Abstract
Previous work has provided strong evidence for a role of peroxisome proliferator-activated receptor β/δ (PPARβ/δ) and transforming growth factor-β (TGFβ) in inflammation and tumor stroma function, raising the possibility that both signaling pathways are interconnected. We have addressed this hypothesis by microarray analyses of human diploid fibroblasts induced to myofibroblastic differentiation, which revealed a substantial, mostly reverse crosstalk of both pathways and identified distinct classes of genes. A major class encompasses classical PPAR target genes, including ANGPTL4, CPT1A, ADRP and PDK4. These genes are repressed by TGFβ, which is counteracted by PPARβ/δ activation. This is mediated, at least in part, by the TGFβ-induced recruitment of the corepressor SMRT to PPAR response elements, and its release by PPARβ/δ ligands, indicating that TGFβ and PPARβ/δ signals are integrated by chromatin-associated complexes. A second class represents TGFβ-induced genes that are downregulated by PPARβ/δ agonists, exemplified by CD274 and IL6, which is consistent with the anti-inflammatory properties of PPARβ/δ ligands. Finally, cooperative regulation by both ligands was observed for a minor group of genes, including several regulators of cell proliferation. These observations indicate that PPARβ/δ is able to influence the expression of distinct sets of both TGFβ-repressed and TGFβ-activated genes in both directions.
INTRODUCTION
Peroxisome proliferator-activated receptors (PPARs) are nuclear receptors that function as ligand-inducible transcription factors (1–3). The three PPAR subtypes (PPARα, PPARβ/δ and PPARγ) activate their target genes through binding to PPAR response elements (PPREs) as heterodimers with members of the retinoid X receptor (RXR) family. PPARs play a central role in lipid metabolism by serving as sensors for fatty acids and fatty acid metabolites with major function as modulators of metabolic and inflammatory processes. Consequently, the transcriptional activity of PPARs is modulated not only by natural fatty acids, but also by lipid-derived metabolites such as prostaglandins J2 and I2, leukotriene A4, 15-hydroxyeicosatetraenoic acid and 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine (4–7). PPARs also play essential roles in developmental processes, wound healing, cell differentiation and proliferation and many associated diseases, including arteriosclerosis, diabetes, fibrosis, inflammatory disorders and cancer (8–12), which led to the development of numerous subtype-selective, high-affinity ligands (13).
We and others have shown that PPARβ/δ plays an essential role in regulating the differentiation, function and proliferation of tumor stroma cells (14–16). Ppard-null mice show gross alterations of tumor endothelial cells and fibroblasts, resulting in a high proportion of immature, dysfunctional microvessels and increased numbers of myofibroblastic cells (14). Consistent with these in vivo data, overexpression of PPARβ/δ inhibited the proliferation of cultured fibroblasts (14). Likewise, the prostacyclin mimetic Treprostinil inhibited the proliferation of lung fibroblasts concomitant with the transcriptional activation of PPARβ/δ (17). A regulatory role for PPARβ/δ in myofibroblasts has also been shown in a cell culture model of cardiac fibrosis, i.e. neonatal rat cardiac fibroblasts induced to myofibroblast transdifferentiation by culturing on a rigid substrate (18). Finally, different PPAR subtypes have been shown to play a role in experimentally induced lung fibrosis, and it has been suggested that PPARβ/δ agonists may attenuate disease progression by inhibiting myofibroblast proliferation and function (19).
A cytokine present in vast amounts in many tumors and playing a pivotal role in both tumor stroma function, inflammation and tissue fibrosis is the transforming growth factor-β (TGFβ) (20), suggesting that PPARβ/δ and TGFβ signaling pathways may functionally interact. To test this hypothesis, we performed microarray analyses of human lung fibroblasts induced to differentiate into myofibroblastic cells by TGFβ and analyzed the influence of PPARβ/δ agonists on the transcriptional profile. This study revealed an extensive, mainly reverse crosstalk of the transcriptional pathways regulated by PPARβ/δ and TGFβ, leading to the definition of distinct classes of genes. Class A genes are repressed by TGFβ, which is, at least in part, due to the induction of the corepressor SMRT and is counteracted by PPARβ/δ agonists. These include many known PPAR target genes with functions in lipid metabolism. A prominent example is the ANGPTL4 gene, which encodes an important regulator of lipid metabolism and presumptive modulator of metastasis (21,22). In contrast, class B genes are induced by TGFβ and downregulated by PPARβ/δ agonists. These genes include IL6, which may be relevant in view of the reported anti-inflammatory and anti-fibrotic properties of PPARβ/δ.
MATERIALS AND METHODS
Chemicals
TGFβ1 was purchased from Sigma-Aldrich (Karlsruhe, Germany), GW501516, GW1929 and GW7647 from Axxora (Lörrach, Germany), and L165,041 from Calbiochem (Merck, Darmstadt, Germany).
Cell culture
WI-38 cells were obtained from the ATCC and maintained in DMEM/MCDB105 (1:1, PAA, Cölbe, Germany/Sigma, Steinheim, Germany) supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin in a humidified incubator at 37°C and 5% CO2. Differentiation by TGFβ1 was carried out in serum-free medium as described (23,24).
Immunostaining and quantification of stress fibers
Cells were fixed with ethanol (70%), stained by indirect immunofluorescence using a polyclonal α-SMA antibody (Sigma, Steinheim, Germany) visualized by a Cy5-labeled secondary antibody (Molecular Probes A11029, Invitrogen, Karlsruhe, Germany), and counterstained with Hoechst 33258 (Invitrogen). Slides were evaluated with a Leica RMB 3 microscope equipped with fluorescence optics. For quantitative evaluation of SMA stress fibers detected by immunofluorescence, cells showing strong, weak or no staining were counted separately. A total of ∼750 cells in eight microscopic fields were counted per sample.
Small-interfering RNA transfections
Cells were seeded at a density of 5 × 105 cells per 6 cm dish in 4 ml DMEM with 10% fetal calf serum (FCS) and cultured for 2 h. 1280 ng small-interfering RNA (siRNA) in 100 µl OptiMEM (Invitrogen) and 20 µl HiPerfect (Qiagen, Hilden, Germany) were mixed and incubated for 5–10 min at room temperature prior to transfection. The cells were replated 24 h post-transfection at a density of 5 × 105 cells per 6 cm dish. Transfection was repeated 48 h after start of the experiment, and cells were passaged after another 24 h. Forty-eight hours following the last transfection, cells were incubated in serum-free medium for 24 h before stimulation. The NCOR2 siRNA pool was composed of the following sequences:
Hs_NCOR2_1: 5′-GGA CGG AGA UCU UCA AUA U;
Hs_NCOR2_2: 5′-GAA CCU CGA UGA GAU CUU G;
Hs_NCOR2_3: 5′-GGA AAA GAC UCA AAG UAA A;
Hs_NCOR2_4: 5′-GCG CAC CUA UGA CAU GAU G;
control siRNA (#1022563, Qiagen, Hilden, Germany).
Quantitative real-time polymerase chain reaction
Complementary DNA (cDNA) was synthesized from 0.1–1 µg of RNA using oligo(dT) primers and the Omniscript kit (Qiagen, Hilden, Germany). Quantitative polymerase chain reaction (qPCR) was performed in a Mx3000P Real-Time PCR system (Stratagene, La Jolla, CA, USA) for 40 cycles at an annealing temperature of 60 °C. PCR reactions were carried out using the Absolute QPCR SYBR Green Mix (Abgene, Hamburg, Germany) and a primer concentration of 0.2 µM following the manufacturer’s instructions. L27 was used as normalizer. Comparative expression analyses were statistically analyzed by Student’s t-test (two-tailed, equal variance) and Bonferroni correction. The sequences of the primers are as follows:
| ANGPTL4fw: | 5′-GATGGCTCAGTGGACTTCAACC; |
| ANGPTL4rv: | 5′-CCCGTGATGCTATGCACCTTC; |
| L27fw: | 5′-AAAGCCGTCATCGTGAAGAAC; |
| L27rv: | 5′-GCTGTCACTTTCCGGGGATAG; |
| PPARDfw: | 5′-TCATTGCGGCCATCATTCTGTGTG; |
| PPARDrv: | 5′-TTCGGTCTTCTTGATCCGCTGCAT; |
| ADRPfw: | 5′-TGTGAGATGGCAGAGAACGGT; |
| ADRPrv: | 5′-CTGCTCACGAGCTGCATCATC; |
| CPT1Afw: | 5′-ACAGTCGGTGAGGCCTCTTATGAA; |
| CPT1Arv: | 5′-TCTTGCTGCCTGAATGTGAGTTGG; |
| PDK4fw: | 5′-TTGAGTGTTCAAGGATGCTCTG; |
| PDK4rv: | 5′-TGCCCGCATTGCATTCTTAAATA; |
| COL4A1fw: | 5′-ACTCTTTTGTGATGCACACCA; |
| COL4A1rv: | 5′-AAGCTGTAAGCGTTTGCGTA; |
| ACTA2fw: | 5′-TGATCACCATCGGAAATGAA; |
| ACTA2rv: | 5′-TGATGCTGTTGTAGGTGGTTTC; |
| SM22Afw: | 5′-TTGAAGGCAAAGACATGGCAG; |
| SM22Arv: | 5′-CCATCTGAAGGCCAATGACAT; |
| CD274fw: | 5′-GGCATCCAAGATACAAACTCAA; |
| CD274rv: | 5′-CAGAAGTTCCAATGCTGGATTA; |
| CLDN1fw: | 5′-CCCTATGACCCCAGTCAATG; |
| CLDN1rv: | 5′-ACCTCCCAGAAGGCAGAGA; |
| IL6fw: | 5′-CAGGAGCCCAGCTATGAACT; |
| IL6rv: | 5′-AGCAGGCAACACCAGGAG; |
| NCOR1fw: | 5′-TCGCTTCCACTGTTTCTGC; |
| NCOR1rv: | 5′-GGGCTTGACAGCTTCAACTT; |
| NCOR2fw: | 5′-CGGAGTGACCACACACTCAC; |
| NCOR2rv: | 5′-GGGTCTGCCAGAGACCTTG. |
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as described (6), except that nuclei were resuspended at 2.5 × 107/ml, and 60 pulses were applied during sonication. The following antibodies were used: IgG pool, I5006 (Sigma-Aldrich, Steinheim, Germany), α-PPARβ/δ, sc-7197 (Santa Cruz, Heidelberg, Germany); α-SMRT, ab24551 (Abcam, Cambridge, UK). Comparative binding analyses were statistically analyzed by Student’s t-test (two-tailed, equal variance) and corrected for multiple hypothesis testing by the Bonferroni method. Primer sequences were as follows:
| ANGPTL4+3500fw: | 5′-CCTTACTGGATGGGAGGAAAG; |
| ANGPTL4+3500rv: | 5′-CCCAGAGTGACCAGGAAGAC; |
| ANGPTL4-12000fw: | 5′-ACCCTGGGTGTTCATGGTAG; |
| ANGPTL4-12000rv: | 5′-CCCAAGGGGTTCAATGTATTC. |
Microarrays
RNA was isolated using the Nucleospin RNA II kit (Macherey-Nagel, Düren, Germany). RNA quality was assessed using the Experion automated electrophoresis station with RNA StdSens chips (Bio-Rad, Munich, Germany). For microarray studies, total RNA samples were amplified and labeled using the Agilent Quick Amp Labeling Kit (Agilent, Santa Clara, CA, USA) according to the manufacturer's instructions. The amplification procedure consists of reverse transcription of total RNA, including spike-in with an oligo(dT) primer bearing a T7 promoter, followed by in vitro transcription of the resulting cDNA with T7 RNA polymerase in the presence of dye labeled CTP to generate multiple fluorescence labeled copies of each messenger RNA (mRNA). After purification, the labeled aRNA was quantified and hybridization samples were prepared according to the manufacturer's instructions. Human Agilent 4-plex Array 44K were used for the analysis of the gene expression of the different samples in a reference-design assay. As a reference, a pool of all samples was used. This reference was labeled with Cy3, while the samples were labeled with Cy5 dye. The hybridization assembly was performed as described in the Agilent Microarray Hybridization Chamber User Guide (G2534-90001). After a 17-h hybridization at 65°C, slides were washed as described by the manufacturer and subsequently scanned using an Agilent DNA Microarray Scanner G2505C; scan software: Agilent Scan Control Version A.8.1.3; quantification software: Agilent Feature Extraction Version 10.5.1.1 (FE Protocol GE_105_Dec08). Raw microarray data were normalized using the ‘loess’ method implemented within the marray package of R/BioConductor (www.bioconductor.org). Regulated probes were selected on the basis that the logarithmic (base 2) average intensity value was ≥6, and that the fluctuation between replicates was ≤50%.
RESULTS
Induction of myofibroblastic differentiation of diploid human fibroblasts
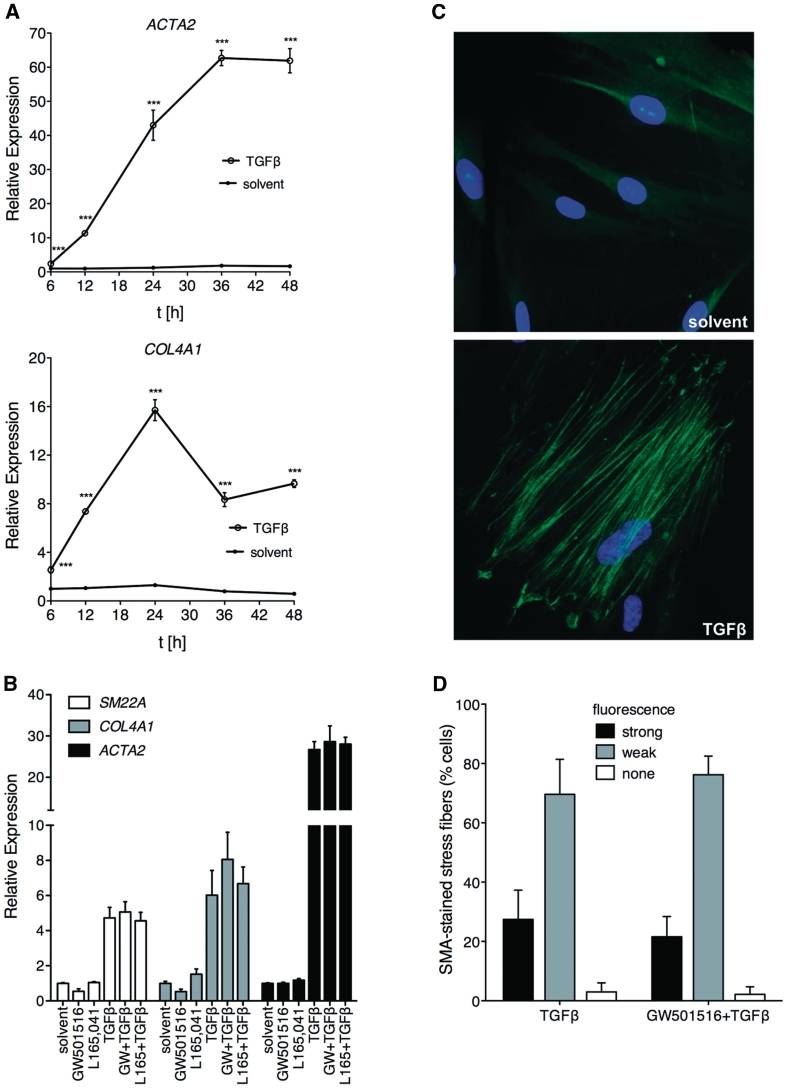
The purpose of the present study was to investigate whether PPARβ/δ and TGFβ signaling pathways functionally interact. As an experimental model, we used diploid human lung fibroblasts (WI38 cells) induced by TGFβ to differentiate into myofibroblast-like cells. In order to characterize this system, we first studied the expression of the myofibroblast marker genes ACTA2 (coding for smooth muscle α-actin; SMA), COL4A1 (encoding collagen type IV α1) and SM22A (coding for smooth muscle protein 22-α). As shown in Figure 1A and B, TGFβ induced the expression all three genes. Increased levels of ACTA2 and COL4A1 mRNA were detectable after 6 h and reached maximum levels after 24–36 h (Figure 1A). In the same experimental setup, no significant effect of the PPARβ/δ agonists GW501516 or L165,041 on the TGFβ-mediated induction of ACTA2, COL4A1 and SM22A was detectable (Figure 1B), suggesting that the ligand-mediated activation of PPARβ/δ does not affect the myofibroblastic differentiation of WI38 cells.
Figure 1.
TGFβ-induced myofibroblast-like differentiation of WI38 cells is not affected by PPARβ/δ ligands. (A) Cells were treated with TGFβ1 (2 ng/ml) or solvent for the indicated times, and the relative expression levels of ACTA2 and COL4A1 were determined by RT-qPCR. ***, significant difference to solvent-treated sample (P < 0.001 by t-test). (B) Expression of ACTA2, COL4A1 and SM22A after 24 h treatment with TGFβ1 (2 ng/ml), GW501516 (0.3 µM), L165,041 (2 µM), TGFβ1 plus PPARβ/δ ligand (as indicated) or solvent determined by RT-qPCR. No significant differences were detectable (t-test, P > 0.1) in PPARβ/δ ligand-treated cells in either the absence or presence of TGFβ. (C) Staining by indirect immunofluorescence of SMA stress fibers (green) in WI38 cells treated with solvent or TGFβ for 24 h as in (A). Nuclei were visualized by Hoechst 33258 staining (blue). (D) Quantitative evaluation of SMA fibers stained by immunofluorescence after treatment of WI38 cells with TGFβ or TGFβ plus GW501516 for 24 h. Cells showing strong, weak or no staining were counted separately. For both samples, a total of 1500 cells in 16 microscopic fields were counted. Error bars represent the standard deviation for individual field counts.
Concomitantly with the induction of these marker genes, SMA-containing stress fibers, a hallmark of differentiating myofibroblasts, were readily detectable after 24 h exposure of WI38 cells to TGFβ (Figure 1C). Consistent with the marker gene expression data in Figure 1B, treatment with the PPARβ/δ agonist GW501516 had no detectable effect on stress fiber formation by TGFβ (Figure 1D).
As the deletion of Ppard in mice has been associated with myofibroblastic differentiation in the tumor stroma, we also investigated whether the inhibition of PPARβ/δ expression in WI38 cells might affect the differentiation status of these cells. Supplementary Figure S1 shows that ACTA2 expression indeed increased after the siRNA-mediated knockdown of PPARβ/δ. Taken together, these observations suggest that PPARβ/δ plays a role in preventing myofibroblastic transdifferentiation under basal conditions, but that its activation by ligands does not prevent TGFβ-induced differentiation.
Genome-wide expression profiling of WI38 cells treated with TGFβ and PPARβ/δ agonist
The fact that PPARβ/δ ligands do not affect the TGFβ-induced differentiation of WI38 cells makes this experimental system suitable to study possible interactions of these signaling pathways in myofibroblasts without interference by an altered differentiation state. Such interactions could, for instance, affect the functional activation or metabolic activity of these cells. We therefore used this model to address two questions: (i) does TGFβ alter the regulation of PPARβ/δ target genes, and (ii) do PPARβ/δ ligands impinge on TGFβ-mediated transcriptional signaling events that are associated with, for instance, inflammatory or fibrotic responses.
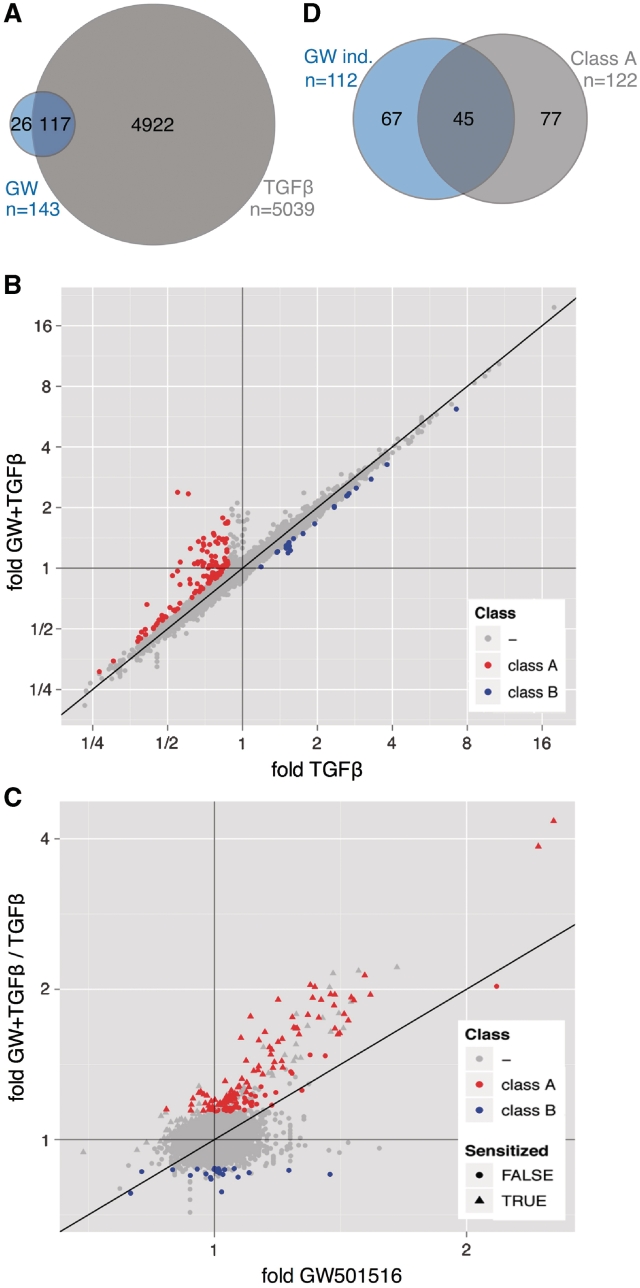
To identify potential functional interactions between TGFβ and PPARβ/δ signaling pathways, we performed microarray analyses of WI38 cells, either untreated (solvent) or treated with GW501516 (0.3 µM), TGFβ1 (2 ng/ml) or both ligands for 24 h (EMBL-EBI ArrayExpress, accession number E-MEXP-2750). As illustrated by the Venn diagram in Figure 2A, 5039 probes indicated regulation by TGFβ and 143 probes regulation by GW501516 (≥1.3-fold change) with an overlap of 117 probes. These correspond to 74 different annotated genes regulated by both ligands.
Figure 2.
Genome-wide expression profiling of WI38 cells treated with TGFβ, PPARβ/δ agonist or both ligands. (A) Venn diagram depicting the numbers of probes showing regulation by TGFβ or GW501516 (≥1.3-fold change). The overlap represents those probes that indicate regulation by both ligands. (B) Dot plot analyzing for individual probes the effect of GW501516 on TGFβ-mediated regulation. Relative expression levels measured after co-treatment of WI38 cells with TGFβ plus GW501516 were plotted against expression levels measured after treatment with TGFβ alone. Red data points represent probes indicating reversion by GW501516 of TGFβ-mediated repression (≥1.3-fold upregulation; class A genes), blue data points represent probes indicating reversion by GW501516 of TGFβ-mediated activation (≥1.3-fold difference; class B genes). (C) Dot plot showing for individual probes a TGFβ-mediated increased GW501516 inducibility. Induction by GW501516 in the presence of TGFβ was plotted against the induction by GW501516 in the absence of TGFβ. The former value was calculated as the ratio of (fold induction by both ligands) / (fold induction by TGFβ). Red data points represent the class A probes defined in panel B. Triangles indicate sensitization by TGFβ, i.e. an increased induction (≥1.3-fold) by GW501516 in the presence of TGFβ (y-value/x-value ≥1.3). (D) Venn diagram illustrating the overlap between class A genes and all genes induced by GW501516 (≥30% induction, n = 112). This analysis includes only those genes, for which the effect of TGFβ could be evaluated in a statistically meaningful way. Therefore, the number of GW501516-induced genes is higher in (A).
To determine cooperative or antagonistic effects exerted by TGFβ and GW501516, we compared for individual genes the transcriptional outcome of exposing WI38 cells to both ligands to that of treatment with either ligand alone, as described in the following sections.
Modulation of TGFβ signaling by PPARβ/δ
The effect of GW501516 on TGFβ-mediated regulation was determined by plotting the relative expression levels measured after co-treatment with both ligands against the expression levels measured after treatment with TGFβ alone. The dot plot in Figure 2B identifies different set of probes showing distinct responses to TGFβ and GW501516.
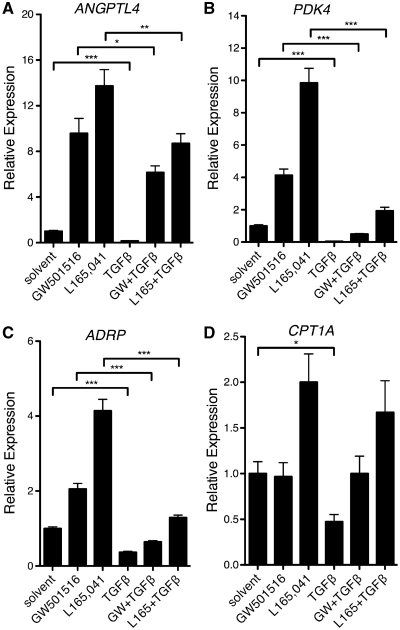
‘Class A’ probes, which represent the major group defined in the present study, indicate repression by TGFβ that is counteracted by GW501516. This pattern was observed for a total of 136 probes, including 122 different annotated genes (cutoff ≥1.3-fold upregulation by GW501516; red data points in Figure 2B; Supplementary Table S1). The characteristic expression pattern of class A genes in response to TGFβ and GW501516 is shown in Figure 3A, and validated by RT-qPCR (Figure 4) for ANGPTL4 (angiopoietin-like 4), PDK4 (pyruvate dehydrogenase kinase 4), CPT1A (carnitine palmitoyltransferase 1A) and ADRP (adipose differentiation-related protein). Several representative genes of this class are listed in Table 1.
Figure 3.
Graphic representation of the reverse effects of GW501516 on TGFβ-mediated gene regulation. The graphics show the expression patterns for the top 10 class A and class B genes identified in Figure 2B. (A) Repression by TGFβ counteracted by GW501516 (class A genes); (B) induction by TGFβ counteracted by GW501516 (class B genes).
Figure 4.
PPARβ/δ counteracts TGFβ-mediated transcriptional repression for a subgroup of target genes. Treatment of WI38 cells with TGFβ and/or PPARβ/δ ligands for 24 h and RT-qPCR analyses of ANGPTL4 (A), PDK4 (B), ADRP (C) and CPT1A (D) expression were performed as in Figure 1B. ***, **, * significant difference (P < 0.001 by t-test, P < 0.01, P < 0.05).
Table 1.
Representative examples of PPARβ/δ target genes regulated by TGFβ-mediated repression and reversal by GW501516 (class A genes)
| Gene | Gene product | TGFβa | GW501516a | (GW501516 + TGFβ) / TGFβb |
|---|---|---|---|---|
| ANGPTL4 | Angiopoietin-like 4 | 0.3 | 6.5 | 18.9 |
| PDK4 | Pyruvate dehydrogenase kinase, isozyme 4 | 0.4 | 4.7 | 4.1 |
| CPT1A | Carnitine palmitoyltransferase 1A | 0.5 | 2.0 | 3.2 |
| DDEF1IT1 | DDEF1 intronic transcript 1 | 0.7 | 1.5 | 2.8 |
| GPR137B | G-protein-coupled receptor 137B | 0.4 | 1.6 | 2.6 |
| SP100 | SP100 nuclear antigen | 0.5 | 1.3 | 2.5 |
| SRGAP1 | SLIT-ROBO Rho GTPase activating protein 1 | 0.5 | 1.4 | 2.3 |
| IMPA2 | Inositol(myo)-1(or 4)-monophosphatase 2 | 0.3 | 1.7 | 2.2 |
| ACAA2 | Acetyl-Coenzyme A acyltransferase 2 | 0.7 | 1.4 | 2.0 |
| ABCA1 | ATP-binding cassette, sub-family A, member 1 (cholesterol transporter) | 0.6 | 1.5 | 1.9 |
| ADRP | Adipose differentiation-related protein | 0.4 | 1.8 | 1.9 |
| CAT | Catalase | 0.4 | 1.5 | 1.8 |
aRelative expression values derived from microarray data (fold induction relative to solvent-treated cells).
bValues reflect GW501516-mediated induction in the presence of TGFβ corrected for the TGFβ effect.
‘Class B’ probes indicate a counteractive effect of GW501516 on TGFβ-mediated activation. This class encompasses 22 probes, representing 21 annotated genes (cutoff ≥1.3-fold difference for TGFβ plus GW501516 relative to TGFβ alone; blue data points in Figure 2B; Supplementary Table S1). Their characteristic expression pattern in response to TGFβ and GW501516 is shown in Figure 3B. The RT-qPCR data in Figure 5 confirm that PPARβ/δ activation counteracts the TGFβ-mediated induction of the class B genes IL6 (interleukin-6), CD274 (B7-H1) and CLDN1 (claudin 1), which was clearly detectable 6 h after application of GW501516, pointing to a direct effect of the PPARβ/δ ligands. No effect on the TGFβ-mediated induction of IL6 was seen with the PPARγ ligand GW1929 or the PPARα agonist GW7647 (Figure 5D), suggesting that the observed effect is PPARβ/δ-specific.
Figure 5.
PPARβ/δ agonists inhibit TGFβ-mediated transcriptional activation for a subgroup of target genes. WI38 cells were treated with the PPARβ/δ ligands GW501516 and L165,041 for 16 h, subsequently stimulated with TGFβ for 6 or 24 h, and CLDN1 (A), CD274 (B) and IL6 (C) expression was analyzed by RT-qPCR. (D) Comparison of the effects of the PPARβ/δ ligand L165, 041, the PPARγ ligand GW1929 and the PPARα agonist GW7647 on TGFβ-mediated induction of IL6. ***, **, * significant difference (P < 0.001 by t-test, P < 0.01, P < 0.05).
Cooperative regulation was also detectable for several probes (Figure 2B; not highlighted; class C and D in Supplementary Table S1), suggesting that GW501516 is able to influence the expression of distinct sets of both TGFβ-repressed and TGFβ-activated genes in both directions. Class C includes KIT, FOXQ1 and TOP2A, which code for the tyrosine kinase receptor KIT, the transcription factor forkhead box Q1 and topoisomerase II, respectively. All three genes have been associated with cell cycle progression and tumorigenesis and may thus be of particular interest with respect to the function of TGFβ and PPARβ/δ in tumor and tumor stroma cells.
Repression of PPARβ/δ target genes by TGFβ and counter-regulation by GW501516
We next determined for individual probes the effect of TGFβ on GW501516 inducibility. This was achieved by plotting the induction by GW501516 in the presence of TGFβ (fold GW501516 plus TGFβ/TGFβ alone) against the induction by GW501516 in the absence of TGFβ (Figure 2C). The predominant probe set identified by this analysis indicates increased induction (≥1.3-fold) by GW501516 in the presence of TGFβ (shown as triangles in Figure 2C). Surprisingly, a substantial number of these probes are identical to those showing repression by TGFβ and counter-regulation by GW501516 (red data points in Figure 2B and C). This overlap (Figure 2D) includes 37% of all class A probes (45/122) and 40% of all GW501516-induced sequences (45/112). The concomitant sensitization by TGFβ to activation by PPARβ/δ agonists and the reversal of the repressive effect of TGFβ by these ligands is also illustrated by the data in Figure 4 and Table 1. These findings suggest that the TGFβ-mediated repression of class A genes and its reversal by PPARβ/δ agonists are functionally linked.
Enhancement of corepressor recruitment to PPAR response elements by TGFβ
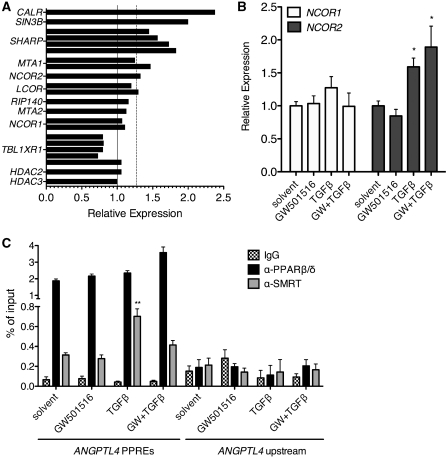
Finally, we addressed the molecular mechanisms that contribute to the regulation of class A genes. The activating and repressive activities of PPARs have been linked to interactions with proteins that serve as coactivators or corepressors, which in turn have profound effects on the local chromatin structure (9,25). Analysis of our microarray revealed a higher expression of several genes encoding corepressors of nuclear receptors in TGFβ-treated cells relative to solvent controls. These include NCOR1 (coding for NCOR), NCOR2 (encoding SMRT), SHARP, LCOR, SIN3B, MTA1 and CALR (Figure 6A). Previous work by several laboratories has established a role for the corepressors NCOR and SMRT in transcriptional repression by unliganded PPARβ/δ in vivo (9,25–28). Upregulation of NCOR2 was observed in RT-qPCR experiments already 6 h after treatment with TGFβ, whereas the induction of NCOR1 was statistically not significant at this time point (Figure 6B).
Figure 6.
TGFβ induces corepressor expression and recruitment to the PPRE enhancer of the ANGPTL4 gene in vivo. (A) Microarray data were analyzed for TGFβ-mediated effects on potential corepressor genes and plotted as relative expression values (TGFβ treatment versus solvent control). The dashed line denotes a threshold of 1.3-fold induction. (B) RT-qPCR analysis of NCOR1 and SMRT expression 6 h following treatment of WI38 cells with GW501516, TGFβ1 or both ligands. (C) TGFβ induces SMRT recruitment to the ANGPTL4 PPRE enhancer in vivo. WI38 cells were treated with 0.3 µM GW501516, 2 ng/ml TGFβ1, or both for 24 h, and ChIP was carried out with antibodies against PPARβ/δ, SMRT or a nonspecific IgG pool, and an ANGPTL4 region containing the PPRE enhancer (+3500 bp relative to the transcription start site) was amplified by qPCR. An ANGPTL4 upstream region was included as a control. Signals were calculated relative to 1% of input DNA. **, * significant difference to solvent-treated sample (P < 0.01 by t-test, P < 0.05).
We therefore analyzed whether TGFβ might influence the recruitment of SMRT to the PPREs of the ANGPTL4 gene in vivo. Figure 6C shows that this is indeed the case. TGFβ treatment induced a 2.2-fold enhanced recruitment relative to solvent-treated cells, which was decreased to 1.3-fold in the presence of GW501516. This correlates well with the observed changes in ANGPTL4 expression, pointing to a causal relationship between the regulation of class A genes and the recruitment of SMRT in response to TGFβ and GW501516.
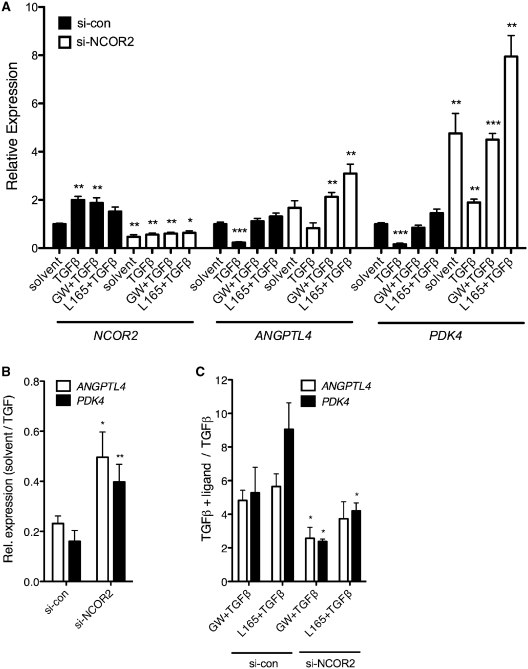
To test this hypothesis, we analyzed the impact of NCOR2 siRNA interference on TGFβ and GW501516-regulated ANGPTL4 and PDK4 gene expression. As shown in Figure 7A (left), the treatment of WI38 cells with NCOR2 siRNA reduced NCOR2 expression to 28–46% relative to cells exposed to control siRNA. The same treatment also attenuated the TGFβ-mediated repression of both PPAR target genes, whose relative expression levels increased in NCOR2 siRNA-treated cells from 0.23 to 0.50 for ANGPTL4, and from 0.16 to 0.40 for PDK4 (Figure 7A and B), respectively. This increased basal level expression was paralleled by a decreased inducibility by PPARβ/δ ligands in the presence of TGFβ, which dropped by ∼50% for both genes (Figure 7A and C). The fact that similar patterns were seen with both ANGPTL4 and PDK4 indicates that the regulatory mechanism identified in this study is not gene-specific. Taken together, these observations clearly establish a functional connection between SMRT, TGFβ and the transcription of PPARβ/δ target genes.
Figure 7.
NCOR2 induction plays a role in the TGFβ-triggered repression of PPARβ/δ target genes. (A) WI38 cells were transfected with NCOR2 or control siRNA as described in ‘Materials and Methods’ section. Twenty-four hours after serum deprivation the cells were treated with TGFβ1 (2 ng/ml), TGFβ1 + GW501516 (0.3 µM), TGFβ1 + L165,041 (2 µM) or solvent for 10 h, and NCOR2, ANGPTL4 and PDK4 mRNA levels were measured by RT-qPCR. The lower inducibility by GW501516 as compared to Figure 4 is presumably due to different cell densities and the siRNA treatment. (B) Effect of siRNA treatment on TGFβ-mediated ANGPTL4 and PDK4 repression. Values represent the ratio of expression levels in solvent-treated cells relative to TGFβ-treated cells. Experimental details as in (A). (C) Effect of siRNA treatment on PPARβ/δ ligand-mediated ANGPTL4 and PDK4 induction in the presence of TGFβ. Values represent the ratio of expression levels in cells treated with PPARβ/δ ligands plus TGFβ relative to TGFβ-treated cells. Experimental details as in (A). ***, **, * significant difference to solvent-treated (A) or si-con (B, C) sample (P < 0.001 by t-test, P < 0.01, P < 0.05).
DISCUSSION
Several lines of evidence strongly suggest that PPARβ/δ plays a role in regulating the differentiation and function of tumor stroma and inflammatory cells, pointing to a crosstalk of PPARβ/δ and cytokine signaling pathways. A cytokine with a pivotal function in inflammation and tumorigenesis is TGFβ. In the present study, we tested this hypothesis by asking whether PPARβ/δ and TGFβ signaling pathways functionally interact and modulate the transcriptional activity of common target genes in diploid human fibroblasts induced to differentiate into myofibroblast-like cells.
Reverse crosstalk of TGFβ and PPARβ/δ signaling
The potential interaction of transcriptional signaling pathways regulated by PPARβ/δ and TGFβ was analyzed by determining the genome-wide transcriptional profile of WI38 cells treated with TGFβ, a PPARβ/δ agonist or both ligands. The data obtained from this analysis point to an extensive crosstalk of the transcriptional signaling pathways regulated by PPARβ/δ and TGFβ (Figures 2 and 3). Bioinformatic analyses identified several classes of genes showing distinct responses to the combined action of TGFβ and PPARβ/δ agonists. Two of these classes that are of particular interest are characterized by the following distinct features (Figures 2B and 3): (i) repression by TGFβ, which is counteracted by PPARβ/δ agonists (class A genes; Table 1), and (ii) induction by TGFβ, which is counteracted by PPARβ/δ agonists (class B genes). In both cases, PPARβ/δ agonists significantly inhibited the effect of TGFβ, indicating that this mode of interaction is a major feature of the interaction of these pathways.
Repression of PPARβ/δ target genes by TGFβ and reversal by GW501516
We also determined for individual probes the effect of TGFβ on ligand-mediated PPARβ/δ activation. This analysis identified a major set of genes, representing mostly classical PPAR target genes, such as ANGPTL4, PDK4, ADRP and CPT1A, which show increased induction by GW501516 in the presence of TGFβ (Figure 2C). These genes overlap to a large extent (37%) with class A genes (red data points in Figure 2B and C), indicating that the enhancement of GW501516 inducibility by TGFβ is functionally linked to their repression by TGFβ.
It has previously been shown that the ANGPTL4 gene is induced by TGFβ in human breast cancer cell lines (21), which is in apparent contrast to the findings reported in the present study. It is, however, well established that TGFβ frequently exerts opposite effects on target gene expression in mesenchymal and epithelial cells, and that neoplastic transformation can subvert TGFβ-mediated transcriptional regulation (29). It would thus be conceivable that the ANGPTL4 gene is also subject to a similarly complex regulatory network. Consistent with this hypothesis is our observation (30) that ANGPTL4 transcription is induced by TGFβ in the epithelial cell line HaCaT (31) and in WPMY-1 cells, which is a SV40-transformed cell line derived from human prostate carcinoma-associated fibroblasts (32). These findings suggest that the ANGPTL4 gene is a useful model to investigate the molecular mechanisms underlying the cell type-specific and transformation-dependent effects of TGFβ-triggered transcriptional signaling pathways.
Correlation of the TGFβ/GW501516-mediated crosstalk with recruitment of the transcriptional corepressor SMRT
In the absence of ligands, PPARβ/δ target genes can be repressed through the recruitment of corepressors to PPRE-bound PPARβ/δ-RXR heterodimers, such as NCOR and SMRT (9,25–28). In the present study, we tested the hypothesis that TGFβ may enhance the formation or function of these repressor complexes. In such a scenario, TGFβ would lead to a decreased transcriptional activity in the absence of ligands, and PPARβ/δ agonists induce the dissociation of corepressors and their replacement with coactivators, thereby counteracting the TGFβ effect. Our data are consistent with this model: (i) the NCOR2 gene (coding for SMRT) is a transcriptional target of TGFβ (Figure 6A and B); (ii) the TGFβ-induced NCOR2 expression leads to an increased recruitment of the SMRT corepressor to the ANGPTL4 PPREs in vivo (Figure 6C); (iii) this enhancement of SMRT recruitment is markedly diminished by the PPARβ/δ agonist GW501516 (Figure 6C); (iv) the siRNA-mediated inhibition of NCOR2 expression leads to a strong derepression of ANGPTL4 transcription and an inhibition of TGFβ-mediated repression (Figure 7A and B); and (v) the same treatment also reduced the inducibility by PPARβ/δ ligands in the presence of TGFβ (Figure 7A and C). These findings provide compelling evidence for a functional link between the TGFβ-induced expression of SMRT, the impact of TGFβ on PPARβ/δ target genes and the counteracting effects of PPARβ/δ ligands. Importantly, similar siRNA effects were also observed with another class A gene, the PPARβ/δ target gene PDK4 (Figures 4 and 7). This suggests that the regulatory mechanism identified here is not a peculiar feature of the ANGPTL4 gene, but appears to a have a broader relevance. Collectively, our findings establish a clear functional connection between the induction of corepressor expression by TGFβ and the transcription of PPARβ/δ target genes, as are illustrated by the model in Figure 8.
Figure 8.
Model illustrating the repression of the PPARβ/δ target genes by TGFβ and its reversion by PPARβ/δ ligands. CoA, coactivator; CoR, corepressor; CoReg, activating or repressing coregulators; orange squares, synthetic PPARβ/δ ligand (GW501516). (Left) the absence of both GW501516 and TGFβ leads to a weak recruitment of positive and negative coregulators, resulting in a low rate of transcription. (Middle) TGFβ induces corepressor genes, including NCOR2, which leads to an enhanced recruitment of SMRT and other corepressors (CoR) to PPRE-bound PPARβ/δ complexes, and consequently an inhibition of transcription. (Bottom) GW501516 induces SMRT dissociation and favors the association with coactivators, leading to transcriptional activation. (Top) Other corepressors (CoR) induced by TGFβ, like those identified in Figure 6A, remain bound to the PPARβ/δ, resulting in a lower level of transcription compared to cells exposed to PPARβ/δ ligands in the absence of TGFβ.
The data in Figure 7A and C indicate that after knockdown of NCOR2 expression, TGFβ still represses ANGPTL4 and PDK4 transcription, albeit to a reduced extent. This suggests that SMRT may not be the only corepressor relevant in this context, and that the PPARβ/δ repressor complex is probably subject to additional regulatory mechanisms triggered by TGFβ. This is supported by the observation that the overall expression level induced by PPARβ/δ ligands is higher than that observed after treatment with ligand plus TGFβ (Figure 4). Consistent with this hypothesis, TGFβ induces several other corepressor genes, such as CALR (calreticulin), LCOR, MTA1, SHARP and SIN3B (Figure 6A), which may play a role in the formation of PPARβ/δ repressor complexes, as previously published for SHARP (9,25–28). The clarification of these questions will be the subject of future studies aiming at a precise dissection of the molecular mechanism involved in the regulation of class A genes by PPARβ/δ and TGFβ.
Inhibition of TGFβ-mediated transcriptional activation by PPARβ/δ ligands
The genes represented by the second group are induced by TGFβ, which is diminished by PPARβ/δ agonists (Figure 2B, blue data points). This group contains several genes that are potentially relevant in view of the known function of PPARβ/δ in modulating the immune responses. One of these is interleukin-6, a cytokine with both pro-inflammatory and anti-inflammatory properties and a vast range of biological and pathophysiological activities, including a role in tissue fibrosis (33). Time course experiments suggest that repression of TGFβ-mediated IL6 induction by PPARβ/δ ligands is a direct event, because it is detectable within 6 h post-treatment (Figure 5C). The IL6 gene is regulated by multiple transcription factors, including NFκB and C/EBP, which have been suggested to interact with PPARs in different experimental systems (34,35). It is possible that the inhibitory effect of PPARβ/δ on TGFβ-induced IL6 transcription is also associated with these transcription factors. Another potentially interesting gene in this context is CD274 coding for B7-H1, a membrane-bound ligand that modulates the activation or inhibition of lymphocytes and myeloid cells (36). Taken together, these data suggest that in differentiating myofibroblasts PPARβ/δ agonists counteract the effects of TGFβ for a subset of target genes with functions in immune regulation, highlighting the relevance of these compounds as potential anti-fibrotic and anti-inflammatory drugs.
Cooperative signaling by TGFβ and PPARβ/δ
We also detected cooperation of the two signaling pathways for several genes (Supplementary Table S1; class C and D). The cooperatively repressed genes (class C) include the cell cycle and tumorigenesis promoting genes KIT, FOXQ1 and TOP2A. This is of potential interest, because we observed cooperative effects of TGFβ and GW5101516 also on cell-cycle regulation. Thus, GW501516 not only inhibited cell-cycle progression in untreated WI38 cells, but also enhanced the inhibitory effect of TGFβ (Figure S2). The cooperative regulation of genes that have been associated with the cell cycle may thus provide an explanation for the cooperation of GW501516 and TGFβ in the inhibition of cell-cycle progression. However, it cannot be ruled out at present that the cell-cycle effects mediated by the two ligands are functionally unrelated. Inhibition of cell proliferation by PPARβ/δ ligands has previously been reported for a number of other cell lines of different origins, but the underlying molecular mechanisms remain largely obscure (10).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Deutsche Forschungsgemeinschaft (Mu601/12-1 and SFB/TR17); Genomics and Bioinformatics core facility of the LOEWE-Schwerpunkt ‘Tumor and Inflammation’. Funding for open access charge: Research grant (DFG).
Conflict of interest statement. None declared.
REFERENCES
- 1.Feige JN, Gelman L, Michalik L, Desvergne B, Wahli W. From molecular action to physiological outputs: peroxisome proliferator-activated receptors are nuclear receptors at the crossroads of key cellular functions. Prog. Lipid Res. 2006;45:120–159. doi: 10.1016/j.plipres.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol. Rev. 2006;86:465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- 3.Barish GD, Narkar VA, Evans RM. PPARδ: a dagger in the heart of the metabolic syndrome. J. Clin. Invest. 2006;116:590–597. doi: 10.1172/JCI27955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forman BM, Tontonoz P, Chen J, Brun RP, Spiegelman BM, Evans RM. 15-Deoxy-delta 12, 14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR gamma. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 5.Chinetti G, Fruchart JC, Staels B. Peroxisome proliferator-activated receptors (PPARs): nuclear receptors at the crossroads between lipid metabolism and inflammation. Inflamm. Res. 2000;49:497–505. doi: 10.1007/s000110050622. [DOI] [PubMed] [Google Scholar]
- 6.Naruhn S, Meissner W, Adhikary T, Kaddatz K, Klein T, Watzer B, Müller-Brüsselbach S, Müller R. 15-hydroxyeicosatetraenoic acid is a preferential peroxisome proliferator-activated receptor β/δ agonist. Mol. Pharmacol. 2010;77:171–184. doi: 10.1124/mol.109.060541. [DOI] [PubMed] [Google Scholar]
- 7.Chakravarthy MV, Lodhi IJ, Yin L, Malapaka RR, Xu HE, Turk J, Semenkovich CF. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. 2009;138:476–488. doi: 10.1016/j.cell.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michalik L, Wahli W. Peroxisome proliferator-activated receptors (PPARs) in skin health, repair and disease. Biochim. Biophys. Acta. 2007;1771:991–998. doi: 10.1016/j.bbalip.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Ricote M, Glass CK. PPARs and molecular mechanisms of transrepression. Biochim. Biophys. Acta. 2007;1771:926–935. doi: 10.1016/j.bbalip.2007.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters JM, Gonzalez FJ. Sorting out the functional role(s) of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) in cell proliferation and cancer. Biochim. Biophys. Acta. 2009;1796:230–241. doi: 10.1016/j.bbcan.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kilgore KS, Billin AN. PPARβ/δ ligands as modulators of the inflammatory response. Curr. Opin. Investig. Drugs. 2008;9:463–469. [PubMed] [Google Scholar]
- 12.Müller R, Rieck M, Müller-Brüsselbach S. Regulation of cell proliferation and differentiation by PPARβ/δ. PPAR Res. 2008;2008:614852. doi: 10.1155/2008/614852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peraza MA, Burdick AD, Marin HE, Gonzalez FJ, Peters JM. The toxicology of ligands for peroxisome proliferator-activated receptors (PPAR) Toxicol. Sci. 2006;90:269–295. doi: 10.1093/toxsci/kfj062. [DOI] [PubMed] [Google Scholar]
- 14.Müller-Brüsselbach S, Kömhoff M, Rieck M, Meissner W, Kaddatz K, Adamkiewicz J, Keil B, Klose KJ, Moll R, Burdick AD, et al. Deregulation of tumor angiogenesis and blockade of tumor growth in PPARβ-deficient mice. EMBO J. 2007;26:3686–3698. doi: 10.1038/sj.emboj.7601803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdollahi A, Schwager C, Kleeff J, Esposito I, Domhan S, Peschke P, Hauser K, Hahnfeldt P, Hlatky L, Debus J, et al. Transcriptional network governing the angiogenic switch in human pancreatic cancer. Proc. Natl Acad. Sci. USA. 2007;104:12890–12895. doi: 10.1073/pnas.0705505104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müller R, Kömhoff M, Peters JM, Müller-Brüsselbach S. A role for PPARβ/δ in tumor stroma and tumorigenesis. PPAR Res. 2008;2008:534294. doi: 10.1155/2008/534294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ali FY, Egan K, FitzGerald GA, Desvergne B, Wahli W, Bishop-Bailey D, Warner TD, Mitchell JA. Role of prostacyclin versus peroxisome proliferator-activated receptor beta receptors in prostacyclin sensing by lung fibroblasts. Am. J. Respir. Cell. Mol. Biol. 2006;34:242–246. doi: 10.1165/rcmb.2005-0289OC. [DOI] [PubMed] [Google Scholar]
- 18.Teunissen BE, Smeets PJ, Willemsen PH, De Windt LJ, Van der Vusse GJ, Van Bilsen M. Activation of PPARδ inhibits cardiac fibroblast proliferation and the transdifferentiation into myofibroblasts. Cardiovasc Res. 2007;75:519–529. doi: 10.1016/j.cardiores.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 19.Lakatos HF, Thatcher TH, Kottmann RM, Garcia TM, Phipps RP, Sime PJ. The role of PPARs in lung fibrosis. PPAR Res. 2007;2007:71323. doi: 10.1155/2007/71323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massague J. TGFβ in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Padua D, Zhang XH, Wang Q, Nadal C, Gerald WL, Gomis RR, Massague J. TGFβ primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133:66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galaup A, Cazes A, Le Jan S, Philippe J, Connault E, Le Coz E, Mekid H, Mir LM, Opolon P, Corvol P, et al. Angiopoietin-like 4 prevents metastasis through inhibition of vascular permeability and tumor cell motility and invasiveness. Proc. Natl Acad. Sci. USA. 2006;103:18721–18726. doi: 10.1073/pnas.0609025103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desmouliere A, Geinoz A, Gabbiani F, Gabbiani G. Transforming growth factor-β1 induces alpha-smooth muscle actin expression in granulation tissue myofibroblasts and in quiescent and growing cultured fibroblasts. J. Cell. Biol. 1993;122:103–111. doi: 10.1083/jcb.122.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serini G, Gabbiana G. Modulation of alpha-smooth muscle actin expression in fibroblasts by transforming growth factor-β isoforms: an in vivo and in vitro study. Wound Repair Regen. 1996;4:278–287. doi: 10.1046/j.1524-475X.1996.40217.x. [DOI] [PubMed] [Google Scholar]
- 25.Shi Y, Hon M, Evans RM. The peroxisome proliferator-activated receptor δ, an integrator of transcriptional repression and nuclear receptor signaling. Proc. Natl Acad. Sci. USA. 2002;99:2613–2618. doi: 10.1073/pnas.052707099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim HJ, Moon I, Han K. Transcriptional cofactors exhibit differential preference toward peroxisome proliferator-activated receptors α and δ in uterine cells. Endocrinology. 2004;145:2886–2895. doi: 10.1210/en.2004-0011. [DOI] [PubMed] [Google Scholar]
- 27.Krogsdam AM, Nielsen CA, Neve S, Holst D, Helledie T, Thomsen B, Bendixen C, Mandrup S, Kristiansen K. Nuclear receptor corepressor-dependent repression of peroxisome-proliferator-activated receptor δ-mediated transactivation. Biochem. J. 2002;363:157–165. doi: 10.1042/0264-6021:3630157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Semple RK, Meirhaeghe A, Vidal-Puig AJ, Schwabe JW, Wiggins D, Gibbons GF, Gurnell M, Chatterjee VK, O'Rahilly S. A dominant negative human peroxisome proliferator-activated receptor (PPAR) α is a constitutive transcriptional corepressor and inhibits signaling through all PPAR isoforms. Endocrinology. 2005;146:1871–1882. doi: 10.1210/en.2004-1405. [DOI] [PubMed] [Google Scholar]
- 29.Massague J, Blain SW, Lo RS. TGFβ signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 30.Kaddatz K, Adhikary T, Finkernagel F, Meissner W, Müller-Brüsselbach S, Müller R. Transcriptional profiling identifies functional interactions of TGFβ and PPARβ/δ signaling: synergistic induction of ANGPTL4 transcription. J. Biol. Chem. 2010 doi: 10.1074/jbc.M110.142018. July 1, 2010 [Epub ahead of print; doi:10.1074/jbc.M110.142018] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell. Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webber MM, Trakul N, Thraves PS, Bello-DeOcampo D, Chu WW, Storto PD, Huard TK, Rhim JS, Williams DE. A human prostatic stromal myofibroblast cell line WPMY-1: a model for stromal-epithelial interactions in prostatic neoplasia. Carcinogenesis. 1999;20:1185–1192. doi: 10.1093/carcin/20.7.1185. [DOI] [PubMed] [Google Scholar]
- 33.Kovalovich K, DeAngelis RA, Li W, Furth EE, Ciliberto G, Taub R. Increased toxin-induced liver injury and fibrosis in interleukin-6-deficient mice. Hepatology. 2000;31:149–159. doi: 10.1002/hep.510310123. [DOI] [PubMed] [Google Scholar]
- 34.Libermann TA, Baltimore D. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol. Cell. Biol. 1990;10:2327–2334. doi: 10.1128/mcb.10.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orjalo AV, Bhaumik D, Gengler BK, Scott GK, Campisi J. Cell surface-bound IL-1alpha is an upstream regulator of the senescence-associated IL-6/IL-8 cytokine network. Proc. Natl Acad. Sci. USA. 2009;106:17031–17036. doi: 10.1073/pnas.0905299106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.