Abstract
It has become increasingly clear that both soluble factors, such as growth factors, and insoluble factors, including the surfaces on which cells grow, can have controlling effects on stem cell behavior and differentiation. While much progress has been made in biomaterial design and application, the rational design of biomaterial cues to direct stem cell behavior and differentiation remains challenging. Recent advances in automated, high-throughput methods for synthesizing and screening combinatorial biomaterial libraries and cellular microenvironments promise to accelerate the discovery of factors that control stem cell behavior. Specific examples include miniaturized, automated, combinatorial material synthesis and extracellular matrix screening methods as well microarrayed methods for creating local microenvironments of soluble factors, such as small molecules, siRNA, and other signaling molecules.
Introduction
Stem cells hold enormous potential for application in human regenerative medicine and tissue engineering [1,2]. There are many types of stem and progenitor cell types that can be derived from human adult and embryonic tissue [3]. Human embryonic stem cells (hESCs) have the unique ability to undergo nearly infinite self-renewal in an undifferentiated state [4] and potential to differentiate into any cell type in the human body [5]; they therefore hold significant promise for cell and tissue replacement. Despite advances made in the molecular and cell biology of stem cells during the past decade, there are still challenges to realizing their application in regenerative therapies. In particular, methods to control stem cell growth and differentiation must be developed before stem cells are used broadly in the clinic.
The controlled expansion and differentiation of stem cells on bioactive degradable scaffolds is one promising tool that may enable scientists to repair, sustain, and improve tissues and organs in vivo [6,7]. The cellular microenvironment is a controlling factor in these systems, and it is composed of both physical and chemical signals, including extracellular matrix proteins, neighboring cells, soluble and immobilized growth factors, and small molecules [8]. Stem cell fate and function are determined by this complex collection of factors present in the local environment of the cell. A number of biomaterials have been employed to regulate a range of cell behaviors for tissue engineering applications, including adhesion, proliferation, and differentiation [9••,10–12].
One emerging trend in biomaterials research is the design of biofunctional materials that can modulate cellular responses via the incorporation of instructive signals into the biomaterials [13]. While much progress has been made in biomaterial design and application, the rational design of biomaterial cues to control stem cell behavior and differentiation remains challenging due to the complexity of cell–material interactions. Furthermore, cells typically discern and react to multiple cues rather than a single signal [14], rendering the rational reconstitution of these signals challenging. The development of high-throughput screening methods has enabled rapid and quantitative methods to study the interaction between cells and their micro-environment.
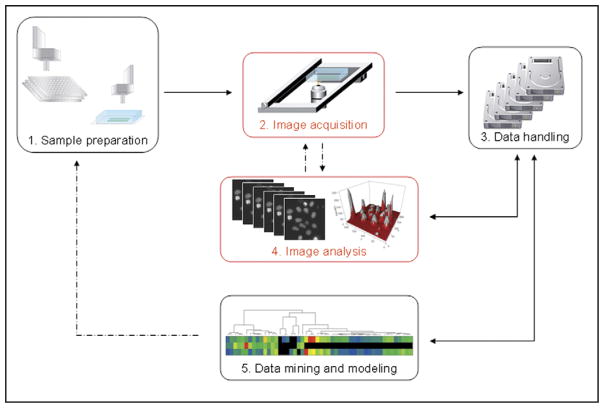
Over the past 20 years, high-throughput approaches have transformed drug discovery in the pharmaceutical industry [15]. These approaches are also being applied to material science [16], and, even more recently, to biomaterial development [17]. The development of high-throughput methods for studying the microenvironment requires, in general, three integrated, high-throughput processes: firstly, library synthesis, secondly, data acquisition, and thirdly, data processing and modeling (see Figure 1) [18,19]. Here, we discuss the emerging trend of high-throughput approaches for cellular microenvironments with an emphasis on biomaterials and applications in stem cell biology and engineering.
Figure 1.
A schematic diagram of high-throughput approaches in biomaterials research (The image is kindly provided by Dr. Ellenberg, and reproduced with permission from [18]).
High-throughput methods for studying the microenvironment
Combinatorial evaluation of synthetic biomaterials
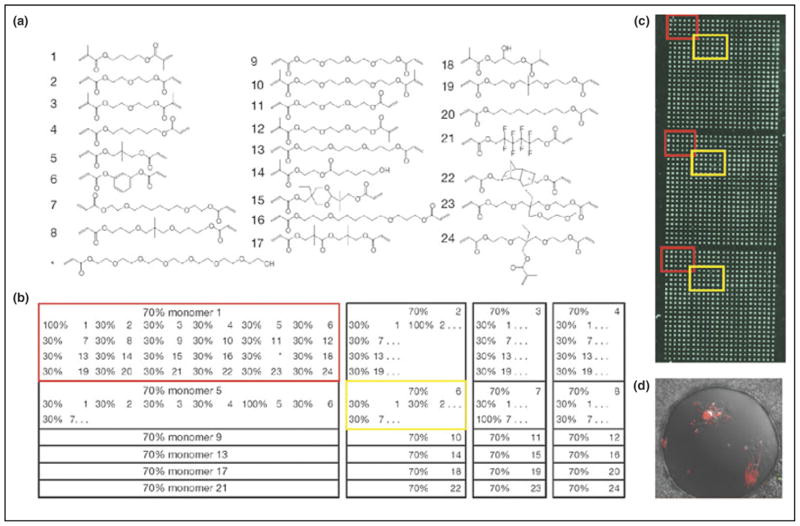
Most of the studies of biomaterial effects on stem cells have focused on the physical surfaces on which the cells are grown. One study investigating the effect of synthetic materials on stem cell differentiation was performed by Anderson et al. [20••] using a library of synthetic polymers. In this study, high-throughput, miniaturized methods for the rapid synthesis and screening of combinatorial libraries of photopolymerizable biomaterials, and the screening of cell–material interactions were developed (see Figure 2). More than 1700 materials were synthesized in nanoliter volumes in microarray format and screened for their affects on hESCs growth and differentiation.
Figure 2.
Biomaterial microarray design. (a) Monomers used for microarray synthesis. (b) Monomers were mixed at a 70:30 ratio pairwise in all possible combinations with the exception of monomer 17, which was substituted with * to increase polymer hydrophilicity. To facilitate analysis, all 24 polymers composed of 70% of a particular monomer were printed as a 6 × 4 group on the array, as highlighted by the red and yellow boxes. Three blocks of 576 polymers were printed on each slide, with a center to center spacing of 740 μm. (c) Printed polymer array imaged by Arrayworx reader. Blocks composed of 70% monomer 1 and 70% monomer 6 are highlighted in red and yellow, respectively. (d) Differential interference contrast light microscopy of typical polymer spot overlaid with a few fluorescent cells (red) (reproduced with permission from [20••]).
Using this method, a number of unexpected cell–polymer interactions were identified, including materials that support high levels of growth and differentiation into epithelial cells. This methodology can be applied to many different adherent cell types, even in the absence of cells, for example to study biofouling. More recently, arrays have been made with roughly 3500 combinations of biodegradable polymers and screened with human mesenchymal stem cells, neural stem cells, and primary articular chondrocytes [21]. Gradient assemblies, such as those described by Mei et al. [22] and Bhat et al. [23], can also be used to characterize biological response to blends of materials and yield information on a cell’s precise point of transition along a gradually changing surface.
Combinatorial extracellular matrix microarrays
Cells have evolved to interact with the materials in their intimate environment. Accordingly, naturally occurring extracellular matrix (ECM) molecules represent a logical source of signaling biomaterials. Initial work on the printing of protein microarrays was performed by Mac-Beath et al. [24], who developed some of the first high-throughput synthesis and screening methodologies for proteins microarrays. This technology was extended by Flaim et al. [25•], who developed a microarrayed system to screen for the effects of ECM molecules on stem cells in a combinatorial, parallel manner. Using a robotic fluid handling device, five natural ECM molecules namely collagen I, collagen III, collagen IV, laminin, and fibronectin were immobilized on a hydrogel surface tested with the two murine cell types. Interestingly, the authors discovered that the precise composition of ECM on which the cells grew affected both mouse embryonic stem cell differentiation and the maintenance of primary rat hepatocyte phenotype. Collagen IV was shown to increase the level of intracellular albumin, a marker of liver-specific function, while collagen III and laminin resulted in decreased albumin levels. The matrix array further revealed that the growth of mouse stem cells on combinations of collagen I and fibronectin led to differentiation into an early hepatic fate.
The technique has several practical advantages. Before this work, photolithographic cell micropatterning tools or custom-built equipment was needed, as conventional microarray spotting methods for cell culture employed process conditions that were incompatible with the ECM protein spotting, required extensive customization of the spotting equipment, or lacked pattern fidelity [25•]. Furthermore, the microarrayed system allowed the testing of materials to be performed using three orders of magnitude less material than required by conventional screens of cell–ECM interaction.
Combinatorial growth factors microarrays
While the work of Flaim et al. focused on natural ECM molecules, Nakajima et al. [26] immobilized both natural and synthetic matrices site-specifically in a combinatorial fashion using photo-assisted patterning. The substrates were loaded with growth factors known to have neurotrophic activities in order to expand undifferentiated neural stem cells or to induce differentiation of neural stem cells into specific lineages. The array-based method identified favorable composite materials. Rat neural stem cells adhered well to laminin-1, fibronectin, ProNectin™, and poly(ethyleneimine) and proliferated most significantly on spots containing epidermal growth factor. Analysis of the matrix-growth factor combinations will facilitate the design of artificial scaffolds for cell therapy-based repair of the central nervous system.
Neural stem cells have the potential to differentiate into several lineages, including neurons or glial cells, of which there are several subtypes. Thus, the incorporation of growth factors with unknown effects on stem cells could be utilized to control the underlying pathway of lineage-specific differentiation. A non-contact, piezoelectric arrayer was employed by Soen et al. [27] to print pre-mixed combinations of signaling molecules — growth factors, ECM components, cell adhesion molecules, and morphogens — onto arrays. Primary bipotent human neural precursor cells were subsequently cultured on the varied natural biomaterials, and proliferation and differentiation were examined by immunostaining and high-throughput quantitative imaging analysis. Whereas the combination of Wnt and Notch was shown to maintain the cells in an undifferentiated state, the presence of bone morphogenic protein 4 notably caused the cells to display an intermediate phenotype characterized by coexpression of both neuronal and glial markers. This work demonstrates the potential of combinatorial studies to assist in the identification of ECM/growth factor microenvironments for the control of stem cell behavior.
Controlled-release microarrays of soluble factors
Another common method of controlling cellular behavior is with small molecules [28•]. The release of small molecules, genetic materials, and growth factors from degradable scaffolds is one important way in which scientists control cellular behavior in the microenvironment [29,30]. High-throughput methods have been developed to screen for soluble factor effects in a microenvironment. For example, Bailey et al. [31] created a controlled release microarray where small molecules were encapsulated in individual spots, and the effects on cells growing locally was studied. The imaging-based readout enables scientists to perform small molecule screens, facilitating the discovery of potential therapeutic agents. Finally, microarrayed DNA and siRNA systems have been developed, creating a high-throughput screen for efficient study of genetic control elements on cell behavior [32]. Other reviews provide a detailed analysis of cell microarrays in drug discovery as well as their design [33].
Methods for optimizing physical cues
In addition to chemical signals, the physical properties of the cell’s microenvironment play a key role in controlling cell behavior, including the mechanical forces exerted on the cell. For example, Taqvi and Roy [34] have studied the effects of physical properties on hematopoietic differentiation of mouse embryonic stem cells. They observed that decreasing the scaffold pore size and increasing the polymer concentration — thereby increasing the scaffold’s compression modulus — resulted in increased differentiation.
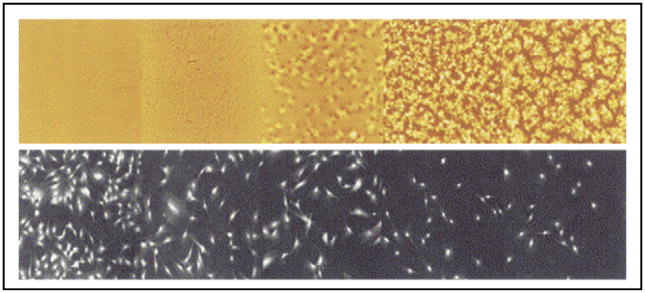
Washburn et al. [35] investigated the influence of polymer crystallinity, specifically nanoscale surface roughness, on the rate of cellular proliferation. Osteoblasts grew at a much greater rate on smooth regions than on rough regions, and a monotonic variation in rate was detected as a function of roughness (see Figure 3). Yang et al. [36] demonstrated that fiber alignment dictates the direction of neural stem cell elongation and that decreased fiber diameters result in increased differentiation. In this way, spatially resolved libraries may be used to complement micropatterning technologies.
Figure 3.
A montage of representative images of PLLA morphology from AFM data (top panels, field of view in each image is 20 μm) and corresponding cell count from fluorescent microscopy (bottom panels, field of view in each image is 1500 μm) (reproduced with permission from [35]).
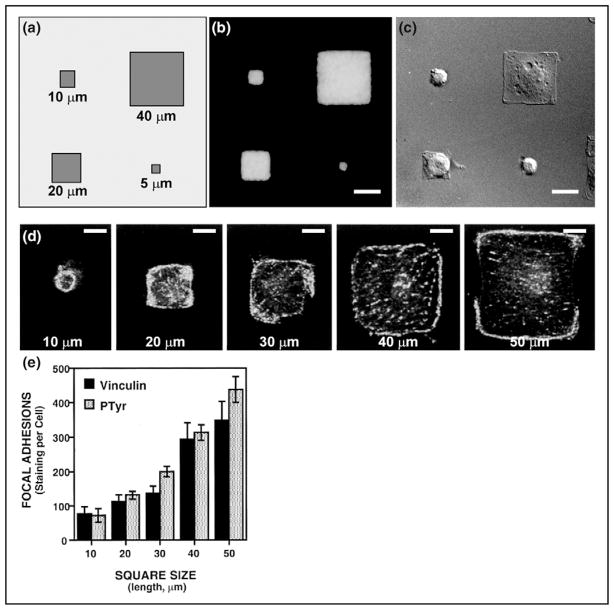
The mechanical forces experienced by a cell are not limited to its contact with surrounding cells, proteins, and surfaces. Nanotechnology [37] and microfabrication technologies [38] both enable the directed study of cell–substrate interactions and contribute to these interactions. McBeath et al. [39••] have shown that the shape of stem cells regulates the differentiation of human mesenchymal stem cells. Flattened, spread cells committed to osteocytes, whereas round cells became adiptocytes. Cellular adhesion and proliferation are also moderated by mechanical cues (see Figure 4). Utilizing microfabrication to control the organization of sheets of cells, Nelson et al. [40] showed that regions of high tractional stress corresponded to regions with high concentrations of growth; thus, tissue form is not only a result but also a modulator of tissue growth.
Figure 4.
Cell shape-dependent control of focal adhesion formation on micropatterned adhesive islands of different size coated with fibronectin. (A) Diagram of an assortment of square adhesive islands with sides ranging from 10 to 50 μm in length that were micropatterned onto substrates using microcontact printing. (B) An immunofluorescence micrograph showing staining for adsorbed fibronectin selectively limited to the square islands. (C) A differential interference contrast micrograph of bovine capillary endothelial cells cultured on different sized, square fibronectin islands. (D) Fluorescent confocal micrographs of individual, vinculin-labeled cells cultured on square islands of different sizes (lengths of sides are indicated). (E) Quantitation of total vinculin and total phosphotyrosine labeling per cell, for cells cultured on different sized squares. Over 30 cells per condition were averaged; error bars indicate standard error of the mean (The image is kindly provided by Dr. Ingber, and reproduced with permission from [43]).
Conclusions
Stem cells have significant potential in regenerative therapies. In order to translate stem cell research into the clinic, a number of challenges must be addressed. One important challenge is the development of methods to provide control of stem cell behaviors, including growth and differentiation. We believe that the high-throughput development of new bioactive materials and microenvironments, as well as the development of in silico modeling systems, will provide important tools in this regard [17]. Furthermore, most high-throughput biomaterial development is limited by robotic throughput. Future studies using materials developed using biological systems, such as through phage display, may greatly expand the diversity and utility of biomaterials [41,42].
Interestingly, most of the high-throughput biomaterials research conducted till date has been limited to the assessment of the interactions between stem cells and material surfaces. This can be attributed, in part, to the fact that most stem cells have been cultured on two-dimensional surfaces, and recent developments in microscopy automation allow stem cell behavior on such surfaces to be evaluated in a high-throughput and high-content manner. We believe the development of automated, high-throughput methods for the study of three-dimensional microenvironment will be important for the identification of medically useful cues. Finally, it is clear that the physical environment surrounding cells is an important controlling feature. We anticipate that future high-throughput methods may be developed to study and optimize the physical cellular microenvironment.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Polak J, Bishop A. Stem cells and tissue engineering: past, present, and future. Ann NY Acad Sci. 2006;1068:352–366. doi: 10.1196/annals.1346.001. [DOI] [PubMed] [Google Scholar]
- 2.Tutter A, Baltus G, Kadam S. Embryonic stem cells: a great hope for a new era of medicine. Curr Opin Drug Discov Dev. 2006;9:169–175. [PubMed] [Google Scholar]
- 3.Verfaillie CM, Pera MF, Lansdorp PM. Stem cells: hype and reality. Hematology. 2002;2002:369–391. doi: 10.1182/asheducation-2002.1.369. [DOI] [PubMed] [Google Scholar]
- 4.Odorico JS, Kaufman DS, Thomson JA. Multilineage differentiation from human embryonic stem cell lines. Stem Cells. 2001;19:193–204. doi: 10.1634/stemcells.19-3-193. [DOI] [PubMed] [Google Scholar]
- 5.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 6.Langer R, Vacanti J. Tissue engineering. Science. 1993;260:920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 7.Griffith LG, Naughton G. Tissue engineering — current challenges and expanding opportunities. Science. 2002;295:1009–1014. doi: 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]
- 8.Jang J-H, Schaffer DV. Microarraying the cellular microenvironment. Mol Syst Biol. 2006;2:39. doi: 10.1038/msb4100079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9••.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. Excellent review on the applications of biomaterials on tissue engineering. A insightful overview of the current status of the field is given, and the future direction is addressed. [DOI] [PubMed] [Google Scholar]
- 10.Elisseeff J, Ferran A, Hwang S, Varghese S, Zhang Z. The role of biomaterials in stem cell differentiation: applications in the musculoskeletal system. Stem Cells Dev. 2006;15:295–303. doi: 10.1089/scd.2006.15.295. [DOI] [PubMed] [Google Scholar]
- 11.Mahoney MJ, Krewson C, Miller J, Saltzman WM. Impact of cell type and density on nerve growth factor distribution and bioactivity in three-dimensional collagen gel cultures. Tissue Eng. 2006;12:1915–1927. doi: 10.1089/ten.2006.12.1915. [DOI] [PubMed] [Google Scholar]
- 12.Prang P, Muller R, Eljaouhari A, Heckmann K, Kunz W, Weber T, Faber C, Vroemen M, Bogdahn U, Weidner N. The promotion of oriented axonal regrowth in the injured spinal cord by alginate-based anisotropic capillary hydrogels. Biomaterials. 2006;27:3560–3569. doi: 10.1016/j.biomaterials.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 13.Pratt AB, Weber FE, Schmoekel HG, Müller R, Hubbell JA. Synthetic extracellular matrices for in situ tissue engineering. Biotechnol Bioeng. 2004;86:27–36. doi: 10.1002/bit.10897. [DOI] [PubMed] [Google Scholar]
- 14.Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- 15.Ramstrom O, Lehn J-M. Drug discovery by dynamic combinatorial libraries. Nat Rev Drug Discov. 2002;1:26–36. doi: 10.1038/nrd704. [DOI] [PubMed] [Google Scholar]
- 16.Amis EJ. Combinatorial materials science: reaching beyond discovery. Nat Mater. 2004;3:83–85. doi: 10.1038/nmat1064. [DOI] [PubMed] [Google Scholar]
- 17.Kohn J. New approaches to biomaterials design. Nat Mater. 2004;3:745–747. doi: 10.1038/nmat1249. [DOI] [PubMed] [Google Scholar]
- 18.Pepperkok R, Ellenberg J. High-throughput fluorescence microscopy for systems biology. Nat Rev Mol Cell Biol. 2006;7:690–696. doi: 10.1038/nrm1979. [DOI] [PubMed] [Google Scholar]
- 19.Jost M, Budde P, Tammen H, Hess R, Kellmann M, Schulte I, Rose H. The concept of functional peptidomics for the discovery of bioactive peptides in cell culture models. Comb Chem High Throughput Screen. 2005;8:767–773. doi: 10.2174/138620705774962445. [DOI] [PubMed] [Google Scholar]
- 20••.Anderson DG, Levenberg S, Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat Biotechnol. 2004;22:863–866. doi: 10.1038/nbt981. A high throughput approach for nanoliter-scale synthesis of biomaterials and characterization of their interactions with cells has been demonstrated. More than 1,700 arrayed biomaterials were generated and their interactions with human embryonic stem cells were simultaneously characterized. [DOI] [PubMed] [Google Scholar]
- 21.Anderson DG, Putnam D, Lavik EB, Mahmood TA, Langer R. Biomaterial microarrays: rapid, microscale screening of polymer–cell interaction. Biomaterials. 2005;26:4892–4897. doi: 10.1016/j.biomaterials.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 22.Mei Y, Elliott JT, Smith JR, Langenbach KJ, Wu T, Xu C, Beers KL, Amis EJ, Henderson L. Gradient substrate assembly for quantifying cellular response to biomaterials. J Biomed Mater Res Part A. 2006;79A:974–988. doi: 10.1002/jbm.a.30883. [DOI] [PubMed] [Google Scholar]
- 23.Bhat RR, Chaney BN, Rowley J, Liebmann-Vinson A, Genzer J. Tailoring cell adhesion using surface-grafted polymer gradient assemblies. Adv Mater. 2005;17:2802–2807. [Google Scholar]
- 24.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 25•.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat Meth. 2005;2:119–125. doi: 10.1038/nmeth736. Protein arrays has been prepared with a standard robotic DNA spotter, and this technology has been applied to study the effects of 32 different combinations of five extracellular matrix molecules on stem cellular differentiations. [DOI] [PubMed] [Google Scholar]
- 26.Nakajima M, Ishimuro T, Kato K, Ko I-K, Hirata I, Arima Y, Iwata H. Combinatorial protein display for the cell-based screening of biomaterials that direct neural stem cell differentiation. Biomaterials. 2007:28. doi: 10.1016/j.biomaterials.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Soen Y, Mori A, Palmer TD, Brown PO. Exploring the regulation of human neural precursor cell differentiation using arrays of signaling microenvironments. Mol Syst Biol. 2006;2:37. doi: 10.1038/msb4100076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Ding S, Schultz PG. A role for chemistry in stem cell biology. Nat Biotechnol. 2004;22:833–840. doi: 10.1038/nbt987. The comprehensive review discusses the use of small molecules to influence the stem cell behaviors. [DOI] [PubMed] [Google Scholar]
- 29.Riddle KW, Kong H-J, Leach JK, Fischbach C, Cheung C, Anseth KS, Mooney DJ. Modifying the proliferative state of target cells to control DNA expression and identifying cell types transfected in vivo. Mol Therapy. 2007;15:361–368. doi: 10.1038/sj.mt.6300017. [DOI] [PubMed] [Google Scholar]
- 30.Hill E, Boontheekul T, Mooney DJ. Regulating activation of transplanted cells controls tissue regeneration. PNAS. 2006;103:2494–2499. doi: 10.1073/pnas.0506004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bailey SN, Sabatini DM, Stockwell BR. Microarrays of small molecules embedded in biodegradable polymers for use in mammalian cell-based screens. PNAS. 2004;101:16144–16149. doi: 10.1073/pnas.0404425101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey SN, Ali SM, Carpenter AE, Higgins CO, Sabatini DM. Microarrays of lentiviruses for gene function screens in immortalized and primary cells. Nat Meth. 2006;3:117–122. doi: 10.1038/nmeth848. [DOI] [PubMed] [Google Scholar]
- 33.Castel D, Pitaval A, Debily M-A, Gidrol X. Cell microarrays in drug discovery. Drug Discov Today. 2006;11:616–622. doi: 10.1016/j.drudis.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 34.Taqvi S, Roy K. Influence of scaffold physical properties and stromal cell coculture on hematopoietic differentiation of mouse embryonic stem cells. Biomaterials. 2006;27:6024–6031. doi: 10.1016/j.biomaterials.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 35.Washburn NR, Yamada KM, Simon CG, Kennedy SB, Amis EJ. High-throughput investigation of osteoblast response to polymer crystallinity: influence of nanometer-scale roughness on proliferation. Biomaterials. 2004;25:1215–1224. doi: 10.1016/j.biomaterials.2003.08.043. [DOI] [PubMed] [Google Scholar]
- 36.Yang F, Murugan R, Wang S, Ramakrishna S. Electrospinning of nano/micro scale poly(L-lactic acid) aligned fibers and their potential in neural tissue engineering. Biomaterials. 2005;26:2603–2610. doi: 10.1016/j.biomaterials.2004.06.051. [DOI] [PubMed] [Google Scholar]
- 37.Sniadecki NJ, Desai RA, Ruiz SA, Chen CS. Nanotechnology for cell–substrate interactions. Ann Biomed Eng. 2006;34:59–74. doi: 10.1007/s10439-005-9006-3. [DOI] [PubMed] [Google Scholar]
- 38.Kane RS, Takayama S, Ostuni E, Ingber DE, Whitesides GM. Patterning proteins and cells using soft lithography. Biomaterials. 1999;20:2363–2376. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 39••.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. A very impressive report illustrate that cell shape regulates commitment of human mesenchymal stem cells (hMSCs) to adipocyte or osteoblast fate. [DOI] [PubMed] [Google Scholar]
- 40.Nelson CM, Jean RP, Tan JL, Liu WF, Sniadecki NJ, Spector AA, Chen CS. Emergent patterns of growth controlled by multicellular form and mechanics. PNAS. 2005;102:11594–11599. doi: 10.1073/pnas.0502575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanghvi AB, Miller KP, Belcher AM, Schmidt CE. Biomaterials functionalization using a novel peptide that selectively binds to a conducting polymer. Nat Mater. 2005;4:496–502. doi: 10.1038/nmat1397. [DOI] [PubMed] [Google Scholar]
- 42.Yoo PJ, Nam KT, Qi J, Lee S-K, Park J, Belcher AM, Hammond PT. Spontaneous assembly of viruses on multilayered polymer surfaces. Nat Mater. 2006;5:234–240. doi: 10.1038/nmat1596. [DOI] [PubMed] [Google Scholar]
- 43.Chen CS, Alonso JL, Ostuni E, Whitesides GM, Ingber DE. Cell shape provides global control of focal adhesion assembly. Biochem Biophys Res Commun. 2003;307:355–361. doi: 10.1016/s0006-291x(03)01165-3. [DOI] [PubMed] [Google Scholar]