Abstract
Dynorphins, the endogenous ligands for the kappa opioid receptor, are implicated in neuropsychiatric disorders through modulation of basal and stimuli-induced dopaminergic, glutamatergic and serotonergic tones. Expression of the prodynorphin gene (PDYN) is critical for rewarding properties of drugs of abuse and stress-induced responses. Epigenetic factors such as DNA methylation play an important role in modulation of gene expression. We analyzed DNA methylation patterns of three CpG-rich regions of PDYN, a CpG island, and cluster A in the proximal promoter, and cluster B in coding exon 4, by bisulfite sequencing of DNA from the caudate and anterior cingulate cortex from post-mortem brain of 35 individuals (22 HIV seropositive), and in peripheral blood mononuclear cells (PBMCs) from 21 of these subjects. We found remarkably similar patterns of methylation across CpG sites in these tissues. However, there were tissue-specific differences in methylation levels (p=0.000001) of the CpG island: higher levels in PBMCs (82%) than in the brain tissues, the caudate (62%), and the anterior cingulate cortex (44%). But there was higher PDYN expression in the caudate than the anterior cingulate cortex. In contrast, cluster A near the transcription start site is hypomethylated. This DNA methylation profile of the PDYN gene is typical for primary responsive genes with regulatory elements for both basal and tissue-specific transcription. Our findings provide a rationale for further studies of the role of other epigenetic factors in regulation of PDYN expression in subjects with psychiatric and neurological disorders.
Keywords: DNA methylation, CpG island, dynorphin, bisulfite sequencing, human post-mortem brain, caudate, anterior cingulate cortex, prodynorphin gene expression
INTRODUCTION
The opioid dynorphin peptides are co-localized with the kappa opioid receptor (OPRK1) in several areas of the dopaminergic nigrostriatal and mesolimbic–mesocortical systems, and play an important role in modulation of opioid, cocaine and other rewarding stimuli, presumably through modulation of basal and drug-induced dopamine-, glutamate- and serotoninergic tones [1–3].
Studies from this and other laboratories have demonstrated that cocaine administration elevates preprodynorphin mRNA levels predominantly in rodent caudate putamen, following a single injection, self-administration, and acute or chronic ”binge” [4–7]. PDYN mRNA levels increased rapidly following acute ”binge” cocaine administration, and this effect persisted with subsequent chronic drug administration [8]. Also, elevated levels of PDYN mRNA and dynorphin peptides were found in the striatum from post-mortem brains of former chronic cocaine users [9,10].
Pretreatment with OPRK1 agonists has been shown to decrease the psychostimulant and conditioned rewarding effects of cocaine in rats, and to decrease the rate of intravenous cocaine self-administration [11,12]. Dynorphin peptides decrease basal and drug-induced dopamine levels in several areas of the striatum (e.g. [12]). The OPRK1–dynorphin system may therefore be considered to be one of the counter-modulatory mechanisms of the brain following direct or indirect drug-induced dopaminergic stimulation [13]. It has been suggested that an activation of the OPRK1–dynorphin system, long understood to cause dysphoria, may also contribute to development of depression (e.g. [14]). Inhibition of dynorphin or OPRK1 functions in rodents was shown to be associated with antidepressant-like activity and block stress-induced behavioral responses [15, 16].
The human prodynorphin gene (hPDYN) is located at chromosome 20pter-p12.2 and spans 15.3 kb. The gene has four exons: exons 1 and 2 contain the 5' untranslated region (UTR); exon 3 encodes a signal peptide; and exon 4 encodes the dynorphin peptides, including α-neoendorphin, β-neoendorphin, dynorphin A, and dynorphin B [17]. In addition to the mRNA with four exons, several splice variants were identified in the embryonic human brain and testis, including forms of mRNA with one, two or three exons [18,19]. PDYN is expressed in many brain regions [18,19], as well in other tissues, including spinal cord [20], and immune cells [21].
Currently, it is well established that epigenetic factors such as DNA methylation and histone modification are critically involved in regulation of gene expression (e.g. [22]). A number of studies have identified differential patterns and tissue-specific DNA methylation across the human genome (e.g. [23–25]). Interestingly, a recent genome-wide analysis of DNA methylation showed that 76% of differential tissue methylation regions (T-DMRs) were not located in CpG islands, but in sequences up to 2 kb distant, termed “CpG island shores” [24].
Infection of human cells by the human immunodeficiency virus type 1 (HIV-1) results in dramatic changes in cellular gene expression, including retroviral-mediated alterations in host DNA methylation [26]. The mechanisms that mediate HIV-induced de novo methylation in vivo remain largely unknown. In vitro studies have shown up-regulation of DNA methyltransferase 1 (DNMT1) in CD4 T cells and HeLa cells after HIV-1 infection, mediated by the transcription factor AP-1 [27]. In relation to HIV-induced neuropathogenesis, researchers found highest concentration of HIV-1 RNA in the caudate [28], and significant decrease in dopamine levels in the caudate, putamen, and substantia nigra in post-mortem brain of HIV-infected subjects [29].
Several potential transcription factor binding sites within the PDYN promoter have been shown to play a role in regulating PDYN expression: a polymorphic 68-bp tandem repeat polymorphism (rs35286281), containing a putative AP-1, located 1250 bp upstream of exon 1 [17, 30]; a site at −156 in the proximal promoter; and −2745 microsatellite [31]. Recently, we reported that the cis-acting polymorphism (rs910079) in the PDYN 3'UTR may be involved in alterations of gene expression in an allele-specific manner [32]. Basal and stimuli-induced PDYN expression of the human PDYN gene is also regulated by the calcium sensitive transcription repressor DREAM (downstream regulatory element antagonist modulator) that binds to the regulatory element (DRE) located in the 5'-untranslated region within exon 1 [33].
Despite existing information on human PDYN expression in brain, little is known about basal, tissue-specific or drug- and stress-induced regulation of the gene. Detailed analysis of epigenetic status of CpG-rich regions of human PDYN may provide insight into the role of epigenetic factors in basal tissue-specific regulation of PDYN, and in response to stress and drug administration.
Our goals in this study were (1) to examine the DNA methylation patterns in the proximal promoter region of hPDYN in two brain regions in post-mortem brain, the caudate and anterior cingulate cortex, and in matched peripheral blood mononuclear cells (PBMCs); (2) to investigate whether PDYN expression levels in these tissues are correlated with DNA methylation level.
We hypothesized that the DNA methylation profile of the hPDYN promoter may lead to the identification of transcriptional cis-regulatory elements and other epigenetic factors (histone modification, DNA CpG binding proteins) that may be involved in the regulation of basal and stimuli-induced PDYN expression in a tissue-specific manner.
MATERIALS AND METHODS
Study subjects, brain and PBMC samples
Brain tissues and PBMCs were obtained from the Manhattan HIV Brain Bank (MHBB, The Mount Sinai Medical Center, New York, NY). The MHBB operates under local IRB-approved ethical guidelines, and individuals or their primary next-of-kin gave consent for collection and use of tissues for medical research and furthering medical knowledge. Specimens from subjects with protracted agonal state, as manifested by extensive anoxic–ischemic damage on histological evaluation, were excluded from this study. Coronal sections of each brain were stored at −80°C. The caudate and anterior cingulate cortex samples were derived from 35 unrelated individuals of different ethnicities (8 Caucasians, 12 African Americans, 14 Hispanics, and 1 Asian) (Table 1). A total of 22 subjects were HIV seropositive and 13 were HIV seronegative. Matched PBMC samples were available for 21 individuals.
Table 1.
Demography and categories of postmortem brain samples
| Subject mhbb ID |
Gender | Ethnicity | Age | PMI (h) | HIV Status |
PBMC available |
|
|---|---|---|---|---|---|---|---|
| 1 | 604 | M | AA | 48 | 9 | − | - |
| 2 | 531 | M | C | 44 | 21.5 | − | - |
| 3 | 553 | M | H | 46 | 16 | − | - |
| 4 | 568 | F | H | 48 | 16 | − | - |
| 5 | 577 | M | C | 65 | 17.5 | − | - |
| 6 | 588 | M | H | 21 | 18.5 | − | - |
| 7 | 589 | M | H | 61 | 6.5 | − | - |
| 8 | 594 | F | C | 57 | 24 | − | - |
| 9 | 601 | F | H | 58 | 16.5 | − | - |
| 10 | 606 | M | H | 50 | 16 | − | - |
| 11 | 610 | F | AA | 54 | 27 | − | - |
| 12 | 611 | M | AA | 59 | 28.4 | − | - |
| 13 | 613 | F | A | 55 | 24 | − | - |
| 14 | 10001 | F | AA | 64 | 4.5 | + | - |
| 15 | 10002 | M | AA | 58 | 15.5 | + | ● |
| 16 | 10013 | M | C | 33 | 17 | + | ● |
| 17 | 10015 | M | C | 33 | 20 | + | ● |
| 18 | 10016 | F | AA | 58 | 17 | + | ● |
| 19 | 10023 | M | AA | 53 | 4.5 | + | ● |
| 20 | 10025 | M | H | 46 | 7.5 | + | ● |
| 21 | 10027 | M | H | 39 | 6 | + | ● |
| 22 | 10034 | M | H | 49 | 5 | + | ● |
| 23 | 10043 | F | AA | 34 | 17 | + | ● |
| 24 | 10045 | F | H | 31 | 9 | + | ● |
| 25 | 10052 | M | AA | 39 | 5 | + | ● |
| 26 | 10063 | M | H | 51 | 5 | + | ● |
| 27 | 10064 | M | C | 49 | 12 | + | ● |
| 28 | 10065 | M | H | 46 | 7 | + | ● |
| 29 | 10066 | F | AA | 54 | 7 | + | ● |
| 30 | 10074 | M | AA | 41 | 17.5 | + | ● |
| 31 | 10086 | F | AA | 48 | 7.5 | + | ● |
| 32 | 10094 | F | H | 41 | 6.5 | + | ● |
| 33 | 10103 | M | H | 40 | 6 | + | ● |
| 34 | 10133 | M | C | 48 | 7.5 | + | ● |
| 35 | 20024 | M | C | 62 | 4 | + | ● |
AA - Afrrican Americans; C - Caucasians; H - Hispanics; A - Asians. PMI -post-mortem interval
DNA Preparation
Tissue (40–60 mg) of each brain sample was homogenized in DNA lysis buffer (Easy-DNA Kit, Invitrogen, Carlsbad, CA, USA) for the isolation of genomic DNA. Genomic DNA was extracted from PBMCs using the Gentra Puregene kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol.
DNA methylation analysis
Genomic DNA was treated with sodium bisulfite using the EZ-96 DNA Methylation Kit (Zymo Research, Orange, CA). Primers for PCR and bisulfite sequencing were custom designed (Supplement Table S1 http://links.lww.com/FPC/A203) based on Methyl Primer Express software (Applied Biosystems, Foster city, CA).
Two methods were employed to determine CpG methylation. The first method employs direct Sanger sequencing of PCR amplicons, which measures the average of methylation of all DNA molecules in a tissue. The second method employs cloning of the PCR-amplified fragments, isolating individual clones, and sequencing of the amplified inserts. This method provides a methylation profile of single DNA molecules. After PCR amplification, unincorporated nucleotides and primers were removed by mixing 4 µl of the final PCR reaction mixture with 1 µL ExoSAP-IT (USB Corp., Cleveland, OH, USA) followed by incubation at 37°C for 30 min and 80°C for 15 min. For sequencing, 1 µL ExoSAP-IT-treated DNA was added to 11 µL of 8 pM forward or reverse primers. Sanger bisulfite DNA sequencing was performed in both forward and reverse directions using the Big Dye Terminator Cycle Sequencing Kit (v3.1, Applied Biosystems) on ABI 3730xl DNA analyzer. Only forward direction produced a good quality sequence which could be used by the Epigenetic Sequencing Methylation Analysis (ESME) for further analysis. The reverse primer sequencing of the PDYN CpG island and cluster A produced highly variable and poor quality sequence traces. The reason for that is a presence of long homogeneous T-nucleotide sequences after conversion C to T nucleotide in bisulfite-treated DNA, particularly in the C-rich promoter region of PDYN, resulting in mispriming and incorrect PCR amplification. Cluster B in PDYN exon 4 is surrounded by heterologous nucleotide sequences, and we obtained good concordance in DNA methylation rate between forward and reverse sequencing. Forward direction sequencing of PDYN CpG island produced a high correlation of the methylation rate of each CpG site with cloning data (r=0.826, p<0.0001).
ESME analysis
For quantification of percent methylation of CpG sites, sequence trace files were analyzed using ESME version 3.2.1 software from Epigenomics AG (Berlin, Germany) [34]. The ESME software was run on a virtual machine (VMware player running on Windows XP) running Red Hat Enterprise Linux 4, which corrects trace files for quality problems, incomplete conversion, imbalanced or overscaled signals, and base caller artifacts to provide quantitative measurement of cytosine methylation. The percent methylation calls by the ESME were reviewed by two independent researchers who visually inspected all the methylation calls using the electropherograms generated by the ESME software. Completeness of conversion C to T in all non-CpG nucleotides (i.e. CpA, CpT, CpC) were confirmed in bisulfite sequence traces of all our samples.
Out of 35, two DNA samples (mhbb # 10001 and 10043) in the caudate and three samples in the anterior cingulate cortex (mhbb # 553, 10001 and 10043) were excluded in ESME analysis due to poor sequence traces. Also, out of PBMCs of 21 individuals, one sample (mhbb #10043) could not be used for analysis because there was no usable methylation data in the brain regions for this subject.
Cloning of PCR Fragments
Amplified bisulfite-modified DNA fragments were cloned using the TA Cloning Kit (Invitrogen) into the pCRII plasmid. DNA from individual clones was sequenced in both directions using M13 forward and reverse primers and the Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems). Samples were run on an ABI Prism 3730xl capillary sequencer (Applied Biosystems). Clones were sequenced for each PCR-amplified region.
Sequence Analysis
The amplified and sequenced region of PDYN was analyzed for predicted transcription factor-binding sites using the Transcription Element Search System [35].
RNA preparation and cDNA synthesis
Additional brain tissues (60–80 mg) from each brain sample were homogenized in RLT buffer (RNeasy Mini Kit, Qiagen) for isolation of total RNA according to the manufacturer’s protocol. RNA samples were treated with RNase-Free DNase (TURBO DNA-free, Ambion, Austin, TX, USA) to remove residual DNA. The RNA integrity number (RIN) and concentration were determined using Agilent 2100 Bioanalyzer (Agilent Technologies, Wilmington, DE, USA). Single strand cDNA was synthesized from 1 µg of total RNA in 20 µL of reaction volume using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) in the presence of random primers and gene-specific antisense primers for PDYN, OPRK1, OPRM1 and CCR5. The antisense PDYN specific primer, 5'-CCTCCCTAAACCCGTCAGA-3', is located in exon 4 at nucleotide position 271–289, relative to the first nucleotide of the exon 4 (GenBank NM_024411). It should be noted that cDNA synthesis was designed to preferentially synthesize cDNAs of PDYN, OPRK1, OPRM1 and CCR5.
Quantitative Real-Time PCR
For quantification of the PDYN mRNA levels in the caudate and anterior cingulate cortex, we used a quantitative real-time polymerase chain reaction (qRT-PCR). cDNA (2 µl) was amplified in a 20 µl solution that contained SYBR® GreenER qPCR SuperMix (Invitrogen) and 10 nM of primers with a PCR condition of 40 cycles of denaturation at 94°C for 30 sec, annealing/extension at 60°C for 1 min. Forward and reverse primers for the PDYN cDNA amplification were located in exon 3 (5'-GGTGCTCCTTGTGTGC TGTAA-3') and exon 4 (5'-GCATCTCTCCCATTCCTCAGA-3'), respectively. The human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primer set (SABiosciences, Frederick, MD, USA) was used for amplification of GAPDH cDNA. Experimental samples were amplified simultaneously with samples that contained serial dilutions of the PDYN or GAPDH cDNA from 101 to 106 copies/2 µl, used to prepare standard curves. qRT-PCR analysis was performed using SDS 2.2 software (ABI) on an ABI Prism® 7900 sequence detection system. The specificity of amplification was confirmed by agarose gel electrophoresis of PCR products and a melting curve profile. PDYN and GAPDH cDNA copy number was quantified by comparing threshold cycles (Ct) of an experimental sample to those in standard curves for PDYN and GAPDH cDNA as described [36]. PDYN cDNA copy number is expressed normalized to copies of GAPDH cDNA copy number.
Statistical Analysis
To determine whether there was a significant difference in the levels of DNA methylation between brain and blood tissues, analyses of variance (ANOVA) with repeated measures were used, followed by Newman-Keuls post hoc tests or planned comparisons as appropriate. The relationship between percent methylation based on cloning or ESME was examined by a Pearson correlation. To examine PDYN expression in two brain regions of the same subjects, a paired t-test was used.
Hierarchical clustering of the methylation profiles of three tissue samples from 20 individuals was performed using the hclust function of R statistical package [37].
RESULTS
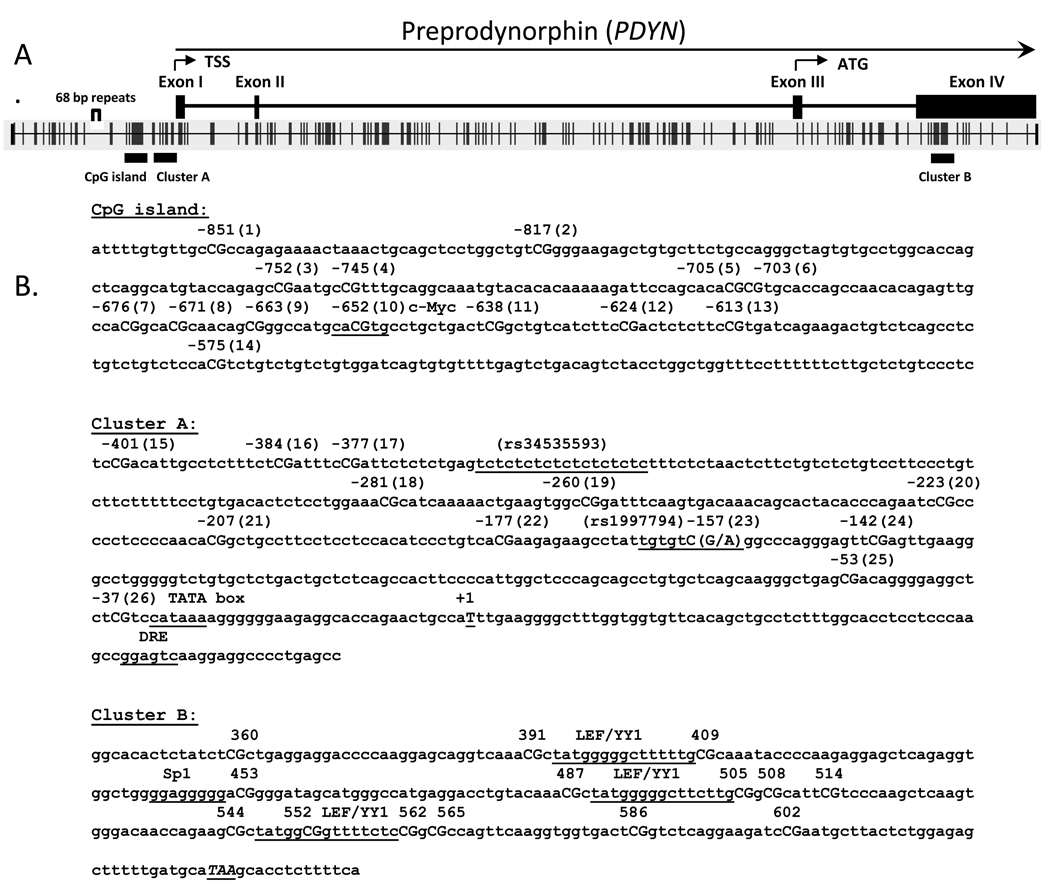
The search for CpG islands in PDYN gene was initially performed using a definition of a CpG island provided by Gardiner-Garden and Frommer [38] implemented in the Methyl Primer Express software (Applied Biosystems). A CpG island is defined as a region of at least 200 bp, with the proportion of Gs or Cs, referred to as “GC content,” greater than 50%, and observed to expected CpG ratio (O/E) greater than 0.6. Analysis of 3 kb upstream of the transcription start site (TSS) of the human PDYN gene showed the presence of a single CpG island which is located from nucleotide −770 to −570 relative to +1 of TSS (Chr 20 19074402-1922702, Build 36.1, GenBank NM_024411) (Fig. 1A). Downstream of the CpG island, a CpG-rich area, which we designated CpG cluster A, in the putative proximal promoter region was detected, located from nucleotides −410 to −37 (Fig. 1B).
Figure 1.
(A) Schematic representation of the human PDYN gene and 5' upstream region (3 kbp from the transcription start site, TSS). ATG is the translation start site. Shown are the distribution of CpG dinucleotides, the location of the CpG island, and the two other CpG-rich regions, cluster A and cluster B. (B) The sequences of the CpG island, cluster A and cluster B that were used for DNA methylation analyses. The position of CpG sites in the CpG island and in cluster A are shown relative to +1 of TSS. The CpG cluster B is located at nucleotides from 346 to 610, relative to the first nucleotide of the exon 4. The TATA-box and putative transcription sites are underlined. The SNP rs1997794 (A to G) affects CpG site #23 and alters the canonical binding site for transcription factor AP-1.
A search for a CpG island in the coding region of the PDYN gene using less stringent parameters (GC percent > 40%, obs/exp >0.6) revealed another CpG-rich area in exon 4 (designated cluster B) located from 346 to 610, relative to the first nucleotide of the exon 4 (Fig. 1B). We examined the DNA methylation status of 12 CpG sites (# 3–14) in the CpG island and two sites (# 1 and 2) upstream of the CpG island, 12 CpG sites (# 15–26) in the CpG cluster A (Fig. 1B), and 14 CpG sites in the CpG cluster B (Fig. 1B). Notably, the region of the 68-base pair nucleotide tandem repeat polymorphism [17], which contains a putative noncanonical AP-1 transcription binding site, does not contain CpG sites.
DNA methylation analysis of the CpG island
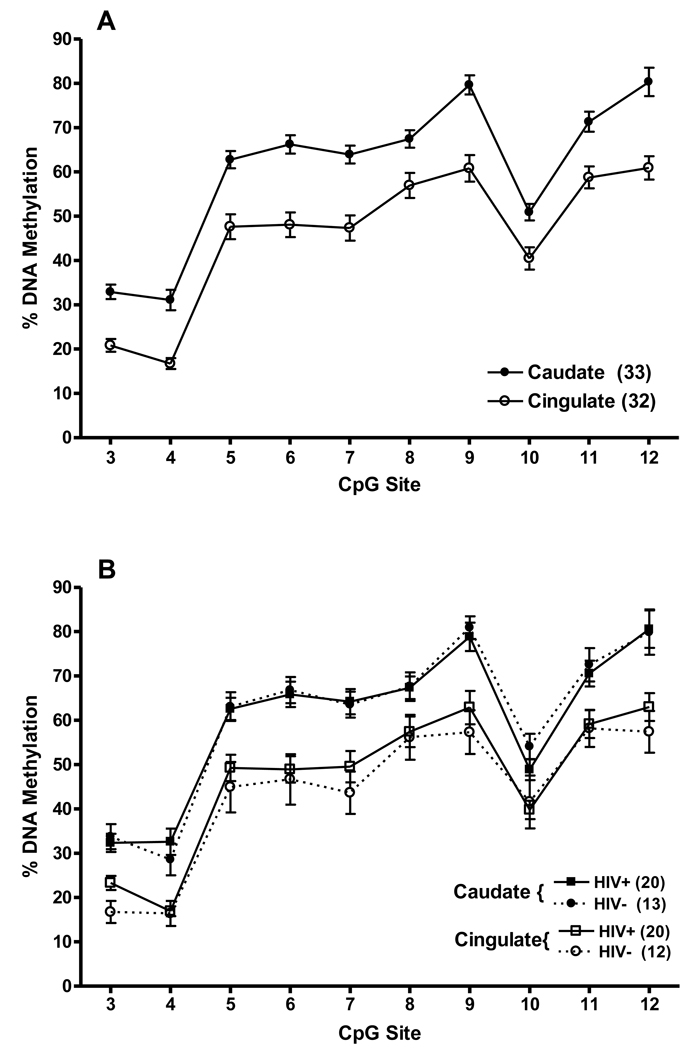
DNA methylation analysis of the CpG island was performed by PCR and direct sequencing of bisulfite-treated DNA obtained from the caudate (n=33), and from the anterior cingulate cortex (n=32), from the post-mortem brains, and PBMCs from 20 of these subjects. Analysis of sequence trace files showed that the ESME software provided reliable percent cytosine methylation calls for 10 CpG sites (# 3–12) in the CpG island. DNA methylation pattern of the CpG island in all samples of two brain regions is shown in Fig.2A. Analysis of variance (ANOVA) showed that there was a significant difference in levels of methylation of DNAs extracted from the caudate and anterior cingulate cortex, with higher levels across the 10 CpG sites in the caudate, F(1, 27) = 32.58, p<0.00001. There were also significant differences among CpG sites, main effect F(9, 243) = 214.19, p<0.000001. However, the pattern of methylation in the two brain regions across the 10 CpG sites in the CpG island in the brain tissues was remarkably similar.
Figure 2.
(A) Percent methylation of 10 CpG dinucleotides (# 3–12) in the PDYN promoter CpG island in two brain regions, the caudate and anterior cingulate cortex. (B) When stratified by HIV status, there was no difference in percent methylation of the CpG sites between HIV positive and HIV negative subjects. Methylation levels at each CpG site in each tissue are shown as mean ± SEM.
Of the 33 individuals whose methylation levels are shown in Fig. 2A, 20 were HIV positive. When the methylation data of the whole sample were examined by ANOVA and stratified by HIV status, there was no difference between HIV positive and HIV negative subjects, F<1.0 (Fig. 2B). Also, no correlation was found between methylation levels of CpG sites in the CpG island and age or PMI.
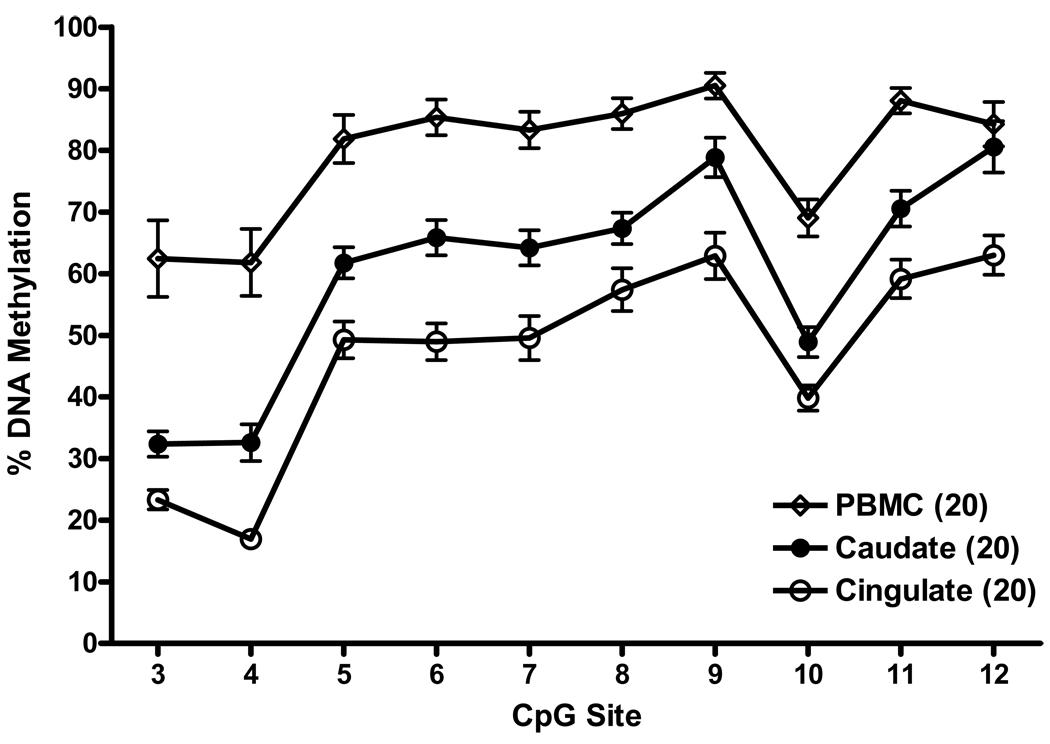
We next asked whether there is a difference in methylation of the PDYN CpG island between PBMCs available for the 20 HIV positive subjects and their two brain regions. The pattern of methylation across the 10 CpG sites in the CpG island in brain tissues and PBMCs was similar (Fig. 3), but the levels of DNA methylation varied significantly among PBMCs and the two brain regions, F(2,34) = 32.01, p<0.000001. DNA methylation levels in PBMCs were significantly greater than in caudate (p<0.0002), which in turn were significantly greater than in anterior cingulate cortex (p<0.02). Also, there was a significant variability of methylation level among CpG sites, F(9,153) = 134.33, p<0.000001, which can clearly be seen in brain regions, from relatively low (CpG sites #3 and #4), to intermediate (#5–8), and high (#9 and #12).
Figure 3.
The pattern of DNA methylation across the 10 CpG sites in the PDYN promoter CpG island in the caudate, anterior cingulate cortex and PBMCs of 20 subjects with data for all three tissues. Methylation levels at each CpG site in each tissue are shown as mean ± SEM.
The levels of methylation for each of 20 subjects in each CpG site in the CpG island in each tissue, caudate, anterior cingulate cortex, and PBMCs are shown in a hierarchical clustering dendrogram (Supplement Fig. S1 http://links.lww.com/FPC/A202). The PBMCs are most clearly heavily methylated and are separate from the two brain regions. Between the two brain regions, the distinction between clustering patterns is not so clear.
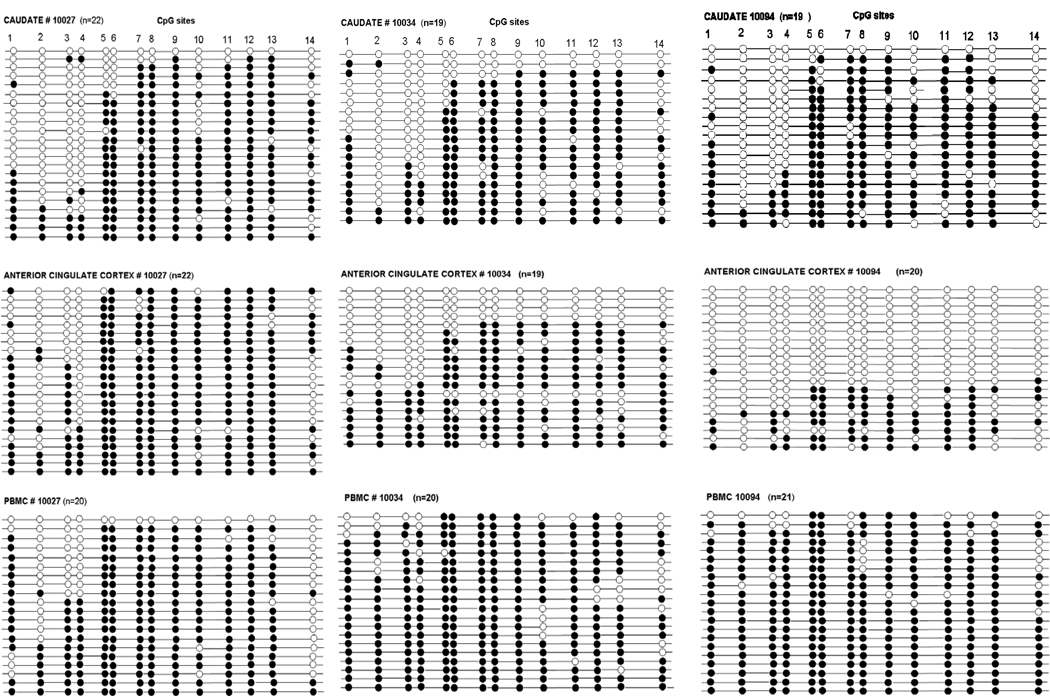
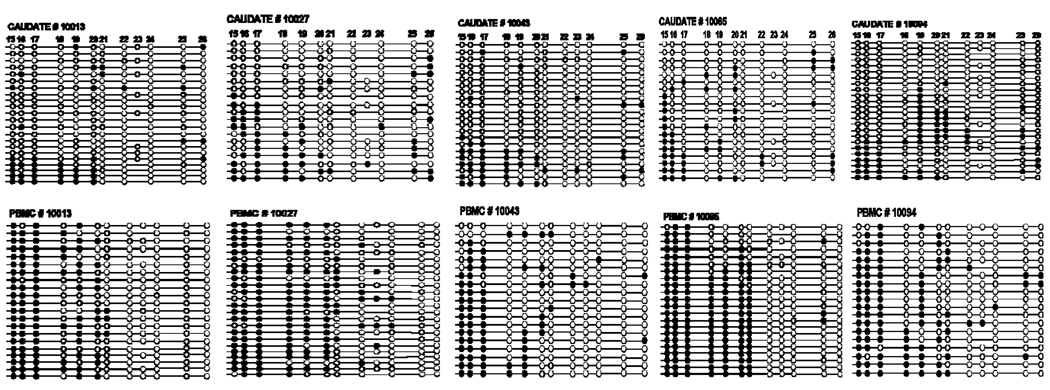
To validate the ESME data, we cloned the PCR amplified bisulfite-treated DNA and sequenced individual clones of the CpG island in the caudate putamen, anterior cingulate cortex, and PBMCs in three subjects (mhbb #10027, 10034, and 10094). The subjects were selected on the basis of differences in methylation extent of CpG sites in ESME data in these subjects, particularly in the anterior cingulate cortex (mean 68%, 51%, and 29%, respectively). Also, while analysis of DNA methylation extent using ESME software provided averaged quantitative values for each CpG site in PCR DNA in each tissue sample, we were interested in the methylation pattern of a single DNA molecule. CpG methylation patterns of 14 CpG sites in 19–22 clones from each region are shown in Fig. 4. With this technique we were able to obtain a methylation pattern or a “methylation haplotype” for all 14 CpG sites in individual DNA strands. Analysis of the CpG site methylation in clones showed (1) low methylation extent of CpG sites # 3 and 4 in brain tissues (consistent with the ESME data), and (2) high variability among clones. The average methylation level was lower in brain tissues, 70% in the caudate and 55% in the anterior cingulate cortex, compared to that in PBMCs (90%). There was a high correlation of the percentage of methylation of CpG sites between ESME and the clonal data for these three subjects (r = 0.826, p<0.0001).
Figure 4.
Methylation status of CpG sites using cloning and sequencing of PCR amplified bisulfite-treated DNA of the CpG island in the caudate, the anterior cingulate cortex and PBMCs from three subjects. Solid circles are methylated CpG sites, open circles are unmethylated CpG sites. Each line represents the result of an independent clone.
DNA methylation analysis of CpG cluster A
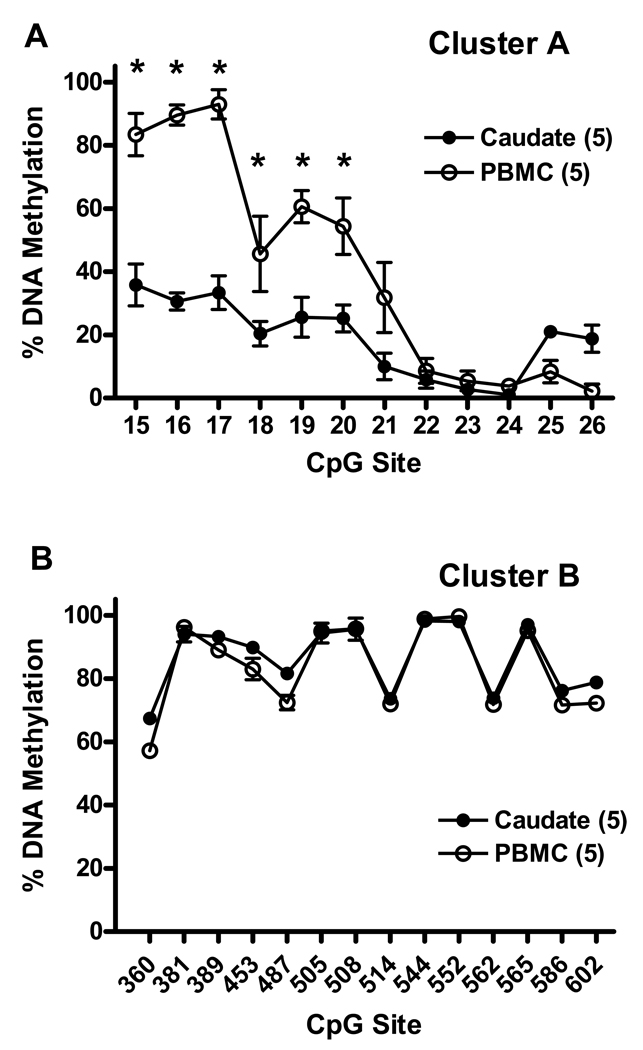
Downstream from the CpG island, there is another CpG-rich region, designated as cluster A, located at nucleotides from −410 to −37 relative to TSS of PDYN (Fig. 1). This region contains 12 CpG sites and is of special interest due to the presence of binding sites for the putative transcription factor AP-1, as well as an immediate proximity to the TATA box. Two polymorphisms in this region, dinucleotide repeats (CT)7–9 (rs34535593) and a single nucleotide polymorphism (SNP) at nucleotide −156 (A>G, rs1997794) in the AP-1 binding site (Fig. 1B), the rs1997794 may alter binding properties of the AP-1 transcription factor. The G-allele of rs1997794 eliminates a putative binding site TGTGTCA for the putative AP-1 transcription factor, and creates a CpG site # 23 (Fig. 1B). The A allele eliminates the CpG site, so that the “C” nucleotide in CpA dinucleotide cannot be methylated. Due to technical problems with direct sequencing of PCR bisulfite-treated DNA (see Methods), the extent of DNA methylation in the caudate and PBMCs in CpG cluster A was analyzed by cloning of PCR amplified bisulfite-treated DNA fragments and subsequent sequencing. We have examined the methylation status of 12 CpG sites in DNA from the caudate and PBMCs of five subjects (mhbb #10013, 10027, 10043, 10065 and 10094) (Fig. 5). The selection of these subjects was based on expression levels of PDYN in the caudate, and the genotype of rs1997794, reported earlier [32]. Among these subjects, four were heterozygous (A/G) and one, #10043, was homozygous (GG) for SNP rs1997794. Of note, CpG site # 23 in heterozygous (A/G) or homozygous (G/G) subjects for SNP rs1997794 is not methylated in most of the clones from both tissues, the caudate and PBMCs. Also, subjects homozygous for the A allele (A/A) do not have this CpG site # 23. The pattern of percent methylation across CpG sites in clones from these five subjects is shown in Fig. 6A. There was a significantly higher methylation level in DNA from PBMCs across CpG sites, F(1,4) = 84.01, p<0.001, a significant difference among individual CpG sites, F(11,44) = 45.58, p<0.000001, and a significantly different pattern of methylation between tissues across sites, interaction F(11, 44) = 13.90, p<0.000001. Across the 12 CpG sites in cluster A there was significantly higher methylation in PBMCs at CpG sites #15–20 (57–82%) than in the caudate (24–38%), with progressively declining methylation at CpG sites #21–24 (0–9%) in both tissues (Fig. 6A).
Figure 5.
Methylation analysis of cluster A in the caudate and matched PBMCs from five subjects (# 10013, 10027, 10043, 10065, and 10094) using cloning and sequencing of PCR amplified bisulfite-treated DNA. Each line represents the result of an independent clone. Solid circles are methylated CpG sites. Open circles are unmethylated CpG sites. Of note, the A allele of the polymorphic CpG site # 23 (rs1997794) eliminates the CpG site # 23.
Figure 6.
(A) DNA methylation pattern of cluster A in the caudate and matched PBMCs shown as mean ± SEM of the percent methylation at each CpG site in individual clones shown in Fig 5 (# 10013, 10027, 10043, 10065, and 10094). (B) DNA methylation pattern of cluster B (in exon 4) in the caudate and matched PBMCs of 5 individuals (# 10001, 10002, 10016, 10045, and 10094).
Interestingly, the clonal data of cluster A show that there is unequal distribution of clones with the A and G alleles, in both the caudate and PBMCs in subjects heterozygous for the SNP rs1997794 in CpG site # 23 (Fig. 5). There is a higher frequency of the G allele in PBMCs (54–86%) compared to the caudate (26–46%). The frequency of the G allele in subject #10013 was 36% in the caudate and 81% in PBMCs; in subject #10027 it was 37% and 54%; in subject #10065 it was 26% and 86%; and in subject # 10094 it was 46% and 79%, respectively. However, due to rare methylation of the CpG site # 23 in both tissues, this difference in the allele distribution does not change the methylation profile of cluster A.
DNA methylation analysis of CpG cluster B
The other CG-rich region, cluster B, is located in exon 4 which encodes preprodynorphin peptides, including α-neoendorphin, β-neoendorphin, dynorphin A, and dynorphin B. Importantly, in band gel-shift experiments it was shown that the conservative sequence coding for [Leu5]enkephalin at the N-terminal of the dynorphin peptides binds the nuclear DNA binding factor (LEF) [39]. One of LEV/YY1 binding sites contains a CpG site at position 552 relative to the first nucleotide of exon 4 (Fig. 1B), a potential site for methylation. In in vitro experiments, LEF inhibited the transcription of reporter genes in the human choriocarcinoma cells, JEG-3. Later, the same group showed that LEF is identical to the multifunctional transcription factor YY1 [40]. It has been shown that the truncated PDYN transcripts, T1 and T2, were initiated within the coding part of exon 4 [18, 19]. Also, several kB elements are present in exon 4, suggesting that NF-kB may be implicated in regulation of transcription of the T1 and T2 PDYN mRNA [41]. Therefore, it was important to elucidate a methylation profile of this region.
The mean methylation status by ESME analysis of 14 CpG sites in cluster B in DNA from the caudate and PBMCs of five subjects (## 10001, 10002, 10016, 10045, and 10094) is shown in Fig. 6B. CpG cluster B is hypermethylated in both the caudate and PBMCs with a remarkably similar pattern. ANOVA showed a significant but negligible difference between tissues (from 86.7% in the caudate to 83.6% in PBMCs), F(1,3) = 13.90, p<0.05. There was a significant difference in methylation among CpG sites, F(13, 39) = 108.71, p<0.000001. For example, methylation at CpG site # 360 was significantly lower than at each of the other sites, p<0.0002 to p<0.05, Newman-Keuls post hoc test.
Quantification of PDYN mRNA levels in brain tissues
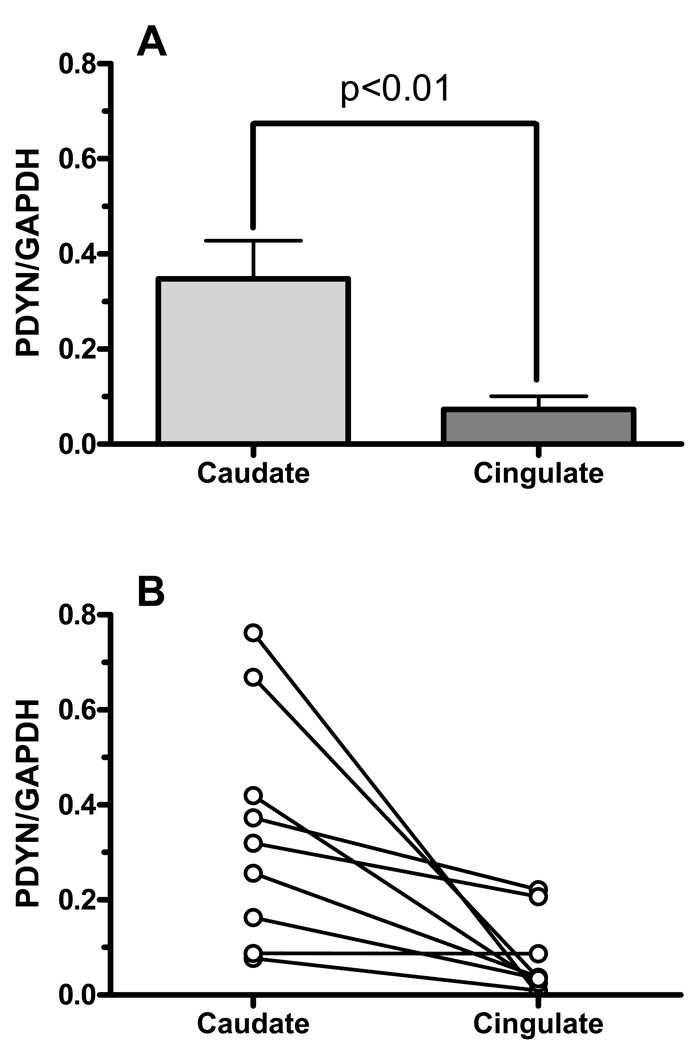
To examine the relationship between the extent of methylation of the CpG island in the caudate and anterior cingulate cortex with PDYN expression, we measured PDYN mRNA levels in those regions from nine subjects (mhbb # 10002, 10016, 10027, 10034, 10052, 10065, 10074, 10094, and 531) using qRT-PCR SYBR assay. These subjects were chosen on the basis of different levels of the PDYN mRNA expression in the caudate, measured quantitatively by the solution hybridization RNase protection assay, reported earlier [29]. Contrary to expectations that a higher level of methylation of the CpG island would lead to lower gene expression, PDYN mRNA levels were significantly higher in the caudate compared to the anterior cingulate cortex, t = 3.10, p<0.02, two-tailed, as can be seen in Fig. 7A. The relationship of PDYN expression in the two brain regions is shown for each individual in Fig. 7B.
Figure 7.
PDYN mRNA levels in the caudate and anterior cingulate in post-mortem brain from nine subjects using qRT-PCR SYBR Green assay. Expression levels were normalized to GAPDH.
DISCUSSION
There is strong evidence of a direct interaction between DNA methylation and specifically modified histones, which plays an important role in modulation of gene expression (e.g. [42]). The methylation of CpG sites attracts methyl-binding proteins, notably the methyl-CpG binding protein MeCP2, which are associated with decreased gene expression and chromatin compaction or may disrupt the binding of transcription factors [43]. Therefore, studies of DNA methylation patterns in genes may lead to the localization and characterization of potential transcription regulatory elements in gene promoters and enhancers. In this study, we found remarkably similar patterns of methylation across CpG sites in the PDYN proximal promoter in brain tissues and PBMCs with high methylation of the CpG island and progressively decreased methylation in a CpG-rich area (cluster A) in immediate proximity to the TATA-like box and the transcription start site. However, the absolute levels of methylation of each CpG site in the CpG island in most subjects differed significantly between brain tissues and PBMCs, and between the caudate and anterior cingulate cortex. The hypermethylation of the CpG island in PBMCs is consistent with low PDYN expression in macrophages, and undetectable expression in lymphocyte cell cultures [21], compared to higher expression of the gene in brain tissues [7]. The methylation extent of the CpG island in the two brain regions varied among subjects. In most cases, there was higher DNA methylation in the caudate than in the anterior cingulate cortex. However, PDYN mRNA levels were significantly higher in the caudate than in the anterior cingulate cortex. Thus, the methylation rate of the CpG island in brain regions was not associated directly with PDYN expression levels. The promoter region of the human PDYN containing the CpG island was shown to be implicated in up-regulation [44] or down-regulation [45] of basal and forskolin-induced PDYN promoter activity in vitro depending on cell culture type. Therefore, the PDYN CpG island may be implicated in a tissue-specific regulation of the gene expression.
Our finding of the similarity of CpG methylation patterns in brain tissues and PBMCs is consistent with results of another study which showed a high correlation of the CpG island methylation profiles among 11 somatic tissues, with no consistent correlation between the CpG island methylation extent and corresponding gene expression [46]. It is possible that CpG methylation status only provides some precondition for the transcription regulatory process, and some other epigenetic factors, such as CpG-binding proteins and histone modifications, play more direct and important roles in tissue-specific gene regulation. For example, CpG site #10 in the PDYN CpG island is located in a putative binding site CACGTG for cMyc or USF1 transcription factors, and shows relatively low methylation in both PBMCs and brain tissues. The cMyc factor may activate or repress transcription at the level of chromatin remodeling in a tissue-specific manner, through its association with histone acetyltransferases or DNA CpG methyltransferases, respectively (e.g. [47]).
We did not find a difference in methylation of the PDYN promoter between HIV-positive and HIV-negative subjects. This result is consistent with a view that only specific genes are susceptible for de novo methylation in HIV-infected cells [48]. It is likely, therefore, that PDYN is not directly involved in HIV pathogenesis.
PDYN mRNA levels in the rat caudate putamen increase rapidly following acute and chronic ”binge” cocaine administration [4, 5, 6, 8]. Therefore, PDYN could be considered as a primary drug- and stress-responsive gene. As shown for other primary responsive genes in lymphocytes and macrophages [49], an unmethylated or hypomethylated state of a comparable region such as the CpG–rich region (cluster A) near the transcription start site in the caudate and PBMCs may be associated with low nucleosome density, active state of chromatin, and greater accessibility of transcription factors. Most CpG-island promoters of the primary responsive genes in un-stimulated macrophages also exhibited higher levels of RNA polymerase II, TATA-binding protein (TBP), and the pre-association of transcription factors [50]. Another feature of primary responsive genes is the regulation of their expression by transcriptional repressors, to counter the active chromatin that is associated with these genes in the basal state [50]. By analogy, basal expression of the human PDYN was reported to be regulated by the calcium-mediated transcriptional repressor, the downstream regulatory element antagonist modulator (DREAM), that binds to the downstream regulatory element (DRE) located within exon 1 [33]. In rodents, an acute cocaine administration was shown to increase the intracellular calcium concentration in brain [51]. Cocaine-induced increased calcium levels have been shown to release DREAM from the DRE site, resulting in the derepression of PDYN transcription.
Cluster B, located in exon 4 of PDYN, is hypermethylated in both the caudate and PBMCs. The sequences coding for [Leu5]enkephalin in dynorphin peptides have been shown to interact with the transcriptional repressor factor LEF/YY1 [39, 40], and in one study, methylation in vitro of DNA coding for the [Leu5]enkephalin interfered with the transcription factor binding. However, we did not find a difference in methylation extent of cluster B between low PDYN-expressing PBMCs and the higher PDYN-expressing caudate.
Although the CpG island and cluster A methylation patterns obtained by both ESME and clonal analysis were remarkably similar among subjects and tissues, the clonal data showed a high diversity in methylation of CpG sites in individual DNA clones. A stochastic model has been suggested for the phenomenon of molecule-to-molecule variation but with stable average methylation of specific sites and stable patterns over larger regions, based on the equilibrium of methylation maintenance and de novo methylation [52]. However, a mosaic-like distribution of the methylated CpG sites or “methylation haplotypes” may also be related to the complexity of the cell type population in tissue samples (e.g. [53]). Indeed, brain tissues contain many types of neuronal and glial cells, and PBMCs are comprised of different subtypes of cells. It has been hypothesized that DNA methylation serves as a molecular mechanism to lock in developmental changes and to determine tissue- and cell-specific functions and gene expression [42].
Our observation of an unequal distribution of alleles of the SNP rs1997794 between the brain tissue and PBMCs in four heterozygous subjects may be important. Currently it is not clear whether this difference is related to technical artifacts like a preferential PCR amplification or cloning [54], or is a real biological phenomenon. If it is a cell-specific phenomenon, then it may lead to differential gene expression. The unequal distribution of alleles observed in this study should be interpreted cautiously, and requires validation by other methods [55].
Unlike animal studies in which investigators may control for a variety of practical aspects of experimental conditions, studies of human post-mortem brain present certain difficulties. Among the factors that may contribute to variability in gene expression and epigenetic profiles are the quality of RNA and DNA, age, post-mortem interval, differences in genetic background, and clinical history of the subjects [56,57]. Furthermore, neuron-to-glia ratios show considerable variation between normal and various pathological conditions, which may lead to differences in methylation profiles in studies of whole tissues [58]. However, other studies have shown that the binding of genomic DNA to the nucleosome core is largely preserved in post-mortem tissues within a wide range of tissue pH and post-mortem intervals [e.g. 59], and in one study no correlation was found between DNA methylation state and pH of cortical post-mortem brain tissues [60].
In conclusion, we have identified DNA methylation profiles in the human PDYN gene which suggest that the CpG island is implicated in tissue- or cell-specific regulation of gene expression, whereas the CpG cluster A may be related to regulation of basal activity of the PDYN promoter. Our findings provide a rationale for further studies of methylated DNA binding proteins, specifically modified histones and associated proteins which are involved in regulation of basal and stimuli-induced PDYN expression in subjects with psychiatric and neurological disorders.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr. Ruijin Shi (Mount Sinai Medical Center, New York) for his help in dissection of brain tissues. The authors would like to acknowledge Susan Russo for her editorial contribution.
Support: NIH-NIDA P60-DA05130 (MJK), NIH-NIMH R01-MH79880 (MJK), NIH-NIMH R24-MH59724 (SM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSE/CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information is available at the website.
REFERENCES
- 1.Bruijnzeel AW. kappa-Opioid receptor signaling and brain reward function. Brain Res Rev. 2009;62:127–146. doi: 10.1016/j.brainresrev.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hauser KF, Aldrich JV, Anderson KJ, Bakalkin G, Christie MJ, Hall ED, et al. Pathobiology of dynorphins in trauma and disease. Front Biosci. 2005;10:216–235. doi: 10.2741/1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kreek MJ, LaForge KS, Butelman E. Pharmacotherapy of addictions. Nat Rev Drug Discov. 2002;1:710–726. doi: 10.1038/nrd897. [DOI] [PubMed] [Google Scholar]
- 4.Daunais JB, McGinty JF. Acute and chronic cocaine administration differentially alters striatal opioid and nuclear transcription factor mRNAs. Synapse. 1994;18:35–45. doi: 10.1002/syn.890180106. [DOI] [PubMed] [Google Scholar]
- 5.Hurd YL, Brown EE, Finlay JM, Fibiger HC, Gerfen CR. Cocaine self-administration differentially alters mRNA expression of striatal peptides. Brain Res Mol Brain Res. 1992;13:165–170. doi: 10.1016/0169-328x(92)90058-j. [DOI] [PubMed] [Google Scholar]
- 6.Spangler R, Unterwald EM, Kreek MJ. 'Binge' cocaine administration induces a sustained increase of prodynorphin mRNA in rat caudate-putamen. Brain Res Mol Brain Res. 1993;19:323–327. doi: 10.1016/0169-328x(93)90133-a. [DOI] [PubMed] [Google Scholar]
- 7.Yuferov V, Zhou Y, LaForge KS, Spangler R, Ho A, Kreek MJ. Elevation of guinea pig brain preprodynorphin mRNA expression and hypothalamic-pituitary-adrenal axis activity by "binge" pattern cocaine administration. Brain Res Bull. 2001;55:65–70. doi: 10.1016/s0361-9230(01)00496-8. [DOI] [PubMed] [Google Scholar]
- 8.Spangler R, Zhou Y, Maggos CE, Schlussman SD, Ho A, Kreek MJ. Prodynorphin, proenkephalin and kappa opioid receptor mRNA responses to acute "binge" cocaine. Brain Res Mol Brain Res. 1997;44:139–142. doi: 10.1016/s0169-328x(96)00249-5. [DOI] [PubMed] [Google Scholar]
- 9.Frankel PS, Alburges ME, Bush L, Hanson GR, Kish SJ. Striatal and ventral pallidum dynorphin concentrations are markedly increased in human chronic cocaine users. Neuropharmacology. 2008;55:41–46. doi: 10.1016/j.neuropharm.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurd YL, Herkenham M. Molecular alterations in the neostriatum of human cocaine addicts. Synapse. 1993;13:357–369. doi: 10.1002/syn.890130408. [DOI] [PubMed] [Google Scholar]
- 11.Shippenberg TS, Zapata A, Chefer VI. Dynorphin and the pathophysiology of drug addiction. Pharmacol Ther. 2007;116:306–321. doi: 10.1016/j.pharmthera.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Butelman ER, Schlussman SD, Ho A, Kreek MJ. Effect of the endogenous kappa opioid agonist dynorphin A(1–17) on cocaine-evoked increases in striatal dopamine levels and cocaine-induced place preference in C57BL/6J mice. Psychopharmacology (Berl) 2004;172:422–429. doi: 10.1007/s00213-003-1688-3. [DOI] [PubMed] [Google Scholar]
- 13.Di Chiara G, Imperato A. Opposite effects of mu and kappa opiate agonists on dopamine release in the nucleus accumbens and in the dorsal caudate of freely moving rats. J Pharmacol Exp Ther. 1988;244:1067–1080. [PubMed] [Google Scholar]
- 14.Knoll AT, Carlezon WA., Jr Dynorphin, stress, and depression. Brain Res. 2010;1314:56–73. doi: 10.1016/j.brainres.2009.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLaughlin JP, Marton-Popovici M, Chavkin C. Kappa opioid receptor antagonism and prodynorphin gene disruption block stress-induced behavioral responses. J Neurosci. 2003;23:5674–5683. doi: 10.1523/JNEUROSCI.23-13-05674.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shirayama Y, Ishida H, Iwata M, Hazama GI, Kawahara R, Duman RS. Stress increases dynorphin immunoreactivity in limbic brain regions and dynorphin antagonism produces antidepressant-like effects. J Neurochem. 2004;90:1258–1268. doi: 10.1111/j.1471-4159.2004.02589.x. [DOI] [PubMed] [Google Scholar]
- 17.Horikawa S, Takai T, Toyosato M, Takahashi H, Noda M, Kakidani H, et al. Isolation and structural organization of the human preproenkephalin B gene. Nature. 1983;306:611–614. doi: 10.1038/306611a0. [DOI] [PubMed] [Google Scholar]
- 18.Nikoshkov A, Hurd YL, Yakovleva T, Bazov I, Marinova Z, Cebers G, et al. Prodynorphin transcripts and proteins differentially expressed and regulated in the adult human brain. FASEB J. 2005;19:1543–1545. doi: 10.1096/fj.05-3743fje. [DOI] [PubMed] [Google Scholar]
- 19.Telkov M, Geijer T, Terenius L. Human prodynorphin gene generates several tissue-specific transcripts. Brain Res. 1998;804:284–295. doi: 10.1016/s0006-8993(98)00706-9. [DOI] [PubMed] [Google Scholar]
- 20.Ji RR, Befort K, Brenner GJ, Woolf CJ. ERK MAP kinase activation in superficial spinal cord neurons induces prodynorphin and NK-1 upregulation and contributes to persistent inflammatory pain hypersensitivity. J Neurosci. 2002;22:478–485. doi: 10.1523/JNEUROSCI.22-02-00478.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun B, Tipton CM, Bidlack JM. The expression of prodynorphin gene is down-regulated by activation with lipopolysaccharide in U-937 macrophage cells. J Neuroimmunol. 2006;174:52–62. doi: 10.1016/j.jneuroim.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Mattick JS, Taft RJ, Faulkner GJ. A global view of genomic information--moving beyond the gene and the master regulator. Trends Genet. 2010;26:21–28. doi: 10.1016/j.tig.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 23.Grunau C, Hindermann W, Rosenthal A. Large-scale methylation analysis of human genomic DNA reveals tissue-specific differences between the methylation profiles of genes and pseudogenes. Hum Mol Genet. 2000;9:2651–2663. doi: 10.1093/hmg/9.18.2651. [DOI] [PubMed] [Google Scholar]
- 24.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ladd-Acosta C, Pevsner J, Sabunciyan S, Yolken RH, Webster MJ, Dinkins T, et al. DNA methylation signatures within the human brain. Am J Hum Genet. 2007;81:1304–1315. doi: 10.1086/524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jähner D, Jaenisch R. Retrovirus-induced de novo methylation of flanking host sequences correlates with gene inactivity. Nature. 1985;315:594–597. doi: 10.1038/315594a0. [DOI] [PubMed] [Google Scholar]
- 27.Youngblood B, Reich NO. The early expressed HIV-1 genes regulate DNMT1 expression. Epigenetics. 2008;3:149–156. doi: 10.4161/epi.3.3.6372. [DOI] [PubMed] [Google Scholar]
- 28.Kumar AM, Borodowsky I, Fernandez B, Gonzalez L, Kumar M. Human immunodeficiency virus type 1 RNA Levels in different regions of human brain: quantification using real-time reverse transcriptase-polymerase chain reaction. J Neurovirol. 2007;13:210–224. doi: 10.1080/13550280701327038. [DOI] [PubMed] [Google Scholar]
- 29.Kumar AM, Fernandez J, Singer EJ, Commins D, Waldrop-Valverde D, Ownby RL, Kumar M. Human immunodeficiency virus type 1 in the central nervous system leads to decreased dopamine in different regions of post-mortem human brains. J Neurovirol. 2009;15:257–274. doi: 10.1080/13550280902973952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimprich A, Kraus J, Woltje M, Mayer P, Rauch E, Hollt V. An allelic variation in the human prodynorphin gene promoter alters stimulus-induced expression. J Neurochem. 2000;74:472–477. doi: 10.1046/j.1471-4159.2000.740472.x. [DOI] [PubMed] [Google Scholar]
- 31.Babbitt CC, Silverman JS, Haygood R, Reininga JM, Rockman MV, Wray GA. Multiple functional variants in cis modulate PDYN expression. Mol Biol Evol. 2010;27:465–479. doi: 10.1093/molbev/msp276. [DOI] [PubMed] [Google Scholar]
- 32.Yuferov V, Ji F, Nielsen DA, Levran O, Ho A, Morgello S, et al. A functional haplotype implicated in vulnerability to develop cocaine dependence is associated with reduced PDYN expression in human brain. Neuropsychopharmacology. 2009;34:1185–1197. doi: 10.1038/npp.2008.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carrion AM, Link WA, Ledo F, Mellstrom B, Naranjo JR. DREAM is a Ca2+regulated transcriptional repressor. Nature. 1999;398:80–84. doi: 10.1038/18044. [DOI] [PubMed] [Google Scholar]
- 34.Lewin J, Schmitt AO, Adorjan P, Hildmann T, Piepenbrock C. Quantitative DNA methylation analysis based on four-dye trace data from direct sequencing of PCR amplificates. Bioinformatics. 2004;20:3005–3012. doi: 10.1093/bioinformatics/bth346. [DOI] [PubMed] [Google Scholar]
- 35.Schug J, Overton G. TESS: Transcription Element Search Software on the WWW. Philadelphia: School of Medicine, University of Pennsylvania; 1977. [Google Scholar]
- 36.Rose'Meyer RB, Mellick AS, Garnham BG, Harrison GJ, Massa HM, Griffiths LR. The measurement of adenosine and estrogen receptor expression in rat brains following ovariectomy using quantitative PCR analysis. Brain Res Brain Res Protoc. 2003;11:9–18. doi: 10.1016/s1385-299x(02)00219-2. [DOI] [PubMed] [Google Scholar]
- 37.Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 38.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 39.Bakalkin G, Telkov M, Yakovleva T, Terenius L. [Leu5]enkephalin-encoding sequences are targets for a specific DNA-binding factor. Proc Natl Acad Sci U S A. 1995;92:9024–9028. doi: 10.1073/pnas.92.20.9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakalkin G, Yakovleva T, Terenius L. The Leu-enkephalin-encoding sequence DNA-binding factor (LEF) is the transcription factor YY1. Biochem Biophys Res Commun. 1997;231:135–139. doi: 10.1006/bbrc.1997.6062. [DOI] [PubMed] [Google Scholar]
- 41.Bakalkin G, Yakovleva T, Terenius L. Prodynorphin gene expression relates to NF-kappa B factors. Brain Res Mol Brain Res. 1994;24:301–312. doi: 10.1016/0169-328x(94)90143-0. [DOI] [PubMed] [Google Scholar]
- 42.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 43.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;(33 Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 44.Carrion AM, Mellstrom B, Naranjo JR. Protein kinase A-dependent derepression of the human prodynorphin gene via differential binding to an intragenic silencer element. Mol Cell Biol. 1998;18:6921–6929. doi: 10.1128/mcb.18.12.6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rouault M, Nielsen DA, Ho A, Kreek MJ, Yuferov V. Cell-specific effects of variants of the 68-base pair tandem repeat on prodynorphin gene promoter activity. Addict Biol. doi: 10.1111/j.1369-1600.2010.00248.x. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan S, Zhang X. CpG island methylation pattern in different human tissues and its correlation with gene expression. Biochem Biophys Res Commun. 2009;383:421–425. doi: 10.1016/j.bbrc.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 47.Brenner C, Deplus R, Didelot C, Loriot A, Vire E, De Smet C, et al. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 2005;24:336–346. doi: 10.1038/sj.emboj.7600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feltus FA, Lee EK, Costello JF, Plass C, Vertino PM. Predicting aberrant CpG island methylation. Proc Natl Acad Sci USA. 2003;100:12253–12258. doi: 10.1073/pnas.2037852100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 51.Du C, Yu M, Volkow ND, Koretsky AP, Fowler JS, Benveniste H. Cocaine increases the intracellular calcium concentration in brain independently of its cerebrovascular effects. J Neurosci. 2006;26:11522–11531. doi: 10.1523/JNEUROSCI.3612-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riggs AD, Xiong Z. Methylation and epigenetic fidelity. Proc Natl Acad Sci U S A. 2004;101:4–5. doi: 10.1073/pnas.0307781100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shiota K. DNA methylation profiles of CpG islands for cellular differentiation and development in mammals. Cytogenet Genome Res. 2004;105:325–334. doi: 10.1159/000078205. [DOI] [PubMed] [Google Scholar]
- 54.Warnecke PM, Stirzaker C, Melki JR, Millar DS, Paul CL, Clark SJ. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 1997;25:4422–4426. doi: 10.1093/nar/25.21.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaminsky ZA, Popendikyte V, Assadzadeh A, Petronis A. Search for somatic DNA variation in the brain: investigation of the serotonin 2A receptor gene. Mamm Genome. 2005;16:587–593. doi: 10.1007/s00335-005-0040-0. [DOI] [PubMed] [Google Scholar]
- 56.Akbarian S, Huang HS. Epigenetic regulation in human brain-focus on histone lysine methylation. Biol Psychiatry. 2009;65:198–203. doi: 10.1016/j.biopsych.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bunney WE, Bunney BG, Vawter MP, Tomita H, Li J, Evans SJ, et al. Microarray technology: a review of new strategies to discover candidate vulnerability genes in psychiatric disorders. Am J Psychiatry. 2003;160:657–666. doi: 10.1176/appi.ajp.160.4.657. [DOI] [PubMed] [Google Scholar]
- 58.Connor CM, Akbarian S. DNA methylation changes in schizophrenia and bipolar disorder. Epigenetics. 2008;3:55–58. doi: 10.4161/epi.3.2.5938. [DOI] [PubMed] [Google Scholar]
- 59.Huang HS, Matevossian A, Jiang Y, Akbarian S. Chromatin immunoprecipitation in post-mortem brain. J Neurosci Methods. 2006;156:284–292. doi: 10.1016/j.jneumeth.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 60.Ernst C, McGowan PO, Deleva V, Meaney MJ, Szyf M, Turecki G. The effects of pH on DNA methylation state: In vitro and post-mortem brain studies. J Neurosci Methods. 2008;174:123–125. doi: 10.1016/j.jneumeth.2008.06.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.