholar]
- 32.Pogozelski WK, Fletcher LD, Cassar CA, Dunn DA, Trounce IA, Pinkert CA. The mitochondrial genome sequence of Mus terricolor: comparison with Mus musculus domesticus and implications for xenomitochondrial mouse modeling. Gene. 2008;418:27–33. doi: 10.1016/j.gene.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonnet C, Kaltimbacher V, Ellouze S, Augustin S, Benit P, Forster V, Rustin P, Sahel JA, Corral-Debrinski M. Allotopic mRNA localization to the mitochondrial surface rescues respiratory chain defects in fibroblasts harboring mitochondrial DNA mutations affecting complex I or v subunits. Rejuvenation Res. 2007;10:127–144. doi: 10.1089/rej.2006.0526. [DOI] [PubMed] [Google Scholar]
- 34.Corral-Debrinski M. mRNA specific subcellular localization represents a crucial step for fine-tuning of gene expression in mammalian cells. Biochim. Biophys. Acta. 2007;1773:473–475. doi: 10.1016/j.bbamcr.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Ellouze S, Augustin S, Bouaita A, Bonnet C, Simonutti M, Forster V, Picaud S, Sahel JA, Corral-Debrinski M. Optimized allotopic expression of the human mitochondrial ND4 prevents blindness in a rat model of mitochondrial dysfunction. Am. J. Hum. Genet. 2008;83:373–387. doi: 10.1016/j.ajhg.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonnet C, Augustin S, Ellouze S, Benit P, Bouaita A, Rustin P, Sahel JA, Corral-Debrinski M. The optimized allotopic expression of ND1 or ND4 genes restores respiratory chain complex I activity in fibroblasts harboring mutations in these genes. Biochim. Biophys. Acta. 2008;1783:1707–1717. doi: 10.1016/j.bbamcr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 37.Claros MG. MitoProt, a Macintosh application for studying mitochondrial proteins. Comput. Appl. Biosci. 1995;11:441–447. doi: 10.1093/bioinformatics/11.4.441. [DOI] [PubMed] [Google Scholar]
- 38.Boer PH, Gray MW. Transfer RNA genes and the genetic code in Chlamydomonas reinhardtii mitochondria. Curr. Genet. 1988;14:583–590. doi: 10.1007/BF00434084. [DOI] [PubMed] [Google Scholar]
- 39.Perez-Martinez X, Vazquez-Acevedo M, Tolkunova E, Funes S, Claros MG, Davidson E, King MP, Gonzalez-Halphen D. Unusual location of a mitochondrial gene. Subunit III of cytochrome C oxidase is encoded in the nucleus of Chlamydomonad algae. J. Biol. Chem. 2000;275:30144–30152. doi: 10.1074/jbc.M003940200. [DOI] [PubMed] [Google Scholar]
- 40.Perez-Martinez X, Antaramian A, Vazquez-Acevedo M, Funes S, Tolkunova E, d'Alayer J, Claros MG, Davidson E, King MP, Gonzalez-Halphen D. Subunit II of cytochrome c oxidase in Chlamydomonad algae is a heterodimer encoded by two independent nuclear genes. J. Biol. Chem. 2001;276:11302–11309. doi: 10.1074/jbc.M010244200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.