Abstract
Bacteriophages are involved in many aspects of the spread and establishment of virulence factors in Staphylococcus aureus, including the mobilization of genetic elements known as pathogenicity islands (SaPIs), which carry genes for superantigen toxins and other virulence factors. SaPIs are packaged into phage-like transducing particles using proteins supplied by the helper phage. We have used cryo-electron microscopy and icosahedral reconstruction to determine the structure of the procapsid and the mature capsid of 80α, a bacteriophage that can mobilize several different SaPIs. The 80α capsid has T = 7 icosahedral symmetry with the capsid protein organized into pentameric and hexameric clusters that interact via prominent trimeric densities. The 80α capsid protein was modeled based on the capsid protein fold of bacteriophage HK97, and fitted into the 80α reconstructions. The models show that the trivalent interactions are mediated primarily by a 22-residue β hairpin structure called the P loop that is not found in HK97. Capsid expansion is associated with a conformational switch in the spine helix that is propagated throughout the subunit, unlike the domain rotation mechanism in phages HK97 or P22.
Keywords: procapsid, structure, assembly, three-dimensional reconstruction, pathogenicity island
Introduction
Staphylococcus aureus is a commensal inhabitant of human skin and mucosal epithelia, but has also been associated with a number of pathogenic conditions 1. Historically, infections occurred mainly in immunocompromised individuals and in hospital settings, but more recently, otherwise healthy individuals have been affected by aggressive strains circulating in the community 2. The emergence of new strains that are resistant to multiple antibiotics, especially methicillin resistant S. aureus (MRSA), has significantly complicated treatment and supportive therapy necessary to clear infection 3.
Bacteriophages are involved in several aspects of S. aureus virulence. Certain bacteriophages are carriers of known virulence genes, including those for Panton-Valentine Leukocidin (PVL), exfoliative toxin A, staphylokinase and enterotoxin A 4,5. Through generalized transduction, bacteriophages can transfer chromosomal and plasmid-encoded virulence and resistance genes between pathogenic and non-pathogenic strains 6,7. Bacteriophages are also involved in the specialized mobilization of S. aureus pathogenicity islands (SaPIs), a family of 14–27 kb genetic elements that are integrated into the host genome and can carry genes for a variety of superantigen toxins, such as the toxic shock syndrome toxin (TSST-1) and a number of enterotoxins 8,9. The SaPIs become mobilized upon infection with certain “helper” bacteriophages or by the induction of endogenous prophages through the SOS response 10. Mobilization involves derepression, excision and replication of the SaPI element 11, which is then packaged into phage-like transducing particles using structural gene products supplied by the helper phage 12,13,14. These transducing particles can then go on to infect naïve hosts, leading to the spread of SaPI-encoded virulence factors through the bacterial population. One striking aspect of the mobilization of many SaPIs is the redirection of capsid assembly to form a smaller particle that accommodates the smaller SaPI genome, but is inadequate for packaging a complete phage genome 15. Understanding the pathway of normal capsid assembly is a prerequisite to elucidating the mechanism by which SaPIs alter capsid size.
S. aureus bacteriophage 80α is a temperate phage that is capable of generalized transduction and also serves as a helper phage for the mobilization of several different SaPIs, including SaPI1, SaPI2, SaPIbov1 and SaPIbov2 9,16. Phage 80α, which is essentially identical to staphylococcal typing phage 53 16, is a double-stranded (ds) DNA phage of the Siphoviridae family. The 80α virion consists of a 63 nm diameter isometric, icosahedrally symmetric capsid attached to a 190 nm long flexuous, non-contractile tail capped with an elaborate baseplate decorated with six tail fibers 13.
Capsids of dsDNA bacteriophages are formed in a stepwise fashion, starting with the initial formation of a roughly spherical precursor capsid called the procapsid, which is subsequently filled with DNA 17. Procapsid formation usually requires a scaffolding protein, which acts as a chaperone for the assembly process 18,19. 80α procapsids are assembled from capsid protein, a product of gene 47 (gp47), scaffolding protein (gp46), a portal protein (gp42) that forms the entry and exit portal for the DNA genome and as a connector between the head and tail, and gp44, a minor capsid protein of unknown function 12,13. The capsid and scaffolding proteins are cleaved between Phe and Ala by an unidentified protease, removing the N-terminal 14 and 13 amino acids, respectively 13. DNA is packaged into the procapsids through the portal vertex by a headful mechanism. This process requires the small (gp40) and large (gp41) terminase subunits (TerS and TerL, respectively), and occurs concomitantly with scaffolding removal and capsid expansion. The organization of the 80α structural gene cluster is shared between several S. aureus phages, and is virtually identical to that of ϕETA2, carrier of the gene for exfoliative toxin A.
The conformational changes that occur in the capsid shells of tailed dsDNA bacteriophages are most extensively described in Escherichia coli bacteriophage HK97, another member of the Siphoviridae family 20. Structures of both the mature capsid and procapsid of HK97 have been solved crystallographically 21,22,23, which identified a unique fold in the gp5 capsid protein that was subsequently observed in many other dsDNA bacteriophages, including P22 and ε15 17,24,25. Studies in HK97 and other bacteriophages have shown that maturation and capsid expansion involve major structural rearrangements and a shift from a thick, rounded shell to a thin, more angular shell, and results in a final structure that is more stable than its precursor 26.
In this paper, we present three-dimensional reconstructions from cryo-electron microscopy (cryo-EM) of procapsids and mature capsids of S. aureus phage 80α. These are the first structures of a bacteriophage that infects S. aureus. The 80α capsid has T = 7 icosahedral symmetry with the gp47 capsid protein organized into hexamers and pentamers that interact at local threefold axes of symmetry. As expected, gp47 has an HK97-like fold, in spite of low sequence similarity. Using HK97 gp5 as template, we generated a model for the gp47 structure. The main differences between gp47 and HK97 gp5 include a 12-residue insertion in gp47 that forms a hairpin loop involved in trivalent interactions, an N-terminal arm that has a helical conformation in the 80α procapsid, and a proline residue in the so-called spine helix of gp47 that acts as a conformational switch during 80α capsid maturation. A kink at this proline leads to a rotation of the N-terminal part of the spine helix that is propagated throughout the subunit and is accompanied by a reorganization of subunit interfaces. The nature of this transition is quite different from the domain movements in either HK97 or P22. Expansion is a key element of capsid maturation in all bacteriophages. The 80α structures show how this complex process can be accomplished via a simple binary switch.
Results
Structure of the 80α mature capsid
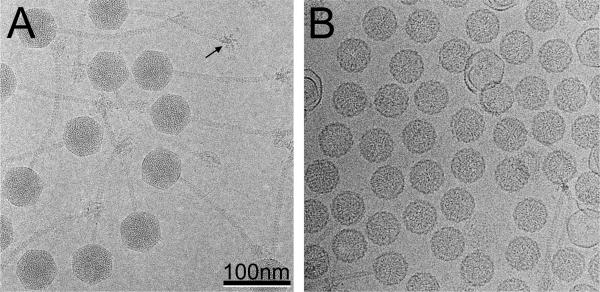
80α virions were produced by induction of the lysogenic strain RN10616 with mitomycin C and purified by CsCl gradient centrifugation, as previously described 13,27. Cryo-EM of 80α virions shows the 57 nm (side-to-side) diameter capsids (63 nm vertex-to-vertex) and 190 nm long, flexuous tails (Fig. 1A)13. The star-shaped, sixfold symmetric baseplates can also be seen. The capsids have a striated or punctate appearance with a spacing of 2.5 nm that is due to the closely packed DNA inside. Only about 1% of the particles in a typical field were empty, presumably having spontaneously ejected their DNA during purification. A total of 8080 individual images of full particles collected from 174 separate micrographs were used to reconstruct the 80α virion structure, using the EMAN program package 28, reaching a resolution of 1.02 nm based on the 0.5 Fourier shell correlation (FSC) criterion.
Figure 1.
Cryo-electron micrographs of 80α. (A) virions and (B) procapsids. A baseplate seen end-on is indicated (arrow), showing the hexagonal star-like structure with six appendages. Scale bar, 100 nm.
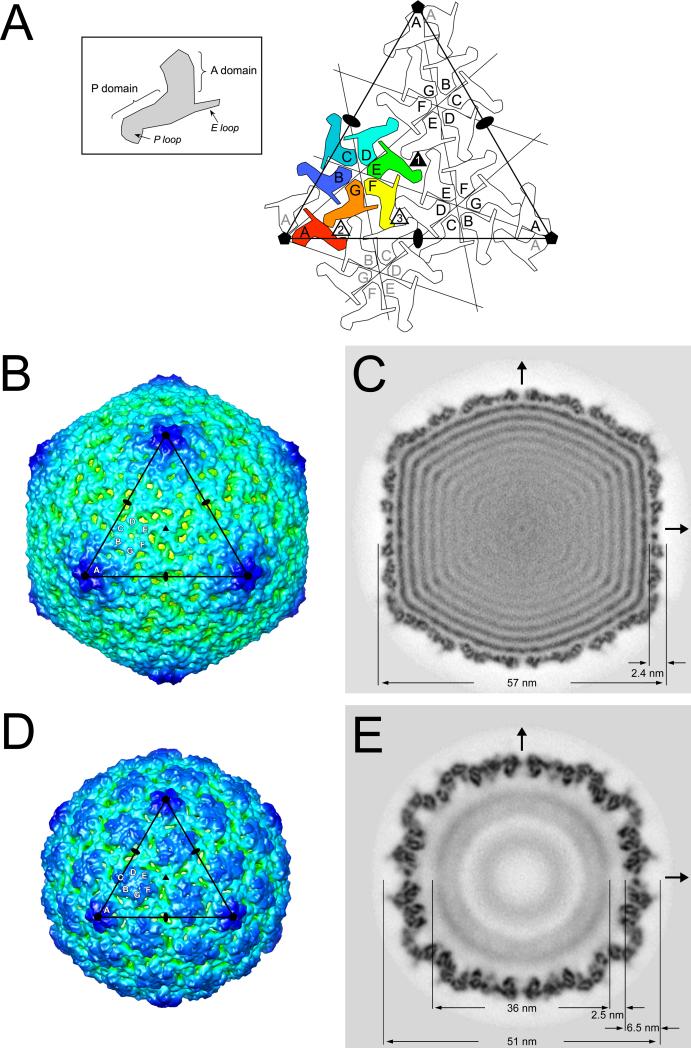
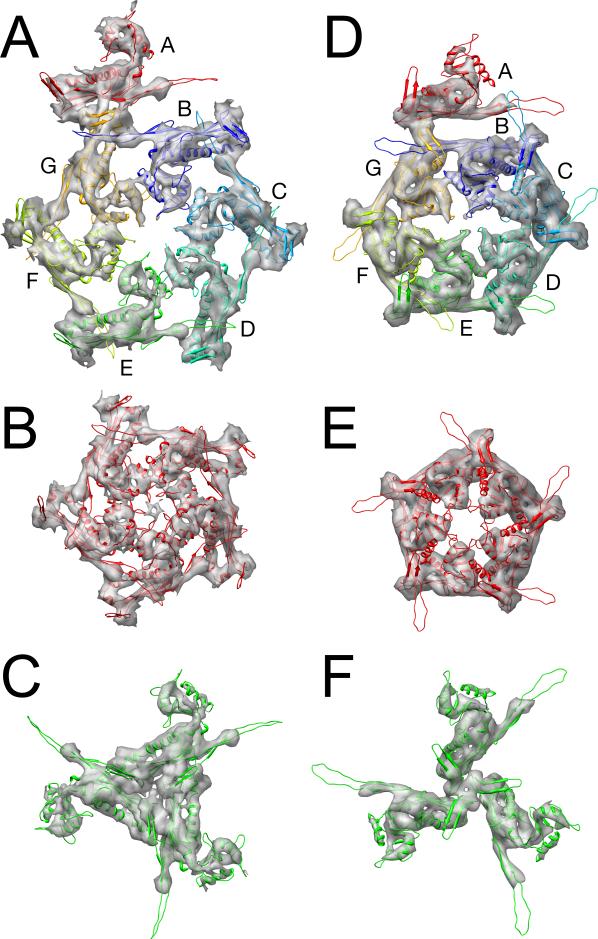
The 80α capsid has an angular appearance with T = 7 laevo icosahedral symmetry, as expected from comparisons of other bacteriophages of similar size, including HK97, λ and P22 21,24,29. The 420 copies of the gp47 capsid protein in the shell are organized into 60 hexamers that sit on the flat triangular faces of the icosahedron and twelve pentamers located on the icosahedral fivefold vertices (Fig. 2A,B). In the asymmetric virion, one pentamer would be replaced by the connector or portal protein at the unique vertex where the tail is attached. There are thus seven non-equivalent gp47 subunits in the 80α virion, arranged into an A5 pentamer and a BCDEFG hexamer (Fig. 2A, B). These capsomers interact via three types of trivalent contacts that we refer to as type 1 (EEE), type 2 (ABG) and type 3 (CDF) (Fig. 2A). Much of the capsid density is clustered in distinct trifoliate densities delineated by numerous 2–3 nm holes at these contact points (Fig. 2B). This appearance is reminiscent of phage λ, in which similar trifoliate densities are made of an additional decoration protein, gpD 29. There is no additional decoration protein in 80α, however, and the trefoils appear to be composed of an extended insertion loop in gp47 (see below). In contrast, hexameric connections are less extensive, characterized by a 1.2 nm diameter hole in the middle of the hexamer (Fig. 2B) when the map is rendered at the cutoff level consistent with the predicted mass of the capsid (420 × 35 kDa = 14,700 kDa).
Figure 2.
Icosahedral reconstructions of 80α capsids. (A) Schematic diagram showing one triangular face (large triangle) delimited by three fivefold and three twofold axes (pentagons and ovals, respectively). The icosahedral threefold (filled triangle) is in the center of the face. Each gp47 subunit is shown as a sphinx-like shape, representing the A and P domains, E loop and P loop as shown in the inset. The seven gp47 subunits corresponding to one asymmetric unit are colored: A, red; B, blue; C, teal; D, cyan; E, green; F, yellow; G, orange. The triangular face has an A5 pentamer at each corner and includes three neighboring BCDEFG hexamers. One neighboring hexamer on an adjacent face is also drawn (gray lettering) to show the three different types of trivalent subunit interactions (numbered triangles). (B) Isosurface representation of the 80α virion reconstruction, viewed down a threefold axis and rendered at a cutoff level consistent with the calculated mass of the capsid. The capsid is radially colored from red (center of capsid) to blue (farthest from center). One icosahedral face corresponding to the triangle in (A) is shown, with twofold, threefold and fivefold symmetry axes indicated. The A, B, C, D, E, F and G subunits in one asymmetric unit are marked. (C) Central section through the virion reconstruction in gray scale representation from white (lowest density) to black (highest density). The arrows indicate the twofold axes. Pertinent dimensions are indicated. (D) Isosurface representation of the 80α procapsid reconstruction, viewed, rendered and labeled as in (B). (E) Central section through the procapsid reconstruction, viewed and labeled as in (C).
The capsid shell is 2.4 nm thick on average, with a flat appearance, except at the fivefold vertices (Fig. 2C). The 2 nm holes are visible in a central section as channels through the shell at the twofold axes. At least 8 concentric layers of density, spaced 2.5 nm apart, are visible inside the shell, corresponding to the near liquid crystal packing of the DNA30,31.
The 80α procapsid reconstruction
Although procapsids form as an intermediate during normal phage production and can be purified on CsCl gradients from wildtype infections, yields are typically low, and contamination with empty, expanded shells is a problem. Thus, a better way to produce procapsids is to block DNA packaging by deleting either of the two terminase genes, which leads to an accumulation of procapsids in the cell lysates. Here, 80α procapsids were produced by induction of a S. aureus 80α lysogen (ST24) that carries a ΔterS deletion, as previously described 13. By cryo-EM, the 80α procapsids appear as 51 nm diameter, rounded particles with a 4 nm thick shell and a characteristic serrated appearance (Fig. 1B). A few thin-walled and aberrant shells are also present in a typical procapsid sample. The 80α procapsid was reconstructed using EMAN from 8029 particles in 106 micrographs to a resolution of 0.88 nm, based on the 0.5 FSC criterion.
The 80α procapsid reconstruction shows a more or less spherical, T = 7 symmetric shell with hexamers and pentamers that protrude much more that those of the virion (Fig. 2D). The hexamers have a characteristically elongated, skewed appearance that has also been described in other bacteriophage procapsids 24,29,32. Compared to the virion, the capsid protein subunits in the procapsid are packed more closely together, and appear to form more extensive intra-capsomeric contacts (Fig. 2D). As in the mature capsid, the hexamers and pentamers interact through trivalent connections at the icosahedral and quasi-threefold axes. In contrast to the mature capsid, there is a 1 nm wide hole in the center of each pentamer in the procapsid, while the hexamers appear completely closed, when the shell is rendered at a cutoff level consistent with the predicted mass.
A central section through the procapsid reconstruction shows that the serrated appearance of the shell results from a tilting of the gp47 subunits relative to the surface of the shell (Fig. 2E). The central section also shows a diffuse central density with a diameter of 36 nm that is assumed to represent the scaffolding core. The capsid and core are separated by a 2.5 nm gap (Fig. 2E), suggesting that there is little direct interaction between the two.
Modeling of the gp47 capsid protein
All capsid proteins of phages belonging to the Caudovirales that have been studied to date conform to a fold known as the HK97 fold, after the first bacteriophage capsid structure to be solved to high resolution 21,26. These structural relationships hold even if sequence relationships are too distant to be detectable. Hence, we expected gp47 to conform to the same general fold. Indeed, inspection of the 80α reconstruction confirmed this assumption. The capsid protein clearly showed the division into an A (axial) and a P (peripheral) domain described in HK97. (The terms axial and peripheral, defined in the original HK97 publication, refer to the proximity of the domain to the fivefold and quasi-sixfold axes.) These domains should perhaps be thought of more as subdomains, since the A domain essentially constitutes an insertion into the P domain with a final, C-terminal arm that enters back into the A domain to complete the fold (Fig. 3).
Figure 3.
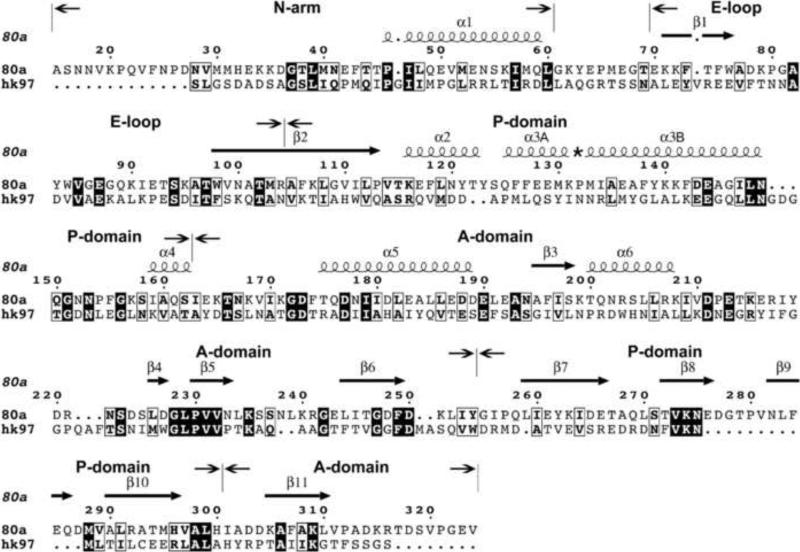
Sequence and secondary structure of 80α gp47 and HK97 gp5. The alignment was made by I-TASSER and edited according to the final model. Filled boxes show sequence identity, while open boxes show residues that share physicochemical properties, as defined in the ESPript program 54. The gp47 sequence starts at Ala 15, after the maturation cleavage site 13. The sequences corresponding to the N-arm, E loop, P domain and A domain are indicated. The secondary structure elements of gp47 according to the final model are shown as spirals (α-helices) and arrows (β-strands) above the alignment. The numbering of the α-helices follows that of HK97 22. The asterisk indicates the kink-forming Pro 132 residue in α3.
Although sequence relationships between bacteriophage capsid proteins are often hard to detect, a search for gp47 structural homologs using the protein homology/analogy recognition server Phyre 33 did return HK97 gp5 as the top homolog, with a sequence identity of 13%. Subsequently, we used the I-TASSER server 34 to produce a gp47 model based on threading into the HK97 gp5 mature capsid structure (PDB ID: 1OHG) as a template. I-TASSER returned a model with a C-score of 0.65 that was used as a starting point for the gp47 model building. The N-terminal 103 residues of HK97 gp5 constitute a “delta” or scaffolding domain that is cleaved from the protein during capsid maturation 32. This sequence is not present in the HK97 crystal structures and was therefore omitted from the comparison. Similarly, the N-terminal 14 residues of gp47, which are cleaved off before or during procapsid assembly 13, were also excluded from the modeling. The resulting gp47 model displays the characteristic feature of the gp5 template, including a long α helix (α3) known as the “spine helix” 23, a protruding β hairpin called the “E loop”, and a three-stranded β sheet in the P domain (Figs. 3, 4A). Most differences were localized to the interspersing loops and in a 12-residue insertion in the P domain.
Figure 4.
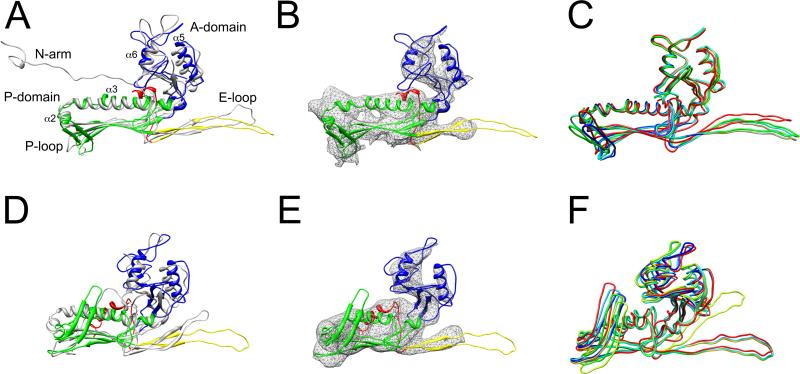
Modeling of gp47. (A) Ribbon representation of the gp47 (subunit B) model in the 80α virion, colored according to structural domains (red, N-arm; yellow, E loop; green, P domain; blue, A domain), and superimposed on HK97 gp5 (PDB ID: 1OHG) in gray. (B) Ribbon diagram of gp47 fitted into corresponding density from the 80α virion reconstruction. (C) Superposition of the models for the seven non-equivalent subunits in the 80α virion, colored as in Fig. 2A. (D) Ribbon representation of the gp47 (subunit B) procapsid model, superimposed on the HK97 gp5 procapsid structure (PDB ID: 3E8K), colored as in (A). (E) The gp47 model fitted into the 80α procapsid reconstruction. (F) Superposition of the seven non-equivalent subunits in the procapsid.
When the I-TASSER model was fitted as a rigid body into the 80α capsid density using cross correlation, it was clear that flexible adjustments would have to be made to the model. As a starting point, the two subdomains (A and P) of the gp47 model were separated at residues 151, 250 and 296. The A domain was then flexibly fitted into the corresponding electron density for subunit B using the MODELLER script Flex-EM 35. Flex-EM was not successful for the P domain, which was fitted as a rigid body with UCSF Chimera 36, upon which the two domains were put back together. The P loop insertion was built manually using the program O 37. The model was then refined through several cycles of manual model building in O, refinement of the local geometry and correlation-based fitting. Molprobity was used to assess the geometric quality of the final model 38. The final gp47 virion model (subunit B) differed from the HK97 template by an average root-mean-square deviation (RMSD) between equivalent Cα atoms of 0.52 nm (Fig. 4A, B). The process was then repeated for all seven non-equivalent subunits in the virion. The seven subunits differed from each other by an average RMSD of 0.14 nm between Cα atoms (Fig. 4C). Pertinent structural features of the gp47 model and the key differences between 80α and the HK97 template are listed below:
(1) The E loop
The E loop of HK97 gp5 constitutes a β hairpin that extends from the body of the P domain by 5.2 nm (Ref. 22; Fig. 4A). The E loop in gp47 is made of β1 and β2 (residues 70–104) and is the same length as in HK97 (Fig. 3). There was only observable density for about half of the loop (70–78 and 96–104) in the 80α reconstruction, possibly due to disorder or flexibility (Fig. 4B). The E loop density extends from the P domain by 2.4 nm and ends with a bulbous feature that matches well with several bulky, aromatic residues in the model, including F73, F75, W76 and W98. Key differences between the seven quasi-equivalent subunits are localized to the E loop and are manifested in the angle between the E loops and the P domain. Subunits B–G are similar, while subunit A (pentamer) is an outlier, with an E loop angled 21° away from the others (Fig. 4C).
(2) The spine helix
The long “spine helix” (α3) is comprised of residues 127–150 of gp47. Together with the shorter α2 helix (117–124), the spine helix forms an L-shaped motif that fits into the 80α density at one end of the P domain (Fig. 4B). The spine helix is interrupted by a proline residue at position 132, which is not present in either HK97, P22 or ε15. Consequently, the spine helix is kinked at an angle of 53° at Pro 132, whereas the spine helices in these other bacteriophages are comparatively straight 22,24. There is a loop immediately following the spine helix that includes a short helix (α4), and that acts as a hinge between the A and P domains.
(3) The A domain
The A domain includes residues 163–254 and 301–324 of gp47 (Fig. 3, 4A). It features two α helices (α5 and α6) and a three-stranded β-sheet that could be confidently fitted into corresponding density in the map (Fig. 4B). The β-sheet is made of β3, β6 and β11, the latter of which comes from the C-terminal arm. The remainder of the A domain was modeled as loops, many of which did not match any density in the map, either due to inherent flexibility or the limited resolution of the reconstruction (Fig. 4B).
(4) The P domain β sheet
The 3.9 nm long, three-stranded β sheet, which together with the spine helix motif makes up the bulk of the P domain, is composed of one strand (β2) that is continuous with the E-loop (residues 105–113), and two strands (β7 and β10) that follow reentry of the sequence into the P domain from the A domain (Fig. 3, 4A). Individual strands cannot be resolved in the map, but the β-sheet can be seen adjacent to the spine helix as a flat density that matches the extent of the modeled sheet well and is very similar to that in HK97.
(5) The P loop
The P domain of gp47 contains a 12-residue insertion (residues 276–287) into the β sheet compared to HK97 (Fig. 3). This insertion was modeled by I-TASSER as part of a 22-residue loop that coincides with a smaller loop in gp5, called the P loop 23. The gp47 P loop was modeled as a β hairpin (β8 and β9) with a classical turn motif (GXP) at its tip (Fig. 4A, B). Bacteriophage ε15 has a long P loop similar to that of gp47 25. This loop fits into the trefoils of density observed at the trivalent contact points between capsomers and is involved in extensive subunit interactions at this point (see below). The tips of the P loops shift over a range of 1.1 nm between the seven non-equivalent subunits, and together with the E loops provide the main source of conformational variation in the shell (Fig. 4C).
(6) N-arm
Residues 15–60 of gp47 correspond to the extended N-terminal “N-arm” (residues 104–132) of gp5 (Fig. 3, 4A)22. In HK97, this arm extends from the body of the P domain and makes twofold contacts with the corresponding arm from an adjacent capsomer 22. However, in the 80α virion reconstruction there was no density corresponding to this sequence (Fig. 4B). This was not entirely surprising, since the N-arm structure is highly divergent between different bacteriophages, such as HK97, P22 and ε15 22,24,25. In the 80α procapsid, a portion of the N-arm forms an ordered α-helical structure (see below).
Capsid protein interactions
The gp47 subunits participate in both intra- and inter-capsomeric interactions. The most prominent intra-capsomeric interactions observed in the map are made between α5 and α6 in adjacent subunits (Fig. 5A, B). These amphipathic helices, which are located at opposite sides of the A domain, interact via several polar and charged residues (D181 and D188 in α5; Q201, S204 and K208 in α6). These contacts are especially prominent between the C and D and the F and G subunits, where the helices make the closest approach, and less prominent in the BC, DE and EF subunit pairs (Fig. 5A). The five A subunits in the pentamer engage in similar α5–α6 interactions; in addition, a loop that extends from the apex of the A domain (residues 201–216) interfaces with fivefold related subunits at the center of the pentamer to form a fivefold lid that effectively closes the central hole (Fig. 5B). Additional contacts are formed by the E loop, in which residues W76 and K79 in the bulbous density are proximal to E129 and P132 in the spine helix of an adjacent subunit in both the hexamers and the pentamers (Fig. 5A, B).
Figure 5.
Capsid protein interactions. Ribbon diagrams of the gp47 model are shown fitted into the capsid electron density in the virion (A–C) and the procapsid (D–F). (A, D) asymmetric unit, comprising one fifth of a pentamer and a BCDEFG hexamer; (B, E) A5 pentamer; and (C, F) the type 1 (EEE) trimer. Subunits are colored as in Fig. 2A.
Inter-capsomeric interactions are primarily mediated via the trefoil formed by the P loop at the true and quasi-threefold axes between capsomers and can be divided into three types, as mentioned above (Figs. 2A and 5C). The structure and organization of the trefoils themselves are very similar in all three types. Instead, the angle between the P loop and the P domains provide the required conformational variation in the shell (Fig. 4C). These interactions might be mediated through a series of polar residues located at the base of the loop. The P loop splays out into the trefoil, but there is no clear density for its apex.
In addition to its role in forming intra-capsomeric contacts with the spine helix, the E loop may also be involved in inter-capsomeric interactions. In HK97, the E loop extends across the P domain of the adjacent subunit to form chemical crosslinks with a P loop in a neighboring capsomer 22. According to the I-TASSER model, the E loop in gp47 is long enough to reach across to neighboring capsomers in the same way even though crosslinking does not occur in 80α (Fig. 4A). The presence of bulky, aromatic residues at the tip of the loop (Y83, W84) would be consistent with a role in protein–protein interactions. However, there is no density for the E loop beyond the bulb, so the structure of the loop is currently speculative (Fig. 4B). Indeed, in the HK97 procapsid, which is not crosslinked, part of the E loop is also disordered 23.
Modeling the procapsid
Inspection of the 80α procapsid density suggested that the organization of gp47 is very different from that of gp5 in the HK97 procapsid (PDB ID: 3E8K)23. Consequently, we used the refined model of gp47 (subunit B) from the virion as a starting point for modeling gp47 subunit B into the 80α procapsid reconstruction (Fig. 2D). The smaller diameter of the procapsid is accommodated by rotating the gp47 subunits so that the A domains protrude more, thereby giving the appearance of a thicker shell (Fig. 2E). This difference is accompanied by major changes both in the conformation of gp47 and its interactions with neighboring subunits. Modeling the procapsid therefore required considerable adjustments (Fig. 4D, E). This was done in a similar fashion to the virion modeling, by splitting the structure into separate A and P subdomains that were rigid body fitted into the procapsid density, followed by cycles of manual manipulation, geometric optimization and correlation fitting. The procapsid model differed from the virion by an RMSD of 0.21 nm (subunit B). After modeling the B subunit in this way, the process was repeated for the remaining six subunits using the B subunit as a template (Fig. 4F). Differences between the procapsid and virion include:
(1) Spine helix
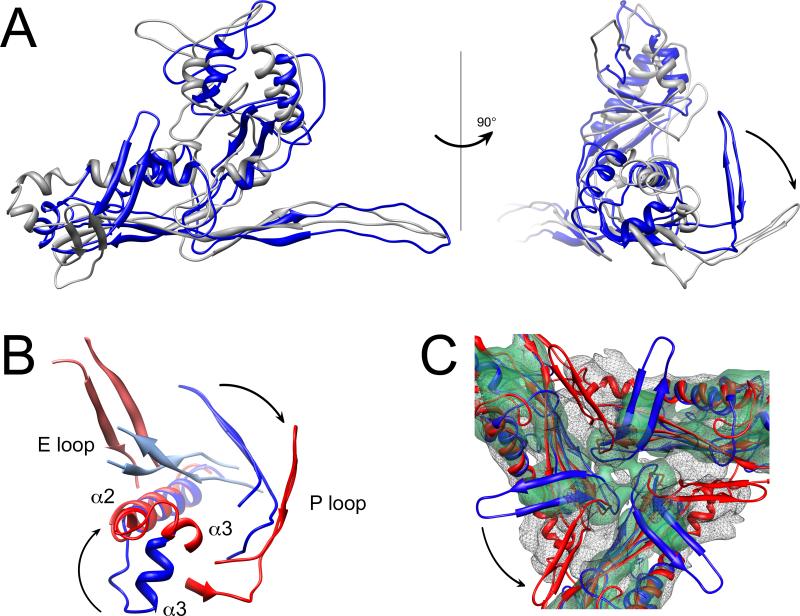
As in the virion, the spine helix (α3) in the procapsid is kinked at Pro 132, this time by about 39°. However, in the procapsid, the N-terminal part of α3 is shifted by 1.9 nm and points in a different direction relative to the virion when the C-terminal part of the helix is superimposed (Fig. 6A). This rotation is accommodated by a bimodal switch in Pro 132, for which the ψ angle changes from +135° to –45° (the two optimal values for Pro residues) between the procapsid and virion. This rotation is propagated from the N-terminal third of α3 through the shorter α2 helix, causing α2 to rotate by 97° and move by around 1.2 nm (Fig. 6A, B). There is bending of the spine helix in the procapsids of HK97 and P22 as well 23,39, but not to the extent that we observe here.
Figure 6.
Comparison of the 80α procapsid and virion. (A) Superposition of the gp47 (subunit B) models in the procapsid (blue) and the virion (gray), aligned on the C-terminal part of the spine helix (residues 133–149). The right panel shows the same superposition rotated by 90° relative to the left panel. (B) Detail of interactions between the E loop and the P domain of an adjacent subunit in the procapsid (blue) and virion (red), aligned as in (A). (C) Superposition of the density for the type 1 trimer in the virion (solid, transparent green) and procapsid (mesh) with the corresponding models of gp47 (subunit E) fitted in (blue, procapsid; red, virion). The arrows indicate domain movements upon capsid expansion.
(2) N-arm
As mentioned above, the N-terminal arm was not observed in the 80α virion reconstruction. In the procapsid map, however, a tubular density, consistent with an α-helix, reaches down behind the P domain towards the interior of the shell. The gp47 I-TASSER model predicted an α-helix in residues 46–60, which fit into the tubular density in the procapsid map (Fig. 4E). Residues 15–45 form an extended arm that might correspond to additional density beneath the “floor” of the P domain where it could form an additional β strand that runs antiparallel to the P domain β sheet. This resembles P22, where the N-arm forms a helix-turn-helix motif that folds underneath the P domain in a similar way 24. However, in P22, the N-arm has a similar conformation in both procapsid and virion. Due to uncertainty about the accuracy of the model in this region that had no equivalent in HK97 and was modeled de novo, as well as the limited resolution of the map, we did not include residues 15–45 in our final model.
(3) E loop
As in the virion, density was only observed for part of the E loop (Fig. 4E). In the virion, the bulbous part of the E loop contacts the spine helix of an adjacent subunit (Fig. 5A, B). In the procapsid, the E loop instead approaches the P loop of an adjacent subunit (Figs. 5D, E and 6B).
(4) P loop
The trivalent interactions formed by the P loop in the procapsid are similar to those in the virion (Fig. 5C, F). However, in the procapsid, the bases of the P loops are more closely packed, while the tips splay out further (Fig. 6C). When the spine helices are superimposed, the P loops in the procapsid are rotated by 63° relative to the virion (Fig. 6A). As in the virion, differences between the seven non-equivalent subunits are accommodated primarily through conformational differences in the E loops and P loops, although the differences are somewhat greater in the procapsid, with an average RMSD of 0.22 nm between subunits (Fig. 4C, F).
Intra-capsomeric interactions are largely preserved between the procapsid and virion structures, and are mediated by α5 and α6 in the A domain and by E loop–P domain contacts (Fig. 5). The smaller radius of the procapsid and the more tightly packed capsomers are mostly accommodated by tilting and rotating the subunits in such a way that the thinner and flatter virion shell has larger gaps between the subunits. When the gp47 subunits flatten out, the α5 and α6 helices move closer (Fig. 5A, D). At the type 1, 2 and 3 trivalent inter-capsomeric interfaces, subunit rotation leads to a more densely packed trefoil in the virion, while preserving the integrity of the P loop contacts (Fig. 5C, F).
Discussion
The HK97 capsid fold has been found in all members of the Caudovirales that have been studied to date, and extends across Gram-positive and Gram-negative hosts, to Archaea and even to eukaryotic species, such as herpesvirus 17,40. Thus, it was expected that the gp47 capsid protein of 80α would conform to the same fold, as we have shown here. Nevertheless, there are differences between gp47 and HK97 gp5 throughout the structure. The biggest difference is in the elongated P-loop of gp47, which forms a trifoliate β hairpin at the trivalent inter-capsomeric contact points in the 80α lattice. This trifoliate density is reflected in a clustering of strong density in the reconstruction, suggesting that this interface is especially critical for capsid stability. The differences between 80α and other phage capsids in this region may reflect alternative strategies for providing capsid stability: phage λ has an additional decoration protein (gpD) at this location that strengthens the threefold interactions 41, while HK97 uses chemical crosslinking to stabilize its shell 21. Like in 80α, the P22 capsid protein has an extended P-loop—albeit shorter than that of gp47— and an additional telokin domain that may also be involved in capsid stabilization 24. The 80α shell has a rather open structure with numerous small holes that appeared when the reconstruction was rendered at a cutoff level consistent with the expected mass of the capsid protein (Fig. 2B, C).
The prominence of the threefold contacts in the 80α lattice may also reflect a important role in assembly. Although HK97 shells can be assembled from hexamers and pentamers, at least under certain conditions 42, P22 assembly proceeds by the addition of monomers 43. HK97 may be unique in this respect, since it does not have a separate scaffolding protein 32. Clearly, the observation of “hexamer/pentamer clustering” in T = 7 bacteriophage shells does not necessarily imply that these clusters represent assembly precursors. Most likely, phage capsids can be assembled via many different and energetically equivalent pathways 44.
Capsid maturation is a dramatic process that involves extensive rearrangement of capsid protein on the surface of the shell, a change from a round to an angular shape and an overall expansion that nearly doubles the volume. Expansion is energetically favored, and occurs spontaneously in many systems as a result of chemical or thermal stress 26. In vivo, however, the expansion needs to be finely coordinated with scaffolding protein release and removal, and DNA packaging.
The expansion in 80α is accompanied by conformational changes in the capsid protein that are reflected in the differences between the virion and procapsid reconstructions observed in this study. One of these differences is the N-arm, which cannot be seen in the virion, but forms an α-helix in the procapsid that extends towards the interior of the capsid. The N-arm might thus be associated with the scaffolding protein in the procapsid. In this case, expansion may be coupled to scaffolding protein release via the observed conformational change in the N-arm. Although both gp47 and gp46 undergo N-terminal processing, this seems to be independent of scaffolding release and capsid expansion, since cleavage appears to occur prior to assembly 13.
The most striking change in the 80α capsid protein during capsid maturation is in the spine helix, and is associated with a switch in the ψ angle of Pro 132 from +135° to –45°, leading to a rotation of the N-terminal half of α3 relative to the rest of the helix (Fig. 6A,B). This change is propagated through α2 to the whole P domain, including the P loop. However, the position of the A domain relative to the P domain remains essentially the same. This differs sharply from HK97, where one of the most striking changes that occur upon capsid maturation is a rotation of the A domain by up to 39° relative to the P domain 23. Likewise, in P22, rotation around the so-called β hinge leads to a jackknifing of the A domain towards the P domain 39. In contrast, the transition in 80α is a more of a concerted movement that is propagated from the α3 kink through the whole structure. This conformational switch is also transmitted to the subunit interactions, specifically the way the E loops make contact with adjacent subunits. In the procapsid, the E loop interacts with the P loop of the adjacent subunit; in the virion, the E loop instead interacts with the spine helix. The trivalent interactions, mediated by the P loops, are mostly preserved (Fig. 5C,F and 6C). The 80α structures thus show how a complex process like capsid expansion can be effected via a simple binary switch. It might have been expected, perhaps, that the capsid protein interactions would be more malleable and thus subject to greater change than the protein conformations. However, in spite of large conformational changes, capsid protein interactions actually tend to be conserved.
When 80α mobilizes SaPI1, the capsids produced are smaller than those normally formed by 80α alone 15. This SaPI1-induced size change is reminiscent of the enterobacterial bacteriophage P2/P4 system, in which P4 can induce a size change of the P2 capsid 45,46,47. In this case, the size change is effected by the P4-encoded protein Sid, which forms an external scaffold around the P4 procapsids 48. We previously showed that while mature SaPI1 virions contain the same proteins as 80α, SaPI1 procapsids contain at least one additional SaPI1-encoded protein, gp6 13. The equivalent protein in the related SaPIbov1 had previously been genetically implicated in size determination 49 and would therefore seem to be a prime candidate for a size determination factor in this system. Work is currently underway to understand whether the SaPI1 size determination process is similar to the P2/P4 system and how additional SaPI1-encoded proteins may lead to reprogramming of the gp47 conformational repertoire to form small capsids.
Materials and methods
Production and purification of 80α virions and procapsids
80α virions and procapsids were produced by induction of the lysogenic S. aureus strains RN10616 and ST24, which carry 80α wild type and 80α ΔterS prophages, respectively 50. The cells were grown at 32 °C in CY broth as previously described 27. The prophages were induced by the addition of 2 mg/L mitomycin C (Sigma) at OD540=0.4-0.5 and grown until lysis, which typically occurred at 2.5 h post induction for 80α virions and 4 h for 80α procapsids.
Lysates were clarified by centrifugation at 5,400g for 20 min. 80α virions were harvested by precipitation of the lysate with 10% (wt/vol) PEG 6,000 and 0.5 M NaCl overnight at 4°C, followed by centrifugation at 5,400g for 20 min. The PEG pellet was resuspended in phage buffer (50 mM Tris pH 7.8, 100 mM NaCl, 1 mM MgSO4 and 4 mM CaCl2), and one-third volume of chloroform was added. The mixture was vortexed and centrifuged at 8,200g for 15 min. The aqueous (top) phase was collected and 0.5 g CsCl were added per ml solution and centrifuged at 339,000g for 20 h at 15°C in a Beckman NVTi 90 rotor. The virion-containing bands were harvested and dialyzed against dialysis buffer (20 mM Tris pH 7.8, 50 mM NaCl, 1 mM MgCl2, 2 mM CaCl2) and used for EM.
80α procapsids were produced similarly, except that 0.3-0.5 g CsCl were added per ml CHCl3-extracted solution. The procapsid-containing bands were collected, dialyzed and loaded onto 10-40% sucrose gradients in phage buffer and centrifuged for 2 h at 110,000g in a Beckman SW41 rotor. Twelve fractions were collected manually from the top of the sucrose gradients and analyzed by SDS-PAGE to check for the presence of procapsid-related proteins. Fractions containing predominantly procapsids were pelleted by centrifugation at 110,000g for 1 hr. The pellet was resuspended in dialysis buffer and used for EM experiments.
Electron microscopy
Cryo-EM was done by standard methods 51: 3μl of sample was applied to non-glow discharged C-flat holey film (Electron Microscopy Sciences, Hatfield, PA), blotted briefly before plunging into liquid ethane, and transferred to a Gatan 626 cryo-sample holder. All samples were observed in an FEI Tecnai F20 electron microscope operated at 200kV, and images were captured on Kodak SO-163 film at a magnification of 62,000×.
Three-dimensional reconstruction
The micrographs were scanned at 4,000 dpi on a Nikon CoolScan 9000 ED film scanner, corresponding to a pixel size of 0.1024 nm on the specimen. The subsequent processing was primarily done using the EMAN program suite 28. The images were binned to a pixel size of 0.2048 nm using the median value as implemented in proc2d, and particles were picked using boxer with a box size of 384×384 pixels for 80α virions and 372×372 pixels for 80α procapsids. The images were low pass filtered with a 0.5 nm resolution Gaussian filter. Contrast transfer function parameters were determined using the program ctfit and phase correction was applied to the images. Classification, generation of class averages and reconstruction by backprojection were done as implemented in EMAN. The resolution was assessed using Fourier shell correlation between two separate reconstructions using half the data set, and the final reconstructions were low pass filtered at 0.6 nm resolution. Visualization of the maps and fitting of the capsid protein structure were done using UCSF Chimera 36,52. The appropriate density cutoff level was determined by first deleting the density attributable to the DNA in the virion and the scaffolding in the procapsid, and then calculating the cutoff level consistent with the predicted 420 × 35.1 kDa mass of the shell. The reconstructions were submitted to the EMDB (http://emdep.rutgers.edu) with accession numbers EMD-5236 and EMD-5237 for the virion and procapsid, respectively.
Modeling
An initial search for structural homologues using the program Phyre 33 identified HK97 gp5 as the top hit, and output a sequence alignment and a gp5-based structural model. An improved model was generated with the program I-TASSER 34, specifying HK97 gp5 (PDB ID: 1OHG) as a template. I-TASSER returned five models with C-scores from – 1.77 to +0.65, all of which followed the fold of the template, but with differences in the connecting loops between secondary structure elements. The highest-scoring model from ITASSER was chosen as a starting point for the fitting of subunit B in the virion map. Initial rigid-body fitting into the 80α virion electron density maps was done using UCSF Chimera36,52. Flexible fitting of the 80α gp47 A domain (residues 164-250) was performed using the simulated annealing molecular dynamics (MD) function of Flex-EM 35. Four MD cycles were done and the resulting model was recombined with the rigid body fitted I-TASSER model for the P domain. This initial fit was followed by several cycles of manual modeling and geometrical optimization in O 37, and cross-correlation fitting using Chimera 36,52. The geometrical quality of the model was assessed using Molprobity 38. Fitting of the procapsid subunits was carried out in a similar manner, using the fitted B subunit from the virion as a starting point for the procapsid B subunit. Subunit B in both the virion and procapsid was then used as a starting point for modeling the other six subunits. Subunit interactions and conformational differences were analyzed by the CCP4 programs contacts and lsqkab, respectively53.
Acknowledgements
This work was funded by NIH grants R21 AI071982 and R01 AI083255 to T.D.; R21 AI067654 and an A.D. Williams Trust and Baruch Foundation Trust Grant-In-Aid to G.E.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers:
Maps have been submitted to EMDep, accession codes EMD-5236 and EMD-5237.
References
- 1.Gordon RJ, Lowy FD. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 2008;46:S350–S359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naber CK. Staphylococcus aureus bacteremia: epidemiology, pathophysiology, and management strategies. Clin. Infect. Dis. 2009;48:S231–S237. doi: 10.1086/598189. [DOI] [PubMed] [Google Scholar]
- 4.Los M, Kuzio J, McConnell MR, Kropinski AM, Wegrzyn G, Christie GE. Lysogenic conversion in bacteria of importance to the food industry. In: Sabour MP, Griffiths M, editors. Bacteriophage in the detection and control of foodborne pathogens. ASM Press; Washington, D.C.: 2010. pp. 157–198. [Google Scholar]
- 5.Brussow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Molec. Biol. Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyer DW, Rock MI, Lee CY, Iandolo JJ. Generation of transducing particles in Staphylococcus aureus. J. Bacteriol. 1985;161:91–95. doi: 10.1128/jb.161.1.91-95.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novick RP, Edelman I, Lofdahl S. Small Staphylococcus aureus plasmids are transduced as linear multimers that are formed and resolved by replicative processes. J. Mol. Biol. 1986;192:209–220. doi: 10.1016/0022-2836(86)90360-8. [DOI] [PubMed] [Google Scholar]
- 8.Novick RP. Mobile genetic elements and bacterial toxinoses: the superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid. 2003;49:93–105. doi: 10.1016/s0147-619x(02)00157-9. [DOI] [PubMed] [Google Scholar]
- 9.Novick RP, Christie GE, Penades JR. The phage-related chromosomal islands of Gram-positive bacteria. Nat. Rev. Microbiol. 2010;8:541–551. doi: 10.1038/nrmicro2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maiques E, Ubeda C, Campoy S, Salvador N, Lasa I, Novick RP, Barbe J, Penades JR. beta-lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J. Bacteriol. 2006;188:2726–2729. doi: 10.1128/JB.188.7.2726-2729.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tormo-Mas MA, Mir I, Shrestha A, Tallent SM, Campoy S, Lasa I, Barbe J, Novick RP, Christie GE, Penades JR. Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands. Nature. 2010;465:779–782. doi: 10.1038/nature09065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tallent SM, Langston TB, Moran RG, Christie GE. Transducing particles of Staphylococcus aureus pathogenicity island SaPI1 are comprised of helper phage-encoded proteins. J. Bacteriol. 2007;189:7520–7524. doi: 10.1128/JB.00738-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poliakov A, Chang JR, Spilman MS, Damle PK, Christie GE, Mobley JA, Dokland T. Capsid size determination by Staphylococcus aureus pathogenicity island SaPI1 involves specific incorporation of SaPI1 proteins into procapsids. J. Mol. Biol. 2008;380:465–475. doi: 10.1016/j.jmb.2008.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tormo MA, Ferrer MD, Maiques E, Ubeda C, Selva L, Lasa I, Calvete JJ, Novick RP, Penades JR. SaPI DNA is packaged in particles composed of phage proteins. J. Bacteriol. 2008;190:2434–2440. doi: 10.1128/JB.01349-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruzin A, Lindsay JA, Novick RP. Molecular genetics of SaPI1 - a mobile pathogenicity island in Staphylococcus aureus. Mol. Microbiol. 2001;41:365–377. doi: 10.1046/j.1365-2958.2001.02488.x. [DOI] [PubMed] [Google Scholar]
- 16.Christie GE, Matthews AM, King DG, Lane KD, Olivarez NP, Tallent SM, Gill SR, Novick RP. The complete genomes of Staphylococcus aureus bacteriophages 80 and 80 alpha - implications for the specificity of SaPI mobilization. Virology. 2010 doi: 10.1016/j.virol.2010.08.036. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson JE, Chiu W. DNA packaging and delivery machines in tailed bacteriophages. Curr. Opin. Struct. Biol. 2007;17:237–243. doi: 10.1016/j.sbi.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Dokland T. Scaffolding proteins and their role in viral assembly. Cell. Mol. Life Sci. 1999;56:580–603. doi: 10.1007/s000180050455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fane BA, Prevelige PE. Mechanism of scaffolding-assisted viral assembly. Adv. Prot. Chem. 2003;64:259–299. doi: 10.1016/s0065-3233(03)01007-6. [DOI] [PubMed] [Google Scholar]
- 20.Hendrix RW. Bacteriophage HK97: assembly of the capsid and evolutionary connections. Adv. Virus Res. 2005;64:1–14. doi: 10.1016/S0065-3527(05)64001-8. [DOI] [PubMed] [Google Scholar]
- 21.Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 22.Helgstrand C, Wikoff WR, Duda RL, Hendrix RW, Johnson JE, Liljas L. The refined structure of a protein catenane: the HK97 bacteriophage capsid at 3.44Å resolution. J. Mol. Biol. 2003;334:865–899. doi: 10.1016/j.jmb.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 23.Gertsman I, Gan L, Guttman M, Lee K, Speir JA, Duda RL, Hendrix RW, Komives EA, Johnson JE. An unexpected twist in viral capsid maturation. Nature. 2009;458:646–650. doi: 10.1038/nature07686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parent KN, Khayat R, Tu LH, Suhanovsky MM, Cortines JR, Teschke CM, Johnson JE, Baker TS. P22 coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks. Structure. 2010;18:390–401. doi: 10.1016/j.str.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1135. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- 26.Johnson JE. Virus particle maturation: insights into elegantly programmed nanomachines. Curr. Opin. Struct. Biol. 2010;20:210–216. doi: 10.1016/j.sbi.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novick RP. Genetic systems in Staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 28.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-partice reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 29.Dokland T, Murialdo H. Structural transitions during maturation of bacteriophage lambda capsids. J. Mol. Biol. 1993;233:682–694. doi: 10.1006/jmbi.1993.1545. [DOI] [PubMed] [Google Scholar]
- 30.Roos WH, Ivanovska IL, Evilevich A, Wuite GJ. Viral capsids: mechanical characteristics, genome packaging and delivery mechanisms. Cell. Mol. Life Sci. 2007;64:1484–1497. doi: 10.1007/s00018-007-6451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrov AS, Boz MB, Harvey SC. The conformation of double-stranded DNA inside bacteriophages depends on capsid size and shape. J. Struct. Biol. 2007;160:241–248. doi: 10.1016/j.jsb.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conway JF, Duda RL, Cheng N, Hendrix RW, Steven AC. Proteolytic and conformational control of virus capsid maturation: the bacteriophage HK97 system. J. Mol. Biol. 1995;253:86–99. doi: 10.1006/jmbi.1995.0538. [DOI] [PubMed] [Google Scholar]
- 33.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Topf M, Baker ML, John B, Chiu W, Sali A. Structural characterization of components of protein assemblies by comparative modeling and electron cryo-microscopy. J. Struct. Biol. 2005;149:191–203. doi: 10.1016/j.jsb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 37.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for the building of protein models in electron density and the location of errors in these models. Acta Cryst. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 38.Chen VB, Arendall W. B. r., Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Cryst. D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teschke CM, Parent KN. ‘Let the phage do the work’: Using the phage P22 coat protein structures as a framework to understand its folding and assembly mutants. Virology. 2010;401:119–130. doi: 10.1016/j.virol.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 2005;15:655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 41.Lander G, Evilevich A, Jeembaeva M, Potter CS, Carragher B, Johnson JE. Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Structure. 2008;16:1399–1406. doi: 10.1016/j.str.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie Z, Hendrix RW. Assembly in vitro of bacteriophage HK97 proheads. J. Mol. Biol. 1995;253:74–85. doi: 10.1006/jmbi.1995.0537. [DOI] [PubMed] [Google Scholar]
- 43.Prevelige P, Thomas D, King J. Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophys. J. 1993;64:824–835. doi: 10.1016/S0006-3495(93)81443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dokland T. Freedom and restraint: themes in virus capsid assembly. Structure. 2000;8:R157–R167. doi: 10.1016/s0969-2126(00)00181-7. [DOI] [PubMed] [Google Scholar]
- 45.Christie GE, Calendar R. Interactions between satellite bacteriophage P4 and its helpers. Annu. Rev. Genet. 1990;24:465–490. doi: 10.1146/annurev.ge.24.120190.002341. [DOI] [PubMed] [Google Scholar]
- 46.Lindqvist BH, Deho G, Calendar R. Mechanisms of genome propagation and helper exploitation by satellite phage P4. Microbiol. Rev. 1993;57:683–702. doi: 10.1128/mr.57.3.683-702.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dokland T, Lindqvist BH, Fuller SD. Image reconstruction from cryo-electron micrographs reveals the morphopoietic mechanism in the P2-P4 bacteriophage system. EMBO J. 1992;11:839–846. doi: 10.1002/j.1460-2075.1992.tb05121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marvik OJ, Dokland T, Nøkling RH, Jacobsen E, Larsen T, Lindqvist BH. The capsid size-determining protein Sid forms an external scaffold on phage P4 procapsids. J. Mol. Biol. 1995;251:59–75. doi: 10.1006/jmbi.1995.0416. [DOI] [PubMed] [Google Scholar]
- 49.Ubeda C, Maiques E, Barry P, Matthews A, Tormo MA, Lasa I, Novick RP, Penades JR. SaPI mutations affecting replication and transfer and enabling autonomous replication in the absence of helper phage. Mol. Microbiol. 2008;67:493–503. doi: 10.1111/j.1365-2958.2007.06027.x. [DOI] [PubMed] [Google Scholar]
- 50.Ubeda C, Olivarez NP, Barry P, Wang H, Kong X, Matthews A, Tallent SM, Christie GE, Novick RP. Specificity of staphylococcal phage and SaPI DNA packaging as revealed by integrase and terminase mutations. Mol. Microbiol. 2009;72:98–108. doi: 10.1111/j.1365-2958.2009.06634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dokland T, Ng ML. Transmission electron microscopy of biological specimens. In: Dokland T, Hutmacher DW, Ng ML, Schantz JT, editors. Techniques in microscopy for biomedical applications. World Scientific Press; Singapore: 2006. [Google Scholar]
- 52.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 53.Collaborative Computational Project, No. 4 The CCP4 suite: programs for protein crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 54.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]