Abstract
Ion chemistry has long played an important role in molecular mass spectrometry (MS), as it is central to the use of MS as a structural characterization tool. With the advent of ionization methods capable of producing gaseous ions from large biomolecules, the chemistry of gaseous bioions has become a highly active area of research. Gas-phase biomolecule-ion reactions are usually driven by interactions with neutral molecules, photons, electrons, ions, or surfaces. Ion dissociation or transformation into different ion types can be achieved. The types of reaction products observed depend on the characteristics of the ions, the transformation methods, and the time frame of observation. This review focuses on the gas-phase chemistries of ions derived from the electrospray ionization of peptides, proteins, and oligonucleotides, with particular emphasis on their utility in bioanalysis. Various ion-transformation strategies, which further facilitate structural interrogation by converting ions from one type to another, are also summarized.
Keywords: tandem mass spectrometry, electrospray ionization, multiply charged biomolecules, dissociation, ion transformation, ion/ion reaction
1. INTRODUCTION
The development of soft-ionization techniques (1–3), which enable the generation of large gaseous ions from biomolecules, has driven research and development in the field of mass spectrometry both in instrumentation and in bioion chemistry. The mass of the intact biomolecule is a particularly important piece of information that has motivated the development of analyzers with both high resolving power and mass-measurement accuracy. However, mass measurements alone provide no information regarding bond connectivity (i.e., primary structure). Ion chemistry has long been used for this purpose in molecular mass spectrometry, and it has been and continues to be heavily explored for the structural characterization of biomolecule ions. Significant advances have been made in structural characterization of biomolecules via tandem mass spectrometry that include both reactions leading to the dissociation of the ions of interest and reactions that transform bioions from one type to another.
Unimolecular dissociation refers to the class of reactions that generally provides the most detailed structural information in tandem mass spectrometry (4, 5). A number of factors may significantly affect the extent of information that can be obtained from dissociation of a given ion. These factors include, for example, the ion type (e.g., radical cation, protonated molecule, deprotonated molecule, multiply protonated molecule, or metal-cationized species), the internal energy distribution of the ion, and the observation window of the experiment. The observation time scale is defined by the instrument and is generally variable to only a limited extent. Beam-type tandem mass spectrometers, such as triple quadrupole instruments and tandem time-of-flight (TOF) instruments, generally allow for observation windows that integrate processes that occur for up to a few tens of microseconds, whereas ion-trapping instruments, such as electrodynamic ion traps and magnetic/electrostatic ion traps, can integrate processes that extend up to many hundreds of milliseconds. Some hybrid instruments, which combine elements of beam-type and ion-trapping instruments, can access both microsecond and millisecond time scales.
For a given instrument, the two main variables available to the analyst for maximizing structural information are the selection of ion type to be probed and the dissociation method. The range of ion types that can be formed in useful abundances from a given analyte is usually constrained by the ionization method, although ion chemistry approaches (discussed below) are being developed to relax this constraint. Often, no single ion type can provide all of the desired information upon dissociation, and complementary information can frequently be obtained from dissociation of alternative ion types derived from the same analyte. A range of dissociation methods have been developed for tandem mass spectrometry, and these approaches are being explored for the structural characterization of bioions. No single ion type or activation method can consistently provide all of the desired structural information across the range of analyte species subjected to tandem mass spectrometry. In fact, given the range of structural and chemical properties that characterize biopolymers, one should be able to (a) form a range of ion types and (b) apply a range of dissociation methods. Thus, due to the important role that mass spectrometry currently plays in the structural characterization of biomolecules, the study and development of dissociation methods, as well as techniques for generating different ion types, are active areas of research. The following sections summarize the dissociation of gaseous peptide, protein, and oligonucleotide ions; ion chemistry approaches to alter the ion types initially formed by the ionization method via proton transfer or metal-ion transfer; and new reactions that covalently modify analyte ions.
2. BIOION-DISSOCIATION METHODS IN TANDEM MASS SPECTROMETRY
The usual objective in tandem mass spectrometry is to generate an intact ion from the molecule (or complex) of interest so that an accurate molecular weight can be determined. A subsequent dissociation step is then taken to generate structural information, which in the case of linear biopolymer ions generally implies fragmentation along the polymer backbone. The most common approach is to add energy into a precursor ion after its mass selection by one of a variety of means, such as collision with a neutral gas, collision with a surface, or photon absorption. An alternative approach is to convert the precursor ion to a less stable ion type that may then fragment either spontaneously or following an activation step. In this section, we review the application of various ion-activation/-dissociation processes for the structural characterization of multiply charged peptide/protein and nucleic acid ions generated via electrospray ionization (ESI).
2.1. Peptide and Protein Characterization
Central to proteomics is the use of tandem mass spectrometry for protein/peptide identification and characterization (6–9), which are usually performed in positive-ion mode. Under collisional-activation conditions, in which accelerated precursor ions can undergo one or more collisions with gaseous neutral target species (usually argon or nitrogen), part of the ion/target relative translational energy is converted to vibrational energy of the ion and is redistributed over the peptide/protein ion. The resulting collision-induced dissociation (CID) occurs at amide bonds along the peptide backbone, either generating b- and y-type fragment ions or leading to losses of small neutral molecules, such as water and/or ammonia or other fragments derived from side chains. CID is, by far, the most commonly used approach for dissociating peptide and protein ions. However, the relative contributions from the various common dissociation channels are highly dependent upon CID conditions (e.g., collision energy, target pressure, and observation time scale). For example, CID conducted in sector or TOF/TOF instruments with kilovolt accelerating voltages and low collision numbers leads to contributions from dissociation of a relatively large number of amide bonds as well as from amino acid side-chain cleavages. However, most tandem mass spectrometers currently in use do not employ precursor-ion accelerating voltages in the kilovolt range. Most triple quadrupole and hybrid instruments that allow for beam-type CID typically employ accelerating voltages of 100 V or less. Combined with a somewhat longer observation window (i.e., up to a millisecond in the lower-energy instruments versus a few microseconds in the TOF/TOF instruments), the lower collision energies give rise to fewer side-chain cleavages and, often, poorer representation from less-favored amide bond cleavages. For an ion-trapping instrument, the dissociation period can be even longer (up to several seconds); therefore, relatively slow dissociation processes (<102 s−1) can be sampled, allowing for use of ion-trap slow-heating methods, such as ion-trap resonance excitation, the sustained off-resonance irradiation approach in Fourier transform ion cyclotron resonance (FT-ICR) instruments (10, 11), black-body infrared radiative dissociation (12), and infrared multiphoton dissociation (IRMPD) (13–20), to efficiently dissociate biomolecules ranging in mass up to tens of kilodaltons. Nozzle-skimmer dissociation (NSD) (21, 22), which is effected in the atmosphere/vacuum region of atmospheric sampling interfaces, is another means for CID, although it is not achieved between stages of mass spectrometry. The sequence characterization of proteins up to 200 kDa has recently been demonstrated via combinations of vibrational-activation methods, including NSD (23), and counterions that inhibit protein folding upon desolvation. CID techniques have also been applied to large complexes, thereby allowing for the determination of protein composition and subunit stoichiometry (24, 25).
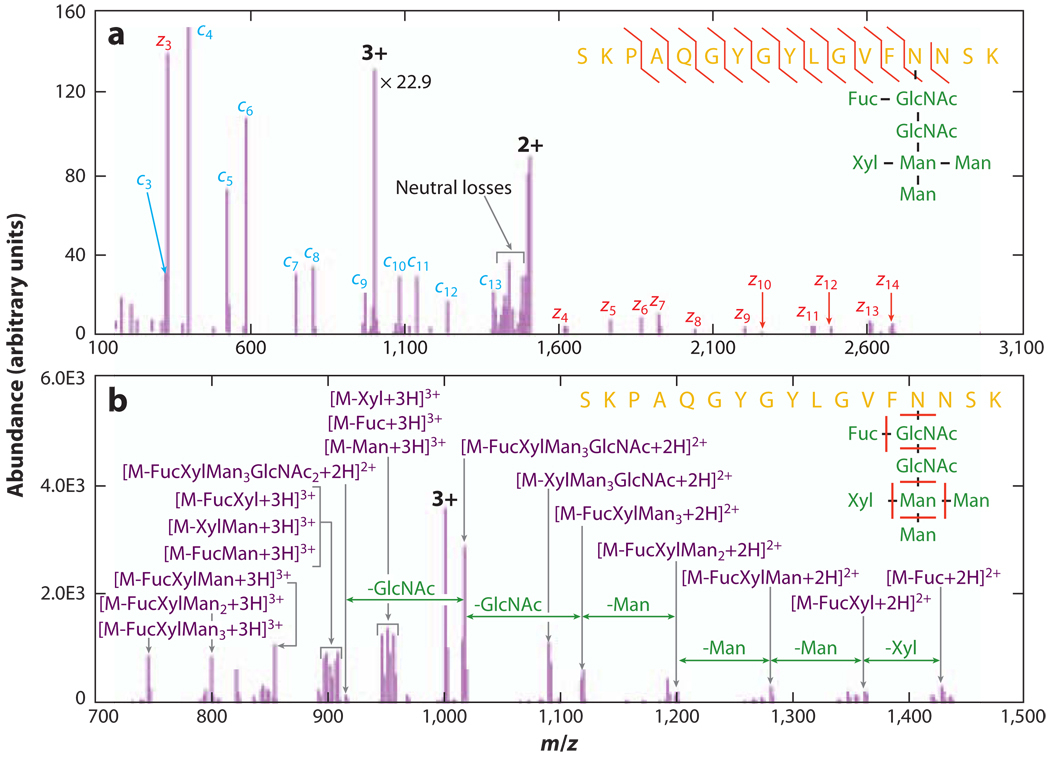
A wide variety of dissociation conditions have also been applied to generate structural information complementary to that provided by vibrational activation of protonated or deprotonated species. For example, dissociation channels that differ from those generated by even-electron precursor ions can dominate for radical species; consequently, complementary sequence information can be obtained. To this end, several gas-phase reactions have been applied to transform electrospray-generated closed-shell ions into open-shell radical ions prior to further tandem mass spectrometry analysis. One of the most common methods for the generation of peptide radical cations is by CID of doubly charged peptide/metal/amine-ligand complexes (26, 27). Additional sequence information can be derived from subsequent dissociation of the peptide radical cations. However, this method is not universally applicable and requires the addition of salts into the ESI solution. Electron-capture dissociation (ECD) is another technique (28–31) that generates radical cations, although hydrogen-rich radicals are generated in this approach. Upon capturing a thermal electron, a multiply protonated peptide or protein is transformed into a hypervalent species. The resulting odd-electron species undergo fragmentation pathways driven by radical chemistry, where the N-Cα backbone bonds are cleaved to generate c- and z-type fragment ions. Usually, more extensive sequence information can be obtained from ECD than from conventional CID. Another important feature is that CID-labile posttranslational modifications (PTMs), such as glycosylation, phosphorylation, and sulfation, are preserved in ECD. In contrast, disulfide linkages, which usually resist dissociation under vibrational-activation conditions, are preferentially dissociated. Therefore, ECD is extremely useful for the analysis of polypeptides with PTMs, providing both the sequence information and the localization of the modification sites. An alternative method for protein/peptide dissociation, referred to as electron-transfer dissociation (ETD), has also been reported (32–34). In the case of ETD, an electron is transferred from a reagent anion to a multiply protonated peptide/protein cation via ion/ion reactions. ETD of multiply protonated peptide/protein ions generally leads to dissociation patterns similar to those of ECD. ECD is generally implemented in the FT-ICR mass spectrometers, whereas ETD is implemented in electrodynamic ion traps. Complementary structural information is usually obtained from the vibrational activation of even-electron ions and from the radical-based ion-dissociation methods. An illustrative example is shown in Figure 1 (34), which summarizes the structural interrogation of a glycopeptide on the basis of ETD and CID. Whereas ETD provides bond connectivity information about the peptide backbone as well as the location of the glycosylation, structural information about the glycan moiety can be derived from CID.
Figure 1.
Tandem mass spectra of a tryptic lectin glycopeptide. (a) Activated electron-transfer dissociation product-ion spectrum of a triply protonated glycopeptide. (b) Beam-type collision-induced dissociation product-ion spectrum of the same glycopeptide ion. Adapted with permission from Reference 34. Copyright 2008, American Chemical Society.
Although studies on the gas-phase dissociation of peptides and proteins are usually carried out on positive ions, the dissociation of multiply deprotonated peptide/protein anions has also been investigated. Such investigations are of interest, especially when the peptide or protein contains more acidic residues than basic amino acid residues. Also, a variety of negatively charged PTMs (e.g., phosphorylation, sulfation, and certain forms of glycosylation) can make the peptide acidic. Such acidic peptides and proteins are more readily ionized as negative ions. However, the interpretation of CID product-ion spectra of deprotonated peptides is not as straightforward as that of protonated peptide ions. CID of deprotonated peptide ions usually generates ions from internal cleavages, losses of amino acid side chains, and consecutive neutral losses from the fragment ions, which complicate the product-ion spectra (35, 36).
Even-electron ESI-generated multiply deprotonated peptide anions have been subjected to various gas-phase ion-transformation methods that lead to radical-ion formation. Zubarev and colleagues (29, 37, 38) and Anusiewicz et al. (39) demonstrated electron-detachment dissociation (EDD) of multiply charged peptide anions. Through the irradiation of peptide anions with high-energy electrons (>10 eV), an electron can be removed from peptide anions. The subsequent dissociation of the resulting peptide radical anions generated the a˙-/x-ion series from preferential Cα-C peptide backbone bond cleavages. Similar results were observed for negative ETD of peptide anions via reactions with xenon radical cations (40). An alternative method for the generation of peptide radical anions, electron-photodetachment dissociation (EPD), has also been demonstrated through the application of ultraviolet radiation on the peptide anions (41). The isolation and dissociation of the resulting peptide radical anions led to fragmentation patterns similar to those observed in ETD or EDD experiments. These methods are especially useful for the characterization of acidic peptides containing PTMs.
2.2. Nucleic Acid Characterization
Sequence characterization of nucleic acids continues to attract interest in the field of genomic research. Compared with other conventional sequencing methods, tandem mass spectrometry of oligonucleotides is rapid and sensitive to modified bases; therefore, it has been widely used for sequencing and structural characterization of natural and chemically modified oligonucleotides. Due to the acidic phosphodiester backbones of oligonucleotides, it is more common to generate oligonucleotide anions than cations via ESI. CID has been the method of choice to cause the dissociation of the phosphodiester backbone bonds and the generation of sequence-specific fragment ions (42–49). Various mechanisms to achieve these ends have been proposed. In general, CID of deprotonated DNA is initiated by loss of neutral or charged bases, followed by subsequent 3′ C-O bond cleavage. The resulting complementary (a–B)-/w-ions provide sequence information. However, the product-ion spectra from CID of DNA usually contain fragment ions from secondary fragmentation, which can complicate spectral interpretation.
IRMPD is another common vibrational-activation method for oligonucleotide analysis (15, 49–57). Preferred dissociation channels similar to CID are usually observed. The dissociation of DNA under CID and that performed under IRMPD conditions have been compared via a quadrupole ion trap (57). For IRMPD, minimal base loss ions were present in the product-ion spectra, presumably due to sequential fragmentation from continued IR irradiation. Furthermore, free-phosphate and nucleobase ions in the low–m/z region, providing modified base information. Using IRMPD, McLafferty and coworkers (52) have demonstrated the dissociation of oligonucleotides in an FT-ICR instrument and obtained extensive sequence information of DNA up to 108 nucleotides long.
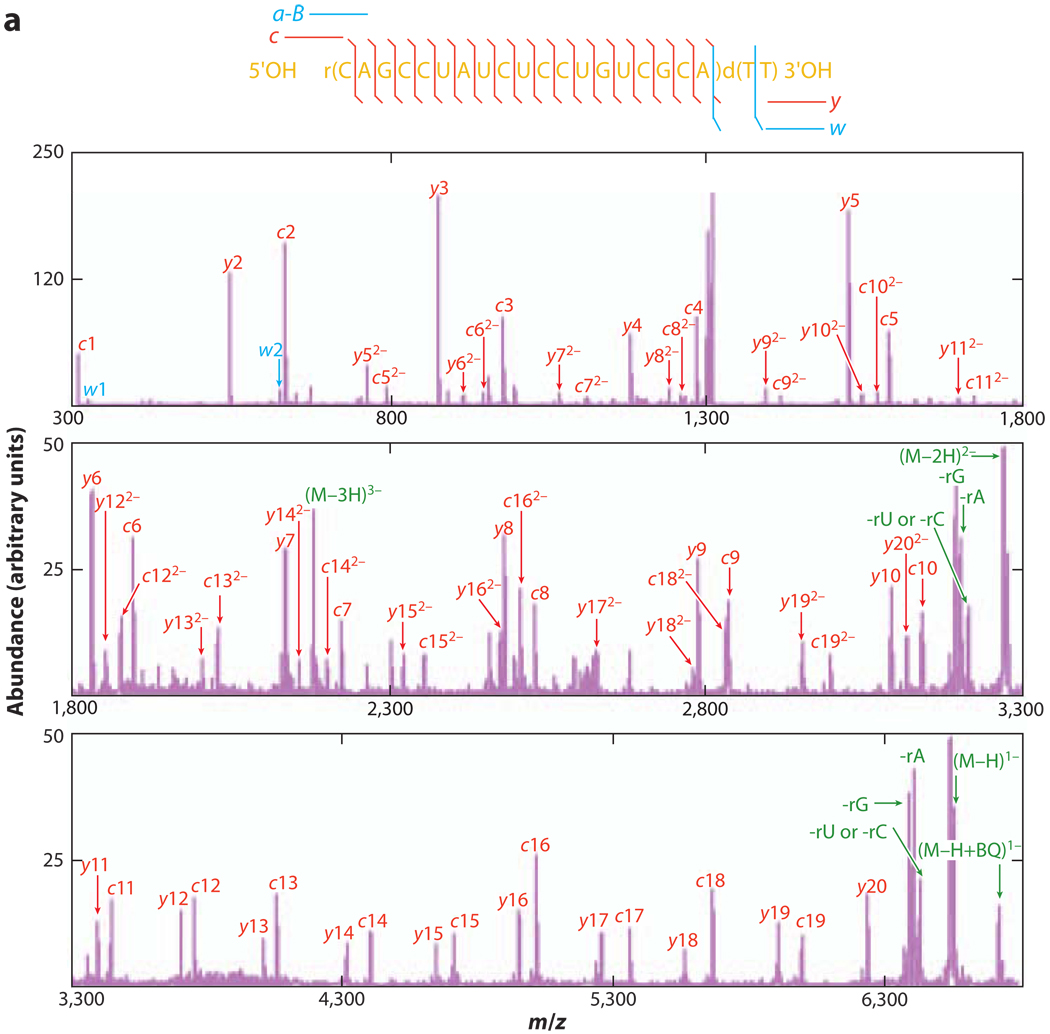
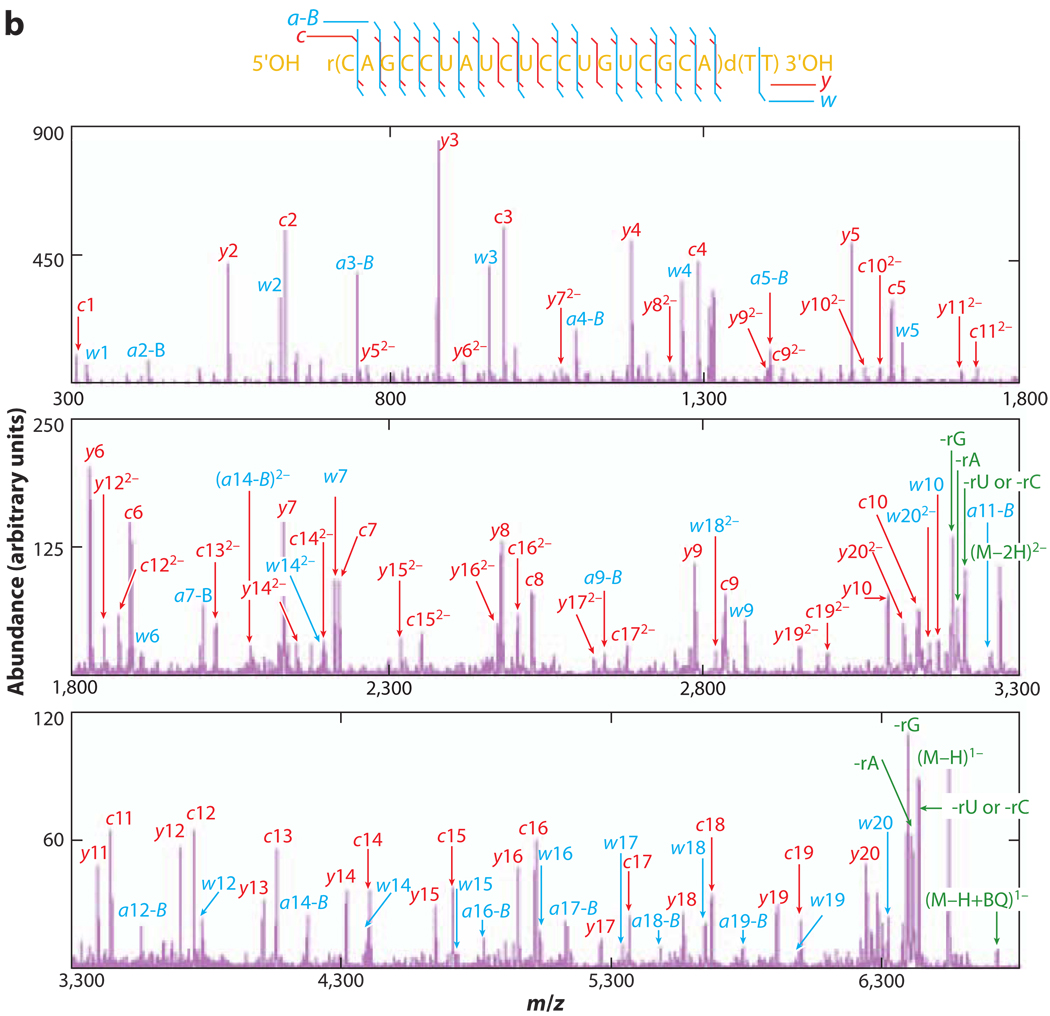
The importance of RNA, especially noncoding RNA, in cellular function has attracted efforts to develop analytical methods for RNA characterization. Dissociation of RNA anions via collisional activation in the gas phase leads to the 5′ P-O backbone cleavage, which gives rise to the characteristic c-/y-ion series (49, 58–61). This dissociation channel is initiated from an intramolecular cyclic intermediate that forms through the bridging between the 2′ OH hydrogen and the 5′ phosphate oxygen. Abstraction of the 2′ OH proton by the 5′ oxygen then leads to the dissociation of the 5′ P-O bond. This cleavage mechanism competes with nucleobase loss and the subsequent backbone cleavage to yield (a–B)-/w-ion series. The characteristic ions from both series can provide primary sequence information and allow the identification of the presence of modified nucleosides. Most RNA dissociation experiments have been conducted under beam-type CID conditions, resulting in multiple dissociation channels together with secondary fragmentations. Recently, the charge state–dependent dissociation of model RNA oligomers and small interfering RNA anions was investigated under ion-trap collisional activation (62, 63). Ion-trap collisional activation, which samples dissociation reactions at lower rates and lower energies than does beam-type collisional activation, shows great potential for RNA de novo sequencing. As shown in Figure 2 (63), complete sequencing of the antisense strand was achieved by selective dissociation of the lower-energy 5′ P-O bonds (c-/y-ion series) or both the 5′ P-O bonds and the higher-energy 3′ C-O bonds [(a–B)-/w-ion series] through variation of the activation conditions. This example illustrates that tuning the activation conditions can allow access to different dissociation channels.
Figure 2.
Dissociation of the antisense strand of a model small interfering RNA (siRNA) molecule under various activation amplitudes. (a) Postion/ion ion-trap collision-induced dissociation (CID) spectrum of [AS siRNA]5− with a relatively low excitation amplitude applied (99.93 kHz, 480 mV). (b) Postion/ion ion-trap CID spectrum of [AS siRNA]5− with a relatively high excitation amplitude applied (99.93 kHz, 800 mV). Adapted with permission from Reference 63. Copyright 2008, American Chemical Society.
The dissociation phenomenology of oligonucleotide radical anions has also been investigated. Reactions of multiply deprotonated DNA anions with various reagent radical cations, such as O2+˙, lead to the generation of radical anions (64–66). The subsequent dissociation of the DNA radical anions generates sequence-specific fragment ions—the a-/w-ion series—that are complementary to those generated by CID. Compared with CID, the cleavage of the N-glycosidic bond is significantly decreased, and therefore fewer base loss ions are observed.
EDD of oligonucleotide anions has also been investigated (67). Reactions of multiply deprotonated DNA and RNA anions with high-energy electrons can also lead to gas-phase dissociation. The observed dissociation behavior varied among different oligonucleotides. The w- and d-type ions are the most prominent fragment ions for both DNA and RNA, but other dissociation channels are also accessible. Also, the noncovalent interactions of DNA duplexes and hairpins are not disrupted by EDD. No nucleobase dependency was observed for EDD, indicating that the dissociation process is initiated via direct electron detachment from the deprotonated phosphate backbone rather than from the nucleobases.
EPD is another dissociation method involving oligonucleotide radical anions (68, 69). Irradiation at 260 nm leads to electron detachment and generates radical anions. The electron-photodetachment yield is inversely correlated to the ionization potential of nucleobases, indicating that the detachment of the electron is initiated at the bases and not at the phosphates. When the resulting DNA radical anions are subjected to CID, w-, d-, a˙-, and z˙-ions, in addition to neutral losses, are observed. As in EDD, the fragile noncovalent interaction is preserved; therefore, EPD can also be used to probe inter- and intramolecular interactions.
The gas-phase dissociation of positive oligonucleotide ions has not been as extensively investigated as the dissociation of negative ions. However, several reports have shown that the major dissociation channels of some model oligonucleotide cations are similar to those of the anions (i.e., nucleobase losses and the preferred 3′ C-O backbone bond cleavages) (57, 70–73). Moreover, gas-phase dissociation methods involving ion/electron reactions (ECD) and ion/ion reactions (ETD) have also been applied to oligonucleotide cations (74–75). ECD of oligonucleotides depends on the nucleotide sequence. Various fragment-ion types have been observed for different model oligonucleotide sequences. ECD of multiply protonated DNA generates radical cations and low-abundance product ions from various backbone bond cleavages; w-/d-ions are the most prominent. However, ECD of multiply protonated RNA generates very limited sequence information. Generally, oligonucleotide sequence information obtained from ECD is not as extensive as that obtained from EDD and vibrational activation. Recently, ETD of oligonucleotide cations has been explored (76). Reaction of the ESI-generated multiprotonated oligonucleotide anions with fluoranthene radical anions leads to efficient electron-transfer charge reduction and limited backbone cleavages to yield sequence ions of low abundance. Subsequent CID of the charge-reduced oligonucleotide radical cations [a process known as electron-transfer collision-activated dissociation (ETcaD)] generates a-/w- and d-/z-ion series. Compared with CID of the even-electron ions, the abundances of base loss ions and internal fragments are significantly lower. ETcaD of an oligonucleotide duplex resulted in specific backbone cleavages, with conservation of weaker noncovalent bonds.
3. BIOMOLECULE ION TRANSFORMATION IN THE GAS PHASE VIA METAL ION– OR PROTON-TRANSFER REACTIONS
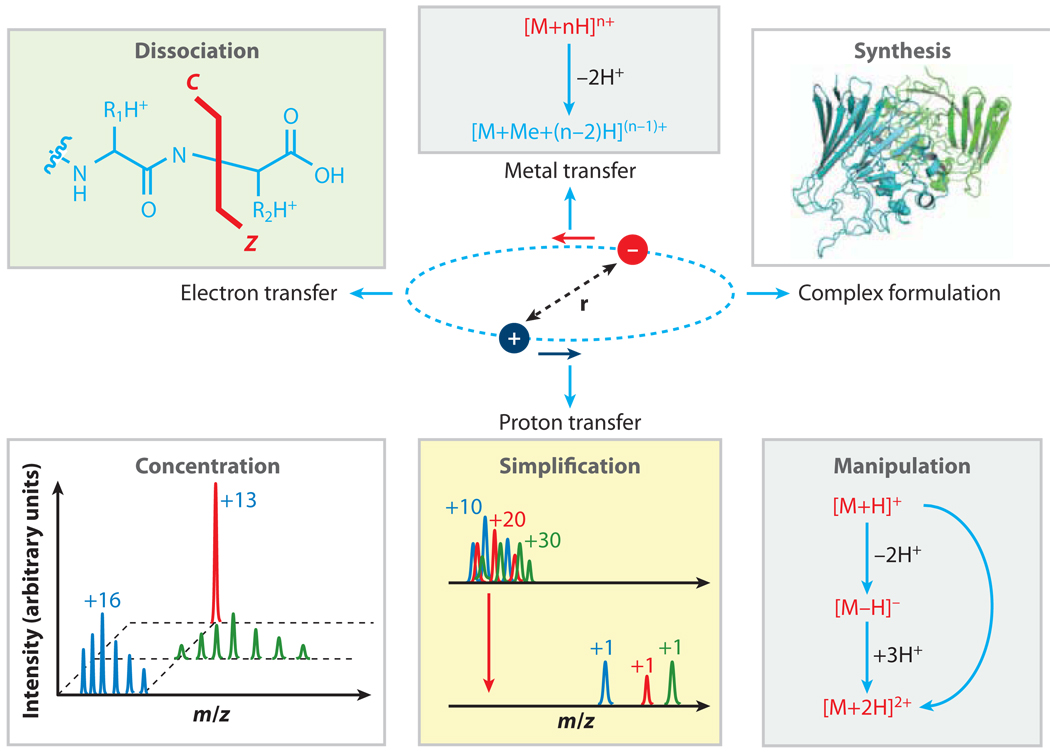
The previous section emphasized dissociation reactions for structural interrogation of ESI-generated multiply charged biomolecules. Although unimolecular dissociation reactions underlie the primary structural characterization of biomolecules in a mass spectrometer, other gas-phase ion chemistries have also been explored for their utility in the field of tandem mass spectrometry. Gas-phase ion/ion reactions (as illustrated in Figure 3) provide means for transforming ion types formed initially by the ionization method, thereby increasing the number of ion types available for structural interrogation by various dissociation methods. Electron transfer or capture provides examples of ion-transformation reactions that can lead directly to precursor-ion dissociation, as discussed above. In this section, we present ion-transformation reactions via either metal-ion transfer or proton transfer to facilitate the tandem mass spectrometry analysis of biomolecules. For example, ESI-generated biomolecule ions can be transformed into other forms before being subjected to mass analysis or dissociation. In some applications, an ion-transformation step is also applied to multiply charged product ions.
Figure 3.
The transformation of various gas-phase biomolecule ions via ion/ion reactions. Abbreviation: r, relative distance between the two charged particles in a stable ion/ion orbit.
3.1. Ion-Transformation Reactions That Lead to Metal Insertion
The effects of cationization by various metal ion types on the gas-phase dissociation of different biomolecules have been an active area of research. Metal-cationized species, including sugars, lipids, peptides/proteins, and oligonucleotides, often show fragmentation patterns that differ from those of their protonated counterparts in tandem mass spectrometry. Dissociation of a metal-cationized, sulfated, heparin-like glycosaminoglycan oligosaccharide such as Arixtra® has been investigated (77). Compared with the extent of losses from multiply deprotonated ions, the extent of neutral sulfur trioxide losses from the sulfated molecules was decreased, and cross-ring cleavages of the sugar molecule were observed for the metal-cationized ions. Also, whereas lipid ions are usually singly protonated, metallation has also been applied to increase the charge states of glycerophosphocholine lipids in order to conduct ETD experiments (78). Although most metallation procedures are carried out by adding metal salts to the solution prior to electrospray, an alternative method involves reacting the ESI-generated multiply charged ions with various metal-containing ion complexes of the opposite polarity in the gas phase (79–85). An example is the transfer of Au(I) to a disulfide-linked peptide via ion/ion reaction with AuCl2− (84). The subsequent dissociation of the disulfide-linked Au(I)-cationized peptides led primarily to the cleavage of the S-S bond, whereas the dissociation of the protonated species led to neutral losses and peptide bond cleavages. The insertion of transition-metal ions into oligonucleotide anions has also been demonstrated via ion/ion reactions between multiply charged oligonucleotide anions and transition-metal complex cations (85). The gas-phase formation of metal-containing species that do not readily form from ESI may benefit structural interrogation in subsequent tandem mass spectrometry experiments. The gas-phase ion/ion metal-insertion approach allows independent optimization of the ESI process and the formation of metal-containing ions while avoiding the detrimental effects of salts in the electrospray process.
3.2. Ion-Transformation Reactions That Lead to Charge Inversion and Charge Increase
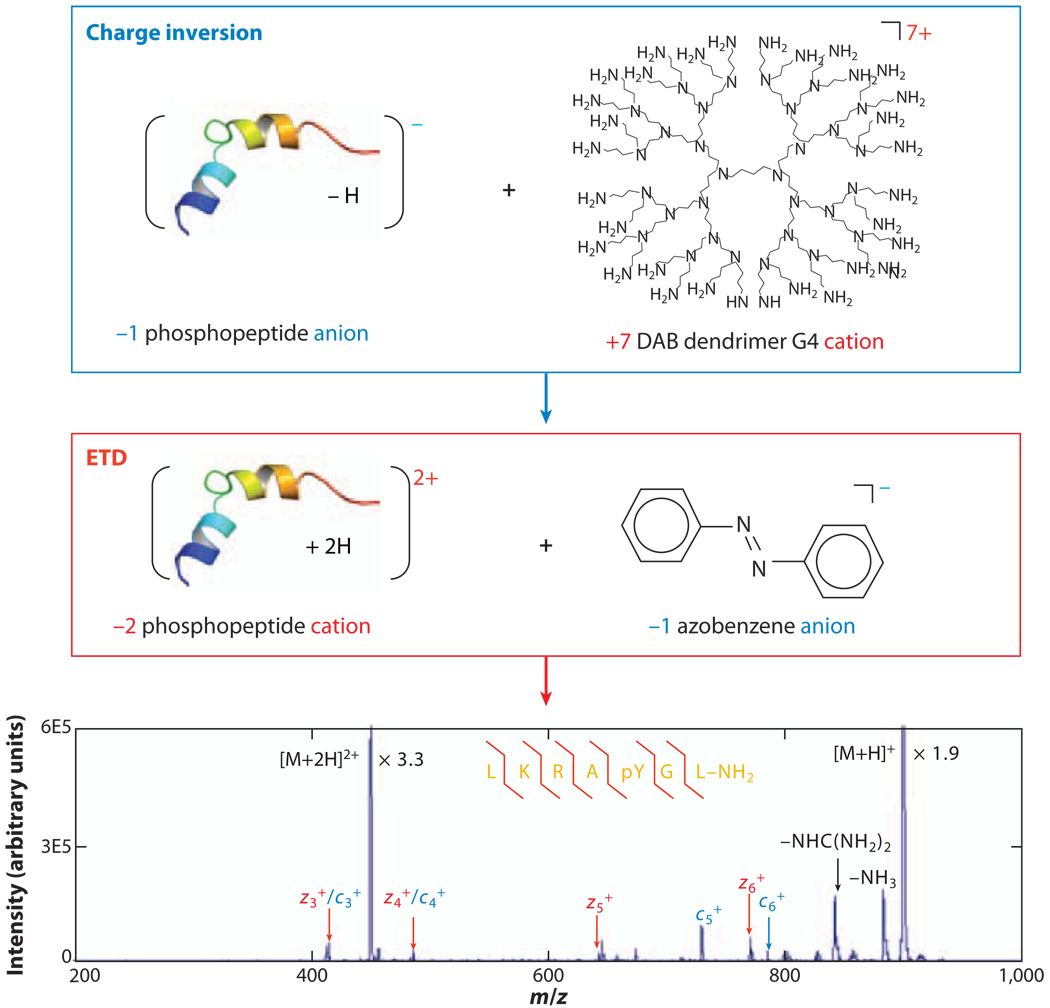
Transformation of ions from one polarity to the opposite polarity has also been demonstrated via ion/ion reactions (86–89). For charge inversion to occur, multiple proton transfers from the multiply charged reagent ions to the singly charged biomolecular ions must take place within a single ion/ion encounter to avoid the neutralization of biomolecular ions. Therefore, reagent ions with large physical cross sections tend to have good charge-inversion efficiency. Reagents such as dendrimers, proteins, and oligonucleotides have been used for the inversion of both cations and anions into the opposite polarities. For example, charge inversion was applied to the analysis of a phosphopeptide in a tryptic digestion mixture through use of ETD, as shown in Figure 4 (89). In this experiment, the acidic phosphopeptide analyte ions were observed in the negative-ion but not the positive-ion mass spectrum of the tryptic peptide mixture. Transformation of the prominent singly deprotonated phosphopeptide ions into doubly protonated ions allowed the subsequent application of ETD to localize the phosphorylation site, which was not possible via CID of the deprotonated peptide. This application is especially useful for the structural characterization of acidic polypeptides via ETD.
Figure 4.
Electron-transfer dissociation (ETD) of doubly protonated tryptic phosphopeptide ions derived from sequential charge inversion of singly deprotonated precursor ions. Adapted with permission from Reference 89. Copyright 2008, American Chemical Society.
Another charge-inversion application involves increasing the analyte-ion charge state by applying two consecutive charge-inversion reactions (86). First, charge inversion of the singly protonated peptide cations to singly deprotonated peptide anions occurs via multiply deprotonated carboxylate-terminated dendrimer ions. Second, the resulting singly deprotonated peptide anions are then transformed into doubly protonated ions via a reaction with multiply protonated amine-terminated dendrimer ions. An increase of the charge states of singly deprotonated ions to doubly deprotonated ions has also been demonstrated (87). The availability of a broader range of analyte-ion charge states allows higher flexibility for downstream ion manipulation and structural interrogation. A scenario in which charge increase is desirable would require an increase in the charge state of matrix-assisted laser desorption/ionization (MALDI)-generated singly charged peptides. Although singly charged ions generated by MALDI usually fragment through neutral losses upon collisional activation, the increase in charge state could result in a greater variety of structurally informative fragmentation. Such results could be of interest specifically for the structural characterization of biomolecules, such as peptides, during MALDI imaging. However, the recent demonstration of the formation of multiply protonated proteins directly via atmospheric-pressure MALDI (90), if generally applicable, may prove to a simpler approach to the generation of multiply charged ions.
3.3. Ion-Transformation Reactions That Lead to Proton-Transfer Charge Reduction
ESI of biomacromolecules usually generates a distribution of charge states. This characteristic, although advantageous in some aspects, can be problematic, especially when a sample of mixture is subjected to analysis. It is sometimes desirable to reduce charge states to simplify the interpretation of the complex electrospray mass spectra of mixtures. Compared with ion/molecule reactions (91–95), ion/ion proton-transfer reactions between multiply charged analyte ions and a singly charged ion of the opposite polarity are more efficient. The multiply protonated cations lose protons to the reagent anions, whereas the multiply deprotonated anions gain protons from the reagent cations. The charge-squared dependency of the reaction kinetics leads to a situation wherein the various charge states of the mixture components are reduced primarily to +1 or −1 charge states with relatively little differential neutralization (98). Ion/ion proton-transfer reactions have been applied to the simplification of (a) electrospray mass spectra of protein mixtures in positive-ion mode and (b) oligonucleotide mixtures in negative-ion mode in an ion trap and at atmospheric pressure (95–101).
Also, the charge-state effects on the gas-phase dissociation of both protein cations and oligonucleotide anions have been investigated. Generally, the collisional activation of precursor ions of lower and medium charge states usually leads to different dissociation patterns, and often greater sequence coverage, than do the higher–charge state ions. Therefore, in those cases it is desirable to expand the available charge states to assess the gas-phase dissociation phenomena. To that end, ion/ion proton-transfer reactions have been applied to transform the charge states of the biomolecular ions into charge states not directly generated from ESI (102, 103). The resulting lower–charge state ions can be used for subsequent structural characterization.
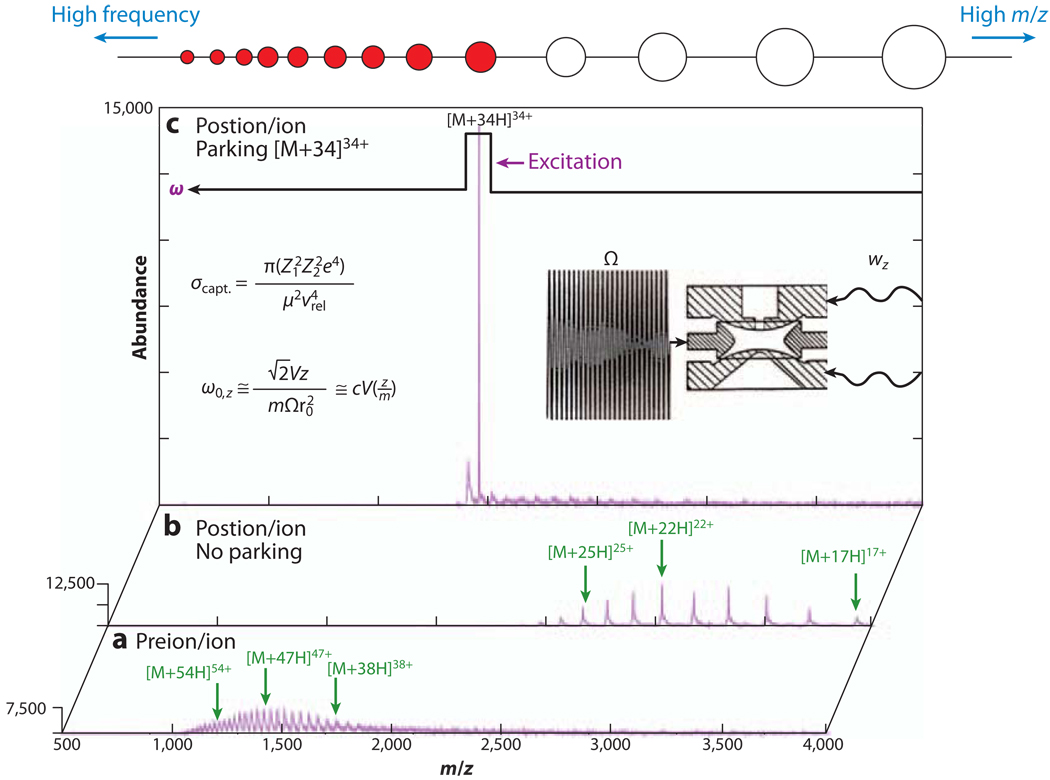
A gas-phase concentration technique that can be performed in conjunction with ion/ion proton-transfer reactions in quadrupolar ion traps is referred to as ion parking (104–109). As shown in Figure 5 (106), in an ion-parking experiment a supplemental dipolar resonant excitation voltage is applied to a pair of opposing electrodes; this voltage matches the fundamental frequency of motion of the ions of interest during ion/ion proton-transfer reactions in ion traps. Ions that match the applied frequency are accelerated, and their overlap with the oppositely charged reagent ions is thereby reduced, inhibiting the ion/ion reaction rate of the selected ion. The ion/ion reaction rates of the other ions are not affected, and they continue to be neutralized. The net result is the accumulation of all higher–charge state ions into the single charge state being “parked.” A similar technique, termed parallel ion parking (109), involves the simultaneous concentration of multiple components by inhibiting the reaction rates of a broad m/z range of ions. Parallel ion parking has also been demonstrated to simplify mixture analyses.
Figure 5.
Gas-phase concentration of bovine serum albumin. (a) Preion/ion reaction electrospray ionization mass spectrum of bovine serum albumin. (b) Postion/ion reaction spectrum after 30 ms of ion/ion proton-transfer reactions with a perfluorocarbon anion (no ion parking). (c) Postion/ion reaction spectrum with acceleration of the [M+34H]34+ ion to effect ion parking. Adapted with permission from Reference 105. Copyright 2003, Elsevier Science.
Moreover, by reducing all product-ion charge states primarily to singly charged ions, ion/ion proton-transfer reactions can resolve the charge-state ambiguity of the product-ion spectra from the unimolecular dissociation of multiply charged ions (108, 110–112) when using mass analyzers of moderate to low resolving powers. This technique has been applied to the sequencing of large, highly charged biomolecule ions, such as transfer RNA anions and protein cations. Consecutive ion/ion proton-transfer reactions and ion-parking steps have also been demonstrated for (a) the concentration and charge-state purification of precursor ions and (b) the identification of product ions in a complex protein mixture from an Escherichia coli cell lysate fraction (108).
4. NOVEL GAS-PHASE ION CHEMISTRY
The range of reactions that ESI-generated biomolecule ions can undergo is limited by the ions’ chemical functionalities and by the range of reagents with which they can react. This section emphasizes reactions with reagent species that allow for phenomena that cannot be produced with most proton-transfer or electron-transfer reagents. The reactions discussed in the above sections involve the transformation of ions via gain or loss of an ion or electron or the cleavage or weakening of certain covalent backbone bonds to generate sequence specific fragmentation; in this section we describe other gas-phase ion chemistries with potentially useful applications.
Many solution-phase derivatization reactions have been used to enhance the ionization, quantification, and structural characterization of biomolecules. Recently, several derivatization reactions were explored for different classes of biomolecules under gas-phase reaction conditions. A nucleophilic substitution reaction, for example, was observed for ion/molecule reactions between multiply charged oligonucleotide anions and trimethylsilyl chloride molecules (113). The SN2 reactions led to the attachment of trimethylsilyl moieties to the phosphodiester backbones of oligonucleotide anions. Another selective ion/molecule reaction involved Schiff base formation through the use of either acetone (114) or 2,5-hexanedione (115) as the neutral reagent. A more recent study (116) demonstrated the selective formation of Schiff base adducts on peptide cations when multiply protonated peptides with primary amine groups reacted with singly deprotonated 4-formyl-1,3-benzenedisulfonic acid. When the resulting derivatized product ions were subjected to collisional activation, either altered dissociation channels or more extensive structural information was generated. The gas-phase derivatization could be further explored via reagent ions with other functionalities to react with other classes of biomolecules. This area of research may provide a general means of modifying analyte ions for enhancing structural characterization that would not require solution-phase reactions.
Ion/ion reactions leading to the attachment of anions or cations have also been observed. For example, protein complexes have been formed following reactions between multiply protonated protein cations and multiply deprotonated protein anions (115–120). The creation of protein heterocomplexes (containing as many as six subunits) from ion/ion reactions of single-subunit reactants of opposite polarities has recently been demonstrated (120), as has the formation of peptide/oligonucleotide complexes under similar reaction conditions (66). However, applications of gas-phase complex formation have been relatively little explored.
Another type of reaction involves the gas-phase ion chemistry of cluster ions. In a recent ion/ion reaction study (121), the authors used protonated water clusters, comprising five or more water molecules, as proton-transfer reagents to reduce the charge state of fragile multiply deprotonated biomolecule anions, achieving significantly less fragmentation. This phenomenon can be attributed to the reaction exothermicities of proton transfer from protonated water clusters, which are lower than those of other proton-transfer reagents, such as protonated strong bases. Also, hydrated cluster ions have been used as chemical thermometers for the determination of energy deposition during gas-phase ion-activation/-transformation processes (122, 123). Following a reaction with electrons or photons in the gas phase, the internal energy deposition from the reaction leads to the evaporation of water molecules from the cluster ions. The degree of water loss, which can be readily determined with mass spectrometry, can reflect the amount of energy deposited.
5. CONCLUSIONS
Ion chemistry plays an important role in modern biological mass spectrometry through its roles in structural characterization and ion transformation. Dissociation of multiply charged ions is useful for determining the primary structure of biomolecules. Dissociation can be induced either via various activation methods or through reactions that lead to the transformation of ESI-generated multiply charged ions into less stable forms. Variations in the derived information (e.g., the identities of the product ions) can result from different activation conditions (e.g., laser wavelength, collision energy, target pressure, time frame of observation) or from examination of different precursor-ion types (e.g., multiply protonated, metal-cationized, and multiply deprotonated ions; radical cations). The availability of numerous dissociation methods and the ability to form a wide range of precursor-ion types maximize the extent of structural information that can be obtained from a biomolecule of interest.
Also, gas-phase ion transformations, such as those arising from proton transfer, have also been applied to analytical mass spectrometry as well as to structural characterization. The manipulation of precursor-ion charge, for example, allows simplification of ESI mass spectra of a complex mixture of biomolecules by reducing spectral overlap and expanding the number of accessible ion species via charge reduction, charge inversions, and the concentration and purification of gas-phase ions through the ion-parking technique. Moreover, simplification of product-ion spectra from gas-phase dissociation of highly charged biomacromolecular ions can also be achieved by reducing the multiply charged product ions to predominantly singly charged ions. Other novel gas-phase ion chemistries, including gas-phase metal ion insertion, selective covalent modification of bioions in the gas phase, and cluster-ion reactions, may provide useful applications in the future.
The field of biological mass spectrometry has seen remarkable advances over the past two decades that have extended the reach of mass spectrometry into many new areas of biological research. The complexities associated with biological systems, however, are daunting, and dramatic improvements are needed in many aspects of mass spectrometry, including sensitivity, dynamic range, and specificity. The developments in bioion chemistry have been impressive, but the ability to completely characterize a large biomolecule, such as a protein, remains elusive, particularly for large molecules. Therefore, we should anticipate further exploration and development of approaches for structural characterization—including methods based on ion chemistry, ion mobility, and ion spectroscopy, among others—over the coming decade.
SUMMARY POINTS
Gas-phase ion chemistry plays an essential role in the structural characterization of biomolecules by mass spectrometry.
Unimolecular dissociation of biomolecular ions, effected by an expanding range of methods, provides structural information for biomolecule characterization. No single dissociation method, however, consistently provides satisfactory structural information for the wide range of bioions of interest.
Structural information can be increased by interrogating the structures of various ion types derived from the biomolecule of interest. The various ion types can be generated either in the ionization process or via gas-phase ion-transformation reactions.
Applications beyond structural characterization are enabled via gas-phase ion-transformation reactions, such as biomolecule mixture analysis, product-ion charge-state determination, and gas-phase concentration of analyte ions largely into a single precursor-ion charge state.
FUTURE ISSUES
The derivation of complete primary sequence information from linear biopolymers becomes increasingly challenging as the size of the polymer increases. So-called top-down approaches that rely on the dissociation of intact precursor ions, as opposed to ions generated from chemical or enzymatic digestion, are currently limited in practice to species below approximately 50 kDa. Even for such ions, sequence coverage can be low. Thus, dissociation methods for moderate-to-large linear biopolymers require significant improvement.
Branched biopolymer (e.g., carbohydrate) structural characterization remains a poorly developed area relative to the characterization of proteins and nucleic acids.
Ion/ion reactions and their application have been largely restricted to single-proton transfer and single-electron transfer. A much wider range of chemistry is possible, in principle. Further exploration of selective chemistries involving covalent bond formation and cleavage will probably enable new analytical strategies.
Gas-phase reactions involving hydrated species promise to enable closer analogies between solution-phase and gas-phase chemistries. The use of hydrated reagent ions may enable well-studied condensed-phase reactions to be implemented within the context of tandem mass spectrometry experiments.
ACKNOWLEDGMENTS
The authors acknowledge support for their laboratory from the Office of Basic Energy Sciences, Office of Sciences, U.S. Department of Energy, under award number DE-FG02-00ER15105 for the study of fundamental aspects of ion/ion chemistry; the National Institutes of Health under grant GM 45372 for the development of instrumentation and applications related to the ion/ion chemistry of peptides and proteins; and the National Science Foundation under CHE-0808380 for their recent ion/ion reaction work with nucleic acids.
Glossary
- ESI
electrospray ionization
- CID
collision-induced dissociation
- IRMPD
infrared multiphoton dissociation
- ECD
electron-capture dissociation
- ETD
electron-transfer dissociation
- EDD
electron-detachment dissociation
- EPD
electron-photodetachment dissociation
Footnotes
DISCLOSURE STATEMENT
S.A.M. is a cofounder of BG Medicine, an early-stage biotechnology company. He and his students and former students either hold or have applied for patents relating to ion/ion chemistry. His group also actively collaborates with MDS Analytical Technologies in various instrumentation and methods-development projects.
LITERATURE CITED
- 1.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10000 daltons. Anal. Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 2.Whitehouse CM, Dreyer RN, Yamashita M, Fenn JB. Electrospray interface for liquid chromatographs and mass spectrometers. Anal. Chem. 1985;57:675–679. doi: 10.1021/ac00280a023. [DOI] [PubMed] [Google Scholar]
- 3.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 4.Cooks RG, Beynon JH, Caprioli RM, Lester GR. Metastable Ions. Amsterdam: Elsevier; 1973. [Google Scholar]
- 5.Cooks RG. Collision-induced dissociation: readings and commentary. J. Mass Spectrom. 1995;30:1215–1221. [Google Scholar]
- 6.Hunt DF, Yates JR, Shabanowitz J, Winston S, Hauer CR. Protein sequencing by tandem mass spectrometry. Proc. Natl. Acad. Sci. USA. 1986;83:6233–6237. doi: 10.1073/pnas.83.17.6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loo JA, Edmonds CG, Smith RD. Primary sequence information from intact proteins by electrospray ionization tandem mass spectrometry. Science. 1990;248:201–204. doi: 10.1126/science.2326633. [DOI] [PubMed] [Google Scholar]
- 8.Wells JM, McLuckey SA. Collision-induced dissociation (CID) of peptide and proteins. Methods Enzymol. 2005;402:148–185. doi: 10.1016/S0076-6879(05)02005-7. [DOI] [PubMed] [Google Scholar]
- 9.Wysocki VH, Resing KA, Zhang Q, Cheng G. Mass spectrometry of peptides and proteins. Methods. 2005;35:211–222. doi: 10.1016/j.ymeth.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 10.Wu Q, Van Orden S, Cheng X, Bakhtiar R, Smith RD. Characterization of cytochrome c variants with high-resolution FT-ICR mass spectrometry: correlation of fragmentation and structure. Anal. Chem. 1995;67:2498–2509. doi: 10.1021/ac00110a027. [DOI] [PubMed] [Google Scholar]
- 11.Senko MW, Speir JP, Mclafferty FW. Collisional activation of large multiply charged ions using Fourier transform mass spectrometry. Anal. Chem. 1994;66:2801–2808. doi: 10.1021/ac00090a003. [DOI] [PubMed] [Google Scholar]
- 12.Jockusch RA, Schnier PD, Price WD, Strittmatter EF, Demirev PA, Williams ER. Effects of charge state of fragmentation pathways, dynamics, and activation energies of ubiquitin ions measured by blackbody infrared radiative dissociation. Anal. Chem. 1997;69:1119–1126. doi: 10.1021/ac960804q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Little DP, Speir JP, Senko MW, O’Connor PB, McLafferty FW. Infrared multiphoton dissociation of large multiply charged ions for biomolecule sequencing. Anal. Chem. 1994;66:2809–2815. doi: 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- 14.Mortz E, O’Connor PB, Roepstorff P, Kelleher NL, Wood TD, et al. Sequence tag identification of intact proteins by matching tandem mass spectral data against sequence data bases. Proc. Natl. Acad. Sci. USA. 1996;93:8264–8267. doi: 10.1073/pnas.93.16.8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W, Hendrickson CL, Emmett MR, Marshall AG. Identification of intact proteins in mixtures by alternated capillary liquid chromatography electrospray ionization and LC ESI infrared multiphoton dissociation Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 1999;71:4397–4402. doi: 10.1021/ac990011e. [DOI] [PubMed] [Google Scholar]
- 16.Masselon C, Anderson GA, Harkewicz R, Bruce JE, Pasa-Tolic L, Smith RD. Accurate mass multiplexed tandem mass spectrometry for high-throughput polypeptide identification from mixtures. Anal. Chem. 2000;72:1918–1924. doi: 10.1021/ac991133+. [DOI] [PubMed] [Google Scholar]
- 17.Flora JW, Muddiman DC. Selective, sensitive, and rapid phosphopeptide identification in enzymatic digests using ESI-FTICR-MS with infrared multiphoton dissociation. Anal. Chem. 2001;73:3305–3311. doi: 10.1021/ac010333u. [DOI] [PubMed] [Google Scholar]
- 18.Hakansson K, Cooper HJ, Emmett MR, Costello CE, Marshall AG, Nilsson CL. Electron capture dissociation and infrared multiphoton dissociation MS/MS of an N-glycosylated tryptic peptide to yield complementary sequence information. Anal. Chem. 2001;73:4530–4536. doi: 10.1021/ac0103470. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Masselon CD, Anderson GA, Pasa-Tolic L, Lee SW, et al. High-throughput peptide identification from protein digests using data-dependent multiplexed tandem FTICR mass spectrometry coupled with capillary liquid chromatography. Anal. Chem. 2001;73:3312–3322. doi: 10.1021/ac010192w. [DOI] [PubMed] [Google Scholar]
- 20.Meng F, Cargile BJ, Patrie SM, Johnson JR, McLoughlin SM, Kelleher NL. Processing complex mixtures of intact proteins for direct analysis by mass spectrometry. Anal. Chem. 2002;74:2923–2929. doi: 10.1021/ac020049i. [DOI] [PubMed] [Google Scholar]
- 21.Loo JA, Udseth HR, Smith RD, Futrell JH. Collisional effects on the charge distribution of ions ffrom large molecules, formed by electrospray-ionization mass spectra. Rapid Commun. Mass Spectrom. 1988;2:207–210. [Google Scholar]
- 22.Zhai H, Han X, Breuker K, McLafferty FW. Consecutive ion activation for top down mass spectrometry: improved protein sequencing by nozzle-skimmer dissociation. Anal. Chem. 2005;77:5777–5784. doi: 10.1021/ac0580215. [DOI] [PubMed] [Google Scholar]
- 23.Han XM, Jin M, Breuker K, McLafferty FW. Extending top-down mass spectrometry to proteins with masses greater than 200 kilodaltons. Science. 2006;314:109–112. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- 24.Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Micelles protect membrane complexes from solution to vacuum. Science. 2008;321:243–246. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- 25.Sharon M, Robinson CV. The role of mass spectrometry in structure elucidation of dynamic protein complexes. Annu. Rev. Biochem. 2007;76:167–193. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]
- 26.Chu IK, Rodriquez CF, Lau TC, Hopkinson AC, Siu KWM. Molecular radical cations of oligopeptides. J. Phys. Chem. B. 2000;104:3393–3397. [Google Scholar]
- 27.Chu IK, Lam CNW, Siu SO. Facile generation of tripeptide radical cations in vacuo via intramolecular electron transfer in Cu(II) tripeptide complexes containing sterically encumbered terpyridine ligands. J. Am. Soc. Mass Spectrom. 2005;16:763–771. doi: 10.1016/j.jasms.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 28.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J. Am. Chem. Soc. 1998;120:3265–3266. [Google Scholar]
- 29.Zubarev RA. Reactions of polypeptide ions with electrons in the gas phase. Mass Spectrom. Rev. 2003;22:57–77. doi: 10.1002/mas.10042. [DOI] [PubMed] [Google Scholar]
- 30.Shi SD, Hemling ME, Carr SA, Horn DM, Lindh I, McLafferty FW. Phosphopeptide/phosphoprotein mapping by electron capture dissociation mass spectrometry. Anal. Chem. 2001;73:19–22. doi: 10.1021/ac000703z. [DOI] [PubMed] [Google Scholar]
- 31.Mirgorodskya E, Roepstoff P, Zubarev RA. Localization of O-glycosylation sites in peptides by electron capture dissociation in a Fourier transform mass spectrometer. Anal. Chem. 1999;71:4431–4436. doi: 10.1021/ac990578v. [DOI] [PubMed] [Google Scholar]
- 32.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coon JJ. Collisions or electrons? Protein sequence analysis in the 21st century. Anal. Chem. 2009;81:3208–3215. doi: 10.1021/ac802330b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han H, Xia Y, Yang M, McLuckey SA. Rapidly alternating transmission mode electron-transfer dissociation and collisional activation for the characterization of polypeptide ions. Anal. Chem. 2008;80:3492–3497. doi: 10.1021/ac7022734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ewing NP, Cassady CJ. Dissociation of multiply charged negative ions for hirudin(54–65), fibrinopeptide B, and insulin A (oxidized) J. Am. Soc. Mass Spectrom. 2000;12:105–116. doi: 10.1016/S1044-0305(00)00195-1. [DOI] [PubMed] [Google Scholar]
- 36.Bowie JH, Brinkworth CS, Dua S. Collision-induced fragmentations of the (M-H)-parent anions of underivatized peptides: an aid to structure determination and some unusual negative ion cleavages. Mass Spectrom. Rev. 2002;21:87–107. doi: 10.1002/mas.10022. [DOI] [PubMed] [Google Scholar]
- 37.Kjeldsen F, Silivra OA, Ivonin IA, Haselmann KF, Gorshkov M, Zubarev RA. Cα-C backbone fragmentation dominates in electron detachment dissociation of gas-phase polypeptide polyanions. Chem. Eur. J. 2005;11:1803–1812. doi: 10.1002/chem.200400806. [DOI] [PubMed] [Google Scholar]
- 38.Budnik B, Haselmann KF, Zubarev RA. Electron detachment dissociation of peptide di-anions: an electron-hole recombination phenomenon. Chem. Phys. Lett. 2001;342:299–302. [Google Scholar]
- 39.Anusiewicz I, Jasionowski M, Skurski P, Simons J. Backbone and side-chain cleavages in electron detachment dissociation (EDD) J. Phys. Chem. A. 2005;109:11332–11337. doi: 10.1021/jp055018g. [DOI] [PubMed] [Google Scholar]
- 40.Coon JJ, Shabanowitz J, Hunt DF, Syka JEP. Electron transfer dissociation of peptide anions. J. Am. Soc. Mass Spectrom. 2005;16:880–882. doi: 10.1016/j.jasms.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 41.Antoine R, Joly L, Tabarin T, Broyer M, Dugourd P, Lemoine J. Photo-induced formation of radical anion peptides. Electron photodetachment dissociation experiments. Rapid Commun. Mass Spectrom. 2007;21:265–268. doi: 10.1002/rcm.2810. [DOI] [PubMed] [Google Scholar]
- 42.McLuckey SA, Van Berkel GJ, Glish GL. Mass spectrometry/mass spectrometry of small multiply charged oligonucleotides. J. Am. Soc. Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 43.McLuckey SA, Habibi-Goudarzi S. Decompositions of multiply-charged oligonucleotide anions. J. Am. Chem. Soc. 1993;115:12085–12095. [Google Scholar]
- 44.Rodgers MT, Campbell S, Marzluff EM, Beauchamp JL. Low-energy collision-induced dissociation of deprotonated dinucleotides: determination of the energetically favored dissociation pathways and the relative acidities of the nucleic acid bases. Int. J. Mass Spectrom. 1994;137:121–149. [Google Scholar]
- 45.Habibi-Ghoudarzi S, McLuckey SA. Ion trap collisional activation of the deprotonated deoxymononucleoside and deoxydinucleoside monophosphates. J. Am. Soc. Mass Spectrom. 1995;6:102–113. doi: 10.1016/S1044-0305(94)00108-C. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z, Wan KX, Ramanatha R, Taylor JS, Gross ML. Structure and fragmentation mechanisms of isomeric T-rich oligodeoxynucleotides: a comparison of four tandem mass spectrometric methods. J. Am. Soc. Mass Spectrom. 1998;9:683–691. doi: 10.1016/S1044-0305(98)00178-0. [DOI] [PubMed] [Google Scholar]
- 47.Wan KX, Gross J, Hillenkamp F, Gross ML. Fragmentation mechanisms of oligodeoxynucleotides studied by H/D exchange and electrospray ionization tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2001;12:193–205. doi: 10.1016/S1044-0305(00)00208-7. [DOI] [PubMed] [Google Scholar]
- 48.Wan KX, Gross ML. Fragmentation mechanisms of oligodeoxynucleotides: effects of replacing phosphates with methylphosphonates and thymines with other bases in T-rich sequences. J. Am. Soc. Mass Spectrom. 2001;12:580–589. doi: 10.1016/S1044-0305(01)00223-9. [DOI] [PubMed] [Google Scholar]
- 49.Wu J, McLuckey SA. Gas-phase fragmentation of oligonucleotide ions. Int. J. Mass Spectrom. 2004;237:197–241. [Google Scholar]
- 50.Little DP, Chorush RA, Speir JP, Senko MW, Kelleher NL, McLafferty FW. Rapid sequencing of oligonucleotides by high-resolution mass spectrometry. J. Am. Chem. Soc. 1994;116:4893–4897. [Google Scholar]
- 51.Little DP, McLafferty FW. Sequencing 50-mer DNAs using electrospray tandem mass spectrometry and complementary fragmentation methods. J. Am. Chem. Soc. 1995;117:6783–6784. [Google Scholar]
- 52.Little DP, Aaserud DJ, Valaskovic GA, McLafferty FW. Sequence information from 42 to 108-mer DNAs (complete for a 50-mer) by tandem mass spectrometry. J. Am. Chem. Soc. 1996;118:9352–9359. [Google Scholar]
- 53.Hofstadler SA, Griffey RH, Pasa-Tolic L, Smith RD. The use of a stable internal mass standard for accurate mass measurements of oligonucleotide fragment ions using electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry with infrared multiphoton dissociation. Rapid Commun. Mass Spectrom. 1998;12:1400–1404. [Google Scholar]
- 54.Hofstadler SA, Sannes-Lowery KA, Griffey RH. Infrared multiphoton dissociation in an external ion reservoir. Anal. Chem. 1999;71:2067–2070. doi: 10.1021/ac990176n. [DOI] [PubMed] [Google Scholar]
- 55.Hofstadler SA, Sannes-Lowery KA, Griffey RH. m/z-Selective infrared multiphoton dissociation in a Penning trap using sidekick trapping and an RF-tickle pulse. Rapid Commun. Mass Spectrom. 2001;15:945–951. [Google Scholar]
- 56.Sannes-Lowery KA, Hofstadler SA. Sequence confirmation of modified oligonucleotides using IRMPD in the external ion reservoir of an electrospray ionization Fourier transform ion cyclotron mass spectrometer. J. Am. Soc. Mass Spectrom. 2003;14:825–833. doi: 10.1016/S1044-0305(03)00335-0. [DOI] [PubMed] [Google Scholar]
- 57.Keller KM, Brodbelt JS. Collisionally activated dissociation and infrared multiphoton dissociation of oligonucleotides in a quadrupole ion trap. Anal. Biochem. 2004;326:200–210. doi: 10.1016/j.ab.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 58.Kirpekar F, Krogh TN. RNA fragmentation studied in a matrix-assisted laser desorption/ionization tandem quadrupole/orthogonal time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 2001;15:8–14. doi: 10.1002/1097-0231(20010115)15:1<8::AID-RCM185>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 59.Schürch S, Bernal-Mendez E, Leumann CJ. Electrospray tandem mass spectrometry of mixed-sequence RNA/DNA oligonucleotides. J. Am. Soc. Mass Spectrom. 2002;13:936–945. doi: 10.1016/S1044-0305(02)00413-0. [DOI] [PubMed] [Google Scholar]
- 60.Tromp JM, Schürch S. Gas-phase dissociation of oligoribonucleotides and their analogues studied by electrospray ionization tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2005;16:1262–1268. doi: 10.1016/j.jasms.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 61.Andersen TE, Kirpekar F, Haselmann KF. RNA fragmentation in MALDI mass spectrometry studied by H/D-exchange: mechanisms of general applicability to nucleic acids. J. Am. Soc. Mass Spectrom. 2006;17:1353–1368. doi: 10.1016/j.jasms.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 62.Huang TY, Kharlamova A, Liu J, McLuckey SA. Ion trap collision–induced dissociation of multiply deprotonated RNA: c/y-ions versus (a–B)/w-ions. J. Am. Soc. Mass Spectrom. 2008;19:1832–1840. doi: 10.1016/j.jasms.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 63.Huang TY, Liu J, Liang X, Hodges BDM, McLuckey SA. Collision-induced dissociation of intact duplex and single-stranded siRNA anions. Anal. Chem. 2008;80:8501–8508. doi: 10.1021/ac801331h. [DOI] [PubMed] [Google Scholar]
- 64.Herron WJ, Goeringer DE, McLuckey SA. Gas-phase electron-transfer reactions from multiply-charged anions to rare-gas cations. J. Am. Chem. Soc. 1995;117:11555–11562. [Google Scholar]
- 65.McLuckey SA, Stephenson JL, O’Hair RAJ. Decompositions of odd- and even-electron anions derived from deoxy-polyadenylates. J. Am. Soc. Mass Spectrom. 1997;8:148–154. [Google Scholar]
- 66.Wu J, McLuckey SA. Ion/ion reactions of multiply charged nucleic acid anions: electron transfer, proton transfer, and ion attachment. Int. J. Mass Spectrom. 2003;228:577–597. [Google Scholar]
- 67.Yang J, Mo J, Adamson JT, Håkansson K. Characterization of oligodeoxynucleotides by electron detachment dissociation Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 2005;77:1876–1882. doi: 10.1021/ac048415g. [DOI] [PubMed] [Google Scholar]
- 68.Gabelica V, Tabarin T, Antoine R, Rosu F, Compagnon I, et al. Electron photodetachment dissociation of DNA polyanions in a quadrupole ion trap mass spectrometer. Anal. Chem. 2006;78:6564–6572. doi: 10.1021/ac060753p. [DOI] [PubMed] [Google Scholar]
- 69.Gabelica V, Tabarin T, Antoine R, Rosu F, Compagnon I, et al. Base-dependent electron photodetachment from negatively charged DNA strands upon 260-nm laser irradiation. J. Am. Chem. Soc. 2007;129:4706–4713. doi: 10.1021/ja068440z. [DOI] [PubMed] [Google Scholar]
- 70.Ni J, Mathews MAA, McCloskey JA. Collision-induced dissociation of polyprotonated oligonucleotides produced by electrospray ionization. Rapid Commun. Mass Spectrom. 1997;11:535–540. doi: 10.1002/(SICI)1097-0231(199704)11:6<535::AID-RCM898>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 71.Wang P, Bartlett MG, Martin LB. Electrospray collision–induced dissociation mass spectra of positively charged oligonucleotides. Rapid Commun. Mass Spectrom. 1997;11:846–856. [Google Scholar]
- 72.Weimann A, Iannitti-Tito P, Sheil MM. Characterization of product ions in high-energy tandem mass spectra of protonated oligonucleotides formed by electrospray ionization. Int. J. Mass Spectrom. 2000;194:269–288. [Google Scholar]
- 73.Vrkic AK, O’Hair RAJ, Foote S, Reid GE. Fragmentation reactions of all 64 protonated trimer oligodeoxynucleotides and 16 mixed base tetramer oligodeoxynucleotides via tandem mass spectrometry in an ion trap. Int. J. Mass Spectrom. 2000;194:145–164. [Google Scholar]
- 74.Håkansson H, Hudgins RR, Marshall AG, O’Hair RAJ. Electron capture dissociation and infrared multiphoton dissociation of oligodeoxynucleotide dications. J. Am. Soc. Mass Spectrom. 2003;14:23–41. doi: 10.1016/S1044-0305(02)00708-0. [DOI] [PubMed] [Google Scholar]
- 75.Schultz KN, Håkansson K. Rapid electron capture dissociation of mass-selectively accumulated oligodeoxynucleotide dications. Int. J. Mass Spectrom. 2004;234:123–130. [Google Scholar]
- 76.Smith SI, Brodbelt JS. Electron transfer dissociation of oligonucleotide cations. Int. J. Mass Spectrom. 2009;283:85–93. doi: 10.1016/j.ijms.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zaia J, Costello CE. Tandem mass spectrometry of sulfated heparin-like glycosaminoglycan oligosaccharides. Anal. Chem. 2003;75:2445–2455. doi: 10.1021/ac0263418. [DOI] [PubMed] [Google Scholar]
- 78.Liang X, Liu J, LeBlanc Y, Covey T, Ptak AC, et al. Electron transfer dissociation of doubly sodiated glycerophosphocholine lipids. J. Am. Soc. Mass Spectrom. 2007;18:1783–1788. doi: 10.1016/j.jasms.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Payne AH, Glish GL. Gas-phase ion/ion interactions between peptides or proteins and iron ions in a quadrupole ion trap. Int. J. Mass Spectrom. 2001;204:47–54. [Google Scholar]
- 80.Newton KA, McLuckey SA. Gas-phase peptide/protein cationizing agent switching via ion/ion reactions. J. Am. Chem. Soc. 2003;125:12404–12405. doi: 10.1021/ja036924e. [DOI] [PubMed] [Google Scholar]
- 81.Newton KA, McLuckey SA. Generation and manipulation of sodium cationized peptides in the gas phase. J. Am. Soc. Mass Spectrom. 2004;15:607–615. doi: 10.1016/j.jasms.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 82.Newton KA, He M, Amunugama R, McLuckey SA. Selective cation removal from gaseous polypeptide ions: proton vs sodium ion abstraction via ion/ion reactions. Phys. Chem. Chem. Phys. 2004;6:2710–2717. [Google Scholar]
- 83.Hodges BDM, Liang X, McLuckey SA. Generation of di-lithiated peptide ions from multiply protonated peptides via ion/ion reactions. Int. J. Mass Spectrom. 2007;267:183–189. [Google Scholar]
- 84.Gunawardena HP, O’Hair RAJ, McLuckey SA. Selective disulfide bond cleavage in gold(I) cationized polypeptide ions formed via gas-phase ion/ion cation switching. J. Proteome Res. 2006;5:2087–2092. doi: 10.1021/pr0602794. [DOI] [PubMed] [Google Scholar]
- 85.Barlow CK, Hodges BDM, Xia Y, O’Hair RAJ, McLuckey SA. Gas-phase ion/ion reactions of transition metal complex cations with multiply charged oligodeoxynucleotide anions. J. Am. Soc. Mass Spectrom. 2008;19:281–293. doi: 10.1016/j.jasms.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 86.He M, McLuckey SA. Two ion/ion charge inversion steps to form a doubly protonated peptide from a singly protonated peptide in the gas phase. J. Am. Chem. Soc. 2003;125:7756–7757. doi: 10.1021/ja0354521. [DOI] [PubMed] [Google Scholar]
- 87.He M, McLuckey SA. Increasing the negative charge of a macroanion in the gas phase via sequential charge inversion reactions. Anal. Chem. 2004;76:4189–4192. doi: 10.1021/ac496087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.He M, Emory JF, McLuckey SA. Reagent anions for charge inversion of polypeptide/protein cations in the gas phase. Anal. Chem. 2005;77:3173–3182. doi: 10.1021/ac0482312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gunawardena HP, Emory JF, McLuckey SA. Phosphopeptide anion characterization via sequential charge inversion and electron-transfer dissociation. Anal. Chem. 2006;78:3788–3793. doi: 10.1021/ac060164j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Trimpin S, Inutan ED, Herath TN, McEwen CN. Matrix-assisted laser desorption/ionization mass spectrometry method for selectively producing either singly or multiply charged molecular ions. Anal. Chem. 2009 doi: 10.1021/ac902066s. doi:10.1021/ac902066s. [DOI] [PubMed] [Google Scholar]
- 91.McLuckey SA, Van Berkel GJ, Glish GL. Reactions of dimethylamine with multiply charged ions of cytochrome c. J. Am. Chem. Soc. 1990;112:5668–5670. [Google Scholar]
- 92.McLuckey SA, Glish GL, Van Berkel GJ. Charge determination of product ions formed from collision-induced dissociation of multiply protonated molecules via ion/molecule reactions. Anal. Chem. 1991;63:1971–1978. doi: 10.1021/ac00018a014. [DOI] [PubMed] [Google Scholar]
- 93.McLuckey SA, Goeringer DE. Ion/molecule reactions for improved effective mass resolution in electrospray mass spectrometry. Anal. Chem. 1995;67:2493–2497. doi: 10.1021/ac00110a026. [DOI] [PubMed] [Google Scholar]
- 94.Cassady CJ, Wronka J, Kruppa GH, Laukien FH. Deprotonation reactions of multiply protonated ubiquitin ions. Rapid Commun. Mass Spectrom. 1994;8:394–400. doi: 10.1002/rcm.1290080511. [DOI] [PubMed] [Google Scholar]
- 95.Loo RRO, Smith RD. Proton-transfer reactions of multiply charged peptide and protein cations and anions. J. Mass Spectrom. 1995;30:339–347. [Google Scholar]
- 96.Scalf M, Westphall MS, Krause J, Kaufma SL, Smith LM. Controlling charge states of large ions. Science. 1999;283:194–197. doi: 10.1126/science.283.5399.194. [DOI] [PubMed] [Google Scholar]
- 97.Scalf M, Westphall MS, Smith LM. Charge reduction electrospray mass spectrometry. Anal. Chem. 2000;72:52–60. doi: 10.1021/ac990878c. [DOI] [PubMed] [Google Scholar]
- 98.Stephenson JL, Jr, McLuckey SA. Ion/ion reactions in the gas-phase: proton transfer reactions involving multiply-charged proteins. J. Am. Chem. Soc. 1996;118:7390–7397. [Google Scholar]
- 99.Stephenson JL, Jr, McLuckey SA. Ion/ion reactions for oligopeptide mixture analysis: application to mixtures comprised of 0.5–100 kDa components. J. Am. Soc. Mass Spectrom. 1998;9:585–596. doi: 10.1016/S1044-0305(98)00025-7. [DOI] [PubMed] [Google Scholar]
- 100.McLuckey SA, Wu J, Bundy JL, Stephenson JL, Hurst GB. Oligonucleotide mixture, analysis via electrospray and ion/ion reactions in a quadrupole ion trap. Anal. Chem. 2002;74:976–984. doi: 10.1021/ac011015y. [DOI] [PubMed] [Google Scholar]
- 101.Stephenson JL, Jr, McLuckey SA. Ion/ion proton transfer reactions for protein mixture analysis. Anal. Chem. 1996;68:4026–4032. doi: 10.1021/ac9605657. [DOI] [PubMed] [Google Scholar]
- 102.Hogan JM, McLuckey SA. Charge state dependent collision-induced dissociation of native and reduced porcine elastase. J. Mass Spectrom. 2003;38:245–256. doi: 10.1002/jms.458. [DOI] [PubMed] [Google Scholar]
- 103.Chanthamontri C, Liu J, McLuckey SA. Charge state dependent fragmentation of gaseous α-synuclein cations via ion trap and beam-type collisional activation. Int. J. Mass Spectrom. 2009;283:9–16. doi: 10.1016/j.ijms.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McLuckey SA, Reid GE, Wells JM. Ion parking during ion/ion reactions in electrodynamic ion traps. Anal. Chem. 2002;74:336–346. doi: 10.1021/ac0109671. [DOI] [PubMed] [Google Scholar]
- 105.Reid GE, Shang H, Hogan JM, Lee GU, McLuckey SA. Gas-phase concentration, purification, and identification of whole proteins from complex mixtures. J. Am. Chem. Soc. 2002;124:7353–7362. doi: 10.1021/ja025966k. [DOI] [PubMed] [Google Scholar]
- 106.Reid GE, Wells JM, Badman ER, McLuckey SA. Performance of a quadrupole ion trap mass spectrometer adapted for ion/ion reaction studies. Int. J. Mass Spectrom. 2003;222:243–258. [Google Scholar]
- 107.Amunugama R, Hogan JM, Newton KA, McLuckey SA. Whole protein dissociation in a quadrupole ion trap: identification of an a priori unknown modified protein. Anal. Chem. 2004;76:720–727. doi: 10.1021/ac034900k. [DOI] [PubMed] [Google Scholar]
- 108.He M, Reid GE, Shang H, Lee GU, McLuckey SA. Dissociation of multiple protein ion charge states following a single gas-phase purification and concentration procedure. Anal. Chem. 2002;74:4653–4661. doi: 10.1021/ac025587+. [DOI] [PubMed] [Google Scholar]
- 109.Chrisman PA, Pitteri SJ, McLuckey SA. Parallel ion parking: improving conversion of parents to first-generation products in electron transfer dissociation. Anal. Chem. 2005;77:3411–3414. doi: 10.1021/ac0503613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Coon JJ, Ueberheide B, Syka JEP, Dryhurst DD, Ausio J, et al. Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc. Natl. Acad. Sci. USA. 102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu J, Huang TY, McLuckey SA. Top-down protein identification/characterization of a priori unknown proteins via ion trap collision-induced dissociation and ion/ion reactions in a quadrupole/time-of-flight tandem mass spectrometer. Anal. Chem. 2009;81:1433–1441. doi: 10.1021/ac802204j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stephenson JL, Jr, McLuckey SA. Simplification of product ion spectra derived from multiply-charged parent ions via ion/ion chemistry. Anal. Chem. 1998;70:3533–3544. doi: 10.1021/ac9802832. [DOI] [PubMed] [Google Scholar]
- 113.O’Hair RAJ, McLuckey SA. Trimethylsilyl derivatization of nucleic acid anions in the gas phase. Int. J. Mass Spectrom. Ion Processes. 1997;162:183–202. [Google Scholar]
- 114.O’Hair RAJ, Reid GE. Derivatization of protonated peptides via gas phase ion-molecule reactions with acetone. J. Am. Soc. Mass Spectrom. 2000;11:244–256. doi: 10.1016/S1044-0305(99)00142-7. [DOI] [PubMed] [Google Scholar]
- 115.Gur EH, de Koning LJ, Nibbering NMM. The bimolecular gas-phase reaction of protonated alkyldipeptides with acetonylacetone. Int J. Mass Spectrom. Ion Processes. 1997;167:135–147. [Google Scholar]
- 116.Han H, McLuckey SA. Selective covalent bond formation in polypeptide ions via gas-phase ion/ion reaction chemistry. J. Am. Chem. Soc. 2009;131:12884–12885. doi: 10.1021/ja904812d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stephenson JL, Jr, McLuckey SA. Gaseous protein cations are amphoteric. J. Am. Chem. Soc. 1997;119:1688–1696. [Google Scholar]
- 118.Wells JM, Chrisman PA, McLuckey SA. Formation of protein-protein complexes in vacuo. J. Am. Chem. Soc. 2001;123:12428–12429. doi: 10.1021/ja0170403. [DOI] [PubMed] [Google Scholar]
- 119.Wells JM, Chrisman PA, McLuckey SA. Formation and characterization of protein-protein complexes in vacuo. J. Am. Chem. Soc. 2003;125:7238–7249. doi: 10.1021/ja035051l. [DOI] [PubMed] [Google Scholar]
- 120.Gunawardena HP, McLuckey SA. Synthesis of multi-unit protein hetero-complexes in the gas phase via ion-ion chemistry. J. Mass Spectrom. 2004;39:630–638. doi: 10.1002/jms.629. [DOI] [PubMed] [Google Scholar]
- 121.Bowers JJ, Hodges BDM, Saad OM, Leary JA, McLuckey SA. Proton hydrates as soft ion/ion proton transfer reagents for multiply deprotonated biomolecules. Int. J. Mass Spectrom. 2008;276:153–159. [Google Scholar]
- 122.Leib RD, Donald WA, Bush MF, O’Brien JT, Williams ER. Internal energy deposition in electron capture dissociation measured using hydrated divalent metal ions as nanocalorimeters. J. Am. Chem. Soc. 2007;129:4894–4895. doi: 10.1021/ja0666607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Prell JS, O’Brien JT, Holm AIS, Leib RD, Donald WA, Williams ER. Electron capture by a hydrated gaseous peptide: effects of water on fragmentation and molecular survival. J. Am. Chem. Soc. 2008;130:12680–12689. doi: 10.1021/ja8022434. [DOI] [PMC free article] [PubMed] [Google Scholar]