Abstract
We report here the complete genome sequence of the squirrel monkey α-herpesvirus saimiriine herpesvirus 1 (HVS1). Unlike the simplexviruses of other primate species, only the unique short region of the HVS1 genome is bounded by inverted repeats. While all Old World simian simplexviruses characterized to date lack the herpes simplex virus RL1 (γ34.5) gene, HVS1 has an RL1 gene. HVS1 lacks several genes that are present in other primate simplexviruses (US8.5, US10–12, UL43/43.5 and UL49A). Although the overall genome structure appears more like that of varicelloviruses, the encoded HVS1 proteins are most closely related to homologous proteins of the primate simplexviruses. Phylogenetic analyses confirm that HVS1 is a simplexvirus. Limited comparison of two HVS1 strains revealed a very low degree of sequence variation more typical of varicelloviruses. HVS1 is thus unique among the primate α-herpesviruses in that its genome has properties of both simplexviruses and varicelloviruses.
Keywords: Herpesvirus, genome structure, squirrel monkey, DNA sequence
Introduction
Herpesvirus saimiri 1 (Saimiriine herpesvirus 1; HVS1) is an α-herpesvirus related to the human herpes simplex viruses (HSV-1, HSV-2). HVS1 was originally isolated from tamarins (Saguinus spp.) that succumbed to lethal generalized disease, hence its original designation as Herpesvirus tamarinus or Herpes T (Holmes et al., 1964; Melnick et al., 1964). Owl monkeys (Aotus trivirgatus) were found to be similarly susceptible to lethal generalized infection by HVS1 (Burkholder and Soave, 1970; Emmons et al., 1968; Leib et al., 1987a) but subsequent studies demonstrated that squirrel monkeys (Saimiri spp.) were the natural host for this virus (Daniel et al., 1967; Emmons et al., 1968; Holmes et al., 1966; King et al., 1967). While there have been several suspected cases, there have been no confirmed cases of human HVS1 infection.
Very little molecular work has been done with HVS1. HVS1 is most closely related to a spider monkey herpesvirus (Ateline herpesvirus 1; HVA1), but also shows limited antigenic cross-reactivity with α-herpesviruses of Old World monkeys as well as the human HSVs (Desrosiers and Falk, 1981; Eberle et al., 1989; Hilliard et al., 1989; Mou et al., 1986). While the HVS1 UL27 (glycoprotein gB) and UL23 [thymidine kinase (TK)] genes have been sequenced (Eberle and Black, 1993; Otsuka and Kit, 1984), the structure of the HVS1 genome has not been resolved. Restriction analysis of the viral genome has shown that HVS1 has both unique long (UL) and unique short (US) regions, but whether or not the UL and US regions are both bounded by repeats and invert during replication as in the other primate α-herpesviruses has not been resolved (Desrosiers and Falk, 1981; Leib et al., 1987a).
The evolution of herpesviruses is a topic of considerable interest in that these viruses appear for the most part to have co-evolved with their hosts (McGeoch and Cook, 1994). In phylogenetic analyses based on coding sequences, the α-herpesviruses of primates [excluding human varicella zoster virus (VZV) and simian varicella virus (SVV)] form a phylogenetic clade (the simplexviruses) separate from most other mammalian α-herpesviruses including VZV and SVV (the varicelloviruses). Within the simplexvirus group, the viruses of humans, Old World monkeys, and New World monkeys each occupy a separate branch (Eberle and Black, 1993; McGeoch et al., 2000, 2006). The complete genome sequences of a number of primate simplexviruses (human, macaque, vervet, and baboon) have been determined (Dolan et al., 1998; McGeoch et al., 1985, 1986, 1988; Perelygina et al., 2003; Tyler et al., 2005; Tyler and Severini, 2006; Szpara et al., 2010), allowing characteristics of the structural layout of these genomes to be compared. Such analyses have identified many conserved features and genes, but also a few features in which these viruses vary. The genome structure of HVS1 has not been determined, but is of interest as a virus of South American primates representing a phylogenetic clade distinct from the α-herpesviruses of humans and cercopithecine monkeys that have been sequenced.
As previously noted (Desrosiers and Falk, 1981), HVS1 is biologically similar to other primate α-herpesviruses that are members of the simplexvirus subgroup. In a serological survey of several hundred squirrel monkeys in a captive breeding colony, it was found that >90% of adults had antibody to HVS1 (unpublished data). Monkeys undergoing primary infections can develop ulcerative oral lesions, and HVS1 has been isolated from tongue lesions, throat swabs, trigeminal ganglia, and brain tissue of infected squirrel monkeys (Daniel et al., 1967; King et al., 1967; unpublished results). Owl monkeys, tamarins, and marmosets all succumb rapidly to HVS1 infection, and lesions are histologically indistinguishable from those produced by HSV-1 (Holmes et al., 1964). Rabbits infected with HVS1 by intradermal inoculation develop latent infections in dorsal root ganglia serving the site of inoculation as does HSV-1 (Leib et al., 1988; McCarthy and Tosolini, 1975). HVS1 also displays aggressive invasion and destruction of the central nervous system in mice after inoculation by skin scarification (Breshears et al., 2001, 2005). Despite these similarities to simplexviruses, some studies suggest that HVS1 may have a genome structure like that of varicelloviruses (which includes most mammalian α-herpesviruses of non-primate species) (Desrosiers and Falk, 1981). Also, it has been noted by several investigators that restriction digests of viral DNA reveals remarkably similar patterns for different HVS1 isolates (Desrosiers and Falk, 1981; Leib et al., 1987a, 1987b), reminiscent of the low genomic sequence variation characteristic of VZV (Peters et al., 2006; Tyler et al., 2007).
In this communication we report the complete genome sequence of HVS1. The results reveal that while in many respects the HVS1 genome is similar to those of other primate simplexviruses in its genetic organization, HVS1 also exhibits a number of properties that are characteristic of varicellovirus genomes.
Materials and Methods
Viral culture and nucleic acid purification
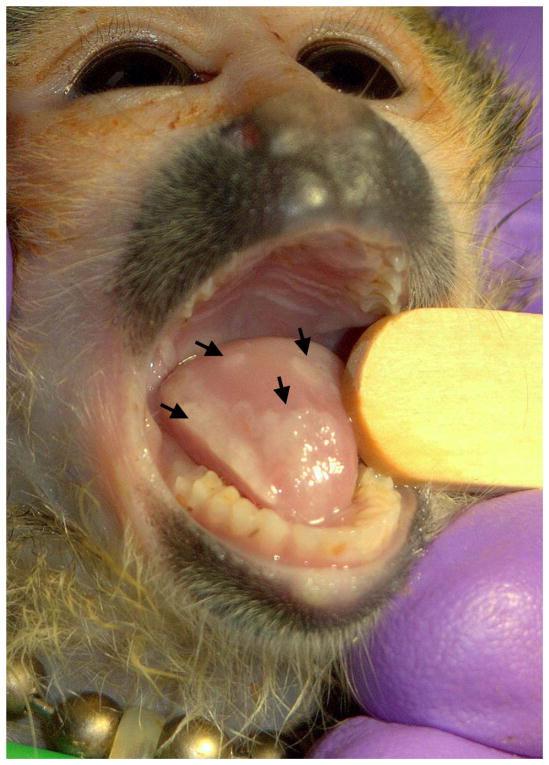
HVS1 strain MV-5-4 was originally isolated from a tamarin (Sanguinus spp.) (Holmes et al., 1964) and was obtained from the American Type Culture Collection (ATCC; Manassas, VA). HVS1 strain 4672 was isolated from an ulcerative lesion on the tongue of a young squirrel monkey (Saimiri sciurens; Figure 1) housed at the Squirrel Monkey Breeding and Research Resource at the University of South Alabama, Mobile, AL (currently located at the Michale E. Keeling Center for Comparative Medicine and Research at the University of Texas M. D. Anderson Cancer Center, Bastrop, TX). Virus stocks were prepared and titers determined in Vero cells as described (Mou et al., 1986).
Figure 1.
HVS1 lesions on the tongue of a squirrel monkey. Shown are multiple consolidated lesions on the tongue of a juvenile squirrel monkey. Multiple juvenile monkeys in the colony developed similar oral lesions over a short time period, and HVS1 strain 4672 was isolated from an oral swab collected during this outbreak.
Viral DNA was purified from infected Vero cells as described (Hilliard et al., 1989). Briefly, 300 cm2 of confluent Vero cells were infected at an MOI of approximately 0.01 PFU/cell and incubated at 37°C until the entire monolayer exhibited CPE. Infected cells were pelleted from the media by centrifugation at 300 x g for 5 min and frozen at −80°C. Infected cell pellets were thawed, resuspended in 4 ml digestion buffer [10 mM Tris (pH 8.0), 0.5% SDS, 5 mM EDTA, 40 μg/ml proteinase K] and incubated approximately 20 hrs at 37°C. EtBr was added to a final concentration of 0.22 μg/ml, 6.0 ml NaI saturated 10 mM Tris (pH 8.0) added, and centrifuged at 132,000 x g for 68 hrs. The cellular DNA band was removed by side puncture of the centrifuge tube before the viral DNA band was removed. EtBr was extracted with chloroform and DNA dialyzed for 24 hrs against 10 mM Tris/ 0.1 mM EDTA at 4°C before being precipitated. Viral DNA was resuspended in 0.1 mM Tris and the concentration determined by spectrophotometric analysis.
Total RNA was prepared from infected Vero cells for transcript analysis. Vero cells were infected at an MOI = 5 PFU/cell and harvested by trypsinization into ice cold PBS at 3 or 8 hrs PI. Cells were pelleted at 300 x g for 5 min, resuspended in 50 μl DEPC-treated water, and 450 μl RNAlater (QIAGEN) added. Samples were stored at −80°C until RNA was extracted using the RNAeasy Mini Kit (Qiagen).
Sequencing
Initial sequencing was performed on a Genome Sequencer 20 (Roche) using the manufacturer’s Shotgun Sequencing protocol. Sequencing was performed in a ¼ region of a picotiter plate to an approximate depth of coverage of 40x and the data assembled with the GS De Novo Assembler (Roche). A number of the resulting contigs were found to be derived from the cell line used to culture the virus or otherwise too small to be of practical use, and a total of 43 contigs were selected as the foundation for the final assembly. The majority of the gaps were subsequently found to be small (often less than 20 bp) with the exception of the RL/RS regions which was highly fragmented and underrepresented in the GS20 sequencing data. In order to facilitate gap closure, a conventional plasmid based shotgun library was also prepared using the TOPO Shotgun Subcloning Kit (Invitrogen). Plasmid DNA was extracted using the Wizard SV or Wizard SV96 kits (Promega) and end sequenced in order to orient them on the genome. Clones of interest were then sequenced with custom primers in order to close sequence gaps. Gaps in regions not covered by spanning clones were closed using conventional PCR with the aid of the FailSafe PCR System (Epicentre). The resulting amplicons were gel purified or purified directly using Microcon PCR filter units (Millipore) and sequenced. Conventional sequencing was performed on an ABI3730XL using BigDye 3.1 chemistry or a 3:1 mix of BigDye 3.1 and dGTP chemistry supplemented with SequenceRx Enhancer Solution A (Invitrogen). The combined data were assembled and edited using the Staden Package (Staden et al., 1999).
The sequences of the genomic termini were determined using a modification of the Rapid Amplification of cDNA Ends (RACE) technique (Davison et al., 2003). In brief, genomic viral DNA was blunt ended using the End-It kit (Epicentre) after which adaptors were ligated on using the Marathon cDNA amplification Kit (Clontech). PCR was then performed using a primer targeting the adaptor and one near the suspected 5′ terminus (nt 180–161; AACTGGCACTGAGCCAAGTG) or one near the suspected 3′ terminus (nt 156244–156263; CCTAGCTGGCGCTCGGCCAA). The resulting amplicons were purified as described above and sequenced.
Transcript verification
The open reading frame (ORF) for a number of genes could not be identified/confirmed based solely on similarity to known genes of other primate simplexviruses. In order to confirm that predicted transcripts from these regions were produced, reverse transcriptase PCR (RT-PCR) was performed on the targeted region using the One-Step RT-PCR kit (Qiagen). RNA was first treated with DNase to ensure a positive result was not due to residual DNA contamination. All reactions were performed in duplicate with one reaction having the RT portion of the thermocycling profile omitted (reactions were subjected to the 95°C denaturing step for 15 min prior to the addition of template to inactivate the reverse transcriptase). This control reaction was done to again ensure that no DNA contamination was present. If the existence of a transcript was confirmed, the resulting amplicon was sequence-verified and RT-PCR was continued until the full transcript sequence had been obtained. These data were supplemented with RACE data using the Marathon cDNA Amplification kit (Clontech) in order to obtain the transcript ends.
Inversion of UL region
Restriction analysis of HVS1 genomic DNA was performed using standard procedures. Briefly, DNA (1 μg) was digested for 3 hrs and restriction fragments separated on a 0.5% agarose gel run overnight at 35 volts. Gels were stained with EtBr, and viewed and photographed under UV light. Restriction fragment sizes for the MV-5-4 strain of HVS1 were predicted using Informax Vector NTI software.
Sequence Analysis
Sequence analysis was facilitated by the tools available via the NCBI web-site. In addition Artemis (Rutherford et al., 2000), Mega (Tamura et al., 2007), and LaserGene (DNAStar) were employed at various stages. Protein sequence comparisons were performed using an in-house perl script implementing the Needleman-Wunsch global alignment algorithm. Transmembrane domains of herpesvirus proteins were predicted using the Web based software TMHMM, from the Centre for Biological Sequence Analysis at the Technical University of Denmark. Amino acid (AA) content, pI, protein charge and prediction of glycosylation sites were determined using Informax Vector NTI and the PredictProtein Web software (http://www.predictprotein.org) (Rost et al., 2004). The complete genome sequence of HVS1 has been deposited in GenBank and assigned the accession number HM625781.
Results and Discussion
General Genome Structure and Organization
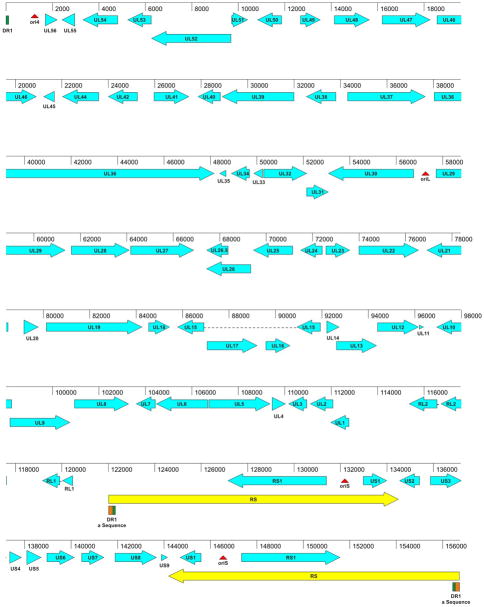
The HVS1 MV-5-4 genome, as sequenced in this study, is 156,742 nt long and is organized as illustrated in Figure 2. It consists of a long unique region (UL; 121,300 nt) and a short unique region (US; 9,650 nt) that is flanked by an inverted repeat sequence (RS; 12,529 nt each) analogous to the “c” repeats (or short repeats; RS) of HSV-1. This is a herpesvirus genomic arrangement of type D structure as defined by Roizman & Pellett (2001) and is similar to that of varicella-zoster virus (VZV) rather than the type E structure typical of other primate simplexviruses. The overall G+C content is 67%, with the RS region having a higher G+C content (77%) than the rest of the genome.
Figure 2.
Genetic organization of the HVS1 genome. The location and orientation of coding sequences are indicated by blue arrows. Coding sequences have been named based on their AA sequence similarity with genes of other primate α-herpesviruses. The location of repeat sequences (yellow) and origins of replication (red) have been identified. The DR1 (green) and “a” repeat sequence regions (orange) are also noted
Unlike the human and Old World monkey viruses, the UL region of the HVS1 genome is not flanked by inverted repeat sequences (analogous to the “b” repeats of HSV-1). Thus, only a single copy of the putative LAT region plus the RL2 (ICP0) and RL1 (γ34.1) ORFs are present in the HVS1 genome. Sequences analogous to the “a” sequence of HSV-1 are present, but the key signal features required for packaging are divided between the 5′ and 3′ termini of the HVS1 genome and are only brought together when the viral genome is circularized (see below). In this respect the HVS1 genome organization is also more like that of varicelloviruses than simplexviruses. The portion of the “a” sequence present at the 3′ (right/RS) terminus is repeated in the internal copy of the RS at the boundary between the UL and RS sequences.
Overall the genes identified in HVS1 are collinear with homologous genes of HSV-1 and other primate simplexviruses, only with the UL1 ORF oriented next to the RS-US region. However, a number of genes in both the UL and US region of HSV-1 that are present in all other primate simplexviruses were found to be absent in HVS1 (see below). Also unlike the other primate simplexviruses, the US1 ORF of HVS1 is located within the short inverted repeats, and therefore is present in two copies.
There is a region in the HVS1 genome between the UL1 and RL2 ORFs that appears to correspond to the LAT region of other primate α-herpesviruses. No ORF likely to encode a protein is apparent in this region, but two poly-adenylation consensus motifs (AATAAA) are present that correspond in position and orientation to those found in the LAT regions of other primate simplexviruses. Although there is not significant sequence homology in this region with other primate α-herpesvirus genomes, HVS1 may express LAT transcripts (and possibly L/STs) analogous to those of other primate α-herpesviruses (Perng and Jones, 2010). However, experimental transcript mapping will be necessary to confirm this.
Genomic Termini and “a” Sequence
In addition to the RACE data identifying the genomic termini, the sequencing library provided further confirmation of the 5′ (left/UL end) terminal sequence. During the course of assembling the sequence data it was observed that one end of several plasmid clones terminated at exactly the same position. The starting DNA for this library was randomly sheared DNA, so it is highly improbable that any two clones would end at exactly the same point unless they were derived from the terminus of the linear DNA. The RACE data confirmed that this termination point was in fact the 5′ terminus of the genome. No clones were identified which correlated with the 3′ terminus; however, this is not surprising as the 5′ terminus would predominate in a replicating viral culture (Severini et al., 1996).
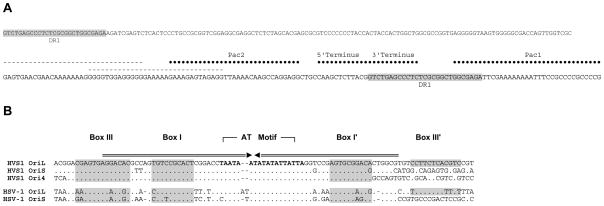
The termini of the HVS-1 genome have different sequences and do not harbor an “a” direct repeat as occurs in other α-herpesviruses. Rather the HVS1 termini structure closely resembles that of HHV-6 with putative Pac1 and Pac2 sequences found on the 5′ (left/UL end) and the 3′ (right/US end) termini of the genome, respectively (Deng and Dewhurst, 1998; Thomson et al., 1994). As shown in Fig. 3A, the joined termini together with 27 bases of the 5′ terminus repeated in the RS region constitute a Psc1/Pac2 packaging signal like that found in HSV (Hodge and Stow, 2001).
Figure 3.
Comparison of Pac1, Pac2, and replication origins of HVS1 and HSV-1. A) HVS1 Pac1 and Pac2 sites are juxtaposed by the joining of the 5′ (left; UL end) and 3′ (right; US end) termini of the genome. The dashed lines indicate possible alternate Pac2 sites. The shaded sequence identifies a repeat sequence (DR1) also found near the ends of the short repeat (RS) sequences. B) Sequences of the replication origins of HVS1 compared to the sequences of OriL and OriS sequences of HSV-1. The arrows above the alignment indicate the inverted repeat and shaded sequences identify the putative binding sites of the origin binding protein as previously defined (Hazuda et al., 1991).
Origins of Replication
Four putative origins of replication (Ori) were identified in the HVS1 genome. As in other α-herpesviruses, one (OriL) is situated in the UL region (centered on nt 57,263) between the UL29 and UL30 ORFs and is a perfect palindrome 146 nt in length. The nucleotide sequence is highly similar to the OriL sequence of HSV-1 and other simplexviruses (Fig. 3B) in the putative binding sites for the origin binding protein. As in other α-herpesviruses, two identical replication origins (OriS) are found in the RS regions. These origins are very similar in sequence to OriL but they lack the rightmost part of the inverted repeat where Box III is situated (Fig. 3B). Interestingly, this is the same structure found in the OriS of HSV-1 and HSV-2, but not of the other primate simplexviruses (SA8, HVP2 and BV) where OriS and OriL are almost identical perfect palindromes (Perelygina et al., 2003; Tyler et al., 2005; Tyler and Severini, 2006). The fourth putative replication origin (Ori4) is situated near the left terminus of the HVS1 genome and is very similar in sequence and structure to OriS (Fig. 3B). The presence of a fourth replication origin is a feature unique to HVS1 among all sequenced α-herpesviruses.
Genome Isomerization
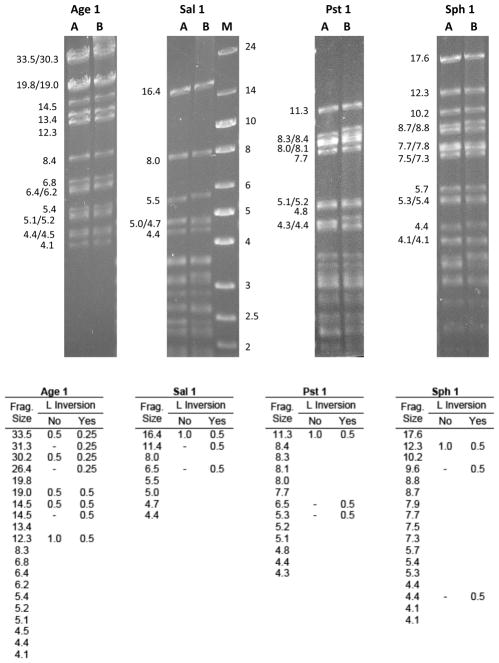
There are two conflicting reports in the literature regarding the structure of the HVS1 genome - whether it is a type D or type E genome (Desrosiers and Falk, 1981; Leib et al., 1987a). Both studies utilized restriction digestion of viral DNA and densitometric analysis of gels to determine the molarity of restriction fragments, with 0.25 M fragments being indicative of a type E genome where four genomic isomers are produced during replication due to inversion of both the UL and US regions. The sequence of the MV-5-4 genome clearly indicates that only the US region is bounded by inverted repeats (a type D genome), and so predicts that there should not be any 0.25 M restriction fragments. Comparing predicted restriction fragments based on the genome sequence with published gels and gel traces in the studies cited above proved difficult due to the size of the fragments of interest (generally >10 Kbp) and the presence of multiple fragments of similar size.
To directly address this discrepancy, we sought experimental evidence for inversion of the L region. Based on the genome sequence, the size and molarity of restriction fragments were predicted for Age1, Sal1, Pst1 and Sph1, assuming either inversion or no inversion of the L region (Fig. 4, bottom). DNA of both the MV-5-4 and 4672 strains of HVS1 were then digested with these restriction enzymes, fragments separated on an agarose gel, and the restriction fragment profile for each enzyme compared to the predicted results (Fig. 4). For each restriction enzyme, the actual restriction profile was consistent with the lack of inversion of the L region with regard to both the presence/absence of certain predicted fragments and the relative molarity of fragments. As one simple and very clear example, if the L region inverts, then Sal1 digestion should produce two fragments representing the two ends of the L region (1.5 and 11.4 Kbp; 0.5 M each) and two fragments representing the L-S joint region (16.4 & 6.5 Kbp; 0.5 M each). If however the L region does not invert, only the 1.5 Kbp L end and the 16.4 L-S joint fragment (both 1.0 M) would result. As shown in Fig. 4, there was no 11.4 Kbp L end or 6.5 Kbp L-S fragment evident for either HVS1 strain, consistent with the lack of L inversion. These experimental data thus support the lack of inversion of the L region of the HVS1 genome as concluded by Desrosier and Falk (1981). However, given the variation in restriction profiles of American and European isolates of HVS1 (Leib et al., 1987b) and the inconsistency of restriction fragments predicted from the genome sequence data with the results of Leib et al. (1987a), it is possible that the European isolate used by these investigators may have a different genome structure from the American HVS1 isolates used in this study. Another (albeit remote) possibility is that the HVS1 isolates used by Leib et al. (1987a) which were isolated from owl monkeys may not be HVS1, but rather a very closely related virus of owl monkeys.
Figure 4.
Restriction analysis confirming the lack of inversion of the L region of the HVS1 genome. Gradient purified DNA of HVS1 strain MV-5-4 (lanes A) and 4672 (lanes B) were digested with restriction enzyme and fragments separated on a 0.5% agarose gel. The sizes (in Kbp) of restriction fragments are shown to the left of each gel and standard size markers (lane M) are shown at right of the Sal1 fragments. All digests were run on the same gel, but have been separated here to accommodate labelling of fragments. The table lists the size (Kbp) and relative molarity (if different from 1.0) of restriction fragments (only those >4.0 Kbp) predicted for each restriction enzyme based on the HVS1 MV-5-4 genome sequence, assuming either inversion or no inversion of the L region. As described in the text, results for all four restriction enzymes tested are consistent with the lack of inversion of the L region of the HVS1 genome.
Open reading frames
As shown in Fig. 2, HVS1 has homologues of most ORFs found in other simplexviruses. Based on AA sequence similarity, the predicted HVS1 proteins all had as their closest homologue the corresponding protein of one of the other primate simplexviruses (Supplemental Table 1). HVS1 predicted proteins have a much lower similarity with homologous proteins of VZV or SVV than with the simplexviruses, indicating that HVS1 is more closely related to the simplexviruses than the varicelloviruses despite its VZV-like type D genome structure.
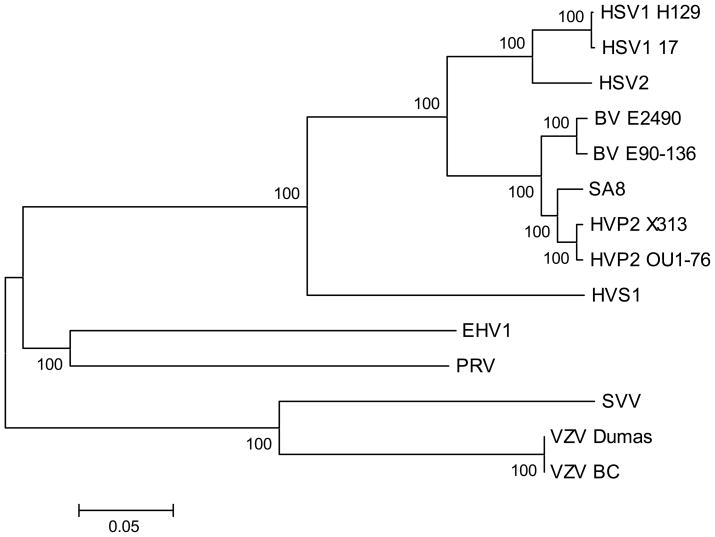
Previous phylogenetic analyses based on single genes (gB or TK) showed that while HVS1 does cluster with anthropoid and simian simplexviruses rather than with varicelloviruses, it occupies a separate branch (together with the other New World simian α-herpesvirus HVA1) within the simplexvirus clade (Eberle and Black, 1993; McGeoch et al., 2000, 2006). To test the veracity of these results with regard to the placement of HVS1, phylogenetic analysis was performed utilizing concatenated AA sequences of 7 conserved proteins [UL2 (uracil DNA glycosylase), UL5 (helicase/primase subunit), UL19 (major capsid protein), UL27 (gB glycoprotein), UL28 (DNA cleavage/packaging protein), UL29 (ssDNA binding protein) and UL30 (DNA polymerase)] as described by McGeoch et al. (1995). As shown in Fig. 5, HVS1 again occupies a separate branch within the simplexvirus clade. These results confirm the inclusion of HVS1 in the simplexvirus group and also confirm that HVS1 occupies the deepest branch within the simplexvirus clade.
Figure 5.
Phylogenetic analysis of HVS1 and related α-herpesviruses. Predicted AA sequences for 7 conserved genes (UL2, UL5, UL19 & UL27–30) were aligned and concatenated as described by McGeoch et al. (1995). Distances were calculated using the MEGA program with Gamma distance and Neighbor Joining for tree construction. Bootstrap values for 500 replications are shown at the root of each major branch. The tree shown is an unrooted tree.
If the hypothesis of co-speciation of herpesviruses is accepted, it would follow that the simplexviruses of Old World primates (BV, HVP2 & SA8) should be more closely related to the human HSVs than HVS1 is. While phylogenetic analyses using conserved genes are consistent with this, the presence of an RL1 gene homologue in HVS1 (see below) while the Old World simian simplexviruses lack one seems contradictory to this. However, there are three other simplexviruses (bovine herpesvirus 2 and parma wallaby herpesviruses 1 & 2) that also have an RL1 gene homologue and (based on phylogenetic analyses) are clearly members of the simplexvirus clade. Like HVS1, these non-primate simplexviruses occupy a separate branch of their own within the simplexvirus tree, with its branchpoint between that of the Old World and New World simian viruses (McGeoch et al., 2000). The inclusion of these non-primate viruses within what is otherwise a clade of primate viruses (the simplexvirus clade) is not consistent with the co-speciation hypothesis unless these non-primate viruses are the result of cross-species transmission of a primate virus. Considering the presence of an RL1 homologue in these viruses, perhaps the absence of an RL1 gene homologue in the Old World simian simplexviruses is more puzzling than the presence of this gene in HVS1. This could represent a loss of the RL1 gene homologue in the Old World monkey herpesvirus lineage, but sequencing of the HVA1 genome to confirm the presence of an RL1 homologue is truly a feature of the New World α-herpesvirus1 lineage.
Divergent Genes
Putative ORFs corresponding in location to the UL56, US4 and US5 genes were evident, but their predicted AA sequences exhibited virtually no similarity to corresponding proteins of other primate simplexviruses. In HSV-1 the UL56 gene codes for a type II membrane protein involved in pathogenicity (Berkowitz et al., 1994), while US4 and US5 code for envelope glycoproteins gG and gJ, respectively (McGeoch et al., 1985, 1987). These proteins are among the least conserved among the primate simplexviruses (Tyler and Severini, 2006), so it is not surprising that homologous ORFs for these genes are not readily identifiable based on sequence similarity. In order to properly identify the correct ORFs for these genes, regions of the HVS1 genome harboring these ORFs were first targeted by RT-PCR to confirm that a transcript was produced from each region. RT-PCR in addition to RACE PCR was continued in order to identify the boundaries of the complete transcripts and to confirm the CDS (Table 1). Analysis of the predicted protein products of these three genes were then compared to the analogous proteins of other primate simplexviruses to determine if the HVS1 protein had similar properties that might suggest functional homology despite the low level of sequence homology.
Table 1.
Genome location of transcripts and coding regions for selected genes.
| Gene1 | CDS Location | Transcript Location |
|---|---|---|
| UL56 | 1667-2230 | 1279-2306 |
| UL49 | 12652-13557 | 12404-13673 |
| RL1 | 120490-120029, 119936-119735, 119178-119174 | 120685-120029, 119936-119735, 119178-119108, 117777-116700, 116579-115215 |
| RL2 | 117642-116700, 116579-115333 | |
| US3 | 135845-137230 | 135598-137959 |
| US4 | 137333-137887 | |
| US5 | 138067-138732 | 138030-140188 |
| US6 | 138938-140161 | |
RL1 and RL2 are spliced genes and are encoded on the negative strand.
In HSV-2, the UL56 protein appears to be a type 2 membrane protein anchored in the membrane by its C-terminal hydrophobic domain (Koshizuka et al., 2002; Davison, 2010). The UL56 protein has several PPxY motifs that facilitate interaction with ubiquitin ligase Nedd4 (Ushijima et al., 2008). Interacting with the myristilated tegument protein encoded by UL11, the UL56 protein also functions in cytoplasmic transport of virions to the plasma membrane and/or release of virions from the infected cell (Ushijima et al., 2009). Alignment of UL56 AA sequences revealed very little sequence identity between the proteins of simian and human viruses (not shown). Although the HVS1 UL56 showed slightly more AA sequence homology with simian virus UL56 proteins than with the HSV UL56 proteins, homology was still very low, there being no stretch of conserved sequence exceeding 4 consecutive AA residues. Despite the lack of AA sequence identity, the UL56 proteins of all the primate simplexviruses including HVS1 have a Leu / Val-rich transmembrane domain of 18–23 AAs at the C-terminus of the protein, suggesting that like the HSV-2 UL56 protein they are all type 2 membrane proteins. While the three PPxY motifs present in the HSV-1 and HSV-2 UL56 proteins are conserved in other simian simplexviruses, only one of these PPxY motifs (the second) is present in the HVS1 UL56 protein. The human and simian simplexviruses also have an Arg-rich domain between the second and third PPxY motifs that is not present in the HVS1 UL56 protein. Despite the lack of extensive AA sequence homology with other α-herpesvirus UL56 proteins, we consider it likely that the HVS1 UL56 protein functions similarly to the HSV UL56 protein.
Although the HSV-1 gJ glycoprotein encoded by the US5 gene has been shown to have anti-apoptotic activity and to induce reactive oxygen species, little else is known about its function (Aubert et al., 2008). The HVS1 US5 protein appears different from the gJ proteins of other primate simplexviruses. It is considerably larger (222 AAs compared to 92–122 AAs for the gJ proteins of other primate simplexviruses). Like the gJ proteins of the other primate simplexviruses, the HVS1 US5 protein has a predicted hydrophobic signal peptide at the N-terminus, an N-linked glycosylation site (NxS/T), a predicted transmembrane domain of 23 AAs, and a highly charged C-terminal domain of 20–22 AA. Sequence homology between the US5 protein of HVS1 and the gJ proteins of other primate simplexviruses is limited to a 4 AA sequence (GGLC) located within the predicted transmembrane domain. The predicted extracellular domain of the HVS1 US5 protein has 5 Cys residues as compared to only one in HSV-2 gJ and none in the HSV-1, BV, SA8 & HVP2 gJ glycoproteins, suggesting additional structure in the HVS1 US5 protein not found in other gJ proteins. None of the primate virus gJ proteins (including HVS1) have the high Pro/Ser/Thr content indicative of addition of O-linked glycosylation. Thus, it does appear that the HVS1 US5 gene encodes a glycoprotein and, despite the lack of extensive AA sequence homology, it may be functionally related to the gJ glycoprotein of other primate simplexviruses.
The HSV-1 and HSV-2 US4 ORFs code for glycoprotein gG, an envelope protein of unknown function but which has been studied extensively for its virus-specific antigenic properties (Ashley et al., 1988; Boucher et al., 1993; Frame et al., 1986; McGeoch et al., 1987). In all primate simplexviruses except HSV-1, gG is a protein of about 600 AA which is cleaved into a secreted portion and a membrane bound portion (mgG; 357 AA in HSV-2) (McGeoch et al., 1987; Ohsawa et al., 2002; Perelygina et al., 2003; Su et al., 1987; Tyler et al., 2005; Tyler and Severini, 2006). The mgG fragment is present in the virion envelope and has a role in viral entry (Balachandran and Hutt-Fletcher, 1985; Su et al., 1987; Tran et al., 2000). In contrast, gG of HSV-1 is a 238 AA envelope protein lacking the secreted fragment (McGeoch et al., 1985). The putative product of the HVS1 US4 ORF is only 182 codons in length and shows no clear AA sequence homology with any other simplexvirus gG protein including that of HSV-1. However, prediction of transmembrane domains shows that the putative HVS1 US4 protein is similar to gG of HSV-1 in that it has an N-terminal hydrophobic signal sequence and a transmembrane domain at the C-terminus (not shown); the mgG from HSV-2 and the simian simplexviruses have a different predicted structure, with the large extracellular domain containing a cleavage site located between the signal sequence and the transmembrane domain at the C-terminus. The gG proteins of the primate simplexviruses (HSV-1 gG and the mgG of HSV-2 and the simian viruses) all have a stretch of acidic AA residues (Asp/Asp), 1–2 N-linked glycosylation sites, and a high concentration of Pro/Ser/Thr residues typical of regions where O-linked glycosylation occurs. The HVS1 US4 protein appears to lack all of these characteristics. Aside from its small size and the presence of two hydrophobic domains, the HVS1 gG protein appears to bear little similarity to the gG proteins of other primate simplexviruses.
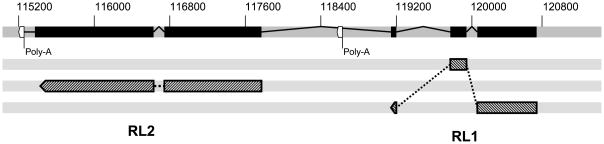
Like all of the other simplexviruses or primates, HVS1 has a homologue of the immediate early RL2 (ICP0) gene, although in the case of HVS1 this gene is present in only a single copy. While HSV-1 and HSV-2 as well as the chimpanzee simplexvirus (unpublished observation) have an RL1 (γ34.5) gene homologue, the cercopithecine monkey viruses (BV, HVP2 & SA8) lack an identifiable RL1 homologue. It is therefore interesting that HVS1 has an RL1 gene homologue. Both the RL1 and RL2 genes of HVS1 are spliced, and the HVS1 RL1 gene possess only minor regions of AA sequence similarity to RL1 genes of other primate herpesviruses such that the predicted AA sequences lack sufficient similarity to accurately identify the HVS1 RL1 ORF. The complete transcripts for the HVS1 RL1 and RL2 genes were amplified and sequenced in order to determine the coding sequence boundaries and splice sites (Table 1). In doing so it was found that the HVS1 RL1 and RL2 genes appear to be transcribed as a single unit as illustrated in Fig. 6. Although a consensus poly-adenylation site (AATAAA) is present downstream of the RL1 ORF, it is located approximately 500 bp 3′ of the RL1 termination codon and does not appear to be utilized (at least in Vero cells in vitro).
Figure 6.
Genetic organization of the RL1 and RL2 genes. The polycistronic RL1/RL2 mRNA is shown at top with introns as black boxes and exons as lines between introns. The locations of consensus poly-adenylation signals in the genome are indicated in white. The reading frame of introns is indicated by hatched boxes below the mRNA map, the three gray bars representing reading frames 1, 2 & 3 in the DNA sequence.
In HSV-1, the RL1 (γ34.5) protein has some AA sequence homology with mammalian protein phosphatase 1 regulatory subunit 15A (PP1r15A), dephosphorylates eIF-2α to prevent shutdown of protein synthesis in the infected cell, facilitates virion egress, and plays a central role in neurovirulence (Cheng et al., 2003; Chou et al., 1990; Chou and Roizman, 1994; He et al., 1997; Jing et al., 2004; MacLean et al., 1991a; Whitley et al., 1993). Unlike the unspliced HSV-1 RL1 gene, the HVS1 RL1 gene is spliced, having three exons. The 852 bp sequence from 90 bp 5′ of exon 1 through exon 2 was 99.9% identical in the 4672 strain of HVS1 (Table 2), indicating that the RL1 gene is not an artifact of one HVS1 isolate (see below). The HVS1 RL1-encoded protein shows sequence homology with PP1r15A that is similar to that between the HSV-1 and HSV-2 γ34.5 proteins and PP1r15A (Fig. 7), suggesting that the RL1 protein of HVS1 may have some of the same functions as γ 34.5 does in HSV-1.
Table 2.
Sequence Variation Between HVS1 Strains MV-5-4 and 4672
| Genome Area Compared | Sequence Differences (MV-5-4 → 4672) | ||||
|---|---|---|---|---|---|
| Genome Coordinates | Length | Genes Covered | Coordinate | Type | Result |
| 23,312 - 24,358 | 1047 nt | UL42 - UL44 | 23,961 | C → A | Noncoding |
| 117,352 - 118,332 | 981 nt | RL2 Exon 1 - RL1/2 IG | 118,302 | Insertion (A) | Noncoding |
| 118,860 - 119,243 | 384 nt | RL1/2 IG - RL1 Intron 2 | None | ||
| 119,727 - 120,894 | 1168 nt | RL1 Intron 2 - RL1/RS1 IG | 120,129 | C → T | RL1 Exon 1 (silent) |
| 120,885 | Deletion (G) | Noncoding | |||
| 136,938 - 137,730 | 793 nt | US3 - US4 | None | ||
| 138,424 - 139,282 | 859 nt | US5 - US6 | 138,757 | C → T | Noncoding |
| 143,015 - 144,686 | 1672 nt | US8 - US1 | 143,260 | C → T | US8 (silent) |
| 144,205 | Insertion (C) | Noncoding | |||
| 144,243 | A → G | Noncoding | |||
| 144,377 | Insertion (G) | Noncoding | |||
| 144,622 | Insertion (T) | Noncoding | |||
Figure 7.
Alignment of the protein phosphatase 1 regulatory subunit 15A (PP1r15A) domain of the predicted HVS1 RL1 protein with analogous domains of HSV-1 and HSV-2 γ34.5 proteins and the murine and human PP1r15A proteins. Accession numbers are AAG39340 (HSV-1 γ34.5), NP044468 (HSV-2 γ34.5), NP032680 (murine PP1r15A) and AAP36135 (human PP1r15A). The darker shaded AA residues represent identity between HVS1 RL1 and the mammalian PP1r15A proteins, and the lighter shaded areas represent identity among the three herpesvirus RL1 proteins.
Adjacent to the RL1 gene is the RL2 gene which in other α-herpesviruses encodes the immediate early (IE) ICP0 protein. In all of the primate simplexviruses except HSV-2, the RL2 gene is spliced and has three exons. The HVS1 RL2 gene is spliced, but has only two exons as does the HSV-2 RL2 gene (McGeoch et al., 1991). The putative HVS1 ICP0 protein has a ring finger domain like ICP0 proteins of other primate simplexviruses, so the HVS1 RL2 protein probably serves a similar function. Transcript mapping revealed that in HVS1 the RL1/RL2 mRNA terminates 3′ of the RL2 ORF. Although northern blots were not performed to detect the synthesis of multiple mRNAs, it is possible that in HVS1 the RL1 and RL2 genes are transcribed as a 3′ co-terminal set of mRNAs.
Missing Genes
ORFs corresponding to UL43/43.5, UL49A, US8.5, US10, US11, and US12 are missing from the HVS1 genome. It is unlikely that these missing genes are the result of sequence assembly errors since sufficient plasmid clones were sequenced in order to establish a tiling path across the entire genome. The sequences obtained from these clones were logically consistent with respect to orientation and insert size range indicating that the unique genome arrangement for HVS1 is not a result of incorrect assembly of the sequence data. In addition to the PCR results (Table 2) confirming this arrangement, the regions found to be lacking particular genes were further scrutinized in order to rule out the possibility that a minor misassembly could have resulted in a localized deletion of a short segment of sequence. All of these regions of missing genes were found to also be covered by several plasmid based sequence reads supporting the 454 sequence data. In addition, in all cases at least one individual sequence read was found to span the entire region in question, further validating the accuracy of the sequence.
The UL43 gene of HSV-1 [and homologues in certain γ-herpesviruses (Tugizov et al., 2003)] codes for a membrane protein involved in infection of polarized epithelial cells, but it is not essential for HSV-1 infection in cell culture or pathogenicity in the mouse model (MacLean et al., 1991b). However, all α-herpesviruses that have been sequenced have a UL43 gene homologue. In the HVS1 genome the UL42 ORF is separated from the UL44 ORF by 357 nucleotides, resulting in insufficient space for an additional ORF of ~1.1 Kbp between them (the average size of other simplexvirus UL43 ORFs); the largest potential ORF in this region is less than 80 bp long. The lack of a UL43 ORF in HVS1 was confirmed by PCR amplification and sequencing of the UL42–UL44 intergenic region in both the MV-5-4 and 4672 strains. A homologue of the HSV-1 ORF 43.5 which is antisense to UL43 is also not present in HVS1.
HVS1 also lacks a homologue of the HSV UL49A gene. In HSV and PRV, the UL49A gene encodes a small membrane protein (glycoprotein gN) that interacts with the gM glycoprotein and appears to function in virion morphogenesis (Adams et al., 1998; Crump et al., 2004). Initial sequence analysis identified candidate ORFs located between UL49 and UL50, but these were substantially shorter than other simplexvirus UL49A ORFs and did not exhibit any AA sequence similarity with UL49A proteins of other simplexviruses. In HSV, UL49 and UL49A as assumed to be co-transcribed. For this reason the HVS1 UL49 transcript was mapped and sequenced (Table 1). The 5′ boundary of the UL49 mRNA was found not to extend into the region where the small potential UL49A ORFs are located. We therefore conclude that it is unlikely that these ORFs encode proteins, and that HVS1 lacks a UL49A gene.
US10, US11 and US12 ORFs are transcribed as three co-terminal mRNAs in HSV-1 (Rixon and McGeoch, 1984), and this entire region is missing from the HVS1 MV-5-4 genome. The absence of the US10/11/12 ORFs was confirmed in the 4672 isolate of HVS1. In HSV-1 US10 encodes a virion tegument phosphoprotein of uncertain function (Nishiyama et al., 1993; Yamada et al., 1997), US11 encodes an RNA binding protein that inhibits protein kinase R and regulates viral gene expression (Attrill et al., 2002; Bryant et al., 2005; Khoo et al., 2002; MacLean et al., 1991b), and US12 encodes an IE protein (ICP47) that is an inhibitor of antigen presentation (Ahn et al., 1996; Hill et al., 1995; York et al., 1994). Deletion of these genes (plus US9) from the HSV-1 genome does not affect viral replication or neurovirulence in mice (Nishiyama et al., 1993). Despite the important role these proteins fulfill in HSV-1, α-herpesviruses of non-primate mammalian species (e.g. bovine, equine and porcine herpesviruses) lack these genes while all other primate simplexviruses have homologues of them. Although VZV and SVV also lack homologues of the US11 and US12 genes, these two viruses have two copies of the US10 gene located within the short repeats adjacent to the US1 gene (Gray et al., 1995). The lack of US10/11/12 homologues represents another feature of the HVS1 genome that is distinct from those of other primate simplexviruses.
Also absent in the HVS1 genome is a homologue of the US8.5 ORF. In other primate simplexviruses this small ORF encodes a type 2 membrane protein and partially overlaps the C-terminus of the US8 ORF which encodes glycoprotein gE (Davison, 2010; Dolan et al., 1998; Georgopoulou et al., 1993; Ohsawa et al., 2002; Perelygina et al., 2003; Tyler et al., 2005; Tyler and Severini, 2006). None of the predicted AA sequences derived from the three reading frames immediately 3′ of the HVS1 US8 ORF (corresponding to sequences encoding the C-terminal half of US8.5 proteins) showed significant homology with any of the simplexvirus US8.5 AA sequences. The US8–US9 intergenic region in other simplexviruses is about ~350 bp, while it is only ~150 bp in HVS1, which also supports the absence of a US8.5 ORF in this region. Again, the MV-5-4 and 4672 strains of HVS1 are consistent. In other primate α-herpesviruses US8, US8.5 and US9 form a single transcriptional unit utilizing a single mRNA poly-adenylation and termination site 3′ of the US9 ORF. Despite the apparent lack of a US8.5 gene, US8 and US9 still appear to constitute a single transcriptional unit in HVS1.
Strain Variation
Since the HVS1 genome is in several ways similar to that of varicelloviruses, the degree of DNA sequence variation between the standard MV-5-4 strain and a recent field isolate of HVS1 was analyzed. In 2007 several young squirrel monkeys in a closed breeding colony developed oral lesions typical of a herpesvirus infection (Figure 1). A total of 3 isolates were obtained from clinical swab specimens, of which strain 4672 was one. The rapid development of rounded cells in Vero cell culture, SDS-PAGE analysis of infected cell proteins, restriction analysis of viral DNA, and PCR/sequencing using primers located in the conserved UL19 gene all indicated that this virus was a strain of HVS1 and that there were no detectable differences that distinguished the three isolates (data not shown).
To assess DNA sequence variation between the standard MV-5-4 and 4672 strains of HVS1, several regions of the genome representing areas that exhibit a comparatively high level of sequence variation among strains of HSV-2 or HVP2 were amplified from strain 4672 by PCR and the products sequenced. The regions analyzed and the sequencing results are summarized in Table 2. Briefly, over a combined 6,904 bp of sequence analyzed, there were a total of only 10 nucleotide differences (0.14% sequence variation) between the two strains. Eight differences were located in noncoding intergenic sequences, and the two that were located in coding sequence were silent (produced no AA change). Of the eight differences in noncoding sequence, five were insertion/deletion differences and occurred within homopolynucleotide tracts that varied in size from 3 to 15 residues. For comparison, alignment of sequences of the conserved glycoprotein gB gene of at least 8 strains each of HSV-1, HSV-2 and VZV were used to estimate the degree of genetic variation for each of these viruses. HSV-1 had the highest value (2.24%) followed by HSV-2 (1.25%) and VZV (0.11%). Sequence variation in the noncoding US3–4 and US4–5 regions of BV and HVP2 is even greater than these HSV gB sequence variation rates (Rogers et al., 2003; Smith et al., 1998). The low level of sequence divergence between HVS1 strains (0.14%) is considerably less than that observed among strains of other simplexviruses but is comparable to the level of sequence divergence among VZV isolates (Norberg et al. 2006; Peters et al., 2006; Tyler et al., 2007).
Conclusions
Biologically, HVS1 is very similar to simplexviruses of other primate species (Eberle and Hilliard, 1995; Hull, 1973; Hunt and Melendez, 1969). Unlike other primate simplexviruses, only the US region of the HVS1 genome is bounded by repeats, resulting in a type D genome with two genomic isomers as found in varicelloviruses rather than the four isomers that occur in the other primate simplexviruses that have type E genomes. While other simian simplexviruses (BV, HVP2 & SA8) all lack a homologue of the RL1 (γ34.5) gene, HVS1 has a gene clearly related to the RL1 genes of HSV-1 and HSV-2 and with limited homology to the mammalian PP1r15A protein. Unlike any other α-herpesvirus sequenced to date, the HVS1 genome has four putative DNA replication origins. The HVS1 US1 gene is present within the RS repeats, but the US10 homologue present in the VZV and SVV short repeats is lacking in HVS1. HVS1 also lacks homologues of several genes that are present in all other primate simplexviruses (US8.5, US10/11/12, UL43/43.5 and UL49A). Despite these differences, HVS1 proteins are clearly more closely related to analogous proteins of the primate simplexviruses than to those of varicelloviruses, such that phylogenetic analyses firmly place HVS1 within the primate simplexvirus clade, albeit on a separate deep branch. The significance of the unique structural characteristics of the HVS1 genome is unclear, and it will be informative to examine the structure of other α-herpesviruses of South American primate species.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the cooperation of Dr. C. Abee and staff of the Michale E. Keeling Center for Comparative Medicine and Research at the University of Texas for tissue samples for virus isolation from squirrel monkeys in this colony and the USPHS grant P40 RR01254 that supports this colony. The assistance of the Bioinformatics Core and DNA Core Facilities of the Canada National Microbiology Laboratory is also acknowledged. This work was supported in part by grants from the USPHS P40 RR12317.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Shaun Tyler, Email: Shaun_Tyler@phac-aspc.gc.ca.
Alberto Severini, Email: Alberto_Severini@phac-aspc.gc.ca.
Darla Black, Email: darla.black@okstate.edu.
R. Eberle, Email: r.eberle@okstate.edu.
References
- Adams R, Cunningham C, Davison MD, MacLean CA, Davison AJ. Characterization of the protein encoded by gene UL49A of herpes simplex virus type 1. J Gen Virol. 1998;79 (Pt 4):813–823. doi: 10.1099/0022-1317-79-4-813. [DOI] [PubMed] [Google Scholar]
- Ahn K, Meyer TH, Uebel S, Sempe P, Djaballah H, Yang Y, Peterson PA, Fruh K, Tampe R. Molecular mechanism and species specificity of TAP inhibition by herpes simplex virus ICP47. EMBO J. 1996;15(13):3247–3255. [PMC free article] [PubMed] [Google Scholar]
- Ashley RL, Militoni J, Lee F, Nahmias A, Corey L. Comparison of Western blot (immunoblot) and glycoprotein G-specific immunodot enzyme assay for detecting antibodies to herpes simplex virus types 1 and 2 in human sera. J Clin Microbiol. 1988;26(4):662–667. doi: 10.1128/jcm.26.4.662-667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attrill HL, Cumming SA, Clements JB, Graham SV. The herpes simplex virus type 1 US11 protein binds the coterminal UL12, UL13, and UL14 RNAs and regulates UL13 expression in vivo. J Virol. 2002;76(16):8090–8100. doi: 10.1128/JVI.76.16.8090-8100.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert M, Chen Z, Lang R, Dang CH, Fowler C, Sloan DD, Jerome KR. The antiapoptotic herpes simplex virus glycoprotein J localizes to multiple cellular organelles and induces reactive oxygen species formation. J Virol. 2008;82(2):617–629. doi: 10.1128/JVI.01341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran N, Hutt-Fletcher LM. Synthesis and processing of glycoprotein gG of herpes simplex virus type 2. J Virol. 1985;54(3):825–832. doi: 10.1128/jvi.54.3.825-832.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz C, Moyal M, Rosen-Wolff A, Darai G, Becker Y. Herpes simplex virus type 1 (HSV-1) UL56 gene is involved in viral intraperitoneal pathogenicity to immunocompetent mice. Arch Virol. 1994;134(1–2):73–83. doi: 10.1007/BF01379108. [DOI] [PubMed] [Google Scholar]
- Boucher FD, Yasukawa LL, Kerns K, Kastelein M, Arvin AM, Prober CG. Detection of antibodies to herpes simplex virus type 2 with a mammalian cell line expressing glycoprotein gG-2. Clin Diagn Virol. 1993;1(1):29–38. doi: 10.1016/0928-0197(93)90031-y. [DOI] [PubMed] [Google Scholar]
- Breshears MA, Eberle R, Ritchey JW. Characterization of gross and histological lesions in Balb/c mice experimentally infected with herpesvirus saimiri 1 (HVS1) J Compar Pathol. 2001;125(1):25–33. doi: 10.1053/jcpa.2001.0473. [DOI] [PubMed] [Google Scholar]
- Breshears MA, Eberle R, Ritchey JW. Temporal progression of viral replication and gross and histological lesions in Balb/c mice inoculated epidermally with Saimiriine herpesvirus 1 (SaHV-1) J Compar Path. 2005;133(2–3):103–113. doi: 10.1016/j.jcpa.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Bryant KF, Cox JC, Wang H, Hogle JM, Ellington AD, Coen DM. Binding of herpes simplex virus-1 US11 to specific RNA sequences. Nucleic Acids Res. 2005;33(19):6090–6100. doi: 10.1093/nar/gki919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkholder CR, Soave OA. Isolation, identification, and experimental transmission of herpesvirus T from an owl monkey (Aotus trivirgatus) Lab Anim Care. 1970;20(2):186–191. [PubMed] [Google Scholar]
- Cheng G, Yang K, He B. Dephosphorylation of eIF-2alpha mediated by the gamma(1)34.5 protein of herpes simplex virus type 1 is required for viral response to interferon but is not sufficient for efficient viral replication. J Virol. 2003;77(18):10154–10161. doi: 10.1128/JVI.77.18.10154-10161.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J, Kern ER, Whitley RJ, Roizman B. Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science. 1990;250(4985):1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- Chou J, Roizman B. Herpes simplex virus 1 gamma(1)34.5 gene function, which blocks the host response to infection, maps in the homologous domain of the genes expressed during growth arrest and DNA damage. Proc Natl Acad Sci USA. 1994;91(12):5247–5251. doi: 10.1073/pnas.91.12.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump CM, Bruun B, Bell S, Pomeranz LE, Minson T, Browne HM. Alphaherpesvirus glycoprotein M causes the relocalization of plasma membrane proteins. J Gen Virol. 2004;85(Pt 12):3517–3527. doi: 10.1099/vir.0.80361-0. [DOI] [PubMed] [Google Scholar]
- Daniel MD, Karpas A, Melendez LV, King NW, Hunt RD. Isolation of herpes-T virus from a spontaneous disease in squirrel monkeys (Saimiri sciureus) Arch Gesamte Virusforsch. 1967;22(3):324–331. doi: 10.1007/BF01242953. [DOI] [PubMed] [Google Scholar]
- Davison AJ, Dolan A, Akter P, Addison C, Dargan DJ, Alcendor DJ, McGeoch DJ, Hayward GS. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J Gen Virol. 2003;84(Pt 1):17–28. doi: 10.1099/vir.0.18606-0. [DOI] [PubMed] [Google Scholar]
- Davison AJ. Herpesvirus sytematics. Vet Microbiol. 2010;143(1):52–69. doi: 10.1016/j.vetmic.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Dewhurst S. Functional identification and analysis of cis-acting sequences which mediate genome cleavage and packaging in human herpesvirus 6. J Virol. 1998;72(1):320–329. doi: 10.1128/jvi.72.1.320-329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers RC, Falk LA., Jr Herpesvirus tamarinus and its relation to herpes simplex virus. J Gen Virol. 1981;56(Pt 1):119–130. doi: 10.1099/0022-1317-56-1-119. [DOI] [PubMed] [Google Scholar]
- Dolan A, Jamieson FE, Cunningham C, Barnett BC, McGeoch DJ. The genome sequence of herpes simplex virus type 2. J Virol. 1998;72(3):2010–2021. doi: 10.1128/jvi.72.3.2010-2021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle R, Black D. Sequence analysis of herpes simplex virus gB gene homologs of two platyrrhine monkey alpha-herpesviruses. Arch Virol. 1993;129(1–4):167–182. doi: 10.1007/BF01316893. [DOI] [PubMed] [Google Scholar]
- Eberle R, Black D, Hilliard JK. Relatedness of glycoproteins expressed on the surface of simian herpes-virus virions and infected cells to specific HSV glycoproteins. Arch Virol. 1989;109(3–4):233–252. doi: 10.1007/BF01311084. [DOI] [PubMed] [Google Scholar]
- Eberle R, Hilliard JK. The simian herpesviruses: a review. Infect Agents Dis. 1995;4:55–70. [PubMed] [Google Scholar]
- Emmons RW, Gribble DH, Lennette EH. Natural fatal infection of an owl monkey (Aotus trivirgatus) with Herpes T virus. J Infect Dis. 1968;118(2):153–159. doi: 10.1093/infdis/118.2.153. [DOI] [PubMed] [Google Scholar]
- Frame MC, Marsden HS, McGeoch DJ. Novel herpes simplex virus type 1 glycoproteins identified by antiserum against a synthetic oligopeptide from the predicted product of gene US4. J Gen Virol. 1986;67 (Pt 4):745–751. doi: 10.1099/0022-1317-67-4-745. [DOI] [PubMed] [Google Scholar]
- Georgopoulou U, Michaelidou A, Roizman B, Mavromara-Nazos P. Identification of a new transcriptional unit that yields a gene product within the unique sequences of the short component of the herpes simplex virus 1 genome. J Virol. 1993;67(7):3961–3968. doi: 10.1128/jvi.67.7.3961-3968.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WL, Gusick NJ, Ek-Kommonen C, Kempson SE, Fletcher TM. The inverted repeat regions of the simian varicella virus and varicella-zoster virus genomes have a similar genetic organization. Virus Res. 1995;39(2–3):181–193. doi: 10.1016/0168-1702(95)00091-7. [DOI] [PubMed] [Google Scholar]
- Hazuda DJ, Perry HC, Naylor AM, McClements WL. Characterization of the herpes simplex virus origin binding protein interaction with OriS. J Biol Chem. 1991;266(36):24621–24626. [PubMed] [Google Scholar]
- He B, Gross M, Roizman B. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1997;94(3):843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. Herpes simplex virus turns off the TAP to evade host immunity. Nature. 1995;375(6530):411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- Hilliard JK, Black DH, Eberle R. Simian alphaherpesviruses and their relation to the human herpes simplex viruses. Arch Virol. 1989;109:83–102. doi: 10.1007/BF01310520. [DOI] [PubMed] [Google Scholar]
- Hodge PD, Stow ND. Effects of mutations within the herpes simplex virus type 1 DNA encapsidation signal on packaging efficiency. J Virol. 2001;75:8977–8986. doi: 10.1128/JVI.75.19.8977-8986.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AW, Caldwell RG, Dedmon RE, Deinhardt F. Isolation and characterization of a new herpes virus. J Immunol. 1964;92:602–610. [PubMed] [Google Scholar]
- Holmes AW, Devine JA, Nowakowski E, Deinhardt F. The epidemiology of a herpes virus infection of New World monkeys. J Immunol. 1966;96(4):668–671. [PubMed] [Google Scholar]
- Hull RN. The simian herpesviruses. In: Kaplan AS, editor. The Herpesviruses. Academic Press; New York: 1973. pp. 389–426. [Google Scholar]
- Hunt RD, Melendez LV. Herpes virus infections of non-human primates: a review. Lab Anim Care. 1969;19(2):221–234. [PubMed] [Google Scholar]
- Jing X, Cerveny M, Yang K, He B. Replication of herpes simplex virus 1 depends on the gamma 134.5 functions that facilitate virus response to interferon and egress in the different stages of productive infection. J Virol. 2004;78(14):7653–7666. doi: 10.1128/JVI.78.14.7653-7666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo D, Perez C, Mohr I. Characterization of RNA determinants recognized by the arginine- and proline-rich region of Us11, a herpes simplex virus type 1-encoded double-stranded RNA binding protein that prevents PKR activation. J Virol. 2002;76(23):11971–11981. doi: 10.1128/JVI.76.23.11971-11981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King NW, Hunt RD, Daniel MD, Melendez LV. Overt herpes-T infection in squirrel monkeys (Saimiri sciureus) Lab Anim Care. 1967;17(4):413–423. [PubMed] [Google Scholar]
- Koshizuka T, Goshima F, Takakuwa H, Nozawa N, Daikoku T, Koiwai O, Nishiyama Y. Identification and characterization of the UL56 gene product of herpes simplex virus type 2. J Virol. 2002;76(13):6718–6728. doi: 10.1128/JVI.76.13.6718-6728.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshizuka T, Kawaguchi Y, Goshima F, Mori I, Nishiyama Y. Association of two membrane proteins encoded by herpes simplex virus type 2, UL11 and UL56. Virus Genes. 2006;32(2):153–163. doi: 10.1007/s11262-005-6871-7. [DOI] [PubMed] [Google Scholar]
- Leib DA, Bradbury JM, Hart CA, McCarthy K. Genome isomerism in two alphaherpesviruses: Herpesvirus saimiri-1 (Herpesvirus tamarinus) and avian infectious laryngotracheitis virus. Arch Virol. 1987a;93(3–4):287–294. doi: 10.1007/BF01310982. [DOI] [PubMed] [Google Scholar]
- Leib DA, Hart CA, McCarthy K. Characterization of four herpesviruses isolated from owl monkeys and their comparison with Herpesvirus saimiri type 1 (Herpesvirus tamarinus) and herpes simplex virus type 1. J Comp Pathol. 1987b;97(2):159–169. doi: 10.1016/0021-9975(87)90036-3. [DOI] [PubMed] [Google Scholar]
- Leib DA, Hart CA, McCarthy K. Alphaherpesvirus saimiri in rabbits: a model for human encephalitis? J Gen Virol. 1988;69 (Pt 7):1609–1615. doi: 10.1099/0022-1317-69-7-1609. [DOI] [PubMed] [Google Scholar]
- MacLean AR, ul-Fareed M, Robertson L, Harland J, Brown SM. Herpes simplex virus type 1 deletion variants 1714 and 1716 pinpoint neurovirulence-related sequences in Glasgow strain 17+ between immediate early gene 1 and the ’a’ sequence. J Gen Virol. 1991a;72 (Pt 3):631–639. doi: 10.1099/0022-1317-72-3-631. [DOI] [PubMed] [Google Scholar]
- MacLean CA, Efstathiou S, Elliott ML, Jamieson FE, McGeoch DJ. Investigation of herpes simplex virus type 1 genes encoding multiply inserted membrane proteins. J Gen Virol. 1991b;72 (Pt 4):897–906. doi: 10.1099/0022-1317-72-4-897. [DOI] [PubMed] [Google Scholar]
- McCarthy K, Tosolini FA. Hazards from simian herpes viruses: reactivation of skin lesions with virus shedding. Lancet. 1975;1(7908):649–650. doi: 10.1016/s0140-6736(75)91756-0. [DOI] [PubMed] [Google Scholar]
- McGeoch DJ, Dolan A, Donald S, Rixon FJ. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J Mol Biol. 1985;181(1):1–13. doi: 10.1016/0022-2836(85)90320-1. [DOI] [PubMed] [Google Scholar]
- McGeoch DJ, Dolan A, Donald S, Brauer DH. Complete DNA sequence of the short repeat region in the genome of herpes simplex virus type 1. Nucleic Acids Res. 1986;14(4):1727–1745. doi: 10.1093/nar/14.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch DJ, Moss HW, McNab D, Frame MC. DNA sequence and genetic content of the HindIII l region in the short unique component of the herpes simplex virus type 2 genome: identification of the gene encoding glycoprotein G, and evolutionary comparisons. J Gen Virol. 1987;68 (Pt 1):19–38. doi: 10.1099/0022-1317-68-1-19. [DOI] [PubMed] [Google Scholar]
- McGeoch DJ, Dalrymple MA, Davison AJ, Dolan A, Frame MC, McNab D, Perry LJ, Scott JE, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69 (Pt 7):1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- McGeoch DJ, Cunningham C, McIntyre G, Dolan A. Comparative sequence analysis of the long repeat regions and adjoining parts of the long unique regions in the genomes of herpes simplex viruses types 1 and 2. J Gen Virol. 1991;72 (Pt 12):3057–3075. doi: 10.1099/0022-1317-72-12-3057. [DOI] [PubMed] [Google Scholar]
- McGeoch DJ, Cook S. Molecular phylogeny of the alphaherpesvirinae subfamily and a proposed evolutionary timescale. J Molec Biol. 1994;238:9–22. doi: 10.1006/jmbi.1994.1264. [DOI] [PubMed] [Google Scholar]
- McGeoch DJ, Cook S, Dolan A, Jamieson FE, Telford EA. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J Mol Biol. 1995;247(3):443–458. doi: 10.1006/jmbi.1995.0152. [DOI] [PubMed] [Google Scholar]
- McGeoch DJ, Dolan A, Ralph AC. Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J Virol. 2000;74(22):10401–10406. doi: 10.1128/jvi.74.22.10401-10406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch DJ, Rixon FJ, Davison AJ. Topics in herpesvirus genomics and evolution. Virus Res. 2006;117(1):90–104. doi: 10.1016/j.virusres.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Melnick JL, Midulla M, Wimberly I, Barrera-Oro JG, Levy BM. A new member of the herpesvirus group isolated from South American marmosets. J Immunol. 1964;92:596–601. [PubMed] [Google Scholar]
- Mou SW, Hilliard JK, Song CH, Eberle R. Comparison of the primate alphaherpesviruses. I. Characterization of two herpesviruses from spider monkeys and squirrel monkeys and viral polypeptides synthesized in infected cells. Arch Virol. 1986;91(1–2):117–133. doi: 10.1007/BF01316733. [DOI] [PubMed] [Google Scholar]
- Nishiyama Y, Kurachi R, Daikoku T, Umene K. The US 9, 10, 11, and 12 genes of herpes simplex virus type 1 are of no importance for its neurovirulence and latency in mice. Virology. 1993;194(1):419–423. doi: 10.1006/viro.1993.1279. [DOI] [PubMed] [Google Scholar]
- Norberg P, Liljeqvist JA, Bergstrom T, Sammons S, Schmid DS, Loparev VN. Complete-genome phylogenetic approach to varicella-zoster virus evolution: genetic divergence and evidence for recombination. J Virol. 2006;80(19):9569–9576. doi: 10.1128/JVI.00835-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa K, Black DH, Sato H, Eberle R. Sequence and genetic arrangement of the unique short region of monkey B virus (Cercopithecine herpesvirus 1) genome and its comparison with other primate herpesviruses. J Virol. 2002;76:1516–1520. doi: 10.1128/JVI.76.3.1516-1520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka H, Kit S. Nucleotide sequence of the marmoset herpesvirus thymidine kinase gene and predicted amino acid sequence of thymidine kinase polypeptide. Virology. 1984;135(2):316–330. doi: 10.1016/0042-6822(84)90189-2. [DOI] [PubMed] [Google Scholar]
- Perelygina L, Zhu L, Zurkuhlen H, Mills R, Borodovsky M, Hilliard JK. Complete sequence and comparative analysis of the genome of herpes B virus (Cercopithecine herpesvirus 1) from a rhesus monkey. J Virol. 2003;77:6167–6177. doi: 10.1128/JVI.77.11.6167-6177.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng GC, Jones C. Towards an understanding of the herpes simplex virus type 1 latency-reactivation cycle. Interdiscip Perspect Infect Dis. 2010;2010:262415. doi: 10.1155/2010/262415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters GA, Tyler SD, Grose C, Severini A, Gray MJ, Upton C, Tipples GA. A full-genome phylogenetic analysis of varicella-zoster virus reveals a novel origin of replication-based genotyping scheme and evidence of recombination between major circulating clades. J Virol. 2006;80:9850–9860. doi: 10.1128/JVI.00715-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rixon FJ, McGeoch DJ. A 3′ co-terminal family of mRNAs from the herpes simplex virus type 1 short region: two overlapping reading frames encode unrelated polypeptide one of which has highly reiterated amino acid sequence. Nucleic Acids Res. 1984;12(5):2473–2487. doi: 10.1093/nar/12.5.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers KM, Ealey KA, Ritchey JW, Black DH, Eberle R. Pathogenicity of different baboon Herpesvirus papio 2 isolates is characterized by either extreme neurovirulence or complete apathogenicity. J Virol. 2003;77:10731–10739. doi: 10.1128/JVI.77.20.10731-10739.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B, Pellett PE. The Family Herpesviridae: A Brief Introduction. In: Knipe DM, Howley PM, editors. Fields Virology. 4. Vol. 2. Lippincott, WIlliams & Wilkins; Philadelphia PA: 2001. pp. 2381–2397. [Google Scholar]
- Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32(Web Server issue):W321–326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16(10):944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Severini A, Scraba DG, Tyrrell DL. Branched structures in the intracellular DNA of herpes simplex virus type 1. J Virol. 1996;70:3169–3175. doi: 10.1128/jvi.70.5.3169-3175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AL, Black D, Eberle R. Molecular evidence for distinct genotypes of monkey B virus (Herpesvirus simiae) which are related to the host macaque species. J Virol. 1998;72:9224–9232. doi: 10.1128/jvi.72.11.9224-9232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpara ML, Parsons L, Enquist LW. Sequence variability in clinical and laboratory isolates of herpes simplex virus 1 reveals new mutations. J Virol. 2010;84(10):5303–5313. doi: 10.1128/JVI.00312-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R, Beal KF, Bonfield JK. The Staden Package 1998. Vol. 132. The Humana Press, Inc; Ttowa, N.J: 1999. [DOI] [PubMed] [Google Scholar]
- Su HK, Eberle R, Courtney RJ. Processing of the herpes simplex virus type 2 glycoprotein gG-2 results in secretion of a 34,000-Mr cleavage product. J Virol. 1987;61(5):1735–1737. doi: 10.1128/jvi.61.5.1735-1737.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thomson BJ, Dewhurst S, Gray D. Structure and heterogeneity of the a sequences of human herpesvirus 6 strain variants U1102 and Z29 and identification of human telomeric repeat sequences at the genomic termini. J Virol. 1994;68(5):3007–3014. doi: 10.1128/jvi.68.5.3007-3014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LC, Kissner JM, Westerman LE, Sears AE. A herpes simplex virus 1 recombinant lacking the glycoprotein G coding sequences is defective in entry through apical surfaces of polarized epithelial cells in culture and in vivo. Proc Natl Acad Sci USA. 2000;97(4):1818–1822. doi: 10.1073/pnas.020510297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugizov SM, Berline JW, Palefsky JM. Epstein-Barr virus infection of polarized tongue and nasopharyngeal epithelial cells. Nat Med. 2003;9(3):307–314. doi: 10.1038/nm830. [DOI] [PubMed] [Google Scholar]
- Tyler SD, Peters GA, Severini A. Complete genome sequence of Cercopithecine herpesvirus 2 (SA8) and comparison with other simplexviruses. Virology. 2005;331:429–440. doi: 10.1016/j.virol.2004.09.042. [DOI] [PubMed] [Google Scholar]
- Tyler SD, Severini A. The complete genome sequence of Herpesvirus papio 2 (Cercopithecine herpesvirus 16) shows evidence of recombination events among various progenitor herpesviruses. J Virol. 2006;80(3):1214–1221. doi: 10.1128/JVI.80.3.1214-1221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler SD, Peters GA, Grose C, Severini A, Gray MJ, Upton C, Tipples GA. Genomic cartography of varicella-zoster virus: a complete genome-based analysis of strain variability with implications for attenuation and phenotypic differences. Virology. 2007;359(2):447–458. doi: 10.1016/j.virol.2006.09.037. [DOI] [PubMed] [Google Scholar]
- Ushijima Y, Koshizuka T, Goshima F, Kimura H, Nishiyama Y. Herpes simplex virus type 2 UL56 interacts with the ubiquitin ligase Nedd4 and increases its ubiquitination. J Virol. 2008;82(11):5220–5233. doi: 10.1128/JVI.02515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima Y, Goshima F, Kimura H, Nishiyama Y. Herpes simplex virus type 2 tegument protein UL56 relocalizes ubiquitin ligase Nedd4 and has a role in transport and/or release of virions. Virol J. 2009;6:168. doi: 10.1186/1743-422X-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley RJ, Kern ER, Chatterjee S, Chou J, Roizman B. Replication, establishment of latency, and induced reactivation of herpes simplex virus gamma 1 34.5 deletion mutants in rodent models. J Clin Invest. 1993;91(6):283728–43. doi: 10.1172/JCI116527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Daikoku T, Yamashita Y, Jiang YM, Tsurumi T, Nishiyama Y. The product of the US10 gene of herpes simplex virus type 1 is a capsid/tegument-associated phosphoprotein which copurifies with the nuclear matrix. J Gen Virol. 1997;78 (Pt 11):2923–2931. doi: 10.1099/0022-1317-78-11-2923. [DOI] [PubMed] [Google Scholar]
- York IA, Roop C, Andrews DW, Riddell SR, Graham FL, Johnson DC. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell. 1994;77(4):525–535. doi: 10.1016/0092-8674(94)90215-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.