Abstract
Rationale
MicroRNAs (miRs) are expanding our understanding of cardiac disease and have the potential to transform cardiovascular therapeutics. One miR can target hundreds of individual mRNAs, but existing methodologies are not sufficient to accurately and comprehensively identify these mRNA targets in vivo.
Objective
To develop methods permitting identification of in vivo miR targets in an unbiased manner, using massively parallel sequencing of mouse cardiac transcriptomes in combination with sequencing of mRNA associated with mouse cardiac RNA-induced silencing complexes (RISCs).
Methods and Results
We optimized techniques for expression profiling small amounts of RNA without introducing amplification bias, and applied this to anti-Argonaute 2 immunoprecipitated RISCs (RISC-Seq) from mouse hearts. By comparing RNA-sequencing results of cardiac RISC and transcriptome from the same individual hearts, we defined 1,645 mRNAs consistently targeted to mouse cardiac RISCs. We employed this approach in hearts overexpressing miRs from Myh6 promoter-driven precursors (programmed RISC-Seq) to identify 209 in vivo targets of miR-133a and 81 in vivo targets of miR-499. Consistent with the fact that miR-133a and miR-499 have widely differing ‘seed’ sequences and belong to different miR families, only 6 targets were common to miR-133a- and miR-499-programmed hearts.
Conclusions
RISC-sequencing is a highly sensitive method for general RISC profiling and individual miR target identification in biological context, and is applicable to any tissue and any disease state.
Summary
MicroRNAs (miRs) are key regulators of mRNA translation in health and disease. While bioinformatic predictions suggest that a single miR may target hundreds of mRNAs, the number of experimentally verified targets of miRs is low. To enable comprehensive, unbiased examination of miR targets, we have performed deep RNA sequencing of cardiac transcriptomes in parallel with cardiac RNA-induced silencing complex (RISC)-associated RNAs (the RISCome), called RISC sequencing. We developed methods that did not require cross-linking of RNAs to RISCs or amplification of mRNA prior to sequencing, making it possible to rapidly perform RISC sequencing from intact tissue while avoiding amplification bias. Comparison of RISCome with transcriptome expression defined the degree of RISC enrichment for each mRNA. The majority of the mRNAs enriched in wild-type cardiac RISComes compared to transcriptomes were bioinformatically predicted to be targets of at least 1 of 139 cardiac-expressed miRs. Programming cardiomyocyte RISCs via transgenic overexpression in adult hearts of miR-133a or miR-499, two miRs that contain entirely different ‘seed’ sequences, elicited differing profiles of RISC-targeted mRNAs. Thus, RISC sequencing represents a highly sensitive method for general RISC profiling and individual miR target identification in biological context.
Keywords: RNA-induced silencing complex, RNA-sequencing, mRNA, miRNA, miR-133a, miR-499
INTRODUCTION
MicroRNAs (miRs) are small, non-coding RNAs that regulate critical aspects of cell function including growth, differentiation, and programmed death. These ~22 nucleotide single-stranded oligomers bind through Watson-Crick base-pairing to complementary sequences in mRNA targets, thereby accelerating mRNA degradation or interfering with protein translation. miRs play essential roles is tissue development and homeostasis, and their capacity to act as mediators of disease have been demonstrated in multiple conditions and organ systems 1–3.
The effects of miRs accrue from their binding to specific mRNA targets. Bioinformatics analysis has suggested that 30% of all mRNAs may be regulated by miRs. Because a single miR can bind to dozens or hundreds of different mRNAs, it has the potential to orchestrate complex gene networks and equally complicated phenotypes. For this reason there is tremendous interest in developing miR antagonists and/or mimetics for clinical therapeutics 4. Before the therapeutic potential of miRs can be fulfilled, however, the full spectrum of their specific mRNA targets needs to be determined in the context of the organ system and pathophysiological processes of interest. Achieving this goal has lagged behind experimental miR manipulation and characterization of resulting phenotypes 5. One approach for identifying miR targets compares miR and mRNA expression signatures to detect reciprocally regulated miR-mRNA pairs 6. This approach is limited because only a fraction of miR effects are mediated through mRNA destabilization (the remainder by translational suppression 7) and because indirect miR effects can regulate mRNAs that are not direct targets 8. Bioinformatics have also been used to identify putative mRNA targets based on miR antisense sequence pairing to mRNA 3′untranslated regions, evolutionary conservation, and on the predicted binding energy of miR-mRNA duplexes. However, different bioinformatics platforms differ markedly in their predictions 9, and even if the predictions are accurate, bioinformatics does not provide information about biological context. Thus, techniques are needed for comprehensive, unbiased in vivo identification of miR targets in health and disease.
An approach of isolating mRNAs sequestered by miRs into RNA-induced silencing complexes (RISCs) has been used to profile miR-targeted mRNAs in cultured cells and mouse brains 10,11. While conceptually attractive, this technique has been limited in practice by use of microarrays requiring RNA amplification, which is a potential source of experimental error. Here, we reveal the extent to which RNA amplification biases transcriptional profiling. To avoid this confounder, we describe a technique for annotation and quantitation of mRNAs in the RNA-induced sequencing complexes (the “RISCome”) of mouse hearts using next generation massively parallel sequencing of unamplified mRNAs isolated from myocardial Argonaute 2 immune complexes. By “programming” the cardiomyocytes with excess miR, we then apply the technique to identification of cardiomyocyte-autonomous mRNA targets for two structurally and functionally distinct cardiac-expressed miRs, miR-133a and miR-499. These procedures are applicable to any miR in any tissue and disease state and will complement mRNA footprinting 11 and proteomics analyses 12 to predict in vivo effects of proposed miR-based therapeutics.
METHODS
Preparation and quantification of total myocardial RNA
Cardiac miR-133a transgenic mice were described previously 13. miR-499 transgenic mice were generated similarly by cloning a 530 bp fragment surrounding the mature mouse miR-499 sequence (chromosome 2, intron 19 of Myh7b) into the Myh6 promoter construct. Total cardiac RNA was isolated from flash-frozen ventricular tissue using Trizol (Invitrogen) and quantified on a UV spectrometer.
Reverse transcription and preparation of cDNA
Preparation of cDNA fragments was modified from previously described protocols 8, 14. For poly(A)+ RNA, 4 μg of total cardiac RNA isolated from ventricular apices were twice oligo(dT) selected using the Dynabead mRNA purification system (Invitrogen). Two hundred ng of cardiac mRNA were fragmented to ~200 nt by heating to 94 C for 2.5 min in 40 mmol/L Tris acetate pH 8.2, 100 mmol/L potassium acetate, 30 mmol/L magnesium acetate, and immediately chilled on ice. After purification on Ambion NucAway columns, 100 ng of fragmented cardiac mRNA were reverse-transcribed using SuperScript III (Invitrogen) with random hexamers as per the manufacturer’s directions (50 min, 50 C), followed by second-strand cDNA synthesis for 2 h, 16 C, in 20 mmol/L Tris.HCl pH 6.9, 90 mmol/L KCl, 4.6 mmol/L MgCl2, 150 mmol/L b-NAD+, 10 mmol/L (NH4)2SO4, 0.067 U/μL E. coli DNA ligase, 0.27 U/μL L E. coli DNA polymerase I, and 0.013 U/μL E. coli RNase H (New England Biolabs).
Integrated reverse-transcription and amplification of low-input RNA
RNA amplification prior to fragmentation and sequencing library construction was performed as per Mitreva and Mardis 15. Briefly, polyA-tailed RNA was selected from total RNA using a primer recognizing 5′ mRNA ends and one containing an oligo(dT) moiety. Following reverse transcription, a third primer recognizing common elements in the first two primers was used to amplify cDNAs and to add biotin tags (primer sequences as described in 15). Optimization of amplification cycles was performed as described 15 and biotin tags were removed on streptavidin beads following restriction enzyme digestion.
Immunoprecipitation of mouse cardiac Ago2-associated RNA and preparation of cDNA
Conditions for Ago2 immunoprecipitation were adapted from Karginov et al. 10. Frozen mouse heart bases were homogenized in 500 μL ice-cold 50 mmol/L Tris.HCl, 5 mmol/L EDTA, 5 mmol/L EGTA, pH 7.5, with Roche Complete protease inhibitors. Yeast tRNA (Invitrogen) and SUPERnase-IN (Ambion) were added to final concentrations of 1 mg/mL and 1 U/μL, respectively, and unbroken cellular material was removed at 100g, 5′, 4 C. Nonidet P-40 was added to a final concentration of 0.5% (w/v) to solubilize proteins (15′, 4 C, rotating) and insoluble material was removed at 10000g, 15′, 4 C. The supernatant was added to 50 μL protein G-coupled Dynabeads (Invitrogen), to which 5 μg anti-mouse Ago2 monoclonal antibody (Wako Pure, clone #2D4, lot PEM0820) had been previously bound according to the Dynabead protocol. Following 1 h rotational incubation at 4 C, the beads were washed 3x with Dynabead washing buffer, transferring the suspension to a fresh tube for the last wash. Beads were pelleted from the remaining suspension, the supernatant was removed, and 500 μL Trizol was added to the immunoprecipitated material to extract RNA. Ago2 immunoprecipitate-associated RNA was fragmented in acetate buffer without poly-A+ selection, purified on NucAway columns, and one half was used for cDNA synthesis using the protocol described above.
Construction of DNA-barcoded short-read libraries for Illumina sequencing
Detailed methods for preparation of Illumina sequencing libraries from mouse cardiac RNA were recently described 8. Briefly, cDNAs were end-repaired and 3′ A-overhangs added. Illumina adapters with T-overhangs and customized to include three nt ‘barcodes’ were ligated to the cDNA at 10:1 molar excess and DNA in the 200–400 bp range was isolated via gel purification (Qiagen) on 2% low-melting agarose. One-third of the gel-purified material was amplified with 12 cycles (total myocardial cDNA) or 16 cycles (RISC-immunoprecipitated cDNA) of Phusion polymerase (New England Biolabs #F531)-mediated PCR (10 sec 98 C, 30 sec 65 C, 30 sec 72 C cycles, followed by final 5 min 72 C), using oligonucleotides complementary to Illumina sequencing adapters. The final, amplified libraries were again column-purified and quantified using PicoGreen (Quant-It, Invitrogen).
Four barcoded libraries were combined in equimolar (10 nmol/L) amounts and diluted to 6 pmol/L for cluster formation on a single Illumina Genome Analyzer II flowcell lane, followed by single-end sequencing. Basecalling, library sorting by barcode and mapping to the transcriptome were performed as previously described 8, using updated versions of the software packages Bowtie (release 0.12.3) 16, TopHat (release 1.0.13) 17, and Cufflinks (release 0.8.1) 18. Cufflinks outputs gene expression values in terms of Fragments Per Kb of exon per Million mapped reads (FPKM) 18. When performing single-end Illumina sequencing, as we have done here, this parameter is equivalent to RPKM (Reads Per Kb of exon per Million mapped reads), in which an RPKM of 3 corresponds to 1 copy/cell in cardiac samples 8, 14. We used the default options supplied with these software packages in our analyses and analyzed only those RNA elements that had expression signals in at least 2 of 4 biological replicates.
Computation of RISC abundance and enrichment scores
Because they are not poly-A+ selected, RISCome sequencing libraries contained a higher proportion of ribosomal RNA (32–42%) than poly-A+ selected transcriptome libraries (3–5%). To permit comparison between the transcriptome and RISCome, measures of RNA abundance expressed as Fragments Per Kb of exon per Million mapped reads (FPKM 18) were adjusted for the proportion of mRNA. RISC enrichment scores to assess RISC enrichment or depletion were defined as (RISCome adjusted FPKM/transcriptome adjusted FPKM) ≥ 2.0 or ≤ 0.5, P<0.0001, FDR<0.01. Scores <1 were plotted as −1/score for clarity.
Luciferase reporter constructs
Full (if ≤ 1 kb) or partial (regions selected to contain miR-133a binding ‘seeds’) 3′ untranslated regions of candidate miR-133a target genes were cloned into the dual-luciferase reporter vector psiCheck2 (Promega). The miR-133a genomic precursor was the same as that used to create miR-133a transgenic mice 13. 100 ng of psiCheck2 construct, together with 100 ng of pcDNA3.1 or pcDNA3.1+miR-133a genomic precursor were transfected in triplicate into HEK293 cells on 24-well plates using Fugene HD (Promega). After 48 h, duplicate determinations of Renilla and firefly luciferase activities were performed after harvesting cells into 120 μL Glo Lysis Buffer (Promega). 30 μL of each lysate were analyzed using Dual-Glo Luciferase reagents (Promega) on a SpectraMax M5e platereader (Molecular Devices). Renilla 3′UTR-coupled luciferase activity was normalized to constitutive firefly luciferase activity for each well.
Statistical analysis
Gene symbols and FPKM values were imported into Partek Genomics Suite v6.5 (Partek, St Louis, MO) for comparison of RISCome and transcriptome expression values, computation of P-values and false discovery rates, and preparation of P-value vs fold-change (volcano) plots. GraphPad Prism was used for chi-square tests. Luciferase activities were compared using Student’s t-test (GraphPad Prism). Gene Ontology over-representation was performed using a hypergeometric test with Benjamini & Hochberg false discovery rate correction 19. Significant differences were defined as P<0.05 unless otherwise described.
RESULTS
RNA amplification biases mRNA expression signatures
miR-mRNA pairings in any tissue depend upon at least three factors: The identity and quantity of expressed miRs, the identity and quantity of expressed mRNAs, and the potential for those miRs and mRNAs to interact via sequence complementarity. The former two factors vary with tissue type and pathological circumstance. It is therefore necessary to profile miR-mRNA interactions in the proper biological context.
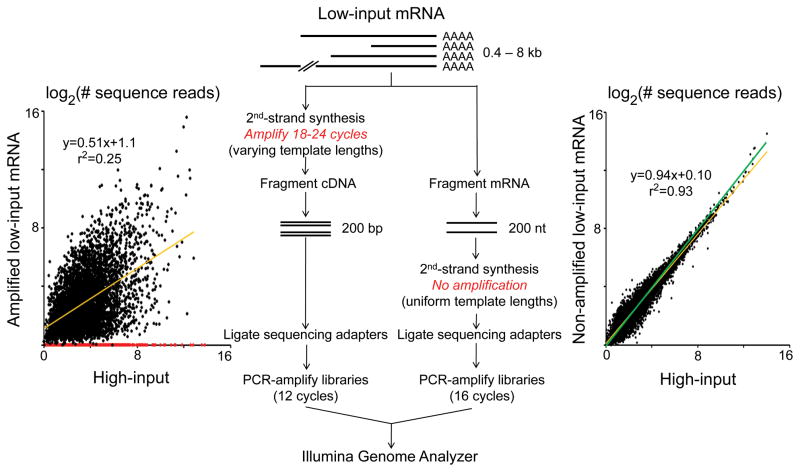
Previous RNA profiling of RISC-associated mRNA used expression microarrays, which require RNA amplification that might alter the apparent transcriptional signature 10, 11. To determine the effect of mRNA amplification on transcript profiling, we compared mRNA signatures of amplified and non-amplified mRNA from the same mouse heart samples. Five μg of unamplified total mouse cardiac RNA underwent two poly-A+ selections to yield 150 ng of mRNA for reverse transcription, sequencing library production, and RNA sequencing on an Illumina Genome Analyzer II 8. In parallel, 50 ng of the same total mouse cardiac RNA (i.e. 1/100th the above initial input amount) were poly-A+ selected, reverse transcribed, and amplified using an integrated multi-stage procedure developed specifically for expression profiling of limited RNA samples, 15 and Illumina sequencing libraries were prepared (Figure 1, left pathway). Sequencing depth was similar in the two samples (9.5×105 mRNA-matching reads in the reference sample vs 8.5×105 in the amplified sample), but whereas 10,501 individual mRNAs were identified from unamplified cardiac RNA, only 5,610 transcripts were detected after RNA amplification. Only 53% of cardiac-expressed mRNAs were represented in sequencing libraries prepared from amplified mRNA (Figure 1 left graph, red data points), and correlation of RNA expression levels between amplified and non-amplified RNAs from the same heart was poor (r2=0.25; Figure 1, left), even compared to RNA sequencing from two separate normal hearts 8 (r2=0.92; Supplemental Figure I). Similar amplification bias was evident when comparing normal and cardiomyopathic hearts (Supplemental Figure II). These results show that RNA amplification is a source of error in mRNA profiling.
Figure 1. Adverse effects of RNA amplification on RNA abundance profiling.
(Center) Schematic diagram outlines methods for RNA sequencing library preparation, with and without RNA amplification. (Left) RNA-Sequencing count data log2(FPKM+1) from the same mouse cardiac mRNA, nonamplified or after amplification. Red dots indicate mRNAs observed in nonamplified but not amplified preparations. (Right) RNA-Sequencing count data from 100 ng (high-input) or 0.5 ng (low-input) polyA+ RNA from the same mouse heart, without amplification. Orange lines are linear regression best fit, green lines are lines of unity.
We considered that small amounts of mRNA might be accurately profiled if the mRNA was fragmented and cDNA sequencing libraries were constructed prior to any amplification (Figure 1, right pathway). To test this, RNA sequencing libraries were prepared from 100 ng or 0.5 ng poly-A+ twice-selected mRNA from the same normal mouse heart. In contrast to amplified mRNA (in which 47% of mRNA transcripts were not represented and the correlation of expression values for represented RNAs was poor), sequencing library preparation from unamplified low-input RNA detected 99.2 % of mRNA transcripts in the high-input library prepared from the same sample (9,336 of 9,564 transcripts) with excellent correlation of RNA expression levels (r2=0.93; Figure 1, right). Thus, next-generation sequencing library production from unamplified RNA generates accurate expression profiles from very small amounts of mRNA.
Defining the cardiac RISCome profiling by deep RNA sequencing of Ago2 complexes
Quantitative analysis of mRNAs sequestered in murine cardiac RISC complexes (the cardiac RISCome) could define the sub-set of cardiac-expressed transcripts targeted by cardiac-expressed miRs. Performing this analysis in vivo is essential to determining how the RISCome is modified in health and disease. As a first step toward using RNA sequencing to define mRNAs recruited into individual mouse cardiac RISComes (a technique that we call RISC-Seq), we developed procedures for isolating the RISC complexes from mouse hearts, capturing the associated mRNAs, and processing the RISC-mRNA libraries for sequencing.
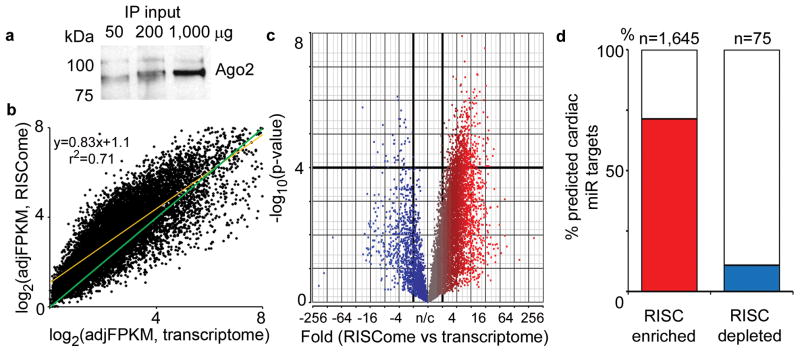
The RISC consists of Argonaute (Ago) 1 and 2 proteins, TNRC6 family members, miRs, and their mRNA targets 20. After identifying an antibody that immunoprecipitates endogenous Ago2 from mouse hearts (Figure 2a), we determined if endogenous cardiac Ago2 immunoprecipitates contained sufficient RISC-associated cardiac mRNA for RNA sequence analysis without sample cross-linking, amplification, or pooling previously used with Ago2 pull-down 10, 11. Ventricular tissue from four 8-week old male FVB/N mouse hearts was transversely sectioned; one third of each heart was used for mRNA sequencing to characterize the overall cardiac transcriptomes, and the remaining two thirds were used for Ago2 immunoprecipitation, RNA extraction, and RISCome RNA sequencing analysis. Although there was 96.5% overlap in the individual mRNAs represented in the cardiac transcriptome and RISCome (9,779 ± 894 transcriptome mRNAs and 10,690 ± 205 RISCome mRNAs; P = 0.31), 1,645 individual mRNAs (~15% of the cardiac transcriptome) were significantly RISC-enriched in the Ago2 immunoprecipitates (Figure 2b). RISC enrichment or depletion was defined as (RISCome expression/transcriptome expression) ≥ 2.0 or ≤ 0.5, P<0.0001, FDR<0.01. The enriched transcripts define the subset of cardiac-expressed mRNAs that are targets of the cardiac-expressed miRs, i.e. the cardiac RISCome (Figure 2c, upper right quadrant; Supplemental Table I).
Figure 2. RISC immunoprecipitation and RISC-Sequencing of normal mouse hearts.
(a) Ago-2 immunoblot of Ago2 immunoprecipitates from 50, 200 and 1000 mg mouse cardiac homogenate. (b) mRNA abundance (log2 adjusted FPKM) in mouse cardiac RISCome and transcriptome. Orange line is best fit linear regression; green is line of unity. (c) Volcano plot (plot of fold-change in expression value vs P-value) for mouse heart RISCome/transcriptome (n/c = no change). Red indicates RISC-enrichment (significant in upper right quadrant); blue indicates RISC-depletion. (d) Proportion of mouse heart RISC-enriched mRNAs (n=1,645) and RISC-depleted mRNAs (n=75) predicted to be targets for any of 139 cardiac-expressed miRs. Red and blue bars indicate mRNAs from the upper right and upper left quadrants of (c), respectively.
Published studies have identified 139 consensus cardiac-expressed miRs (Supplemental Table II) 4, 21. Since the mouse cardiac RISCome should consist of mRNAs that interact with these (and any other) cardiac-expressed miRs, we compared our results with RISC-Seq to the targets predicted for these 139 cardiac-expressed miRs by the TargetScan algorithm 12, 22. 1,171 of 1,645 RISC-enriched mRNAs (71.2%) were predicted targets of one or more cardiac-expressed miRs (Figure 2d), compared to only 45.7% of all cardiac-expressed mRNAs. By contrast, only 8 of 75 RISC-depleted mRNAs (10.7%) are predicted cardiac miR targets (Figure 2d; P=0.0003, Fisher’s exact test). Thus, our RISC-Seq results are supported by bioinformatic predictions based on established cardiac miR expression signatures.
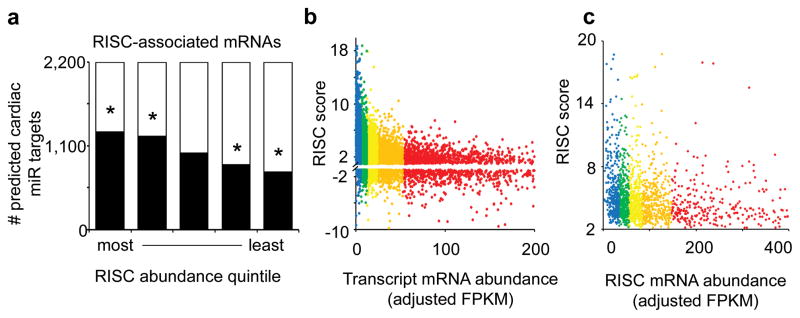
RISC enrichment scoring ranks transcripts by abundance in RISCome relative to transcriptome (simply: [RISC-mRNA]/[transcript mRNA] for each individual mRNA). Thus, reciprocal regulation of mRNAs in the RISCome and transcriptome, as expected with miR-induced mRNA destabilization, produces a higher score. Because this ranking method does not account for absolute mRNA expression, a transcript expressed at 1 copy per cell and enriched two-fold in the RISC is scored the same as a transcript having 50 copies per cell with similar RISC enrichment. To determine if the absolute number of mRNA copies in the RISCome provided critical information about miR targets, we ranked the RISC-associated transcripts according to their absolute abundance, and determined the proportion of TargetScan-predicted cardiac miR targets by quintile. RISC-associated mRNA abundance and the likelihood of being a predicted cardiac miR target were associated (P<0.01, χ2 test; Figure 3a), demonstrating that absolute RISC mRNA abundance is a factor in miR targeting. Surprisingly however, the association between RISC enrichment score and either transcriptome mRNA abundance (Figure 3b) or RISC-associated mRNA abundance (Figure 3c) is inverted, revealing that the most RISC-enriched targets tend to be the rarer mRNA transcripts. This finding is consistent with previous observations that most abundant cardiac mRNAs, encoding myofilament and other constitutively expressed proteins, tend to undergo the least regulation 8, 23. These results indicate that the analytical approach of relating RISC mRNA abundance to transcript mRNA abundance, i.e. RISC enrichment, is superior to just identifying mRNAs in the RISCome.
Figure 3. Comparison of RISC score and RISC abundance for miR target prediction.
(a) Number of RISC-associated mRNAs predicted to be targets for any of 139 cardiac-expressed miRs in each RISCome abundance quintile. * = P<0.01 vs same transcriptome abundance quintile by χ2 test. (b) RISC mRNA enrichment score versus transcript mRNA abundance (FPKM) vs, for the cardiac transcriptome (only transcripts with FPKM to 200 are shown). Colors indicate different transcript abundance quintiles. (c) RISC mRNA enrichment score versus RISCome mRNA abundance for 1,645 significantly RISC-enriched transcripts) vs RISC enrichment score (only transcripts with FPKM to 400 are shown). Colors indicate different RISC-enriched mRNA abundance quintiles.
Programming of the cardiac RISCome to identify individual miR targets
A thorough understanding of the pathological roles of dysregulated miRs requires delineating individual in vivo miR-mRNA functional pairs, which has been problematic using bioinformatics predictions and tissue culture experimentation 13. We considered that the cardiomyocyte RISC could be programmed to retain specific miR-targeted mRNAs 10 by cardiomyocyte-specific overexpression of a miR precursor. RISC-Seq comparison of miR-programmed and un-programmed cardiac RISComes would then identify mRNAs targeted by the programming miR. We tested this notion using RISC-Seq of our miR-133a transgenic mice 13. miR-133a is a muscle-specific miR essential for cardiac development that is dysregulated in cardiac disease 24–27. Importantly, miR-133a transgenic mice have no basal cardiac phenotype to confound miR target analysis by dysregulating cardiac gene expression.
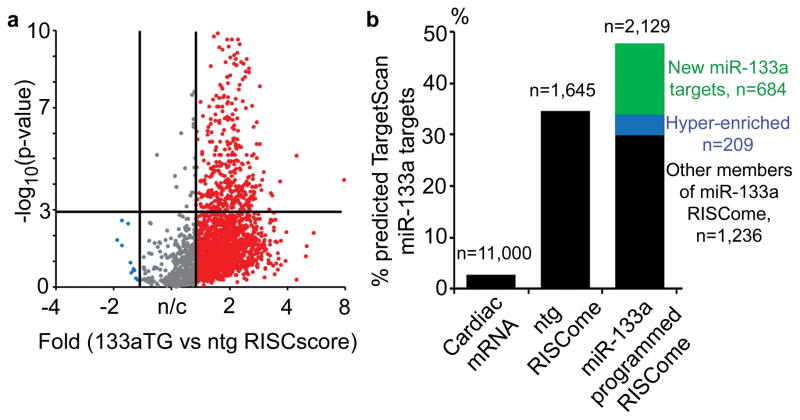
RNA sequencing of the miR-133a programmed transcriptome and RISCome was performed as described above. Sequencing read depth (1.5 to 2.0×106 mRNA-matching reads per barcoded sample), alignment to the mouse genome, and the identities of detected transcripts were similar for all 16 libraries (one RNA-seq and one RISC-seq library from each of four nontransgenic and four miR-133a transgenic mice). RISC scoring identified 2,149 RISC-enriched mRNAs in miR-133a programmed hearts, 684 of which had not met criteria for being RISC-enriched in nontransgenic hearts (“new miR-133a targets”), and 209 of which were enriched to a significantly greater extent (>1.3 fold greater RISC score, P<0.001, χ2 test) in the miR-133a cardiac RISCome (“hyper-enriched miR-133a targets”) (Figure 4a and Supplemental Table III). TargetScan reports only 435 predicted mouse miR-133a targets among 17,315 total mouse transcripts in the database: 310 of these mRNAs are among the ~10,000 mRNAs present in the cardiac transcriptome (~2.8% of all cardiac-expressed mRNAs). The 1,645 mRNAs enriched in the normal cardiac RISCome contain 107 (34.5%) of the 310 TargetScan predicted cardiac miR-133a targets, consistent with miR-133a being one of the more highly expressed cardiac miRs 6, 25. Finally, 147 (47.4%) of the 310 predicted targets were present in miR-133a programmed transcriptomes (Figure 4b). Thus, bioinformatics analysis shows a progressive increase in the proportion of predicted miR-133a targets from the cardiac transcriptome to the cardiac RISCome, and to the miR-133a programmed cardiac RISCome.
Figure 4. RISC-Sequencing of miR-133a transgenic mouse hearts.
(a) Volcano plot of RISC score differences and associated p-values between miR-133a transgenic and normal hearts. Red is hyper-enriched, blue is depleted; grey indicates less than 1.3-fold change. Upper right quadrant includes genes with RISC scores ≥ 1.3-fold higher in miR-133a heart, P<0.001 (χ2 test). (b) Proportion of TargetScan-predicted miR-133a targets in the cardiac transcriptome, nontransgenic RISCome, and miR-133a transgenic RISCome.
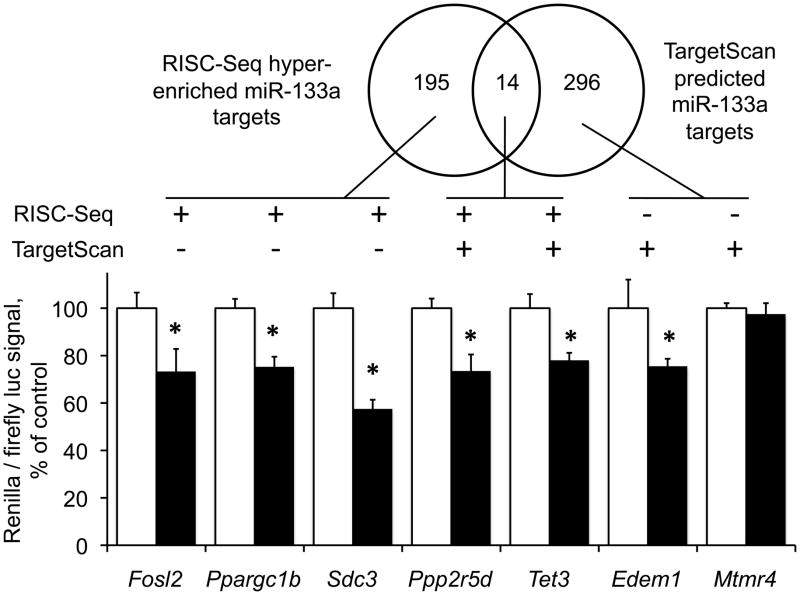
Enrichment of miR-133a targets in programmed cardiac RISCs and the presence of miR-133a programmed RISC mRNAs previously shown to be miR-133a targets (Whsc2 25, Hcn4 28, Ccnd2 27, Casp9 29, and Ctgf 28; only Whsc2 and Ctgf predicted by TargetScan) validated our method of individual miR target analysis by programmed RISC-Seq. However, a number of miR-133a programmed RISC enriched mRNAs identified experimentally were not predicted by TargetScan. To determine the reason for this discrepancy, we characterized miR-133a effects on 7 miR-133a programmed RISC-associated mRNAs using standard luciferase reporter assays 3, 10. All five miR-133a RISC-enriched mRNA 3′ UTRs were significantly suppressed by miR-133a (Figure 5), including three (Fosl2, Ppargc1b, and Sdc3) that were not identified by TargetScan as miR-133a targets. Only one of the two TargetScan-predicted/RISC-Seq not predicted miR-133a targets was suppressed (Edem1, which was indeed increased 2.5-fold in miR-133a programmed RISComes, but with a p-value [0.016] that did not meet our selection criteria) (Figure 5). Based on these results and the five previously validated miR-133a targets, RISC-Seq exhibits a sensitivity of 91% for target identification, versus 45% for TargetScan.
Figure 5. Luciferase reporter assays for putative miR-133a target 3′UTRs.
(Top) 3′ UTRs of putative miR-133a target genes were cloned into a dual Renilla/firefly luciferase reporter vector and co-transfected with miR-133a precursor expression plasmid. (Bottom) Luciferase signals for miR-133a (black bars) as percentage of the Renilla/firefly ratio observed with co-transfection empty expression plasmid (white bars). * = P<0.05 vs control by Student’s t-test.
RISC-Seq comparison of miR-133a and miR-499 programmed cardiac RISComes
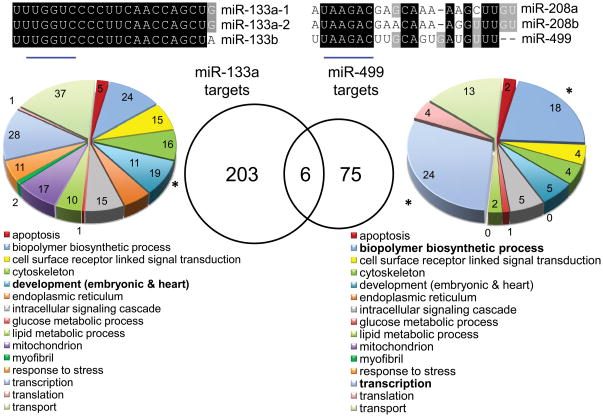
The true applicability of any research technique is its ability to answer a real-world question. Accordingly, we used RISC-Seq to compare in vivo mRNA targeting of two cardiac-expressed, but functionally divergent miRs. miR-133a and miR-499 are both muscle-specific miRs that are expressed at high levels in the heart and are regulated in cardiac disease 30. Importantly, they belong to different miR-subfamilies (Figure 6a). The distinct functions and lack of sequence homology of these two miRs suggest that they target different mRNAs. Indeed, the population of hyper-enriched mRNAs in miR-133a programmed RISComes has little in common with that from miR-499 transgenic mouse hearts (Figure 6b, Supplemental Table IV); gene-ontology classification 19 of the miR-133a and miR-499 programmed RISComes showed significant over-representation (see Methods) of cardiac development genes for miR-133a, and myofibrillar transcriptional control (biopolymer biosynthesis) genes for miR-499. Thus, the results of RISC-Seq are consistent with the previously established biological effects of these two functionally divergent cardiac-expressed miRs 27, 31.
Figure 6. Gene Ontology (GO) analysis of miR-133a and miR-499 RISCome hyper-enriched mRNAs.
(a) Sequence homology of members of the miR-133 and miR-208/499 families. (b) Venn diagram of hyper-enriched mRNAs in mir-133a (left) and mir-499 (right) programmed cardiac RISComes with respective GO categories. * = over-represented category compared to distribution of all genes across the transcriptome (see Methods).
DISCUSSION
Here, we describe RISC-Seq, a method that marries RNA-sequencing to Ago2 immunoprecipitation for in vivo RISCome profiling and identification of specific miR targets. Our method avoids experimental bias introduced by RNA amplification and takes advantage of superior mRNA profiling results from RNA sequencing, compared to expression arrays 8. Because our technique works with endogenous Ago2 protein and does not require crosslinking or sample pooling, it is broadly applicable to miR target analysis in genetic- and physiologically-modeled in vivo models.
The cardiac system is especially well suited for analyis of endogenous miR targets because miR effects are central to embryonic heart formation, reactive cardiac hypertrophy, excitation-contraction coupling, and programmed cardiomyocyte death 4. miR expression is dynamically regulated in heart disease 6, and so miRs are being evaluated as diagnostic tools and therapeutic targets 6, 32, 33. The complement of individual cardiac-expressed mRNAs that are potentially regulated by cardiac-expressed miRs (i.e. the cardiac RISCome) was not previously known. Here, RISC-Seq defined the normal cardiac RISCome consisting of ~2,000 of ~10,000 cardiac-expressed mRNAs. Thus, approximately 20% of cardiac mRNAs are regulated by miRs under normal conditions. RISC-Seq should be equally useful to define RISC-associated mRNAs in pathological conditions, such as cardiac hypertrophy or heart failure, where the miRome and transcriptome are jointly dysregulated.
Our studies show that absolute RISC-associated mRNA abundance is inferior to RISC enrichment in identifying miR targets. Loosely-associated, highly abundant transcriptome mRNAs are unavoidably captured during Ago2 immunoprecipitation, explaining the nearly complete representation of cardiac-expressed mRNAs in the RISC complexes. We controlled for non-specific representation of highly-expressed transcripts in Ago2 immunoprecipitates by relating their abundance in the RISCome to that in the transcriptome, setting a threshold level of 2-fold enrichment. We found that lower abundance mRNAs were disproportionately recruited to the RISCome, consistent with recent results demonstrating that the most abundant mRNA transcripts are also subjected to less transcriptional regulation in disease 8, 23.
To attack the question of individual miR targeting in the heart, we used cardiomyocyte-specific transgenic expression of miR-133a and miR-499 precursors to “program” the cardiomyocyte RISCome 10. These two well-studied miRs have central roles in cardiac development and myofilament identity, respectively 30, and are regulated in cardiac disease to approximately the levels at which they were overexpressed. In both instances we identified mRNA targets that were not predicted by TargetScan. These findings demonstrate a significant impact of other determinants of miR-mRNA interactions on RISCome recruitment, suggesting roles for non-seed sequence complementation, the number of complementary miR sequences in a given transcript, and the cumulative effects of other miRs on the same transcript.
Programming cardiomyocytes with transgenic miRs was used to hyper-enrich the RISCome with strong miR targets (which are already present in the RISCome due to effects of endogenous miRs), and to identify weaker miR targets (new RISCome mRNAs). We took advantage of the Myh6 promoter to drive miR expression so that miR content, and RISCome mRNA profiles, were selectively altered in cardiac myocytes of the heart. Although non-myocytes comprise ~30% of ventricular mass, only the transcriptomes and RISComes of cardiomyocytes are affected by our genetic manipulations. Thus, the comparison between non-transgenic and transgenic RISC-Seq subtracts non-myocyte information.
The RISCome programming approach raises the possibility of antithetic experiments to “de-program” the RISCome, i.e. RISC-Seq of miR knockout models. In the case of miR-133, the severe pathology resulting from miR-133 ablation (embryonic lethality or aggressive cardiomyopathy 27) precludes a meaningful analysis; secondary effects of altered miR and mRNA expression in the diseased hearts would obfuscate any primary change in RISCome mRNA content caused by absent miR-133. Furthermore, we think that RISC de-programming is likely to be of limited utility even in the absence of a confounding phenotype because legitimate mRNA targets of the absent miR will continue to be recruited to the RISC by miR family members having similar sequences (see Figure 6).
In addition to providing comprehensive and unbiased analysis of RISComes in pathophysiological context, we believe RISC-Seq in tissues programmed with endogenous miRs will be useful to identify unsuspected mRNA targets having imperfect seed sequence complementarity or extra-seed sequence binding, which are generally overlooked by bioinformatics. mRNA targets with perfect seed sequence complementation to a highly expressed miR are likely to already be present in the cardiac RISCome, and may be less enriched by the presence of more miR than weaker targets. We observed this with Srf, an experimentally validated miR-133a target 27 that is 2.4-fold RISC-enriched in miR-133a programmed hearts, but did not meet our criteria for statistical significance (P<0.0001), and with Sox6, a validated 31 miR-499 target that is already 5.1-fold RISC-enriched in nontransgenic hearts and only increases a further 1.2-fold (below our threshold for 1.3-fold change) in miR-499 programmed hearts. Thus, RISC programming with endogenously expressed miRs may have the greatest proportional impact on the less obvious, and most difficult to define, mRNA targets. Perhaps in combination with RNA footprinting to define miR-mRNA binding domains 11 and proteomics studies to elucidate the biology of miR effects 12, RISC-Seq will be especially useful to define the in vivo targets of synthetic “designer” miRs and miR-mimetics undergoing evaluation as therapeutics.
Novelty and Significance.
What is known?
MicroRNAs (miRs) are important regulators of gene expression, play roles in cardiovascular development and disease and may serve as novel cardiovascular therapeutics.
While a single miR can potentially suppress hundreds of mRNAs, comprehensive identification of in vivo targets is lacking.
What new information does this article contribute?
Methods were developed for deep sequencing of mRNAs targeted by miRs into RNA-induced silencing complexes (RISCs). The method avoids bias introduced by RNA amplification, cross-linking of RNA to RISCs, and use of expression arrays.
Identification of mRNAs enriched in wild-type mouse cardiac RISCs was performed by comparing expression levels in the RISCome (RISC-associated RNAs) to that in the transcriptome (poly(A+)-selected RNA).
Programming cardiomyocyte RISCs via transgenic overexpression of miRs reveals specific profiles of RISC-enriched mRNAs in intact mouse hearts, and thus the in vivo targets of miRs in cardiomyocytes.
Supplementary Material
Acknowledgments
SOURCES OF FUNDING
Supported by NIH RC2 HL102222, R01 HL59888, and UL1 RR024992.
NON-STANDARD ABBREVIATIONS AND ACRONYMS
- Ago2
Argonaute 2
- FPKM
fragments per kb of exon per million mapped reads
- RISC
RNA-induced silencing complex
- 3′ UTR
3′ untranslated region
Footnotes
DISCLOSURES
None.
References
- 1.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 2.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, Olson EN. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Latronico MV, Condorelli G. MicroRNAs and cardiac pathology. Nat Rev Cardiol. 2009;6:419–429. doi: 10.1038/nrcardio.2009.56. [DOI] [PubMed] [Google Scholar]
- 5.Abdellatif M. The role of microRNA-133 in cardiac hypertrophy uncovered. Circ Res. 2010;106:16–18. doi: 10.1161/CIRCRESAHA.109.212183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matkovich SJ, Van Booven DJ, Youker KA, Torre-Amione G, Diwan A, Eschenbacher WH, Dorn LE, Watson MA, Margulies KB, Dorn GW., 2nd Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation. 2009;119:1263–1271. doi: 10.1161/CIRCULATIONAHA.108.813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 8.Matkovich SJ, Zhang Y, Van Booven DJ, Dorn GW., 2nd Deep mRNA sequencing for in vivo functional analysis of cardiac transcriptional regulators: application to Gaq. Circ Res. 2010;106:1459–1467. doi: 10.1161/CIRCRESAHA.110.217513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Verbeek FJ. Comparison and integration of target prediction algorithms for microRNA studies. J Integr Bioinf. 2010;7 doi: 10.2390/biecoll-jib-2010–2127. [DOI] [PubMed] [Google Scholar]
- 10.Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci USA. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matkovich SJ, Wang W, Tu Y, Eschenbacher WH, Dorn LE, Condorelli G, Diwan A, Nerbonne JM, Dorn GW., 2nd MicroRNA-133a protects against myocardial fibrosis and modulates electrical repolarization without affecting hypertrophy in pressure-overloaded adult hearts. Circ Res. 2010;106:166–175. doi: 10.1161/CIRCRESAHA.109.202176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Meth. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 15.Mitreva M, Mardis ER. Large-scale sequencing and analytical processing of ESTs. Methods Mol Biol. 2009;533:153–187. doi: 10.1007/978-1-60327-136-3_8. [DOI] [PubMed] [Google Scholar]
- 16.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and abundance estimation from RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 20.Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Condorelli G, Latronico MV, Dorn GW., 2nd microRNAs in heart disease: putative novel therapeutic targets? Eur Heart J. 2010;31:649–658. doi: 10.1093/eurheartj/ehp573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman RC, Farh KK, Burge CB, Bartel D. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim JB, Porreca GJ, Song L, Greenway SC, Gorham JM, Church GM, Seidman CE, Seidman JG. Polony multiplex analysis of gene expression (PMAGE) in mouse hypertrophic cardiomyopathy. Science. 2007;316:1481–1484. doi: 10.1126/science.1137325. [DOI] [PubMed] [Google Scholar]
- 24.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MV, Hoydal M, Autore C, Russo MA, Dorn GW, 2nd, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 26.Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Path. 2007;170:1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van dMI, Herias V, van Leeuwen RE, Schellings MW, Barenbrug P, Maessen JG, Heymans S, Pinto YM, Creemers EE. miR–133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res. 2009;104:170–178. doi: 10.1161/CIRCRESAHA.108.182535. [DOI] [PubMed] [Google Scholar]
- 29.Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, Xiao J, Shan H, Wang Z, Yang B. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120:3045–3052. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- 30.Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18:510–525. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, Kok WE, Pinto YM. MiR423-5p as a circulating biomarker for heart failure. Circ Res. 2010;106:1035–1039. doi: 10.1161/CIRCRESAHA.110.218297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.