Abstract
GroEL is a group I chaperonin that facilitates protein folding and prevents protein aggregation in the bacterial cytosol. Mycobacteria are unusual in encoding two or more copies of GroEL in their genome. While GroEL2 is essential for viability and likely functions as the general housekeeping chaperonin, GroEL1 is dispensable but its structure and function remain unclear.
Here we present the 2.2 Å resolution crystal structure of a 23 kDa fragment of Mycobacterium tuberculosis GroEL1 consisting of an extended apical domain. Our X-ray structure of the GroEL1 apical domain closely resembles those of Escherichia coli GroEL and M. tuberculosis GroEL2; thus, highlighting the remarkable structural conservation of bacterial chaperonins. Notably, in our structure, the proposed substrate-binding site of GroEL1 interacts with the N-terminal region of a symmetry related, neighboring GroEL1 molecule. The latter is consistent with the known GroEL apical domain function in substrate binding, and is supported by results obtained from using peptide array technology. Taken together, we show that the apical domains of M. tuberculosis GroEL paralogs are conserved in three-dimensional structure, suggesting that GroEL1, like GroEL2, is a chaperonin.
Keywords: Molecular chaperones, Hsp60, apical domain, protein folding, KasA
Introduction
Molecular chaperones assist protein folding by facilitating the productive folding of newly synthesized polypeptides and by preventing protein aggregation in the crowded environment of the cell.1 Escherichia coli GroEL is a group I chaperonin that assembles into an 800 kDa homo-tetradecamer composed of two heptameric rings that are stacked back-to-back.2; 3 Each GroEL subunit has a molecular weight of 57 kDa and consists of an equatorial, an intermediate, and an apical domain.3 The equatorial domain contains the ATP-binding site and mediates contacts between subunits in the cis and trans rings. The intermediate domain functions as a hinge that connects the equatorial domain to the apical domain. The latter forms the entrance to the GroEL cavity and is involved in GroES binding4 as well as polypeptide recognition.5; 6 It has been suggested from X-ray crystallographic studies that helices H and I of the E. coli GroEL apical domain form the substrate binding site.7; 8; 9
Interestingly, many mycobacteria contain genes encoding two or more GroEL paralogs.10 GroEL1 and GroEL2 from the human pathogen Mycobacterium tuberculosis11; 12 are of particular interest as both proteins are involved in the host immune response to M. tuberculosis infection.13; 14 While GroEL2 is essential and likely functions as the principal housekeeping chaperonin,10; 11 GroEL1 is non-essential and is dispensable for viability. It has been proposed that M. tuberculosis GroEL1 is a nucleoid-associated protein,15 and that the closely related GroEL1 ortholog from M. smegmatis plays a role in biofilm formation by modulating mycolic acid biosynthesis through direct interaction with the β-ketoacyl ACP synthase KasA.10
Like other bacterial chaperonins, M. tuberculosis GroEL1 and GroEL2 are up-regulated upon heat shock16 as well as in response to oxidative stress,17 indicating that both copies may have chaperone activity inside cells. In contrast, recombinant GroEL1 and GroEL2 overexpressed in E. coli exist as dimers, and exhibit low ATPase and no folding activities.18 Since native GroEL1 forms higher-order oligomers in M. tuberculosis cells,19 lack of chaperone activity might be attributed to the inability of the recombinant proteins to self-assemble.
Consistent with its essential cellular role, the X-ray structure of a M. tuberculosis GroEL2 dimer20 showed that the GroEL2 monomer has the same fold as E. coli GroEL,20 supporting the notion that GroEL2 is a chaperonin. However, at present, no high-resolution structural information is available for M. tuberculosis GroEL1, and its structure-function relationship remains unclear.
Here we present the 2.2 Å resolution crystal structure of a 23 kDa M. tuberculosis GroEL1 fragment consisting of the GroEL1 apical domain flanked by flexible segments that are part of the intermediate domain. This structure is hereafter referred to as the GroEL1 apical domain. We found that the atomic structure of the GroEL1 apical domain is very similar to those of M. tuberculosis GroEL220 and E. coli GroEL.7; 8 Fortuitously, in our crystal structure, the N-terminus of one molecule interacts with the putative GroEL substrate-binding site of a symmetry related molecule. This interaction is reminiscent of the X-ray structures of E. coli chaperonin-substrate peptide complexes.7; 8; 9 Moreover, we found using peptide array technology that both full-length M. tuberculosis GroEL1 and the isolated GroEL1 apical domain recognize the same peptide motifs present in the M. tuberculosis KasA sequence, which resemble binding motifs reported for E. coli GroEL.21 Thus, our combined structural and functional data suggest that M. tuberculosis GroEL1, like GroEL2, is a chaperonin and support the notion that the apical domain is sufficient for substrate interaction.
Results and Discussion
Crystal Structure of the M. tuberculosis GroEL1 Apical Domain
Crystals of the GroEL1 apical domain (residues 184–377) diffracted to 2.2 Å resolution on a home X-ray source, and belonged to the orthorhombic space group P21212 with one molecule in the asymmetric unit. After rigid body refinement of the initial model in CNS22 the resulting electron density map was readily interpretable. Several rounds of model building, simulated annealing, least-squares positional, and individual B-factor refinement were performed resulting in a final model with a Rfactor and Rfree 23 of 21.0% and 23.2%, respectively. The final model consists of 194 residues and 65 water molecules. The Ramachandran plot showed that 91.1% of all residues were in the most favored regions and 8.9% in additional allowed regions. No residue was in a generously allowed or disallowed region. The refinement statistics are summarized in Table 1.
Table 1.
Summary of Data Collection and Refinement Statistics
| M. tuberculosis GroEL1 apical domain | |
|---|---|
| Diffraction data statistics | |
| Resolution (Å) | 40.0 – 2.2 (2.28 – 2.2) |
| Space group | P 21212 |
| Unit cell parameters | a = 75.47 Å, b = 78.65 Å, c = 34.89 Å |
| α = β = γ = 90° | |
| No. of unique reflections | 10,994 |
| Completeness (%) | 98.7 (91.9) |
| Redundancy | 4.3 (2.6) |
| I/sigma (I) | 12.3 (2.4) |
| Rsym (%)b | 10.1 (36.7) |
| Refinement statistics | |
| Resolution (Å) | 20.0 – 2.2 |
| Rfactor / Rfree (%)c | 21.0 / 23.2 |
| Average B-factor (Å2) | 30.3 |
| Number of atoms | |
| Protein | 1461 |
| Water | 65 |
| RMSD of ideal bond length/angle | |
| Bond length (Å) | 0.006 |
| Bond angle (°) | 1.2 |
Values in parentheses are for the highest resolution shell.
Rsym = Σ |I − (I)| / Σ (I), where I is the observed intensity and (I) is the average intensity.
Rfactor Σ |Fobs − Fcalc| / Σ |Fobs|, where Fobs are the observed structure factors and Fcalc are the calculated structure factors. The crystallographic Rfactor is based on 95% of the data used in refinement, and the Rfree is based on 5% of the data withheld for cross-validation test.
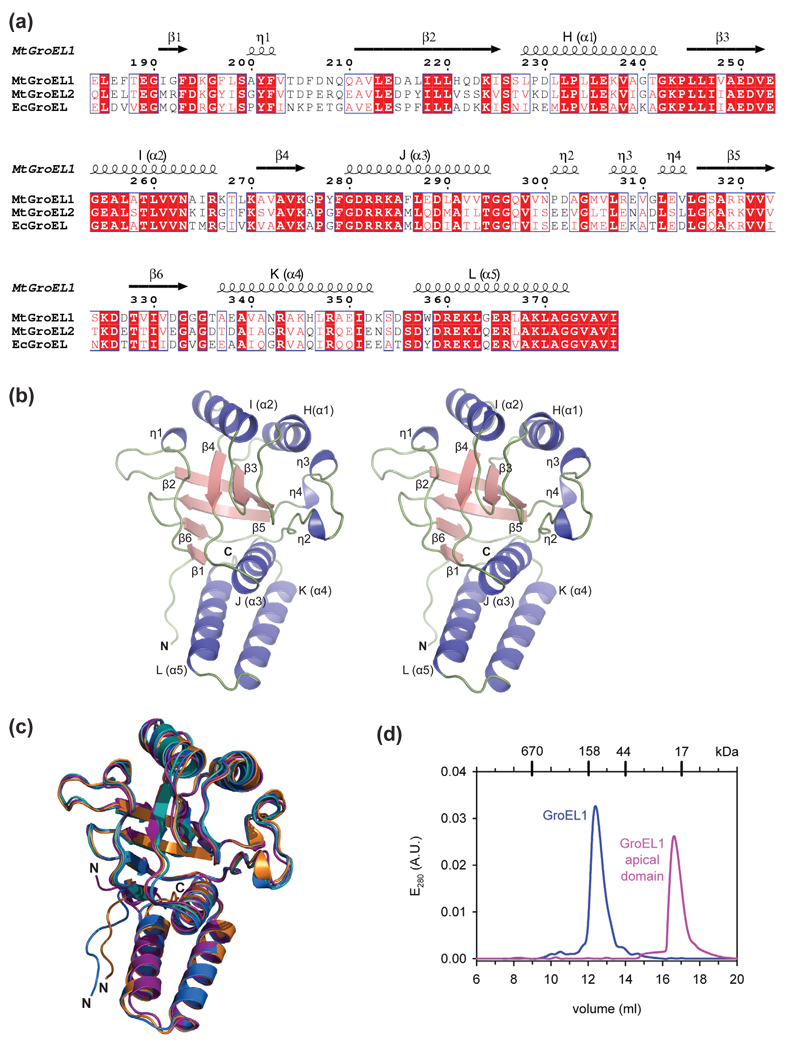
The structure of the M. tuberculosis GroEL1 apical domain consists of a β-sandwich scaffold flanked by several α-helices and loops (Fig. 1a and b). Structural comparison of the M. tuberculosis GroEL1 apical domain with those of M. tuberculosis GroEL2 (PDB ID: 1SJP-A)20 and E. coli GroEL (PDB ID: 1KID and 1DKD-A)7; 8 showed that they are very similar (Fig. 1c). The Cα atoms of the refined M. tuberculosis GroEL1 apical domain structure (residues 191–372) superimposed pairwise with those of M. tuberculosis GroEL2 residues 190–371 (PDB ID: 1SJP-A) 20 with a root mean square deviation (RMSD) of 0.82 Å, and with those of E. coli GroEL residues 193–375 (PDB ID: 1KID)7 and residues 193–336 (PDB ID: 1DKD-A)8 with a RMSD of 0.63 Å and 0.75 Å, respectively.
Fig. 1.
Bacterial chaperonins are structurally conserved. (a) Sequence alignment of the apical domains of M. tuberculosis (Mt) GroEL1, Mt GroEL2, and E. coli (Ec) GroEL. Secondary structure elements of M. tuberculosis GroEL1 are represented by helices (α- and η [310]-helices) and arrows (β-strands). Conserved residues are boxed. Nomenclature of α-helices (H–L) is the same as previously described for the E. coli GroEL apical domain.8 (b) Stereo view of the M. tuberculosis GroEL1 apical domain. The crystal structure is shown as ribbon diagram with α-helices colored blue, β-strands red, and the N- and C-terminal extensions and loops green. η denotes a 310 helix. (c) Superposition of the apical domain structures from M. tuberculosis GroEL1 (orange), M. tuberculosis GroEL2 (magenta),20 and E. coli GroEL residues 191–376 (blue)7 and residues 191–336 (cyan).8 (d) Analysis of full-length GroEL1 (blue) and GroEL1 apical domain (magenta) by size-exclusion chromatography. The chromatogram shows that the full-length protein elutes as a dimer, whereas the apical domain is a monomer.
Protein-protein interactions between symmetry related GroEL1 molecules
Our crystal structure showed two types of protein interfaces with crystallographic symmetry related GroEL1 apical domain molecules in the crystal lattice. While the observed contacts may hint at the existence of higher oligomers, none of the observed interactions are consistent with the known interfaces in the E. coli GroEL tetradecamer structure.2; 3 It has been proposed that both recombinant, full-length M. tuberculosis GroEL1 and GroEL2 form dimers18 through inter-subunit contacts between apical domains.20 While our biochemical results support the notion that recombinantly expressed, full-length GroEL1 is a dimer, we found that the isolated GroEL1 apical domain is a monomer in solution (Fig. 1d).
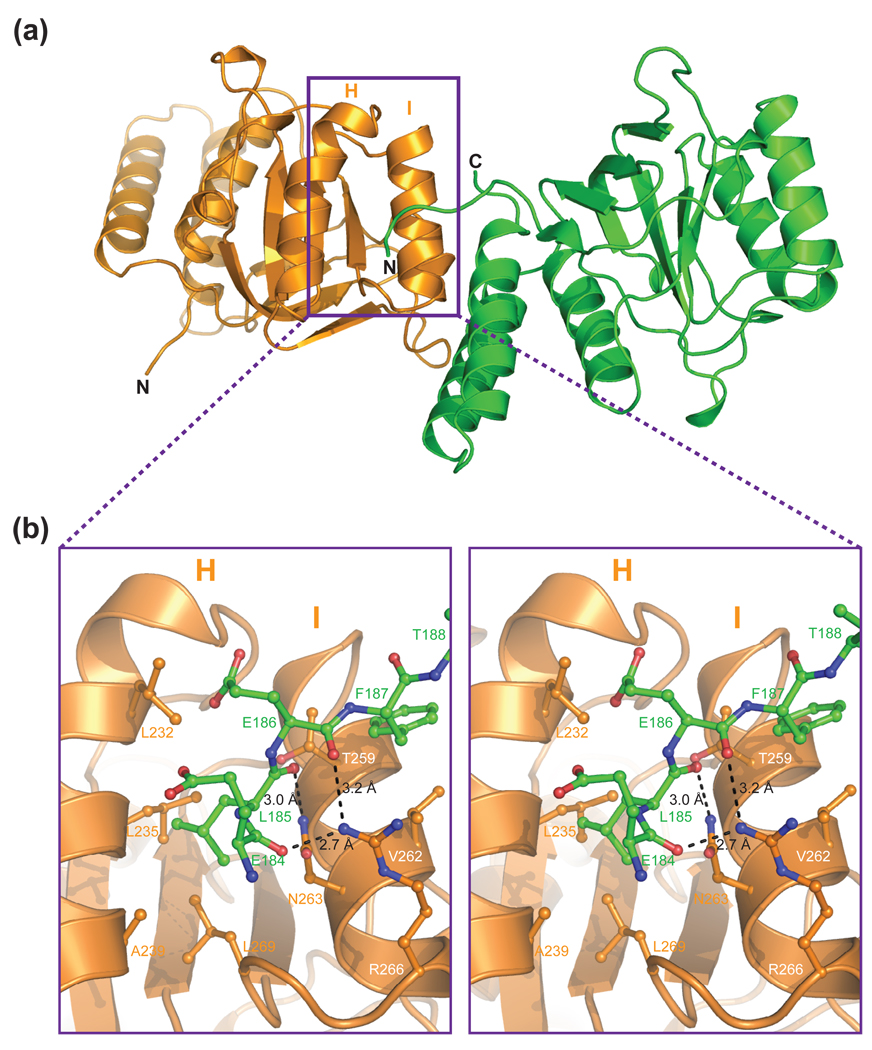
In our structure, the N-terminal residues 184 to 187 with sequence Glu-Leu-Glu-Phe interact with helices H and I of a symmetry-related neighboring molecule (Fig. 2a and b). Notably, the Leu185 side-chain is in van der Waals contact with the side-chains of Leu232, Leu235, Ala239, and Leu269, which form a hydrophobic pocket (Fig. 2b and 3a). In addition, there is a network of hydrogen bond interactions between the polar side chains of Asn263 and Arg266 and the main chain carbonyl oxygens of Glu184, Leu185, and Glu186 (Fig. 2b and 3a). These interactions are reminiscent of the previously observed E. coli GroEL apical domain contacts with different peptides (Fig. 3b–c).7; 8; 9
Fig. 2.
Structure of the GroEL1 substrate-binding site. (a) Ribbon diagram of the GroEL1 apical domain (orange) and that of a crystallographic symmetry related molecule (green). The putative substrate binding site is indicated by a purple box. (b) Close-up stereo view of the substrate binding site in GroEL1. Interacting residues from helices H and I (orange) and from the N-terminus of a crystallographic symmetry related molecule (green) are depicted as ball-and-stick models. Hydrogen bonds are indicated by dashed lines and are shown together with the corresponding bond lengths.
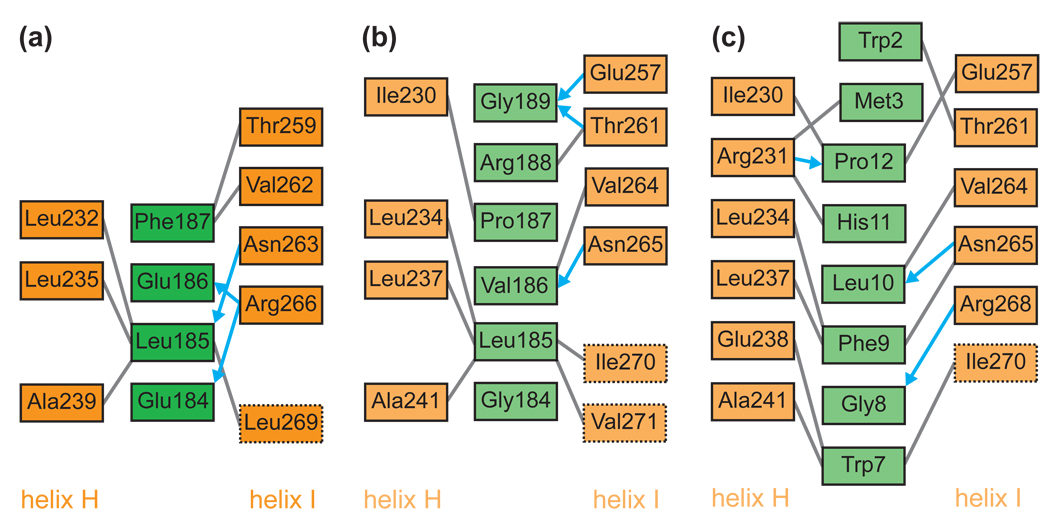
Fig. 3.
Schematic representation of molecular interactions at the putative chaperonin-substrate peptide interface in M. tuberculosis GroEL1 and E. coli GroEL. (a) M. tuberculosis GroEL1 apical domain bound to an N-terminal segment of GroEL1, (b) E. coli GroEL apical domain bound to a non-native peptide,7 and (c) E. coli GroEL apical domain bound to a strong binding peptide selected by phage display.8 Gray lines indicate van der Waals contacts, blue arrows represent hydrogen bonds. Residues surrounded by a dotted box are located on a loop immediately downstream of helix I.
GroEL1 substrate binding site
It has been proposed that the apical domain of E. coli GroEL contains the substrate binding site.5; 6 This is supported by crystal structures of the E. coli GroEL apical domain bound to a strong binding peptide selected by phage-display,8 and to an extended N-terminal segment of a symmetry related molecule,7 respectively. In the latter two structures, protein-protein interactions were mediated by both non-polar and hydrogen bond interactions with helices H and I of the GroEL apical domain.7; 8 Although the sequence of the bound peptides differed in each case, the two previously reported X-ray structures of the E. coli GroEL apical domain7; 8 and our atomic structure of the M. tuberculosis GroEL1 apical domain display common binding features. First, the hydrophobic pocket is lined by the conserved residues Leu232, Leu235, and Ala239 from helix H and the less conserved Leu269, which correspond to Leu234, Leu237, Ala241, and Val271 in E. coli GroEL (Fig. 3a–c). Second, hydrogen bond interactions mainly involve residues from helix I (Fig. 3a–c). Third, hydrogen bonds are exclusively formed between the side chains of GroEL and the main chain of the bound peptide (Fig. 3a–c). Notably, in our structure, only four residues (Glu-Leu-Glu-Phe) contact the substrate binding site of the apical domain, which may represent the minimal binding region.
GroEL1-KasA peptide interaction
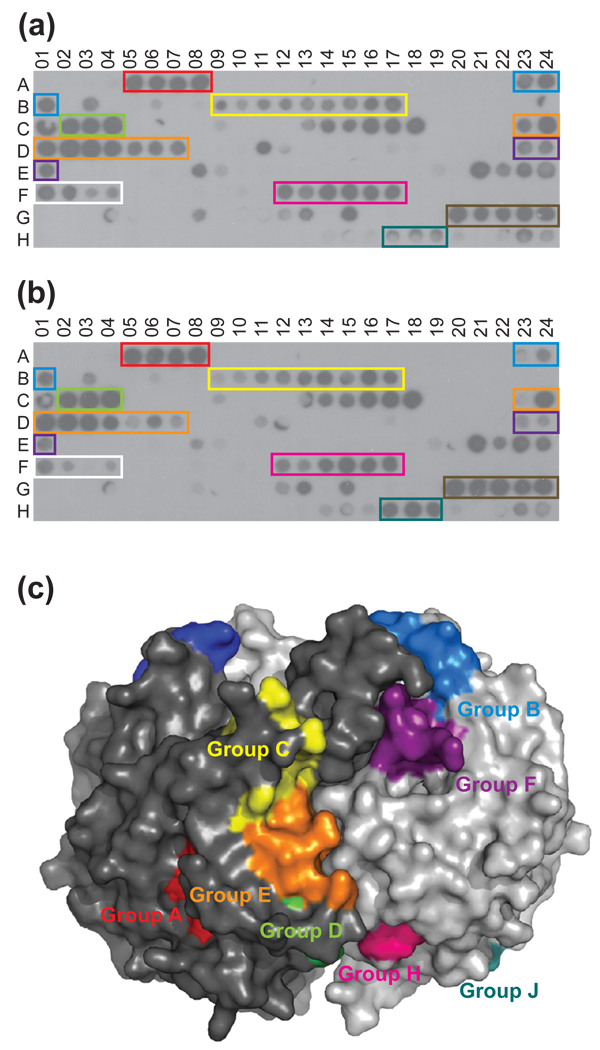
To gain further insight into substrate recognition by M. tuberculosis GroEL1, we synthesized a miniaturized peptide array of overlapping 12-mer peptides derived from the M. tuberculosis KasA amino acid sequence. This work followed from an earlier report demonstrating a direct interaction between KasA and GroEL1 in M. smegmatis cells,10 which serves as a model organism for different mycobacterial species, including M. tuberculosis. Probing our KasA-derived peptide array with full-length GroEL1 showed binding to 67 out of 192 peptides (Fig. 4a). Eleven of those peptides (C01, C14, C16 – C18, E21 – E24, G13, and G15) also bound to a control protein and, therefore, were excluded from further analysis (data not shown). Remarkably, we found that the isolated GroEL1 apical domain bound to the same peptides as full-length GroEL1, suggesting that the apical domain is sufficient for substrate binding, although, some spots differed in intensity indicating different binding affinities (Fig. 4a and b). Moreover, the same KasA peptides, in addition to other KasA motifs, were also recognized by full-length GroEL2 in a control experiment (Fig. S1).
Fig. 4.
GroEL1 apical domain is sufficient for substrate recognition. Binding of (a) full-length M. tuberculosis GroEL1 or (b) the isolated GroEL1 apical domain to a miniaturized peptide array derived from the M. tuberculosis KasA sequence. Peptides belonging to the same group are boxed together. Groups are differentiated by the color of boxes: Group A (red), Group B (blue), Group C (yellow), Group D (green), Group E (orange), Group F (purple), Group G (white), Group H (pink), Group I (brown), and Group J (teal). (c) Surface representation of the KasA dimer structure (PDB ID: 2WGF)37 with the KasA monomers displayed in different hues. Consensus motifs are mapped onto the KasA structure using the same color scheme as in Fig. 4a and b.
Consecutive binding peptides with at least three members were grouped together to identify consensus motifs within each group (Table 2). A consensus motif was defined as an amino acid sequence that was present in most members of a group, and was at least four amino acid residues long. Only one acidic residue (Asp) was found in all consensus motifs (Table 2; Group B), which is very low compared to the abundance of negatively charged residues (14 %) in the KasA sequence. On the other hand, positively charged (Arg), and hydrophobic residues (Val and Met) were enriched 2 to 2.6-fold in the consensus motifs relative to the full-length sequence. The latter is in good agreement with previous studies of peptide sequences bound by E. coli GroEL, which also revealed a strong preference for hydrophobic and positively charged residues.21; 24; 25; 26
Table 2.
KasA peptides bound by GroEL1
| Group | Spots | Sequencea |
|---|---|---|
| A | A05-A08 | 9GGFPSVVVTAVTATTSIS26 |
| B | A23-B01 | 49EFVTKWDLAVKIGGHL64 |
| C | B09-B17 | 68DSHMDGRLDMRRMSYVQRMGKLLGGQLWE86 |
| D | C02-C04 | 103VDPDRFAVVVGTGLGG118 |
| E | C23-D07 | 145IMPNGAAAVIGLQLGARAGVMTPVSACS172 |
| F | D23-E01 | 205LPIAAFSMMRAMSTRN220 |
| G | F01-F04 | 257RGAKPLARLLGAGITSDA274 |
| H | F12-F17 | 279APAADGVRAGRAMTRSLELAGL300 |
| I | G20-G24 | 349AVGALESVLTVLTLRDGVIP368 |
| J | H17-H19 | 391YGDYRYAVNNSFGFGG406 |
Consensus motifs are depicted in bold. Lower case numbers indicate the position in the M. tuberculosis KasA sequence.
To provide a spatial explanation for the location of our identified peptides, we mapped our consensus motifs onto the available X-ray structure of M. tuberculosis KasA27 (Fig. 4c). We found that six of our consensus motifs (Groups A, D, G, H, I, J) are mostly buried in the native protein structure, as it might be expected for a chaperonin substrate.28 However, other consensus motifs (Groups B, C, E, and F) are solvent-exposed (Fig. 4c), suggesting that GroEL1 may also interact with native, folded KasA. Taken together, our findings suggest that M. tuberculosis GroEL1 and GroEL2 are chaperonins that recognize both distinct and overlapping KasA peptide motifs. How GroEL1 modulates fatty-acid synthesis through interaction with KasA remains unclear and is subject to further investigation.
Materials and Methods
Protein preparation and analysis
Cloning, expression, and purification of full-length GroEL1 from M. tuberculosis H37Rv, as well as the crystallization and preliminary crystallographic analysis of the GroEL1 fragment have been described.29 For peptide array analysis, the apical domain of GroEL1 (Glu188 to Val373) was cloned by PCR into pProEx Htb generating a plasmid that harbors the GroEL1 apical domain with a Tobacco Etch Virus protease-cleavable N-terminal His6-tag. Expression and purification of the GroEL1 apical domain was performed as previously described for full-length GroEL1.29 GroEL2 was cloned, overexpressed, and purified in a similar manner, except for addition of a complete EDTA-free protease inhibitor cocktail (Roche Diagnostics) in the lysis buffer.
To determine the oligomeric state, purified full-length GroEL1 and GroEL1 apical domain were analyzed at 4 °C by size-exclusion chromatography on a Superdex 200 HR 10/30 column (GE Healthcare) in 50 mM Tris pH 7.5 and 150 mM NaCl. Molecular weight protein standards (BioRad) were run in the same buffer in order to correlate elution volume with protein size.
X-ray crystallographic analysis and structure refinement
The structure of the GroEL1 apical domain was determined by the molecular replacement technique using the apical domain (residues 188 to 372) of a M. tuberculosis GroEL2 monomer (PDB ID: 1SJP-A)20 as search model. This model shares 64% sequence identity with the M. tuberculosis GroEL1 apical domain, distributed evenly over the amino acid sequence (Fig. 1a). 5% of the observed data were randomly chosen and excluded from refinement for cross-validation purposes. The model was refined in CNS 1.2.22 Model refinement was interspersed by manual rebuilding of the atomic structure using COOT.30 Water molecules were selected automatically in CNS 1.2,22 and confirmed manually according to the peak height and distance criteria in the calculated Fo-Fc and 2Fo-Fc maps. The stereochemistry of the final model was analyzed using PROCHECK.31
Superpositioning of molecules was done in PyMol32, the SSAP Server33 was used for RMSD calculations, and ESPript34 was used for preparing the secondary structure alignment.
Peptide array synthesis
A peptide array of 12-mer overlapping peptides derived from the amino acid sequence of M. tuberculosis KasA was prepared by the SPOT synthesis technique using a semi-automated ASP 222 peptide synthesis robot (Intavis) essentially as described.35 The sequence was walked through by advancing two to three amino acids at each position in order to fit two complete sets of peptides onto one membrane. The membrane was cut into halves, blocked for 2 h in 1× Pierce Superblock in TBS-1 (20 mM Tris-HCl pH 7.5, 137 mM NaCl, and 0.1 % Tween-20), and washed for 10 min in blocking buffer consisting of 10% Superblock and 5% sucrose in TBS-2 (20 mM Tris-HCl pH 7.5, 137 mM NaCl, and 0.05 % Tween-20). Next, each membrane half was incubated in blocking buffer for 1 h in the presence of either 500 nM His6-tagged GroEL1 or 500 nM His6-tagged GroEL1 apical domain. After washing the membranes three times in TBS-1, bound protein was electro-transferred to a Hybond-ECL nitrocellulose membrane (GE Healthcare), and probed and detected as previously described.35 In addition, membranes were also probed directly with 50 ng/ml of a commercially available, anti-His6 monoclonal antibody-horse radish peroxidase conjugate (BD Biosciences) for 1 h in blocking buffer without sucrose. Despite slight differences in spot intensities, both methods generated identical results. After detection, bound protein and antibody were stripped off the membrane by washing the membrane three times in 8 M urea, 1% SDS, 0.5% 2-mercaptoethanol for 30 min each, followed by three times in 20% acetic acid and 50% ethanol for 15 min each, and three times in 100% ethanol for 10 min each. A negative control was performed with His6-tagged GrpE which was expressed and purified as described.36 After stripping the membrane, another control was performed with His6-tagged GroEL2. All incubations were carried out under gentle rocking at room temperature.
PDB accession code
The atomic coordinates and corresponding structure factors have been deposited in the Protein Data Bank under PDB ID: 3M6C.
Supplementary Material
Acknowledgments
We thank Dr. T. Palzkill for advice and suggestions, and Dr. A. Reger for help with X-ray data collection. Work in F.T.F.T.'s laboratory is supported by grants from the National Institutes of Health (R01-AI076239), the Welch Foundation (Q-1530), the Department of Defense, and the American Cancer Society. B.S. was a Welch postdoctoral fellow and a recipient of a training fellowship from the Pharmacoinformatics Training Program of the Keck Center of the Gulf Coast Consortia (NIH Grant No. R90-DK071505).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
B.S. designed and performed research, analyzed data, and wrote the paper. K.S.L. performed research, analyzed data, and wrote the paper. F.T.FT. designed research, analyzed data, and wrote the paper.
References
- 1.Houry WA, Frishman D, Eckerskorn C, Lottspeich F, Hartl FU. Identification of in vivo substrates of the chaperonin GroEL. Nature. 1999;402:147–154. doi: 10.1038/45977. [DOI] [PubMed] [Google Scholar]
- 2.Bartolucci C, Lamba D, Grazulis S, Manakova E, Heumann H. Crystal structure of wild-type chaperonin GroEL. J Mol Biol. 2005;354:940–951. doi: 10.1016/j.jmb.2005.09.096. [DOI] [PubMed] [Google Scholar]
- 3.Braig K, Otwinowski Z, Hegde R, Boisvert DC, Joachimiak A, Horwich AL, Sigler PB. The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature. 1994;371:578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 4.Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 5.Chen S, Roseman AM, Hunter AS, Wood SP, Burston SG, Ranson NA, Clarke AR, Saibil HR. Location of a folding protein and shape changes in GroEL-GroES complexes imaged by cryo-electron microscopy. Nature. 1994;371:261–264. doi: 10.1038/371261a0. [DOI] [PubMed] [Google Scholar]
- 6.Fenton WA, Kashi Y, Furtak K, Horwich AL. Residues in chaperonin GroEL required for polypeptide binding and release. Nature. 1994;371:614–619. doi: 10.1038/371614a0. [DOI] [PubMed] [Google Scholar]
- 7.Buckle AM, Zahn R, Fersht AR. A structural model for GroEL-polypeptide recognition. Proc Natl Acad Sci U S A. 1997;94:3571–3575. doi: 10.1073/pnas.94.8.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Sigler PB. The crystal structure of a GroEL/peptide complex: plasticity as a basis for substrate diversity. Cell. 1999;99:757–768. doi: 10.1016/s0092-8674(00)81673-6. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Chen L. Domain motions in GroEL upon binding of an oligopeptide. J Mol Biol. 2003;334:489–499. doi: 10.1016/j.jmb.2003.09.074. [DOI] [PubMed] [Google Scholar]
- 10.Ojha A, Anand M, Bhatt A, Kremer L, Jacobs WR, Jr, Hatfull GF. GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell. 2005;123:861–873. doi: 10.1016/j.cell.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Hu Y, Henderson B, Lund PA, Tormay P, Ahmed MT, Gurcha SS, Besra GS, Coates AR. A Mycobacterium tuberculosis mutant lacking the groEL homologue cpn60.1 is viable but fails to induce an inflammatory response in animal models of infection. Infect Immun. 2008;76:1535–1546. doi: 10.1128/IAI.01078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong TH, Coates AR, Butcher PD, Hickman CJ, Shinnick TM. Mycobacterium tuberculosis expresses two chaperonin-60 homologs. Proc Natl Acad Sci U S A. 1993;90:2608–2612. doi: 10.1073/pnas.90.7.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewthwaite JC, Coates AR, Tormay P, Singh M, Mascagni P, Poole S, Roberts M, Sharp L, Henderson B. Mycobacterium tuberculosis chaperonin 60.1 is a more potent cytokine stimulator than chaperonin 60.2 (Hsp 65) and contains a CD14-binding domain. Infect Immun. 2001;69:7349–7355. doi: 10.1128/IAI.69.12.7349-7355.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orme IM, Roberts AD, Griffin JP, Abrams JS. Cytokine secretion by CD4 T lymphocytes acquired in response to Mycobacterium tuberculosis infection. J Immunol. 1993;151:518–525. [PubMed] [Google Scholar]
- 15.Basu D, Khare G, Singh S, Tyagi A, Khosla S, Mande SC. A novel nucleoid-associated protein of Mycobacterium tuberculosis is a sequence homolog of GroEL. Nucleic Acids Res. 2009;37:4944–4954. doi: 10.1093/nar/gkp502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart GR, Wernisch L, Stabler R, Mangan JA, Hinds J, Laing KG, Young DB, Butcher PD. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology. 2002;148:3129–3138. doi: 10.1099/00221287-148-10-3129. [DOI] [PubMed] [Google Scholar]
- 17.Dosanjh NS, Rawat M, Chung JH, Av-Gay Y. Thiol specific oxidative stress response in Mycobacteria. FEMS Microbiol Lett. 2005;249:87–94. doi: 10.1016/j.femsle.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Qamra R, Srinivas V, Mande SC. Mycobacterium tuberculosis GroEL homologues unusually exist as lower oligomers and retain the ability to suppress aggregation of substrate proteins. J Mol Biol. 2004;342:605–617. doi: 10.1016/j.jmb.2004.07.066. [DOI] [PubMed] [Google Scholar]
- 19.Kumar CM, Khare G, Srikanth CV, Tyagi AK, Sardesai AA, Mande SC. Facilitated oligomerization of mycobacterial GroEL: evidence for phosphorylation-mediated oligomerization. J Bacteriol. 2009;191:6525–6538. doi: 10.1128/JB.00652-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qamra R, Mande SC. Crystal structure of the 65-kilodalton heat shock protein, chaperonin 60.2, of Mycobacterium tuberculosis. J Bacteriol. 2004;186:8105–8113. doi: 10.1128/JB.186.23.8105-8113.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coyle JE, Jaeger J, Gross M, Robinson CV, Radford SE. Structural and mechanistic consequences of polypeptide binding by GroEL. Fold Des. 1997;2:R93–R104. doi: 10.1016/S1359-0278(97)00046-1. [DOI] [PubMed] [Google Scholar]
- 22.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Gao X, Chen L. GroEL Recognizes an Amphipathic Helix and Binds to the Hydrophobic Side. J Biol Chem. 2009;284:4324–4331. doi: 10.1074/jbc.M804818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Preuss M, Hutchinson JP, Miller AD. Secondary structure forming propensity coupled with amphiphilicity is an optimal motif in a peptide or protein for association with chaperonin 60 (GroEL) Biochemistry. 1999;38:10272–10286. doi: 10.1021/bi990342l. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Feng H, Landry SJ, Maxwell J, Gierasch LM. Basis of substrate binding by the chaperonin GroEL. Biochemistry. 1999;38:12537–12546. doi: 10.1021/bi991070p. [DOI] [PubMed] [Google Scholar]
- 27.Luckner SR, Machutta CA, Tonge PJ, Kisker C. Crystal structures of Mycobacterium tuberculosis KasA show mode of action within cell wall biosynthesis and its inhibition by thiolactomycin. Structure. 2009;17:1004–1013. doi: 10.1016/j.str.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stan G, Brooks BR, Lorimer GH, Thirumalai D. Residues in substrate proteins that interact with GroEL in the capture process are buried in the native state. Proc Natl Acad Sci U S A. 2006;103:4433–4438. doi: 10.1073/pnas.0600433103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sielaff B, Lee KS, Tsai FT. Crystallization and preliminary X-ray crystallographic analysis of a GroEL1 fragment from Mycobacterium tuberculosis H37Rv. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:418–420. doi: 10.1107/S1744309110004409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paul E, Kevin C. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Laskowski R, MacArthur M, Moss D, Thornton J. Procheck - a program to check the stereochemical quality of protein structures. J Appl Cryst. 1996;26:283–291. [Google Scholar]
- 32.DeLano W. The PYMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientic LLC; 2002. ( http://www.pymol.org) [Google Scholar]
- 33.Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. CATH--a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 34.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 35.Rees I, Lee S, Kim H, Tsai FT. The E3 ubiquitin ligase CHIP binds the androgen receptor in a phosphorylation-dependent manner. Biochim Biophys Acta. 2006;1764:1073–1079. doi: 10.1016/j.bbapap.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 36.Sielaff B, Tsai FT. The M-Domain Controls Hsp104 Protein Remodeling Activity in an Hsp70/Hsp40-Dependent Manner. J Mol Biol. 2010;402:30–37. doi: 10.1016/j.jmb.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.