Abstract
Influenza viruses infect a wide range of avian and mammalian host species including humans. Infections with influenza viruses are responsible for major causes of human respiratory infections and mortality. Influenza viruses are recognized by the innate immune system through multiple mechanisms. These include endosomal recognition through the Toll-like receptor 7 (TLR7) and cytosolic recognition through the retinoic acid inducible gene I (RIG-I). Recent studies also identified the role of NOD-like receptors (NLR) in innate detection of influenza viruses leading to the activation of the inflammasomes. Here, we review the cellular and molecular mechanisms by which influenza virus infection leads to inflammasome activation, and discuss the consequences of such activation in innate and adaptive immune defense against influenza viruses.
Infection and innate recognition of influenza virus
Influenza A virus is a negative single-strand RNA virus that is responsible for the annual seasonal epidemics worldwide, and novel virus strains emerge sporadically as pandemic viruses. The unpredictable nature of emergent pandemic strains requires an urgent need to develop vaccines and therapeutics that can be rapidly prepared and deployed to prevent or treat the next influenza pandemic. The outcome of influenza A virus infection is determined by the host’s immune response against the virus. Thus, understanding the mechanism by which influenza A virus infection is detected and cleared by the immune system provides a useful basis for the development of effective vaccines.
The innate immune system is essential for the rapid and effective control of viral infections as well as for initiation of adaptive immunity. The primary target of influenza virus infection is the airway epithelial cells lining the respiratory mucosa. Once these cells are infected and lysed, influenza A virus can also infect alveolar macrophages and dendritic cells (DCs) that reside in the airway. Upon sensing infection, DCs that have taken up viral antigens play a key role in priming effector T cell responses [1–3]. Once activated, influenza virus-specific CD4 and CD8 T cells act in concert with antibody-producing B cells to promote viral clearance in the lung [4].
Influenza infection can be recognized by the innate immune system in multiple ways. These include endosomal recognition through the Toll-like receptor 7 (TLR7) and cytosolic recognition through the retinoic acid inducible gene I (RIG-I). Recent studies also identified the role of NOD-like receptors (NLR) in innate recognition of influenza virus infection. Interestingly, the innate system sensing of the influenza infection by the NLR depends not on the pathogen-associated molecular patterns (PAMPs) but also the activity of the viral replication strategy. In this article, we focus on the role of the NLR inflammasomes in innate and adaptive immune defense against influenza A virus infection. The underlying mechanism for virus-mediated inflammasome activation is also considered.
Viral sensing by innate recognition receptors direct the development of adaptive immunity
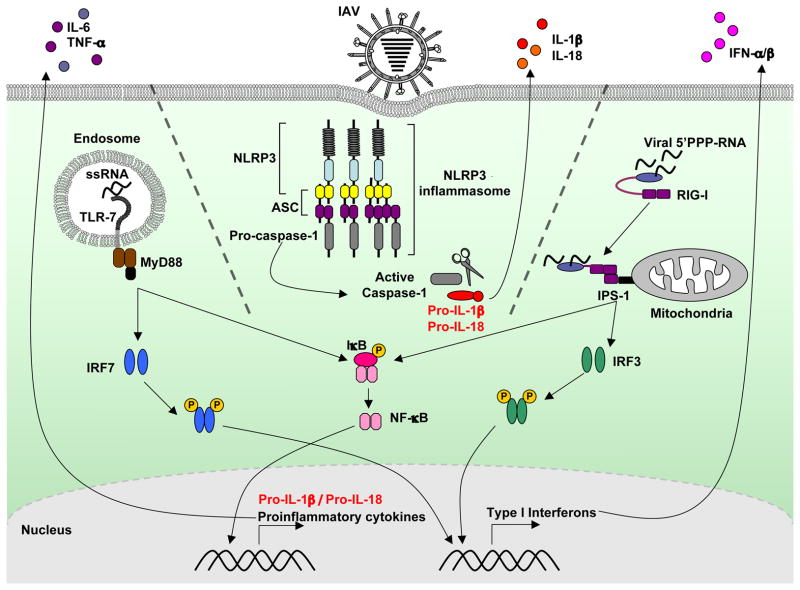
Influenza A virus is recognized by innate immune cell types through distinct mechanisms that utilize different germ line-encoded pathogen-recognition receptors (PRRs) (Figure 1). In infected cells, the cytosolic sensor retinoic acid inducible gene I (RIG-I) detects influenza virus through recognition of 5′-triphosphates on genomic single stranded (ss)RNA, which is revealed after viral fusion and replication [5]. In specialized sentinel cells, such as the plasmacytoid dendritic cells (pDCs), influenza ssRNA, upon its release in the acidified endosome during infection is recognized by Toll-like receptor (TLR)-7 in the endosome [6,7]. Both RIG-I and TLR7 pathways induce production of proinflammatory cytokines and type I interferons (IFNs), which elicit an anti-viral response and induce viral resistance in neighboring uninfected cells. RIG-I is critical for viral detection and type I IFN production in infected fibroblasts and conventional dendritic cells (cDCs), whereas pDCs use the TLR7 pathway for innate influenza A virus recognition [8]. TLR3 also recognizes double stranded (ds)RNA in the endosomes and mediates a proinflammatory response to influenza A virus infection leading to pathology [9–11].
Figure 1. Innate recognition of influenza A virus.
Influenza A virus is sensed by at least three different types of PRRs. First, viral single-stranded RNA is recognized by TLR7 in the endosome. Second, the cytosolic sensor RIG-I detects viral ssRNA bearing 5′ triphosphates. TLR7 signals through the adaptor protein, MyD88, which leads to the downstream activation of IRF7 and NF-κB. In contrast, RIG-I signals through the adaptor IPS-I located on the mitochondria for the activation of IRF3 and NF-kB. Activated IRF-7 and IRF-3 can translocate to the nucleus to activate the production of type I interferons while NF-κB acts as a transcription factor for the induction of a proinflammatory cytokines including IL-6, TNF-α, and the pro-form IL-1β. The TLR7 pathway is critical for the production of type I interferons in specialized sentinel cells, pDCs, while most other virally infected cells utilize the RIG-I pathway. Third, influenza A virus activates the NLRP3 inflammasome in dendritic cells and macrophages leading to the activation of caspase-1, which mediates the processing of pro-IL-1β to mature IL-1β and its subsequent release.
The importance of TLRs and RIG-I for induction of adaptive anti-viral immunity has also been studied, with somewhat conflicting results. In one study, in response to influenza A virus infection, neither T cell nor antibody (Ab) responses were dependent on MyD88, an adaptor protein required for TLR signaling [12]. In contrast, a different group found that TLR7 and MyD88 were required for virus-specific IgG responses, but dispensable for CD4 and CD8 T cell activation and effector function [13]. It was also reported that TLR7 and MyD88 signaling is required for the induction of CD4 T cell and B cell anti-hemagglutinin Ab responses, but not Ab responses to whole influenza A/PR8 virions [14]. In contrast, TLR3 and IPS-1, the adaptor protein that mediates RIG-I signaling, do not appear to play a role in the induction of CD4, CD8 or Ab responses [13,14]. It is possible that the dose of virus used to infect the mice, the strain of influenza A virus used, and the mode of virus delivery (aerosolized [12] vs. solution [13,14]) might explain these different findings. Interestingly, these studies all demonstrate that the activation of primary CD8 T cells during an anti-influenza immune response relies on mechanisms other than the MyD88-dependent TLR7 pathway and the IPS-1-dependent RIG-I pathway.
More recent studies have revealed that cytoplasmic complexes called inflammasomes play an essential role in influenza virus recognition and defense [15–17]. The inflammasomes are multiprotein complexes that acts as a platform for caspase-1 activation, which in turn cleaves immature (‘pro’) forms of cytokines such as pro-interleukin-1β (IL-1β) and pro-IL-18, resulting in their secretion into the extracellular space [18,19] (Box 1). The inflammasomes come in a variety of flavors. The best studied are those that contain members of the NLRs, which are a family of proteins that contain three major domains: an N-terminal protein-interacting effector domain, a central nucleotide-binding domain, and a C-terminal leucine-rich repeat (LRR) domain [20]. NLRP3 (NALP3/Cryopyrin/NACHT- LRR- Pyrin (PYD)- containing protein 3) is one of the best characterized NLRs. NLRP3 recruits the adaptor ASC (apoptosis-associated speck-like protein containing a caspase recruitment domain) also known as PYCARD, which consists of an N-terminal PYD domain and a C-terminal caspase activation and recruitment domain (CARD) that is essential for the binding and recruitment of caspase-1 to the inflammasome [21]. The NLRP3 inflammasome has been linked to innate recognition of both DNA and RNA viruses [22–24].
Box 1. IL-1β and IL-18 in innate and adaptive immunity.
Even before the discovery of the inflammasome complex, the roles of IL-1β and IL-18 in orchestrating an effective innate and adaptive immune response was known. IL-1β induces the expression of hundreds of genes, including cytokines (IL-6 and TNF-α), proinflammatory mediators (iNOS, COX2, PLA2), and adhesion molecules that are important for leukocyte trafficking [51]. In adaptive immunity, IL-1β plays a central role in the induction of Th17 cells in humans [52] and the antigen-driven expansion and differentiation of CD4 T cells [53]. While IL-1R is widely expressed by many cell types, IL-1β is mainly produced by the sentinel cells of the innate immune system including macrophages and DCs, although fibroblasts and keratinocytes can also synthesize this cytokine in response to tissue injury or stress signals [51,54]. On the other hand, IL-18, which is expressed mainly by macrophages and DCs, work alone or in combination with IL-15 prime natural killer (NK) cells for the production of IFN-γ and enhance their cytolytic activity [55–57]. IL-18 in combination with IL-12 or IL-2 can also induce differentiation of either Th1 or Th2 cell types depending on the cytokine milieu [58,59].
In response to influenza A virus infection in IL-1R−/− mice, which are unable to respond to IL-1β and IL-1α, impaired leukocyte recruitment to the lung after intranasal influenza A virus infection was observed [60]. This study also found that IL-1R−/− mice had diminished CD4 and CD8 T cell responses, impaired IgM responses and increased incidences of virus-induced fatality. In a separate study, mice deficient in IL-18 were found to have reduced cellular infiltration into the lung, and decreased level of IFN-γ in the alveolar space early after influenza A virus infection [61]. Further analysis revealed that natural killer cell cytolytic activity was compromised, which could explain the significantly higher viral titer in these mice in the first three days post infection [61]. IL-18−/− mice have normal number of epitope-specific CD8 T cells but these CD8 T cells are less capable of secreting IFN-γ, TNF-α and IL-2, indicating that IL-18 is required for optimal cytokine production by CTLs [62]. Collectively, these studies indicate that IL-1 and IL-18 have non-redundant roles in triggering different aspects of innate and adaptive immunity to generate a protective anti-influenza response. This requirement for IL-1β and IL-18, but not IL-12, is consistent with the ability of the host animal to recognize influenza A virus and trigger the activation of the inflammasomes [15–17].
In addition to the NLRs, other cytoplasmic PRRs can also act as initiators of the inflammasome in virus-infected cells. RIG-I recruits caspase-1 independent of IPS-1 and induces inflammasome activation by directly engaging the adaptor ASC in cells infected with vesicular stomatitis virus (VSV) or transfected with 5′ triphosphates [25]. In contrast, the AIM2 binds cytosolic DNA and engages ASC to form a caspase-1 activation complex that is critical for host defense against certain DNA viruses and intracellular bacteria [26–31]. Whether the RIG-I or AIM2 inflammasome is required for host defense against influenza A virus infection remains to be investigated.
Influenza virus infection activates inflammasome-dependent innate and adaptive responses
Evidence for the ability of influenza virus to trigger the inflammasome was published more than a decade ago, in a study showing that human macrophages produce IL-1β and IL-18 upon influenza virus infection; a response that can be blocked by a caspase-1 specific inhibitor [32]. A more recent study showed that caspase-1 activation in response to influenza virus infection requires NLRP3 [22] Three recent reports provide further insight into the role of inflammasomes in the host immune response and survival after influenza A virus infection [15–17]. Increased production of IL-1β and IL-18 was detected in cultured bone marrow derived dendritic cells (BMDCs) and bone marrow derived macrophages (BMMs) and in the bronchoalveolar lavage (BAL) after influenza A virus infection in mice in vivo [15–17]. However, mice that are deficient in NLRP3, ASC or caspse-1 fail to secrete IL-1β or IL-18 into the alveolar space in response to influenza A virus infection. All three studies found that early viral clearance, up to 7 days after infection, was not affected in the inflammasome-deficient mice [15–17]. But late stage viral clearance (examined between 7–10 days post infection) was dependent on the inflammasome complex [15,17]. While all three studies attribute these findings to different mechanisms, the data are consistent with a requirement for inflammasome-dependent responses in antiviral defense against respiratory influenza A virus infection.
Innate defense responses against influenza A virus requires NLRP3 inflammasome
Two groups [17][16] have used high dose viral challenge (3,000–8,000 PFU of A/PR8 influenza A virus per mouse) to probe the importance of inflammasomes in innate antiviral responses against influenza A virus infection. These studies found that NLRP3 is essential for the innate cellular responses to influenza A virus, and for the production of chemokines including KC and MIP-2α after 3 days post infection. Both studies found that in the absence of NLRP3 or caspase-1, cellular infiltration into the BAL is significantly decreased after intranasal influenza A virus infection. Consequently, only 20% [17] or 40% [16] of mice deficient in NLRP3 survived high dose A/PR8 challenge compared to 70% [17] or 60% [16] of WT mice. Thus, inflammasome activation and IL-1β secretion leads to the production of chemokines that enhance the recruitment of inflammatory cells including neutrophils and monocytes and/or their maintenance in the lung following influenza A virus infection. One of these studies showed that diminished production of cytokines and chemokines after influenza A virus infection enhanced the severity of influenza pneumonia, which is characterized by increased pulmonary necrosis and reduced respiratory function without affecting early viral clearance (on days 3 and 6 of infection) [16]. This suggests that the NLRP3-dependent caspase-1 activation and IL-1β secretion promote tissue repair after infection-induced injury rather than causing immunopathology in the infected host. A contrasting study, however, has reported that reduced cellular recruitment in NLRP3– deficient mice results in impaired antiviral defense, as indicated by sustained viral burden in the Nlrp3−/− mice compared to WT counterpart 7 days after infection [17]. Combined, these results demonstrate the importance of NLRP3, ASC and caspase-1 dependent inflammasome activation in innate antiviral defense, through cellular recruitment [16,17] or induction of tissue repair [16] in the respiratory tract after influenza A virus infections. However, to what extent these different mechanisms account for the lethality in the respective knockout mice remains to be resolved.
Adaptive immune defense against influenza A virus requires the ASC inflammasome
In response to influenza A virus infection, both CD4 and CD8 T cells can develop into effector T cells that migrate to the lung to promote viral clearance. In addition, B cells producing antibodies specific to influenza antigens are critical for neutralizing and clearing virions throughout the body. One study reported, using a low sublethal dose of viral challenge (10 PFU of A/PR8, or 0.4 LD50 per mouse), that IL-1β secretion in the BAL occurs in a manner dependent on NLRP3, ASC and caspase-1 [15]. In contrast, in the lung parenchyma, IL-1β secretion and cellular recruitment remained intact in Nlrp3−/−, but not ASC-knockout (pycard−/−) or caspase-1−/−, mice. Caspase-1 activation within hematopoietic cells, but not the stromal compartment, was responsible for IL-1β production and subsequent cellular infiltration into the lung after influenza A virus infection [15]. Caspase-1−/− and pycard−/− mice, but not Nlrp3−/− mice, failed to activate virus-specific IFN-γ secreting CD4 and CD8 T cells, and to generate of nasal IgA and serum IgG isotypes. These mice succumbed to virus-induced mortality. Thus, this study indicates that ASC inflammasomes are required for the development of adaptive immune responses to influenza A virus infection. The requirement for inflammasomes to generate adaptive immune responses was attributed to impaired secretion of IL-1β since Il1r1−/− mice suffered from a similar lack of immune responses and viral disease. In contrast, a different study found that antibody production, as well as tetramer+ CD8 T cell number in the BAL, was intact in both Nlrp3−/− and caspase-1−/− mice between 7–11 days of infection [16]. Thus, although NLRP3 is required for inflammasome activation in certain cell types such as DCs and macrophages, NLRP3 deficiency does not impact the generation of adaptive immunity against influenza A virus infection [15,16]. The two studies differ, however, in their reported requirement for caspase-1 in adaptive immunity. In addition, our study [15] and others [16,17] differed with respect to the role of NLRP3 in the clinical outcome of the influenza A virus challenge. The reason for the discrepancies remains unclear, but might be related to: 1) the different viral challenge doses used to infect the mice; 2) a difference in methods of virus propagation and maintainance; 3) inclusion of alum, a NLRP3 agonist, in viral inoculum [17]; 4) the Nlrp3−/− mouse strain used; and 5) environmental factors, including possible differences in the commensal microbiota in different animal facilities. While these issues remain to be resolved, these studies collectively highlight the importance of the NLRP3 inflammasome in mediating innate immunity to influenza A virus and the NLRP3-independent activation of ASC inflammasomes in both innate and adaptive immune defense against influenza A virus.
Influenza A virus infection triggers both signal 1 and signal 2 for inflammasome activation
The activation of the NLRP3 inflammasome and production of IL-1β usually requires two signals. The first signal can be triggered by TLR agonists, including viral genomic RNA and synthetic dsRNA, poly(I:C) [33]. The priming signal leads to transcriptional activation of the genes encoding pro-IL-1β, pro-IL-18 and NLRP3 [34]. The second signal is triggered in response to various stress signals associated with damaged-self and non-self molecules [34,35]. Three models – that are not necessarily exclusive – have been proposed to explain the second signal for NLRP3 inflammasome activation [36–39]. One model is the binding of extracellular ATP – to and subsequent activation of – the cell surface receptor P2X7. Activation of this ATP-gated ion channel triggers K+ efflux and recruitment of pannexin 1 to form a large non-selective pore, which might enable entry of NLRP3 agonists into the cell. In a second model, after phagocytosis of large crystals such as monosodium urate (MSU), silica, asbestos and aluminum salts, lysosomal damage and rupture of lysosomal content might activate NLRP3. Finally, production of reactive oxygen species (ROS) is proposed to lead to the formation of the NLRP3 inflammasome. It remains unclear, however, how these diverse signals might converge to activate the NLRP3 inflammasome.
In response to influenza virus infection, one report showed that influenza A virus infected macrophages required NLRP3 to activate caspase-1 when pulsed transiently with ATP (a NLRP3 agonist) for 30 minutes, demonstrating that infection is sufficient to trigger signal 1 for the activation of the NLRP3 inflammasome. However, the nature of the two signals, and the underlying mechanism by which influenza A virus infection triggers the activation of NLRP3 inflammasome remained elusive. It was reported that influenza A virus-mediated activation of the NLRP3 inflammasome depends on lysosomal maturation and the production of ROS [17]. Furthermore, intranasal delivery of poly (I:C) induces NLRP3-dependent airway inflammation in vivo and the transfection of viral genomic RNA alone activated NLRP3 inflammasome in cultured BMDCs in vitro, suggesting that viral RNA acts as both signal 1 and signal 2 to trigger the activation of the NLRP3 inflammasome [16,17]. These results seemingly differed from reports that infection of influenza A virus inactivated by UV light or transfection of cultured BMDCs with viral RNA, poly (I:C), or infection with reovirus or VSV failed to elicit caspase-1 activation [15,24].
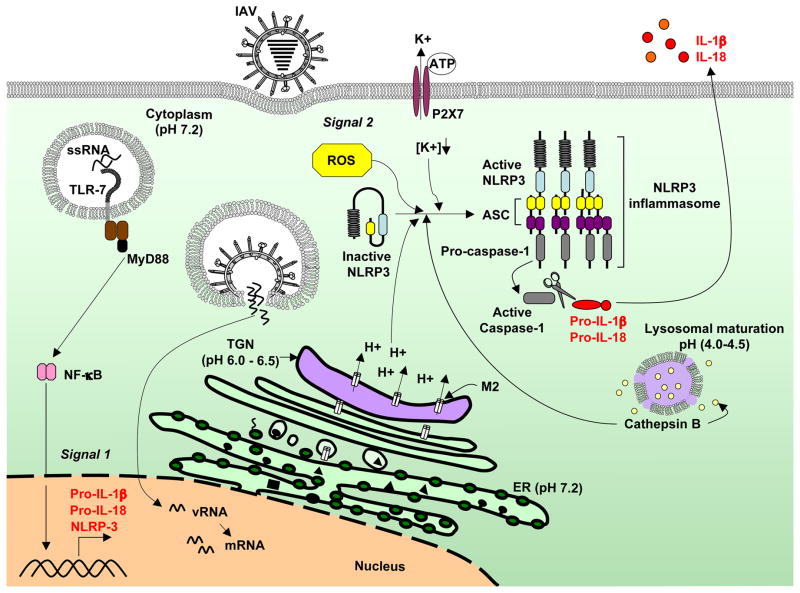
Recently, a molecular mechanism for activation of the NLRP3 inflammasome by influenza A virus has been provided [33]. Infection by influenza virus, but not other viruses, is sufficient to trigger both signals 1 and 2 required for NLRP3 inflammasome activation. It was found that delivery of viral RNA into WT but not Tlr7−/− BMMs induced transcriptional activation of pro-IL-1β, indicating that TLR7 senses viral genomic RNA in the endosome and provides signal 1. However, TLR7-dependent recognition alone was insufficient to trigger the activation of caspase-1 and cleavage of IL-1β. Influenza A virus fusion and replication were required to stimulate NLRP3 inflammasome activation and this was mediated by the virally-encoded ion channel M2. The M2 channel of influenza A virus is a homotetrameric integral membrane protein that associates to form a highly specific proton channel [40]. It is essential for influenza A virus infection and replication [41]. M2 protein allows influenza viral entry into the cell by equilibrating the pH of the virus within the acidifying endosome. This step is critical as it facilitates uncoating (release of genetic material in to the cytoplasm) of the viral genome by disruption of the interactions between the viral ribonucleoprotein (vRNP) complex and the matrix protein, M1, which coats the vRNP [42,43]. In addition, the M2 channel exports protons in the acidic trans-Golgi network (TGN) to neutralize the pH of the lumen of the TGN in order to prevent the premature maturation of hemagglutinin to its low-pH fusogenic form [44]. While the fusogenic property of HA is required during viral entry for the fusion of viral and host intracellular membranes, it must be avoided during virion production [45].
Ectopic expression of the M2 channel restored the production of IL-1β from BMMs and BMDCs that were infected with an influenza A virus that encoded a mutant M2 protein that lacks the H+ transport activity [33]. This indicates that the M2 ion channel activity is both required and sufficient to trigger signal 2 for inflammasome activation and IL-1β production. After infection, virus-encoded M2 is expressed in the secretory pathway and trafficked from the Golgi apparatus to the plasma membrane within 12 hours [33]. Pharmacological inhibitors and biochemical agents that neutralize the pH of the TGN mimic the activity of M2 and activate the inflammasome in cultured BMMs, suggesting that ionic perturbation in the TGN by M2 channel activity triggers inflammasome activation. Moreover, M2-dependent IL-1β production by influenza A virus-infected BMMs can be blocked by high concentration of extracellular K+ or by adding ROS inhibitor, consistent with a previous report [17]. How M2 affects ROS production and K+ efflux remains unclear. M2 has an anti-porter like property by facilitating cation efflux when protons flow down a concentration gradient into vesicles [46], which suggests that M2 can directly modulate intracellular K+ concentration. Further studies are required to characterize the downstream pathway leading to NLRP3 activation by influenza M2 protein and to determine whether K+ efflux and ROS production are cross-regulated or independently required for NLRP3 activation in influenza A virus infected cells.
Future perspective.
Several studies now reveal the importance of inflammasomes in innate viral defense and for the development of adaptive immune responses. Furthermore, we have begun to learn about the underlying mechanisms of NLRP3-dependent inflammasome activation following influenza A virus infections. Although some progress has been made in this regard, additional studies are required to fully understand how and why the inflammasomes are required to mediate protection against influenza A virus infections (see Box for Outstanding Questions).
Many viruses are known to encode proteins that facilitate ion transport across host cell membranes [47]. It is tempting to speculate that sensing of cellular stress imposed by imbalances in ionic concentrations in intracellular vesicles is a general pathogen-recognition pathway used by the eukaryotic host to signal the activation of the NLR inflammasomes. For instance, picornaviruses, which engage the NLRP3 inflammasome and the cytosolic sensor Mda5 to activate caspase-1, encode the ion channel protein 2B that is mainly localized to the endoplasmic reticulum and Golgi [25,47]. Whether the ion channel activity of Picornaviruses is required to trigger the NLRP3 inflammasome remains to be investigated. Likewise, the HIV accessory protein Vpu, which is required for the assembly and release of the virus, has ion channel activity [48]. A recent study has demonstrated association between NLRP3 single nucleotide polymorphisms (SNP) and susceptibility to HIV infection, although the mechanism behind this association remains unknown (Box 2) [49].
Mutant influenza virus lacking the M2 protein (M2KO) has been used as live attenuated vaccine in experimental studies, and mice intranasally infected with M2KO virus generated a protective immune response against influenza A virus [50]. This is consistent with the observation that NLRP3 inflammasome activation is dispensable for the development of adaptive immunity [15,16]. The induction of protective immunity against influenza A virus, however, requires the activation of caspase-1 by binding to the adaptor protein, ASC [15]. This underscores the importance of further studies to identify additional caspase-1-activating mechanisms that are critical for the establishment of anti-influenza immunity. Addressing these and other questions regarding influenza A virus recognition through NLRs and potentially other inflammasome complexes will likely provide clues to making more effective vaccines and therapeutics against impending flu pandemics.
Box 2. Genetic evidence for inflammasomes in human antimicrobial defense.
Polymorphisms or mutations in human NLRs have been known to associate with autoinflammatory disorders such as Muckle-Wells syndrome and Crohn’s disease, but epidemiological studies of genetic polymorphisms in human populations have only recently begun to provide us with a better understanding of the role of NLRs in the antiviral response and immune activation against microbial infection [63]. Several single-nucleotide polymorphisms (SNPs) have been identified in the gene encoding NLRP3 that result either in heightened or reduced inflammasome activation. For instance, the NLRP3 inflammasome is critical for host defense against the fungal pathogen Candida albicans [64]. Consistently, in women with vulvar vestibulitis syndrome, polymorphism in the NLRP3 gene is associated with reduced levels of IL-1β production and heightened susceptibility to a recurrent form of vaginal C. albicans infections [65]. Interestingly, the same group identified that among women undergoing in vitro fertilization, those carrying this NLRP3 SNP are more likely to acquire cervical mycoplasma infection-associated infertility [66]. Other genetic factors can also have a substantial influence on the production of IL-1β. Caspase-12 is thought to negatively regulate IL-1β production by antagonizing the inflammasome and NF-κB signaling. Most healthy individuals express a truncated form of caspase-12 that is biologically inactive. However, expression of the full-length caspase-12 protein in humans is associated with an enhanced susceptibility to bacterial sepsis, suggesting that these individuals are prone to infections [67]. In addition, polymorphisms in the IL-1 gene family are known to affect the susceptibility to and the outcome of both viral and bacterial infections [68,69]. Taken together, not only do NLR mutations predispose humans to autoinflammatory disorders, emerging studies on humans carrying mutations or polymorphisms in the NLRs support the vital role of inflammasome activation in antimicrobial defense.
Box for Outstanding Questions.
What is being sensed by NLRP3 to activate the inflammasome?
How does ion transport activity at the TGN membrane translate to the activation of cytosolic NLRP3 – are adaptor molecules involved?
Does the sensing of ion channel activity represent a general mechanism used by the host to activate the NLRP3 inflammasome?
Does the NLRP3 inflammasome share redundant roles with other inflammasome receptors to generate a protective immunity response against influenza A virus? This type of cooperativity between two NLRs have been seen for salmonella infection, in which mice lacking both the NLRP3 and NLRC4 are markedly more susceptible compared to mice lacking only NLRP3 or NLRC4 [70]?
Do other members of the NLRP family form an ASC-dependent caspase-1 activating complex in response to influenza A virus infection? In addition to NLRP3, NLRP6 and NLRP12 have been shown to engage ASC to activate caspase-1 [71,72].
Figure 2. Proposed mechanism of NLRP3 inflammasome activation by influenza A virus.
Influenza A virus can trigger both signal 1 and signal 2 for the NLRP3 inflammasome activation. Sensing of influenza ssRNA in the endosome by TLR7 induces the transcription of pro-IL-1β and NLRP3. Within the TGN, the influenza-encoded M2 ion channel protein transports protons (H+) out of the lumen, leading to the neutralization of the TGN pH. Ionic imbalance in the Golgi compartment triggers M2-mediated inflammasome activation. The P2X7 receptor, an ATP-gated ion channel that causes potassium (K+) efflux when activated, is partially required for M2-induced inflammasome activation. Lysosomal maturation and the activity of cathepsin B and ROS also play a role in influenza-induced inflammasome activation but the underlying mechanisms remain to be defined.
Acknowledgments
We thank Dr. Takeshi Ichinohe for his contributions that are discussed in this review and for his help with the figures. This work was supported by National Institutes of Health (NIH) (AI062428, AI064705 and AI083242 to A.I.). A.I. holds an Investigators in Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. I.K.P. was supported by the NIH National Research Service Award (T32AI07019) from the Interdisciplinary Immunology Training Program in Yale University, Department of Immunobiology. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ballesteros-Tato A, et al. Temporal changes in dendritic cell subsets, cross-priming and costimulation via CD70 control CD8+ T cell responses to influenza. Nat Immunol. 2010;11 (3):216–224. doi: 10.1038/ni.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGill J, et al. Protective influenza-specific CD8 T cell responses require interactions with dendritic cells in the lungs. J Exp Med. 2008;205 (7):1635–1646. doi: 10.1084/jem.20080314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roman E, et al. CD4 effector T cell subsets in the response to influenza: heterogeneity, migration, and function. J Exp Med. 2002;196 (7):957–968. doi: 10.1084/jem.20021052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohlmeier JE, Woodland DL. Immunity to Respiratory Viruses. Annual Review of Immunology. 2009;27 (1):61–82. doi: 10.1146/annurev.immunol.021908.132625. [DOI] [PubMed] [Google Scholar]
- 5.Pichlmair A, et al. RIG-I-Mediated Antiviral Responses to Single-Stranded RNA Bearing 5′-Phosphates. Science. 2006;314 (5801):997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 6.Lund JM, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci U S A. 2004;101 (15):5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diebold SS, et al. Innate Antiviral Responses by Means of TLR7-Mediated Recognition of Single-Stranded RNA. Science. 2004;303 (5663):1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 8.Kato H, et al. Cell Type-Specific Involvement of RIG-I in Antiviral Response. Immunity. 2005;23 (1):19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 9.Le Goffic R, et al. Cutting Edge: Influenza A virus activates TLR3-dependent inflammatory and RIG-I-dependent antiviral responses in human lung epithelial cells. J Immunol. 2007;178 (6):3368–3372. doi: 10.4049/jimmunol.178.6.3368. [DOI] [PubMed] [Google Scholar]
- 10.Guillot L, et al. Involvement of toll-like receptor 3 in the immune response of lung epithelial cells to double-stranded RNA and influenza A virus. J Biol Chem. 2005;280 (7):5571–5580. doi: 10.1074/jbc.M410592200. [DOI] [PubMed] [Google Scholar]
- 11.Le Goffic R, et al. Detrimental contribution of the Toll-like receptor (TLR)3 to influenza A virus-induced acute pneumonia. PLoS Pathog. 2006;2 (6):e53. doi: 10.1371/journal.ppat.0020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez CB, et al. TLR-Independent Induction of Dendritic Cell Maturation and Adaptive Immunity by Negative-Strand RNA Viruses. J Immunol. 2004;173 (11):6882–6889. doi: 10.4049/jimmunol.173.11.6882. [DOI] [PubMed] [Google Scholar]
- 13.Heer AK, et al. TLR Signaling Fine-Tunes Anti-Influenza B Cell Responses without Regulating Effector T Cell Responses. J Immunol. 2007;178 (4):2182–2191. doi: 10.4049/jimmunol.178.4.2182. [DOI] [PubMed] [Google Scholar]
- 14.Koyama S, et al. Differential Role of TLR- and RLR-Signaling in the Immune Responses to Influenza A Virus Infection and Vaccination. J Immunol. 2007;179 (7):4711–4720. doi: 10.4049/jimmunol.179.7.4711. [DOI] [PubMed] [Google Scholar]
- 15.Ichinohe T, et al. Inflammasome recognition of influenza virus is essential for adaptive immune responses. The Journal of Experimental Medicine. 2009;206 (1):79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas PG, et al. The Intracellular Sensor NLRP3 Mediates Key Innate and Healing Responses to Influenza A Virus via the Regulation of Caspase-1. 2009;30(4):566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allen IC, et al. The NLRP3 Inflammasome Mediates In Vivo Innate Immunity to Influenza A Virus through Recognition of Viral RNA. Immunity. 2009;30 (4):556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinarello CA. Immunological and Inflammatory Functions of the Interleukin-1 Family. Annual Review of Immunology. 2009;27 (1):519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 19.Schroder K, Tschopp J. The Inflammasomes. Cell. 2010;140 (6):821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 20.Yu HB, Finlay BB. The Caspase-1 Inflammasome: A Pilot of Innate Immune Responses. Cell Host & Microbe. 2008;4 (3):198–208. doi: 10.1016/j.chom.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Martinon F, Tschopp Jr. NLRs join TLRs as innate sensors of pathogens. Trends in Immunology. 2005;26 (8):447–454. doi: 10.1016/j.it.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Kanneganti TD, et al. Critical Role for Cryopyrin/Nalp3 in Activation of Caspase-1 in Response to Viral Infection and Double-stranded RNA. Journal of Biological Chemistry. 2006;281 (48):36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 23.Kanneganti TD, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440 (7081):233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 24.Muruve DA, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452 (7183):103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 25.Poeck H, et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1[beta] production. Nat Immunol. 2010;11 (1):63–69. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- 26.Rathinam VAK, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11 (5):395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323 (5917):1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 28.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458 (7237):514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandes-Alnemri T, et al. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458 (7237):509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burckstummer T, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10 (3):266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 31.Fernandes-Alnemri T, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11 (5):385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pirhonen J, et al. Virus Infection Activates IL-1{beta} and IL-18 Production in Human Macrophages by a Caspase-1-Dependent Pathway. J Immunol. 1999;162 (12):7322–7329. [PubMed] [Google Scholar]
- 33.Ichinohe T, et al. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 2010;11 (5):404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bauernfeind FG, et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol. 2009;183 (2):787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franchi L, et al. Cutting edge: TNF-alpha mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J Immunol. 2009;183 (2):792–796. doi: 10.4049/jimmunol.0900173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinon F, et al. The Inflammasomes: Guardians of the Body. Annual Review of Immunology. 2009;27 (1):229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 37.Tschopp J, Schroder K. NLRP3 inflammasome activation: the convergence of multiple signalling pathways on ROS production? Nat Rev Immunol. 2010;10 (3):210–215. doi: 10.1038/nri2725. [DOI] [PubMed] [Google Scholar]
- 38.Schroder K, et al. The NLRP3 Inflammasome: A Sensor for Metabolic Danger? Science. 2010;327 (5963):296–300. doi: 10.1126/science.1184003. [DOI] [PubMed] [Google Scholar]
- 39.Hornung V, Latz E. Critical functions of priming and lysosomal damage for NLRP3 activation. European Journal of Immunology. 2010;40 (3):620–623. doi: 10.1002/eji.200940185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinto LH, Lamb RA. The M2 Proton Channels of Influenza A and B Viruses. Journal of Biological Chemistry. 2006;281 (14):8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 41.Takeda M, et al. Influenza a virus M2 ion channel activity is essential for efficient replication in tissue culture. J Virol. 2002;76 (3):1391–1399. doi: 10.1128/JVI.76.3.1391-1399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhirnov OP. Solubilization of matrix protein M1/M from virions occurs at different pH for orthomyxo- and paramyxoviruses. Virology. 1990;176 (1):274–279. doi: 10.1016/0042-6822(90)90253-n. [DOI] [PubMed] [Google Scholar]
- 43.Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67 (1):117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 44.Ciampor F, et al. Evidence that the amantadine-induced, M2-mediated conversion of influenza A virus hemagglutinin to the low pH conformation occurs in an acidic trans Golgi compartment. Virology. 1992;188 (1):14–24. doi: 10.1016/0042-6822(92)90730-d. [DOI] [PubMed] [Google Scholar]
- 45.Sakaguchi T, et al. The ion channel activity of the influenza virus M2 protein affects transport through the Golgi apparatus. The Journal of Cell Biology. 1996;133 (4):733–747. doi: 10.1083/jcb.133.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leiding T, et al. Proton and cation transport activity of the M2 proton channel from influenza A virus. Proceedings of the National Academy of Sciences. 2010;107 (35):15409–15414. doi: 10.1073/pnas.1009997107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang K, et al. Viral proteins function as ion channels. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbamem.2010.05.006. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schubert U, et al. Identification of an ion channel activity of the Vpu transmembrane domain and its involvement in the regulation of virus release from HIV-1-infected cells. FEBS Letters. 1996;398 (1):12–18. doi: 10.1016/s0014-5793(96)01146-5. [DOI] [PubMed] [Google Scholar]
- 49.Pontillo A, et al. A 3′UTR SNP in NLRP3 gene is associated with susceptibility to HIV-1 infection. J Acquir Immune Defic Syndr. 2010;54 (3):236–240. doi: 10.1097/QAI.0b013e3181dd17d4. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe S, et al. Influenza A virus lacking M2 protein as a live attenuated vaccine. J Virol. 2009;83 (11):5947–5950. doi: 10.1128/JVI.00450-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dinarello CA. Biologic basis for interleukin-1 in disease. Blood. 1996;87 (6):2095–2147. [PubMed] [Google Scholar]
- 52.Acosta-Rodriguez EV, et al. Interleukins 1[beta] and 6 but not transforming growth factor-[beta] are essential for the differentiation of interleukin 17-producing human T helper cells. Nat Immunol. 2007;8 (9):942–949. doi: 10.1038/ni1496. [DOI] [PubMed] [Google Scholar]
- 53.Ben-Sasson SZ, et al. IL-1 acts directly on CD4 T cells to enhance their antigen-driven expansion and differentiation. Proceedings of the National Academy of Sciences. 2009;106 (17):7119–7124. doi: 10.1073/pnas.0902745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sims JE, Smith DE. The IL-1 family: regulators of immunity. Nat Rev Immunol. 2010;10 (2):89–102. doi: 10.1038/nri2691. [DOI] [PubMed] [Google Scholar]
- 55.French AR, et al. IL-18 acts synergistically with IL-15 in stimulating natural killer cell proliferation. Cytokine. 2006;35 (5–6):229–234. doi: 10.1016/j.cyto.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 56.Takeda K, et al. Defective NK cell activity and Th1 response in IL-18-deficient mice. Immunity. 1998;8 (3):383–390. doi: 10.1016/s1074-7613(00)80543-9. [DOI] [PubMed] [Google Scholar]
- 57.Hyodo Y, et al. IL-18 up-regulates perforin-mediated NK activity without increasing perforin messenger RNA expression by binding to constitutively expressed IL-18 receptor. J Immunol. 1999;162 (3):1662–1668. [PubMed] [Google Scholar]
- 58.Hoshino T, et al. Cutting edge: IL-18-transgenic mice: in vivo evidence of a broad role for IL-18 in modulating immune function. J Immunol. 2001;166 (12):7014–7018. doi: 10.4049/jimmunol.166.12.7014. [DOI] [PubMed] [Google Scholar]
- 59.Okamura H, et al. Cloning of a new cytokine that induces IFN-gamma production by T cells. Nature. 1995;378 (6552):88–91. doi: 10.1038/378088a0. [DOI] [PubMed] [Google Scholar]
- 60.Schmitz N, et al. Interleukin-1 is responsible for acute lung immunopathology but increases survival of respiratory influenza virus infection. J Virol. 2005;79 (10):6441–6448. doi: 10.1128/JVI.79.10.6441-6448.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu B, et al. Interleukin-18 improves the early defence system against influenza virus infection by augmenting natural killer cell-mediated cytotoxicity. J Gen Virol. 2004;85 (2):423–428. doi: 10.1099/vir.0.19596-0. [DOI] [PubMed] [Google Scholar]
- 62.Denton A, et al. IL-18, but not IL-12, is required for optimal cytokine production by influenza virus-specific CD8+ T cells. European Journal of Immunology. 2007;37 (2):368–375. doi: 10.1002/eji.200636766. [DOI] [PubMed] [Google Scholar]
- 63.Rodrigue-Gervais IG, Saleh M. Genetics of inflammasome-associated disorders: A lesson in the guiding principals of inflammasome function. European Journal of Immunology. 2010;40 (3):643–648. doi: 10.1002/eji.200940225. [DOI] [PubMed] [Google Scholar]
- 64.Hise AG, et al. An Essential Role for the NLRP3 Inflammasome in Host Defense against the Human Fungal Pathogen Candida albicans. Cell Host & Microbe. 2009;5 (5):487–497. doi: 10.1016/j.chom.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lev-Sagie A, et al. Polymorphism in a gene coding for the inflammasome component NALP3 and recurrent vulvovaginal candidiasis in women with vulvar vestibulitis syndrome. Am J Obstet Gynecol. 2009;200(3):303, e301–306. doi: 10.1016/j.ajog.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 66.Witkin SS, et al. Genetic polymorphism in an inflammasome component, cervical mycoplasma detection and female infertility in women undergoing in vitro fertilization. J Reprod Immunol. 2010;84 (2):171–175. doi: 10.1016/j.jri.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 67.Saleh M, et al. Differential modulation of endotoxin responsiveness by human caspase-12 polymorphisms. Nature. 2004;429 (6987):75–79. doi: 10.1038/nature02451. [DOI] [PubMed] [Google Scholar]
- 68.Wilkinson RJ, et al. Influence of Polymorphism in the Genes for the Interleukin (IL)-1 Receptor Antagonist and IL-1β on Tuberculosis. The Journal of Experimental Medicine. 1999;189 (12):1863–1874. doi: 10.1084/jem.189.12.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Witkin SS, et al. Influence of interleukin-1 receptor antagonist gene polymorphism on disease. Clin Infect Dis. 2002;34 (2):204–209. doi: 10.1086/338261. [DOI] [PubMed] [Google Scholar]
- 70.Broz P, et al. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. The Journal of Experimental Medicine. 2010;207 (8):1745–1755. doi: 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang L, et al. PYPAF7, a Novel PYRIN-containing Apaf1-like Protein That Regulates Activation of NF-κB and Caspase-1-dependent Cytokine Processing. Journal of Biological Chemistry. 2002;277 (33):29874–29880. doi: 10.1074/jbc.M203915200. [DOI] [PubMed] [Google Scholar]
- 72.Martinon F, et al. The Inflammasome: A Molecular Platform Triggering Activation of Inflammatory Caspases and Processing of proIL-[beta] Molecular Cell. 2002;10 (2):417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]