Abstract
DNA methyltransferases (DNMTs) play an important role in establishing and maintaining DNA methylation. Aberrant expression of DNMTs and their isoforms has been found in many types of cancer, and their contribution to aberrant DNA methylation has been proposed. Here, we generated HEK 293T cells stably transfected with each of 13 different DNMTs (DNMT1, two DNMT3A isoforms, nine DNMT3B isoforms and DNMT3L) and assessed the DNA methylation changes induced by each DNMT. We obtained DNA methylation profiles of DNA repetitive elements and 1505 CpG sites from 808 cancer-related genes. We found that DNMTs have specific and overlapping target sites and their DNA methylation target profiles are a reflection of the DNMT domains. By examining H3K4me3 and H3K27me3 modifications in the 808 gene promoter regions using promoter ChIP-on-chip analysis, we found that specific de novo DNA methylation target sites of DNMT3A1 are associated with H3K4me3 modification that are transcriptionally active, whereas the specific target sites of DNMT3B1 are associated with H3K27me3 modification that are transcriptionally inactive. Our data suggest that different DNMT domains are responsible for targeting DNA methylation to specific regions of the genome, and this targeting might be associated with histone modifications.
INTRODUCTION
DNA methyltransferases can transfer a methyl group from S-adenosylmethionine to cytosine at CpG dinucleotides in mammalian genomes (1). There are three enzymatically active DNA methyltransferases (DNMTs), DNMT1, DNMT3A and DNMT3B, in addition to an enzymatically inactive regulatory factor, DNMT3L (DNA methyltransferase3-like). Furthermore, at least two DNMT3A isoforms and more than 20 DNMT3B isoforms have been identified (2). DNMT1 is essential for cell survival and mammalian development (3). It preferentially methylates hemimethylated CpG palindromes in the DNA and is referred to as a ‘maintenance’ DNA methylation enzyme that faithfully copies the DNA methylation pattern from the parent to the daughter strand of DNA after replication. DNMT3A and DNMT3B are structurally similar and play essential roles in establishing de novo DNA methylation, as well as maintaining DNA methylation (4). In addition to the catalytically active DNMTs, DNMT3L plays an important role in establishing DNA methylation by recruiting or activating de novo DNMTs (5–7). Although many studies have shown differential roles for DNMTs in DNA methylation during development, the roles of DNMTs and their isoforms in aberrant DNA methylation remain to be studied (4,8).
The causal roles of DNMTs and their isoforms in aberrant DNA methylation have been studied. Overexpression of DNMT1 induces hypermethylation of the imprinted Igf2 gene and leads to its biallelic expression (9). Overexpression of Dnmt3b1 induces a loss of imprinting and increases the number of colon tumors in heterozygous mice carrying a mutant adenomatus polyposis coli (Apc) tumor suppressor gene (10). Altered expression levels of DNMT3B have also been observed during oncogenic transformation induced by SV40T antigen and activated Ras (11). Overexpression of the DNMT3B4 isoform, a splice variant of DNA methyltransferase 3B, is associated with DNA hypomethylation in pericentromeric satellite regions and affects specific gene expression during hepatocarcinogenesis (12,13). DNMTΔ3B isoforms, which lack the N-terminal domain of DNMT3B, are predominantly expressed in non-small cell lung cancer and are associated with RASSF1A promoter methylation (14–16). The DNMT3B7 isoform encodes truncated proteins lacking a catalytic methyltransferase domain and alters both gene expression and DNA methylation (2). The roles of DNMT isoforms in gene-specific DNA methylation have been studied previously; however, the profiling of de novo DNA methylation target sites for each DNMT isoform is necessary to understand the role of each DNMT isoform.
Hypermethylation of CpG islands in cancer cells is found to be linked to the gain of repressive histone marks, such as methylation of histone H3 on lysine 9 (H3K9) and lysine 27 (H3K27). H3K9 methylation, a repressive mark, is associated with hypermethylated genes, and H3K9 di- and tri-methylation (H3K9me2 and H3K9me3) are found in hypermethylated genes in embryonic carcinoma cells (17,18). Cancer-specific hypermethylated genes are also packaged with polycomb-mediated H3K27me3 in both embryonic stem (ES) cells and cancer cells (18–20). The interaction of EZH2 (Enhancer of Zeste homolog 2), which is a component of polycomb complex 2/3, with DNMTs provides a mechanism for crosstalk between the polycomb complex and DNA methylation (21). Most of the genes marked with H3K27me3, however, also possess H3K4me3, which is known to be ‘bivalent chromatin structure’ in ES cells (22,23). This bivalent state usually commit to either H3K4me3 or H3K27me3 during differentiation (23), suggesting that both H3K4me3 and H3K27me3 marked regions in somatic cells might be vulnerable to the aberrant DNA methylation observed in cancer cells. The mechanism of how these different histone marked genes become methylated in cancer, however, is not clear.
In this study, we successfully identified de novo DNA methylation target sites of specific DNMTs for the first time by mimicking cancer cell overexpression of DNMTs. Furthermore, we characterized the relationship of histone modifications at de novo DNA methylation target sites of DNMTs in 808 cancer-related genes.
MATERIALS AND METHODS
DNA methyltransferase isoform constructs
pIRESpuro3 vector (Clontech), a stable integrating plasmid, was modified by ligating a myc epitope tag sequence (forward 5′-CTAGCCCACCATGGAGCAGAAGCTGATCTCAG-AGGAGGACCTGG-3′ and reverse 5′-AATTCCAG-GTCCTCCTCTGAGATCAG-CTTCTGCTCCATGGTGGG-3′) to the NheI-EcoRI sites of the pIRESpuro3 vector. pcDNA3/Myc-human DNMT1, DNMT3A1, DNMT3A2, DNMT3B1, DNMT3B2, DNMT3B3 and DNMT3L constructs were kindly provided by Dr. Arthur D. Riggs from the City of Hope Cancer Center (24). These DNMTs were subcloned into the EcoR1–NotI sites of the pIRESpuro/Myc vector to facilitate selection of functional clones stably overexpressing the N-terminal myc-tagged DNMT isoforms. The C-terminus of DNMT3B4 was amplified from a pcDNA/Myc DNMT3B2 construct using Pfu Taq polymerase (Stratagene) with primers (forward 5′-TGGGTCCAGTGGTTTGGCGATG-3′ and reverse 5′-GAGCGGCCGCTTAACTGTTCA-TCCCGGGTAGG -3′) and ligated into the BstXI–NotI sites of the pIRESpuro/Myc DNMT3B2 vector. The C-terminus of the DNMT3B5 isoform was amplified with primers (forward 5′-AACTCGAGCTGCAGGACTGCTTGGAATACAATAGGA-TAGCCAAGGATCTTTGGCTTTCCTGTGC-3′ and reverse 5′-TTCTCGAGCTATT-CACATGCAAAGTAGTC-3′), ligated to the XhoI site of pcDNA3/Myc DNMT 3B2, and subcloned into the pIRESpuro/Myc vector. Human DNMTΔ3B1or DNMTΔ3B2 cDNA was amplified from pcDNA3/Myc-DNMT 3B1 with primers (forward; 5′-TCGAATTCATGGAGTCCCCGCAGGTGGAG-3′ and reverse; 5′- CATCGCCAA-ACCACTGGACCCA-3′) and ligated into the EcoRI–BstX1 sites of pIRESpuro/Myc-DNMT3B1 or DNMT3B2, respectively. To generate DNMTΔ3B3 and DNMTΔ3B4, the N-terminal sequence of DNMTΔ3B3 was amplified with primers (forward; 5′- CGAATTCATGGAGTCCCCGCAGGTGGAGGCAGACAGTGGAGATGGAGACAGTTCAGAGTATCAGGTCTCTGC-3′ and reverse; 5′-CTTTCTCCAGAGCATGGTAC-3′) from a DNMTΔ3B3 cDNA, which was amplified from HCT 116 cDNA using RT-PCR and ligated into the EcoRI–MscI sites of IRESpuro/Myc-DNMT3B1 or DNMT3B2, respectively.
Generation of stable cell lines overexpressing DNA methyltransferases
Expression vectors encoding N-terminal myc-tagged DNMT isoforms were transfected into HEK 293T cells using LipofectamineTM 2000 (Invitrogen), and cells were subsequently selected in medium containing Dulbecco’s modified Eagle's medium (DMEM), 10% fetal bovine serum, 1% penicillin/streptomycin and 3 ug/ml puromycin (Clontech) for 30 days. Two independent polyclonal cell lines, indicated as cell line 1 and cell line 2, were established by performing two independent transfection and selection experiments. Cell line 1 was subjected to Illumina DNA methylation analysis and mRNA expression microarray analysis. Both cell line 1 and cell line 2 were subjected to bisulfite PCR pyrosequencing analysis.
mRNA expression by quantitative real time polymerase chain reaction
Total RNA was extracted from puromycin-resistant polyclonal cells stably transfected with DNMT isoforms using the RNeasy Mini kit (Qiagen). Reverse transcription was performed using a first strand cDNA synthesis kit (NEB), according to the manufacturer’s recommended protocol. To quantify mRNA expression, real-time polymerase chain reaction (PCR) was performed with Brilliant SYBR Green QPCR Master Mix, according to the manufacturer’s instructions (Stratagene). For each sample, an RT reaction under the same conditions but without reverse transcriptase was used as a negative control. All data were converted to relative values based on a standard curve and normalized to the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) values obtained for the same sample. All primers are listed in Supplementary Table S1.
Illumina mRNA expression microarray
mRNA expression microarray experiments were performed using a whole-genome expression array (Sentrix Human-6 Expression BeadChip version 3, Illumina) at the University of Southern California Epigenome Center. This array chip consists of about 48 000 probe sequences derived from RefSeq genes and the latest Unigene release (http://www.switchtoi.com/annotationfiles.ilmn). Data were analyzed using the BeadStudio software provided by Illumina.
Protein expression of DNA methyltransferases
The level of ectopically expressed DNMT isoforms was examined by immunoblotting. Briefly, whole-cell extracts from puromycin-resistant polyclonal cell line 1 stably expressing DNMT isoforms were prepared using 2xSDS sample buffer (Bio-Rad). After boiling, cell extracts were separated in 6% and 8% SDS-PAGE gels and transferred onto PVDF membranes (GE Healthcare). DNMT isoforms were detected using an anti-Myc antibody (Invitrogen), and the level of GAPDH in the same samples was determined using an anti-GAPDH antibody (Research Diagnostics Inc.) as a loading control. Alternatively, the percentage of cells overexpressing DNMT proteins was assessed by immunocytochemistry, comparing anti-Myc antibody staining and DAPI staining (Supplementart Figure S3).
DNA methylation measurements by bisulfite PCR pyrosequencing
DNA from cell lines was extracted as described previously (25). Sodium bisulfite conversion of genomic DNA was performed using the EZ DNA Methylation-Gold KitTM (Zymo Research), according to the manufacturer’s recommended protocol. All DNA repetitive elements, including LINE1, Alu Yb8, Sat-α and NBL2 were measured in duplicate, as described previously (26,27). For validation of the Illumina DNA methylation data, DNA methylation at the same CpG site measured by the Illumina DNA methylation assay was quantitatively assessed by measuring the ratio of unmethylated CpG to methylated CpG using bisulfite PCR pyrosequencing (28). All primers used for bisulfite PCR and pyrosequencing primers are listed in Supplementary Table S1. DNA from HEK 293T cells treated with 5-aza-2′-deoxycytidine (DAC)- or M.SssI-treated DNA was used as a decreased or fully methylated control, respectively, for bisulfite PCR pyrosequencing. DAC-treated DNA was extracted from HEK 293T cells treated with DAC daily for 3 days. M.SssI-treated DNA was obtained as described previously (29).
Illumina DNA methylation analysis
The Illumina GoldenGate Methylation Cancer Panel I was used to measure de novo DNA methylation induced by each DNMT isoform. The Illumina DNA methylation assay was performed on sodium bisulfite-converted DNA from cell line 1 overexpressing each DNMT isoform by the University of Southern California Epigenome Center. Sodium bisulfite conversion for the Illumina assay was performed using the EZ-96 DNA Methylation Kit (Zymo Research). Illumina GoldenGate methods and reagents have been previously described (30). In brief, this assay employs fluorescently tagged competitive primers to amplify methylated (red) or unmethylated (green) alleles at specific CpG sites. The ratio of these competitive PCR primers is scored as a β-value that correlates with the level of DNA methylation (30). In total, 1505 CpG sites from 808 genes are interrogated for DNA methylation using the Illumina GoldenGate Methylation Cancer Panel I (Illumina, GM-17-211) (31). All information from the 1505 loci is listed in Supplementary Table S2. The 1505 sequences on the GoldenGate Methylation Cancer Panel I are fully described at www.illumina.com. Hierarchical cluster analysis of the 1505 loci was performed using the software at www.discover.nci.nih.gov/cimminer/, and the average linkage and Euclidean distance were used in the hierarchical clustering algorithm.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed largely as described previously (32). In brief, HEK 293T cells were grown in 150-mm dishes to 90% confluence. Cells were fixed with 1% formaldehyde for 10 min and sonicated to an average DNA size of 200–1000 bp in nuclear lysis buffer. Lysates were diluted 10 times with the dilution buffer and pre-cleared with 30 µl of a mixture of salmon sperm DNA–protein A agarose slurry (Upstate Biotechnology) to reduce non-specific background. Immunoprecipitation was conducted with normal rabbit IgG (Santa Cruz Biotechnology, sc-2027), anti-H3K4me3 (Abcam, ab8580), anti-H3K9me3 (Abcam, ab8898) or anti-H3K27me3 (Upstate, 07-449) with rotation overnight at 4°C. We added 50 µl of protein A-agarose (Upstate Biotechnology) to the lysates for 2 h, and then washed one time each with low salt wash buffer (150 mM NaCl), high salt wash buffer (300 mM NaCl), and LiCl wash buffer, followed by two times with TE buffer. The immunoprecipitated DNA was purified using a PCR purification kit (Qiagen) and quantified by qPCR using Brilliant SYBR Green QPCR Master Mix or FullVelocity™ SYBR® Green QPCR Master Mix according to the manufacturer's instructions (Stratagene). All data were converted to relative values based on a standard curve and normalized to 1% input values of the same sample. All primers are listed in Supplementary Table S1.
ChIP-on-chip assay
ChIP DNA was amplified using the WGA2 Kit (Sigma), according to manufacturer’s recommended protocol. Labeling of ChIP DNA and input, hybridization to NimbleGen 385K RefSeq promoter arrays, and scanning were performed using NimbleGen protocols in their service laboratory in Iceland. To detect the peaks for histone modification, the data were processed using a false discovery rate (FDR < 0.2) by NimbleGen (www.nimblegen.com).
RESULTS
Generation of cell lines stably expressing DNMT isoforms
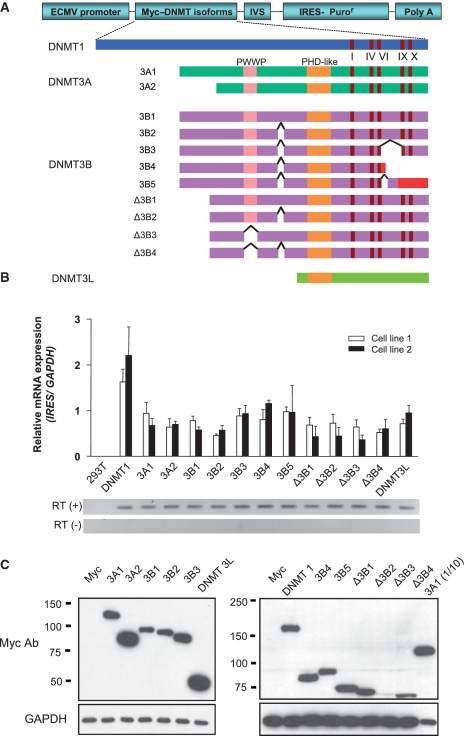
We established cell lines that stably express each of 13 different DNMTs in HEK 293T cells. Endogenous expression of these DNMTs, except DNMT3A2, was detected in HEK 293T cells, as shown in Supplementary Figure S1. At 30 days post-transfection, we were able to obtain polyclonal cells stably expressing various levels of different DNMTs (Figure 1A). The level of ectopically overexpressed DNMTs was compared with the endogenous expression of DNMT using real-time RT-PCR (Supplementary Figure S2). We examined the ectopic mRNA expression of each DNMT by real time RT-PCR of the IRES (internal ribosomal entry site), which is transcribed with DNMT (Figure 1B). We found that different DNMTs and their isoforms are expressed at similar levels in two different polyclonal cells by independent transfection experiments. We also confirmed the protein expression levels of DNMTs and their isoforms using immunoblotting with a myc antibody (Figure 1C). In contrast to mRNA expression, the protein expression levels of DNMTs varied. Although it is not completely clear, differences in protein expression levels might be attributable to differences in the stability of the DNMT3B isoforms. We compared the level of endogenous and exogenous DNMT1 and DNMT3A1 proteins (Supplementary Figure S1-C). For DNMT3B1, we could detect exogenous DNMT3B1 but not the endogenous DNMT3B1, possibly due to low endogenous expression, as seen in Supplementary Figure S1-A. Importantly, we later found that the protein expression levels of different DNMTs were not correlated with the DNA methylation changes shown in Figure 3A. Finally, the transfection efficiency was confirmed using immunocytochemistry with a myc antibody (Supplementary Figure S3). Protein expression levels obtained by immunocytochemistry were concordant with the immunoblotting results and showed that all cells express each DNMT. Therefore, the transfection efficiency of different DNMTs did not influence the level of DNA methylation changes detected later.
Figure 1.
Generation of cell lines stably overexpressing DNA methyltransferases (DNMTs). (A) Schematic structure of DNMT isoforms and expression vector constructs. The conserved PWWP and PHD-like domains, DNMT catalytic motifs (I, IV, VI, IX and X), and alternative splicing sites are indicated. The coding regions of DNMT 3B4 and DNMT 3B5 in red represent frame shift mutations generated by alternative splicing. The expression vectors encode N-terminal myc-tagged DNMT isoforms. (B) mRNA expression of exogenous DNMTs. mRNA expression of each DNMT isoform was assessed by real time RT-PCR with primers designed for the IRES region located on a bicistronic transcript and normalized to the expression of GAPDH. (C) Protein expression levels of exogenous DNMT isoforms by western blotting using a myc antibody. The expression level of DNMTs shown in the right panel was lower than those in the left panel, such that a 10-fold higher amount of protein was loaded in the left panel, as shown by the GAPDH control. Note the 10-fold higher amount of DNMT 3A1 in the left panel was compared with 3A1 (1/10) in the right panel.
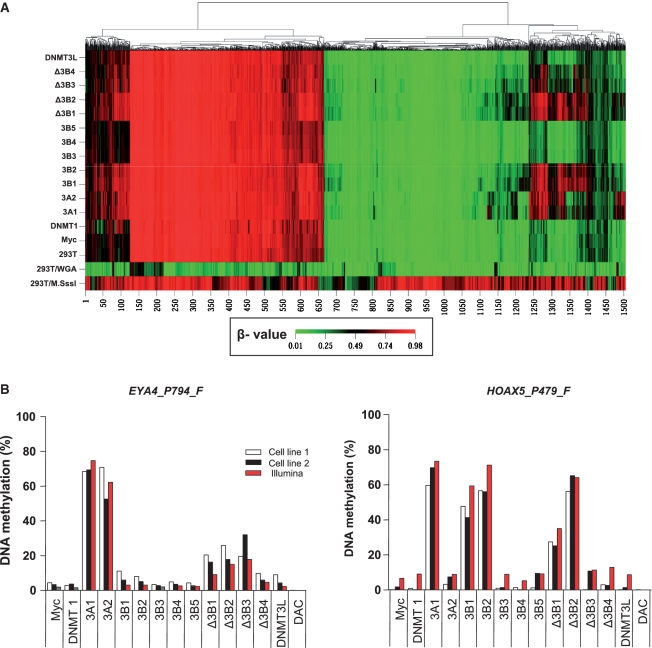
Figure 3.
Gene-specific DNA methylation changes induced by DNMT isoforms. (A) Illumina DNA methylation assay for gene-specific DNA methylation. DNA methylation profiles were one-dimensionally clustered by methylation level at 1505 loci based on the β-value. The β-value, a continuous measure of DNA methylation levels, is represented in colors ranging from green (β-value 0, unmethylated sites) to red (β-value 1, methylated sites) (30). Each column represents 1 of 1505 CpG sites, and each row represents the DNA methylation profile from a stably transfected cell line that expresses each of the different DNMTs indicated on the left. The relatedness of the DNA methylation patterns is represented by a tree (top), with the lengths of the branches representing the degree of similarity based on the average linkage and Euclidean distance. 293T/M.SssI and 293T/WGA (whole genome amplification) were used as methylated and unmethylated controls. (B) Validation of the Illumina DNA methylation assay using bisulfite PCR pyrosequencing. Absolute DNA methylation at the EYA4_P794_F and HOXA5_P479_F loci was directly measured by bisulfite PCR pyrosequencing and compared with the percent methylation extrapolated from the β-value in Illumina DNA methylation assay. Two independent stable transfection experiments are indicated as cell line 1 and cell line 2.
DNA methylation of repetitive elements induced by DNMT isoforms
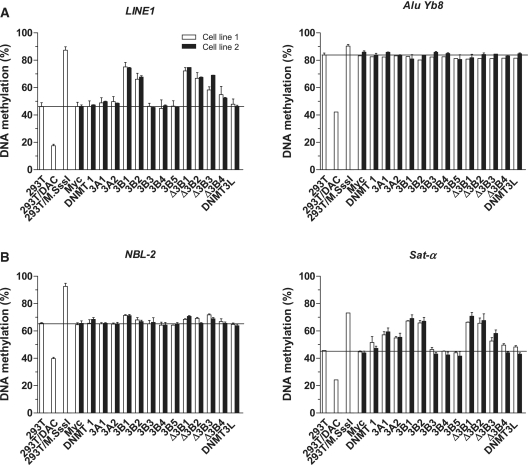
DNA methylation of repetitive elements can be unequally restored by different DNMTs in mouse ES cells (8), raising the possibility that different DNMTs and their isoforms may play distinct roles in methylating different types of repetitive elements in somatic cells as well. To investigate whether different DNMTs could change the DNA methylation of different repetitive elements, we measured the DNA methylation of both interspersed DNA repeats and tandem repeats using a previously described bisulfite PCR pyrosequencing assay (27) (Figure 2). Interspersed DNA repeats can act as a surrogate for global DNA methylation changes through the analysis of the methylation of a pool of repetitive elements dispersed throughout the genome. LINE1, a subfamily of LINEs (Long Interspersed Nuclear Elements), is relatively hypomethylated in HEK 293T cells compared to normal tissues (76%, data not shown); its methylation was increased by DNMT3B1, DNMT3B2 and DNMTΔ3B isoforms, but not de novo DNMT3A isoforms (Figure 2A, left). Another interspersed repeat, AluYb8, a subfamily of SINEs (Short Interspersed Nuclear Elements), did not change its DNA methylation levels significantly with the expression of any isoform. Methylation levels of this element, however, were high to begin with, and this may reflect a technical limitation of our AluYb8 assay (Figure 2A, right).
Figure 2.
DNA methylation changes of DNA repetitive elements induced by DNMT isoforms. (A) DNA methylation changes of two interspersed DNA repeats, LINE1 and AluYb8. (B) DNA methylation changes of two tandem repeats, Sat-α and NBL-2. DNA methylation in HEK 293T cells treated with 5-aza-2′-deoxycytidine, a DNA methylation inhibitor (decreased methylation control), or M.SssI, a CpG methyltransferase (fully methylated control), is shown as 293T/DAC or 293T/M.SssI, respectively. Two independent transfection experiments are indicated as cell line 1 and cell line 2.
We also measured DNA methylation of Sat-α and NBL-2 tandem repeats. Satellite-α (Sat-α) repeats are known to be hypomethylated in cancer and NBL-2 repeats are known to be hypomethylated in ICF (Immunodeficiency, Centromeric instability and Facial abnormalities) syndrome patients who have a germline mutation of DNMT3B. Tandem repeats showed similar, but slightly different, results than the interspersed repeats (Figure 2B). Both DNMT3A and DNMT3B isoforms could increase the DNA methylation of Sat-α, but DNMT3B isoforms (except the catalytically inactive forms) were more efficient in methylating Sat-α repeats. As expected, the maintenance form of the enzyme, DNMT1, in addition to the catalytically inactive DNMT3B isoforms (DNMT3B3, DNMT3B4 and DNMT3B5) and DNMT3L did not change the methylation level of all DNA repeats studied. The role of DNMT3B3 in DNA methylation is controversial. In vitro assays have shown that DNMT3B3 purified from mammalian cells has the same DNA methylation activity as DNMT3B1 (11), but DNMT3B3 in vivo does not affect episome DNA methylation (24). Our data are consistent with the latter finding and support the idea DNMT3B3 might be an inactive DNMT, with impairment of its methyltransferase catalytic motifs VIII and IX (33). Overexpression of DNMT3B4 has been suggested to lead to hypomethylation of pericentromeric satellite regions in human hepatocellular carcinoma (13). According to our data, however, cells with overexpressed DNMT3B4 showed no methylation changes in the DNA repeats studied. The results of two independent stable transfection experiments showed consistent DNA methylation changes for all four different repetitive elements; thus, different DNMTs and their isoforms are responsible for the methylation of different DNA repetitive elements.
High-throughput gene-specific DNA methylation analysis
To identify multiple target sites of different DNMTs and their isoforms in the human genome, we studied the methylation of specific gene promoters using the Illumina GoldenGate Cancer Methylation Panel I, which evaluates the methylation status at 1505 CpG sites of 808 gene promoters. A one-dimensional supervised cluster analysis of all 1505 loci using correlation to measure the distance between loci illustrates groups of genes that are de novo methylated by each DNMT isoform (Figure 3A and Supplementary Table S2). Four de novo methylation sites detected by the Illumina DNA methylation assay were randomly selected, and the methylation changes at these loci were validated using bisulfite PCR pyrosequencing, a more precise and quantitative way to assess DNA methylation (Figure 3B and Supplementary Figure S4). DNA methylation, as determined by the Illumina DNA methylation assay, was highly concordant with the methylation status determined by bisulfite PCR pyrosequencing for the four selected loci in 15 different groups, with the Spearman’s rank correlation coefficient rs = 0.71 (P < 0.0001). These results confirmed the overall validity of the Illumina DNA methylation assay. In addition, two independent transfection experiments (cell line 1 and cell line 2) resulted in the same methylation levels, confirming that DNA methylation induced by different DNMTs is not a stochastic event and is independent of clonal selection.
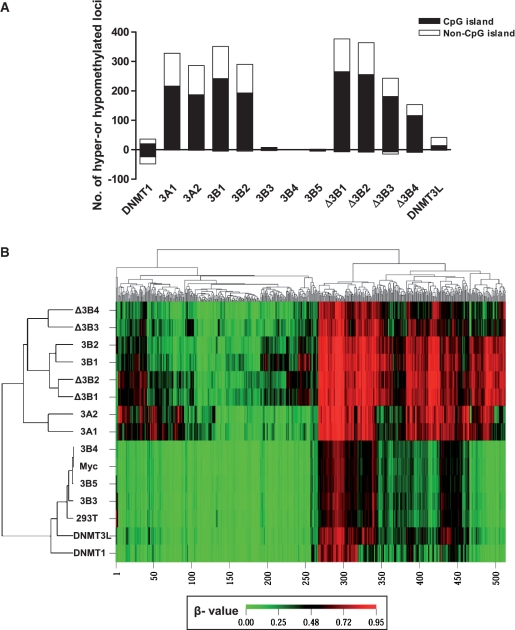
In the Illumina DNA methylation assay, the level of DNA methylation was expressed as a β-value, ranging from 0 (unmethylated sites) to 1 (methylated sites) (30). To determine the loci with changed DNA methylation status induced by DNMTs and their isoforms, we used the absolute difference in β-value (|Δβ| > 0.1843) with two-sided 99% confidence intervals between the non-transfected and the mock vector-transfected HEK 293T control groups (R2 = 0.98). This difference in β-value is comparable to a value previously described as the technical variability of the assay (30). At a change in β > 0.1843, we found that different DNMTs and their isoforms change a variable number of CpG sites (Figure 4A). Overexpression of DNMT3A and DNMT3B isoforms increased DNA methylation at 153–377 loci and both CpG islands and non-CpG islands were hypermethylated. As expected, the catalytically inactive DNMT3B3, 3B4 and 3B5 isoforms did not change the DNA methylation. Interestingly, DNMT1 (36 loci) and DNMT3L (42 loci) induced hypermethylation at a relatively small number of sites compared to the de novo DNMT3A and DNMT3B isoforms, suggesting that DNMT1 and DNMT3L might play a regulatory role in DNA methylation at certain CpG sites through cooperation with endogenous de novo DNMTs (24).
Figure 4.
Distribution of CpG islands and histone marks at loci with changes in DNA methylation. (A) Distribution of CpG islands (black) and non-CpG islands (white) at hyper- and hypomethylated loci induced by each DNMT isoform. Hyper-and hypomethylated CpG sites induced by each DNMT were selected by an absolute difference in the beta value (|Δβ|-value > 0.1843). DNMT1 equally induced hyper and hypomethylated loci. Most isoforms induced exclusively hypermethylation at both CpG islands and non-CpG islands, except 3B3, 3B4 and 3B5, which are known to be catalytically inactive. (B) Unsupervised two-dimensional clustering analysis with all 514 de novo methylated loci induced by any DNMT isoform shows that de novo DNA methylation profiles induced by DNMT isoforms were clustered according to the structural similarity of the DNMT isoforms.
Specific and overlapping target sites of DNMTs
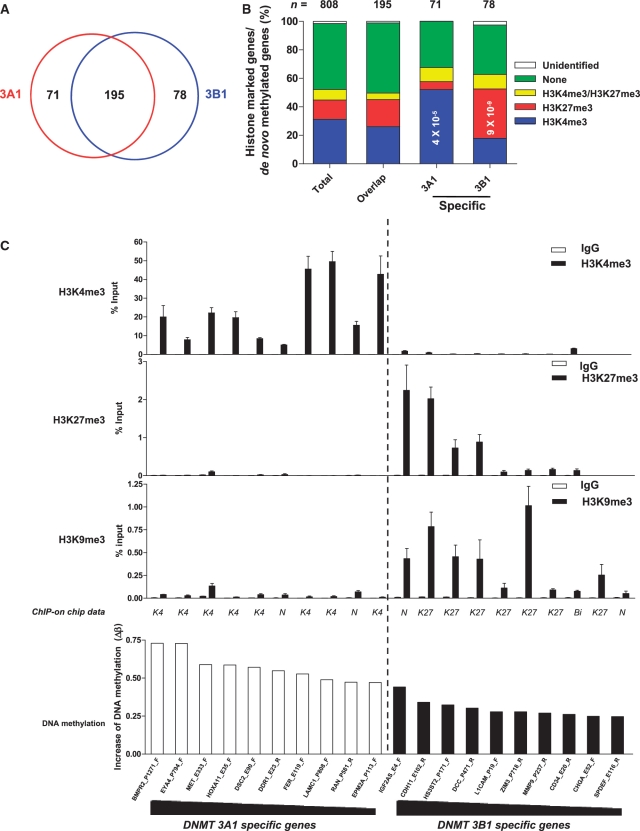
To identify specific and overlapping target sites of DNMTs, 514 de novo methylated sites (β-value increase >0.1843) for any of DNMTs were used in cluster analysis (Figure 4B). Among 514 loci, 304 loci that were not methylated or only slightly methylated in HEK 293T cells (β-value < 0.1843) became methylated exclusively in a subset of DNMT-overexpressing cells. In addition, 210 loci that were already methylated to some extent in HEK 293T cells (β-value ≥ 0.1843) became heavily methylated by a subset of DNMTs. Two-dimensional unsupervised hierarchical cluster analysis showed a correlation of DNA methylation profiles induced by structurally similar DNMTs rather than by the level of DNMT expression, as shown in Figure 1C. We compared the specific and overlapping target sites of pairs of DNMTs that were closely clustered. With regard to DNA methylation target sites, DNMT3A1 overlapped with DNMT3A2 by 93%; DNMT3B1 overlapped with DNMT3B2 by 93%; DNMTΔ3B1 overlapped with DNMTΔ3B2 by 87%; and DNMTΔ3B3 overlapped with DNMTΔ3B4 by 83%. DNMT3A1 and DNMT3B1, which showed the most distinct methylation pattern, made up 90.8% of all 514 de novo methylated loci. The 514 de novo methylated loci corresponded to 397 gene promoter regions, as individual CpG sites were grouped into gene promoter regions. We next compared specific and overlapping target gene promoter regions for DNMT3A1 and DNMT3B1. The DNMT3A1 target genes overlapped with the DNMT3B1 target genes in 195 gene promoter regions; DNMT3A1 had 71 specific target genes, and DNMT3B1 had 78 specific target genes (Figure 5A).
Figure 5.
Histone modifications of DNMT3A1- and DNMT3B1-specific target genes. (A) The Venn diagram depicts the number of specific and overlapping target gene promoter regions between DNMT3A1 and DNMT3B1. DNMT3A1 and DNMT3B1 have 195 overlapping target genes. DNMT3A1 has 71 specific target genes, and DNMT3B1 has 78 specific target genes. (B) The association between DNMT target genes and histone modifications. Cross-application analysis between DNA methylation measurement and ChIP-on-chip analysis of histone modifications (H3K4me3 and H3K27me3) showed that specific target genes of DNMT3A1 are associated with H3K4me3 modifications, whereas the specific target genes of DNMT3B1 are associated with H3K27me3. The total represents the percentage of histone modifications for all 808 genes. Overlapping target genes and specific target genes of DNMTs are shown Figure 5A. The association between de novo methylated sites and histone marks was calculated using a Chi-squared test. (C) Validation of the association between DNA methylation and histone modifications. Chromatin from HEK 293T cells was immunoprecipitated using antibodies against H3K4me3, H3K27me3 or H3K9me3 and analyzed by real-time PCR for 20 loci that showed the greatest increase in de novo DNA methylation changes specific for DNMT3A1 (left 10 loci) and DNMT3B1 (right 10 loci). The increase in DNA methylation was calculated by the difference in the β-value between mock vector-transfected cell lines and DNMT3A1- or DNMT3B1-transfected cell lines. ChIP-on-chip histone data are shown as K4: H3K4me3, K27: H3K27me3, Bi: H3K4me3/H3K27me3, and N: neither H3K4me3 nor H3K27me3. ChIP data represent the means ± SD. of three independent experiments. The white bar represents the IgG control, and the black bar represents enrichment of histone modifications. All data were converted to relative values based on a standard curve and normalized to 1% input values. Real-time PCR primers for 20 loci were designed within ±500 bp at the CpG sites analyzed in the Illumina DNA methylation assay, except the HS3ST2 genes (+1359 bp). See details for the primers in Supplementary Table S1.
Association between DNA methylation and histone modifications
To understand the characteristics of the specific and overlapping target genes for DNMT3Al and DNMT3B1, we studied the association between de novo methylated genes and histone modifications in HEK 293T cells (Figure 5B). We examined H3K4me3 and H3K27me3 modifications of all known well-characterized RefSeq genes using promoter ChIP-on-chip experiments. ChIP DNA for H3K4me3 and H3K27me3 was hybridized to NimbleGen 385K RefSeq promoter arrays. We identified an initial list of 8527 and 3133 genomic regions for H3K4me3 and for H3K27me3, respectively, from 2.2 kb upstream to 0.5 kb downstream of the transcription start sites (Supplementary Tables S3 and S4). By cross-application analysis between the ChIP-on-chip and Illumina DNA methylation assays, we identified histone modifications of 808 of the genes studied for DNA methylation analysis (Figure 5B and Supplementary Table S5).
The overlapping genes between DNMT3A1 and DNMT3B1 had no preference for histone marks; however, we found that specific target genes of DNMT3A1 were associated with H3K4me3 modification (P = 4 × 10−5), while specific target genes of DNMT3B1 were associated with H3K27me3 modification (P = 9 × 10−9). To validate the ChIP-on-chip data, as well as the association between histone modification and DNA methylation, we performed ChIP combined with real-time PCR for the 20 gene promoter regions that showed the greatest increase in de novo DNA methylation specific for DNMT3A1 and for DNMT3B1 (Figure 5C). By comparing the ChIP-on-chip data to the ChIP data for these 20 genes, we found that histone modifications were concordant between the two assays at 17 of the 20 gene promoter regions. In the ChIP experiment, we detected histone modifications at three gene promoter regions (DDR1; H3K4me3, RAN; H3K4me3 and IGF2AS; H3K27me3) for which histone modifications were not detected in the ChIP-on-chip experiment. In the ChIP experiment, all specific DNMT3A1 target genes were marked by H3K4me3; whereas specific DNMT3B1 target genes were marked with H3K27me3 and/or H3K9me3. These data demonstrated that specific target sites of DNMT3A1 and DNMT3B1 are associated with active (H3K4me3) and repressive histone modifications (H3K27me3 and H3K9me3), respectively.
Changes in mRNA expression and histone modification by DNMTs
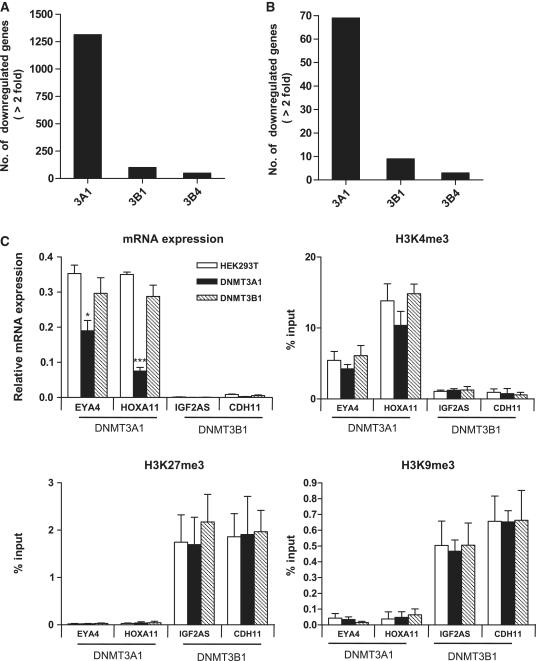
To examine the different effects of DNMT3A1 and DNMT3B1 on gene expression, we have conducted expression array using the Illumina mRNA expression array (HumanWG-6 v3.0 containing 38 276 transcripts) (Figure 6A). Genes with expression levels that were downregulated more than 2-fold with two-sided 99.9% confidence intervals between HEK 293T cells and mock vector-transfected cells were selected (R2 = 0.99). We found that 1314 genes (3.4%) were downregulated in DNMT3A1-overexpressing cells compared to parent cells; whereas a relatively small number of 101 genes (0.26%) was downregulated in DNMT3B1-overexpressing cells, which is comparable to the 49 genes (0.12%) downregulated in catalytically inactive DNMT3B4-overexpressing cells. The same pattern of mRNA downregulation by DNMT3A1 and DNMT3B1 was exhibited in the analysis of 808 genes studied for DNA methylation (Figure 6B). We also found that histone modifications are tightly associated with transcription activity of genes as previously reported (23) (Supplementary Figure S5). Thus, these data suggest that DNMT3A1 can methylate genes that are transcriptionally active with H3K4me3, while DNMT3B1 can methylate genes that are silenced by repressive histone modifications (H3K27me3/H3K9me3). Furthermore, we examined whether this gene repression was coupled to histone modification changes by performing qRT-PCR to evaluate mRNA expression and ChIP experiments to determine the levels of H3K4me3, H3K9me3 and H3K27me3 for DNMT3A1- and DNMT3B1-specific target genes. From the mRNA expression microarray data, we chose EYA4 and HOXA11, which showed both increased DNA methylation and gene downregulation, as DNMT3A1-specific target genes. We found that the basal expression levels of DNMT3A1 target genes (EYA4 and HOXA11) were relatively higher than those of DNMT3B1 target genes (IGF2AS and CDH11), which is consistent with the histone modifications of these genes (Figure 6C). The mRNA expression of the DNMT3A1 target genes was significantly decreased in DNMT3Al overexpressed cells, whereas DNMT3B1 target gene expression at baseline was too low to measure decreases in DNMT3B1 overexpressed cells. However, H3K4me3 was slightly decreased or remained unchanged in DNMT3A1 or DNMT3B1 overexpressed cells, compared to HEK293T cells. We also did not see significant changes in the repressive histone marks, H3K27me3 and H3K9me3 in DNMT3A1 or DNMT3B1 overexpressed cells (Supplementary Figure S6).
Figure 6.
Changes in mRNA expression and histone modification by overexpression of DNMT3A1 and DNMT3B1. Number of downregulated (>2-fold) genes in DNMT3A1- and DNMT3B1-overexpressing cells. The number of downregulated genes in whole genome mRNA representing 8276 transcripts (A) and in the 808 genes studied for DNA methylation (B). (C) Changes in mRNA expression and histone modification. Real-time PCR to measure the mRNA expression of DNMT3A1 target genes (EYA4 and HOXA11) and DNMT3B1 target genes (IGF2AS and CDH11), and ChIP combined with real-time PCR for histone medications (H3K4me3, H3K27me3 and H3K9me3) in HEK 293T cells (white bar), DNMT3A1 cells (black bar) and DNMT3B1 cells (gray bar). Real-time RT-PCR and ChIP data represent the means ± SD. For mRNA expression, the primer efficiency for all four genes was ∼95%, and comparison of the fold changes of the standard curve was consistent with the ΔCt-calculated fold changes. (t-test; *P < 0.05 and ***P < 0.001 versus HEK 293T cells).
The roles of DNMT domains in targeting DNMT isoforms
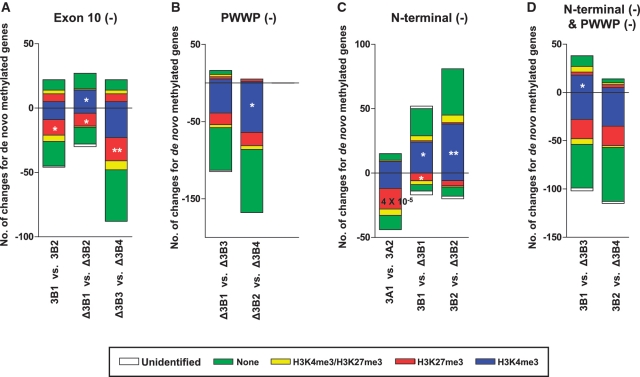
Clustering analysis showed that DNA methylation profiles were dependent on the structural similarity of DNMT isoforms in Figure 4B, suggesting a functional role of DNMT domains in targeting DNMT isoforms to specific sites. To study the role of each domain, we dissected DNMT isoforms by directly comparing isoforms that contained or lacked specific domains. The de novo methylated sites of DNMT isoforms that contained specific domains were directly compared to the de novo methylated gene promoter regions of DNMT isoforms that were identical, with the exception that they lacked the specific domain of interest (Figure 7 and Supplementary Table S6). DNMT3B exon10, which is commonly removed from some DNMT3B isoforms, seemed to be important for de novo DNA methylation, given that overexpression of DNMT3B2 and DNMT Δ3B2, which lack this exon, led to fewer de novo DNA methylation sites than did the overexpression of their counterparts (Figure 7A). The PWWP domain of DNMT3B is known to play an important role in DNA binding (34), and our results support this role given that the lack of the PWWP domain led to fewer de novo methylation events compared to when this domain was present (Figure 7B). Interestingly, the N-terminal domain appeared to play different roles in DNMT3A and DNMT3B. Overexpression of the DNMT3A2 isoform, which lacks 220 N-terminal amino acids of DNMT3A1, led to the de novo methylation of 29 fewer genes (+15 genes and −44 genes) than did overexpression of DNMT3A1. Conversely, the loss of 199 amino acids from the DNMT3B1 N-terminal domain, which defines the delta forms of the DNMT3B isoforms, led to a greater number of de novo methylation sites than did its counterparts (Figure 7C).
Figure 7.
Changes in DNA methylation target sites of DNMT isoforms in the absence of specific DNMT domains. DNA methylated loci were compared between DNMT isoforms that were identical, with the exception that they lacked specific domains. (A) DNMT3B exon10, (B) DNMT3B PWWP, (C) N-terminus of DNMT3A or DNMT3B and (D) both DNMT3B PWWP and the N-terminus. The association between de novo methylated loci and histone marks was calculated using a Chi-squared test; *P < 0.05 and **P < 0.01. All genes are listed in Supplementary Table S6.
The loss of each domain led to changes in DNMT targeting for histone modification (Figure 7). The lack of DNMT3B exon 10 was associated with a decrease in the preference for H3K27me, but the lack of the PWWP domain was related to a decrease in the preference for H3K4me. The loss of the DNMT3A1 N-terminus was associated with a decrease in the preference for H3K27me, while the loss of the DNMT3B1 N-terminus was associated with an increase in the preference for H3K4me3. These results suggest that specific domains can play a role in targeting DNMT isoforms to specific genomic sites and that this targeting may be related to specific histone modifications.
DISCUSSION
While there are numerous studies on DNA methyltransferase enzymes, the target sites of individual DNMT enzymes are poorly understood due to the lack of an efficient cell model system. We generated a cell model system that overexpresses each of 13 different DNMTs and comprehensively identified the de novo methylation sites. Overexpression of catalytically active DNMT3A and DNMT3B isoforms increased de novo DNA methylation, but inactive forms of DNMT3B could not increase DNA methylation. In addition, DNMT1 and DNMT3L induced hypermethylation at a relatively small number of sites compared to isoforms of DNMT3A and DNMT3B, as might be expected. We found that de novo DNA methylation induced by different DNMTs was tightly related to the structure of DNMTs and that their specific target sites were associated with specific histone modifications.
Here, we propose a possible mechanism by which de novo DNA methylation is targeted to different DNA sites in somatic cells (Supplementary Figure S7). By studying the association between unique de novo methylated sites of each DNMT isoform with specific histone modifications, we found that DNMT3A1 was preferentially targeted to H3K4me3-marked genes, while DNMT3B1 was preferentially targeted to H3K27me3- and/or H3K9me3-marked genes. Previous studies have shown a relationship between DNA methylation and H3K4me3 and H3K27me3. The polycomb repressor complexes can directly recruit de novo methyltransferases to H3K27me3-marked genes and lead to specific gene hypermethylation (21). Furthermore, recent genome-wide mapping of histone modifications indicates that bivalent histone marks (H3K4me3/H3K27me3) are more widespread than H3K27me3 alone in ES cells (22). Therefore, frequently hypermethylated genes in malignant cells are significantly enriched with regard to ‘bivalent chromatin structure’ in ES cells, which are usually committed to either H3K4me3 or H3K27me3 in somatic cells, suggesting that not only H3K27me3-marked genes, but also H3K4me3-marked genes, can be methylated in cancer cells. Kondo et al. (35) also showed that the majority of gene silencing induced by DNA hypermethylation in prostate cancer is independent of polycomb-mediated gene silencing, suggesting that the majority of gene silencing caused by DNA hypermethylation in cancer might occur via a mechanism involving more than the polycomb repressive complex. It is not clear, however, how active genes marked with H3K4me3 can be hypermethylated in cancer cells. H3K4me3-marked genes are largely protected from DNA methylation in ES cells since DNMT3L recruits de novo DNMTs to genomic regions devoid of H3K4me3 modifications (5), but an increase in DNA methylation is detected with the loss of H3K4 methylation during differentiation (36). Okitsu et al. (37) reported that DNA methylation plays a role in the exclusion of H3K4 methylation and leads to gene silencing. Here, we show that overexpression of DNMT3A1 or DNMT3B1 could induce de novo DNA methylation at different histone-marked loci, suggesting a possible mechanism for how H3K4me3- or H3K27me3-marked loci could acquire DNA methylation.
It is important to underscore that de novo DNA methylation targeting by DNMT3A1 and DNMT3B1 was not absolutely associated with H3K4me3 and H3K27me3, respectively. Clearly, genes that are associated with H3K4me3 could be de novo methylated by DNMT3B1 and the opposite was true as well. Although it is attractive to think of DNMT targeting to specific histone marks, it is also possible that other factors such as transcriptionally active versus inactive sites could be the determining factor. We attempted to delineate the relationship of DNMT3A1 and DNMT3B1 with either histone mark or transcription by performing global ChIP-on-chip experiments and comparing to expression array profiles; however, due to the tight association of H3K4me3 with transcriptionally active and H3K27me3 with inactive genes, we were unable to determine the exact mechanism of targeting in this work.
It remains to be seen whether these DNA methylation patterns shown in our cell model system can occur in ES cells and primary tumor tissues. Unique cellular environments in different cell types might generate DNA methylation patterns that differ from those observed in our studies. For example, DNMT3L-deficient female mice show DNA methylation defects in maternally imprinted genes, but DNMT3L-deficient male germ cells abolish the methylation of interspersed DNA repeats (6,7). The bivalent chromatin structure and the unique expression patterns of DNMTs might provide a different environment for de novo DNA methylation in ES cells. Another limitation of our cell model system is that target sites already methylated by endogenous DNMTs could not be identified. Nonetheless, the importance of the observed DNA methylation changes might be directly translatable to the epigenetic changes found in cancer and in other diseases. In the present study, de novo methylated genes by different DNMTs coincided with genes that are known to be aberrantly methylated in different types of cancer (Supplementary Table S7). Of the 397 de novo methylated genes, 84 are known to be aberrantly hypermethylated in cancer (http://www.pubmeth.org/). Aberrant DNA methylation via overexpression of DNMT isoforms or changes in DNMT isoform expression patterns could also reflect an important cancer mechanism. The expression of DNMT3A1 and DNMT3B1 is low in normal tissues, but the expression of DNMT3B isoforms is significantly increased in cancer (15,38). DNMTΔ3B isoforms are the predominant forms of DNMT3B expressed in human lung cancer, and one of these isoforms, DNMTΔ3B4, is responsible for promoter-specific methylation of RASSF1A in an NSCLC cell line (14). Recent studies have shown that overexpression of Dnmt3b in conjunction with Apc gene mutations promotes tumorigenesis in mice (10). It is possible that specific aberrant DNA methylation patterns are associated with the overexpression of specific DNMT isoforms detected in different types of cancer.
We believe our model system can be used as a valuable tool to understand how different DNMT isoforms are involved in establishing normal and aberrant DNA methylation patterns. Using this system, we demonstrated that overexpression of DNMT isoforms induces aberrant de novo DNA methylation at specific loci, and analysis of these de novo targets revealed that DNMT isoforms have specific genomic preferences, possibly directed by histone modifications. The knowledge that DNMT isoforms can have specific genomic targets based on specific isoform domains and histone medications suggests a possible mechanism by which normal and abnormal DNA methylation patterns are established.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
A.S.Y. is a Franklin T. Williams scholar and the recipient of an American Society of Clinical Oncology-Association of Subspecialty Professors Career Development Award in Geriatric Oncology. The Southern California Environmental Health Sciences Center pilot grant funded by the National Institute of Environmental Health Sciences (Grant number 5P30ES07048 to S.H.C and the project). S.H.C is the recipient of California Institute for Regenerative Medicine training grant. Funding for open access charge: American Society of Clinical Oncology-Association of Subspecialty Professors Career Development Award in Geriatric Oncology.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Arthur D. Riggs and Zhao-Xia Chen at the City of Hopes Cancer center for generously providing several DNA methyltransferase constructs. We also thank Peter A. Jones, Gangning Liang and Shinwu Jeong for critical reading of the manuscript and Peter W. Laird and Kwang-Ho Lee for helpful discussions regarding the present work.
REFERENCES
- 1.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 2.Ostler KR, Davis EM, Payne SL, Gosalia BB, Exposito-Cespedes J, Le Beau MM, Godley LA. Cancer cells express aberrant DNMT3B transcripts encoding truncated proteins. Oncogene. 2007;26:5553–5563. doi: 10.1038/sj.onc.1210351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, Ueda Y, Li E. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat. Genet. 2007;39:391–396. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- 4.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 5.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 7.Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 8.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biniszkiewicz D, Gribnau J, Ramsahoye B, Gaudet F, Eggan K, Humpherys D, Mastrangelo MA, Jun Z, Walter J, Jaenisch R. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol. Cell. Biol. 2002;22:2124–2135. doi: 10.1128/MCB.22.7.2124-2135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linhart HG, Lin H, Yamada Y, Moran E, Steine EJ, Gokhale S, Lo G, Cantu E, Ehrich M, He T, et al. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soejima K, Fang W, Rollins BJ. DNA methyltransferase 3b contributes to oncogenic transformation induced by SV40T antigen and activated Ras. Oncogene. 2003;22:4723–4733. doi: 10.1038/sj.onc.1206510. [DOI] [PubMed] [Google Scholar]
- 12.Kanai Y, Saito Y, Ushijima S, Hirohashi S. Alterations in gene expression associated with the overexpression of a splice variant of DNA methyltransferase 3b, DNMT3b4, during human hepatocarcinogenesis. J. Cancer Res. Clin. Oncol. 2004;130:636–644. doi: 10.1007/s00432-004-0586-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H, Hirohashi S. Overexpression of a splice variant of DNA methyltransferase 3b, DNMT3b4, associated with DNA hypomethylation on pericentromeric satellite regions during human hepatocarcinogenesis. Proc. Natl Acad. Sci. USA. 2002;99:10060–10065. doi: 10.1073/pnas.152121799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Bhutani M, Pathak AK, Lang W, Ren H, Jelinek J, He R, Shen L, Issa JP, Mao L. Delta DNMT3B variants regulate DNA methylation in a promoter-specific manner. Cancer Res. 2007;67:10647–10652. doi: 10.1158/0008-5472.CAN-07-1337. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Wang J, Sun S, Rodriguez M, Yue P, Jang SJ, Mao L. A novel DNMT3B subfamily, DeltaDNMT3B, is the predominant form of DNMT3B in non-small cell lung cancer. Int. J. Oncol. 2006;29:201–207. [PubMed] [Google Scholar]
- 16.Wang J, Walsh G, Liu DD, Lee JJ, Mao L. Expression of {Delta}DNMT3B variants and its association with promoter methylation of p16 and RASSF1A in primary non-small cell lung cancer. Cancer Res. 2006;66:8361–8366. doi: 10.1158/0008-5472.CAN-06-2031. [DOI] [PubMed] [Google Scholar]
- 17.Kondo Y, Shen L, Yan PS, Huang TH, Issa JP. Chromatin immunoprecipitation microarrays for identification of genes silenced by histone H3 lysine 9 methylation. Proc. Natl Acad. Sci. USA. 2004;101:7398–7403. doi: 10.1073/pnas.0306641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. Epigenetic stem cell signature in cancer. Nat. Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 20.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 21.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 22.Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 24.Chen ZX, Mann JR, Hsieh CL, Riggs AD, Chedin F. Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J. Cell. Biochem. 2005;95:902–917. doi: 10.1002/jcb.20447. [DOI] [PubMed] [Google Scholar]
- 25.Laird PW, Zijderveld A, Linders K, Rudnicki MA, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi SH, Byun HM, Kwan JM, Issa JP, Yang AS. Hydroxycarbamide in combination with azacitidine or decitabine is antagonistic on DNA methylation inhibition. Br. J. Haematol. 2007;138:616–623. doi: 10.1111/j.1365-2141.2007.06707.x. [DOI] [PubMed] [Google Scholar]
- 27.Choi SH, Worswick S, Byun HM, Shear T, Soussa JC, Wolff EM, Douer D, Garcia-Manero G, Liang G, Yang AS. Changes in DNA methylation of tandem DNA repeats are different from interspersed repeats in cancer. Int. J. Cancer. 2009;125:723–729. doi: 10.1002/ijc.24384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uhlmann K, Brinckmann A, Toliat MR, Ritter H, Nurnberg P. Evaluation of a potential epigenetic biomarker by quantitative methyl-single nucleotide polymorphism analysis. Electrophoresis. 2002;23:4072–4079. doi: 10.1002/elps.200290023. [DOI] [PubMed] [Google Scholar]
- 29.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, Doucet D, Thomas NJ, Wang Y, Vollmer E, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Houshdaran S, Cortessis VK, Siegmund K, Yang A, Laird PW, Sokol RZ. Widespread epigenetic abnormalities suggest a broad DNA methylation erasure defect in abnormal human sperm. PLoS ONE. 2007;2:e1289. doi: 10.1371/journal.pone.0001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huebert DJ, Kamal M, O'Donovan A, Bernstein BE. Genome-wide analysis of histone modifications by ChIP-on-chip. Methods. 2006;40:365–369. doi: 10.1016/j.ymeth.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 33.Aoki A, Suetake I, Miyagawa J, Fujio T, Chijiwa T, Sasaki H, Tajima S. Enzymatic properties of de novo-type mouse DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 2001;29:3506–3512. doi: 10.1093/nar/29.17.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen T, Tsujimoto N, Li E. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol. Cell. Biol. 2004;24:9048–9058. doi: 10.1128/MCB.24.20.9048-9058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat. Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 36.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okitsu CY, Hsieh CL. DNA methylation dictates histone H3K4 methylation. Mol. Cell. Biol. 2007;27:2746–2757. doi: 10.1128/MCB.02291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.