Abstract
The viral determinants of Alphavirus-induced rheumatic disease have not been elucidated. We identified an RRV strain (DC5692) which, in contrast to the T48 strain, does not induce musculoskeletal inflammation in a mouse model of RRV disease. Substitution of the RRV T48 strain nonstructural protein 1 (nsP1) coding sequence with that from strain DC5692 generated a virus that was attenuated in vivo despite similar viral loads in tissues. In contrast, substitution of the T48 PE2 coding region with the PE2 coding region from DC5692 resulted in attenuation in vivo and reduced viral loads in tissues. In gain of virulence experiments, substitution of the DC5692 strain nsP1 and PE2 coding regions with those from the T48 strain was sufficient to restore full virulence to the DC5692 strain. These findings indicate that determinants in both nsP1 and PE2 have critical and distinct roles in the pathogenesis of RRV-induced musculoskeletal inflammatory disease in mice.
Keywords: Alphavirus, pathogenesis, virulence, inflammation, rheumatic disease
Introduction
Ross River virus (RRV) is a positive-sense, single-stranded RNA virus in the Alphavirus genus of the family Togaviridae (Powers et al., 2001). RRV is among a group of related mosquito-transmitted alphaviruses, which includes chikungunya virus, Mayaro virus, o’nyong ‘nyong virus, and others, that typically cause a debilitating musculoskeletal inflammatory disease in humans. These viruses are an emerging disease threat due to their ability to cause explosive epidemics. Past epidemics include a 1979-to-1980 epidemic of RRV disease in the South Pacific that involved more than 60,000 patients (Harley, Sleigh, and Ritchie, 2001) and a 1959-to-1962 epidemic of o’nyong-nyong fever in Africa that involved at least 2 million infections (Williams, Woodall, and Gillett, 1965). Since 2004, chikungunya virus has caused major epidemics in multiple countries in the Indian Ocean region with estimates on the order of 1–6 million cases (Weaver and Reisen, 2010).
Clinical manifestations following infection with arthritis/myositis-associated alphaviruses develop following an incubation period ranging from 2–12 days (Harley, Sleigh, and Ritchie, 2001; Pialoux et al., 2007). The disease is most commonly characterized by fever, intense pain in the peripheral joints, myalgias, and an impaired ability to ambulate (Harley, Sleigh, and Ritchie, 2001; Pinheiro et al., 1981; Staples, Breiman, and Powers, 2009). A number of studies indicate that musculoskeletal pain lasts for months to years in a subset of persons infected with RRV or CHIKV, however, the cause of these long lasting symptoms is unclear (Borgherini et al., 2008; Brighton, Prozesky, and de la Harpe, 1983; Harley, Sleigh, and Ritchie, 2001; Larrieu et al., 2010; Simon et al., 2007; Sissoko et al., 2009). There are currently no licensed antivirals or vaccines for any of the arthritis/myositis-associated alphaviruses. Treatment is limited to supportive care with analgesics and anti-inflammatory drugs (Harley, Sleigh, and Ritchie, 2001; Pialoux et al., 2007).
To study the pathogenesis of arthritis/myositis-associated alphaviruses, we have developed a mouse model of RRV-induced disease, based on subcutaneous inoculation of 1000 pfu of the T48 strain of RRV into the footpad of 3–4 week old C57BL/6 mice, that recapitulates many aspects of the human disease (Morrison et al., 2007; Morrison, Simmons, and Heise, 2008; Morrison et al., 2006). Studies with the RRV mouse model demonstrated that, following a high titer serum viremia, bone/joint-associated tissues and skeletal muscle tissue are the primary targets of RRV replication (Morrison et al., 2006). RRV replication at these sites results in a severe inflammatory response with abundant tissue damage leading to deficits in grip strength and an altered gait. This model recapitulates many aspects of human RRV and CHIKV infection, including i) high titer serum viremia in people infected with RRV or CHIKV (Hoarau et al.; Parola et al., 2006), ii) the detection of RRV RNA and antigen in human joint tissue (Fraser et al., 1981; Soden et al., 2000), iii) the detection of CHIKV antigen in quadriceps muscle biopsies (Ozden et al., 2007), iv) the presence of mononuclear inflammatory infiltrates in joints and muscle tissue of RRV or CHIKV infected patients (Fraser et al., 1981; Hazelton, Hughes, and Aaskov, 1985; Hoarau et al.; Ozden et al., 2007), and v) the disease signs experienced by infected patients. Thus, understanding the host and viral factors that contribute to pathogenesis in this mouse model may aid the development of therapies and vaccines to treat or prevent human disease.
A number of studies have identified genomic regions, genes, or specific molecular determinants that contribute to Alphavirus neurovirulence (Aguilar et al., 2008; Bernard, Klimstra, and Johnston, 2000; Davis et al., 1991; Frolova et al., 2002; Gardner et al., 2009; Glasgow et al., 1994; Grieder et al., 1995; Kobiler et al., 1999; Powers et al., 2000; Russell, Dalrymple, and Johnston, 1989; Santagati et al., 1995; Schoepp and Johnston, 1993; Suthar et al., 2005; Tucker and Griffin, 1991; Tuittila et al., 2000; White et al., 2001). However, no studies have identified viral determinants important for Alphavirus-induced musculoskeletal inflammatory disease. Utilizing our RRV disease model, we report here the identification of a murine-attenuated RRV strain (DC5692). To identify genetic determinants associated with virulence, chimeric viruses composed of various portions of the mouse virulent T48 strain and the DC5692 strain were constructed and analyzed in vitro and in vivo. These studies identified critical, yet distinct, roles for determinants encoded in the nonstructural protein 1 (nsP1) and the PE2 coding regions in the pathogenesis of RRV-induced musculoskeletal inflammatory disease.
Results
RRV strain DC5692 and virus derived from a molecular clone of RRV strain DC5692 (RR87) replicate like the T48 strain in Vero cells, but do not cause musculoskeletal inflammatory disease in mice
We have developed a mouse model of RRV-induced rheumatic disease that has been utilized to identify host factors that promote inflammation and damage of musculoskeletal tissues following RRV infection (Lidbury et al., 2008; Morrison et al., 2007; Morrison, Simmons, and Heise, 2008; Morrison et al., 2006; Rulli et al., 2009). In contrast, the viral genetic determinants of Alphavirus-induced musculoskeletal inflammatory disease are unknown. In an effort to identify such determinants, we first sought to identify RRV strains that were defective for the induction of inflammatory disease in vivo, but which replicated like the virulent RR64 (virus derived from the molecular clone of the T48 strain) in vitro. This strategy was adopted to eliminate viruses that may be attenuated in vivo due to intrinsic replication differences in simple mammalian cell culture systems. Screening of multiple RRV isolates resulted in the identification of one candidate strain (RRV DC5692), which replicated with similar kinetics to RR64-derived virus in Vero cells (Fig.1A and 1B), but unlike RR64, DC5692 did not cause overt disease signs in infected mice (Fig. 2A). At 10 dpi, RR64-infected mice had severe inflammation and tissue damage in musculoskeletal tissues such as the quadriceps muscles (Fig. 2B). Similar to control mice, mice inoculated with 103 pfu of RRV DC5692 did not have evidence of inflammation or damage of quadriceps muscle tissue at 10 dpi (Fig. 2B).
Figure 1. Multi-step replication analysis.
(A) Schematic representations of the genomes of the T48 strain molecular clone RR64 (white), RRV strain DC5692 (grey), and the DC5692 molecular clone RR87. (B) Vero cells were inoculated with 0.01 pfu/cell of RR64, RRV strain DC5692, or RR87. At the times indicated, 100 μl of supernatant was harvested and virus titers were determined by plaque assay on BHK-21 cell monolayers. The dashed line indicates the limit of detection. No statistically significant differences were detected.
Figure 2. The RRV DC5692 strain and virus derived from a molecular clone of the DC5692 strain (RR87) are attenuated in vivo.
(A) Twenty-four-day-old C57BL/6J mice were inoculated with PBS (n = 3), or 103 PFU of RR64 (n = 4), RRV strain DC5692 (n = 5), or the molecular clone of DC5692, RR87 (n = 5) by injection in the left rear footpad. Mice were scored for the development of disease signs including loss of gripping ability, hind-limb weakness, and altered gait. Each data point represents the arithmetic mean ± standard deviation (SD). Data was evaluated for statistically significant differences by ANOVA (P < 0.0001) followed by Bonferroni’s Multiple Comparison Test. (B) At 10 dpi, mice were sacrificed and perfused with 4% paraformaldehyde. 5 micron-thick paraffin-embedded sections generated from quadriceps muscles were stained with H&E. Images are representative of three mice per group.
The fact that the DC5692 strain showed similar replication to the T48 strain in Vero cells, but a defect in the induction of virus-induced disease, suggested that this virus might be a useful tool for identifying critical viral determinants of T48 strain-induced musculoskeletal inflammatory disease. Therefore, as a first step in this process, we sought to create a full length molecular clone of DC5692 with the goal of using this clone to create chimeric viruses with RR64 to map viral virulence determinants. To establish this system, the complete genome sequence, except the 5′ untranslated region (UTR), of RRV strain DC5692 was determined (Genbank HM234643) and compared to the sequence of RR64. This analysis revealed a total of 356 nucleotide differences (96.8% identity) within the coding region of the genomes (nucleotides 79-11,330). As shown in Table 1, 47 of the 356 nucleotide changes resulted in an amino acid difference. Due to the existence of only 2 synonymous nucleotide differences within the first 200 nucleotides of the nsP1 coding sequence of these two RRV strains, a molecular clone that encodes nucleotides 272-11329 of RRV strain DC5692 (pRR87; Fig. 1A) was constructed as described in Materials and Methods. Infectious virus derived from the molecular clone of strain DC5692 (RR87) replicated like the biological isolate and RR64 in Vero cells (Fig. 1B). Furthermore, inoculation of mice with 103 pfu of RR87 did not result in detectable disease signs or evidence of inflammation of quadriceps muscle tissue (Fig. 2B).
Table 1.
Amino acid differences of RRV T48 (RR64) and RRV DC5692.
| nsP1 |
| S79C |
| A112S |
| L224I |
| C416F |
| S424N |
| L463I |
| nsP2 |
| R11K |
| A31T |
| E116D |
| M140T |
| T175A |
| S406T |
| R464K |
| nsP3 |
| I192V |
| S330P |
| T331M |
| V362L |
| V412I |
| K427T |
| A431T |
| A435del. |
| V461A |
| K494E |
| I510V |
| S518P |
| nsP4 |
| T7I |
| V29A |
| T89A |
| V99A |
| A359V |
| E399D |
| V438I |
| V459I |
| K514N |
| Capsid |
| K73Q |
| N84K |
| P89S |
| E3 |
| R59G |
| E2 |
| Y18H |
| I67M |
| H94R |
| R251K |
| H255Q |
| E302V |
| E1 |
| S120L |
| V304A |
| L413M |
A chimeric virus that encodes the RRV DC5692 3′ UTR in the genetic background of the T48 strain (RR67) caused disease in mice that was indistinguishable from RRV T48 (RR64)-induced disease
Sequence analysis revealed a number of differences in the nucleotide sequences of the RRV DC5692 3′ UTR compared to the 3′ UTR of the T48 strain (Fig. 3A). These differences include a 79 nucleotide deletion within the DC5692 3′ UTR, which eliminates most of repeat element 2, a 34 bp deletion, which eliminates a large region of repeat element 3, an 11 nucleotide deletion, and numerous other nucleotide changes. Thus, the RRV DC5692 strain 3′ UTR (not including the poly A tail) is 398 nucleotides in length with 2 intact repeat elements (1 and 4) whereas the 3′ UTR of the T48 strain is 523 nucleotides in length and encodes 4 repeat elements (1–4). To test whether the 3′ UTR of the T48 strain contributed to virulence in the mouse model, a chimeric virus encoding the RRV DC5692 strain 3′ UTR in the T48 strain genetic background (RR67) was generated (Fig 3B). Inoculation of 103 pfu of either RR64 or RR67 resulted in disease signs (Fig. 3C) and inflammation of quadriceps muscles (Fig. 3D) that were indistinguishable. These data, together with the data that indicated that the 5′ and 3′ UTRs of the T48 strain were not sufficient to enhance the virulence of the DC5692 strain (Fig. 2), suggest that the 3′ UTRs of these RRV strains do not encode major virulence determinants in this mouse model of inflammatory rheumatic disease.
Figure 3. A chimeric virus that encodes the RRV DC5692 3′UTR in the T48 genetic background (RR67) caused disease in mice that was indistinguishable from RRV T48-induced disease.
(A) Alignment of the 3′ UTR of the RRV strains DC5692 and T48. The underlined TAA (first row) is the E1 stop codon. The italicized and underlined sequences are the repeat elements present in the T48 3′ UTR. (B) Schematic representations of the T48 derived (white) and DC5692 derived (grey) sequence within the genomes of RR64 and RR67. (C) Twenty-four-day-old C57BL/6J mice were inoculated with 103 PFU of RR64 (n = 4), RRV strain DC5692 (n = 4), or RR67 (n = 4) by injection in the left rear footpad. Mice were scored for the development of disease signs including loss of gripping ability, hind-limb weakness, and altered gait. Each data point represents the arithmetic mean ± SD. No statistically significant differences were detected. (D) At 10 dpi, mice were sacrificed and perfused with 4% paraformaldehyde. 5 micron-thick paraffin-embedded sections generated from quadriceps muscles were stained with H&E. Images are representative of four mice per group.
Chimeric viruses that encode the RRV DC5692 nsP1 coding region in the T48 strain genetic background are attenuated in vivo
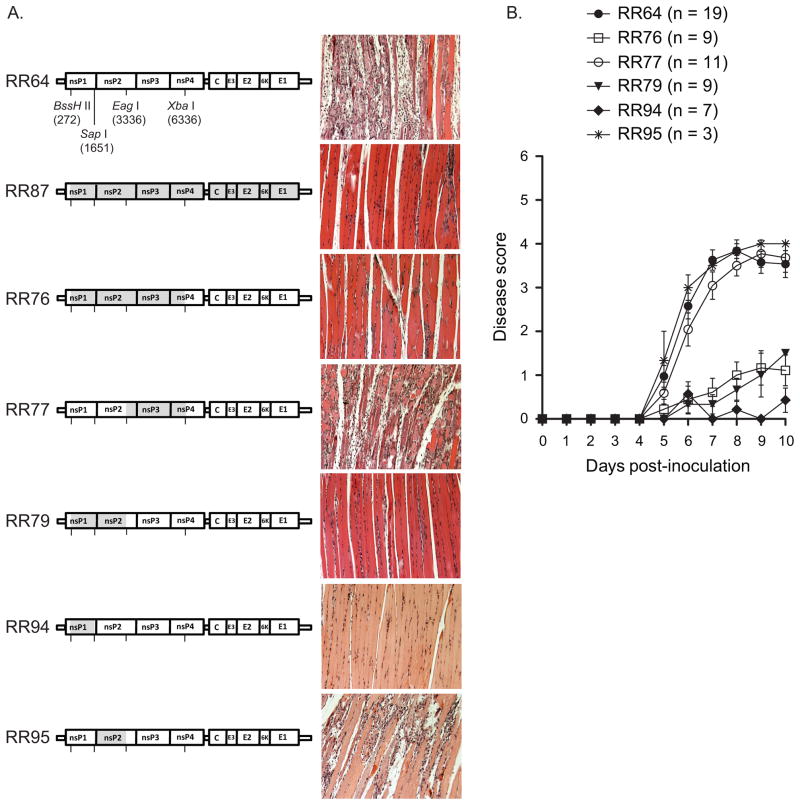
To test whether important virulence determinants of T48 strain-induced musculoskeletal inflammatory disease are encoded in the nonstructural region of the genome, a panel of chimeric virus cDNAs was constructed (Fig. 4). Full-length RNAs encoding the parental and chimeric virus genomes formed plaques with similar efficiency after transfection into BHK-21 cells (Fig. S1A), whereas all of the chimeric viruses that encode the nsP1 coding region from the DC5692 strain had a small plaque phenotype on BHK-21 cells compared to RR64 (Fig. S1B). Similar to virus-derived from the DC5692 molecular clone, RR87, inoculation of mice with RR76, which encodes nsP1, nsP2, nsP3, and the N-terminal portion of nsP4 from RRV DC5692 strain in the T48 strain genetic background resulted in severely diminished inflammation of quadriceps muscle tissue and disease signs compared to RR64 (Fig. 4A and 4B; P ≤ 0.05). To further map the determinants underlying this phenotype, we compared chimeras encoding either nsP1-nsP2 (RR79) or nsP3 and the N-terminal portion of nsP4 (RR77) from RRV DC5692 in the T48 genetic background. As shown in Fig. 4A and 4B, while RR77 elicited disease signs identical to the virulent RR64 virus, RR79 was significantly attenuated (P≤ 0.01). Additional chimeras encoding either nsP1 or nsP2 from the RRV DC5692 strain in the T48 strain genetic background (RR94 and RR95, respectively) demonstrated that the presence of the nsP1, but not the nsP2, coding region of the DC5692 strain resulted in diminished inflammation in skeletal muscle tissues and significantly less severe disease signs (Fig. 4A and 4B; P≤0.001). To investigate whether the attenuated phenotype of RR94 was due to inefficient processing of the chimeric P12 protein, we compared nonstructural protein processing kinetics of the parental viruses, RR64 and RR87, to RR94. As shown in Fig. S2, no differences in nonstructural protein processing were observed in vitro. Taken together, these data suggest that a determinant(s) in nsP1 plays a critical role in the pathogenesis of the musculoskeletal inflammatory disease caused by the mouse virulent T48 strain (RR64).
Figure 4. Chimeric viruses that encode the RRV DC5692 nsP1 coding region in the T48 strain genetic background are attenuated in vivo.
(A) Twenty-four-day-old C57BL/6J mice were inoculated with 103 PFU of RR64, RR76, RR79, RR77, RR94, or RR95 by injection in the left rear footpad. At 10 dpi, mice were sacrificed and perfused with 4% paraformaldehyde. 5 micron-thick paraffin-embedded sections generated from the quadriceps muscle were stained with H&E. Images are representative of three mice per group. (B) Twenty-four-day-old C57BL/6J mice were inoculated with 103 PFU of RR64 (n = 19), RR76 (n = 9), RR77 (n = 11), RR79 (n = 9), RR94 (n = 7), or RR95 (n = 3) by injection in the left rear footpad. Mice were scored for the development of disease signs including loss of gripping ability, hind-limb weakness, and altered gait. Each data point represents the arithmetic mean ± standard error of the mean (SEM). Data are combined from four independent experiments and were evaluated for statistically significant differences by ANOVA (P < 0.0001) followed by Bonferroni’s Multiple Comparison Test (see text).
Chimeric viruses that encode the RRV DC5692 PE2 coding region in the T48 genetic background are attenuated in vivo
While the data presented above demonstrated that determinants within nsP1 are required for the induction of inflammatory disease by the T48 strain (RR64), we also sought to assess the relative contribution of the viral structural genes to disease. Therefore, a second panel of chimeric viruses was constructed to test whether important virulence determinants of T48 strain-induced inflammatory disease are encoded in the structural region of the genome (Fig. 5). Full-length RNAs encoding these chimeric virus genomes formed plaques with similar efficiency compared to the T48 strain after transfection into BHK-21 cells (Fig. S1A). Interestingly, similar to the chimeric viruses encoding nsP1 from the DC5692 strain, all of the chimeric viruses that encode PE2 from the DC5692 strain had a small plaque phenotype on BHK-21 cells compared to the T48 strain (Fig. S1B). Inoculation of mice with RR73, which encodes the very C-terminus of E2, 6K, and E1 from the RRV DC5692 strain in the T48 strain genetic background resulted in musculoskeletal inflammation and disease signs indistinguishable from those observed following inoculation of RR64 (Fig. 5). In contrast, inoculation of mice with a chimera that encodes the C-terminal of nsP4, capsid, E3, and the majority of E2 from RRV DC5692 (RR78) or a chimera that encodes E3, E2, 6K, and E1 from RRV DC5692 (RR101) resulted in an attenuated phenotype (Fig. 5A and 5B; P ≤ 0.01). Taken together, these findings suggested that an important determinant(s) was located in the PE2 coding region. To confirm these finding, two additional chimeras, RR102 and RR100, were constructed. Inoculation of mice with RR102, which encodes portions of RRV DC5692 nsP4 and capsid in the T48 strain genetic background, resulted in musculoskeletal inflammation and disease signs indistinguishable from RR64-inoculated mice (Fig. 5A and 5B). Inoculation of mice with RR100, which encodes RRV DC5692 E3 and the majority of E2 in the genetic background of the T48 strain resulted in an attenuated phenotype (Fig. 5A and 5B; P≤0.05). Thus, all chimeras that encode the PE2 coding region of the RRV DC5692 strain in the T48 strain genetic background (RR78, RR101, and RR100) were attenuated when inoculated into mice. These findings suggest that a determinant(s) in the PE2 coding region also plays a critical role in the pathogenesis of RRV-induced musculoskeletal inflammatory disease.
Figure 5. Chimeric viruses that encode the RRV DC5692 PE2 coding region in the T48 genetic background are attenuated in vivo.
(A) Twenty-four-day-old C57BL/6J mice were inoculated with 103 PFU of RR64, RR73, RR78, RR101, RR102, or RR100 by injection in the left rear footpad. At 10 dpi, mice were sacrificed and perfused with 4% paraformaldehyde. 5 micron-thick paraffin-embedded sections generated from quadriceps muscles were stained with H&E. Images are representative of three mice per group. (B) Twenty-four-day-old C57BL/6J mice were inoculated with 103 PFU of RR64 (n = 10), RR73 (n = 7), RR78 (n = 4), RR101 (n = 3), RR102 (n = 3), or RR100 (n =7) by injection in the left rear footpad. Mice were scored for the development of disease signs including loss of gripping ability, hind-limb weakness, and altered gait. Each data point represents the arithmetic mean ± SEM. Data are combined from three independent experiments and were evaluated for statistically significant differences by ANOVA (P < 0.0001) followed by Bonferroni’s Multiple Comparison Test (see text).
Attenuating determinants in PE2, but not nsP1, regulate viral tissue titers in mice
To investigate whether the attenuation of RR94 and RR100 chimeric viruses in vivo was associated with differences in viral loads in tissues, mice were inoculated with RR64, RR87, RR94, or RR100 and the amounts of infectious virus present in serum, joint, and skeletal muscle tissues at 1, 3, and 5 dpi were quantified by plaque assays (Fig. 6). The viral loads in tissues of RR87-inoculated mice were significantly lower compared to RR64-inoculated mice at all time points examined (Fig. 6). Interestingly, the amounts of infectious virus in tissues of RR94-inoculated mice were similar to the amounts in tissues of RR64-inoculated mice, suggesting that a determinant(s) in RRV nsP1 regulates activation of host inflammatory responses in musculoskeletal tissues independent of affects on viral titers in these tissues (Fig. 6). In contrast, the pattern of tissue titers detected in mice inoculated with RR100 was very similar to that detected in mice inoculated with RR87, with the exception of serum titers at 3 dpi (Fig. 6). At 1 dpi, RR87 and RR100 titers were dramatically lower than RR64 and RR94 titers in serum, ankle joints, and quadriceps muscles. Whereas RR64 and RR94 peak viral titers occurred by 1 dpi, RR87 and RR100 tissue titers reached a lower peak by 3 dpi and then returned to lower levels by 5 dpi. These findings indicate that the attenuating determinant(s) in the RRV DC5692 PE2 coding region regulates efficient viral replication in tissues.
Figure 6. Attenuating determinants in PE2, but not nsP1, regulate viral tissue titers in mice.
Twenty-four-day-old C57BL/6J mice were inoculated with 103 PFU of RR64 (filled circles), RR87 (open circles), RR94 (filled squares), or RR100 (open squares) by injection in the left rear footpad. At 1, 3, and 5 dpi mice (n = 3–6) were sacrificed, blood was collected via cardiac puncture, and mice were perfused by intracardial injection with 1X PBS. Tissues were dissected, weighed, homogenized and the amount of infectious virus present was quantified by plaque assay on BHK-21 cells. Dashed lines indicate the limit of detection. *P≤0.05, **P≤0.01, ***P≤0.001 as determined by ANOVA.
A chimeric virus that encodes the RRV T48 strain nsP1 and PE2 coding regions in the RRV DC5692 genetic background caused disease in mice that was indistinguishable from RRV T48 (RR64)-induced disease
We next constructed a panel of chimeric viruses to test whether the nsP1 and PE2 coding regions derived from the T48 strain were sufficient to convert the DC5692 strain, which is highly attenuated in mice (Fig. 2), into a virus that causes musculoskeletal inflammatory disease (Fig. 7). Each of these chimeric virus genomes had similar RNA infectivities compared to the T48 strain (Fig. S1A). Inoculation of mice with RR106, which encodes the T48 nsP1 coding region in the DC5692 strain genetic background (the reciprocal virus to RR94), resulted in severely diminished inflammation of quadriceps muscle tissue and disease signs compared to RR64 (Fig. 7A and 7B; P ≤ 0.01). Similarly, inoculation of mice with RR108, which encodes the T48 PE2 coding region in the DC5692 strain genetic background (the reciprocal virus to RR100), resulted in an attenuated disease phenotype (Fig. 8A and 8B; P≤0.05). These results indicated that neither the T48 nsP1 nor PE2 coding regions alone were sufficient to enhance the virulence of the DC5692 strain to that of the T48 strain. Inoculation of mice with RR109, which encodes both the T48 nsP1 and PE2 coding regions in the DC5692 genetic background, resulted in inflammation of musculoskeletal tissue and disease signs indistinguishable from RR64-inoculated mice (Fig. 8A and 8B). Interestingly, although neither the T48 nsP1 (RR106) nor the PE2 coding regions alone (RR108) were sufficient to reverse the small plaque phenotype of the DC5692 strain (RR87) on BHK-21 cells, RR109 formed plaques on BHK-21 cells that were indistinguishable from those formed by the T48 strain (Fig. S1B). These findings indicate that both the T48 nsP1 and PE2 coding regions are necessary and sufficient to convert strain DC5692 into a virus that caused musculoskeletal inflammatory disease in mice indistinguishable from that caused by the T48 strain.
Figure 7. Substitution of the DC5692 strain nsP1 and PE2 coding regions with those from the T48 strain is sufficient to enhance the virulence of the DC5692 strain.
(A) Twenty-four-day-old C57BL/6J mice were inoculated with 103 PFU of RR64, RR106, RR108, or RR109 by injection in the left rear footpad. At 10 dpi, mice were sacrificed and perfused with 4% paraformaldehyde. 5 micron-thick paraffin-embedded sections generated from quadriceps muscles were stained with H&E. Images are representative of three mice per group. (B) Twenty-four-day-old C57BL/6J mice were inoculated with 103 PFU of RR64 (n = 10), RR106 (n = 5), RR108 (n = 7), or RR109 (n = 9) by injection in the left rear footpad. Mice were scored for the development of disease signs including loss of gripping ability, hind-limb weakness, and altered gait. Each data point represents the arithmetic mean ± SEM. Data are combined from three independent experiments and were evaluated for statistically significant differences by ANOVA (P < 0.0001) followed by Bonferroni’s Multiple Comparison Test (see text).
Discussion
A large number of studies, utilizing mouse models of Sindbis virus (SINV), Semliki Forest virus (SFV), Eastern equine encephalitis virus (EEEV), or Venezuelan equine encephalitis virus (VEEV) neuropathogenesis, have shed light on the genetic determinants of Alphavirus neurovirulence. In contrast, no studies have identified viral genetic determinants that promote Alphavirus-induced rheumatic disease. We identified a strain of RRV (RRV strain DC5692) that replicates like the mouse virulent T48 strain (RR64) in Vero cells but fails to initiate musculoskeletal inflammation following inoculation into mice. These findings suggested that RRV strain DC5692 could be used to identify RRV genetic determinants critical for the development of disease in the mouse model. Thus, we generated a molecular clone of RRV strain DC5692 and a panel of T48-DC5692 chimeric viruses and used them to identify specific genes associated with the musculoskeletal inflammatory disease.
The 3′ UTRs of Alphaviruses encode a number of sequence elements. These elements include a 19 nucleotide sequence adjacent to the poly A tail that is highly conserved among alphaviruses and functions in RNA replication (Strauss and Strauss, 1994). In addition, Alphavirus 3′ UTRs contain 40–60 nucleotide repeat sequence elements that vary in sequence and length among different alphaviruses (Ou, Trent, and Strauss, 1982). The biologic function of the repeat sequence elements is unclear, although recent evidence suggests these elements may regulate viral RNA deadenylation (Garneau et al., 2008). Alignment of the DC5692 and T48 genome sequences revealed a number of differences in the 3′ UTRs of these two viruses. Extensive variability in 3′ UTRs among RRV isolates has been previously reported (Faragher and Dalgarno, 1986), however, whether these differences impact the development of disease is not known. To test whether the differences in the 3′ UTRs contributed to pathogenesis in the mouse model, we generated a chimeric virus that encodes the DC5692 3′ UTR in the T48 strain genetic backbone (RR67). Inoculation of mice with RR67 resulted in disease indistinguishable from mice inoculated with the T48 strain, suggesting that the 3′ UTR of the DC5692 strain is not sufficient to attenuate T48 strain-induced disease in this model of musculoskeletal inflammatory disease. Furthermore, RR87, which is phenotypically indistinguishable in vitro and in vivo from the DC5692 biological isolate, encodes the complete DC5692 coding region bracketed by the 5′ and 3′ UTRs of the T48 strain. Our experiments indicated that replacing the 3′ UTR of the DC5692 strain with the T48 strain 3′ UTR did not result in a gain of virulence.
To investigate whether genes within the nonstructural coding region of the genome contributed to pathogenesis in the mouse model, a series of chimeric viruses was constructed and tested in vivo for their ability to cause disease. From these experiments, we discovered that substitution of the T48 nsP1coding region with the nsP1 coding region from strain DC5692, which differ by six amino acids, generated a virus (RR94) that failed to cause disease in mice. In contrast to RR94, chimeric viruses encoding DC5692 nsP2, nsP3, or various portions of nsP4 in the T48 strain genetic background caused disease like the T48 strain. Interestingly, viral loads of RR94 in serum and musculoskeletal tissues were indistinguishable from those in mice inoculated with the T48 strain (RR64) from 1-5 dpi, suggesting that a determinant(s) in nsP1 modulates the host inflammatory response independent of effects on RRV tissue titers during the initiation phase of the inflammatory disease in mice.
Alphavirus nsP1 is a replicase protein that functions in viral RNA replication and viral RNA capping (Strauss and Strauss, 1994). In studies of temperature sensitive (ts) mutants of SFV and SINV, only three ts mutations have been mapped to nsP1 and each of these mutants is specifically defective in minus-strand synthesis (Lulla et al., 2008; Wang, Sawicki, and Sawicki, 1991). In addition, nsP1 encodes guanine-7-methyltransferase and guanyltransferase activities which are essential for capping and cap methylation of viral genomic and subgenomic RNAs (Ahola and Kaariainen, 1995; Ahola et al., 1997; Mi et al., 1989; Scheidel, Durbin, and Stollar, 1989). The structure of nsP1 is unknown and deletion studies have failed to define specific enzymatic subdomains. However, specific mutations that affect the binding of enzymatic substrates have been located in the first 310 amino acids of nsP1 (Lulla et al., 2008). For example, two conserved acidic residues, D62/D88 in RRV nsP1, are essential for S-adenosyl-L-methionine binding and residues S23 and V302 may function in GTP binding (Lulla et al., 2008).
Previous studies have identified determinants within nsP1 critical for Alphavirus neurovirulence. In all alphaviruses, nsP1 contains 1–3 cysteine residues at positions 418-420 (SFV numbering) and the nsP1 protein of Semliki Forest virus (SFV) and Sindbis virus (SINV) is palmitoylated at these conserved cysteines (Ahola et al., 2000; Laakkonen, Ahola, and Kaariainen, 1996). Mutation of the SFV nsP1 palmitoylation site (CCC to AAA at position 418 to 420) generated a virus that was avirulent in mice (Ahola et al., 2000). Interestingly, the mouse attenuated RRV strain DC5692 encodes a phenylalanine instead of a cysteine at RRV nsP1 position 416 (C416F). However, in contrast to the T48-nsP15692 chimera (RR94), the SFV mutant had dramatically altered growth in BHK-21 cells and dramatically reduced tissue titers in mice at all time points examined (Ahola et al., 2000), suggesting there is a different mechanism of attenuation for RR94.
Our studies suggest that nsP1 plays a critical role in the initiation of musculoskeletal inflammation and are the first to directly implicate a nonstructural protein in the pathogenesis of Alphavirus-induced inflammatory rheumatic disease. The divergent in vivo phenotypes of RR64 and RR94, highly inflammatory vs. noninflammatory, respectively, suggest that a specific determinant(s) within nsP1 of the T48 strain may promote the severe host inflammatory response, independent of viral titers at the normal sites of inflammation. Comparison of the T48 and RRV DC5692 nsP1 sequences revealed a total of 36 nucleotide changes. Six of these changes resulted in an amino acid coding change (S79S, A112S, L224I, C416F, S424N, and L463I) (Table 1). Future experiments will assess whether one or a combination of these amino acid changes is responsible for the attenuated phenotype.
In this study, we also investigated whether determinants within the structural region of the genome contributed to the pathogenic versus nonpathogenic phenotypes of the T48 strain and the DC5692 strain in the mouse model. Our experiments identified a critical role for a determinant(s) in the PE2 coding region, as substitution of the T48 PE2 coding region with that from the DC5692 strain, which differ by 7 amino acids, resulted in an attenuated phenotype. In contrast to the RR94 chimera, tissue titers in mice inoculated with RR100 (which encodes the DC5692 PE2 coding region in the T48 strain genetic background), were dramatically altered compared to mice inoculated with the T48 strain. These findings support a model in which determinants within PE2 and nsP1 regulate distinct steps within the pathogenic sequence that leads to musculoskeletal inflammation and disease.
The mechanism(s) by which the DC5692 PE2 protein leads to attenuation will require further study. PE2 and the mature E2 form of the protein have a number of roles in the viral life cycle, including roles in virus attachment and entry, virion assembly, and budding. In addition, the mature E2 glycoprotein is the major target of neutralizing antibodies. A number of studies have implicated determinants in the E2 glycoprotein in Alphavirus neurovirulence. For example, alternating intracranial passage of a laboratory-adapted SINV in neonatal and weanling mice led to the selection of a SINV strain with increased neurovirulence (Griffin and Johnson, 1977). A glutamine to histidine change at E2 position 55, which enhances binding and entry into neurons as well as virus-induced apoptosis of neurons, was identified as the primary determinant associated with the neurovirulent phenotype (Lee et al., 2002; Tucker et al., 1997). It will be of interest to test whether the amino acid changes present in the DC5692 E2 protein regulate RRV binding and entry of specific cell types associated with disease, such as connective tissue fibroblasts, osteoblasts, myofibers, and/or macrophages. In other studies, serial passaging of SINV or VEEV in BHK-21 cells resulted in attenuating mutations associated with adaptation to use of heparan sulfate as a receptor by the acquisition of positively charged amino acid residues in E2 (Bernard, Klimstra, and Johnston, 2000; Davis et al., 1986). It remains to be formally tested whether the DC5692 PE2 protein confers enhanced binding to heparan sulfate.
To test whether the nsP1 and PE2 coding regions could increase the virulence of the DC5692 strain, we constructed an additional set of chimeric viruses. Our results indicated the neither the T48 nsP1 nor PE2 coding regions alone were sufficient to enhance the virulence of the DC5692 strain to that of the T48 strain. However, a chimeric virus that encodes both the T48 strain nsP1 and PE2 coding regions in the DC5692 genetic background caused disease in mice that was indistinguishable from that caused by the T48 strain. These findings provide further evidence that mutations in nsP1 and PE2 are critical determinants of RRV-induced musculosketelal inflammatory disease.
In summary, utilizing pathogenic and apathogenic strains of RRV and a chimeric virus approach, we have identified critical, yet distinct, roles for nsP1 and PE2 in the pathogenesis of RRV-induced rheumatic disease in mice. These are the first studies to identify viral gene regions that promote alphavirus-induced rheumatic disease. Future studies will investigate the specific determinants within nsP1 and PE2 that impact disease outcomes and the mechanism(s) by which these determinants regulate pathogenesis.
Materials and Methods
Viruses
The T48 stain of RRV was isolated from Aedes vigilax mosquitoes in Queensland, Australia. Prior to cDNA cloning, the virus was passaged 10 times in suckling mice followed by two passages on Vero cells. Thus, this strain may be adapted for enhanced virulence in mice. RRV strain DC5692 was isolated in 1995 from Aedes camptorhynchus mosquitoes at Dawesville Cut in the Peel region of Western Australia. The virus was passaged 1 time in C6/36 cells, 1 time in Vero cells, and 1 time in BHK-21 cells prior to cDNA cloning. Trizol reagent (Invitrogen) was added to an aliquot of infected BHK-21 cell supernatant and RRV DC5692 RNA was purified using a Mini RNA isolation kit (Invitrogen) according to the manufacturer’s instructions. RNA was reverse-transcribed and the RRV DC5692 genome was amplified in a series of overlapping fragments by PCR using primers based on the T48 strain sequence and Deep Vent DNA polymerase (NEB). Amplicons were cloned into pCR-Blunt using the Zero Blunt PCR cloning kit (Invitrogen). Forward and reverse sequencing of at least 2 independent amplicons per region was performed at the complete genome sequence , except the 5′ UTR, was determined (Genbank HM234643).
Viral stocks were generated from full-length and chimeric virus cDNAs as previously described (Morrison et al., 2006). Briefly, plasmids encoding virus cDNAs were linearized by digestion with Sac I. 5’-capped full-length RNA transcripts were generated in vitro using SP6-specific mMessage mMachine in vitro transcription kits (Ambion). Full-length transcripts were electroporated into BHK-21 cells using a Bio-Rad Gene pulser electroporator . Culture supernatants were harvested at 24 hours after electroporation, centrifuged for 20 min at 3,000 rpm, aliquoted, and stored at −80°C. Viral stocks were titrated by plaque assay on BHK-21 cells.
Construction of virus cDNAs
Plasmids encoding full-length and chimeric viral cDNAs are designated by the prefix "p". Infectious virus derived from the cDNA clone does not contain the prefix "p". Plasmid pRR64 (provided by Richard Kuhn, Purdue University) encodes the full-length cDNA of the T48 strain of RRV. To generate a chimeric virus cDNA that encodes the RRV DC5692 3’UTR in the T48 genetic background, a shuttle vector (pRR65) was created by religation of pRR64 that had been digested with Afl II and Psh AI followed by a klenow fill in reaction. This resulted in a plasmid with a unique Hind III site at RRV position 11329, which is at the junction of the E1 gene and the 3’UTR and a unique Sac I site at the 3’ end of the RRV poly A tail. The 3’UTR of RRV DC5692 was PCR amplified with a 3’ primer that engineered a Sac I restriction site downstream of the poly A tail. The RRV DC5692 3’UTR was inserted into the Hind III-Sac I site of pRR65 to create pRR66. The Xma I-Sac I fragment of pRR66 was inserted into the Xma I-Sac I site of pRR64 to generate pRR67 which encodes a chimeric virus genome composed of the RRV DC5692 3’UTR in the T48 genetic background.
To generate pRR77, the RRV DC5692 Eag I-XbaI fragment was inserted into the Eag I-Xba I site of pRR64. To generate plasmids pRR76 and pRR79, the RRV DC5692 BssH II-Eag I fragment was inserted into the BssH II-Eag I site of pRR77 and pRR64, respectively. To generate pRR94 and pRR95, pRR64 and pRR79 were digested with Eco RI and the ~8300 bp fragment was gel isolated and religated to generate pRR90 and pRR91, respectively. This eliminated nucleotides 6407-11884 of the RRV sequence and generated plasmids with a unique Sap I site just 20 bp upstream of the junction of the nsP1 and nsP2 coding regions. Next, the BssH II-Sap I fragments were swapped between pRR90 and pRR91 to create pRR92, which encodes RRV DC5692 strain sequence from BssH II-Sap I and T48 strain sequence from Sap I-Eag I, and pRR93, which encodes T48 strain sequence from BssH II-Sap I and RRV DC5692 strain sequence from Sap I-Eag I. Finally, the BssH II-Eag I fragment of pRR92 and pRR93 was inserted in the BssH II-Eag I site of pRR64 to create pRR94 and pRR95, respectively.
To generate plasmid pRR73, the RRV DC5692 Xma I-Hind III fragment was inserted into the Xma I-Hind III site of pRR65 to generate pRR71. The Hind III-Sac I fragment of pRR71was inserted into the Hind III-Sac I site of pRR64 to generate pRR72. Finally, the RRV DC5692 Rsr II-Xma I fragment was cloned into the Rsr II-Xma I site of pRR72 to generate pRR73. Plasmid pRR78 was generated by inserting the RRV DC5692 Xba I-Rsr II fragment into the Xba I-Rsr II site of pRR64. To generate plasmids pRR100, pRR101, and pRR102, pRR64 and pRR73 were digested with Age I, to remove the Age I-Age I fragment, and then religated at the Age I site to create pRR96 and pRR97, respectively. The RRV DC5692 Age I-Rsr II fragment was inserted into the Age I-Rsr II site pf pRR96 and pRR97 to yield pRR98 and pRR99, respectively. Next, the pRR64 Age I-Age I fragment was inserted into the Age I site of pRR98 and pRR99 to generate pRR100 and pRR101, respectively. The RRV DC5692 Age I-Age I fragment was inserted into pRR96 to yield pRR102. Comparison of the RR64 and DC5692 genome sequences revealed only 2 synonymous nucleotide differences within the first 200 nucleotides of the nsP1 coding sequence of these two RRV strains. Therefore, pRR87 encodes the entire coding region of RRV DC5692 except for the first 193 nucleotides (nucleotides 272–11329). To generate pRR87, the RRV DC5692 BssH II-Xba I fragment was inserted into pRR72 to create pRR82. Next, the RRV DC5692 Rsr II-Xma I fragment was inserted into pRR78 to create pRR85. Finally, The Xba I-Xma I fragment of pRR85 was inserted into pRR82 to create pRR87. To generate plasmid pRR106, pRR87 was digested with Eco RI and the ~8300 bp fragment was gel isolated and religated to generate pRR104. This eliminated nucleotides 6407-11884 of the RRV sequence and generated a plasmid with a unique Sap I site just 20 bp upstream of the junction of the nsP1 and nsP2 coding regions. Next, the BssH II-Sap I fragment from pRR64 was cloned into pRR104 to create pRR105, which encodes RRV T48 strain sequence from BssH II-Sap I and DC5692 strain sequence from Sap I-Eag I. The BssH II-Xba I fragment of pRR105 was inserted in the BssH II-Eag I site of pRR87 to create pRR106. To create plasmid pRR108 and pRR109, the Age I-Xma I fragment from pRR102 was inserted into the Rsr II-Xma I site of pRR87 to create pRR107. Finally, the Xba I-Xma I fragment of pRR107 was inserted into the Xba I-Xma I site of pRR87 or pR106 to create pRR108 and pRR109, respectively.
Cells
BHK-21 cells (ATCC CCL10) were grown in α-minimal essential medium (Gibco) supplemented with 10% bovine calf serum (Hyclone), 10% tryptose phosphate broth, penicillin and streptomycin, and 0.29 mg/ml L-glutamine. Vero cells (ATCC CCL81) were grown in DMEM/F12 (Gibco) supplemented with 10% fetal bovine serum (Lonza), non-essential amino acids (Gibco), sodium bicarbonate, penicillin and streptomycin, and 0.29 mg/ml L-glutamine.
RNA infectivity assays
BHK-21 cells were plated at 5.0 × 104 cells/well in 12-well dishes. Cells were transfected with 100 ng, 10 ng, 1 ng, and 0.1 ng of 5′-capped full-length virus RNA transcripts using Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen). At 1.5 hours post-transfection, growth media was removed and cell monolayers were overlaid with 0.5% immunodiffusion agarose. Forty hours posttransfection, cells were stained with neutral red, plaques were enumerated, and the PFU per microgram of RNA was determined.
Plaque Assays
BHK-21 cells were seeded into 6 well dishes. Growth medium was removed and cell monolayers were inoculated with serial 10-fold dilutions of virus-containing samples. Following a one hour adsorption at 37°C, cell monolayers were overlaid with 0.5% immunodiffusion agarose (MP Biomedicals) in medium for 38 to 40 h and then stained with neutral red (Sigma). Plaque sizes were measured (n = 25–30) and total plaques were enumerated to determine the number of PFU per milliliter of culture supernatants and mouse serum or PFU per gram of tissue.
In vitro replication
Vero cells were plated at 105 cells/well in 24-well dishes. Growth medium was removed, and wells were inoculated with virus in triplicate at a multiplicity of infection of 0.01 PFU/cell. Viruses were adsorbed to cells for 1 h at 37°C. Wells were then washed three times with 1 ml of room-temperature phosphate-buffered saline (PBS). One milliliter of growth medium was then added to each well, and cells were incubated at 37°C. For cumulative growth analysis, 100 μl samples of culture supernatants were removed at various times post-inoculation and an equal volume of fresh medium was added to maintain a constant volume within each well. Samples were frozen at −80°C until analysis by plaque assay on BHK-21 cells.
In vitro nonstructural protein processing
5′-capped full-length RNA transcripts were generated in vitro using SP6-specific mMessage mMachine in vitro transcription kits (Ambion). 0.64 μg of each viral RNA was added to a 100-μl reticulocyte lysate reaction supplemented with [35S]-methionine (Heise et al., 2003). Reaction mixtures were also supplemented with KCl to a final concentration of 0.05 M, and then incubated at 30°C for 40 min, at which point the chase was initiated by adding unlabeled methionine to a final concentration of 1 mM and cycloheximide at a final concentration of 0.6 mg/ml. Samples were then incubated at 37°C for the duration of the chase. Five-microliter samples were removed at 0, 20, 40, 60, and 80 min into the chase and placed into 20 μl of gel loading buffer. Samples were heat denatured at 95°C for 5 min. and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide). Gels were analyzed on a phosphorimager.
Mouse experiments
C57BL/6J were obtained from The Jackson Laboratory (Bar Harbor, Maine) and bred in house. Animal husbandry and experiments were performed in accordance with all University of Colorado School of Medicine Institutional Animal Care and Use Committee guidelines. Although RRV is classified as a biosafety level 2 pathogen, due to its exotic nature all mouse studies were performed in a biosafety level 3 laboratory. Twenty-four day old mice were used for all in vivo studies. Mice were inoculated in the left rear footpad with 10 3 pfu of virus in diluent (PBS/1% bovine calf serum) in a 10 μl volume. Mock-infected animals received diluent alone. RRV inoculation resulted in 0% mortality. Mice were monitored for disease signs and weighed at 24 hour intervals. Disease scores were determined by assessing grip strength, hind limb weakness, and altered gait as previously described using the following system: 1 = very mild deficit in hind paw gripping ability of injected foot only; 2 = very mild deficit in bilateral hind paw gripping ability; 3 = bilateral loss of gripping ability, mild bilateral hind limb paresis; altered gait not readily observable; 4 = Bilateral loss of gripping ability, moderate bilateral hind limb paresis, observable altered gait; difficulty righting self; 5 = bilateral loss of gripping ability, severe bilateral hind limb paresis, altered gait, unable to right self; 6 = moribund. Consistent with previous studies which indicated that there is little to no evidence of virus-induced pathology in the central nervous system in this model, no RRV-inoculated mouse displayed evidence of paralysis, no mouse developed a disease score > 5, and no mouse was euthanized based on the criteria of weight loss or a moribund state during the course of these studies.
To determine viral titers in tissues, mice were sacrificed by exsanguinations, blood was collected, and mice were perfused by intracardial injection of 1x PBS. The right and left ankles and right and left quadriceps muscles were removed by dissection and weighed. Tissues were homogenized in 1x PBS supplemented with 1% bovine calf serum, Ca2+, and Mg2+ with a MagNA Lyser (Roche) and stored at −80°C until the amount of virus present was quantified by a standard plaque assay on BHK-21 cells.
Histological analysis
At the times indicated, mice were sacrificed and perfused by intracardial injection of 4% paraformaldehyde, pH 7.3. Tissues were embedded in paraffin and 5-μm sections were prepared. To assess histopathological changes such as tissue inflammation and damage, tissue sections were stained with hematoxylin and eosin (H&E) and evaluated by light microscopy.
Statistical analysis
Viral replication and tissue titers were evaluated for statistically significant differences using either the Student's t test or one-way analysis of variance (ANOVA) followed by Dunn's multiple comparison test. Disease scores were evaluated for statistically significant differences by repeated measures ANOVA followed by Bonferroni’s multiple comparison test.
Supplementary Material
Acknowledgments
This research was supported by NIH research grant K22 AI079163 awarded to T.E.M. and R01 AR047190 awarded to M.T.H. We thank Drs. Cheryl Johansen and David Smith (University of Western Australia) who were instrumental in the isolation of RRV strain DC5692 from mosquitos.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguilar PV, Adams AP, Wang E, Kang W, Carrara AS, Anishchenko M, Frolov I, Weaver SC. Structural and nonstructural protein genome regions of eastern equine encephalitis virus are determinants of interferon sensitivity and murine virulence. J Virol. 2008;82(10):4920–30. doi: 10.1128/JVI.02514-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahola T, Kaariainen L. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc Natl Acad Sci U S A. 1995;92(2):507–11. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahola T, Kujala P, Tuittila M, Blom T, Laakkonen P, Hinkkanen A, Auvinen P. Effects of palmitoylation of replicase protein nsP1 on alphavirus infection. J Virol. 2000;74(15):6725–33. doi: 10.1128/jvi.74.15.6725-6733.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahola T, Laakkonen P, Vihinen H, Kaariainen L. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J Virol. 1997;71(1):392–7. doi: 10.1128/jvi.71.1.392-397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard KA, Klimstra WB, Johnston RE. Mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus confer heparan sulfate interaction, low morbidity, and rapid clearance from blood of mice. Virology. 2000;276(1):93–103. doi: 10.1006/viro.2000.0546. [DOI] [PubMed] [Google Scholar]
- Borgherini G, Poubeau P, Jossaume A, Gouix A, Cotte L, Michault A, Arvin-Berod C, Paganin F. Persistent arthralgia associated with chikungunya virus: a study of 88 adult patients on reunion island. Clin Infect Dis. 2008;47(4):469–75. doi: 10.1086/590003. [DOI] [PubMed] [Google Scholar]
- Brighton SW, Prozesky OW, de la Harpe AL. Chikungunya virus infection. A retrospective study of 107 cases. S Afr Med J. 1983;63(9):313–5. [PubMed] [Google Scholar]
- Davis NL, Fuller FJ, Dougherty WG, Olmsted RA, Johnston RE. A single nucleotide change in the E2 glycoprotein gene of Sindbis virus affects penetration rate in cell culture and virulence in neonatal mice. Proc Natl Acad Sci U S A. 1986;83(18):6771–5. doi: 10.1073/pnas.83.18.6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis NL, Powell N, Greenwald GF, Willis LV, Johnson BJ, Smith JF, Johnston RE. Attenuating mutations in the E2 glycoprotein gene of Venezuelan equine encephalitis virus: construction of single and multiple mutants in a full-length cDNA clone. Virology. 1991;183(1):20–31. doi: 10.1016/0042-6822(91)90114-q. [DOI] [PubMed] [Google Scholar]
- Faragher SG, Dalgarno L. Regions of conservation and divergence in the 3′ untranslated sequences of genomic RNA from Ross River virus isolates. J Mol Biol. 1986;190(2):141–8. doi: 10.1016/0022-2836(86)90287-1. [DOI] [PubMed] [Google Scholar]
- Fraser JR, Cunningham AL, Clarris BJ, Aaskov JG, Leach R. Cytology of synovial effusions in epidemic polyarthritis. Aust N Z J Med. 1981;11(2):168–73. doi: 10.1111/j.1445-5994.1981.tb04226.x. [DOI] [PubMed] [Google Scholar]
- Frolova EI, Fayzulin RZ, Cook SH, Griffin DE, Rice CM, Frolov I. Roles of nonstructural protein nsP2 and Alpha/Beta interferons in determining the outcome of Sindbis virus infection. J Virol. 2002;76(22):11254–64. doi: 10.1128/JVI.76.22.11254-11264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner CL, Yin J, Burke CW, Klimstra WB, Ryman KD. Type I interferon induction is correlated with attenuation of a South American eastern equine encephalitis virus strain in mice. Virology. 2009;390(2):338–47. doi: 10.1016/j.virol.2009.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Sokoloski KJ, Opyrchal M, Neff CP, Wilusz CJ, Wilusz J. The 3′ untranslated region of sindbis virus represses deadenylation of viral transcripts in mosquito and Mammalian cells. J Virol. 2008;82(2):880–92. doi: 10.1128/JVI.01205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow GM, Killen HM, Liljestrom P, Sheahan BJ, Atkins GJ. A single amino acid change in the E2 spike protein of a virulent strain of Semliki Forest virus attenuates pathogenicity. J Gen Virol. 1994;75 ( Pt 3):663–8. doi: 10.1099/0022-1317-75-3-663. [DOI] [PubMed] [Google Scholar]
- Grieder FB, Davis NL, Aronson JF, Charles PC, Sellon DC, Suzuki K, Johnston RE. Specific restrictions in the progression of Venezuelan equine encephalitis virus-induced disease resulting from single amino acid changes in the glycoproteins. Virology. 1995;206(2):994–1006. doi: 10.1006/viro.1995.1022. [DOI] [PubMed] [Google Scholar]
- Griffin DE, Johnson RT. Role of the immune response in recovery from Sindbis virus encephalitis in mice. J Immunol. 1977;118(3):1070–5. [PubMed] [Google Scholar]
- Harley D, Sleigh A, Ritchie S. Ross River virus transmission, infection, and disease: a cross-disciplinary review. Clin Microbiol Rev. 2001;14(4):909–32. doi: 10.1128/CMR.14.4.909-932.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelton RA, Hughes C, Aaskov JG. The inflammatory response in the synovium of a patient with Ross River arbovirus infection. Aust N Z J Med. 1985;15(3):336–9. doi: 10.1111/j.1445-5994.1985.tb04048.x. [DOI] [PubMed] [Google Scholar]
- Heise MT, White LJ, Simpson DA, Leonard C, Bernard KA, Meeker RB, Johnston RE. An attenuating mutation in nsP1 of the Sindbis-group virus S.A.AR86 accelerates nonstructural protein processing and up-regulates viral 26S RNA synthesis. J Virol. 2003;77(2):1149–56. doi: 10.1128/JVI.77.2.1149-1156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoarau JJ, Jaffar Bandjee MC, Trotot PK, Das T, Li-Pat-Yuen G, Dassa B, Denizot M, Guichard E, Ribera A, Henni T, Tallet F, Moiton MP, Gauzere BA, Bruniquet S, Jaffar Bandjee Z, Morbidelli P, Martigny G, Jolivet M, Gay F, Grandadam M, Tolou H, Vieillard V, Debre P, Autran B, Gasque P. Persistent chronic inflammation and infection by Chikungunya arthritogenic alphavirus in spite of a robust host immune response. J Immunol. 184(10):5914–27. doi: 10.4049/jimmunol.0900255. [DOI] [PubMed] [Google Scholar]
- Kobiler D, Rice CM, Brodie C, Shahar A, Dubuisson J, Halevy M, Lustig S. A single nucleotide change in the 5′ noncoding region of Sindbis virus confers neurovirulence in rats. J Virol. 1999;73(12):10440–6. doi: 10.1128/jvi.73.12.10440-10446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laakkonen P, Ahola T, Kaariainen L. The effects of palmitoylation on membrane association of Semliki forest virus RNA capping enzyme. J Biol Chem. 1996;271(45):28567–71. doi: 10.1074/jbc.271.45.28567. [DOI] [PubMed] [Google Scholar]
- Larrieu S, Pouderoux N, Pistone T, Filleul L, Receveur MC, Sissoko D, Ezzedine K, Malvy D. Factors associated with persistence of arthralgia among Chikungunya virus-infected travellers: report of 42 French cases. J Clin Virol. 2010;47(1):85–8. doi: 10.1016/j.jcv.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Lee P, Knight R, Smit JM, Wilschut J, Griffin DE. A single mutation in the E2 glycoprotein important for neurovirulence influences binding of sindbis virus to neuroblastoma cells. J Virol. 2002;76(12):6302–10. doi: 10.1128/JVI.76.12.6302-6310.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidbury BA, Rulli NE, Suhrbier A, Smith PN, McColl SR, Cunningham AL, Tarkowski A, van Rooijen N, Fraser RJ, Mahalingam S. Macrophage-derived proinflammatory factors contribute to the development of arthritis and myositis after infection with an arthrogenic alphavirus. J Infect Dis. 2008;197(11):1585–93. doi: 10.1086/587841. [DOI] [PubMed] [Google Scholar]
- Lulla V, Sawicki DL, Sawicki SG, Lulla A, Merits A, Ahola T. Molecular defects caused by temperature-sensitive mutations in Semliki Forest virus nsP1. J Virol. 2008;82 (18):9236–44. doi: 10.1128/JVI.00711-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Durbin R, Huang HV, Rice CM, Stollar V. Association of the Sindbis virus RNA methyltransferase activity with the nonstructural protein nsP1. Virology. 1989;170(2):385–91. doi: 10.1016/0042-6822(89)90429-7. [DOI] [PubMed] [Google Scholar]
- Morrison TE, Fraser RJ, Smith PN, Mahalingam S, Heise MT. Complement contributes to inflammatory tissue destruction in a mouse model of Ross River virus-induced disease. J Virol. 2007;81(10):5132–43. doi: 10.1128/JVI.02799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison TE, Simmons JD, Heise MT. Complement receptor 3 promotes severe ross river virus-induced disease. J Virol. 2008;82(22):11263–72. doi: 10.1128/JVI.01352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison TE, Whitmore AC, Shabman RS, Lidbury BA, Mahalingam S, Heise MT. Characterization of Ross River virus tropism and virus-induced inflammation in a mouse model of viral arthritis and myositis. J Virol. 2006;80(2):737–49. doi: 10.1128/JVI.80.2.737-749.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou JH, Trent DW, Strauss JH. The 3'-non-coding regions of alphavirus RNAs contain repeating sequences. J Mol Biol. 1982;156(4):719–30. doi: 10.1016/0022-2836(82)90138-3. [DOI] [PubMed] [Google Scholar]
- Ozden S, Huerre M, Riviere JP, Coffey LL, Afonso PV, Mouly V, de Monredon J, Roger JC, El Amrani M, Yvin JL, Jaffar MC, Frenkiel MP, Sourisseau M, Schwartz O, Butler-Browne G, Despres P, Gessain A, Ceccaldi PE. Human muscle satellite cells as targets of Chikungunya virus infection. PLoS One. 2007;2(6):e527. doi: 10.1371/journal.pone.0000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parola P, de Lamballerie X, Jourdan J, Rovery C, Vaillant V, Minodier P, Brouqui P, Flahault A, Raoult D, Charrel RN. Novel chikungunya virus variant in travelers returning from Indian Ocean islands. Emerg Infect Dis. 2006;12(10):1493–9. doi: 10.3201/eid1210.060610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pialoux G, Gauzere BA, Jaureguiberry S, Strobel M. Chikungunya, an epidemic arbovirosis. Lancet Infect Dis. 2007;7(5):319–27. doi: 10.1016/S1473-3099(07)70107-X. [DOI] [PubMed] [Google Scholar]
- Pinheiro FP, Freitas RB, Travassos da Rosa JF, Gabbay YB, Mello WA, LeDuc JW. An outbreak of Mayaro virus disease in Belterra, Brazil. I. Clinical and virological findings. Am J Trop Med Hyg. 1981;30(3):674–81. doi: 10.4269/ajtmh.1981.30.674. [DOI] [PubMed] [Google Scholar]
- Powers AM, Brault AC, Kinney RM, Weaver SC. The use of chimeric Venezuelan equine encephalitis viruses as an approach for the molecular identification of natural virulence determinants. J Virol. 2000;74(9):4258–63. doi: 10.1128/jvi.74.9.4258-4263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers AM, Brault AC, Shirako Y, Strauss EG, Kang W, Strauss JH, Weaver SC. Evolutionary relationships and systematics of the alphaviruses. J Virol. 2001;75 (21):10118–31. doi: 10.1128/JVI.75.21.10118-10131.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulli NE, Guglielmotti A, Mangano G, Rolph MS, Apicella C, Zaid A, Suhrbier A, Mahalingam S. Amelioration of alphavirus-induced arthritis and myositis in a mouse model by treatment with bindarit, an inhibitor of monocyte chemotactic proteins. Arthritis Rheum. 2009;60(8):2513–23. doi: 10.1002/art.24682. [DOI] [PubMed] [Google Scholar]
- Russell DL, Dalrymple JM, Johnston RE. Sindbis virus mutations which coordinately affect glycoprotein processing, penetration, and virulence in mice. J Virol. 1989;63 (4):1619–29. doi: 10.1128/jvi.63.4.1619-1629.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagati MG, Maatta JA, Itaranta PV, Salmi AA, Hinkkanen AE. The Semliki Forest virus E2 gene as a virulence determinant. J Gen Virol. 1995;76 ( Pt 1):47–52. doi: 10.1099/0022-1317-76-1-47. [DOI] [PubMed] [Google Scholar]
- Scheidel LM, Durbin RK, Stollar V. SVLM21, a Sindbis virus mutant resistant to methionine deprivation, encodes an altered methyltransferase. Virology. 1989;173(2):408–14. doi: 10.1016/0042-6822(89)90553-9. [DOI] [PubMed] [Google Scholar]
- Schoepp RJ, Johnston RE. Directed mutagenesis of a Sindbis virus pathogenesis site. Virology. 1993;193(1):149–59. doi: 10.1006/viro.1993.1111. [DOI] [PubMed] [Google Scholar]
- Simon F, Parola P, Grandadam M, Fourcade S, Oliver M, Brouqui P, Hance P, Kraemer P, Ali Mohamed A, de Lamballerie X, Charrel R, Tolou H. Chikungunya infection: an emerging rheumatism among travelers returned from Indian Ocean islands. Report of 47 cases. Medicine (Baltimore) 2007;86(3):123–37. doi: 10.1097/MD/0b013e31806010a5. [DOI] [PubMed] [Google Scholar]
- Sissoko D, Malvy D, Ezzedine K, Renault P, Moscetti F, Ledrans M, Pierre V. Post-Epidemic Chikungunya Disease on Reunion Island: Course of Rheumatic Manifestations and Associated Factors over a 15-Month Period. PLoS Negl Trop Dis. 2009;3 (3):e389. doi: 10.1371/journal.pntd.0000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soden M, Vasudevan H, Roberts B, Coelen R, Hamlin G, Vasudevan S, La Brooy J. Detection of viral ribonucleic acid and histologic analysis of inflamed synovium in Ross River virus infection. Arthritis Rheum. 2000;43(2):365–9. doi: 10.1002/1529-0131(200002)43:2<365::AID-ANR16>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Staples JE, Breiman RF, Powers AM. Chikungunya fever: an epidemiological review of a re-emerging infectious disease. Clin Infect Dis. 2009;49(6):942–8. doi: 10.1086/605496. [DOI] [PubMed] [Google Scholar]
- Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58(3):491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthar MS, Shabman R, Madric K, Lambeth C, Heise MT. Identification of adult mouse neurovirulence determinants of the Sindbis virus strain AR86. J Virol. 2005;79(7):4219–28. doi: 10.1128/JVI.79.7.4219-4228.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker PC, Griffin DE. Mechanism of altered Sindbis virus neurovirulence associated with a single-amino-acid change in the E2 Glycoprotein. J Virol. 1991;65(3):1551–7. doi: 10.1128/jvi.65.3.1551-1557.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker PC, Lee SH, Bui N, Martinie D, Griffin DE. Amino acid changes in the Sindbis virus E2 glycoprotein that increase neurovirulence improve entry into neuroblastoma cells. J Virol. 1997;71(8):6106–12. doi: 10.1128/jvi.71.8.6106-6112.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuittila MT, Santagati MG, Roytta M, Maatta JA, Hinkkanen AE. Replicase complex genes of Semliki Forest virus confer lethal neurovirulence. J Virol. 2000;74 (10):4579–89. doi: 10.1128/jvi.74.10.4579-4589.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YF, Sawicki SG, Sawicki DL. Sindbis virus nsP1 functions in negative-strand RNA synthesis. J Virol. 1991;65(2):985–8. doi: 10.1128/jvi.65.2.985-988.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Reisen WK. Present and future arboviral threats. Antiviral Res. 2010;85 (2):328–45. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White LJ, Wang JG, Davis NL, Johnston RE. Role of alpha/beta interferon in Venezuelan equine encephalitis virus pathogenesis: effect of an attenuating mutation in the 5′ untranslated region. J Virol. 2001;75(8):3706–18. doi: 10.1128/JVI.75.8.3706-3718.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MC, Woodall JP, Gillett JD. O’nyong-Nyong Fever: An Epidemic Virus Diesease in East Africa. Vii. Virus Isolations from Man and Serological Studies up to July 1961. Trans R Soc Trop Med Hyg. 1965;59:186–97. doi: 10.1016/0035-9203(65)90080-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.