Abstract
Patients with extensive ulcerative colitis (UC) have an increased risk of colorectal cancer. Although UC patients generally undergo lifelong colonoscopic surveillance to detect dysplasia or cancer in the colon, detection of cancer in this manner is expensive and invasive. An objective biomarker of dysplasia would vastly improve the clinical management of cancer risk in UC patients. In the current study, accurate mass and time methods with ion intensity-based label-free proteomics are applied to profile individual rectal and colon samples from UC patients with dysplasia or cancer (UC progressors) compared to rectal samples from patients that are dysplasia/cancer free (UC non-progressors) in order to identify a set of proteins in the rectum mucosa that differentiate the two groups. In addition to the identification of proteins in UC dysplastic colon tissue, we for the first time identified differentially expressed proteins in non-dysplastic rectal tissue from UC progressors. This provides a candidate pool of biomarkers for dysplasia/cancer that could be detected in a random non-dysplastic rectal biopsy. Mitochondrial proteins, cytoskeletal proteins, RAS superfamily, proteins relating to apoptosis, and metabolism were important protein clusters differentially expressed in the non-dysplastic and dysplastic tissues of UC progressors, suggesting their importance in the early stages of UC neoplastic progression. Among the differentially expressed proteins, immunohistochemistry analysis confirmed that TRAP1 displayed increased IHC staining in UC progressors, in both dysplastic and non-dysplastic tissue, and CPS1 showed a statistically significant difference in IHC staining between the non-progressor and progressor groups. Furthermore, rectal CPS1 staining could be used to predict dysplasia or cancer in the colon with 87% sensitivity and 45% specificity, demonstrating the feasibility of using surrogate biomarkers in rectal biopsies to predict dysplasia and/or cancer in the colon.
Keywords: Proteomics, ulcerative colitis, dysplasia, colorectal cancer, biomarker, label-free, mass spectrometry, AMT
Introduction
Ulcerative colitis (UC) is an inflammatory bowel disease affecting approximately half a million patients in the United States. Patients with extensive UC of more than eight years duration have an increased risk of colorectal cancer, approximating 0.5–1% per year of colitis1,2. For example, a patient with 20 years of UC will have a neoplastic risk of approximately 10–20%. These patients are usually advised to undergo lifelong colonoscopic surveillance to detect the presence of dysplasia (pre-cancer) or cancer in the colon. There are major challenges with UC cancer surveillance. Unlike sporadic colon cancer, dysplasia or cancer frequently occurs in benign-appearing colonic mucosa of UC patients without evidence of a polyp. Moreover, the large surface area of the colon, which ranges up to one square meter, makes it difficult to find endoscopically-invisible focal cancer or pre-cancer, and therefore numerous random biopsies are usually taken throughout the colon for evaluation by a pathologist3. Dysplasia is graded and ranges from indefinite for dysplasia to low-grade dysplasia (LGD) to high-grade dysplasia (HGD) to cancer. Colectomy is recommended for all patients with HGD or cancer. The histologic diagnosis of dysplasia, however, is highly subjective and requires an experienced pathologist for optimum accuracy4. An objective biomarker of dysplasia would vastly improve the clinical management of cancer risk in UC patients. Of even greater utility would be biomarkers present in the normal-appearing (i.e., non-dysplastic) rectum for prediction of dysplasia in the colon, since a random rectal biopsy could be performed in a short clinic visit and would not need prior colon preparation or sedation as is required in colonoscopy surveillance. Patients who have a positive biomarker detected in the rectal mucosa could then be scheduled for the full colonoscopy with biopsy. Because the biomarker test would be used for decision analysis for colonoscopic surveillance, the sensitivity must be close to 100%, i.e., no dysplasia or cancer should be missed. However, the requirement for specificity of the biomarker would not need to be as high since the patients who are false positive for the biomarker would need to undergo full colonoscopy to confirm the presence of dysplasia. Cost savings will increase with specificity, but even if only one third of true negative patients were spared colonoscopy (specificity 33%) the health care cost reduction would be substantial.
To identify dysregulated proteins in the rectum, the current study uses a comparative label-free proteomics approach to investigate the non-dysplastic and normal-appearing rectal tissue from UC patients who have dysplasia or cancer (progressors) versus UC patients who are dysplasia/cancer-free (non-progressors). As noted above, a cancer biomarker that could be found in the rectum would be ideal for testing, as the colonoscopic procedure could be avoided. The label-free approach allows us to perform global quantitative proteome comparison using individual samples, thus allowing assessment of the variation between samples within the same experimental group. By analyzing protein abundance differences between samples in the same experimental group and between groups, we can estimate not only the magnitude of any differential abundance of proteins, but also the probability that the differential observation is due to random experimental variation. A label-free methodology based on LC-MS ion intensity was chosen over the more popular MS/MS spectral counting approach5,6,7 because an approach that is based on high-resolution ion abundance data has the potential for higher sensitivity and precise quantitation for less-abundant proteins8,9,5. The proteomics data analysis described here was performed using the freely available, open-source msInspect software platform. In our related work, we investigated dysregulated proteins in the non-dysplastic colon tissue immediately surrounding the dysplasia as well as the dysplastic colon tissue10. Our current work is highly complementary to our previous work, and, more importantly, the current work focuses on discovering dysregulated proteins in the rectal tissue, which has not been investigated previously.
We present ranked lists of differential proteins identified by label-free comparative proteomics in non-dysplastic rectal tissue and dysplastic colon tissue from UC progressors as compared with non-progressors. The list is restricted both by effect size (i.e., fold change) and by approximate statistical significance (i.e., p-value). This discovery work was followed by a confirmation study, selecting two of the top-ranked differential proteins for validation by immunohistochemistry (IHC). These ranked lists are utilized in functional analysis and pathway analysis to explore the mechanisms that may underlie UC neoplastic progression. Our results have identified protein markers that may prove useful in segregating dysplasia and cancer in the colon, and, after suitable validation studies are conducted, might be used in a variety of clinical applications, including predictive risk assessment and/or early detection diagnostics.
Methods and Materials
Patients and Tissue Specimens
Tissue specimens from patients with ulcerative colitis were collected in accordance with approved Human Subjects guidelines at the University of Washington and the Cleveland Clinic via institutional internal tissue banks. Once procured, all specimens were assigned study IDs and specimen IDs. These specimens, obtained at the time of colonoscopy or from surgical resections, were placed in frozen media containing Minimal Essential Medium with 10% DMSO and kept frozen at −70°C until use. For proteomic analysis, colonic epithelial samples from 5 subjects (patients) from each of the following three categories were used: 1) Non-dysplastic rectal tissue from non-progressors (NP); 2) Non-dysplastic rectal tissue from progressors (P-NEG); 3) Tissue with high-grade dysplasia from UC progressors: (P-HGD). The P-NEG specimens were from UC patients who had high-grade dysplasia or colon cancer, but the particular specimens chosen were histologically negative for dysplasia in the rectum. The histological grades for each specimen were assigned by co-author (MPB) who has extensive experience in evaluating the pathology of IBD (inflammatory bowel disease) samples. The clinical characteristics of these patients are listed in supplemental Table 1. Progressor and non-progressor specimens were matched on gender, patient age, duration of disease, and degree of inflammation. The degree of inflammation was assessed by pathologist and scored from 0 to 4, with 4 indicating the most severe inflammation: 0 = inactive disease, 1 = mild activity with cryptitis; 2 = crypt abscess; 3 = 3 or more crypt abscesses per high power field; and 4 = ulceration and granulation tissue.
Sample Preparation
Colonic epithelial cells were isolated from specimens by EDTA shake-off, which provides over 90% purity of epithelial cells, as previously described11. Protein lysates were obtained by lysing the epithelial cells in T-Per (Pierce, Rockford, IL) or CHAPS (Millipore, Billerica, MA) with 1× Protease Inhibitor Cocktail (Pierce, Rockford, IL)10. Protein lysates (20μg) were reduced with 20 mM DTT and blocked with 25 mM iodoacetamide. The proteins were then digested with trypsin (trypsin to protein ratio: 1/50) overnight (16–18 hours) and purified by C18 desalting column. 1 μg of each purified peptide sample was injected into the mass spectrometer for analysis.
Mass Spectrometry
An LTQ-Orbitrap hybrid mass spectrometer (Thermo Fisher Scientific, Waltham, MA) coupled with nano-flow HPLC was used in this study. The liquid chromatography/mass spectrometry setup consisted of a trap column (100μm × 1.5cm) made from an IntegraFrit (New Objective, Woburn, MA) packed with Magic C18AQ resin (5μm, 200Å particles; Michrom Bioresources, Auburn CA), followed by an analytical column (75μm × 27cm) made from a PicoFrit (New Objective) packed with Magic C18AQ resin (5μm, 100Å particles; Michrom Bioresources). The columns were connected in-line to an Eksigent 2D nano-HPLC (Eksigent Technologies, Dublin, CA) in a vented column configuration to allow fast sample loading at 3uL/min12. The peptide samples were analyzed by LC-MS/MS using a 90-minute non-linear gradient as follows: start at 5% acetonitrile with 0.1% formic acid (against water with 0.1% formic acid), change to 7% over 2 minutes, then to 35% over 90 minutes, then to 50% over 1 minute, hold at 50% for 9 minutes, change to 95% over 1 minute, hold at 95% for 5 minutes, drop to 5% over 1 minute and recondition at 5%. The flow rate for the peptide separation was 300 nL/min. A spray voltage of 2.25 kV was applied to the nanospray tip. The mass spectrometry experiment consisted of a full MS scan in the Orbitrap (AGC target value 1e6, resolution 60K, and one microscan, FT preview scan on) followed by up to 5 MS/MS spectra acquisitions in the linear ion trap. The five most intense ions from the Orbitrap scan were selected for MS/MS using collision induced dissociation (isolation width 2 m/z, target value 1e4, collision energy 35%, max injection time 100 ms). Lower abundance peptide ions were interrogated using dynamic exclusion (repeat count 1, repeat duration 30 sec., exclusion list size 100, exclusion time 45 sec., exclusion mass width −0.55 m/z low to 1.55 m/z high). Charge state screen was used, allowing for MS/MS of any ions with identifiable charge states +2, +3, and +4 and higher.
Peptide and Protein Identification from Tandem MS Data
Raw machine output files from all MS runs were converted to mzXML files and searched with X!Tandem13 configured with the k-score scoring algorithm14, against version 3.65 of the human International Protein Index (IPI) database. The search parameters were as follows: enzyme, trypsin; maximum missed cleavages, 1; fixed modification, carboxamidomethylation on cysteine; potential modification, oxidization on methionine; parent monositopic mass error, 2.5Da. Peptide identifications were assigned probability by PeptideProphet15, and all identifications assigned probability <0.95 (estimated false discovery rate varied per experimental run in the range 0.006–0.007) were discarded. The msInspect/AMT tools16 were used to create an Accurate Mass and Time (AMT) database containing all passing peptide identifications from all sample runs that were observed in at least two runs independently. These stringent filtration steps ensured that AMT matching occurs with a minimum of background noise due to false identifications.
Proteomics Data Analysis Pipeline
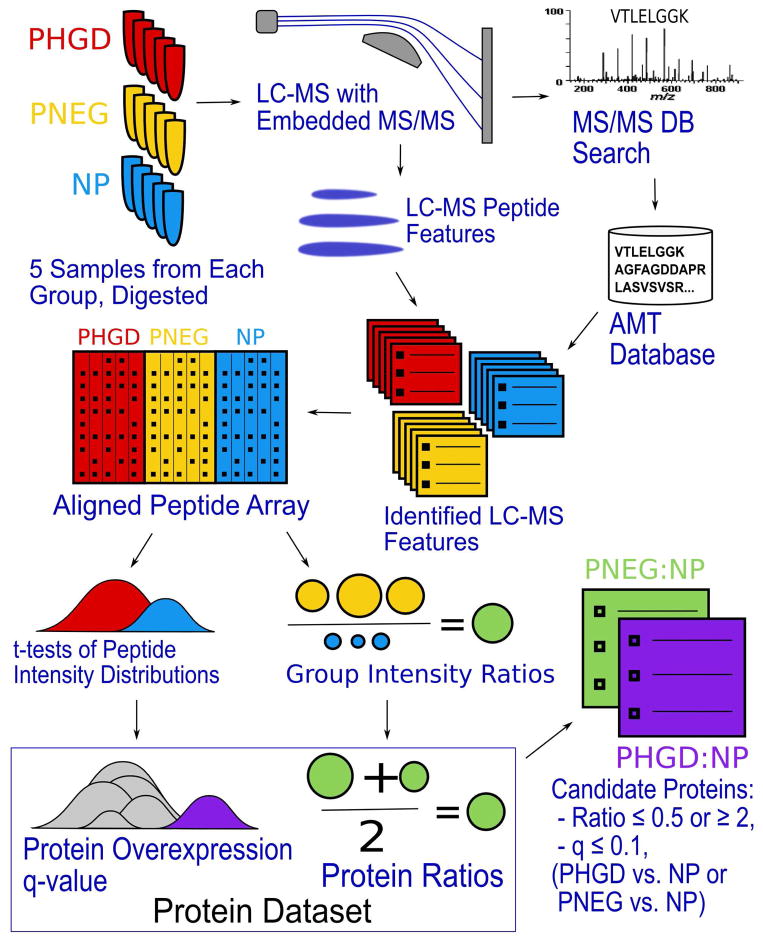
In brief, 5 samples were analyzed from each of the P-HGD, P-NEG and NP experimental groups. For each sample, LC-MS peptide ions were located and assigned peptide identifications using AMT database matching, and this information was combined across replicate sample runs. Peptide identification and intensity information from all samples was assembled into a single “peptide array”, and peptide abundance ratios between the P-HGD and NP groups and between the P-NEG and NP groups were calculated using the geometric mean peptide intensity value from all samples in each group. Protein inference was performed using ProteinProphet, and abundance ratios for each protein identified were obtained by combining peptide information. Where possible, q-values were calculated for each protein describing the probability of observing the observed peptide information for that protein under the null hypothesis that the protein was not differentially expressed. This pipeline is described in further detail below and visualized in Figure 1.
Figure 1.
Experimental workflow. 5 samples from each experimental group were processed. Samples were run in replicate on LTQ-Orbitrap. All runs were searched with X!Tandem against a human IPI database; LC-MS feature-finding was performed. AMT database matching assigned peptide IDs to LC-MS features. Replicate run information was combined for each sample (not shown). LC-MS features were aligned across all samples by mass and RT. Peptide intensity ratios were calculated between the P-NEG (yellow) and NP (blue) groups, and between the P-HGD (red) and NP groups; these ratios were combined for each protein (PNEG:NP in green, PHGD:NP in purple). Peptide t-scores were calculated to assess group differences; t-scores were summarized for each protein (purple) and compared with the t-scores from all other peptides (gray). p-values and q-values were calculated for each protein. Candidate proteins chosen based on q-value missing or ≤ 0.1, and ratio ≤ 0.5 or ≥ 2.
1. LC-MS Feature Detection, Identification and Peptide Array Creation
msInspect17 version 2.3 located LC-MS peptide features in each run using the default settings. AMT matching assigned peptide identifications to LC-MS features. Match probabilities were calculated based on mass and RT error18, and only matches with probability ≥ 0.95 were retained. LC-MS data from the two replicate runs of each sample were combined into a single dataset per sample. The replicates were combined by creating a “peptide array” from the two replicate datasets as previously described17. Briefly, a “peptide array” associates features across multiple machine runs by mass and retention time; each row of the array represents a feature associated across one or more runs, and the array contains columns describing peptide intensity within each run. Feature intensities are normalized based on ion intensity distribution as previously described17, 19 to eliminate the effect of differences between machine runs. In the replicate peptide array dataset for each sample, features observed in only one replicate were retained as-is; features observed in both replicates were collapsed into a single feature and assigned the geometric mean intensity (i.e., log intensities were averaged) of the constituent features. The resulting combined datasets for each sample were, in turn, assembled into a single peptide array containing all 15 samples from all three groups.
2. Peptide Abundance Comparison
Next, we created a dataset describing peptide abundance ratios between the experimental groups. All peptide identifications across the 5 P-HGD and the 5 NP samples were assembled into a single pepXML file20 with a quantitative “ratio” (where available) representing the ratio of geometric mean abundance in the P-HGD samples to the mean abundance of matching peptides in the NP samples. This process was then repeated comparing the P-NEG and NP samples. This representation allowed us to use standard tools for further data processing, treating the dataset as though it were the result of a single LC-MS run using isotopic labeling for quantitation. Peptides observed in one of the compared groups, but not the other, were assigned a ratio of 0 or infinity accordingly, but those peptides were removed from the current analysis, which focuses on quantifiable variation and can thereby use more powerful statistical tests to compare the two groups.
For each of the two-group comparisons (P-HGD:NP and P-NEG:NP) a t-test compared the peptide ion intensities observed in the two groups for each peptide that was observed in at least two patient samples in each group and generated a t-statistic.
3. Protein Abundance Comparison
The peptide-level datasets were processed to create datasets comparing protein abundance between the two groups. For each of the two group-comparison pepXML files (P-HGD:NP and P-NEG:NP), ProteinProphet inferred protein identifications from the identified peptide evidence. Protein probabilities calculated by ProteinProphet were ignored, except to exclude protein identifications subsumed by other proteins that were explained by more peptide evidence. Protein ratios were calculated using the geometric mean of all associated peptide ratios and the results retained in a protXML file. The two two-group-comparison protXML files were combined into a single dataset by associating protein identifications across the two comparisons based on peptide identification information, as described previously21.
For each protein having two or more peptides with t-scores, we performed an overrepresentation analysis, comparing the t-statistics from all of the protein’s peptides with the t-statistics from all other peptide t-statistics observed using a Wilcox test. The p-values calculated from this analysis represented the probability of observing the observed data if the protein was not differentially abundant between groups. Finally, to correct for multiple testing error, q-values (false discovery rates calculated commonly in gene expression array experiments, i.e., FDR in the sense of Storey et al.22) were calculated from those p-values. Differential proteins were chosen from this dataset based on both ratio and q-value as described below.
Immunohistochemistry Analysis
Paraffin-embedded, formalin-fixed tissue blocks were sectioned into 5μM on charge slides. The deparaffinized sections were processed for antigen retrieval using Heat Induced Epitope Retrieval (HIER) Techniques in EDTA buffer (PH 8.0) and microwaved for 18 minutes, followed by cooling to room temperature and then primary antibody incubation. Dilution of the primary antibody TRAP1 (Abcam Cambridge, MA) was 1:50. Biotinylated rabbit IgG made in Goat was used as secondary antibody at 1:500 dilution. IHC staining for CPS1 was performed as described previously10. CPS1 staining was graded semiquantitatively for intensity (0–4) and percentage of cells staining (0=no staining, A=approximately 25%, B=approximately 50%, C=approximately 75%, and D=approximately 100%). Positive staining was defined as cytoplasmic staining of epithelial cells. A combined numeric score was calculated as the product of staining intensity and percentage of staining cells.
Statistical analyses of the CPS1 IHC staining were performed using GraphPad Prism (La Jolla, CA). Differences in the CPS1 level between non-progressors and progressors were tested for statistical significance using the Mann-Whitney test. Empirical receiver operating characteristic (ROC) curves were used to determine the sensitivity and specificity of CPS1 in separating non-dysplastic rectal tissues in progressors from non-dysplastic tissues in non-progressors. Statistical significance was defined as P < 0.05.
Results and Discussion
Proteomics Data Summary
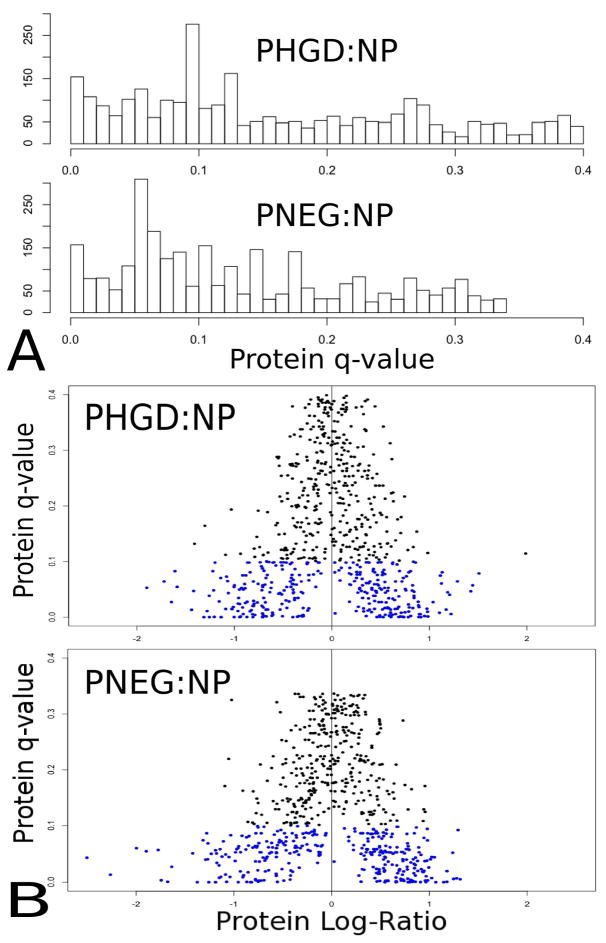
An average of 3,142 unique peptide sequences were identified by MS/MS database search in each of the original 30 runs from all 15 samples and assigned probability ≥ 0.95 by PeptideProphet. The AMT database created from these runs contained 9,679 unique peptides observed independently in at least two different runs. msInspect located an average of 35,562 ions per run, and AMT matching identified an average of 4,461 of these ions per run with probability ≥ 0.95, representing 4,353 unique peptides per run. Sample replicate runs were combined; each sample contained an average of 5,028 unique sequences. Peptide information was combined to infer proteins and compare protein abundances; 1,703 protein group ratios were calculated between the P-HGD and NP groups and 1,705 protein group ratios were calculated between the P-NEG and NP groups. The t-statistics comparing P-HGD and NP groups were calculated for 3,851 peptides observed in at least two samples per group, and for 3,908 peptides comparing P-NEG and NP groups. Protein group-level q-values were calculated as described above for 715 protein groups in the P-HGD:NP comparison and for 708 protein groups in the P-NEG:NP comparison. Figure 2 describes the distribution of q-values and the relationship between protein ratio and protein q-value.
Figure 2.
Figures 2A and 2B contain separate charts comparing the P-HGD group with the NP group and the P-NEG group with the NP group. A. Histogram of q-values for quantitated proteins with sufficient peptide evidence. B. A scatter plot relating the log of the protein ratio (horizontal axis) to the protein q-value (vertical axis). Blue dots indicate proteins with q-value ≤ 0.1.
We compared the current label-free experiment with our previous iTRAQ-based quantitative proteomics study, which used different samples, instrumentation and experimental procedures. In the current study, we quantified 761 of the original 1,106 proteins quantified in the earlier iTRAQ analysis, a 68% overlap based on the total proteins quantified in the iTRAQ study. In addition, an additional 1,165 proteins that were not identified in the iTRAQ analysis were also identified and quantified. The pooling of protein identification data from the analysis of 15 individual samples and the use of AMT database search led to significantly increased protein identification yield in the current study. The differences between the iTRAQ study and the current investigation in sample types (pooled colon specimens vs. individual rectal specimens), quantitative techniques (iTRAQ vs. label-free) and mass spectrometers used (QTOF vs. OrbiTrap) all contribute to the differences in protein identification and quantification. Nonetheless, the high degree of overlap between the very different experiments and the larger number of proteins quantified in the label-free experiment are encouraging, and they are consistent with conclusions reached in other comparisons of labeled and label-free methods23.
Reproducibility
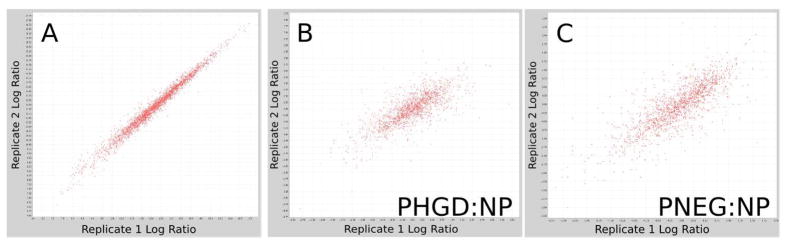
We evaluated the reliability of our approach in two complementary ways using replicate experiments; first the reproducibility of ion intensities across technical replicates was assessed, and second the reproducibility of between group protein-ratios was examined by comparing results when using only the first of the technical replicates to the results when using only the second technical replicates. Peptide-level ion intensity was quite reproducible between the replicates (Figure 3a). The median peptide intensity coefficient of variation between replicates ranged from 0.076 to 0.152 across all 15 samples, with a mean of 0.099. The (less informative) correlation coefficient ranged between 0.961 and 0.995 across all 15 samples, with a mean of 0.987.
Figure 3.
A. A scatter plot of peptide log-intensity values for LC-MS features with the same peptide identification and charge state, for one sample. Horizontal axis: log-intensity in replicate run 1. Vertical axis: log-intensity in replicate run 2. B. A scatter plot of protein log-ratios calculated between the P-HGD and NP groups using only the data from the first replicate run from each sample (horizontal axis) vs. the second replicate run (vertical axis). C. As B, but with data from the comparison between P-NEG and NP.
We evaluated protein-ratio reproducibility in a manner that includes effects from the entire analytic platform, including instrument variation, peptide intensity assessment variability, and peptide identification variability. Sample runs were divided into two groups, one containing only the first replicates of each sample and another with all of the second replicates; the same automated analysis workflow described above was performed separately on each replicate group and the protein ratios calculated by the two analyses were compared. The correlation coefficient of the replicate protein log ratios was 0.75 for the P-HGD:NP comparison and 0.80 for the P-NEG:NP comparison (see Figure 3b). Note that the correlations of peptide ion intensity relationship and the protein log-ratio relationship should not be compared directly since they reflect very different measurement scales: Figure 3a correlates peptide raw intensities, but the protein-level comparison compares aggregated differences of those intensities, as well as additional sources of computational variation that could vary from experiment to experiment (e.g., protein-level inference). Figure 3b shows that, when all these sources of variation are taken together, the reproducibility of the analytic platform is adequate to identify systematic changes across experimental conditions. Moreover, the protein ratios calculated for our study make use of both replicates, and so the reproducibility of protein ratios should be greater in our experiment than demonstrated here.
Differential Proteins
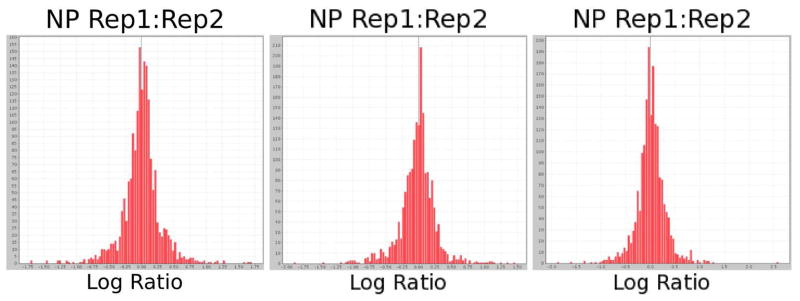
We chose to limit our attention to proteins with ratio ≤ 0.5 or ≥ 2.0, a threshold based on the replicate analysis described above. Figure 4 shows the distribution of (log-transformed) protein ratios calculated between two replicate analyses for each experimental group. In all three groups only 4% of proteins show a variation greater than twofold between the two replicate analyses. Many of those apparently differential proteins in the replicate comparison were identified with few peptides, potentially resulting in poor quantification. Therefore, our list of candidate proteins was further restricted to those proteins with at least two unique peptides observed. Proteins without q-values were considered, as these could be proteins with very low abundance in one experimental group but not another (and therefore insufficient in-common peptide identifications for a q-value calculation); these proteins were evaluated on a case-by-case basis. For proteins with q-values, however, only proteins with q-value ≤ 0.1 were considered. This combination of large effect size of observed dysregulation with low probability of observation by chance provided a list of candidates whose effects are likely to be observable and reproducible in subsequent experiments.
Figure 4.
Log-ratio distributions comparing abundance calculated using only replicate 1 vs. abundance calculated using only replicate 2 of each sample, for each of the three experimental groups. For all three sets of replicate experiments, 96% of the proteins display less than 2-fold change between the replicates.
In the P-HGD vs. NP comparison, 1,249 proteins had at least 2 unique peptides; 1,178 of these had quantitative ratios, of which 294 showed a twofold or greater change; of those changing proteins, 147 also had q-values, 126 of which were ≤ 0.1. In the P-NEG vs. NP comparison, 1,245 proteins had at least 2 unique peptides; 1,160 of these had quantitative ratios, of which 310 showed a twofold or greater change; of those changing proteins, 164 also had q-values, 144 of which were ≤ 0.1. Restricting the protein candidate list using the criteria described above, 303 and 273 differential proteins were identified in P-HGD and P-NEG, respectively. Of these proteins, 155 were observed as differentially expressed in both P-HGD and P-NEG, implying a possible correlation of proteome changes in a field effect of non-dysplasia tissues related to cancer. These findings provide the feasibility of detecting colon dysplasia or cancer using a surrogate biomarker in non-dysplasia rectal biopsies. When compared with the differential proteins identified in the colon tissues from our previous study10, the differential proteins identified in the rectal tissue from the current study using label-free proteomics provide a complementary pool of candidates for UC cancer biomarker development, especially rectal biomarker development. The differential proteins identified in this study are listed in supplemental Table 2.
Functional annotation of differential proteins in UC progressors
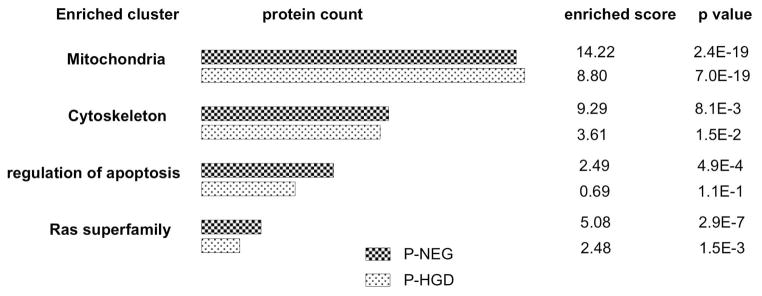
To identify and interpret the potential biological groups and pathways associated with UC neoplastic progression, the genes associated with the two lists of differential proteins in progressor groups (P-NEG and P-HGD) were uploaded separately to the DAVID online database for functional enrichment analysis24,25. These enrichment analyses were based on both up-regulated and down-regulated proteins. Selected enriched groups are presented in Figure 5 and discussed below. The top 10 clusters of the DAVID enrichment analysis are presented in supplemental Tables 3 and 4.
Figure 5.
Selected enriched clusters of the differential proteins in UC progressors. Functional enrichments were analyzed by the DAVID online database. A full list of enrichment clusters is presented in supplemental Tables 3 and 4.
Mitochondrial proteins
One of the groups most enriched for differentially expressed proteins was the mitochondrion. This group was highly enriched by 14.22 fold and 8.8 fold, and accounted for 24% and 28% of the entire list of differential proteins, for progressors P-NEG and P-HGD respectively. The majority of these mitochondrial proteins were mitochondrial inner membrane proteins (36 and 38 for P-NEG and P-HGD respectively), mitochondrial matrix proteins (23 and 19 for P-NEG and P-HGD respectively) and oxidoreductase (40 and 47 for P-NEG and P-HGD respectively). Proteins in the oxidative phosphorylation pathway, especially in Complexes I, III and VI were particularly enriched (15 and 17 for P-NEG and P-HGD respectively). In our previous iTRAQ-based quantitative proteomics study, we also discovered that mitochondrial proteins were enriched among the differentially expressed proteins10. The results from our current study consistently suggest the possible involvement of mitochondrial dysregulation in UC tumorigenesis, i.e., alterations in mitochondrial proteome may be associated with precursor lesions during UC neoplastic progression to cancer.
Cytoskeletal proteins
Cytoskeletal proteins are also enriched in UC progressors. There are 44 and 40 cytoskeletal proteins dysregulated in UC P-NEG and P-HGD respectively, or 9.29 fold and 3.6 fold enrichment respectively. Sixteen of these were actin cytoskeleton or/and actin binding proteins. The pathways involved regulation of actin cytoskeleton (8 proteins), tight junction pathway (6 proteins), and vascular smooth muscle contraction (5 proteins). The cytoskeleton is not simply a structural framework playing a role in cell shape and motile events. Recent studies show that the cytoskeleton also plays critical roles in the regulation of various cellular processes relating to proliferation, contact inhibition, anchorage-independent cell growth, and apoptosis26. Identification of cytoskeletal proteins associated with UC progressors suggests that alterations in the cytoskeleton might be important in UC neoplastic progression. Moreover, the observation of such cytoskeleton changes in non-dysplastic regions implies that cytoskeletal changes may be occurring at a very early stage of UC neoplastic progression, i.e., before cells become dysplastic.
RAS superfamily
Several members of RAS oncogene family were differentially expressed in UC progressors: 14 and 9 in P-NEG and P-HGD, enriched by 5.08 fold and 2.48 fold, respectively. The pathways included the MAPK signaling pathway, chemokine signaling pathway, focal adhesion pathway, VEGF signaling, and RAS signaling pathway. The RAS superfamily of GTPases has been implicated in the regulation of proliferation, cell migration, adhesion, apoptosis, and differentiation27. RAS signaling pathways are well known for their involvement in tumor initiation and cellular transformation. Alterations in the pathways of RAS superfamily in the non-dysplastic epithelium of UC progressors indicate that these epithelial cells are undergoing molecular events required for tumor progression even when they are still morphologically normal.
Apoptosis and regulation of apoptosis
Proteins relating to apoptosis and regulation of apoptosis were also enriched for differentially expressed proteins. Nineteen proteins involved in apoptosis and 32 proteins involved in regulation of apoptosis were differentially expressed in P-NEG, with 2.49-fold enrichment (p<0.05). In progressor P-HGD, 16 proteins involved in apoptosis and 22 proteins involved in regulation of apoptosis were differentially expressed, although the enrichment did not reach statistical significance (p=0.2). Apoptosis is a critical mechanism that allows multicellular organisms to maintain tissue integrity and function and to eliminate damaged or unwanted cells28. Alterations in proteins relating to and regulate apoptosis enable UC epithelial cells to eventually to evade programmed cell death, a trait that is critical in cancer development and progression.
Canonical pathway analysis of differential proteins in UC progressors
Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com) was used to further explore the well-defined canonical pathways involved in UC progressors. The whole datasets were imported into Ingenuity and 2-fold change was used as cut-off value for focus genes. The top five canonical pathways for UC P-NEG were: Mitochondrial Dysfunction (p=6.11E-11), Valine, Leucine and Isoleucine Degradation (p=1.4E-8), Fatty Acid Metabolism (p=2.14E-8), Fatty Acid Elongation in Mitochondria (p=9.38E-8), and Inositol Metabolism (p=2.15E-7). Among the 169 proteins (genes) in mitochondrial dysfunction pathway, 20 proteins (genes) were differentially expressed in UC P-NEG. Moreover, 19 of these 20 proteins (genes) were under-expressed. The top five canonical pathways for UC P-HGD were: Mitochondrial Dysfunction (p=3.93E-14), Oxidative Phosphorylation (p=9.36E-12), Valine, Leucine and Isoleucine Degradation (p=2.38E-8), Arginine and Proline Metabolism (p=1.14E-7), and Fatty Acid Metabolism (p=6.62E-7). Twenty-two proteins (genes) of the total 169 proteins (genes) in the mitochondrial dysfunction pathway were differentially expressed in P-HGD. All expect one protein were under-expressed. The canonical pathway analysis confirms that mitochondrial dysfunction is involved in UC neoplastic progression. In addition, metabolism (including energy, carbohydrate, lipid and amino acid metabolism) is also important in UC neoplastic progression. The pathway analysis results are available in supplemental Tables 5 and 6.
IHC analysis of selected differential proteins
Two mitochondrial proteins CPS1 and TRAP1 were chosen for IHC analysis based on their abundance in the analysis, as well as potential functional relevance to UC neoplastic progression. The current study and our previous study10 both suggest that mitochondrial dysfunction is involved in UC neoplastic progression. CPS1 is a mitochondrial enzyme involved in the urea cycle, and we have previously identified its overexpression in the random colon tissue from progressors. The current study again reveals its overexpression in the rectal tissue. TRAP1 has previously been showed to have a protective role for oxidative stress. Thus TRAP1 might have a functionally relevant role in ulcerative colitis.
CPS1 (carbamoyl-phosphate synthase 1)
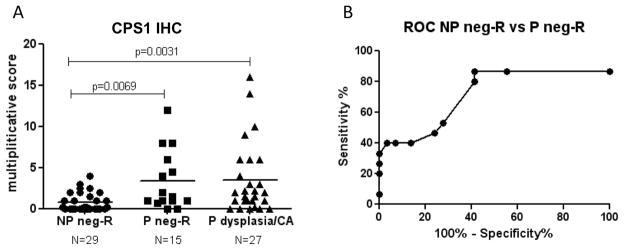
We have previously found up-regulation of CPS1 in the UC dysplasia and its surrounding non-dysplastic colon mucosa10. In the current study, CPS1 was found to be up-regulated in the non-dysplastic rectal tissues from progressors as well as in the dysplastic colon tissues. In individual samples, CPS1 was not observed by MS/MS in any of the five non-progressors, but it was observed in 7 out of the 10 progressors (5 P-NEG and 5 P-HGD). To evaluate the utility of CPS1 as a potential tissue marker for UC progression, IHC analysis was performed on a tissue microarray containing multiple tissues from 15 progressors and 30 non-progressors. As shown in Figure 6A, CPS1 staining in the progressors is significantly increased in the non-dysplastic rectal tissue (p=0.0069), as well as the dysplastic colon tissue (p=0.0031). Further ROC analysis suggested that CPS1 can distinguish non-dysplastic rectal tissues of progressors from rectal tissue from UC non-progressors with statistical significance (AUC=0.75, p=0.008, see Figure 6B). Rectal CPS1 staining could achieve 87% sensitivity and 45% specificity in predicting current dysplasia or cancer in the colon, demonstrating the feasibility of using surrogate biomarkers in rectal biopsies to identify dysplasia and/cancer in UC.
Figure 6.
IHC analysis of CPS1. A. CPS1 IHC scores in non-progressor non-dysplastic rectal tissues (NP-neg-R), progressor non-dysplastic rectal tissues (P neg-R), and progressor dysplastic or cancerous tissues (P dysplasia/CA). The IHC scores are available in supplemental Table 7. B. ROC analysis of CPS1 staining in rectal tissues of progressors and non-progressors.
TRAP1 (TNF receptor-associated protein 1)
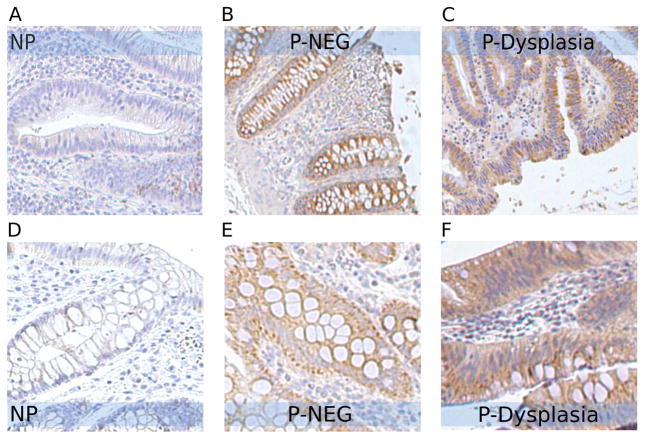
TRAP1 is a mitochondrial heat shock protein involved in protection against oxidative stress and apoptosis. In our label-free proteomic profiling study, it was increased by 2.41-fold in progressor P-NEG (q=0.02) and 1.74-fold in progressor P-HGD (q=0.03). In our MS/MS analysis, TRAP1 was observed in all 15 samples. It was observed in all 10 progressor samples with high spectral count, and in four of the five non-progressor samples with much lower counts. IHC analysis was then performed on tissue sections from 15 independent samples. Only one of five NP samples had moderate staining; the other four NP samples had no TRAP1 signal (Figure 7). In contrast, all 10 progressors (including 5 non-dysplastic tissues and 5 dysplastic tissues) exhibited moderate to strong TRAP1 staining. IHC data thus confirmed the proteomics finding that increased expression of TRAP1 is associated with UC neoplastic progression. UC mucosa constantly undergoes cycles of inflammation, resulting in extensive reactive oxygen species and causing oxidative stress in UC mucosa. The increased expression of TRAP1 in UC progressors detected in the current study is consistent with its protective role in cells under oxidative stress. TRAP1 might play a role in UC tumorigenesis. Other studies have also showed increased expression of TRAP1 in colorectal carcinomas29, in cisplatin-resistant ovarian tumors and ovarian carcinoma cell lines30, and in localized and metastatic prostate cancer31.
Figure 7.
IHC analysis of TRAP1 in UC non-progressors and progressors. Moderate to strong staining of TRAP1 was observed in non-dysplastic tissues (B and E) and dysplastic tissues (C and F) from progressors compared to the minimal staining in the non-dysplastic tissues from non-progressors (A and D).
Summary
Patients with ulcerative colitis would benefit immensely from objective molecular biomarkers of the development of colon cancer. In this study, we use label-free comparative proteomics to expand our earlier isotopically-labeled proteomics biomarker discovery work to develop biomarkers for UC dysplasia detection. A list of differential proteins associated with dysplasia in UC progressors was identified. Moreover, for the first time, a group of differential proteins in non-dysplastic rectal tissue from UC progressors was identified, providing important candidates for developing surrogate rectal biomarkers for predicting dysplasia or cancer in colon. Mitochondrial proteins, cytoskeletal proteins, RAS superfamily, proteins relating to apoptosis, and metabolism were the important protein pathways differentially expressed in the non-dysplastic and dysplastic tissues of UC progressors, suggesting their importance in UC neoplastic progression. One of the differential proteins, TRAP1, displayed increased IHC staining in both dysplastic and non-dysplastic tissues from UC progressors compared to UC non-progressors. Another differential protein, CPS1, also showed a statistically significant difference in IHC staining between the non-progressor and progressor groups. More interestingly, rectal CPS1 staining detected dysplasia or cancer in the colon with 87% sensitivity and 45% specificity, demonstrating the feasibility of using surrogate biomarkers in rectal biopsies to predict dysplasia and cancer in colon.
Supplementary Material
Acknowledgments
We thank Josephine Maurer, Allyn Stevens, and Yasuko Tamura for technical support and study coordination. This work was supported by grants from Crohn’s & Colitis Foundation (RC) and NIH NCI R01CA068124 (TB).
Footnotes
Supporting Information Available. Sample descriptions, a ranked list of all candidate proteins considered, a summary of our functional annotation clustering analysis and pathway analysis, and list of CPS1 IHC scores are provided as supporting information.
References
- 1.Ekbom A, Helmick C, Zack M, Adami HO. Ulcerative colitis and colorectal cancer. A population-based study. N Engl J Med. 1990;323(18):1228–33. doi: 10.1056/NEJM199011013231802. [DOI] [PubMed] [Google Scholar]
- 2.Prior P, Gyde SN, Macartney JC, Thompson H, Waterhouse JA, Allan RN. Cancer morbidity in ulcerative colitis. Gut. 1982;23(6):490–7. doi: 10.1136/gut.23.6.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubin CE, Haggitt RC, Burmer GC, Brentnall TA, Stevens AC, Levine DS, Dean PJ, Kimmey M, Perera DR, Rabinovitch PS. DNA aneuploidy in colonic biopsies predicts future development of dysplasia in ulcerative colitis. Gastroenterology. 1992;103(5):1611–20. doi: 10.1016/0016-5085(92)91185-7. [DOI] [PubMed] [Google Scholar]
- 4.Riddell RH, Goldman H, Ransohoff DF, Appelman HD, Fenoglio CM, Haggitt RC, Ahren C, Correa P, Hamilton SR, Morson BC, et al. Dysplasia in inflammatory bowel disease: standardized classification with provisional clinical applications. Hum Pathol. 1983;14(11):931–68. doi: 10.1016/s0046-8177(83)80175-0. [DOI] [PubMed] [Google Scholar]
- 5.America AH, Cordewener JH. Comparative LC-MS: a landscape of peaks and valleys. Proteomics. 2008;8(4):731–49. doi: 10.1002/pmic.200700694. [DOI] [PubMed] [Google Scholar]
- 6.Mueller LN, Brusniak MY, Mani DR, Aebersold R. An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J Proteome Res. 2008;7(1):51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- 7.Choi H, Fermin D, Nesvizhskii A. Significance Analysis of Spectral Count Data in Label-free Shotgun Proteomics. Mol Cell Proteomics. 2008;7(12):2373–85. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryu S, Gallis B, Goo Y, Shaffer SA, Radulovic D, Goodlett D. Comparison of a Label-Free Quantitative Proteomic Method Based on Peptide Ion Current Area to the Isotope Coded Affinity Tag Method. Cancer Inform. 2008;6:243–255. doi: 10.4137/cin.s385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Old W, Meyer-Arendt K, Aveline-Wolf L, Pierce K, Mendoza A, Sevinsky J, Resing K, Ahn N. Comparison of Label-free Methods for Quantifying Human Proteins by Shotgun Proteomics. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Brentnall TA, Pan S, Bronner MP, Crispin DA, Mirzaei H, Cooke K, Tamura Y, Nikolskaya T, Jebailey L, Goodlett DR, McIntosh M, Aebersold R, Rabinovitch PS, Chen R. Proteins That Underlie Neoplastic Progression of Ulcerative Colitis. Proteomics Clin Appl. 2009;3(11):1326–1337. doi: 10.1002/prca.200900061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabinovitch PS, Dziadon S, Brentnall TA, Emond MJ, Crispin DA, Haggitt RC, Bronner MP. Pancolonic chromosomal instability precedes dysplasia and cancer in ulcerative colitis. Cancer Res. 1999;59(20):5148–53. [PubMed] [Google Scholar]
- 12.Licklider LJ, Thoreen CC, Peng J, Gygi SP. Automation of nanoscale microcapillary liquid chromatography-tandem mass spectrometry with a vented column. Anal Chem. 2002;74:3076–3083. doi: 10.1021/ac025529o. [DOI] [PubMed] [Google Scholar]
- 13.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20(9):1466–7. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 14.Keller A, Eng JK, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Molecular Systems Biology. 2005 August; doi: 10.1038/msb4100024. EPub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 16.May D, Fitzgibbon M, Liu Y, Holzman T, Eng J, Kemp CJ, Whiteaker J, Paulovich A, McIntosh M. A Platform for Accurate Mass and Time Analyses of Mass Spectrometry Data. J Proteome Res. 2007 doi: 10.1021/pr070146y. [DOI] [PubMed] [Google Scholar]
- 17.Bellew M, Coram M, Fitzgibbon M, Igra M, Randolph T, Wang P, May D, Eng J, Fang R, Lin C, Chen J, Goodlett D, Whiteaker J, Paulovich A, McIntosh M. A suite of algorithms for the comprehensive analysis of complex protein mixtures using high-resolution LC-MS. Bioinformatics. 2006;22(15):1902–9. doi: 10.1093/bioinformatics/btl276. [DOI] [PubMed] [Google Scholar]
- 18.May D, Liu Y, Law W, Fitzgibbon M, Wang H, Hanash S, McIntosh M. Peptide Sequence Confidence in Accurate Mass and Time Analysis and Its Use in Complex Proteomics Experiments. J Proteome Res. 2008 doi: 10.1021/pr8004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang P, Tang H, Zhang H, Whiteaker J, Paulovich AG, McIntosh M. Normalization regarding non-random missing values in high-throughput mass spectrometry data. Proceedings of the Pacific Symposium on Biocomputing. 2006;11:315–326. [PubMed] [Google Scholar]
- 20.Deutsch EW, Mendoza A, Shteynberg D, Farrah T, Lam H, Tasman N, Sun Z, Nilsson E, Pratt B, Prazen B, Eng J, Martin D, Nesvizhskii A, Aebersold R. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10:1–10. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang Q, Strand A, Law W, Faca VM, Fitzgibbon MP, Hamel N, Houle B, Liu X, May DH, Poschmann G, Roy L, Stuhler K, Ying W, Zhang J, Zheng Z, Bergeron JJ, Hanash S, He F, Leavitt BR, Meyer HE, Qian X, McIntosh MW. Brain-specific proteins decline in the cerebrospinal fluid of humans with Huntington’s disease. Mol Cell Proteomics. 2008 doi: 10.1074/mcp.M800231-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel VJ, Thalassinos K, Slade SE, Connolly JB, Crombie A, Murrell JC, Scrivens JH. A comparison of labeling and label-free mass spectrometry-based proteomics approaches. J Proteome Res. 2009;8(7):3752–9. doi: 10.1021/pr900080y. [DOI] [PubMed] [Google Scholar]
- 24.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):3. [PubMed] [Google Scholar]
- 25.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Pawlak G, Helfman DM. Cytoskeletal changes in cell transformation and tumorigenesis. Curr Opin Genet Dev. 2001;11(1):41–7. doi: 10.1016/s0959-437x(00)00154-4. [DOI] [PubMed] [Google Scholar]
- 27.Goldfinger LE. Choose your own path: specificity in Ras GTPase signaling. Mol Biosyst. 2008;4(4):293–9. doi: 10.1039/b716887j. [DOI] [PubMed] [Google Scholar]
- 28.Ghobrial IM, Witzig TE, Adjei AA. Targeting apoptosis pathways in cancer therapy. CA Cancer J Clin. 2005;55(3):178–94. doi: 10.3322/canjclin.55.3.178. [DOI] [PubMed] [Google Scholar]
- 29.Costantino E, Maddalena F, Calise S, Piscazzi A, Tirino V, Fersini A, Ambrosi A, Neri V, Esposito F, Landriscina M. TRAP1, a novel mitochondrial chaperone responsible for multi-drug resistance and protection from apoptotis in human colorectal carcinoma cells. Cancer Lett. 2009;279(1):39–46. doi: 10.1016/j.canlet.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 30.Macleod K, Mullen P, Sewell J, Rabiasz G, Lawrie S, Miller E, Smyth JF, Langdon SP. Altered ErbB receptor signaling and gene expression in cisplatin-resistant ovarian cancer. Cancer Res. 2005;65(15):6789–800. doi: 10.1158/0008-5472.CAN-04-2684. [DOI] [PubMed] [Google Scholar]
- 31.Leav I, Plescia J, Goel HL, Li J, Jiang Z, Cohen RJ, Languino LR, Altieri DC. Cytoprotective mitochondrial chaperone TRAP-1 as a novel molecular target in localized and metastatic prostate cancer. Am J Pathol. 176(1):393–401. doi: 10.2353/ajpath.2010.090521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.