Abstract
RNA-binding proteins (RBPs) play a major role in many post-transcriptional processes, including mRNA stability, alternative splicing and translation. PCBP4, also called MCG10, is an RBP belonging to the poly(C)-binding protein family and a target of p53 tumor suppressor. Ectopic expression of PCBP4 induces cell-cycle arrest in G2 and apoptosis. To identify RNA targets regulated by PCBP4 and further decipher its function, we generated multiple cell lines in which PCBP4 is either inducibly over-expressed or knocked down. We found that PCBP4 expression decreases cyclin-dependent kinase inhibitor p21 induction in response to DNA damage. We also provided evidence that PCBP4 regulates p21 expression independently of p53. In addition, we showed that a deficiency in PCBP4 enhances p21 induction upon DNA damage. To validate PCBP4 regulation of p21, we made PCBP4-deficient mice and showed that p21 expression is markedly increased in PCBP4-deficient primary mouse embryo fibroblasts compared to that in wild-type counterparts. Finally, we uncovered that PCBP4 binds to the 3′-UTR of p21 transcript in vitro and in vivo to regulate p21 mRNA stability. Taken together, we revealed that PCBP4 regulates both basal and stress-induced p21 expression through binding p21 3′-UTR and modulating p21 mRNA stability.
INTRODUCTION
Poly(C)-binding proteins (PCBPs), also known as αCPs or hnRNP Es, are ubiquitously expressed RNA-binding proteins (RBPs) involved in many biological processes including gene expression, mRNA stabilization and translation (1–4). The PCBP family is composed of five major proteins: hnRNP K, PCBP1, PCBP2, PCBP3 and PCBP4 (5–9). All PCBP family members possess several hnRNP K homology (KH) domains, which consist of 70 amino-acid motifs essential for binding to poly(C)-rich elements in DNA or RNA targets (10,11). Indeed, PCBPs bind pyrimidine-rich elements in the promoter of various genes to regulate transcription. For example, hnRNP K binds c-myc and eIF4E promoters and recruits the RNA polymerase II machinery to promote transcription (3,12). hnRNP K also acts as co-activator of tumor suppressor p53 to induce p53 target genes and mediate cell-cycle arrest in response to DNA damage (13). In addition, several PCBPs bind the mu opioid receptor (MOR) gene promoter and can either activate (hnRNP K, PCBP1 and PCBP2) or repress (PCBP3) MOR transcription in neuronal cells (4,14,15). Notably, PCBP1 and PCPB2 promote the stabilization of important long-lived transcripts, such as α-globin, collagen α 1 and tyrosine hydroxylase through binding cytosine-rich elements in their 3′ untranslated region (3′-UTR) (16–18). Through mRNA binding, PCBP1, PCBP2 and hnRNP K also regulate the translation of several cellular and viral mRNAs, such as c-myc, 15-lipoxygenase, poliovirus and HPV 16 L2 mRNAs (2,19–21). To this day, specific DNA or RNA targets regulated by PCBP4 have not been identified.
The PCBP4 gene encodes at least four variants: MCG10, MCG10a, αCP-4 and αCP-4a. MCG10 is a 424-amino-acid protein containing two KH domains, several proline-rich domains, a nuclear export signal and a nuclear import signal (5). MCG10a is a variant of MCG10 lacking 55 residues in the first KH domain due to alternative splicing in exon 4. αCP-4 is similar to MCG10a but has 34 unique residues at the NH2-terminus, which encode an additional KH domain (6). αCP-4a is an alternative splice variant of αCP-4 with a shorter and distinct COOH-terminus that lacks proline-rich domains, nuclear export signal and nuclear import signal. The physiological significance of each PCBP4 isoform is not yet fully understood. Interestingly, MCG10, a target of tumor suppressor p53, can be induced in response to DNA damage (5). In addition, MCG10 mediates cell-cycle arrest in G2-M and apoptosis in lung cancer cell lines. Likewise, MCG10a and αCP-4 trigger cell-cycle arrest in G2-M but αCP-4a does not (5,22,23). Notably, the PCBP4 gene is located at 3p21, a chromosomal region highly susceptible to alterations in lung cancers (24). Indeed, loss of αCP-4 expression is common in squamous cell carcinomas and is associated with poorly differentiated and highly proliferative tumors (23). In view of this, PCBP4 is suggested to be a candidate lung tumor suppressor gene and a potential target in lung carcinogenesis.
p21 is a cyclin-dependent kinase inhibitor and a major mediator of p53 to induce G1 arrest in response to stress signals. In addition, p21 plays a role in other cellular processes, including cell differentiation and senescence (25–28). Due to these important functions, p21 is tightly regulated at transcriptional and post-transcriptional levels (29,30). Indeed, various RBPs are implicated in p21 regulation. For example, HuD, HuR and RNPC1, which possess one or several RNA recognition motifs (RRMs), regulate p21 mRNA stability through binding AU-rich elements (AREs) in the 3′-UTR of p21 transcript (31–34). HuR is an important mediator of p21 stabilization in response to UV light, gamma radiation and other stress signals (32,33,35). Similarly, the p53 target RNPC1 is involved in maintaining the stability of basal and stress-induced p21 transcript (36). HuR and RNPC1 bind diverse AREs on p21 3′-UTR but function cooperatively to stabilize p21 mRNA (37). In contrast, PCBP1, PCBP2 and hnRNP K bind CU-rich elements in the p21 3′-UTR to negatively regulate p21 expression (38,39). Indeed, co-depletion of PCBP1 and PCBP2 trigger p21 mRNA stabilization and G1 arrest (38). Instead, hnRNP K acts through inhibiting p21 mRNA translation (39). Interestingly, it is believed that HuR, through antagonizing the regulation of p21 by hnRNP K, controls the switch between neuronal cell proliferation and differentiation (39).
In this study, we aimed to identify novel PCBP4 RNA targets and further decipher PCBP4 physiological functions. We generated multiple cell lines in which PCBP4 is either inducibly expressed or knocked down and found that PCBP4 regulates basal and DNA damage-induced p21 mRNA and protein levels. Consistent with this, we showed that basal p21 mRNA and protein levels were increased in PCBP4−/– MEFs compared to wild-type MEFs. Furthermore, we provided evidence that PCBP4 binds to CU-rich regions in the 3′-UTR of p21 transcript in vitro and in vivo. Finally, we uncovered that PCBP4 regulates p21 mRNA stabilization. Therefore, we suggest that PCBP4 plays a role in the regulation of p21 mRNA stability in normal and cellular stress conditions.
MATERIALS AND METHODS
Reagents
Anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH), anti-p53, anti-p21 and anti-MDM2 (SMP14) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-hemagglutinin (HA) and anti-MDM2 (AB-2) were from Covance (Berkeley, CA, USA) and EMB Biosciences (San Diego, CA, USA), respectively. Anti-PCBP4 was from ProSci Incorporated (Poway, CA, USA). Actinomycin D and other reagents were from Sigma (St. Louis, MO, USA).
Plasmids
To generate PCBP4 shRNA vector, oligonucleotides (5′-AGC TTT TCC AAA AAG AGC GAG CTG TTA CGG TAT CTC TTG AAT ACC GTA ACA GCT CGC TCT GGG-3′ and 5′-GAT CCC CAG AGC GAG CTG TTA CGG TAT TCA AGA GAT ACC GTA ACA GCT CGC TCT TTT TTG GAA A-3′) were designed to target nucleotides 643–656 (shown in boldface) of PCBP4 mRNA (GenBank accession number NM_033008.2). Notably, this target sequence is also present in MCG10 and MCG10a. Oligonucleotides were annealed and cloned into pTER, a PolIII promoter-driven shRNA expression vector. The resulting plasmid was named pTER/siPCBP4.
To generate a pcDNA3 vector expressing C-terminally HA-tagged PCBP4, a cDNA fragment encoding amino acids 1–306 of PCBP4 was amplified using primers 355F (5′-ACA CAC TCG CAG GTC GCT GT-3′) and 1704R (5′-GCA GTG ATG AGG TAC TGG GC-3′). This NH2-terminal PCBP4 fragment was then cloned between EcoRI and KasI sites in the pcDNA3/MCG10HA vector described previously (5). The resulting plasmid was named pcDNA3/PCBP4HA. To generate a pcDNA4 vector expressing C-terminally HA-tagged PCBP4, an EcoRI fragment containing HA-tagged PCBP4 was obtained from pcDNA3/PCBP4HA and cloned into a pcDNA4 vector. The resulting plasmid was named pcDNA4/PCBP4HA.
Cell culture
MCF7 breast adenocarcinoma, RKO colon carcinoma and H1299 lung carcinoma cells were obtained from ATCC (Manassas, VA, USA) and cultured in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 8% fetal bovine serum (Hyclone, Logan, UT, USA). Parental MCF7-pTR-7, RKO-pTR-13 and H1299-pTR-8 cell lines were generated by transfection with a pcDNA6 vector that expresses a tetracycline repressor as previously described (40,41). To generate cell lines that inducibly express PCBP4 shRNA, parental cell lines were transfected with pTER/siPCBP4 and the pBabe vector for puromycin selection. To generate cell lines that inducibly express C-terminally HA-tagged PCBP4, parental cell lines were transfected with pcDNA4/PCBP4HA and the pBabe vector for puromycin selection. Individual clones were screened for inducible knockdown or expression of PCBP4 by Western blot analysis and two representative clones were chosen for subsequent studies.
Generation of heterozygous PCBP4 knockout mice and isolation of MEFs
To generate PCBP4 knockout mice, TBV-2 embryonic stem cells (ES) containing a gene trap rFRosaβgeo+1s vector inserted into PCBP4 first intron were obtained from the German Gene Trap Consortium (clone ID D136D11) (42). These ES cells were microinjected into C57BL/6 blastocysts at the UC Davis murine targeted genomics laboratory. Resulting male chimeras were bred to C57BL/6 females (Jackson Laboratory, Bar Harbor, ME, USA) and agouti pups were tested for germline transmission. Genotyping was performed by PCR on DNA extracted from toe clipping samples. Mice heterozygous for PCBP4 were bred and murine embryonic fibroblasts (MEFs) were isolated from 13.5-day-old embryos. Primary MEFs were cultured in DMEM supplemented with 10% fetal bovine serum. All animals were housed at UC Davis CLAS vivarium facility. All animal care and use protocols were approved by the UC Davis Institutional Animal Care and Use Committee.
Western blot analysis
Cells were washed twice with PBS, re-suspended with 2X SDS sample buffer, incubated at 95°C for 5 min, and used for Western blot analysis as previously described (43).
RNA isolation, RT-PCR and quantitative PCR
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and cDNA was synthesized using iScript (Biorad, Life Science Research, Hercules, CA, USA). To determine which PCBP4 variant is predominantly expressed in MCF7, RKO and H1299 cells, RT-PCR experiments were performed using primers 355F, 885R (5′-GCA CAA AGG TCC TCA TCC AG-3′), 769F (5′-GAT CAC CAT CTC CGA GGG CT-3′) and 1222R (5′-TGT GGA GTT GGG GAG CAG GT-3′). To determine p21 mRNA levels and stability, quantitative RT-PCR was performed on a Mastercycler® ep Realpex (Eppendorf, Hauppauye, NY, USA) using a total volume of 20 µl containing 200 nM primers and 1X Absolute™ Blue QPCR SYBR® Green Mix (ABgene, Thermo Fisher Scientific, Rockford, IL, USA). For GAPDH amplification, primers GAPDH-F (5′-AGC CTC AAG ATC ATC AGC AAT G-3′) and GAPDH-R (5′-ATG GAC TGT GGT CAT GAG TCC TT-3′) were used. For p21 amplification, primers p21-F (5′-TGA GCC GCG ACT GTG ATG-3′) and p21-R (5′-GTC TCG GTG ACA AAG TCG AAG TT-3′) were used.
To assess p21 and GAPDH transcript levels in wild-type, PCBP4+/– and PCBP4−/– MEFs, quantitative RT-PCR experiments were performed. For mouse p21 amplification, primers mp21F (5′-GCC TTA GCC CTC ACT CTG TG-3′) and mp21R (5′-AGC TGG CCT TAG AGG TGA CA-3′) were used. For mouse GAPDH amplification, primers mGAPDHF (5′-AAC TTT GGC ATT GTG GAA GG-3′) and mGAPDHR (5′-ACA CAT TGG GGG TAG GAA CA-3′) were used.
RNA immunoprecipitation and RT-PCR
RNA immunoprecipitation and RT-PCR (RIP) was performed as previously described (44). Briefly, cells (2 × 107) were uninduced or induced to express PCBP4 for 24 h, collected in immunoprecipitation buffer, and then incubated with 2 µg of anti-HA or mouse IgG antibody at 4°C overnight. RNA–protein immunocomplexes were precipitated using protein A/G beads and subjected to RT-PCR.
Luciferase assay
Dual-luciferase reporter assay was done according to the manufacturer’s instructions (Promega, Madison, WI, USA). Briefly, cells (4 × 104) were plated on a 24-well plate, untreated or treated with tetracycline for 24 h, and then transfected with control pGL3 reporter vector or pGL3 vector containing full-length p21 3′-UTR (200 ng) and a Renilla luciferase vector (5 ng). Luciferase activity was measured on triplicate wells using a Turner Designs luminometer 24 h post-transfection.
RNA electrophoretic mobility shift assay
Recombinant HA-tagged PCBP4-GST and GST proteins expressed in E. coli BL21 were purified using glutathione sepharose beads. Various regions in the 3′-UTR of p21 transcript were PCR-amplified using primers containing the T7 promoter. p21 RNA probes were made by in vitro transcription using T7 RNA polymerase in the presence of α-32P-UTP. RNA electrophoretic mobility shift assays (REMSAs) were performed by combining indicated amounts of HA-tagged PCBP4 protein, 1 mg/ml of yeast tRNA and 50 000 CPM 32P-labeled p21 RNA probe in reaction buffer, followed by incubation at 25°C for 20 min as previously described (37). Next, RNA–protein complexes were digested with RNaseT1 and separated on a 7% native acrylamide gel. Supershift assays were performed by pre-incubating 3 µg of anti-HA antibody with HA-tagged PCBP4 protein for 30 min on ice before incubation with p21 RNA probes.
RESULTS
αCP-4 is the main PCBP4 variant expressed in MCF7, RKO and H1299 cells
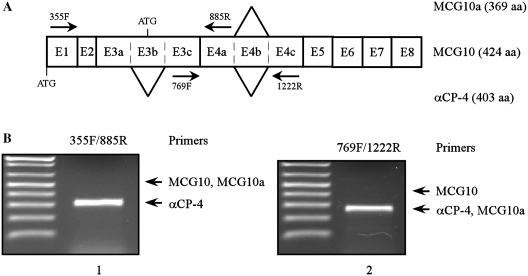
The PCBP4 gene encodes four known transcripts: MCG10, MCG10a, αCP-4 and αCP-4a. MCG10 and MCG10a variants use a start ATG codon located in exon 3b and encode proteins with two KH domains (Figure 1A). αCP-4 and αCP-4a variants lack exon 3b due to alternative splicing and utilize an upstream start ATG to encode proteins with three KH domains (Figure 1A). In addition, MCG10a, αCP-4 and αCP-4a lack exon 4b due to alternative splicing, which results in a deletion of 55 amino acids in the first KH domain (MCG10a) or second KH domain (αCP-4 and αCP-4a). To determine which PCBP4 variant is mainly expressed in MCF7 cells, RT-PCR was performed to amplify exon 3 (Figure 1B, lane 1) and exon 4 (Figure 1B, lane 2). Here, we found that a major PCR product corresponding to αCP-4 was detected upon amplification of exons 3 and 4. Similarly, αCP-4 was the main PCBP4 variant detected in RKO and p53-deficient H1299 cells (data not shown). In view of this, this study will focus on three KH domains-containing αCP-4 protein. As αCP-4 is the major variant of the PCBP4 gene, we will name it PCBP4 from here on.
Figure 1.
αCP-4 is the main PCBP4 variant expressed in MCF7, RKO and H1299 cells. (A) Schematic representation of PCBP4 transcripts. Exons are shown as numbered boxes. Locations of two alternative splice sites are indicated by lines. MCG10a does not contain exon 4b. αCP-4 does not contain exons 3b and 4b. Primers used for RT-PCR experiments are shown. (B) αCP-4 is the main isoform expressed in MCF7 cells. Levels of MCG10, MCG10a and αCP-4 transcripts were analyzed by RT-PCR using primers 355F and 885R (left), 769F and 1222R (right). Positions of expected PCR products for MCG10, MCG10a and αCP-4 are indicated by arrows.
PCBP4 regulates the induction of p21 by DNA damage in a p53-independent manner
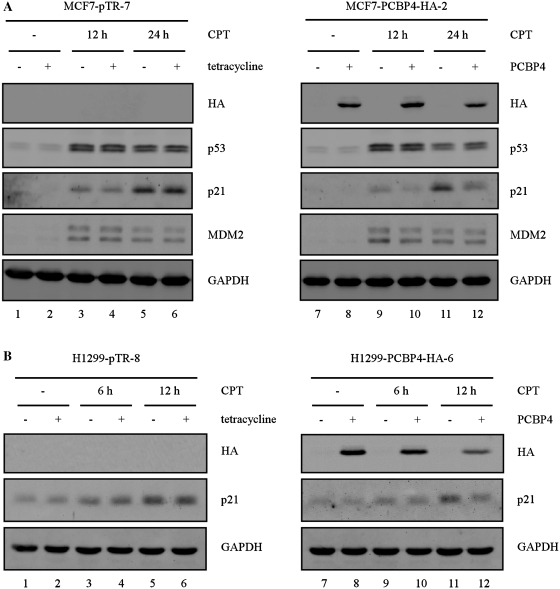
To examine a potential role for PCBP4 in the p53 pathway, we generated MCF7 cell lines in which C-terminally HA-tagged PCBP4 is inducibly expressed under the control of a tetracycline-regulated promoter. Next, parental MCF7-pTR-7 and MCF7-PCBP4-HA-2 cell lines were pretreated with tetracycline for 2 days, followed by treatment with DNA damage agent camptothecin (CPT) for 12 or 24 h (Figure 2). In parental MCF7 cells, camptothecin treatment stabilized endogenous p53, which resulted in induction of p53 targets p21 and MDM2 after 12 or 24 h (Figure 2A, compare lane 1 with lanes 3 and 5). Importantly, we showed that tetracycline had no effect on p53 stabilization and induction of p21 or MDM2 by camptothecin (Figure 2A, compare lanes 1, 3 and 5 with lanes 2, 4 and 6, respectively). In MCF7-PCBP4-HA-2 cells, HA-tagged PCBP4 was detected upon tetracycline treatment for 2 days (Figure 2A, compare lanes 7, 9 and 11 with lanes 8, 10 and 12, respectively). In addition, camptothecin treatment stabilized endogenous p53, which led to induction of p21 and MDM2. Here, we found that PCBP4 had no significant effect on p53 stabilization 12 h after camptothecin treatment (Figure 2A, compare lanes 9 and 10). However, PCBP4 slightly enhanced p53 stabilization 24 h after camptothecin treatment (Figure 2A, compare lanes 11 and 12). Interestingly, we showed that PCBP4 markedly inhibited p21 induction in cells treated with camptothecin for 12 or 24 h (Figure 2A, compare lanes 9 and 11 with lanes 10 and 12, respectively). By contrast, PCBP4 had no effect on MDM2 induction by camptothecin. To verify that the regulation of p21 by PCBP4 is not cell-type specific, we generated RKO cell lines in which C-terminally HA-tagged PCBP4 can be inducibly expressed (Supplementary Figure S1). Next, parental RKO-pTR-13 and RKO-PCBP4-HA-2 cell lines were pretreated with tetracycline for 2 days, followed by treatment with camptothecin for 3, 6 or 12 h. We found that, similar to that in MCF7 cells, PCBP4 decreased p21 levels induced in RKO cells treated with camptothecin for 6 or 12 h (Supplementary Figure S1). Taken together, these results suggest that PCBP4 modulates the induction of p21 by DNA damage.
Figure 2.
PCBP4 regulates the induction of p21 by DNA damage in a p53-independent manner. (A) PCBP4 inhibits p21 induction by DNA damage. Cell extracts were prepared from MCF7-pTR-7 and MCF7-PCBP4-HA-2 cells uninduced (–) or induced (+) to express PCBP4 for 2 days, and then untreated or treated with 200 nM CPT for 12 or 24 h. Levels of HA-tagged PCBP4, p53, p21, MDM2 and GAPDH were detected by Western blot analysis. (B) PCBP4 regulates stress-induced p21 levels in a p53-independent manner. Cell extracts were prepared from H1299-pTR-8 and H1299-PCBP4-HA-6 cells uninduced (–) or induced (+) to express PCBP4 for 2 days, and then untreated or treated with 200 nM CPT for 6 or 12 h. The data are representative of two independent experiments.
To determine whether the regulation of p21 by PCBP4 is dependent on p53, we generated p53-null H1299 cell lines in which HA-tagged PCBP4 can be inducibly expressed. Next, parental H1299-pTR-8 and H1299-PCBP4-HA-6 cell lines were pretreated with tetracycline for 2 days, followed by treatment with camptothecin for 6 or 12 h (Figure 2B). In parental H1299 cells, camptothecin treatment led to a slight increase in p21 at 6 or 12 h (Figure 2B, compare lanes 1 with lanes 3 and 5). In addition, tetracycline had no effect on p21 induction by camptothecin. In H1299-PCBP4-HA-6 cells, HA-tagged PCBP4 was detected upon tetracycline treatment for 2 days (Figure 2B, compare lanes 7, 9 and 11 with lanes 8, 10 and 12, respectively). Most importantly, we found that PCBP4 inhibited p21 induction in p53-null cells treated with camptothecin for 6 or 12 h (Figure 2B, compare lanes 9 and 11 with lanes 10 and 12, respectively). This suggests that PCBP4 regulates p21 levels in a p53-independent manner.
Deficiency in PCBP4 enhances the induction of p21 by DNA damage
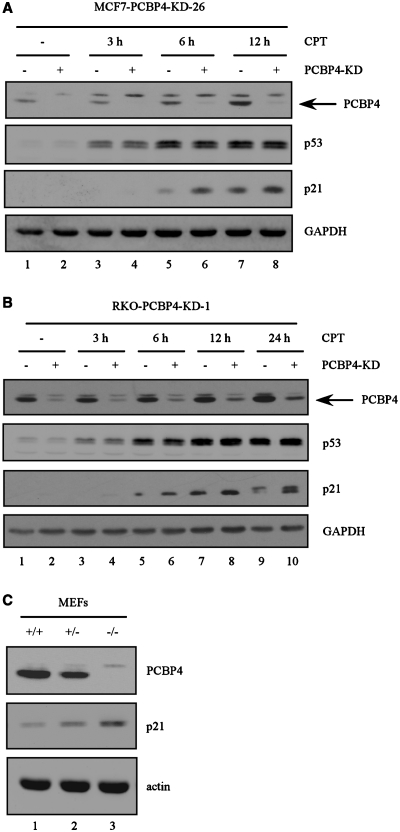
To assess whether a lack of PCBP4 expression has any effect on p21 levels, we generated MCF7 cell lines in which endogenous PCBP4 can be inducibly knocked down. In a representative PCBP4-KD cell line, MCF7-PCBP4-KD-26, we showed that induction of PCBP4 shRNA for 3 days elicited a significant decrease in PCBP4 levels (Figure 3A, compare lanes 1 and 2). Next, MCF7-PCBP4-KD-26 cells uninduced or induced to express PCBP4 shRNA for 3 days were treated with camptothecin for 3, 6 or 12 h. We showed that p53 was stabilized after treatment with camptothecin, which resulted in PCBP4 and p21 inductions 6 or 12 h post-treatment (Figure 3A, compare lanes 1 with lanes 5 and 7). Furthermore, we found that induction of p21 by DNA damage was further enhanced upon PCBP4 knockdown 6 or 12 h post-treatment with camptothecin (Figure 3A, compare lanes 5 and 7 with lanes 6 and 8, respectively). Taken together, our data suggest that PCBP4 modulates p21 induction in response to stress signals.
Figure 3.
Deficiency in PCBP4 increases basal and DNA damage-induced p21 levels. (A) Knockdown of PCBP4 enhances p21 induction upon DNA damage. Cell extracts were prepared from MCF7-PCBP4-KD-26 uninduced (–) or induced (+) to express PCBP4 shRNA for 3 days, and then untreated or treated with 200 nM CPT for 3, 6 or 12 h. Levels of PCBP4, p53, p21 and GAPDH were detected by Western blot analysis. (B) Cell extracts were prepared from RKO-PCBP4-KD-1 uninduced (–) or induced (+) to express PCBP4 shRNA for 3 days, and then untreated or treated with 200 nM CPT for 3, 6, 12 or 24 h. The data are representative of two independent experiments. (C) Deficiency in PCBP4 increases basal p21 expression. Cell extracts were prepared from PCBP4+/+, PCBP4+/− and PCBP4−/− MEFs. Levels of PCBP4, p21 and actin were detected by Western blot analysis.
To verify that the effect of PCBP4 on p21 expression is not cell-type specific, we generated RKO cell lines in which endogenous PCBP4 can be inducibly knocked down by the tetracycline-inducible shRNA expression system. In a representative PCBP4-KD cell line, RKO-PCBP4-KD-1, we showed that induction of PCBP4 shRNA for 3 days elicited a significant decrease in PCBP4 levels (Figure 3B, compare lanes 1 and 2). Next, RKO-PCBP4-KD-1 cells uninduced or induced to express PCBP4 shRNA were treated with camptothecin for 3, 6, 12 or 24 h. We showed that camptothecin stabilized endogenous p53, which resulted in induction of PCBP4 and p21 (Figure 3B, compare lanes 1 with lanes 3, 5, 7 and 9). Similarly, we found that PCBP4 knockdown enhanced p21 induction following camptothecin treatment for 6, 12 or 24 h (Figure 3B, compare lanes 5, 7 and 9 with lanes 6, 8 and 10, respectively).
To determine whether PCBP4 regulates basal p21 expression in a physiologically relevant condition, we created PCBP4+/– mice and intercrossed those mice to obtain primary mouse embryonic fibroblasts (MEFs) of three genotypes: wild-type, PCBP4+/– and PCBP4−/–. Next, Western blot analyses were performed (Figure 3C). We showed that PCBP4 protein levels were reduced by 50% in PCBP4+/– MEFs and were not detectable in PCBP4−/– MEFs (Figure 3C, compare lane 1 with lanes 2 and 3). Importantly, basal p21 protein levels were increased in PCBP4+/– MEFs and increased further in PCBP4−/– MEFs (Figure 3C, compare lane 1 with lanes 2 and 3).
PCBP4 regulates p21 mRNA level and stability
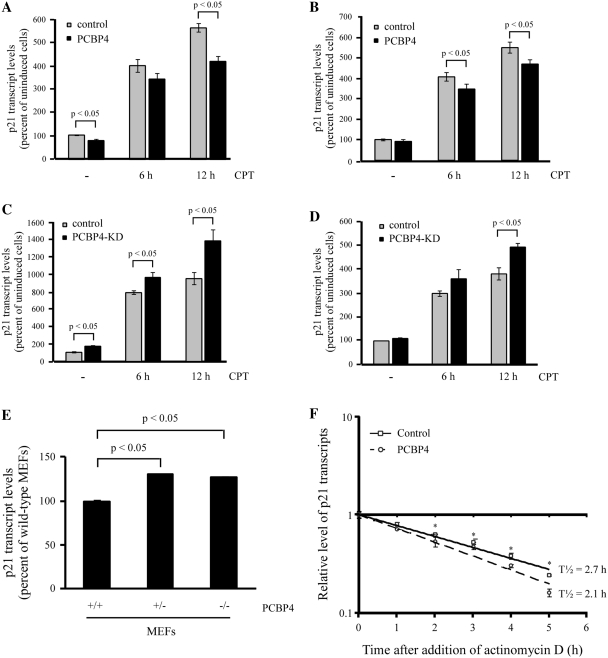
To assess how PCBP4 regulates p21 expression, we examined whether the decrease in p21 protein by PCBP4 is correlated with a decrease in p21 transcript. To this end, quantitative RT-PCR was performed on MCF7 cells that were uninduced or induced to express PCBP4 for 2 days, followed by treatment with camptothecin for 6 or 12 h (Figure 4A). In uninduced cells, we showed that camptothecin treatment led to an increase in p21 mRNA levels by 4-fold and 5.5-fold after 6 and 12 h, respectively (Figure 4A). Interestingly, we found that PCBP4 markedly reduced basal p21 transcript levels by 20%. Furthermore, PCBP4 decreased the induction of p21 upon camptothecin treatment by 17% and 25% after 6 and 12 h, respectively (Figure 4A). To exclude potential cell-type specific effects, p21 transcript levels were examined in RKO cells. We found that, similar to MCF7 cells, PCBP4 reduced p21 transcript levels by 15% in untreated and camptothecin-treated cells (Figure 4B).
Figure 4.
PCBP4 regulates p21 mRNA level and stability. (A and B) PCBP4 decreases p21 mRNA levels induced by DNA damage. Quantitative RT-PCR was performed on cDNA samples from MCF7-PCBP4-HA-2 cells (A) or RKO-PCBP4-HA-2 cells (B) uninduced (–) or induced (+) to express PCBP4 for 2 days, and then untreated or treated with 200 nM CPT for 6 or 12 h. Upon normalization to GAPDH transcript levels, percentage of p21 transcript levels compared to untreated cells was plotted as mean ± SD from triplicate samples. (C and D) Knockdown of PCBP4 increases p21 mRNA levels induced upon DNA damage. Quantitative RT-PCR was performed on cDNA samples from MCF7-PCBP4-KD-26 cells (C) or RKO-PCBP4-KD-1 cells (D) uninduced (–) or induced (+) to express PCBP4 shRNA for 3 days, and then untreated or treated with 200 nM CPT for 6 or 12 h. Percentage of normalized p21 transcript levels compared to untreated cells was plotted as mean ± SD from triplicate samples. (E) Deficiency in PCBP4 increases basal p21 mRNA level. Quantitative RT-PCR was performed on cDNA samples from PCBP4+/+, PCBP4+/– and PCBP4−/– MEFs. (F) PCBP4 expression decreases p21 mRNA half-life. Levels of p21 and GAPDH transcripts were measured by quantitative RT-PCR on cDNA samples from RKO cells that were uninduced or induced to express PCBP4 for 2 days, and then treated with 5 µg/ml actinomycin D for 0, 1, 2, 3, 4 or 5 h. Upon normalization to GAPDH transcript levels, the mean ± SD from triplicate samples was plotted and the relative half-life of p21 transcript was calculated. *, P < 0.05.
To examine whether a lack of PCBP4 expression has any effect on p21 mRNA levels, quantitative RT-PCR was performed on MCF7 cells uninduced or induced to express PCBP4 shRNA for 3 days, followed by treatment with camptothecin for 6 or 12 h. Here, we found that p21 transcript levels were increased by 70% in PCBP4-deficient cells (Figure 4C). In addition, PCBP4 knockdown increased p21 induction after 6 and 12 h camptothecin treatment by 30% and 50%, respectively (Figure 4C). To further confirm the above findings, similar experiments were performed in RKO cells. We found that, similar to MCF7 cells, a deficiency in PCBP4 led to a 20% increase in p21 mRNA levels in untreated and camptothecin-treated cells (Figure 4D).
To determine whether the increase in p21 protein upon PCBP4 knockout is correlated with an increase in p21 transcript, we performed quantitative RT-PCR on wild-type, PCBP4+/– and PCBP4−/– MEFs (Figure 4E). We showed that p21 mRNA levels were increased by 25% in PCBP4+/– and PCBP4−/– MEFs compared to wild-type MEFs. Taken together, our data suggest that PCBP4 modulates p21 expression at the mRNA level in normal and DNA damage conditions.
To further examine the mechanism through which PCBP4 regulates p21, quantitative RT-PCR was performed to measure p21 mRNA half-life in RKO cells that were uninduced or induced to express PCBP4 for 2 days, followed by treatment with transcriptional inhibitor actinomycin D for 0, 1, 2, 3, 4 or 5 h (Figure 4F). In uninduced RKO cells, we found that p21 mRNA levels were gradually decreased upon actinomycin D treatment. The relative p21 mRNA half-life was calculated to be 2.7 h. Notably, in PCBP4-expressing RKO cells, p21 mRNA levels were rapidly decreased upon actinomycin D treatment (Figure 4F). As a result, the relative p21 mRNA half-life (2.1 h) was reduced by 20%. To verify that this effect is not cell-type specific, we examined whether PCBP4 regulates p21 mRNA half-life in MCF7 cells. Quantitative RT-PCR was performed in MCF7 cells uninduced or induced to express PCBP4 for 2 days, and then treated with actinomycin D for various times (Supplemenary Figure S2). In uninduced MCF7 cells, we found that the relative p21 mRNA half-life was 4.1 h. Notably, in PCBP4-expressing MCF7 cells, p21 mRNA levels were decreased faster upon actinomycin D treatment and the resulting relative p21 mRNA half-life (3.4 h) was reduced by 17%. Therefore, we uncovered that PCBP4 plays a role in the regulation of p21 mRNA stability.
PCBP4 binds to the 3′-UTR of p21 transcript
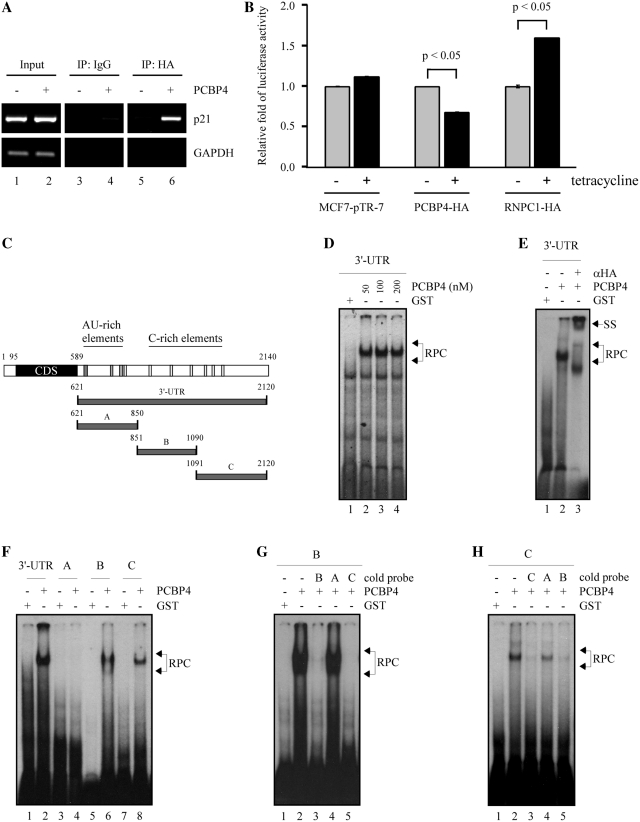
The post-transcriptional regulation of p21 is commonly mediated through RBPs binding to p21 3′-UTR. To assess whether PCBP4 binds to p21 3′-UTR in vivo, RIP assays were performed on RKO cells uninduced or induced to express HA-tagged PCBP4 for 24 h (Figure 5A). Importantly, we found that p21 transcripts were detected upon PCBP4 induction in PCBP4 but not IgG immunocomplexes (Figure 5A, p21 panel, compare lanes 4 and 6). As a control, GAPDH transcripts were not detected upon PCBP4 induction (Figure 5A, GAPDH panel, compare lanes 4 and 6).
Figure 5.
PCBP4 binds to the 3′-UTR of p21. (A) PCBP4 interacts with p21 3′-UTR in vivo. RKO cells were uninduced (–) or induced (+) to express PCBP4 for 24 h, followed by immunoprecipitation with control IgG or anti-HA antibody. RT-PCR was performed to detect p21 and GAPDH transcript levels in control and PCBP4 immunocomplexes. (B) PCBP4 regulates mRNA stability through p21 3′-UTR. Parental MCF7-pTR-7, MCF7-PCBP4-HA-2 and MCF7-RNPC1-HA-16 cells were uninduced (–) or induced (+) for 2 days and then co-transfected with pGL3 reporter vector containing p21 3′-UTR (nt 571 to 2121) and control Renilla luciferase vector. Luciferase assays were performed and relative luciferase activity ± SD of triplicate samples was plotted. The data are representative of two independent experiments. (C) Schematic representation of p21 3′-UTR along with the location of AU-rich elements, C-rich elements and RNA probes used for REMSAs. (D) PCBP4 binds to p21 3′-UTR. REMSA was performed by mixing 32P-labeled full-length p21 3′-UTR with recombinant GST or various amounts of GST-PCBP4-HA proteins. RPC, RNA–protein complexes. (E) Supershift assay was performed by adding 3 µg of anti-HA antibody to a reaction mixture containing full-length p21 3′-UTR and GST-PCBP4-HA. (F) PCBP4 binds to two regions in p21 3′-UTR. REMSA was performed using 32P-labeled p21 full-length, A, B or C probes. (G) Competition assay was done by adding an excess amount of unlabeled p21 probe (B, A or C) to compete the binding of PCBP4 to radiolabeled p21 probe B. (H) Competition assay was performed by adding an excess amount of unlabeled p21 probe (C, A or B) to compete the binding of PCBP4 to radiolabeled p21 probe C.
Next, to determine whether PCBP4 regulates p21 mRNA stability through the 3′-UTR, we performed luciferase assays on parental, PCBP4-expressing or RNPC1-expressing MCF7 cells uninduced or induced to express PCBP4 or RNPC1 for 2 days, followed by transfection with a pGL3 reporter vector that contains p21 3′-UTR (nucleotides 571–2121) (34). Here, we showed that tetracycline had no effect on luciferase activity (Figure 5B). In contrast, PCBP4 markedly reduced luciferase activity by 25%. As a control, known p21 regulator RNPC1 increased luciferase activity by 50%. Taken together, these findings suggest that the binding of PCBP4 to p21 3′-UTR in vivo regulates p21 mRNA stability.
To further decipher the region in p21 3′-UTR that is bound by PCBP4, REMSA was performed using 50, 100 or 200 nM HA-tagged PCBP4 protein and a radiolabeled full-length p21 3′-UTR RNA probe (nucleotides 621–2120, GenBank accession number NM_000389) as described in Figure 5C. Here, we found that there was a strong binding of PCBP4 to full-length p21 3′-UTR (Figure 5D, compare lane 1 with lanes 2–4). Next, to verify the specificity of PCBP4 binding to p21 3′-UTR, a supershift assay was performed by adding anti-HA antibody to the reaction mixture (Figure 5E). Indeed, we showed that anti-HA antibody recognized HA-tagged PCBP4 protein in the PCBP4–RNA complex, which led to a supershift of the complex on the gel (Figure 5E, compare lanes 2 and 3). The specificity of PCBP4 binding to p21 3′-UTR was further confirmed by a competition assay in which unlabeled p21 3′-UTR probe (cold probe) was added to the reaction mixture (Supplementary Figure S3). We found that the binding of PCBP4 to radiolabeled p21 3′-UTR probe was reduced in the presence of cold probe (Supplementary Figure S3, compare lanes 2 and 3).
To identify the region in p21 3′-UTR bound by PCBP4, three p21 RNA probes (A, B and C) were made as described in Figure 5C. Probe A (nucleotides 621–850) contains ARE elements for binding of RRM-containing RBPs, such as HuD, HuR and RNPC1, and a potential poly(C)-rich element for PCBP1 binding (31,37). Probe B (nucleotides 851–1090) contains several poly(C)-rich elements for PCBP1 and PCBP2 binding (38). Probe C (nucleotides 1091–2120) also has several poly(C)-rich elements. Next, REMSAs were performed and showed that PCBP4 bound strongly to regions B and C but not to region A (Figure 5F, compare lane 2 with lanes 4, 6 and 8). To confirm the specificity of PCBP4 binding to regions B and C, competition assays were performed (Figure 5G and H). Here, we showed that the binding of PCBP4 to radiolabeled probe B was inhibited by unlabeled probe B or C, but not probe A (Figure 5G, compare lane 2 with lanes 3–5). Similarly, the binding of PCBP4 to radiolabeled probe C was inhibited by unlabeled probe C or B, but not probe A (Figure 5H, compare lane 2 with lanes 3–5). Taken together, these results imply that PCBP4 specifically binds to two poly(C)-rich regions in the 3′-UTR of p21 transcript to regulate its stability.
DISCUSSION
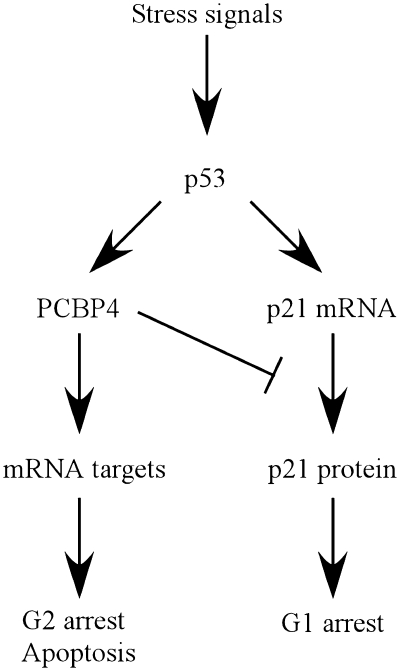
RBPs are increasingly recognized for their importance in many steps of RNA metabolism and as key regulators of gene expression. In view of this, alterations in RBPs are implicated in many human diseases ranging from fragile X syndrome to cancer (45). In this study, we showed that poly(C)-binding protein PCBP4 plays a role in the regulation of cyclin-dependent kinase inhibitor p21. Indeed, we found that the basal level of p21 was decreased by overexpressed PCBP4 but increased by knockdown or knockout of PCBP4 (Figure 4). Consistent with this, it was recently reported that PCBP1 and PCBP2 also regulate p21 (38). These findings uncover p21 as a common RNA target regulated by the PCBP family. However, in response to various stimuli, only PCBP4 and p21 were found to be induced in a p53-dependent manner (5,26). p21 is the major p53 mediator of G1 arrest, whereas PCBP4 induces G2 arrest and apoptosis (5,22). Unexpectedly, we showed here that PCBP4 decreases p21 levels induced by DNA damage. To reconcile these findings, we hypothesize that PCBP4 may have a unique function in the p53 pathway through controlling p21-mediated G1 arrest and facilitating G2 arrest and/or apoptosis depending on the type or level of cellular stress (Figure 6). Interestingly, we found that p53 activation mediates a rapid p21 induction and a more gradual PCBP4 induction in response to DNA damage (data not shown). In view of this, prolonged stress signals can potentially mediate a sustained PCBP4 induction and ultimately switch the cellular response from G1 arrest to G2 arrest and apoptosis. Similarly, studies have revealed that low or high doses of cellular stress, such as UV irradiation, can produce specific dynamics of p53 activation to ultimately implement G2 arrest or G2 arrest release and apoptosis (46,47). In addition, it is expected that PCBP4 regulates additional RNA targets, especially cell-cycle related targets.
Figure 6.
A model for the role of p21 regulation by PCBP4 in the p53 tumor suppressor pathway.
p21 expression is tightly controlled at transcriptional and post-transcriptional levels (29). The major regulator of p21 gene expression is tumor suppressor p53 (26). Here, we provided evidence that PCBP4 modulates p21 expression in a p53-independent manner. In addition, PCBP4 had no major effect on levels of p53 and MDM2. Finally, we showed that PCBP4 binds to p21 3′-UTR, a known site for post-transcriptional control by RBPs (31,32). Interestingly, PCBP4 can bind in two regions containing several poly(C)-rich sequences in p21 3′-UTR. Notably, the region between nt 851 and 1090 was recently found to contain PCBP1 and PCBP2 binding sites (38). Interestingly, the region between nt 1091 and 2120 was reported as a binding site for Musashi-1, a RRM-containing RBP involved in the regulation of p21 translation (48). Therefore, it is likely that the binding sites for PCBP1, PCBP2 and PCBP4 are the same or clustered between nt 851 and 1090 of p21 3′-UTR. In view of this, it is possible that p21 mRNA stability is coordinately regulated by the PCBP family, which merits further investigation. Of note, PCBP family members were reported to regulate the translation of mRNA targets, such as c-myc and 15-lipoxygenase. Therefore, it remains possible that PCBP4 binds p21 3′-UTR to regulate p21 mRNA stability as well as p21 translation.
Interestingly, p21 3′-UTR contains a region from nt 621 to 850 with several AU-rich elements recognized by RRM-containing RBPs (37). From this and previous studies, we conclude that p21 3′-UTR is composed of two domains for p21 post-transcriptional regulation, a first domain mainly bound by RRM-containing RBPs for p21 mRNA stabilization/destabilization and a second domain primarily bound by KH-containing RBPs for p21 mRNA destabilization. Future studies will likely reveal some interesting crosstalks between the PCBP family and other RBPs, such as HuR and RNPC1, essential for appropriate p21 expression at normal and stress conditions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: The National Institutes of Health (CA121137, CA076069).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr. Giles (University of Western Australia) for providing pGL3 reporter constructs. We thank all members of the Chen laboratory for helpful discussions and technical advice.
REFERENCES
- 1.Kiledjian M, Wang X, Liebhaber SA. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. EMBO J. 1995;14:4357–4364. doi: 10.1002/j.1460-2075.1995.tb00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collier B, Goobar-Larsson L, Sokolowski M, Schwartz S. Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogenous ribonucleoprotein K and poly(rC)-binding proteins 1 and 2. J. Biol. Chem. 1998;273:22648–22656. doi: 10.1074/jbc.273.35.22648. [DOI] [PubMed] [Google Scholar]
- 3.Michelotti EF, Michelotti GA, Aronsohn AI, Levens D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell Biol. 1996;16:2350–2360. doi: 10.1128/mcb.16.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi HS, Hwang CK, Song KY, Law PY, Wei LN, Loh HH. Poly(C)-binding proteins as transcriptional regulators of gene expression. Biochem. Biophys. Res. Commun. 2009;380:431–436. doi: 10.1016/j.bbrc.2009.01.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu J, Chen X. MCG10, a novel p53 target gene that encodes a KH domain RNA-binding protein, is capable of inducing apoptosis and cell cycle arrest in G(2)-M. Mol. Cell. Biol. 2000;20:5602–5618. doi: 10.1128/mcb.20.15.5602-5618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makeyev AV, Liebhaber SA. Identification of two novel mammalian genes establishes a subfamily of KH-domain RNA-binding proteins. Genomics. 2000;67:301–316. doi: 10.1006/geno.2000.6244. [DOI] [PubMed] [Google Scholar]
- 7.Makeyev AV, Chkheidze AN, Liebhaber SA. A set of highly conserved RNA-binding proteins, alphaCP-1 and alphaCP-2, implicated in mRNA stabilization, are coexpressed from an intronless gene and its intron-containing paralog. J. Biol. Chem. 1999;274:24849–24857. doi: 10.1074/jbc.274.35.24849. [DOI] [PubMed] [Google Scholar]
- 8.Matunis MJ, Michael WM, Dreyfuss G. Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol. Cell. Biol. 1992;12:164–171. doi: 10.1128/mcb.12.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makeyev AV, Liebhaber SA. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA. 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson TJ, Thompson JD, Heringa J. The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett. 1993;324:361–366. doi: 10.1016/0014-5793(93)80152-k. [DOI] [PubMed] [Google Scholar]
- 11.Valverde R, Edwards L, Regan L. Structure and function of KH domains. FEBS J. 2008;275:2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- 12.Lynch M, Chen L, Ravitz MJ, Mehtani S, Korenblat K, Pazin MJ, Schmidt EV. hnRNP K binds a core polypyrimidine element in the eukaryotic translation initiation factor 4E (eIF4E) promoter, and its regulation of eIF4E contributes to neoplastic transformation. Mol. Cell. Biol. 2005;25:6436–6453. doi: 10.1128/MCB.25.15.6436-6453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moumen A, Masterson P, O'Connor MJ, Jackson SP. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–1078. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 14.Choi HS, Song KY, Hwang CK, Kim CS, Law PY, Wei LN, Loh HH. A proteomics approach for identification of single strand DNA-binding proteins involved in transcriptional regulation of mouse mu opioid receptor gene. Mol. Cell Proteomics. 2008;7:1517–1529. doi: 10.1074/mcp.M800052-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi HS, Kim CS, Hwang CK, Song KY, Law PY, Wei LN, Loh HH. Novel function of the poly(C)-binding protein alpha CP3 as a transcriptional repressor of the mu opioid receptor gene. FASEB J. 2007;21:3963–3973. doi: 10.1096/fj.07-8561com. [DOI] [PubMed] [Google Scholar]
- 16.Paulding WR, Czyzyk-Krzeska MF. Regulation of tyrosine hydroxylase mRNA stability by protein-binding, pyrimidine-rich sequence in the 3′-untranslated region. J. Biol. Chem. 1999;274:2532–2538. doi: 10.1074/jbc.274.4.2532. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Kiledjian M, Weiss IM, Liebhaber SA. Detection and characterization of a 3′ untranslated region ribonucleoprotein complex associated with human alpha-globin mRNA stability. Mol. Cell. Biol. 1995;15:1769–1777. doi: 10.1128/mcb.15.3.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stefanovic B, Hellerbrand C, Holcik M, Briendl M, Aliebhaber S, Brenner DA. Posttranscriptional regulation of collagen alpha1(I) mRNA in hepatic stellate cells. Mol. Cell. Biol. 1997;17:5201–5209. doi: 10.1128/mcb.17.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ostareck DH, Ostareck-Lederer A, Shatsky IN, Hentze MW. Lipoxygenase mRNA silencing in erythroid differentiation: the 3′UTR regulatory complex controls 60S ribosomal subunit joining. Cell. 2001;104:281–290. doi: 10.1016/s0092-8674(01)00212-4. [DOI] [PubMed] [Google Scholar]
- 20.Evans JR, Mitchell SA, Spriggs KA, Ostrowski J, Bomsztyk K, Ostarek D, Willis AE. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene. 2003;22:8012–8020. doi: 10.1038/sj.onc.1206645. [DOI] [PubMed] [Google Scholar]
- 21.Parsley TB, Towner JS, Blyn LB, Ehrenfeld E, Semler BL. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- 22.Castano Z, Vergara-Irigaray N, Pajares MJ, Montuenga LM, Pio R. Expression of alpha CP-4 inhibits cell cycle progression and suppresses tumorigenicity of lung cancer cells. Int. J. Cancer. 2008;122:1512–1520. doi: 10.1002/ijc.23236. [DOI] [PubMed] [Google Scholar]
- 23.Pio R, Zudaire I, Pino I, Castano Z, Zabalegui N, Vicent S, Garcia-Amigot F, Odero MD, Lozano MD, Garcia-Foncillas J, et al. Alpha CP-4, encoded by a putative tumor suppressor gene at 3p21, but not its alternative splice variant alpha CP-4a, is underexpressed in lung cancer. Cancer Res. 2004;64:4171–4179. doi: 10.1158/0008-5472.CAN-03-2982. [DOI] [PubMed] [Google Scholar]
- 24.Angeloni D. Molecular analysis of deletions in human chromosome 3p21 and the role of resident cancer genes in disease. Brief Funct. Genomic Proteomic. 2007;6:19–39. doi: 10.1093/bfgp/elm007. [DOI] [PubMed] [Google Scholar]
- 25.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 26.el-Deiry WS, Harper JW, O'Connor PM, Velculescu VE, Canman CE, Jackman J, Pietenpol JA, Burrell M, Hill DE, Wang Y, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 27.Dulic V, Drullinger LF, Lees E, Reed SI, Stein GH. Altered regulation of G1 cyclins in senescent human diploid fibroblasts: accumulation of inactive cyclin E-Cdk2 and cyclin D1-Cdk2 complexes. Proc. Natl Acad. Sci. USA. 1993;90:11034–11038. doi: 10.1073/pnas.90.23.11034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erhardt JA, Pittman RN. Ectopic p21(WAF1) expression induces differentiation-specific cell cycle changes in PC12 cells characteristic of nerve growth factor treatment. J. Biol. Chem. 1998;273:23517–23523. doi: 10.1074/jbc.273.36.23517. [DOI] [PubMed] [Google Scholar]
- 29.Jung YS, Qian Y, Chen X. Examination of the expanding pathways for the regulation of p21 expression and activity. Cell Signal. 22:1003–1012. doi: 10.1016/j.cellsig.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorospe M, Wang X, Holbrook NJ. p53-dependent elevation of p21Waf1 expression by UV light is mediated through mRNA stabilization and involves a vanadate-sensitive regulatory system. Mol. Cell. Biol. 1998;18:1400–1407. doi: 10.1128/mcb.18.3.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joseph B, Orlian M, Furneaux H. p21(waf1) mRNA contains a conserved element in its 3′-untranslated region that is bound by the Elav-like mRNA-stabilizing proteins. J. Biol. Chem. 1998;273:20511–20516. doi: 10.1074/jbc.273.32.20511. [DOI] [PubMed] [Google Scholar]
- 32.Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook N, Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 2000;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang X, Wang W, Fan J, Lal A, Yang D, Cheng H, Gorospe M. Prostaglandin A2-mediated stabilization of p21 mRNA through an ERK-dependent pathway requiring the RNA-binding protein HuR. J. Biol. Chem. 2004;279:49298–49306. doi: 10.1074/jbc.M407535200. [DOI] [PubMed] [Google Scholar]
- 34.Giles KM, Daly JM, Beveridge DJ, Thomson AM, Voon DC, Furneaux HM, Jazayeri JA, Leedman PJ. The 3′-untranslated region of p21WAF1 mRNA is a composite cis-acting sequence bound by RNA-binding proteins from breast cancer cells, including HuR and poly(C)-binding protein. J. Biol. Chem. 2003;278:2937–2946. doi: 10.1074/jbc.M208439200. [DOI] [PubMed] [Google Scholar]
- 35.Lafarga V, Cuadrado A, Lopez de Silanes I, Bengoechea R, Fernandez-Capetillo O, Nebreda AR. p38 Mitogen-activated protein kinase- and HuR-dependent stabilization of p21(Cip1) mRNA mediates the G(1)/S checkpoint. Mol. Cell. Biol. 2009;29:4341–4351. doi: 10.1128/MCB.00210-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shu L, Yan W, Chen X. RNPC1, an RNA-binding protein and a target of the p53 family, is required for maintaining the stability of the basal and stress-induced p21 transcript. Genes Dev. 2006;20:2961–2972. doi: 10.1101/gad.1463306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho SJ, Zhang J, Chen X. RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability. Nucleic Acids Res. 38:2256–2267. doi: 10.1093/nar/gkp1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waggoner SA, Johannes GJ, Liebhaber SA. Depletion of the poly(C)-binding proteins alphaCP1 and alphaCP2 from K562 cells leads to p53-independent induction of cyclin-dependent kinase inhibitor (CDKN1A) and G1 arrest. J. Biol. Chem. 2009;284:9039–9049. doi: 10.1074/jbc.M806986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yano M, Okano HJ, Okano H. Involvement of Hu and heterogeneous nuclear ribonucleoprotein K in neuronal differentiation through p21 mRNA post-transcriptional regulation. J. Biol. Chem. 2005;280:12690–12699. doi: 10.1074/jbc.M411119200. [DOI] [PubMed] [Google Scholar]
- 40.Liu G, Chen X. DNA polymerase eta, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the DNA damage checkpoint and p53 activation. Mol. Cell. Biol. 2006;26:1398–1413. doi: 10.1128/MCB.26.4.1398-1413.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scoumanne A, Chen X. The epithelial cell transforming sequence 2, a guanine nucleotide exchange factor for Rho GTPases, is repressed by p53 via protein methyltransferases and is required for G1-S transition. Cancer Res. 2006;66:6271–6279. doi: 10.1158/0008-5472.CAN-06-0121. [DOI] [PubMed] [Google Scholar]
- 42.Schnutgen F, De-Zolt S, Van Sloun P, Hollatz M, Floss T, Hansen J, Altschmied J, Seisenberger C, Ghyselinck NB, Ruiz P, et al. Genomewide production of multipurpose alleles for the functional analysis of the mouse genome. Proc. Natl Acad. Sci. USA. 2005;102:7221–7226. doi: 10.1073/pnas.0502273102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu J, Zhou W, Jiang J, Chen X. Identification of a novel p53 functional domain that is necessary for mediating apoptosis. J. Biol. Chem. 1998;273:13030–13036. doi: 10.1074/jbc.273.21.13030. [DOI] [PubMed] [Google Scholar]
- 44.Peritz T, Zeng F, Kannanayakal TJ, Kilk K, Eiriksdottir E, Langel U, Eberwine J. Immunoprecipitation of mRNA-protein complexes. Nat. Protoc. 2006;1:577–580. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
- 45.Lukong KE, Chang KW, Khandjian EW, Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 46.Hamada H, Tashima Y, Kisaka Y, Iwamoto K, Hanai T, Eguchi Y, Okamoto M. Sophisticated framework between cell cycle arrest and apoptosis induction based on p53 dynamics. PLoS One. 2009;4:e4795. doi: 10.1371/journal.pone.0004795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Batchelor E, Loewer A, Lahav G. The ups and downs of p53: understanding protein dynamics in single cells. Nat. Rev. Cancer. 2009;9:371–377. doi: 10.1038/nrc2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Battelli C, Nikopoulos GN, Mitchell JG, Verdi JM. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Mol. Cell. Neurosci. 2006;31:85–96. doi: 10.1016/j.mcn.2005.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.