Abstract
In this study we report a novel use for the iTRAQ® reagent combined with a peptide mass inclusion list to enhance the signal of low-abundance proteins during analysis by mass spectrometry. C-tagged-SH-EGFR was retrovirally-transduced into two mutant lung cancer cell lines (HCC827 and PC9) and the core protein complexes enriched by tandem affinity purification. Tryptically-digested peptides were derivatised with iTRAQ® and analysed by higher-energy collision-induced dissociation mass spectrometry. The data revealed that UBS3B is a member of the EGFR core complex in the HCC827 cell line, that was not apparent by standard, unbiased one-dimensional shotgun analysis and collision-induced dissociation. The expression level of UBS3B, however, was 6 to 10 times lower than that observed in the PC9 cell line. Thus, using iTRAQ® in this fashion allows the identification of low-abundance interactors when combined with samples where the same protein has a higher abundance. Ultimately, this approach may uncover proteins that were previously unknown or only suspected as members of core protein complexes.
Keywords: EGFR, iTRAQ®, acid elution, strep-HA TAP, orbitrap, shotgun
Introduction
The combination of affinity purification with mass spectrometric-based proteomics is a powerful methodology that enables the discernment of proteins involved in forming stable, non-covalent functional complexes. Large-scale comprehensive protein-protein interaction networks of entire organisms;1–4 disease-associated proteins5 and pathways6–9 have shown the value of combining affinity purification with mass spectrometry to deepen our understanding of systems biology. On a more focussed level, TAP combined with LCMS has resulted in the successful charting of selected molecular machines.10–13 Such studies primarily utilised a gel-based LCMS strategy. Thus the majority of the abundant bait protein was physically separated by SDS-PAGE from low-abundance interacting proteins, thereby increasing the chances of detecting the weaker proteins by mass spectrometry. The introduction of the streptavidin-haemagglutinin-(SH)-TAP tag8 combined with one-dimensional shotgun LCMS markedly improved the TAP-LCMS workflow with respect to quantitity of required input material and throughput speed. A drawback of this approach, however, is that peptides from any abundant bait protein can potentially mask the selection, fragmentation and subsequent identification of peptides from low-abundance interacting proteins. If there was a means by which: (i) the signal intensity of peptides from low-abundance proteins could be enhanced in the mass spectrometer; and/or (ii) the selection of abundant bait peptides could be reduced; such a combined methodology has the potential to uncover previously unknown, low-abundance interacting proteins.
The EGFR tyrosine kinase can drive numerous downstream signalling pathways responsible for cancer growth and survival. Inhibition of EGFR by the small molecule inhibitor erlotinib can extend survival in patients with advanced lung cancer refractory to chemotherapy.14 Activating somatic mutations in EGFR enhance receptor signalling and predict sensitivity to tyrosine kinase inhibitors (TKI’s, e.g., erlotinib and gefitinib14–16) that target EGFR. Unfortunately resistance mechanisms such as secondary mutations in EGFR or MET amplification can rapidly lead to drug resistance and tumor growth.17–19
Mutant EGFR leads to constitutive activation of downstream ERK, PI3K/Akt, and STAT signalling resulting in ‘oncogene addiction’ and tumour cell growth and survival.20 Mutant EGFR proteins not only possess constitutive kinase activity but are also known to be defective in internalisation leading to increased membrane signalling.21–23 Understanding the process specific to mutant EGFR proteins could offer novel therapeutic strategies that may overcome resistance mechanisms. A global understanding of this process, however, is daunting. Numerous mechanisms have been proposed in different cell types, but most of these studies examined wild type EGFR proteins. Redundancy or robustness of these mechanisms is still unclear.
A deeper and more thorough understanding of the molecular network of EGFR could lead to novel therapeutic targets and/or strategies. One of the major strengths of such a network approach is the ability to identify proteins whose expression level, or complex formation with other proteins, affects survival of cells with mutant EGFR. Importantly, this ‘EGFR network’ can be considered as a target for therapeutic intervention. The distinction here is that rather than targeting EGFR only, the focus would be on targeting the network of proteins used by EGFR to activate downstream signalling. This network view also produces insights into the redundancy of mechanisms that allow the maintenance of EGFR signalling, thereby enabling approaches that can disable these mechanisms and result in EGFR degradation. Overall, such knowledge has the potential to lead to therapeutic approaches to attack both sensitive and resistant forms of EGFR resulting in cell death and regression in patients.
In this study, SH-tagged versions of mutant EGFR were expressed in two lung cancer cell lines via retroviral transduction. A tandem affinity purification strategy coupled to LCMS was then used to identify the EGFR core protein complexes (Figure 1). To compare the complexes from the two distinct cell lines, we developed an iTRAQ®-based strategy to identify low-abundance peptides that were not observed using an unbiased, one-dimensional shotgun top 6 collision-induced dissociation (1D-SG-CID) data-dependent acquisition. Normally, the iTRAQ® reagent is used to determine the relative quantities of a protein between samples.24 Here we show a novel use of iTRAQ® to identify a low-abundance protein (UBS3A) when combined with a sample where the same protein is present in higher quantities. This approach enhances the weaker peptide signals in the mass spectrometer due to the additive effect from the more abundant signals. Analysis of the iTRAQ® reporter ions indicated that indeed UBS3A was present in both the HCC827 and PC9 cell lines, but in markedly reduced quantities in the former and thus not observed by standard 1D-SG-CID mass spectrometry. Combining this with a peptide m/z plus retention time inclusion list, we also show preferential selection of certain peptides thereby alleviating the dynamic range issue of abundant bait proteins. This approach offers the possibility of not only quantitatively comparing data between TAP samples, but also to identify low-abundance proteins that are normally not observed by conventional mass spectrometric methods.
Figure 1.
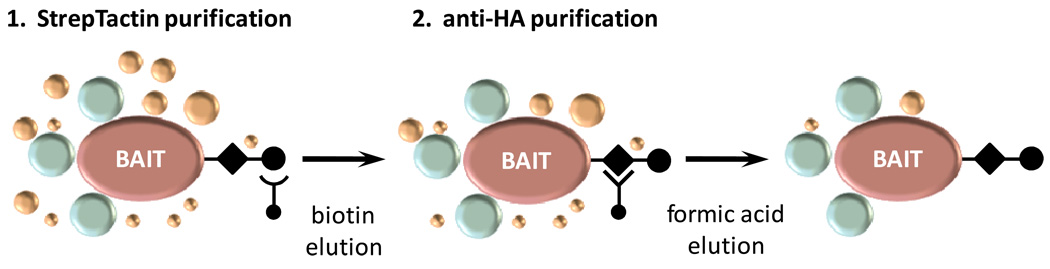
Schematic representation of the tandem affinity purification (TAP) procedure (adapted from Gstaiger et al.8). HCC827 or PC9 cells expressing SH-tagged proteins were lysed and purified from total protein extracts using streptavidin sepharose (StrepTactin beads). After several wash steps, tagged proteins were eluted with 2.5 mM d-biotin for subsequent immunoaffinity purification using anti-HA agarose. After further wash steps, protein complexes were eluted with 100 mM formic acid (pH 2.5) and processed for mass spectrometric analysis.
Material and Methods
Materials
Iodoacetamide, dithiothreitol, 1 M triethylammonium bicarbonate (TEAB), protease inhibitor cocktail, anti-HA agarose, polybrene (SIGMA-Aldrich, St. Louis, MO); trypsin (Promega Corp., Madison, WI); formic acid (HCOOH) (MERCK, Darmstadt, Germany); Strep-Tactin sepharose (IBA TAGnologies, Götingen, Germany); d-biotin (Alfa Aesar, Karlsruhe, Germany); 4-plex iTRAQ® reagent (ABI, Framingham, MA); micro Bio-Spin chromatography columns (Bio-Rad, Hercules, CA); Gateway LR Clonase™ II Enzyme Mix Kit, foetal bovine serum, lipofectamine 2000 (Invitrogen, Carlsbad, CA).
Retroviral Infection
cDNA for EGFR Del was provided by Dr. William Pao (Vanderbilt University, Nashville, TN). The design of PCR primers, amplification and pENTR TOPO cloning of EGFR Del were performed as previously described.25 EGFR Del was inserted into pfMSCV-C-SH IRES GFP gateway vector from pENTR TOPO vector by Gateway LR Clonase™ II Enzyme Mix Kit. The retroviral expression clone was verified by DNA sequencing using an Applied Biosystems 3130X1 Genetic analyser (HITACHI) with data analysis performed using Lasergene software V7.2. Phoenix HEK293 cells were obtained from ATCC (Manassas, VA) and grown in DMEM medium containing 10% foetal bovine serum (FBS). On day one, 8 × 105 Phoenix cells per well were seeded in a 6-well plate. On day two, cells were transfected with 3 µg VSV-G and 5 µg retroviral plasmids using lipofectamine 2000. Six hours after transfection, the supernatant was replaced with 2 mL DMEM 20% FBS and the cells incubated in a 5% CO2 incubator at 32°C for 48 h. The supernatant (viruses) was collected by centrifugation at 4°C; either stored at −80°C, or used immediately to infect the target cells. PC9 and HCC827 cells were maintained in RPMI-1640 medium supplemented with 10% FBS. For retroviral transduction, 2 × 105 cells per well were seeded in a 6-well plate. After overnight incubation, cells were infected with 800 µL of the virus supernatant plus 6 µg/mL polybrene for 24 h and then supplemented with 4 mL media per well. Cells were grown continuously until cell sorting. One week after infection, GFP-positive cells were sorted by FACSVantage from BD Biosciences (San Diego, CA). GFP positivity and HA expression were assessed by flow cytometry and immunoblot, respectively, before expanding the cells to 10 × 15 cm dishes. When approximately 90% confluent, the EGFR Del-tagged cells were washed with ice-cold PBS containing 1 mM sodium orthovanadate and scraped with a cell lifter on ice. Two cell pellets (each consisting of 5 × 15 cm dishes) were collected in 15 mL conical tubes by centrifugation at 129 × g at 4°C and stored at −80°C until required.
SH-Tandem-Affinity Purification (adapted from Gstaiger et al.).8
EGFR- and GFP-expressing PC9 or HCC827 cells were lysed in TNN-HS buffer (50 mM HEPES pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 50 mM NaF, 1.5 mM Na3VO4, 1.0 mM PMSF and protease inhibitor cocktail). Insoluble material was removed by centrifugation at 39,443 × g for 15 min at 4°C. 200 µL StrepTactin sepharose (400 µL slurry/pulldown) was transferred to a 14 mL dust-free Falcon tube and washed with 2 × 1 mL TNN-HS buffer. The lysates (approximately 50 mg total protein from 5 × 15 cm plates) were added to the washed StrepTactin sepharose and rotated for 20 min at 4°C. The sepharose beads and supernatant were transferred to a spin column and gravity drained. The sepharose was washed with 4 × 1 mL TNN-HS buffer, and the bound proteins eluted with 3 × 300 µL freshly-prepared 2.5 mM d-biotin in TNN-HS buffer into a fresh dust-free 1.5 mL eppendorf tube. 100 µL anti-HA agarose beads (200 µL slurry/pulldown) were transferred into a 1.5 mL eppendorf tube, washed with 1 × 1 mL TNN-HS buffer, and centrifuged at 200 × g for 1 min at 4°C. The supernatant was removed and the agarose resuspended in 100 µL TNN-HS buffer. The anti-HA agarose beads were added to the biotin eluate and rotated for 1 h at 4°C. The samples were centrifuged at 200 × g for 1 min at 4°C and the supernatant removed. The agarose beads were resuspended in 1 mL TNN-HS buffer, the washed beads and buffer loaded into a fresh dust-free Biospin column and gravity drained. The anti-HA agarose was washed with 3 × 1 mL TNN-HS buffer and then with 2 × 1 mL TNN-HS buffer consisting of only HEPES, NaCl and EDTA. Retained proteins were eluted from the column directly into a glass HPLC vial with 500 µL 100 mM HCOOH and immediately neutralised with 125 µL 1 M TEAB.26 200 µL were removed for silver-stain one-dimensional gel electrophoresis and/or immunoblot analysis as required. The remaining sample was frozen at −20°C until further processing.
Solution Tryptic Digestion and iTRAQ® Derivatisation
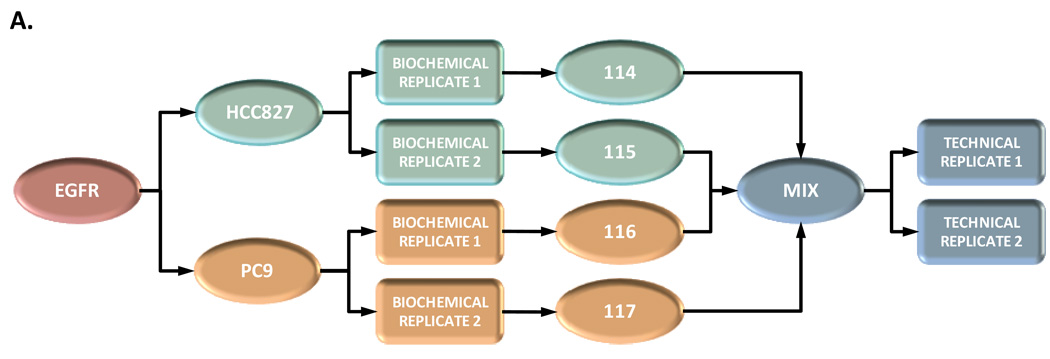
Proteins from the TEAB-neutralised acid eluate were reduced with dithiothreitol, alkylated with iodoacetamide and digested with trypsin. Multiples of 3% of the total eluate volume were desalted and concentrated with customised reversed-phase stage tips.27 The volume of the eluted sample was reduced to approximately 2 µL in a vacuum centrifuge and reconstituted to 8 µL with 5% formic acid and multiples thereof. Tryptically-digested samples were derivatised with 4-plex iTRAQ® reagent24 according to the instructions provided. The biochemical replicates of the strep-HA tandem affinity purifications of EGFR from the HCC827 and PC9 cell lines were labelled with iTRAQ® 114 and 115; and 116 and 117, respectively (Figure 2A).
Figure 2.
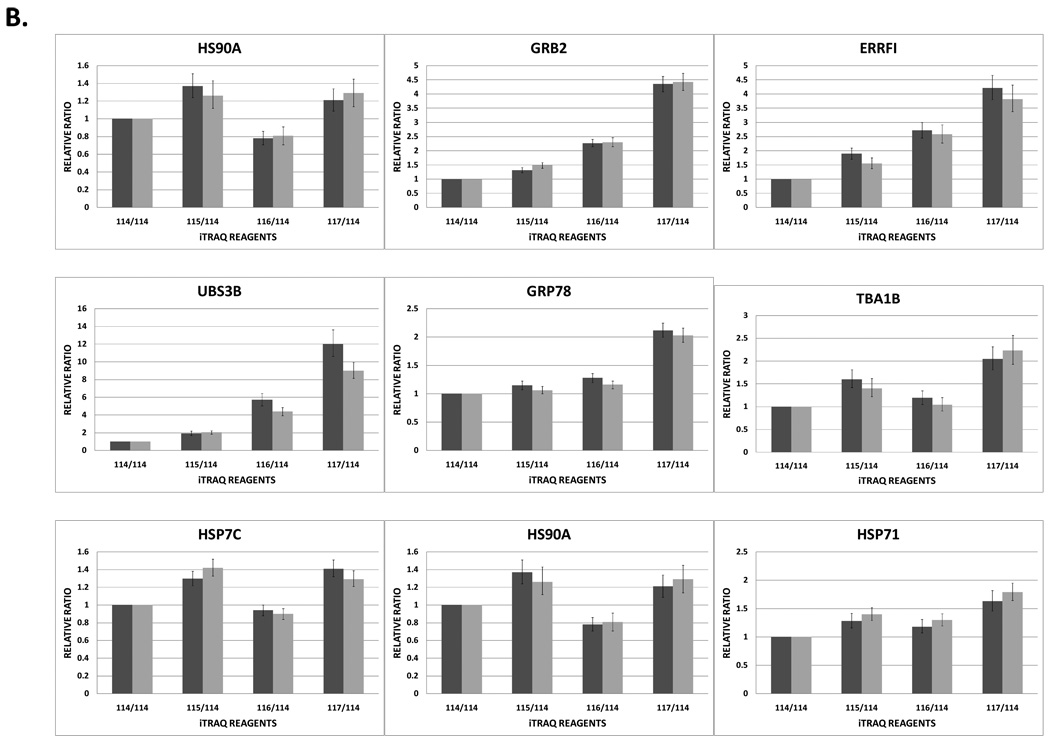
Relative iTRAQ® ratios for the four iTRAQ®-labelled and pooled TAP experiments analysed as technical replicates by higher-energy collision-induced dissociation (HCD). Mass spectrometry method (C), i.e., global inclusion list plus the most intense ions, was used for the analyses. (A) Schematic overview of the iTRAQ® labelling strategy. The biochemical replicates of the EGFR TAP performed from the HCC827 and PC9 cell lines are labelled with 114, 115 and 116, 117 reagents, respectively. (B) Relative quantitation of EGFR and eight interacting proteins. The 114:114 ratio (always equal to one) is included for clarity.
Liquid Chromatography and Mass Spectrometry
Mass spectrometry was performed on a hybrid LTQ-Orbitrap XL mass spectrometer (ThermoFisher Scientific, Waltham, MA) using the Xcalibur version 2.0.7 coupled to an Agilent 1200 HPLC nanoflow system (dual pump system with one precolumn and one analytical column) (Agilent Biotechnologies, Palo Alto, CA) via a nanoelectrospray ion source using liquid junction (Proxeon, Odense, Denmark). Solvents for LCMS separation of the digested samples were as follows: solvent A consisted of 0.4% formic acid in water and solvent B consisted of 0.4% formic acid in 70% methanol and 20% isopropanol. From a thermostatted microautosampler, 8 µL of the tryptic peptide mixture were automatically loaded onto a trap column (Zorbax 300SB-C18 5 µm, 5 × 0.3 mm, Agilent Biotechnologies, Palo Alto, CA) with a binary pump at a flow rate of 45 µL/min. 0.1% TFA was used for loading and washing the pre-column. After washing, the peptides were eluted by back-flushing onto a 16 cm fused silica analytical column with an inner diameter of 50 µm packed with C18 reversed phase material (ReproSil-Pur 120 C18-AQ, 3 µm, Dr. Maisch GmbH, Ammerbuch-Entringen, Germany). The peptides were eluted from the analytical column with a 27 minute gradient ranging from 3 to 30 percent solvent B, followed by a 25 minute gradient from 30 to 70 percent solvent B and, finally, a 7 minute gradient from 70 to 100 percent solvent B at a constant flow rate of 100 nL/min. The analyses were performed in a data-dependent acquisition mode using a top 6 collision-induced dissociation (CID) method for peptide identification alone; or a top 4 high-energy collision-induced dissociation (HCD) method for peptide identification plus relative quantitation of iTRAQ® reporter ions. Dynamic exclusion for selected ions was 60 seconds. No lock masses were employed. Maximal ion accumulation time allowed on the LTQ Orbitrap in CID mode was 150 ms for MSn in the LTQ and 1,000 ms in the C-trap. Automatic gain control was used to prevent overfilling of the ion traps and were set to 5,000 (CID) in MSn mode for the LTQ, 106 ions for a full FTMS scan and 105 ions for HCD. Maximum ion time for HCD was set to 1,000 ms for acquiring 1 microscan at a resolution of 7,500. Injection waveforms were activated for both LTQ and Orbitrap. Intact peptides were detected in the Orbitrap at 100,000 resolution for CID fragmentation and 60,000 for HCD fragmentation experiments. The threshold for switching from MS to MSMS was 2000 counts.
Generation of a Global Inclusion List
The iTRAQ®-labelled biochemical replicate of the EGFR complex purified from the PC9 cell line was analysed as a 1D shotgun experiment by CID. A global m/z precursor ion list was generated from the identified proteins. The list was comprised of 53 tryptic peptides plus retention time (Tr). A window of approximately ± 30 s was set for each Tr. To activate targeted analysis, the MS method parameters were as follows. Preview mode was enabled, the precursor ion mass width was ±5 ppm relative to the mass, and the use of m/z as masses was deselected. If a precursor mass on the inclusion list is not observed, the mass spectrometer selects the four ions with highest intensity for fragmentation.
Data Analysis
The acquired data were processed with Bioworks v3.3.1 SP1 (ThermoFisher, Waltham, MA, USA), dta files merged with an internally-developed program, and searched against the human SwissProt database version v57.4 (34,579 sequences, including isoforms as obtained from varsplic.pl) with the search engines MASCOT (v2.2.03, MatrixScience, London, UK) and Phenyx (v2.5.14, GeneBio, Geneva, Switzerland). 28 Submission to the search engines was via a Perl script that performs an initial search with relatively broad mass tolerances (MASCOT only) on both the precursor and fragment ions (±10 ppm and ±0.6 Da, respectively). High-confidence peptide identifications are used to recalibrate all precursor and fragment ion masses prior to a second search with narrower mass tolerances (±4 ppm and ±0.3 Da for CID and ±4 ppm and ±0.025 Da for HCD, respectively). One missed tryptic cleavage site was allowed. Carbamidomethyl cysteine and iTRAQ® 4-plex (N-terminii and lysine) were set as fixed modifications, and oxidised methionine was set as a variable modification. To validate the proteins, MASCOT and Phenyx output files were processed by internally-developed parsers. For MASCOT, two unique peptides with an ion score >18 (plus additional peptides from proteins fulfilling the criteria with an ion score >10) are required. For Phenyx, two unique peptides with a z-score >4.5 and a P-value <0.001 are required (plus additional peptides from proteins fulfilling the criteria with a z-score >3.5 and a P-value <0.001).
The validated proteins retrieved by the two algorithms are merged, any spectral conflicts discarded and grouped according to shared peptides. A false positive detection rate (FDR) of <0.25 % and <0.1 % (including the peptides exported with lower scores) was determined for proteins and peptides, respectively, by applying the same procedure against a reversed database. Comparisons between analytical methods involved comparisons between the corresponding sets of identified proteins. This was achieved by an internally-developed program that simultaneously computes the protein groups in all samples and extracts statistical data such as the number of distinct peptides, number of spectra, and sequence coverage.
iTRAQ® Relative Quantitation
Identified peptides that are unique to a specific protein were used to determine relative quantitation of a protein between the four samples. The intensities of the iTRAQ® reporter ion m/z values (114.1112, 115.1083, 116.1116 and 117.1150) for these peptides were extracted from the centroided data in the merged.mgf peak lists. The computation and analysis of the relative iTRAQ® ratios were performed with an internally-developed Perl script. The data was corrected for reagent impurity (according to the values supplied by the manufacturer). By using a sliding window approach, noise levels were calculated relative to the retention time and subtracted. Only iTRAQ® reporter ion intensities > 2000 counts were considered for the calculation of the ratios. For the final relative quantitation of a protein, the median ratio of the relative intensities of the reporter ions of all (specific) spectra for a protein are used.
Results and Discussion
One-Dimensional Shotgun Analysis of C-tagged-SH-EGFR
Mutant EGFR protein complexes from two cell lines were examined in this study. The PC9 cell line harbours the E746-A750 EGFR deletion mutant and has 5 copies per cell of the EGFR gene, whilst the HCC827 cell line harbours the E746-A750 EGFR deletion mutant and has 34 copies per cell of the EGFR gene.29 Both cells show preferential amplification of mutant EGFR alleles over wild type EGFR. Taking into consideration the differences between the endogenous EGFR expression levels, it was hypothesised that a certain proportion of the EGFR core complex proteins in the HCC827 cells could be potentially sequestered from the tagged EGFR by endogenous EGFR. The tryptic digests of the acid-eluted and neutralised protein complexes purified from N-terminally-tagged green fluorescent protein (GFP) and C-terminally-tagged EGFR were analysed by standard, unbiased 1D-SG-CID LCMS.
Analysis of the EGFR complexes purified from the PC9 lung cancer cells resulted in the identification of 15 protein groups (n = 4, Table 1). This was following subtraction of the proteins observed from the GFP purification in the same cell line and removal of non-specifically interacting proteins. As is usually observed with SH purifications, the bait (EGFR) was the most abundant protein with a maximum sequence coverage of 54% (61 unique peptides identified from 3,916 spectra). Proteins identified as part of the EGFR core complex were: GRB2 (64% SC, 13 PCT from 332 SCT); ERRFI (34% SC, 15 PCT from 225 SCT); ERBB2 (4% SC, 4 PCT from 207 SCT); UBS3B (41% SC, 21 PCT from 198 SCT); HS90A (11% SC, 9 PCT from 55 SCT); HS90B (9% SC, 7 PCT from 46 SCT); and CDC37 (8% SC, 2 PCT from 10 SCT). AP2M1 and AP2A2 proteins involved in EGFR endocytocytosis; the adaptor protein SHC1; and the chaperone GRP75 were also identified. GRP78, HSP7C, HSP71 and HS71L were observed in both the EGFR and GFP purifications. The spectral counts were markedly increased when EGFR was the bait, indicating enrichment of heat shock proteins in the mutant EGFR complex. For example, from the GFP TAP there were a total of 31 spectra matching HSP7C. This increased by 91% to 345 spectral counts when EGFR was the bait (Table 1). In addition to the core complex proteins, a number of cytoplasmic and ribosomal proteins were observed (TBA1B, TBA1C, TBB5, TBB2C, TBA4A, RL23, RS27, RS27L). Some proteins that are not known to interact with EGFR (ADT3, EFTU, 1433S, 1433T, UBIQ, HNRPU, LMNB1 and MPCP) were also apparent and are most likely indicative of non-specifically-interacting proteins.
Table 1.
Proteins identified by 1D-SG-CID in the EGFR TAP from PC9 (n = 4). Proteins groups identified in the GFP negative control are subtracted from the data set. Proteins shown in bold italics were observed in the GFP TAP, but were enhanced in the EGFR TAP (1D-SG-CID, one-dimensional shotgun collision-induced dissociation; PCT, peptide counts; SCT, spectral counts, SCV, sequence coverage).
| ACCESSION CODE |
SWISSPROT IDENTIFIER |
PCT | SCT | SCV | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TOTAL | AVERAGE | STD. DEV. | TOTAL | AVERAGE | STD. DEV. | TOTAL | AVERAGE | STD. DEV. | ||
| P00533-1 | EGFR | 61 | 56.75 | 3.4 | 3916 | 979 | 110.21 | 0.54 | 0.52 | 0.02 |
| P11021 | GRP78 | 27 | 22 | 2.94 | 382 | 95.5 | 8.06 | 0.44 | 0.37 | 0.03 |
| P11142-1 | HSP7C | 20 | 18.75 | 0.96 | 345 | 86.25 | 14.2 | 0.38 | 0.36 | 0.03 |
| P62993-1 | GRB2 | 13 | 11.5 | 1 | 332 | 83 | 10.42 | 0.64 | 0.5 | 0.09 |
| Q9UJM3 | ERRFI | 15 | 13.5 | 0.58 | 225 | 56.25 | 7.97 | 0.34 | 0.31 | 0.02 |
| P04626 | ERBB2 | 4 | 2.75 | 0.96 | 207 | 51.75 | 6.02 | 0.04 | 0.03 | 0.01 |
| P08107 | HSP71 | 19 | 17 | 1.41 | 206 | 51.5 | 9.47 | 0.39 | 0.35 | 0.03 |
| Q8TF42 | UBS3B | 21 | 16.25 | 2.22 | 198 | 49.5 | 16.62 | 0.41 | 0.29 | 0.06 |
| P34931 | HS71L | 10 | 9.25 | 0.96 | 150 | 37.5 | 7 | 0.21 | 0.19 | 0.02 |
| P68363 | TBA1B | 10 | 9 | 1.41 | 91 | 22.75 | 4.65 | 0.31 | 0.29 | 0.04 |
| Q9BQE3 | TBA1C | 10 | 8.25 | 1.71 | 86 | 21.5 | 4.43 | 0.32 | 0.27 | 0.05 |
| P07900-1 | HS90A | 9 | 8 | 0.82 | 55 | 13.75 | 2.63 | 0.11 | 0.1 | 0.01 |
| P07437 | TBB5 | 10 | 7 | 0.82 | 49 | 12.25 | 1.5 | 0.33 | 0.21 | 0.03 |
| P68371 | TBB2C | 9 | 6.25 | 0.96 | 46 | 11.5 | 1.73 | 0.3 | 0.18 | 0.04 |
| P08238 | HS90B | 7 | 6.75 | 0.5 | 46 | 11.5 | 1.29 | 0.09 | 0.08 | 0 |
| P68366 | TBA4A | 7 | 6 | 1.15 | 43 | 10.75 | 2.63 | 0.23 | 0.2 | 0.04 |
| P12236 | ADT3 | 6 | 4.25 | 0.96 | 36 | 9 | 1.15 | 0.21 | 0.14 | 0.04 |
| P49411 | EFTU | 7 | 3.75 | 2.5 | 17 | 4.25 | 2.99 | 0.18 | 0.09 | 0.06 |
| Q96CW1-1 | AP2M1 | 3 | 3 | 0 | 14 | 3.5 | 0.58 | 0.06 | 0.06 | 0 |
| P62829 | RL23 | 2 | 2 | 0 | 13 | 3.25 | 0.96 | 0.21 | 0.21 | 0 |
| P38646 | GRP75 | 4 | 2 | 1.41 | 10 | 2.5 | 1.73 | 0.08 | 0.04 | 0.03 |
| P27348 | 1433T | 4 | 2.25 | 0.5 | 10 | 2.5 | 0.58 | 0.16 | 0.08 | 0.03 |
| Q16543 | CDC37 | 2 | 1.5 | 1 | 10 | 2.5 | 2.08 | 0.08 | 0.06 | 0.04 |
| P62988 | UBIQ | 2 | 1 | 1.15 | 8 | 2 | 2.31 | 0.29 | 0.14 | 0.17 |
| P29353-1 | SHC1 | 3 | 1.5 | 1.73 | 6 | 1.5 | 1.73 | 0.05 | 0.03 | 0.03 |
| Q00839-1 | HNRPU | 2 | 1 | 1.15 | 5 | 1.25 | 1.5 | 0.03 | 0.01 | 0.02 |
| P31947-1 | 1433S | 2 | 1 | 1.15 | 5 | 1.25 | 1.5 | 0.06 | 0.03 | 0.04 |
| P42677 | RS27 | 2 | 1 | 1.15 | 4 | 1 | 1.15 | 0.29 | 0.14 | 0.16 |
| P20700 | LMNB1 | 2 | 0.5 | 1 | 3 | 0.75 | 1.5 | 0.03 | 0.01 | 0.02 |
| Q00325-1 | MPCP | 2 | 0.5 | 1 | 3 | 0.75 | 1.5 | 0.05 | 0.01 | 0.03 |
| O94973-1 | AP2A2 | 2 | 0.5 | 1 | 3 | 0.75 | 1.5 | 0.02 | 0.01 | 0.01 |
| Q71UM5 | RS27L | 2 | 0.5 | 1 | 2 | 0.5 | 1 | 0.29 | 0.07 | 0.14 |
For the HCC827 cells, subtraction of the protein groups observed in the GFP negative control and exclusion of proteins likely to act as non-specific binders (EFTU, RS27 and MPCP) resulted in 10 proteins that were assumed to interact with C-tagged SH-EGFR (n = 4, Table 2). Again, the EGFR bait was the most abundant protein with a maximum sequence coverage of 51% (58 PCT from 3,308 SCT). The proteins identified as part of the core complex of EGFR purified from the HCC827 lung cancer cell line were: GRB2 (35% SCV, 8 PCT from 55 SCT); ERRFI (25% SCV, 10 PCT from 45 SCT); HS90B (20% SCV, 14 PCT from 88 SCT); HS90A (17% SCV, 12 PCT from 80 SCT); CDC37 (10% SCV, 3 PCT from 15 SCT); and STAT3 (8% SCV, 4 unique PCT from 6 SCT). As for the EGFR-TAP from the PC9 cell line, HSP7C, GRP78, HSP71 and HS71L were identified in both the EGFR and GFP experiments, and again the spectral counts were markedly increased in the former.
Table 2.
Proteins identified by 1D-SG-CID in the EGFR TAP from HCC827 (n = 4). Proteins groups identified in the GFP negative control are subtracted from the data set. Proteins shown in bold italics were observed in the GFP TAP, but were enhanced in the EGFR TAP (EGFR, epidermal growth factor receptor; TAP, tandem affinity purification; GFP, green fluorescent protein; 1D-SG-CID, one-dimensional shotgun collision-induced dissociation; PCT, peptide counts; SCT, spectral counts, SCV, sequence coverage).
| ACCESSION CODE |
SWISSPROT IDENTIFIER |
PCT | SCT | SCV | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TOTAL | AVERAGE | STD. DEV. | TOTAL | AVERAGE | STD. DEV. | TOTAL | AVERAGE | STD. DEV. | ||
| P00533-1 | EGFR | 58 | 56.5 | 0.58 | 3308 | 827 | 36.89 | 0.51 | 0.51 | 0 |
| P11142-1 | HSP7C | 20 | 18 | 0.82 | 312 | 78 | 6.48 | 0.38 | 0.35 | 0.03 |
| P11021 | GRP78 | 17 | 13.75 | 0.5 | 177 | 44.25 | 3.4 | 0.33 | 0.28 | 0.01 |
| P08107 | HSP71 | 10 | 7.75 | 2.06 | 98 | 24.5 | 4.8 | 0.19 | 0.14 | 0.04 |
| P34931 | HS71L | 6 | 5.25 | 0.5 | 90 | 22.5 | 3 | 0.12 | 0.1 | 0.01 |
| P08238 | HS90B | 14 | 12 | 0.82 | 88 | 22 | 2 | 0.2 | 0.17 | 0.01 |
| P07900-1 | HS90A | 12 | 9.5 | 0.58 | 80 | 20 | 2.58 | 0.17 | 0.13 | 0.01 |
| P62993-1 | GRB2 | 8 | 6.5 | 0.58 | 55 | 13.75 | 1.71 | 0.35 | 0.3 | 0.02 |
| Q9UJM3 | ERRFI | 10 | 7.75 | 0.5 | 45 | 11.25 | 1.5 | 0.25 | 0.21 | 0.02 |
| Q16543 | CDC37 | 3 | 2.25 | 0.5 | 15 | 3.75 | 1.5 | 0.1 | 0.09 | 0.01 |
| P40763-1 | STAT3 | 4 | 1.5 | 1.91 | 6 | 1.5 | 1.91 | 0.08 | 0.03 | 0.04 |
| P49411 | EFTU | 3 | 1.25 | 1.5 | 5 | 1.25 | 1.5 | 0.06 | 0.02 | 0.03 |
| P42677 | RS27 | 2 | 0.5 | 1 | 2 | 0.5 | 1 | 0.29 | 0.07 | 0.14 |
| Q00325-1 | MPCP | 2 | 0.5 | 1 | 2 | 0.5 | 1 | 0.05 | 0.01 | 0.03 |
GRB2 and ERRFI had a lower sequence coverage than that observed in the PC9 cell line while HS90A, HS90B and CDC37 were higher in the HCC827 cells. A number of proteins observed in the PC9 cells were not identified in the EGFR complex isolated from the HCC827 cell line. This included ERBB2, UBS3B, AP2M1, SHC1, and AP2A2. It was hypothesised that these proteins are indeed part of the EGFR complex in the HCC827 cell line. The abundance, however, may be low and therefore the proteins are not identified using an unbiased one-dimensional shotgun data-dependent CID method. To examine this in more detail, we developed an alternative use for the iTRAQ® reagent that resulted in the identification of a lower-abundance EGFR interacting protein purified from the HCC827 cell line.
One-Dimensional Shotgun CID Analysis of iTRAQ®-labelled C-tagged-SH-EGFR
The 117-iTRAQ®-labelled biochemical replicate of C-tagged SH-EGFR purified from the PC9 cell line was analysed by a data-dependent top 6 CID method to identify the peptides in the sample (Table 1). This is an important step to generate a global inclusion list with corresponding retention time windows. iTRAQ® labelling results in a shift in retention times of the peptides as compared to the unlabelled counterparts, i.e., the addition of an iTRAQ® label leads to increased peptide hydophobicity. As shown in Table 1, back-to-back analyses of EGFR purifications from PC9 cells resulted in the identification of a total of 11 protein groups from the unlabelled sample (GFP subtracted, enriched heat shock proteins included and non-specific proteins removed). Analysis of the 117-labelled iTRAQ® counterpart resulted in the identification of 9 protein groups. Note that the quantity of starting material used to generate the 117-iTRAQ®-labelled reference sample was quite low. Thus, a loss of information especially for low-abundance peptides may be due to sample processing during labelling that undoubtedly culminates in incomplete peptide recovery. In addition, altered retention time behaviour may also change ion suppression effects, particularly for low-abundance peptides. Compared to the initial analyses (Table 1), AP2A2 and SHC1 were not identified in the unlabelled sample. Following iTRAQ®-labelling, ERBB2, AP2M1 and HS90B were no longer identified, however, SHC1 was apparent.
A complete and comprehensive list of all the m/z values selected for fragmentation plus retention time (Tr) windows was extracted from the 117-iTRAQ®-labelled raw data MS file. Peptides were chosen for addition on the inclusion list according to the following criteria. For the abundant bait protein EGFR, ions with a high intensity and a Mascot peptide score ≥ 55 were selected. Additional peptides with a lower Mascot peptide score were also added based on the retention time value such that all peptides chosen were evenly-distributed across the entire time range of the LCMS chromatogram. The rationale for this approach is to provide sufficient peptides to clearly identify the protein, but to also minimise the selection of over-abundant EGFR peptides and ‘force’ the mass spectrometer to choose an alternative ion in an attempt to identify other peptides from the interacting proteins in the EGFR core complex. For the heat shock proteins GRP78, HSP7C and HSP71, peptides with a Mascot peptide score ≥ 35 were chosen plus additional peptides based on retention time. For the weaker, less-abundant proteins, a minimum of two unique peptides identified by Mascot with a peptide score ≥ 18 were included. The final inclusion list consisted of 53 ions as follows: EGFR (7), GRP78 (6), HSP7C (7), HSP71 (3), GRB2 (7), UBS3B (6), HS90A (6), ERRFI (4), SHC1 (4) and UBIQ (3). Nonspecific proteins and known contaminants such as keratin and trypsin were excluded.
iTRAQ®-labelled biochemical replicates of the EGFR purifications from the two cells lines were mixed in equal volumes and analysed by a top 4 HCD fragmentation method to identify the peptides (Table 4) and provide relative quantitation on the iTRAQ® reporter ions (Figure 2B). It is noteworthy to add at this point that we (unpublished observations) and others30,31 have observed that one-dimensional shotgun analyses of iTRAQ®-labelled complex peptide mixtures (e.g., body fluids, whole cell lysates) can lead to problems with accurate quantitation. Co-elution of peptides from the separation column results in mixed MSMS spectra and subsequent false augmentation of the iTRAQ reporter ions. For the most part, this is overcome by fractionation of the mixtures. For reduced complexity samples such as a SH-TAP this caveat does not appear to affect the quantitation as the total number of proteins (and thus peptides) is less than in a body fluid or whole cell lysate. Hence the probability of co-eluting peptides, mixed MSMS spectra and skewed iTRAQ® reporter ions is markedly reduced.
Table 2.
Proteins identified by 1D-SG-HCD in the combined iTRAQ®-labelled EGFR TAP from HCC827 (n = 2) and PC9 (n = 2) with (A) no inclusion list; (B) global inclusion list only; and (C) global inclusion list plus most intense ions (1D-SG-HCD, one-dimensional shotgun higher-energy collision-induced dissociation; PCT, peptide counts; SCT, spectral counts, SCV, sequence coverage).
| ACCESSION CODE | SWISSPROT NAME | PCT | SCT | SCV | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NO INCLUSION LIST | GLOBAL INCLUSION LIST | GLOBAL INCLUSION LIST + MOST INTENSE IONS | NO INCLUSION LIST | GLOBAL INCLUSION LIST | GLOBAL INCLUSION LIST + MOST INTENSE IONS | NO INCLUSION LIST | GLOBAL INCLUSION LIST | GLOBAL INCLUSION LIST + MOST INTENSE IONS | ||||||||
| TOTAL | TOTAL | TOTAL | AVERAGE | STD. DEV. | TOTAL | TOTAL | TOTAL | AVERAGE | STD. DEV. | TOTAL | TOTAL | TOTAL | AVERAGE | STD. DEV. | ||
| P00533-1 | EGFR | 47 | 7 | 54 | 51.5 | 2.12 | 244 | 18 | 345 | 172.5 | 6.36 | 0.46 | 0.08 | 0.48 | 0.46 | 0.03 |
| P62993-1 | GRB2 | 8 | 6 | 8 | 7.5 | 0.71 | 13 | 8 | 33 | 16.5 | 2.12 | 0.32 | 0.26 | 0.34 | 0.32 | 0.03 |
| P11142-1 | HSP7C | 7 | 7 | 14 | 12 | 2.83 | 14 | 9 | 29 | 14.5 | 2.12 | 0.14 | 0.12 | 0.23 | 0.19 | 0.05 |
| P11021 | GRP78 | 8 | 5 | 14 | 12.5 | 0.71 | 12 | 5 | 34 | 17 | 0 | 0.14 | 0.1 | 0.24 | 0.22 | 0 |
| P08107 | HSP71 | 4 | 4 | 7 | 6 | 1.41 | 8 | 4 | 15 | 7.5 | 2.12 | 0.06 | 0.08 | 0.11 | 0.09 | 0.02 |
| Q8TF42 | UBS3B | 4 | 4 | 5 | 4.5 | 0.71 | 5 | 5 | 12 | 6 | 1.41 | 0.07 | 0.07 | 0.09 | 0.08 | 0.02 |
| Q9UJM3 | ERRFI | 6 | 2 | 6 | 5 | 1.41 | 6 | 3 | 12 | 6 | 0 | 0.17 | 0.05 | 0.15 | 0.12 | 0.05 |
| P07900-1 | HS90A | 2 | 4 | 6 | 4.5 | 2.12 | 2 | 4 | 10 | 5 | 1.41 | 0.03 | 0.05 | 0.09 | 0.07 | 0.03 |
| P68363 | TBA1B | 4 | 0 | 5 | 4 | 0 | 4 | 0 | 8 | 4 | 0 | 0.12 | 0 | 0.15 | 0.11 | 0.01 |
| P05141 | ADT2 | 2 | 0 | 2 | 1 | 1.41 | 2 | 0 | 2 | 1 | 1.41 | 0.05 | 0 | 0.05 | 0.03 | 0.04 |
| P81605 | DCD | 0 | 0 | 2 | 1 | 1.41 | 0 | 0 | 2 | 1 | 1.41 | 0 | 0 | 0.15 | 0.08 | 0.11 |
The samples were analysed by three alternative mass spectrometric methods: (A) standard data-dependent acquisition, i.e., no inclusion list; (B) global inclusion list only; and (C) global inclusion list plus the most intense ions. The third method initially favours peptides on the inclusion list, regardless of intensity. If none of the peptides on the list are observed, the mass spectrometer selects and fragments the peptide with the highest intensity. Using method (A), 11 proteins (including trypsin) were identified (n = 1). This decreased to 8 proteins when the inclusion list was activated (n = 1); and increased to 12 proteins (including trypsin) when a combined method incorporating the inclusion list plus most intense ions was used (n = 2). With method (A), EGFR was identified with a sequence coverage of 46% (47 PCT from 244 SCT). Not surprisingly, the sequence coverage decreased to 8% (7 PCT from 18 SCT) when only the global inclusion list was used (only 7 peptides from EGFR were on the inclusion list); and increased again to 48% (54 PCT from 354 SCT) when the global inclusion list plus most intense ions was used. GRB2, HSP7C and GRP78 followed a similar trend to EGFR. HSP71 and HS90A all increased in sequence coverage with each MS method. The sequence coverage for ERRFI was 17% for method (A); decreased to 5% with method (B) and increased to 15% with method (C). UBS3B had the same sequence coverage with methods (A) and (B), but increased from 7% to 9% with method (C). SHC1 and UBIQ were not identified via any of the approaches, however, this is reasonable as the intensities of these particular ions were quite low in the original unlabelled eluate. In addition, the cycle time for HCD fragmentation is slower than for CID, thus there are always fewer proteins identified with lower spectral counts than a comparative analysis with CID. Overall these data showed that method (C) provided the best compromise between total number of proteins identified without over-selection of the abundant peptides from the EGFR bait.
Relative Quantitation of iTRAQ®-labelled C-tagged-SH-EGFR
Analysis of the relative quantitative results from the technical replicates generated using mass spectrometric method (C) are shown in Figure 2B. The ratio of EGFR varied across the 4 iTRAQ® channels, although the relative ratio of EGFR in the HCC827 cell lines was close to equimolar. With the PC9 cell line, the ratio of EGFR from the first biochemical replicate was lower than the second replicate. This may have been a consequence of inefficient elution of the protein by formic acid (Figure 2B). Overall, the second replicate from the PC9 cell line yielded approximately 1.5 time more bait protein than EGFR from the HCC827 cell line. The relative ratios of EGFR-interacting proteins GRB2, ERRFI, UBS3B and GRP78 were higher in the PC9 cell line compared to HCC827 (Figure 2B). UBS3B was not identified in the experiments conducted in the HCC827 cell line and analysed by 1D-SG-CID (see Table 2), however, the combined iTRAQ®-labelled peptides from UBS3B that is more abundant in the PC9 (or less sequestered by endogenous EGFR compared to HCC827) indicated that UBS3B was actually present at low levels in the EGFR TAP from the HCC827 cells (Figure 2B). The relative quantitation between the channels showed that 6 to 10 times more UBS3B was incorporated in the EGFR core complex purified from the PC9 cell line than that observed with the HCC827 cells. The relative ratios of TBA1B, HSP7C, HS90A and HSP71 followed a similar trend to that observed with EGFR. Namely, that the second biochemical replicate of the EGFR purification from either cell line had slightly higher protein ratios as compared to the first biochemical replicates (Figure 2B).
Thus, here we show a new approach to identify a protein in a system that normally would not be observed because of low-abundance due to sequestration by endogenous bait protein and/or suppression by the high quantities of the bait. When combined with an inclusion list, low-abundance peptides have an improved chance of detection and fragmentation in the mass spectrometer. Therefore we show a novel application of a current methodology to identify low-abundance (or weakly-expressed) proteins. This approach may also be beneficial in detecting transient proteins that only remain in minute quantities after the tandem affinity purification. The entire methodology as described in this study, can also be extended to a range of other applications, e.g., pooling of samples can generically provide a means to increase the detection of low-abundance species due to the additive effect of the iTRAQ®-modified proteins.
Conclusion
Using the iTRAQ® reagent combined with an inclusion list of known specific peptides we have shown a novel application to enhance the intensity of peptides from low-abundance proteins that are otherwise not observed via standard one-dimensional shotgun approaches with faster collision-induced dissociation fragmentation method in the hybrid LTQ-Orbitrap XL mass spectrometer. We have shown that UBS3B is a member of the EGFR core complex in HCC827 cell line, that was not apparent by standard methodology. Using iTRAQ® in this fashion, will allow the identification of low-abundance interacting proteins when combined with samples where the protein has a higher abundance.
Alternatively, this approach could be used to spike peptides into samples where it is believed the protein is present but is too weak to be identified by conventional techniques. Ultimately, this should lead to confirmation of interactors that are suspected and may also result in the identification of transiently-interacting (and thus low-abundance) proteins.
Table 1.
Proteins identified by 1D-SG-CID in the EGFR TAP from PC9 cell line, unlabelled and 117-iTRAQ®-labelled. Proteins groups identified in the GFP negative control are subtracted from the data set. Proteins shown in bold italics were observed in the GFP TAP, but were enhanced in the EGFR TAP (1D-SG-CID, one-dimensional shotgun collision-induced dissociation; PCT, peptide counts; SCT, spectral counts, SCV, sequence coverage).
| ACCESSION CODE |
SWISSPROT IDENTIFIER |
PCT | SCT | SCV | |||
|---|---|---|---|---|---|---|---|
| UNLABELLED | iTRAQ®-LABELLED | UNLABELLED | iTRAQ®-LABELLED | UNLABELLED | iTRAQ®-LABELLED | ||
| P00533-1 | EGFR | 53 | 48 | 832 | 586 | 0.47 | 0.5 |
| P11021 | GRP78 | 21 | 18 | 98 | 61 | 0.37 | 0.32 |
| P62993-1 | GRB2 | 9 | 8 | 82 | 63 | 0.41 | 0.35 |
| P11142-1 | HSP7C | 18 | 16 | 71 | 64 | 0.34 | 0.32 |
| Q8TF42 | UBS3B | 14 | 7 | 65 | 18 | 0.25 | 0.14 |
| P08107 | HSP71 | 14 | 8 | 43 | 44 | 0.25 | 0.14 |
| Q9UJM3 | ERRFI | 11 | 3 | 35 | 10 | 0.31 | 0.11 |
| P07900-1 | HS90A | 6 | 6 | 19 | 11 | 0.08 | 0.09 |
| P34931 | HS71L | 7 | 6 | 34 | 36 | 0.13 | 0.1 |
| P68363 | TBA1B | 8 | 7 | 15 | 17 | 0.24 | 0.22 |
| P08238 | HS90B | 5 | 0 | 18 | 0 | 0.07 | 0 |
| P07437 | TBB5 | 5 | 4 | 10 | 6 | 0.14 | 0.11 |
| P68371 | TBB2C | 4 | 0 | 7 | 0 | 0.1 | 0 |
| P05141 | ADT2 | 2 | 3 | 4 | 5 | 0.06 | 0.09 |
| P29353-1 | SHC1 | 0 | 4 | 0 | 5 | 0 | 0.08 |
| P49411 | EFTU | 4 | 0 | 5 | 0 | 0.08 | 0 |
| P62988 | UBIQ | 3 | 3 | 4 | 12 | 0.5 | 0.41 |
| P12236 | ADT3 | 3 | 0 | 5 | 0 | 0.1 | 0 |
| Q96CW1-1 | AP2M1 | 3 | 0 | 3 | 0 | 0.06 | 0 |
| P04626 | ERBB2 | 3 | 0 | 60 | 0 | 0.03 | 0 |
| P20700 | LMNB1 | 2 | 0 | 2 | 0 | 0.03 | 0 |
| P23396 | RS3 | 0 | 2 | 0 | 2 | 0 | 0.09 |
| P42677 | RS27 | 2 | 0 | 3 | 0 | 0.23 | 0 |
| P25705 | ATPA | 2 | 0 | 3 | 0 | 0.04 | 0 |
| O75369-1 | FLNB | 2 | 0 | 3 | 0 | 0.01 | 0 |
| P62829 | RL23 | 2 | 0 | 2 | 0 | 0.21 | 0 |
Acknowledgement
The authors wish to thank Giulio Superti-Furga at the Center for Molecular Medicine (CeMM) for supporting the visit of EBH; William Pao for providing cDNA for EGFR; Matthias Gstaiger for supplying the original SH constructs and providing valuable advice in establishing SH-TAP protocols at CeMM, and Fumi Kinose for assistance with cell culture. Work in our laboratory is funded by the Austrian Academy of Sciences, the Austrian Federal Ministry for Science and Research (Gen-Au projects, APP and BIN) and by the Austrian Science Fund FWF (P22282-B11, FG). The work was also financed in part by NIH 5P50CA119997 (EBH).
References
- 1.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415(6868):141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 2.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415(6868):180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 3.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440(7084):631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 4.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MH, Butland G, Altaf-Ul AM, Kanaya S, Shilatifard A, O'Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440(7084):637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 5.Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y, Vasilescu J, Abu-Farha M, Lambert JP, Duewel HS, Stewart II, Kuehl B, Hogue K, Colwill K, Gladwish K, Muskat B, Kinach R, Adams SL, Moran MF, Morin GB, Topaloglou T, Figeys D. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blagoev B, Kratchmarova I, Ong SE, Nielsen M, Foster LJ, Mann M. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat Biotechnol. 2003;21(3):315–318. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- 7.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti-Furga G. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6(2):97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 8.Glatter T, Wepf A, Aebersold R, Gstaiger M. An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol Syst Biol. 2009;5:237. doi: 10.1038/msb.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466(7302):68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertwistle D, Sugimoto M, Sherr CJ. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol Cell Biol. 2004;24(3):985–996. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. Plos Biology. 2005;3(6):986–997. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Galova M, Petronczki M, Gregan J, Cetin B, Mudrak I, Ogris E, Mechtler K, Pelletier L, Buchholz F, Shirahige K, Nasmyth K. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441(7089):53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- 13.Brehme M, Hantschel O, Colinge J, Kaupe I, Planyavsky M, Kocher T, Mechtler K, Bennett KL, Superti-Furga G. Charting the molecular network of the drug target Bcr-Abl. Proc Natl Acad Sci U S A. 2009;106(18):7414–7419. doi: 10.1073/pnas.0900653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S, Smylie M, Martins R, van Kooten M, Dediu M, Findlay B, Tu D, Johnston D, Bezjak A, Clark G, Santabarbara P, Seymour L. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353(2):123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 15.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 16.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 17.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, Chitale D, Motoi N, Szoke J, Broderick S, Balak M, Chang WC, Yu CJ, Gazdar A, Pass H, Rusch V, Gerald W, Huang SF, Yang PC, Miller V, Ladanyi M, Yang CH, Pao W. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104(52):20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C, Johnson BE, Cantley LC, Janne PA. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 20.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305(5687):1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 21.Yang S, Qu S, Perez-Tores M, Sawai A, Rosen N, Solit DB, Arteaga CL. Association with HSP90 inhibits Cbl-mediated down-regulation of mutant epidermal growth factor receptors. Cancer Res. 2006;66(14):6990–6997. doi: 10.1158/0008-5472.CAN-06-1042. [DOI] [PubMed] [Google Scholar]
- 22.Shtiegman K, Kochupurakkal BS, Zwang Y, Pines G, Starr A, Vexler A, Citri A, Katz M, Lavi S, Ben-Basat Y, Benjamin S, Corso S, Gan J, Yosef RB, Giordano S, Yarden Y. Defective ubiquitinylation of EGFR mutants of lung cancer confers prolonged signaling. Oncogene. 2007;26(49):6968–6978. doi: 10.1038/sj.onc.1210503. [DOI] [PubMed] [Google Scholar]
- 23.Padron D, Sato M, Shay JW, Gazdar AF, Minna JD, Roth MG. Epidermal growth factor receptors with tyrosine kinase domain mutations exhibit reduced Cbl association, poor ubiquitylation, and down-regulation but are efficiently internalized. Cancer Res. 2007;67(16):7695–7702. doi: 10.1158/0008-5472.CAN-07-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3(12):1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Rix U, Fang B, Bai Y, Edwards A, Colinge J, Bennett KL, Gao J, Song L, Eschrich S, Superti-Furga G, Koomen J, Haura EB. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat Chem Biol. 2010;6(4):291–299. doi: 10.1038/nchembio.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernbach NV, Planyavsky M, Muller A, Breitwieser FP, Colinge J, Rix U, Bennett KL. Acid elution and one-dimensional shotgun analysis on an Orbitrap mass spectrometer: an application to drug affinity chromatography. J Proteome Res. 2009;8(10):4753–4765. doi: 10.1021/pr900455x. [DOI] [PubMed] [Google Scholar]
- 27.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75(3):663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 28.Colinge J, Masselot A, Giron M, Dessingy T, Magnin J. OLAV: towards high-throughput tandem mass spectrometry data identification. Proteomics. 2003;3(8):1454–1463. doi: 10.1002/pmic.200300485. [DOI] [PubMed] [Google Scholar]
- 29.Gandhi J, Zhang J, Xie Y, Soh J, Shigematsu H, Zhang W, Yamamoto H, Peyton M, Girard L, Lockwood WW, Lam WL, Varella-Garcia M, Minna JD, Gazdar AF. Alterations in genes of the EGFR signaling pathway and their relationship to EGFR tyrosine kinase inhibitor sensitivity in lung cancer cell lines. PLoS One. 2009;4(2):e4576. doi: 10.1371/journal.pone.0004576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bantscheff M, Boesche M, Eberhard D, Matthieson T, Sweetman G, Kuster B. Robust and sensitive iTRAQ quantification on an LTQ Orbitrap mass spectrometer. Mol Cell Proteomics. 2008;7(9):1702–1713. doi: 10.1074/mcp.M800029-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ow SY, Salim M, Noirel J, Evans C, Rehman I, Wright PC. iTRAQ underestimation in simple and complex mixtures: "the good, the bad and the ugly". J Proteome Res. 2009;8(11):5347–5355. doi: 10.1021/pr900634c. [DOI] [PubMed] [Google Scholar]