Abstract
Research in the areas of drug delivery and tissue engineering has witnessed tremendous progress in recent years due to their unlimited potential to improve human health. Meanwhile, the development of nanotechnology provides opportunities to characterize, manipulate and organize matter systematically at the nanometer scale. Biomaterials with nano-scale organizations have been used as controlled release reservoirs for drug delivery and artificial matrices for tissue engineering. Drug-delivery systems can be synthesized with controlled composition, shape, size and morphology. Their surface properties can be manipulated to increase solubility, immunocompatibility and cellular uptake. The limitations of current drug delivery systems include suboptimal bioavailability, limited effective targeting and potential cytotoxicity. Promising and versatile nano-scale drug-delivery systems include nanoparticles, nanocapsules, nanotubes, nanogels and dendrimers. They can be used to deliver both small-molecule drugs and various classes of biomacromolecules, such as peptides, proteins, plasmid DNA and synthetic oligodeoxynucleotides. Whereas traditional tissue-engineering scaffolds were based on hydrolytically degradable macroporous materials, current approaches emphasize the control over cell behaviors and tissue formation by nano-scale topography that closely mimics the natural extracellular matrix (ECM). The understanding that the natural ECM is a multifunctional nanocomposite motivated researchers to develop nanofibrous scaffolds through electrospinning or self-assembly. Nanocomposites containing nanocrystals have been shown to elicit active bone growth. Drug delivery and tissue engineering are closely related fields. In fact, tissue engineering can be viewed as a special case of drug delivery where the goal is to accomplish controlled delivery of mammalian cells. Controlled release of therapeutic factors in turn will enhance the efficacy of tissue engineering. From a materials point of view, both the drug-delivery vehicles and tissue-engineering scaffolds need to be biocompatible and biodegradable. The biological functions of encapsulated drugs and cells can be dramatically enhanced by designing biomaterials with controlled organizations at the nanometer scale. This review summarizes the most recent development in utilizing nanostructured materials for applications in drug delivery and tissue engineering.
Keywords: Nanomaterials, biomaterials, drug delivery, tissue engineering, nanoparticles, nanocapsules, nanotubes, nanogels, dendrimers, nanofibril, network, hydrogel, electrospinning, self-assembly, nanocomposites
NANOSTRUCTURED MATERIALS AS DRUG-DELIVERY SYSTEMS
Introduction
The interdisciplinary field of nanobiotechnology, which combines chemistry, biology, engineering and medicine, is revolutionizing the development of drug-delivery systems and devices. Novel materials and formulations are enabling the site-specific targeting and controlled release of traditional pharmaceuticals, recombinant proteins, vaccines and nucleic acids. Nano-scale drug-delivery systems can be devised to tune release kinetics, to regulate biodistribution and to minimize toxic side effects, thereby enhancing the therapeutic index of a given drug [1]. Future generation systems will include biosensing functionalities with in vivo feedback that will permit “smart” drug delivery [2, 3].
The goals of drug delivery are (1) to overcome the inherent limitations associated with biomacromolecular therapeutics, which include a short plasma half-life, poor stability and potential immunogenicity, and (2) to maximize the therapeutic activity while minimizing the toxic side effects of drugs [4]. Current drug-delivery systems are effective at releasing drugs in a controlled fashion to produce a high local concentration; however, the scope is limited to targeting tissues rather than individual cells.
At the expense of lower drug loading capacity, the nanometer size range enhances the ability of drug-delivery carriers to cross cell membranes, reduces the risk of undesired clearance from the body through the liver or spleen and minimizes their uptake by the reticuloendothelial system [4, 5]. Smaller particles have greater surface area-to-volume ratios, which increase the particles’ dissolution rate, enabling them to overcome solubility-limited bioavailability [6]. Particle size is extremely important to the biological properties and, hence, function, of nano-scale drug-delivery systems [7].
Even within the nano-scale range, size variation strongly affects bioavailability and blood circulation time [8, 9]. Following systemic administration, particles with diameters less than 10 nm are rapidly removed through extravasation and renal clearance [7]; particles with diameters ranging from 10 to 70 nm can penetrate even very small capillaries [10]; particles with diameters ranging from 70 to 200 nm demonstrate the most prolonged circulation times [8]; and particles with diameters greater than 200 nm are usually sequestered by the spleen and eventually removed by phagocytes [11].
Combining this information with the knowledge that particles smaller than 100 nm can be enclosed within endocytic vesicles [12] leads to the conclusion that the preferred size range for nanoparticle drug delivery is 10 to 100 nm. Interestingly, nano-scale drug-delivery systems display not only decreased size but also altered attributes.
Nano-scale drug-delivery systems take advantage of the fact that nano-scaled materials (10−9 to 10−7 m) can exhibit distinctive physical properties, electrical, mechanical and optical, that differ from those observed in the macroscopic and atomic realms [13]. Through rational design, nano-scale drug-delivery systems can be manufactured to combine desirable modules, both biological and synthetic, for various applications, including implantable, inhalable, injectable, oral, topical and transdermal drug delivery.
Many properties of nano-scale drug-delivery systems can be tailored for specific applications (controlling factors listed in parentheses): solubility (inherent hydrophilicity of the material, addition of solubilizing moiety); biodistribution (molecular weight, addition of targeting group); biocompatibility (electrical charge, addition of bioinert functionality); biodegradability (backbone, spacer); drug release (physical interaction between drug and carrier, chemical cleavage of covalent spacer); drug encapsulation (physical interaction between drug and carrier); and shape (materials and chemistry employed) [4].
Nanoparticles can be functionalized with biomolecules through various techniques, including physical adsorption, electrostatic binding, complementary recognition and covalent coupling [14]. The conjugation of targeting moieties enhances receptor-mediated delivery [15]. Nanoparticles can even be modified to achieve efficient intracellular targeting to specific organelles. For example, altering the nanoparticles’ surface charge can dictate their location within the cell; specifically, anionic particles remain in lysosomes, while cationic particles become localized in the cytoplasm and within mitochondria [16].
Drug delivery can be localized or systemic. Parenteral drug delivery is typically achieved using liposomal or polymeric carriers [17]. There is no universal platform, as each system has its advantages and disadvantages. While liposomes have a high drug-carrying capacity, their release profiles are more difficult to regulate. In contrast, polymeric drug-delivery systems can be synthesized to generate specific molecular weights and compositions, but their drug-carrying capacity is relatively low [18]. While an ideal general drug-delivery platform may never be realized, hybrid drug-delivery systems that incorporate the benefits of various approaches will be tailored to address the needs of specific applications.
This section examines nanobiomaterial structures developed for drug delivery. While spherical nanoparticles are the simplest to create, other shapes and constructions offer advantages for certain applications. These supramolecular systems include nanocapsules, nanotubes, nanogels and dendrimers (Fig. 1). The sections are intended to characterize each morphology, describing structural features, fabrication methods, materials and recent applications. The final sections consider the growing field of nucleic-acid delivery, as well as novel nano-scale drug-delivery systems that have been invented recently.
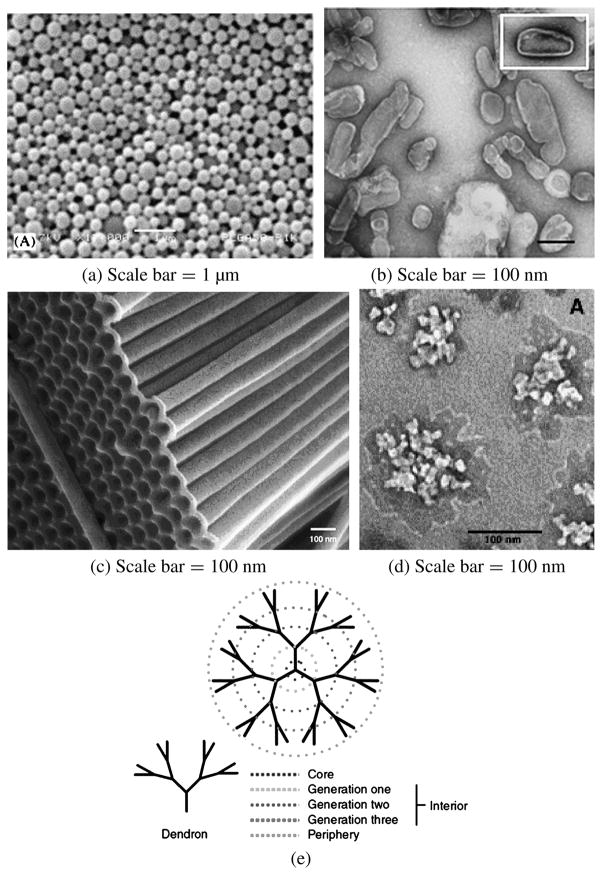
Figure 1.
Representative nano drug-delivery systems. (a) Nanoparticles (reprinted from Ref. [40] with permission), (b) nanocapsules (reprinted from Ref. [46] with permission), (c) nanotubes (reprinted from Ref. [58] with permission), (d) nanogels (reprinted from Ref. [73] with permission) and (e) dendrimers (reprinted from Ref. [79] with permission).
Polymers
Polymer therapeutics comprise rationally-designed macromolecular drugs, polymer–drug and polymer–protein conjugates, polymeric micelles and polyplexes for DNA delivery [18]. The greatest advantage of these species is their amenability to chemical modification, resulting in defined chemical composition, customized surface functionality and the potential for defined three-dimensional structures [18].
Several polymers are widely used in clinical therapies: synthetic polymers (poly(ethylene glycol), N-(2-hydroxypropyl) methacrylamide co-polymers, poly(vinylpyrrolidone), poly(ethyleneimine) and linear polyamidoamines); natural polymers (dextran (α-1,6 polyglucose), dextrin (α-1,4 polyglucose), hyaluronic acid and chitosans); and pseudosynthetic polymers (the man-made poly(amino acids) poly(L-lysine), poly(L-glutamic acid), poly(malic acid) and poly(aspartamides)) [19].
Polymers offer effectively unlimited diversity in chemistry, dimensions and topology, rendering them a class of materials that is particularly suitable for applications in nano-scale drug-delivery systems [4]. Insights into the structure–function relationships of polymers continue to advance their utility. The range of polymer architectures is very wide: linear, branched, cross-linked, block, graft, multivalent, dendronized and star-shaped polymers [4, 18].
Interestingly, polymer architecture can be as significant to the effectiveness of drug-delivery carriers as chemical composition (polyester, polyanhydride, polyamide); backbone stability (biodegradable, non-biodegradable); and water solubility (hydrophilic, hydrophobic) [4]. That is, for the same monomer ratios, copolymer topology dictates the material’s physicochemical properties [20]. Polymer architecture affects not only the carrier’s physicochemical properties but also drug-loading efficiency, drug-release rate and biodistribution [4].
Drugs can be physically entrapped within polymer shells and matrices, or they can be covalently attached to the polymer backbone. Polymer–drug conjugates, which contain a systemically-stable, bioresponsive polymer–drug linker, alter the pharmacokinetics of the drug by increasing the drug’s effective molecular weight [21]. The linker ensures that the pro-drug remains inactive in circulation until it is specifically liberated at the target site by the appropriate enzyme or pH.
Unlike drugs, which are homogeneous defined species, polymers are inherently heterogeneous mixtures of chains of varying lengths [18]. Polydispersity is of particular importance because biological properties are molecular weight dependent [22]. Because polymers are often internalized by cells, biocompatibility is another area of concern [23]. Polycations are often cytotoxic, hemolytic and complement-activating, while polyanions are less cytotoxic but can induce anticoagulant activity and cytokine release [18]. Extensive research has identified polymers that are well-suited to in vivo applications [24].
Novel and improved polymeric nano-scale drug-delivery systems will be realized through a combination of biological rationale and advanced synthetic chemistries. To be suitable for therapeutic applications, such chemistries must be amenable to industrial-scale development [18]. Scientists continue to explore new biodegradable polymers that exhibit more sophisticated three-dimensional structures [25] and are better suited to frequent parenteral administration [26].
Nanoparticles
While all of the nano-scale drug-delivery systems to be discussed, nanocapsules, nanotubes, nanogels and dendrimers, could be considered “nanoparticles”, this section refers specifically to polymer-based matrix particulate systems. Specifically, nanoparticles have been defined as “submicron-sized polymeric colloidal particles with a therapeutic agent of interest encapsulated within their polymeric matrix or adsorbed or conjugated onto the surface” [16].
In general, drug-delivery systems can be administered locally or systemically, with the potential for the attachment of targeting moieties. The drug payload can be released outside of or within the target cells. While larger drug-delivery systems can create high local drug concentrations, smaller drug-delivery systems can be endocytosed directly. Subsequent to their uptake, nanoparticles are trafficked from early endosomes to sorting endosomes. Some of the nanoparticles are exported from the cell, while others are transported to secondary endosomes/lysosomes, from which they can escape into the cytoplasm to serve as intracellular drug reservoirs [16].
In addition to the standard advantages of polymeric and liposomal drug-delivery systems, including sustained and controlled release, as well as the potential for targeted delivery, nano-scale dimensions confer improved bioavailability by increasing the payload’s solubility and stability. Smaller particle sizes facilitate deeper penetration into capillaries and through fenestrations and, ultimately, enhanced cellular uptake. Indeed, 100 nm size particles exhibit in situ uptake efficiencies 15–250-fold of those of small microparticles (1 and 10 μm) [27]. Nanoparticles have even been shown to cross the blood–brain barrier [28].
Different formulation methodologies have been explored not only to control the dimensions of drug-delivery systems but also to optimize drug loading and release profiles. High entrapment efficiencies (>80%) can be achieved with poly(D,L-lactic-co-glycolic acid) (PLGA) when sonication is used for both emulsification steps in the water/oil/water double-emulsion method, independent of the length and intensity of mixing [29]. In contrast, vortexing to achieve the primary emulsion yields poor loading (approx. 25%). PLGA exhibits inherently good loading because of its high molecular weight, high hydrophilicity and free carboxylate end-groups. Other factors that increase drug entrapment efficiency are low drug loading, a large volume of the inner, primary water phase and the use of methylene chloride as the organic solvent; entrapment efficiency can be made to approach 100% [29]. Methylene chloride does, however, increase the mean particle size substantially, as does increasing the polymer concentration in the organic phase [29].
A variety of materials can be used to synthesize nanoparticles and more advanced chemistries are pending [30]. Recent novel examples, many of which are not polymer-based, include calcium carbonate [31], calcium-deficient hydroxyapatite [32], chitosan [33], oligo-3-hydroxybutyrates [34] and porous hollow silica [35]. Polymer micelles have emerged as a very important system for targeted drug delivery. Self-assembly of amphiphilic block or graft co-polymers in aqueous media leads to nanoparticles with hydrophobic cores for drug encapsulation and hydrophilic shells for stabilization and specific targeting [36]. Using prostate cancer as a model, Farokhzad et al. demonstrate the potential utility of polymer micelle–aptamer bioconjugates that encapsulate docetaxel for therapeutic applications [37]. To improve the structure integrity of polymer micelles, the hydrophilic corona has been selectively cross-linked, leading to static entities that are not easily disrupted [38]. Intelligent core–shell nanoparticles have also been produced: thermo-responsive, pH-responsive and biodegradable nanoparticles were made from poly(D,L-lactide)-graft-poly(N-isopropyl acrylamide-co-methacrylic acid) (PLA-g-P(NIPAm-co-MAA)) to yield a hydrophilic outer shell and a hydrophobic inner core that exhibited a phase transition temperature above 37°C, rendering them apposite for biomedical applications [39].
Material selection is vital because the matrix interacts with both the encapsulated drug and the external environment. Surface modification can alter a nanoparticle’s effective exterior [40]. PEGylation is a strategy used to solubilize the carrier, to protect the payload from plasma enzymes, to prevent an immune response and to hinder renal excretion [41, 42]. Targeting ligands can similarly be added to increase the drug’s effective concentration at a desired site. Thus, targeting can be achieved both passively (via enhanced permeation and retention) and actively (via the conjugation of molecular homing devices) [43].
Nanocapsules
Lipid and polymeric nanocapsules are nano-scale drug-delivery systems that can provide controlled release and efficient targeting [44–46]. The composition of the outer coating, in particular, dictates their dispersion stability and the primary physiological response [44]. The fabrication of nanocapsules can be achieved by interfacial deposition, interfacial polymerization, interfacial precipitation, layer-by-layer deposition and self-assembly procedures. Important variables include capsule size, radius distribution, capsule thickness, membrane decomposition and surfactant type [44].
Lipid-based nanocapsules can be modified to alter membrane permeability via channel insertion and to target specific tissues or cells via antibody attachment. Lipid-based nanocapsules have been shown to stabilize their cargo. Cisplatin nanocapsules display an unprecedented cisplatin-to-lipid molar ratio and exhibit greatly improved cytotoxicity against tumor cells in vitro relative to free drugs [47]. Still, the use of lipids can be limited by their instability in biological media and by their sensitivity to many external parameters, including temperature and osmotic pressure [48].
The stability of lipid-based nanocapsules can be improved by synthesizing lipid–polymer-conjugate nanocapsules. Strategies include polymerizing a two-dimensional network in the membrane’s hydrophobic core, adding surface-active polymers to create mixed vesicular structures and coating the liposome with a polyelectrolyte shell [48].
Polyelectrolyte shells, created via layer-by-layer deposition, offer several advantages, including control of surface properties, membrane thickness and release kinetics [49]. Because these shells can adopt both open and closed states, in response to environmental conditions such as temperature and pH, various materials can be readily loaded and released [50]. Examples include drugs, enzymes, nucleic acids and dyes [51–53].
A recent advance in the pursuit of a biocompatible nanocapsule has been the use of “the vault”, a naturally occurring cellular nanoparticle thus named for its morphology [54]. The internal cavity of these 13-MDa ribonucleoprotein particles can accommodate hundreds of protein molecules. Attaching a vault-targeting peptide to any protein of interest sequesters the protein within the vault cavity, enabling the engineering of vault particles with contrived properties and functionalities [54].
Another interesting development was the synthesis of disulfide cross-linked polymer capsules [55]. The disulfide bonds confer enhanced stability to hydrogen-bonded multilayer thin films at physiological pH. They also render the system susceptible to disassembly in the presence of thiol–disulfide exchange reagents. Accordingly, these nanocapsules have the potential to be used as “biodeconstructible” nano-scale drug-delivery systems, as intracellular proteins, such as glutathione, will promote in vivo capsule deconstruction [56, 57].
Nanotubes
Nanotubes, structures that resemble tiny drinking straws, offer advantages over spherical nanoparticles for some applications [58]. Their large inner volumes can be filled with sundry chemicals and biomolecules, ranging in size from small molecules to proteins [59, 60]. Because the inner and outer surfaces of certain types of nanotubes are distinct, they can be differentially modified to encapsulate specific drugs internally and to evade immunogenic response externally [60]. Finally, the open-mouthed structure of nanotubes renders loading especially simple.
Nanotubes can be fabricated from many materials and via distinct routes, ranging from self-assembly to deliberate deposition. Examples include fullerene carbon nanotubes, cyclic peptide nanotubes and template-synthesized nanotubes [58]. Polymeric nanotubes can be synthesized via in-pore polymerization; metal nanotubes can be produced by electroless deposition; and inorganic nanotubes can be created by sol–gel chemistry [61].
The template approach is the most versatile route to nanotube fabrication [58]. The nanotube is made by depositing the material (polymer, silica, metal, or carbon) within the cylindrical pores of a solid surface [61]. The outer diameter is determined by the diameter of the template, while the inner diameter is determined by the deposition time [58]. Composite nanostructures, such as concentric tubular structures [62] and segmented nanowires [63], can also be prepared by this method. Future work on nanotubes will focus on manipulating the caps to control drug release.
Recently, nanotube spearing was used to achieve extremely high transfection efficiency with high post-induction cell viability in hard-to-transfect cells, including B cells and primary neurons [64]. This technique uses nickel-embedded nanotubes to penetrate cell membranes via magnetic field driving, thereby delivering macromolecules that are immobilized on the nanotubes. This approach offers several benefits, including increased control, decreased cytotoxicity and greater efficiency than standard transfection reagents [65, 66].
Penetration efficiency can be tuned by altering magnetic field strength, nanotube speed and duration of spearing. Carbon nanotubes do not cause cytotoxicity at low nanotube concentrations (<10 μM), perhaps owing to the nano-scale penetration, which results in less perturbation than other, larger mechanical delivery systems [64]. Because the DNA payload does not rely on lysosomal escape and can even be deposited directly into the nucleus, nanotube spearing is a much more efficient transduction method than standard magnetofection or endocytosis [67].
Nanotube spearing, which works optimally at only 100 fM nanotubes, is also much more amenable to high-throughput biochemical assays than other membrane-penetrating tools [64]. While a silicon nanoneedle attached to an atomic force microscopy tip may be able to deliver macromolecules to an individual targeted cell, it will not be suitable for in vivo gene therapy applications [68].
Nanogels
In addition to exhibiting high biocompatibility and tunable properties, hydrogel matrices are advantageous for use in drug delivery because of their ability to prevent payload aggregation [69]. Perhaps the greatest advantage of hydrogels as drug carriers is that they can be synthesized in the absence of drugs, ameliorating potential drug inactivation [70]. Generally, the drug is subsequently loaded via self-assembly processes based on non-covalent interactions. Both charged and hydrophobic biomolecules can be incorporated into hydrogel networks, which are themselves characterized by the physical properties of their constituent polymers [7].
Nano-scale hydrogels, or “nanogels”, offer straightforward synthesis and relatively high drug-loading capacity [7]. Hydrogels are hydrophilic, three-dimensional cross-linked polymer networks that swell in the presence of water [7, 71]. They can be designed to respond to many physiological stimuli, including ionic strength, pH and temperature [7, 71]. Hybrid polymerizable nanogels that incorporate both physical and chemical cross-linking structures have been synthesized [69]. Nanogel particles combine the properties of gels with the properties of colloids: high surface-to-volume ratio, microheterogeneous structure and small size.
Polymeric nanogel vectors display extended stability, sustained release, low cytotoxicity and protection from enzymatic degradation [72, 73]. However, their widespread use in nano-scale drug-delivery systems has been limited by the dearth of a general methodology for hydrogel surface coating [74]. While hydrogels can been coated with lipids to preserve colloidal stability, these procedures are limited by low coating efficiency [75, 76].
“Self-exploding” microgels were recently created for the purpose of pulsed drug delivery [74, 77]. A degradable gel is surrounded by a membrane that is permeable to water but impermeable to the drug and the gel-degradation products. Gel degradation increases swelling pressure, resulting in membrane rupture and drug release. Distinct microgel populations could potentially be synthesized with variable degradation rates to yield pulsed explosions from a single injection [74, 77]. Perhaps this novel technique will be modified in the future to allow for the use of nanogels in place of microgels.
It was recently shown that well-defined, stable, cross-linked, amphiphilic hydrogel nanoparticles can be prepared by inverse emulsion photo-polymerization [78]. The size of the colloidal material, which can be loaded with hydrophobic drugs, can be tuned by varying polymerization conditions. While the hydrophobic core solubilizes lipophilic molecules, the hydrophilic corona prevents aggregation, protein adsorption and immunogenic response [78]. This platform, amenable to many amphiphilic precursors, may give rise to a new class of colloidal nano-scale drug-delivery systems.
Dendrimers
The architectural design of dendrimers can be greatly controlled, yielding well-defined shapes, sizes, branching length and density and surface functionality [5, 79]. Accordingly, dendrimers are attractive candidates as nano-scale drug-delivery systems. The drug payload can be physically entrapped within the dendrimer or chemically attached to the surface. Dendrimers are characterized by their high density of exo-presented surface groups, facilitating targeting and biocompatibility [5].
Dendrimer synthesis, which has recently been simplified, can be achieved by the divergent ‘lego’ approach [80] or by the convergent ‘click’ approach [81]. Both strategies permit easy purification and produce environmentally-safe byproducts. The former route requires only one step per generation [82]. The latter route, based on the Cu(I)-catalyzed synthesis of 1,2,3-triazoles from azides and alkynes, yields dendrimers with various surface groups in high purity and excellent yield [83].
The core–shell architecture of dendrimers, such as poly(amidoamine) (PAMAM), grows linearly in diameter as a function of added generations and exponentially with respect to the surface groups. As a result, the branches become entwined as multiplicities increase, producing geometrically-closed nanostructures that can encapsulate small molecule drugs [5]. Notably, drug encapsulation is not limited to physical entrapment; many dendrimers are comprised of internal amines that complex negatively-charged drugs through electrostatic interactions [84], increasing loading capacity relative to poly-alcohols that can only participate in hydrogen bonding [85].
Most dendrimeric drug delivery has focused on chemotherapeutic agents, including cisplatin [86], methotrexate [87] and 5-fluorouracil [88], affording slower release, greater accumulation in solid tumors and decreased systemic toxicity than free drugs, particularly when the dendrimer is PEGylated.
PAMAM dendrimers have been shown to cross cell membranes through a combination of paracellular transport and adsorptive endocytosis [89]. Dendrimer solubility increases with size and, for a given number of surface groups, ester-terminated dendrimers are more bioavailable than their amino-terminated analogues [90]. Unfortunately, along with solubility, dendrimer-induced cytotoxicity increases with generation number [91, 92].
Dendrimers with positively-charged surface groups are apt to destabilize cell membranes, causing lysis. Amino-terminated PAMAM dendrimers do, however, exhibit lower toxicity than more flexible amino-functionalized linear polymers [5]. Primary amines are more toxic than secondary or tertiary amines [92]. As mentioned, surface functionalization, particularly with PEG, can reduce cytotoxicity. Still, dendrimers can only be regarded as “safe” in relation to a specific application: limited clinical experience with dendrimers render safety generalizations about given chemistries difficult [93].
Novel dendrimeric systems for applications as nano-scale drug-delivery systems include biaryl-based dendrimeric micelles that exhibit environment-dependent conformations [94], ‘bow-tie’ architectures that covalently connect a drug-loaded dendron to a PEGylated, solubilizing dendron [25] and hybrid dendrimer-based microcapsules that enable a dual release scheme [95]. In addition to serving as drug-delivery vectors, dendrimers can act as inherently-active antitumor, antiviral and antibacterial agents [96] and as permeability enhancers that can promote oral and transdermal drug delivery [93, 97].
Nucleic acid delivery
Gene therapy and RNA interference (RNAi) are two therapies of notable interest: the former (gain-of-function) eliminates the need for subsequent drug administration, while the latter (loss-of-function) confers highly-specific gene silencing. Gene expression can be controlled by transcriptional (incorporation of genetic regulatory elements) and transductional (conjugation of targeting ligands) targeting.
While extremely efficient, viral delivery poses safety concerns. In addition to being much safer and less expensive, non-viral vectors can be synthesized easily and reproducibly in large quantities and afford much greater nucleic acid carrying capacity [98]. To achieve non-viral delivery of nucleic acids, DNA and RNA are typically condensed by polycationic lipids or polycationic polymers to form nanocomplexes.
In order to generate protein expression, DNA payloads must first enter the cell, escape the endosome, dissociate from their carrier, cross the nuclear membrane and be transcribed by host machinery [99]. Extracellular interactions between the complexes and anionic glycosaminoglycans (GAGs) located on the cell surface inhibit gene transfer by nano-scale drug-delivery systems [100]. If the complex is taken up by the target cell, it should promote endosomolysis via a pH-responsive functionality in the carrier backbone, such as a “proton sponge” [101].
Poly(ethylene imine) (PEI) is the most widely characterized polymeric vector for nucleic acid delivery. Linear polymers with a molecular mass of 22 kDa overcome the nuclear barrier most effectively [102], resulting in the highest transfection rates [103]; however, in vivo use of PEI is limited by its relatively high toxicity.
Alternative classes of endosomolytic polymers have been synthesized, including amphoteric poly(amido amines) [104], poly(acrylic acids) [105], poly(lysine imidazoles) [106] and poly(β-amino esters) [107]. Other materials include PLGA [108], poly(lysine) [109], protamine [110], chitosan [111], calcium phosphate [112], cationized gelatin [113] and carbonate apatite [114]. Recently, several stimuli-responsive polymers have been synthesized to provide site-, timing- and duration period-specific gene expression [98].
Novel systems
Novel nano-scale drug-delivery systems that explore original applications of structures, shapes, materials and phase boundaries are being developed. The following systems represent some recent advances from the literature.
A silicon-based nanochannel was constructed to deliver anti-tumor compounds locally to unresectable tumors with zero-order kinetics [115]. The device can be implanted using a minimally-invasive procedure to overcome the inconvenience of frequent local injections. Biodegradable implantable nano-scale drug-delivery systems have also been fashioned. Polymeric nanofibers with controlled surface and internal molecular structures can be produced by electrospinning to yield scaffolds for controlled drug release [116]. Polypeptide nanofilms can be used as nano-scaled devices with defined structural properties or to customize the surfaces of other materials [117].
Various physical forms have been explored for nano drug delivery. Self-assembled, peptidic doughnut-shaped nanoreactors have been presented as potential nano-scale drug-delivery systems [118]. ‘Nanoeggs’ were prepared from inorganic minerals using boundary-organized reaction droplets as templates based on the interfacial properties of organic architectures [119]. Drug delivery can be achieved through systems that do not even rely upon polymer matrices or lipid enclosures. Hydrophobic drug nanoaggregates can be prepared directly by adding an excess of the hydrophobic drug to a solution of the PEGylated, protonated form of the same drug in an aqueous phase [120].
Furthermore, standard nanoparticles can be altered to create more complex structures. Specifically, nanoparticles can be coated with polymeric nanoshells via layer-by-layer stepwise self-assembly [121]. The nanoshell provides a template upon which surface modifications can be made to create stealth or targeted nano-scale drug-delivery systems. Nanoparticles can also be formulated to address delivery route-specific needs. Nanoparticles have been formulated as microparticle agglomerates with cleavable in vivo chemical cross-linkages to achieve post-inhalation modulation of pulmonary drug-release rates [122].
Nano-scale drug-delivery systems based on phase separation are receiving considerable attention. Polymeric nanomicelles can be created from amphiphilic copolymers that self-assemble in aqueous solution, yielding highly efficient drug-delivery vehicles for hydrophobic and partially hydrophilic drugs, both transdermally and orally [123]. Nanoemulsions are a group of dispersed particles that can exist as either a water-in-oil or an oil-in-water form [124]. The interface between the dispersed droplet and the dispersion medium influences the partitioning and extraction of droplet contents. Low surface tension results in thermodynamically-stable particles with large surface areas [124].
Unlike nanoemulsions, which are characterized by two liquid phases, nanosuspensions are dispersions of solid drug particles in a liquid that are stabilized by surfactants. These simple formulations have the advantage of being amenable to many drugs and are especially useful for drugs with high crystal energies, which render them insoluble in both hydrophilic and lipophilic carriers and solvents [6]. This attribute is particularly pertinent to the modern drug discovery landscape, in which most candidates fit into the pockets of hydrophobic receptors and have limited solubility. Crystalline nano-suspensions not only overcome solubility issues but also afford pharmacokinetic benefits and high weight per loading volume [6].
Finally, external influences can also modulate drug delivery from nano-scale drug-delivery systems. Ultrasound [125] and magnetism [126] have been used to accumulate chemotherapeutic drugs selectively at tumor sites. Progress in the manipulation of traditional nano-scale drug-delivery systems, combined with the ability to control architectures, to create novel materials and to customize formulations, will enable engineers, scientists and physicians to exploit nanotechnology for advanced applications in drug delivery.
NANOSTRUCTURED MATERIALS AS TISSUE-ENGINEERING SCAFFOLDS
Introduction
The field of tissue engineering has witnessed great progress over the past few decades [127–129]. The underlying principle is that the dissociated cells have the ability to reassemble into structures that resemble the original tissue. In order to control and direct cell behavior, a defined biomimetic environment which surrounds the cells and promotes specific cell interactions is necessary. Critical environmental parameters include a scaffolding material, soluble factors and external physical stimulations [130]. The three-dimensional artificial scaffold is designed to not only provide the initial structural integrity for cells, but also direct cell differentiation and proliferation, ultimately leading to the assembly of functional tissue. Obviously, the chemical, physical and mechanical properties of the scaffolds are essential to the functional tissue regeneration.
Historical aspects
Both natural and synthetic materials have been evaluated as scaffolds for tissue engineering. Natural materials include collagen [131], silk protein [132], Matrigel [133], small intestinal submucosa (SIS) [134], agarose [135], alginate [136] and chitosan [137]. Although these materials show promise in tissue repair, critical issues regarding biocompatibility, mechanical properties and degradation cannot be neglected. On the other hand, synthetic materials can be created with improved biocompatibility, controlled degradation and tunable mechanical properties [138, 139]. Furthermore, bioactive moieties and functional groups can be readily incorporated into the polymeric system, giving rise to smart and responsive materials [140–144]. Synthetic polymers such as poly(lactic acid), poly(glycolic acid) and their co-polymers, poly(lactic acid-co-glycolic acid), poly(anhydride), poly(4-hydroxybutyrate), poly(urethane), polyphosphoesters and polyphosphazenes have been employed as degradable scaffolds for a variety of tissues and organs [145]. To this end, macroporous scaffolds were generated by melt-spinning, particulate leaching and the scaffolds were eroded by hydrolytic degradation [127]. Despite the expectations raised by tissue engineering, it has only achieved moderate success so far. Considering the fact that it takes 15 years or so for the human body to mature, generating a functional tissue that can rival nature’s design using isolated cells and artificial scaffolds under in vitro conditions can be a daunting task. The macroporous synthetic polymers listed above do not recapitulate the hierarchical organization and biological functions of the natural ECM [146–148]. Since tissue-engineering scaffolds are designed to serve as temporary, artificial ECM to accommodate cells and to guide 3D tissue formation, materials that most closely mimic the critical features of the natural ECM are the most promising candidates.
The need for the development of nanobiomaterials
The native ECM is a dynamic and hierarchically organized nanocomposite that not only provides mechanical support for embedded cells, but also interacts with cells and promotes and regulates cellular functions such as adhesion, migration, proliferation, differentiation and morphogenesis [146]. Extracellular molecules exhibit complex supramolecular structures and they are linked to one another by multiple binding domains that lead to a stable multifunctional matrix. In a typical connective tissue, structural protein fibers, such as collagen fibers and elastin fibers, have dimensions ranging from 10 to several hundred nanometers. The nano-scaled protein fibers entangle with each other to form a non-woven mesh that provides tensile strength and elasticity for the tissue. Adhesive proteins, such as fibronectin and laminin, which provide specific binding sites for cell adhesion, also exist as nano-scaled fibers in the ECM. The amorphous components of the ECM (such as GAGs) are both structural and functional. They serve as a space filler that resists compression, giving rise to a composite material with desired materials properties. They also play an important role in maintaining normal cellular activities [147–149]. In tendons (and possibly in most other tissues), proteoglycans tie adjoining collagen fibrils together and seem to have a definite role in guaranteeing the mechanical coupling of fibrils and ultimately for distributing the mechanical stress within the whole tissue (Fig. 2).
Figure 2.
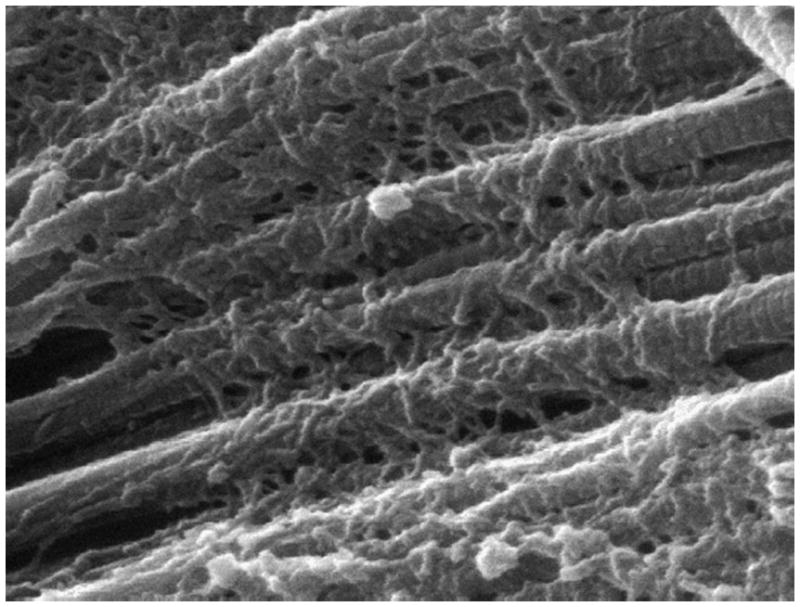
Scanning electron micrograph of rat tail tendon showing collagen fibrils and collagen-bound proteoglycans. The horizontal field of view spans 2 μm (reprinted from Ref. [149] with permission).
On the other hand, nature tends to assemble structures with the minimum amount of materials necessary. The natural ECM consists of less than 1% solid materials, yet they are mechanically robust and functionally diverse. Obviously, building hierarchically organized structures from molecular level up to macroscopic scale is the optimum solution. Nature modulates the mechanical properties of biological tissue by subtle adjustments of its composition with a perceivable alteration of its nano-scale organization.
Cells respond to the ECM through plasma membrane receptors that tie the matrix to the cell cytosketleton. The principle receptors for binding most ECM proteins, such as collagen, laminin and fibronectin, are the integrins. These proteins possess an arginine–glycine–aspartic (RGD) acid sequence that is recognized by integrins [148]. The anisotropic fibrillar architecture of natural ECMs has apparent consequences for cell behaviors. Because of a tight connection between the cytoskeleton and the ECM through cell-surface receptors, cells sense and respond to the mechanical properties of their environment by converting mechanical signals into chemical signals. Consequently, the biophysical properties of ECM influence various cell functions, including adhesion and migration. Moreover, the fibrillar structure of matrix components brings about adhesion ligand clustering, which has been demonstrated to alter cell behavior. Structural ECM features, such as fibrils and pores, are often of a size compatible with cellular processes involved in migration, which may influence the strategy by which cells migrate through ECM [150]. As a result, cells can respond not only to micrometer-scale topography, but also to nanometer-scale topography. Figure 3 shows a comparison between cells attached to macrofibrous and nanofibrous scaffolds. Studies on 2D surfaces with various types of nanotopography have been used to elucidate the mechanism by which cells respond to nano-scale features [151, 152]. Dably et al. investigated the fibroblast responses to 2D nano-islands produced by polymer demixing. The results show that the cells respond to the islands by broad gene up-regulation, notably in the areas of cell signaling, proliferation, cytoskeleton and production of ECM proteins [153]. It has been suggested that topography alone can be used to elicit different responses from the same cell phenotype [154].
Figure 3.
Comparison of chondrocytes on macrofibrous scaffold (PGA) (a) and nanofiberous scaffold (PLGA) (b) (images reprinted from Refs [130] and [163], respectively, with permission).
The unique biochemical composition, structural organization and viscoelastic properties of different types of tissues pose significant challenges for their regeneration using “universal” biodegradable polymers with macroscopic features. The attainment of the understanding of the hierachical tissue organization from moleuclar level up to the macroscopic scale will likely guide rational design of the synthetic ECM substitutes. In order for tissue engineering to be successful, one should strive to engineer biomaterials that recapitulate the hierarchical organization of natural ECM. With the advance of nanotechnology, critical insight on nano-scale organization of ECM components and how they interact with one another to organize a functional ECM are being accumulated. Atomic force microscopy has become particularly important for the study of biological systems. Its major advantage is that it can produce high-resolution topographic images in aqueous and physiologically relevant environments without the need to stain the specimen [155]. More importantly, when operated in the force mode, AFM can reveal molecular level information that is critical for the proper functioning of the native tissue [156].
Taking lessons from the nature, researchers have started to engineer artificial scaffolds that mimic the nano-scale morphological features of the natural ECM. To this end, both nanofibrous and nanocomposite scaffolds have been investigated.
Nanofibrous scaffolds by electrospinning
Electrospinning aims at producing ECM-mimetics that exhibit a physical structure similar to that of the fibrous proteins in native ECM albeit their completely different chemical compositions. The principle of electrospinning is to use an electric field to draw a polymer solution from an orifice to a collector, producing submicron polymer fiber mesh with fiber diameter of several hundred nanometers [157, 158]. High voltages, usually 10–20 kV, are applied to generate a sufficient surface charge to overcome the surface tension in a pendant drop of the polymer fluid, resulting in a 2D membrane.
Due to the simplicity of this method, electrospinning has been widely used by a variety of research groups to prepare nanofibrillar matrices. Many studies have been carried out in the application of polymer nanofibers in the tissue engineering of bone [159], blood vessel [160], cartilage [161–163], cardiac tissue [164], peripheral nerve system [165], ligaments [166], liver [167] and skin [162]. In most of these studies, biodegradable polymer materials such as PCL, PLA, PGA and PLGA were used. In addition, naturally occurring macromolecules, such as collagen [168], silk protein [169, 170], fibrinogen [171], elastin mimetic polypeptides [172–174], chitosan [175], dextran [176] and hyaluronic acid [177], have been fabricated into nanofibers by electrospinning. Nanofibers thus produced are expected to possess high axial strength combined with extreme flexibility [178]. The high surface area, high porosity and high spatial interconnectivity maximize the cell–ECM interaction and promote tissue regeneration. The porosity may play a significant role in the transport of nutrients and cell migration during tissue regeneration.
In general, the electrospun nanofibrous matrices are found to support cell adhesion and proliferation and cells tend to maintain their phenotypic shape and guided growth according to the fiber orientation. Xu et al. studied the potential of non-woven and aligned nanofiber matrices made from PLA-PCL and PLLA-PCL/collagen as a scaffold for blood-vessel regeneration [160]. A favorable interaction between the scaffold and human coronary artery smooth muscle cells (SMCs) was demonstrated. The SMCs attached and migrated along the axis of the aligned nanofibers and expressed a spindle-like contractile phenotype; the distribution and organization of smooth muscle cytoskeleton proteins inside SMCs were parallel to the direction of the nanofibers; the adhesion and proliferation rate of SMCs on the aligned nanofibrous scaffold was significantly improved than on the polymer films. The above results strongly suggest that this synthetic aligned matrix combines with the advantages of synthetic biodegradable polymers, nanometer-scale dimension mimicking the natural ECM and a defined architecture replicating the in vivo-like vascular structure, may represent an ideal tissue-engineering scaffold, especially for blood-vessel engineering.
Although robust nano-fibrous matrices can be easily generated by electrospinning, they lack the biological recognition signals [179] and cell-responsive domains that are conducive to tissue growth [140, 150]. Cells attach to these substrates through non-specific adhesion. These problems can be overcome by surface immobilization of natural materials on the synthetic scaffolds or simply by blending the two materials, which may improve the biocompatibility of the synthetic nanofibers while preserving the desired mechanical strength [180]. The applicability of this method has been demonstrated by culturing human coronary artery endothelial cells on electrospun collagen-blended poly(L-lactic acid)-co-poly(ε-caprolactone) nanofibers with diameters between 100 and 200 nm. The blending electrospinning technique shows potential in refining the composition of polymer nanofibers by adding various ingredients (e.g., growth factors) according to cell types to fabricate a tissue-engineering scaffold, particularly blood-vessel-engineering scaffold [181]. Layer-by-layer deposition provides an alternative strategy to fine tune the surface properties of polymeric scaffolds on the nanometer scale [182–184]. Recent studies demonstrate that one can easily control the cell behaviors on these self-assembled multilayers by simple manipulation of the deposition process [185–190]. However, polyelectrolyte multilayer studies are limited to planner surfaces and spherical particles. Given the versatility of this coating process, its combination with the electrospinning technique is likely to provide promising methods for the fabrication of bioactive nanofibers.
When natural proteins are used, the electrospinning process can lead to denaturing or disruption of their native structure, defeating the purpose of utilizing the natural polymers as conducive scaffolds for tissue engineering. Collagen nanofibrous matrices obtained by electrospinning from 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) solution have been shown to have relatively low cell adhesion in normal human keratinocytes and the fibers have to be coated with native ECM proteins in order to restore collagen’s biological and structural properties [191]. The denaturing problems can be overcome by electrospinning collagen in the presence of GAG [192]. To produce stable nanofibers from water soluble precursors, chemical cross-linking has to be introduced after the spinning process in order to prevent fiber dissolution in the aqueous media. In the case of electrospun collagen fibers, the nanofibrillar structures are mostly lost when the scaffold is partially degraded by the enzymes, exposing the cells to macroscopic features that are not originally intended [192]. Moreover, experimental parameters such as flow rate, applied electric field, collecting distance and composition of the starting solutions determined the morphology of the obtained fibers and often fibers with the same geometry cannot be prepared reproducibly. Studies of electrospinning collagen nanofibers have found that the structural properties of electrospun collagen vary with the tissue of origin, the isotope and the concentration of the collagen solution [168]. Finally, challenges exist in utilizing this technique to fabricate complex 3D scaffolds [193]. Cells cultured on the fibrous membrane can penetrate into the scaffolds only after the scaffolds have been degraded to a large extent. It is difficult to produce nanofibers with diameter smaller than 50 nm using electrospinning.
Nanofibrous scaffolds by self-assembly
Molecular self-assembly has recently emerged as a new approach in engineering artificial scaffolding materials that emulate natural ECM both structurally and functionally. Unlike electrospinning, this technology not only incorporates specific biological components of the ECM, but also mimics the process of ECM assembly from the bottom up [194]. Self-assembly involves concerted action of weak and non-covalent interactions that result in hierarchical structures [195].
Stupp and co-workers designed small, self-assembling molecules with a hydrophobic alkyl tail and a hydrophilic oliogopeptide head. The amphiphilicity of these molecules creates cylindrical nanofibers with well-defined diameters, which subsequently induce a liquid-to-gel transformation [196–198]. In one demonstration, the de novo designed peptide amphiphile contains five key structural features which are conducive to in situ directed self-assembly, reversible cross-linking, mineralization and cell adhesion. Upon acidification of the peptide solution below pH 4, cylindrical micelles form, in which the alkyl tails pack on the inside of the fiber and the peptide segments are displayed on the fiber surface. Upon cross-linking of the cysteine residues on adjacent molecules, the stable nanofibers are able to direct the mineralization of hydroxyapatite. The resultant mineral is aligned with the direction of the self-assembled fibrils [196]. These mineralized nanofibers resemble the lowest level of hierachical organization of bone; their mechanical properties, however, are likely to be inferior to those of the native bone. Since the fibrillar hydrogel formation is triggered at relatively low pH, it is not applicable for in situ encapsulation of cells. Therefore, no further study regarding the potential of this material as tissue engineering scaffolds for bone can be found.
A slight modification of the peptide amphiphilic architecture, nanofibrillar hydrogel formation is possible at physiological condition [199]. Neural progenitor cells were encapsulated in vitro within a three-dimensional network of nanofibers formed by self-assembly of peptide amphiphile molecules. The self-assembly is triggered by mixing cell suspensions in media with dilute aqueous solutions of the molecules and cells survive the growth of the nanofibers around them. These nanofibers were designed to present to cells the neurite-promoting laminin epitope IKVAV at nearly van der Waals density. Relative to laminin or soluble peptide, the artificial nanofiber scaffold induced very rapid differentiation of cells into neurons, while discouraging the development of astrocytes. This rapid selective differentiation is linked to the amplification of bioactive epitope presentation to cells by the nanofibers. This is a powerful example of directing the differentiation of neutral progenitor cells to a specific linage without the use of growth factors.
Zhang and co-workers developed another class of nanofibrillar hydrogels with very high water content through self-assembly of self-complementary amphiphilic peptides in physiological conditions [195, 200]. They are characterized by an alternating sequence of hydrophobic and hydrophilic residues, in which the hydrophilic residues, in turn, alternate between being positively and negatively charged. The alternation between polar and non-polar residues promotes the formation of a β-strand building block with hydrophobic and hydrophilic faces. The blocks stack with the charged side chains facing each other in a complementary “lego-like” manner. The self-assembly process can be triggered by adjusting the ionic strength and the solution pH. The resulting hydrogels contain nanofibers of 7–10 nm in diameter and 50–400-nm-sized nanopores, closely mimicking the morphological characteristics of natural ECM. The mechanical strength, however, is relatively low, which has been attributed to the ease with which cells migrate through the matrix. These types of nanofibrillar hydrogels have been shown to be non-cytotoxic. Although these peptides do not contain RGD sequence, they can organize cells in a 3D fashion. They have been shown to support a wide spectrum of mammalian cells [201], maintain the functions of differentiated neural cells [202] and chondrocytes [203], promote the differentiation of liver progenitor cells [204] and even foster functional return of vision [205]. The underlying mechanism for these observations, however, has not been elucidated.
A higher level of organizational control was demonstrated using a de novo designed peptide that forms a pH-responsive self-assembled β-hairpin [206]. The sequence consists of alternating hydrophobic (valine) and hydrophilic (lysine) residues of high β-sheet propensity flanking a four-residue segment incorporated to promote the formation of a type II′ β-turn, as in (VK)4-V-DPPT-(KV)4. This system is unique in that a nanofibrillar network forms only after intramolecular folding. The network is branched with fibrils 3 nm in diameter and can extend for over several hundred nanometers in length [207]. Due to the non-covalent nature of the hydrogel, it is responsive to external stimuli, including addition of salt, pH and temperature changes and mechanical shear [208], making it an ideal matrix for in situ cell encapsulation. The hydrogel construct has been shown to be non-toxic and cytocompatible [209]. The application of this type of peptides as tissue-engineering scaffolds has not fully been explored.
To further develop the self-assembled nanofibrous scaffolds for tissue-engineering applications, several technical hurdles need to be addressed. First, the solid-phase peptide synthesis cannot be readily scaled up, thereby limiting their applicability. Secondly, their degradation profiles have not been systematically addressed. Unlike the electrospun nanofibers that are continuous, the self-assembled nanofibers are fragmented and may be susceptible to endocytosis [210]. Further, the peptide backbone can be easily degraded by a variety of enzymes, making it difficult to adjust their degradability accordingly. Due to the dynamic nature of the self-assembly, uncontrolled matrix erosion is possible. Finally, most of these hydrogels are mechanically weak and they do not effectively sustain and transfer mechanical loadings to the cells.
Nanocomposite scaffolds for hard-tissue engineering
In the field of hard-tissue engineering, traditional bio-inert materials such as metals, alloys, ceramics and composites have been widely used, with the goal of achieving a suitable combination of physical properties to match those of the replaced tissue with a minimal toxic response in the host [211]. However, most implanted materials elicit an unnatural healing process that eventually leads to fibrous capsule formation around the implant [212]. The ideal situation for hard tissue regeneration is to utilize “bioactive” materials that stimulate active tissue regeneration. The mechanism of bonding of bioglass to living tissue was shown to involve multiple reaction steps that ultimately led to osteoblasts colonization and proliferation and differentiation of the cells to form new bone that had a mechanically strong bonding to the implant surface [213]. There are opportunities for the development of tissue engineered nanocomposites that emulate the structural, compositional and biological characteristics of natural bone. Most of the nanocomposites studied so far for bone regeneration are based primarily on nano-hydroxyapatite (HA) and collagen [214, 215]. In one example, a 3D nanocomposite matrix based on nano-HA/collagen and PLA was used to culture osteoblasts [216]. Due to its compositional and structural similarity to natural bone, the nanocomposite was shown to support osteoblast adhesion, proliferation and migration. In another demonstration, nano-HA/collagen/alginate was used as the matrix with the goal of enhancing the composite strength and mimicking the GAG components found in the natural bone. Fibroblasts were co-cultured with osteoblasts in the matrix and were found to attach and proliferate well on the scaffold [217].
CONCLUSIONS
Drug delivery is becoming an increasingly important aspect of medicine, as more potent and specific drugs are being developed – particularly with the increased understanding of disease pathways generated by the Human Genome Project. New opportunities to prevent and to treat diseases are emerging. Many biomaterials, primarily polymer- or lipid-based, can be used to this end, offering extensive chemical diversity and the potential for further modification. Drug delivery is no longer restricted to small-molecule drugs. The inherent limitations of therapeutic biomacromolecules, such as proteins and nucleic acids, are being addressed through rationally-designed delivery vehicles.
Drug delivery has matured from its original goal of prolonging the duration of drug release; it now involves customized systems that are designed to achieve specific spatial and temporal control; future generations of drug-delivery systems will incorporate “smart” biosensing functionalities that will enable unaided in vivo feedback control.
Tissue engineering is an exciting multidisciplinary research that promises to bring exciting discoveries over time. Advances in characterization of materials and tissues at nanometer scale will likely stimulate rational material design. For example, recent studies of single molecule biomechanics using AFM [218, 219] demonstrate that biological assemblies are dynamic entities whose transitions may include the disassembly and re-assembly of some of the subunits to the accomplishment of local and global conformation changes. The presence of folded domains held together by secondary forces in natural polymers give rise to combined strength, toughness and elasticity. The folded domain structures also provide the natural ECM molecules multifaceted biological functions. The development of complex nanobiomaterials that resemble the native ECM is of critical importance in achieving this goal.
Footnotes
This paper is part of the Special Issue devoted to Nanobiomaterials (Guest Editors: Vasif Hasırcıand Karel Petrak). The other papers of this Special Issue have been published in J. Biomater. Sci. Polymer Edn, Volume 17, No. 11 (2006).
References
- 1.Kayser O, Lemke A, Hernandez-Trejo N. Curr Pharm Biotechnol. 2005;6:3. doi: 10.2174/1389201053167158. [DOI] [PubMed] [Google Scholar]
- 2.Anderson DG, Burdick JA, Langer R. Science. 2004;305:1923. doi: 10.1126/science.1099987. [DOI] [PubMed] [Google Scholar]
- 3.LaVan DA, McGuire T, Langer R. Nature Biotechnol. 2003;21:1184. doi: 10.1038/nbt876. [DOI] [PubMed] [Google Scholar]
- 4.Qiu LY, Bae YH. Pharm Res. 2006;23:1. doi: 10.1007/s11095-005-9046-2. [DOI] [PubMed] [Google Scholar]
- 5.Svenson S, Tomalia DA. Adv Drug Deliv Rev. 2005;57:2106. doi: 10.1016/j.addr.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 6.Rabinow BE. Nature Rev Drug Discov. 2004;3:785. doi: 10.1038/nrd1494. [DOI] [PubMed] [Google Scholar]
- 7.Vinogradov SV, Bronich TK, Kabanov AV. Adv Drug Deliv Rev. 2002;54:135. doi: 10.1016/s0169-409x(01)00245-9. [DOI] [PubMed] [Google Scholar]
- 8.Ishida O, Maruyama K, Sasaki K, Iwatsuru M. Int J Pharm. 1999;190:49. doi: 10.1016/s0378-5173(99)00256-2. [DOI] [PubMed] [Google Scholar]
- 9.Kong G, Braun RD, Dewhirst MW. Cancer Res. 2000;60:4440. [PubMed] [Google Scholar]
- 10.Hawley AE, Davis SS, Illum L. Adv Drug Deliv Rev. 1995;17:129. [Google Scholar]
- 11.Stolnik S, Illum L, Davis SS. Adv Drug Deliv Rev. 1995;16:195. [Google Scholar]
- 12.Ogawara K, Yoshida M, Furumoto K, Takakura Y, Hashida M, Higaki K, Kimura T. J Drug Target. 1999;7:213. doi: 10.3109/10611869909085504. [DOI] [PubMed] [Google Scholar]
- 13.Hu E. Annu Rev Mater Res. 2004;34:151. [Google Scholar]
- 14.Katz E, Willner I. Angew Chem Int Edn Engl. 2004;43:6042. doi: 10.1002/anie.200400651. [DOI] [PubMed] [Google Scholar]
- 15.Allen TM. Nature Rev Drug Discov. 2002;2:750. [Google Scholar]
- 16.Panyam J, Labhasetwar V. Adv Drug Deliv Rev. 2003;55:329. doi: 10.1016/s0169-409x(02)00228-4. [DOI] [PubMed] [Google Scholar]
- 17.Langer R. Nature. 1998;392:5. [PubMed] [Google Scholar]
- 18.Duncan R. Nature Rev Drug Discov. 2003;2:347. doi: 10.1038/nrd1088. [DOI] [PubMed] [Google Scholar]
- 19.Brocchini S, Duncan R. In: Encyclopaedia of Controlled Drug Delivery. Mathiowitz E, editor. Wiley; New York, NY: 1999. p. 786. [Google Scholar]
- 20.Peng T, Cheng Y-L. Polymer. 2001;42:2091. [Google Scholar]
- 21.Duncan R, Spreafico F. Clin Pharmacokinet. 1994;27:290. doi: 10.2165/00003088-199427040-00004. [DOI] [PubMed] [Google Scholar]
- 22.Seymour LW, Duncan R, Strohalm J, Kopecek J. J Biomed Mater Res. 1987;21:1341. doi: 10.1002/jbm.820211106. [DOI] [PubMed] [Google Scholar]
- 23.Wissing SA, Kayser O, Muller RH. Adv Drug Deliv Rev. 2004;56:1257. doi: 10.1016/j.addr.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y-X, Robertson JL, Spillman WB, Claus RO. Pharm Res. 2004;21:1362. doi: 10.1023/b:pham.0000036909.41843.18. [DOI] [PubMed] [Google Scholar]
- 25.Gillies ER, Frechet JMJ. J Am Chem Soc. 2002;124:14137. doi: 10.1021/ja028100n. [DOI] [PubMed] [Google Scholar]
- 26.Tomlinson R, Klee M, Garrett S, Heller J, Duncan R, Brocchini S. Macromolecules. 2002;35:473. [Google Scholar]
- 27.Desai MP, Labhasetwar V, Amidon GL, Levy RJ. Pharm Res. 1996;13:1838. doi: 10.1023/a:1016085108889. [DOI] [PubMed] [Google Scholar]
- 28.Roney C, Kulkarni P, Arora V, Antich P, Bonte F, Wu A, Mallikarjuana NN, Manohar S, Liang H-F, Kulkarni AR. J Control Rel. 2005;108:193. doi: 10.1016/j.jconrel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 29.Bilati U, Allemann E, Doelker E. J Microencapsul. 2005;22:205. doi: 10.1080/02652040400026442. [DOI] [PubMed] [Google Scholar]
- 30.Hawker CJ, Wooley KL. Science. 2005;309:1200. doi: 10.1126/science.1109778. [DOI] [PubMed] [Google Scholar]
- 31.Ueno Y, Futagawa H, Takagi Y, Ueno A, Mizushima Y. J Control Rel. 2005;103:93. doi: 10.1016/j.jconrel.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 32.Liu T-Y, Chen S-Y, Liu D-M, Liou S-C. J Control Rel. 2005;107:112. doi: 10.1016/j.jconrel.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 33.Bodnar M, Hartmann JF, Borbely J. Biomacromolecules. 2005;6:2521. doi: 10.1021/bm0502258. [DOI] [PubMed] [Google Scholar]
- 34.Piddubnyak V, Kurcok P, Matuszowicz A, Glowala M, Fiszer-Kierzkowska A, Jedlinski Z, Juzwa M, Krawczyk Z. Biomaterials. 2004;25:5271. doi: 10.1016/j.biomaterials.2003.12.029. [DOI] [PubMed] [Google Scholar]
- 35.Chen J-F, Ding H-M, Wang J-X, Shao L. Biomaterials. 2004;25:723. doi: 10.1016/s0142-9612(03)00566-0. [DOI] [PubMed] [Google Scholar]
- 36.Nasongkla N, Shuai X, Ai H, Weinberg BD, Pink J, Boothman DA, Gao JM. Angew Chem Int Edn Engl. 2004;43:6323. doi: 10.1002/anie.200460800. [DOI] [PubMed] [Google Scholar]
- 37.Farokhzad OC, Cheng JJ, Teply BA, Sherifi I, Jon S, Kantoff PW, Richie JP, Langer R. Proc Natl Acad Sci USA. 2006;103:6315. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan D, Turner JL, Wooley KL. Chem Commun. 2003:2400. doi: 10.1039/b307878g. [DOI] [PubMed] [Google Scholar]
- 39.Lo CL, Lin KM, Hsiue GH. J Control Rel. 2005;104:477. doi: 10.1016/j.jconrel.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 40.Win KY, Feng SS. Biomaterials. 2005;26:2713. doi: 10.1016/j.biomaterials.2004.07.050. [DOI] [PubMed] [Google Scholar]
- 41.Veronese FM, Pasut G. Drug Discov Today. 2005;10:1451. doi: 10.1016/S1359-6446(05)03575-0. [DOI] [PubMed] [Google Scholar]
- 42.Gref R, Minamitake Y, Peracchia MT, Trubetskoy V, Torchilin V, Langer R. Science. 1994;263:1600. doi: 10.1126/science.8128245. [DOI] [PubMed] [Google Scholar]
- 43.Yokoyama M. J Artif Organs. 2005;8:77. doi: 10.1007/s10047-005-0285-0. [DOI] [PubMed] [Google Scholar]
- 44.Mayer C. Int J Artif Organs. 2005;28:1163. doi: 10.1177/039139880502801114. [DOI] [PubMed] [Google Scholar]
- 45.Krol S, Diaspro A, Magrassi R, Ballario P, Grimaldi B, Filetici P, Ornaghi P, Ramoino P, Gliozzi A. IEEE Trans Nanobiosci. 2004;3:32. doi: 10.1109/tnb.2004.824279. [DOI] [PubMed] [Google Scholar]
- 46.Burger KNJ, Staffhorst RWHM, de Vijlder HC, Velinova MJ, Bomans PH, Frederik PM, de Kruijff B. Nature Med. 2002;8:81. doi: 10.1038/nm0102-81. [DOI] [PubMed] [Google Scholar]
- 47.de Kroon AIPM, Staffhorst RWHM, de Kruijff B, Burger KNJ. Methods Enzymol. 1995;391:118. doi: 10.1016/S0076-6879(05)91006-9. [DOI] [PubMed] [Google Scholar]
- 48.Ruysschaert T, Germain M, Gomes JF, Fournier D, Sukhorukov GB, Meier WMW. IEEE Trans Nanobiosci. 2004;3:49. doi: 10.1109/tnb.2004.824273. [DOI] [PubMed] [Google Scholar]
- 49.Ai H, Pink J, Shuai X, Boothman D, Gao J. J Biomed Mater Res. 2005;73A:303. doi: 10.1002/jbm.a.30289. [DOI] [PubMed] [Google Scholar]
- 50.Peyratout CS, Dahne L. Angew Chem Int Edn Engl. 2004;43:3762. doi: 10.1002/anie.200300568. [DOI] [PubMed] [Google Scholar]
- 51.Dahne L, Leporatti S, Donath E, Mohwald H. J Am Chem Soc. 2001;123:5431. doi: 10.1021/ja002911e. [DOI] [PubMed] [Google Scholar]
- 52.Shchukin DG, Patel AA, Sukhorukov GB, Lvov YM. J Am Chem Soc. 2004;126:3374. doi: 10.1021/ja036952x. [DOI] [PubMed] [Google Scholar]
- 53.Tiourina OP, Antipov AA, Sukhorukov GB, Larionova NI, Lvov Y, Möhwald H. Macromol Biosci. 2001;1:209. [Google Scholar]
- 54.Kickhoefer VA, Garcia Y, Mikyas Y, Johansson E, Zhou JC, Raval-Fernandes S, Minoofar P, Zink JI, Dunn B, Stewart PL, Rome LH. Proc Natl Acad Sci USA. 2005;102:4348. doi: 10.1073/pnas.0500929102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haynie DT, Palath N, Liu Y, Li BY, Pargaonkar N. Langmuir. 2005;21:1136. doi: 10.1021/la047833d. [DOI] [PubMed] [Google Scholar]
- 56.Sukhishvili SA, Granick S. Macromolecules. 2002;35:301. [Google Scholar]
- 57.Zelikin AN, Quinn JF, Caruso F. Biomacromolecules. 2006;7:27. doi: 10.1021/bm050832v. [DOI] [PubMed] [Google Scholar]
- 58.Martin CR, Kohli P. Nature Rev Drug Discov. 2003;2:29. doi: 10.1038/nrd988. [DOI] [PubMed] [Google Scholar]
- 59.Lee SB, Mitchell DT, Trofin L, Nevanen TK, Soderlund H, Martin CR. Science. 2002;296:2198. doi: 10.1126/science.1071396. [DOI] [PubMed] [Google Scholar]
- 60.Mitchell DT, Lee SB, Trofin L, Li N, Nevanen TK, Soderlund H, Martin CR. J Am Chem Soc. 2002;124:11864. doi: 10.1021/ja027247b. [DOI] [PubMed] [Google Scholar]
- 61.Martin CR. Science. 1994;266:1961. doi: 10.1126/science.266.5193.1961. [DOI] [PubMed] [Google Scholar]
- 62.Cepak VM, Hulteen JC, Che GL, Jirage KB, Lakshmi BB, Fisher ER, Martin CR. J Mater Res. 1998;13:3070. [Google Scholar]
- 63.Nicewarner-Pena SR, Freeman RG, Reiss BD, He L, Pena DJ, Walton ID, Cromer R, Keating CD, Natan MJ. Science. 2001;294:137. doi: 10.1126/science.294.5540.137. [DOI] [PubMed] [Google Scholar]
- 64.Cai D, Mataraza JM, Qin ZH, Huang Z, Huang J, Chiles TC, Carnahan D, Kempa K, Ren Z. Nature Methods. 2005;2:449. doi: 10.1038/nmeth761. [DOI] [PubMed] [Google Scholar]
- 65.Pantarotto D, Briand JP, Prato M, Bianco A. Chem Commun. 2004:16. doi: 10.1039/b311254c. [DOI] [PubMed] [Google Scholar]
- 66.ShiKam NW, Jessop TC, Wender PA, Dai H. J Am Chem Soc. 2004;126:6850. doi: 10.1021/ja0486059. [DOI] [PubMed] [Google Scholar]
- 67.Scherer F, Anton M, Schillinger U, Henke J, Bergemann C, Kruger A, Gansbacher B, Plank C. Gene Ther. 2002;9:102. doi: 10.1038/sj.gt.3301624. [DOI] [PubMed] [Google Scholar]
- 68.Obataya I, Nakamura C, Han SW, Nakamura N, Miyake J. Nano Lett. 2005;5:27. doi: 10.1021/nl0485399. [DOI] [PubMed] [Google Scholar]
- 69.Morimoto N, Endo T, Ohtomi M, Iwasaki Y, Akiyoshi K. Macromol Biosci. 2005;5:710. doi: 10.1002/mabi.200500051. [DOI] [PubMed] [Google Scholar]
- 70.Shin Y, Chang JH, Liu J, Williford R, Shin Y-K, Exarhos GJ. J Control Rel. 2001;73:1. doi: 10.1016/s0168-3659(01)00247-4. [DOI] [PubMed] [Google Scholar]
- 71.Peppas NA, Bures P, Leobandung W, Ichikawa H. Eur J Pharm Biopharm. 2000;50:27. doi: 10.1016/s0939-6411(00)00090-4. [DOI] [PubMed] [Google Scholar]
- 72.McAllister K, Sazani P, Adam M, Cho MJ, Rubinstein M, Samulski RJ, DeSimone JM. J Am Chem Soc. 2002;124:15198. doi: 10.1021/ja027759q. [DOI] [PubMed] [Google Scholar]
- 73.Vinogradov SV, Zeman AD, Batrakova EV, Kabanov AV. J Control Rel. 2005;107:143. doi: 10.1016/j.jconrel.2005.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.De Geest BG, Stubbe BG, Jonas AM, Van Thienen T, Hinrichs WL, Demeester J, De Smedt SC. Biomacromolecules. 2006;7:373. doi: 10.1021/bm0507296. [DOI] [PubMed] [Google Scholar]
- 75.Kiser PF, Wilson G, Needham D. Nature. 1998;394:459. doi: 10.1038/28822. [DOI] [PubMed] [Google Scholar]
- 76.Kraft ML, Moore JS. J Am Chem Soc. 2001;123:12921. doi: 10.1021/ja016975g. [DOI] [PubMed] [Google Scholar]
- 77.De Geest BG, Déjugnat C, Sukhorukov GB, Braeckmans K, De Smedt SC, Demeester J. Adv Mater. 2005;17:2357. [Google Scholar]
- 78.Missirlis D, Tirelli N, Hubbell JA. Langmuir. 2005;21:2605. doi: 10.1021/la047367s. [DOI] [PubMed] [Google Scholar]
- 79.Lee CC, MacKay JA, Frechet JMJ, Szoka FC. Nature Biotechnol. 2005;23:1517. doi: 10.1038/nbt1171. [DOI] [PubMed] [Google Scholar]
- 80.Tomalia DA. Macromol Symp. 1996;101:243. [Google Scholar]
- 81.Hawker CJ, Frechet JMJ. J Am Chem Soc. 1990;112:7638. [Google Scholar]
- 82.Maraval V, Pyzowski J, Caminade A-M, Majoral J-P. J Org Chem. 2003;68:6043. doi: 10.1021/jo0344438. [DOI] [PubMed] [Google Scholar]
- 83.Wu P, Feldman AK, Nugent AK, Hawker CJ, Scheel A, Voit B, Pyun J, Fréchet JMJ, Sharpless KB, Fokin VV. Angew Chem Int Edn Engl. 2004;43:3928. doi: 10.1002/anie.200454078. [DOI] [PubMed] [Google Scholar]
- 84.Beezer AE, King ASH, Martin IK, Mitchel JC, Twyman LJ, Wain CF. Tetrahedron. 2003;59:3873. [Google Scholar]
- 85.Kolhe P, Misra E, Kannan RM, Kannan S, Lieh-Lai M. Int J Pharm. 2003;259:143. doi: 10.1016/s0378-5173(03)00225-4. [DOI] [PubMed] [Google Scholar]
- 86.Malik N, Evagorou EG, Duncan R. Anti-Cancer Drugs. 1999;10:767. [PubMed] [Google Scholar]
- 87.Kojima C, Kono K, Maruyama K, Takagishi T. Bioconjug Chem. 2000;11:910. doi: 10.1021/bc0000583. [DOI] [PubMed] [Google Scholar]
- 88.Bhadra D, Bhadra S, Jain S, Jain NK. Int J Pharm. 2003;257:111. doi: 10.1016/s0378-5173(03)00132-7. [DOI] [PubMed] [Google Scholar]
- 89.El-Sayed M, Rhodes CA, Ginski M, Ghandehari H. Int J Pharm. 2003;265:151. doi: 10.1016/s0378-5173(03)00391-0. [DOI] [PubMed] [Google Scholar]
- 90.Devarakonda B, Hill RA, de Villiers MM. Int J Pharm. 2004;284:133. doi: 10.1016/j.ijpharm.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 91.Jevprasesphant R, Penny J, Jalal R, Attwood D, McKeown NB, D’Emanuele A. Int J Pharm. 2003;252:263. doi: 10.1016/s0378-5173(02)00623-3. [DOI] [PubMed] [Google Scholar]
- 92.Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. Biomaterials. 2003;24:1121. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- 93.Duncan R, Izzo L. Adv Drug Deliv Rev. 2005;57:2215. doi: 10.1016/j.addr.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 94.Ambade AV, Savariar EN, Thayumanavan S. Mol Pharm. 2005;2:264. doi: 10.1021/mp050020d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Khopade AJ, Caruso F. Biomacromolecules. 2002;3:1154. doi: 10.1021/bm025562k. [DOI] [PubMed] [Google Scholar]
- 96.Boas U, Heegaard PMH. Chem Soc Rev. 2004;33:43. doi: 10.1039/b309043b. [DOI] [PubMed] [Google Scholar]
- 97.Chauhan AS, Sridevi S, Chalasani KB, Jain AK, Jain SK, Jain NK, Diwan PV. J Control Rel. 2003;90:335. doi: 10.1016/s0168-3659(03)00200-1. [DOI] [PubMed] [Google Scholar]
- 98.Dincer S, Turk M, Piskin E. Gene Ther. 2005;12:S139. doi: 10.1038/sj.gt.3302628. [DOI] [PubMed] [Google Scholar]
- 99.Luo D, Saltzman WM. Nature Biotechnol. 2000;18:33. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- 100.Ruponen M, Honkakoski P, Ronkko S, Pelkonen J, Tammi M, Urtti A. J Control Rel. 2003;93:213. doi: 10.1016/j.jconrel.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 101.Remy J-S, Abdallah B, Zanta MA, Boussif O, Behr J-P, Demeneix B. Adv Drug Deliv Rev. 1998;30:85. doi: 10.1016/s0169-409x(97)00109-9. [DOI] [PubMed] [Google Scholar]
- 102.Brunner S, Furtbauer E, Sauer T, Kursa M, Wagner E. Mol Ther. 2002;5:80. doi: 10.1006/mthe.2001.0509. [DOI] [PubMed] [Google Scholar]
- 103.Wightman L, Kircheis R, Rössler V, Carotta S, Ruzicka R, Kursa M, Wagner E. J Gene Med. 2001;3:362. doi: 10.1002/jgm.187. [DOI] [PubMed] [Google Scholar]
- 104.Ferruti P, Marchisio MA, Duncan R. Macromol Rapid Commun. 2002;23:332. [Google Scholar]
- 105.Stayton PS, Hoffman AS, Murthy N, Lackey C, Cheung C, Tan P, Klumb LA, Chilkoti A, Wilbur FS, Press OW. J Control Rel. 2000;65:203. doi: 10.1016/s0168-3659(99)00236-9. [DOI] [PubMed] [Google Scholar]
- 106.Putnam D, Gentry CA, Pack DW, Langer R. Proc Natl Acad Sci USA. 2001;98:1200. doi: 10.1073/pnas.031577698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Akinc A, Anderson DG, Lynn DM, Langer R. Bioconjug Chem. 2003;14:979. doi: 10.1021/bc034067y. [DOI] [PubMed] [Google Scholar]
- 108.Kumar MN, Mohapatra SS, Kong X, Jena PK, Bakowsky U, Lehr CM. J Nanosci Nanotechnol. 2004;4:990. doi: 10.1166/jnn.2004.130. [DOI] [PubMed] [Google Scholar]
- 109.Lee H, Jeong JH, Park TG. J Control Rel. 2002;79:283. doi: 10.1016/s0168-3659(02)00002-0. [DOI] [PubMed] [Google Scholar]
- 110.Zhang J-S, Li S, Huang L. Methods Enzymol. 2003;373:332. doi: 10.1016/S0076-6879(03)73021-3. [DOI] [PubMed] [Google Scholar]
- 111.Ravi Kumar M, Hellermann G, Lockey RF, Mohapatra SS. Expert Opin Biol Ther. 2004;4:1213. doi: 10.1517/14712598.4.8.1213. [DOI] [PubMed] [Google Scholar]
- 112.Fu H, Hu Y, McNelis T, Hollinger JO. J Biomed Mater Res. 2005;74A:40. doi: 10.1002/jbm.a.30267. [DOI] [PubMed] [Google Scholar]
- 113.Kushibiki T, Tabata Y. J Biomater Sci Polymer Edn. 2005;16:1447. doi: 10.1163/156856205774472326. [DOI] [PubMed] [Google Scholar]
- 114.Chowdhury EH, Akaike T. Biotechnol Bioeng. 2005;90:414. doi: 10.1002/bit.20398. [DOI] [PubMed] [Google Scholar]
- 115.Lesinski GB, Sharma S, Varker KA, Sinha P, Ferrari M, Carson WE., 3rd Biomed Microdevices. 2005;7:71. doi: 10.1007/s10544-005-6174-8. [DOI] [PubMed] [Google Scholar]
- 116.Venugopal J, Ramakrishna S. Appl Biochem Biotechnol. 2005;125:147. doi: 10.1385/abab:125:3:147. [DOI] [PubMed] [Google Scholar]
- 117.Haynie DT, Zhang L, Rudra JS, Zhao WH, Zhong Y, Palath N. Biomacromolecules. 2005;6:2895. doi: 10.1021/bm050525p. [DOI] [PubMed] [Google Scholar]
- 118.Djalali R, Samson J, Matsui H. J Am Chem Soc. 2004;126:7935. doi: 10.1021/ja0319691. [DOI] [PubMed] [Google Scholar]
- 119.Yamaguchi Y, Nagasawa T, Nakamura N, Takenaga M, Mizoguchi M, Kawai S-I, Mizushima Y, Igarashi R. J Control Rel. 2005;104:29. doi: 10.1016/j.jconrel.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 120.Yoo HS, Park TG. J Control Rel. 2004;100:247. doi: 10.1016/j.jconrel.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 121.Zahr AS, de Villiers M, Pishko MV. Langmuir. 2005;21:403. doi: 10.1021/la0478595. [DOI] [PubMed] [Google Scholar]
- 122.Karathanasis E, Ayyagari AL, Bhavane R, Bellamkonda RV, Annapragada AV. J Control Rel. 2005;103:159. doi: 10.1016/j.jconrel.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 123.Kumar R, Chen MH, Parmar VS, Samuelson LA, Kumar J, Nicolosi R, Yoganathan S, Watterson AC. J Am Chem Soc. 2004;126:10640. doi: 10.1021/ja039651w. [DOI] [PubMed] [Google Scholar]
- 124.Sarker DK. Curr Drug Deliv. 2005;2:297. doi: 10.2174/156720105774370267. [DOI] [PubMed] [Google Scholar]
- 125.Larina IV, Evers BM, Ashitkov TV, Bartels C, Larin KV, Esenaliev RO. Technol Cancer Res Treat. 2005;4:217. doi: 10.1177/153303460500400211. [DOI] [PubMed] [Google Scholar]
- 126.Hafeli UO. Int J Pharm. 2004;277:19. doi: 10.1016/j.ijpharm.2003.03.002. [DOI] [PubMed] [Google Scholar]
- 127.Langer R, Vacanti JP. Science. 1993;260:920. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 128.Cima LG, Langer R. Chem Eng Prog. 1993;6:46. [Google Scholar]
- 129.Langer R, Tirrell DA. Nature. 2004;428:487. doi: 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]
- 130.Freed LE, Vunjak-Novakovic G. Adv Drug Deliv Rev. 1998;33:15. doi: 10.1016/s0169-409x(98)00017-9. [DOI] [PubMed] [Google Scholar]
- 131.Freyman TM, Yannas IV, Yokoo R, Gibson LJ. Biomaterials. 2001;22:2883. doi: 10.1016/s0142-9612(01)00034-5. [DOI] [PubMed] [Google Scholar]
- 132.Wang YZ, Kim UJ, Blasioli DJ, Kim HJ, Kaplan DL. Biomaterials. 2005;26:7082. doi: 10.1016/j.biomaterials.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 133.Nahmias Y, Schwartz RE, Verfaillie CM, Odde DJ. Biotechnol Bioeng. 2005;92:129. doi: 10.1002/bit.20585. [DOI] [PubMed] [Google Scholar]
- 134.Badylak SF, Record R, Lindberg K, Hodde J, Park K. J Biomater Sci Polymer Edn. 1998;9:863. doi: 10.1163/156856298x00208. [DOI] [PubMed] [Google Scholar]
- 135.Mauck RL, Yuan X, Tuan RS. Osteoarthritis Cartil. 2006;14:179. doi: 10.1016/j.joca.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 136.Marijnissen W, van Osch G, Aigner J, van der Veen SW, Hollander AP, Verwoerd-Verhoef HL, Verhaar JAN. Biomaterials. 2002;23:1511. doi: 10.1016/s0142-9612(01)00281-2. [DOI] [PubMed] [Google Scholar]
- 137.Ciardelli G, Chiono V. Macromol Biosci. 2006;6:13. doi: 10.1002/mabi.200500151. [DOI] [PubMed] [Google Scholar]
- 138.Wang Y, Ameer GA, Sheppard BJ, Langer R. Nature Biotechnol. 2002;20:602. doi: 10.1038/nbt0602-602. [DOI] [PubMed] [Google Scholar]
- 139.Lendlein A, Langer R. Science. 2002;296:1673. doi: 10.1126/science.1066102. [DOI] [PubMed] [Google Scholar]
- 140.Lutolf MP, Raeber GP, Zisch AH, Tirelli N, Hubbell JA. Adv Mater. 2003;15:888. [Google Scholar]
- 141.Barrera DA, Zylstra E, Lansbury PT, Langer R. J Am Chem Soc. 1993;115:11010. [Google Scholar]
- 142.Cook AD, Hrkach JS, Gao NN, Johnson IM, Pajvani UB, Cannizzaro SM, Langer R. J Biomed Mater Res. 1997;35:513. doi: 10.1002/(sici)1097-4636(19970615)35:4<513::aid-jbm11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 143.Parrish B, Emrick T. Macromolecules. 2004;37:5863. [Google Scholar]
- 144.Parrish B, Breitenkamp RB, Emrick T. J Am Chem Soc. 2005;127:7404. doi: 10.1021/ja050310n. [DOI] [PubMed] [Google Scholar]
- 145.Ma PX. Mater Today. 2004;7:30. [Google Scholar]
- 146.Zagris N. Micron. 2001;32:427. doi: 10.1016/s0968-4328(00)00011-1. [DOI] [PubMed] [Google Scholar]
- 147.Aumailley M, Gayraud B. J Mol Med. 1998;76:253. doi: 10.1007/s001090050215. [DOI] [PubMed] [Google Scholar]
- 148.Gullberg D, Ekblom P. Int J Dev Biol. 1995;39:845. [PubMed] [Google Scholar]
- 149.Ottani V, Martini D, Franchi M, Ruggeri A, Raspanti M. Micron. 2002;33:587. doi: 10.1016/s0968-4328(02)00033-1. [DOI] [PubMed] [Google Scholar]
- 150.Lutolf MP, Hubbell JA. Nature Biotechnol. 2005;23:47. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 151.Diehl KA, Foley JD, Nealey PF, Murphy CJ. J Biomed Mater Res. 2005;75A:603. doi: 10.1002/jbm.a.30467. [DOI] [PubMed] [Google Scholar]
- 152.Curtis A. IEEE Trans Nanobiosci. 2004;3:293. doi: 10.1109/tnb.2004.837898. [DOI] [PubMed] [Google Scholar]
- 153.Dalby MJ, Yarwood SJ, Riehle MO, Johnstone JH, Affrossman S, Curtis ASG. Exp Cell Res. 2002;276:1. doi: 10.1006/excr.2002.5498. [DOI] [PubMed] [Google Scholar]
- 154.Stevens MM, George JH. Science. 2005;310:1135. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- 155.Horber JKH, Miles MJ. Science. 2003;302:1002. doi: 10.1126/science.1067410. [DOI] [PubMed] [Google Scholar]
- 156.Fantner GE, Hassenkam T, Kindt JH, Weaver JC, Birkedal H, Pechenik L, Cutroni JA, Cidadem GAG, Stucky GD, Morse DE, Hansma PK. Nature Mater. 2005;4:612. doi: 10.1038/nmat1428. [DOI] [PubMed] [Google Scholar]
- 157.Formhals A. 1,975,504. US Patent. 1934
- 158.Reneker DH, Chun I. Nanotechnology. 1996;7:216. [Google Scholar]
- 159.Yoshimoto H, Shin YM, Terai H, Vacanti JP. Biomaterials. 2003;24:2077. doi: 10.1016/s0142-9612(02)00635-x. [DOI] [PubMed] [Google Scholar]
- 160.Xu CY, Inai R, Kotaki M, Ramakrishna S. Biomaterials. 2004;25:877. doi: 10.1016/s0142-9612(03)00593-3. [DOI] [PubMed] [Google Scholar]
- 161.Li WJ, Tuli R, Okafor C, Derfoul A, Danielson KG, Hall DJ, Tuan RS. Biomaterials. 2005;26:599. doi: 10.1016/j.biomaterials.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 162.Li WJ, Laurencin CT, Caterson EJ, Tuan RS, Ko FK. J Biomed Mater Res. 2002;60:613. doi: 10.1002/jbm.10167. [DOI] [PubMed] [Google Scholar]
- 163.Shin HJ, Lee CH, Cho IH, Kim YJ, Lee YJ, Kim IA, Park KD, Yui N, Shin JW. J Biomater Sci Polymer Edn. 2006;17:103. doi: 10.1163/156856206774879126. [DOI] [PubMed] [Google Scholar]
- 164.Shin M, Ishii O, Sueda T, Vacanti JP. Biomaterials. 2004;25:3717. doi: 10.1016/j.biomaterials.2003.10.055. [DOI] [PubMed] [Google Scholar]
- 165.Bini TB, Gao SJ, Tan TC, Wang S, Lim A, Hai LB, Ramakrishna S. Nanotechnology. 2004;15:1459. [Google Scholar]
- 166.Lee CH, Shin HJ, Cho IH, Kang YM, Kim IA, Park KD, Shin JW. Biomaterials. 2005;26:1261. doi: 10.1016/j.biomaterials.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 167.Chua KN, Lim WS, Zhang PC, Lu HF, Wen J, Ramakrishna S, Leong KW, Mao HQ. Biomaterials. 2005;26:2537. doi: 10.1016/j.biomaterials.2004.07.040. [DOI] [PubMed] [Google Scholar]
- 168.Matthews JA, Wnek GE, Simpson DG, Bowlin GL. Biomacromolecules. 2002;3:232. doi: 10.1021/bm015533u. [DOI] [PubMed] [Google Scholar]
- 169.Li CM, Jin HJ, Botsaris GD, Kaplan DL. J Mater Res. 2005;20:3374. [Google Scholar]
- 170.Kim KH, Jeong L, Park HN, Shin SY, Park WH, Lee SC, Kim TI, Park YJ, Seol YJ, Lee YM, Ku Y, Rhyu IC, Han SB, Chung CP. J Biotech. 2005;120:327. doi: 10.1016/j.jbiotec.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 171.Wnek GE, Carr ME, Simpson DG, Bowlin GL. Nano Lett. 2003;3:216. [Google Scholar]
- 172.Li M, Mondrinos MJ, Gandhi MR, Ko FK, Weiss AS, Lelkes PI. Biomaterials. 2005;26:5999. doi: 10.1016/j.biomaterials.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 173.Boland ED, Matthews JA, Pawlowski KJ, Simpson DG, Wnek GE, Bowlin GL. Front Biosci. 2004;9:1422. doi: 10.2741/1313. [DOI] [PubMed] [Google Scholar]
- 174.Huang L, McMillan RA, Apkarian RP, Pourdeyhimi B, Conticello VP, Chaikof EL. Macromolecules. 2000;33:2989. [Google Scholar]
- 175.Ohkawa K, Cha DI, Kim H, Nishida A, Yamamoto H. Macromol Rapid Commun. 2004;25:1600. [Google Scholar]
- 176.Jiang HL, Fang DF, Hsiao BS, Chu B, Chen WL. Biomacromolecules. 2004;5:326. doi: 10.1021/bm034345w. [DOI] [PubMed] [Google Scholar]
- 177.Um IC, Fang DF, Hsiao BS, Okamoto A, Chu B. Biomacromolecules. 2004;5:1428. doi: 10.1021/bm034539b. [DOI] [PubMed] [Google Scholar]
- 178.Dzenis Y. Science. 2004;304:1917. doi: 10.1126/science.1099074. [DOI] [PubMed] [Google Scholar]
- 179.Kim BS, Mooney DJ. Trends Biotechnol. 1998;16 doi: 10.1016/s0167-7799(98)01191-3. [DOI] [PubMed] [Google Scholar]
- 180.Kwon IK, Matsuda T. Biomacromolecules. 2005;6:2096. doi: 10.1021/bm050086u. [DOI] [PubMed] [Google Scholar]
- 181.He W, Yong T, Teo WE, Ma ZW, Ramakrishna S. Tissue Eng. 2005;11:1574. doi: 10.1089/ten.2005.11.1574. [DOI] [PubMed] [Google Scholar]
- 182.Hammond PT. Adv Mater. 2004;16:1271. [Google Scholar]
- 183.Shi XY, Shen MW, Mohwald H. Prog Polym Sci. 2004;29:987. [Google Scholar]
- 184.Ai H, Jones SA, Lvov YM. Cell Biochem Biophys. 2003;39:23. doi: 10.1385/CBB:39:1:23. [DOI] [PubMed] [Google Scholar]
- 185.Mendelsohn JD, Yang SY, Hiller J, Hochbaum AI, Rubner MF. Biomacromolecules. 2003;4:96. doi: 10.1021/bm0256101. [DOI] [PubMed] [Google Scholar]
- 186.Olenych SG, Moussallem MD, Salloum DS, Schlenoff JB, Keller TCS. Biomacromolecules. 2005;6:3252. doi: 10.1021/bm050298r. [DOI] [PubMed] [Google Scholar]
- 187.Richert L, Lavalle P, Vautier D, Senger B, Stoltz JF, Schaaf P, Voegel JC, Picart C. Biomacromolecules. 2002;3:1170. doi: 10.1021/bm0255490. [DOI] [PubMed] [Google Scholar]
- 188.Salloum DS, Olenych SG, Keller TCS, Schlenoff JB. Biomacromolecules. 2005;6:161. doi: 10.1021/bm0497015. [DOI] [PubMed] [Google Scholar]
- 189.Richert L, Arntz Y, Schaaf P, Voegel JC, Picart C. Surface Sci. 2004;570:13. [Google Scholar]
- 190.Zhu HG, Ji J, Shen JC. Biomacromolecules. 2004;5:1933. doi: 10.1021/bm049753u. [DOI] [PubMed] [Google Scholar]
- 191.Rho KS, Jeong L, Lee G, Seo BM, Park YJ, Hong SD, Roh S, Cho JJ, Park WH, Min BM. Biomaterials. 2006;27:1452. doi: 10.1016/j.biomaterials.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 192.Zhong SP, Teo WE, Zhu X, Beuerman R, Ramakrishna S, Yung LYL. Biomacromolecules. 2005;6:2998. doi: 10.1021/bm050318p. [DOI] [PubMed] [Google Scholar]
- 193.Van Lieshout MI, Vaz CM, Rutten MCM, Peters GWM, Baaijens FPT. J Biomater Sci Polymer Edn. 2006;17:77. doi: 10.1163/156856206774879153. [DOI] [PubMed] [Google Scholar]
- 194.Fairman R, Akerfeldt KS. Curr Opin Struct Biol. 2005;15:453. doi: 10.1016/j.sbi.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 195.Zhang S, Zhao X. J Mater Chem. 2004;14:2082. [Google Scholar]
- 196.Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294:1684. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 197.Hartgerink JD, Beniash E, Stupp SI. Proc Natl Acad Sci USA. 2002;99:5133. doi: 10.1073/pnas.072699999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Stupp SI, Donners J, Li LS, Mata A. MRS Bull. 2005;30:864. [Google Scholar]
- 199.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Science. 2004;303:1352. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 200.Zhang S. Nature Biotechnol. 2003;21:1171. doi: 10.1038/nbt874. [DOI] [PubMed] [Google Scholar]
- 201.Zhang S, Holmes TC, DiPersio CM, Hynes RO, Su X, Rich A. Biomaterials. 1995;16:1385. doi: 10.1016/0142-9612(95)96874-y. [DOI] [PubMed] [Google Scholar]
- 202.Holmes TC, de Lacalle S, Su X, Liu GS, Rich A, Zhang SG. Proc Natl Acad Sci USA. 2000;97:6728. doi: 10.1073/pnas.97.12.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kisiday J, Jin M, Kurtz B, Hung H, Semino C, Zhang S, Grodzinsky AJ. Proc Natl Acad Sci USA. 2002;99:9996. doi: 10.1073/pnas.142309999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Semino CE, Merok JR, Crane GG, Panagiotakos G, Zhang SG. Differentiation. 2003;71:262. doi: 10.1046/j.1432-0436.2003.7104503.x. [DOI] [PubMed] [Google Scholar]
- 205.Ellis-Behnke RG, Liang Y-X, You S-W, Tay DKC, Zhang S, So K-F, Schneider GE. Proc Natl Acad Sci USA. 2006;103:5054. doi: 10.1073/pnas.0600559103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Schneider JP, Pochan DJ, Ozbas B, Rajagopal K, Pakstis L, Kretsinger J. J Am Chem Soc. 2002;124:15030. doi: 10.1021/ja027993g. [DOI] [PubMed] [Google Scholar]
- 207.Ozbas B, Rajagopal K, Schneider JP, Pochan DJ. Phys Rev Lett. 2004;93:268106. doi: 10.1103/PhysRevLett.93.268106. [DOI] [PubMed] [Google Scholar]
- 208.Ozbas B, Kretsinger J, Rajagopal K, Schneider JP, Pochan DJ. Macromolecules. 2004;37:7331. [Google Scholar]
- 209.Kretsinger JK, Haines LA, Ozbas B, Pochan DJ, Schneider JP. Biomaterials. 2005;26:5177. doi: 10.1016/j.biomaterials.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 210.Beniash E, Hartgerink JD, Storrie H, Stendahl JC, Stupp SI. Acta Biomater. 2005;1:387. doi: 10.1016/j.actbio.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 211.Hench LL, Polak JM. Science. 2002;295:1014. doi: 10.1126/science.1067404. [DOI] [PubMed] [Google Scholar]
- 212.Ratner BD, Bryant SJ. Annu Rev Biomed Eng. 2004;6:41. doi: 10.1146/annurev.bioeng.6.040803.140027. [DOI] [PubMed] [Google Scholar]
- 213.Hench LL. J Am Ceram Soc. 1998;81:1705. [Google Scholar]
- 214.Murugan R, Ramakrishna S. Compos Sci Technol. 2005;65:2385. [Google Scholar]
- 215.Du C, Cui FZ, Zhu XD, de Groot K. J Biomed Mater Res. 1999;44:407. doi: 10.1002/(sici)1097-4636(19990315)44:4<407::aid-jbm6>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 216.Liao SS, Cui FZ, Zhu XD. J Bioactive Compat Polym. 2004;19:117. [Google Scholar]
- 217.Zhang SM, Cui FZ, Liao SS, Zhu Y, Han L. J Mater Sci Mater Med. 2003;14:641. doi: 10.1023/a:1024083309982. [DOI] [PubMed] [Google Scholar]
- 218.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Science. 1997;276:1109. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 219.Li HB, Linke WA, Oberhauser AF, Carrion-Vazquez M, Kerkviliet JG, Lu H, Marszalek PE, Fernandez JM. Nature. 2002;418:998. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]