Abstract
Background
Protein prenylation is a common post-translational modification in metazoans, protozoans, fungi, and plants. This modification, which mediates protein-membrane and protein-protein interactions, is characterized by the covalent attachment of a fifteen-carbon farnesyl or twenty-carbon geranylgeranyl group to the cysteine residue of a carboxyl terminal CaaX motif. In Arabidopsis, era1 mutants lacking protein farnesyltransferase exhibit enlarged meristems, supernumerary floral organs, an enhanced response to abscisic acid (ABA), and drought tolerance. In contrast, ggb mutants lacking protein geranylgeranyltransferase type 1 exhibit subtle changes in ABA and auxin responsiveness, but develop normally.
Results
We have expressed recombinant Arabidopsis protein farnesyltransferase (PFT) and protein geranylgeranyltransferase type 1 (PGGT1) in E. coli and characterized purified enzymes with respect to kinetic constants and substrate specificities. Our results indicate that, whereas PFT exhibits little specificity for the terminal amino acid of the CaaX motif, PGGT1 exclusively prenylates CaaX proteins with a leucine in the terminal position. Moreover, we found that different substrates exhibit similar Km but different kcat values in the presence of PFT and PGGT1, indicating that substrate specificities are determined primarily by reactivity rather than binding affinity.
Conclusions
The data presented here potentially explain the relatively strong phenotype of era1 mutants and weak phenotype of ggb mutants. Specifically, the substrate specificities of PFT and PGGT1 suggest that PFT can compensate for loss of PGGT1 in ggb mutants more effectively than PGGT1 can compensate for loss of PFT in era1 mutants. Moreover, our results indicate that PFT and PGGT1 substrate specificities are primarily due to differences in catalysis, rather than differences in substrate binding.
Background
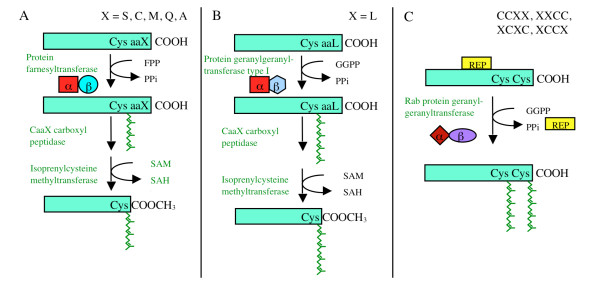
Protein farnesylation is the process by which proteins bearing a carboxyl terminal CaaX motif (C = Cys; a = aliphatic; X = Ser, Cys, Met, Gln, Ala) are post-translationally modified by the covalent attachment of a fifteen-carbon farnesyl group [1-4]. This modification results in the formation of a stable thioether bond between the cysteine of the CaaX motif and the farnesyl moiety, with farnesyl diphosphate serving as the farnesyl donor (Figure 1). This lipidation reaction is catalyzed by protein farnesyltransferase (PFT), which is a cytosolic enzyme consisting of α- and β-subunits [1-4]. In a similar process, proteins bearing a carboxyl terminal CaaX motif with Leu, Ile, Met, or Phe in the terminal position are modified by the covalent attachment of a twenty-carbon geranylgeranyl group to the cysteine of the CaaX motif. This modification is catalyzed by protein geranylgeranyltransferase type I (PGGT1), which is a cytosolic enzyme consisting of an α-subunit identical to that of PFT and a distinct β-subunit [1-5]. A third enzyme, protein geranylgeranyltransferase type II (PGGT II), also called RAB geranylgeranyltransferase (RAB GGT), catalyzes the geranylgeranylation of RAB proteins bound to the RAB ESCORT PROTEIN (REP). All three enzymes have been found in protozoans, metazoans, fungi, and plants, including peas [6,7], tomato [8,9], and Arabidopsis [10-14].
Figure 1.
Protein prenylation and processing in eukaryotes. A, prenylation and processing of farnesylated CaaX proteins. B, prenylation and processing of geranylgeranylated CaaX proteins. C, prenylation of RAB GTPases.
In Arabidopsis, a single gene encodes the common α-subunit of PFT and PGGT1 (PLURIPETALA, PLP, At3g59380) [14], a second gene encodes the β-subunit of PFT (ENHANCED RESPONSE TO ABA1, ERA1, At5g40280) [11,13], and a third gene encodes the β-subunit of PGGT1 (GERANYLGERANYLTRANSFERASE BETA, GGB, At2g39550) [10,12]. The ERA1 gene was so named because knockout mutations in this gene cause an enhanced response to abscisic acid (ABA) in both seed germination and stomatal closure assays. Consequently, era1 mutants exhibit increased seed dormancy and stomatal closure in response to ABA, and are drought tolerant [11,13,15-17]. These observations suggest that at least one farnesylated protein functions as a negative regulator of ABA signaling. However, to date, a farnesylated negative regulator of ABA signaling has not been definitively identified. era1 plants also exhibit enlarged meristems and supernumerary floral organs, especially petals, and this phenotype is greatly exaggerated in plp mutants lacking the common α-subunit of PFT and PGGT1 [14,18-21]. The more severe developmental phenotype of plp mutants compared to era1 mutants suggests that PGGT1 partially compensates for loss of PFT in era1 mutants [14]. Plants with defects in the GGB gene exhibit increased ABA-induced stomatal closure and auxin-induced lateral root formation [12], but without significant developmental phenotypes. These observations suggest that at least one geranylgeranylated protein functions as a negative regulator of ABA signaling and at least one functions as a negative regulator of auxin signaling. Indeed, ROP2 and ROP6, which are geranylgeranylated small GTPases [22,23], have been shown to function as negative regulators of ABA signaling, and ROP2 and AUX 2-11 (a geranylgeranylated member of the AUX/IAA family) have been shown to function as negative regulators of auxin signaling [10,24]. Moreover, Arabidopsis plants possess two genes encoding G protein γ-subunits, both of which are geranylgeranylated, and mutants lacking either of these genes exhibit an enhanced response to auxin-induced lateral root formation [25]. Prenylated proteins have also been implicated in a plethora of other processes, including calcium signal transduction [26,27], response to heat and heavy metal stress [28-30], cytokinin biosynthesis [31], and regulation of the cell division cycle [6,32,33]. Given these multiple roles, it is surprising that, unlike other organisms, Arabidopsis plants survive without the shared α-subunit of PFT and PGGT1 [14].
Proteins that are prenylated by either PFT or PGGT1 are further modified. First, the aaX portion of the CaaX motif is proteolytically removed by specific CaaX proteases (AtSTE24, At4g01320 and AtFACE-2, At2g36305 in Arabidopsis) [34-36] and, second, the isoprenylcysteine at the newly formed carboxyl terminus is methylated (Figure 1) [37-43]. Two distinct isoprenylcysteine methyltransferase (ICMT) enzymes, encoded by the AtSTE14A (At5g23320) and AtSTE14B (ICMT, At5g08335) genes, catalyze the methylation of carboxyl terminal isoprenylcysteines in Arabidopsis [41,43-46]. Demethylation of isoprenylcysteine methyl esters is catalyzed by isoprenylcysteine methylesterase (ICME), which is encoded by the ICME gene (At5g15860) [46,47].
As described above, two geranylgeranylated proteins (ROP2 and ROP6) and at least one farnesylated protein negatively regulate ABA signaling in Arabidopsis. However, it is not clear at the present time how these proteins function in ABA signaling. The stomata of ggb plants were found to exhibit an enhanced response to ABA, consistent with the known role of ROP6 in negative regulation of ABA-induced stomatal closure [12,23], but the response of ggb seeds to ABA was normal, despite a report that ROP2 is involved in negative regulation of ABA signaling in seeds [22]. While this may seem like a contradiction, it is possible that PFT activity in ggb plants is sufficient for the prenylation and function of certain prenylated proteins, such as ROP2 (i.e., PFT compensates for loss of PGGT1 in ggb mutants). Indeed, numerous reports exist of prenylated proteins that are substrates of both PFT and PGGT1 and others that are substrates of either PFT or PGGT1 [48,49]. Given this heterogeneity in the specificity of PFT and PGGT1 for certain CaaX proteins, deconvoluting the complex roles of protein prenylation in negative regulation of ABA signaling, meristem development, and other fundamental processes poses a significant challenge. Nevertheless, to address this problem, we characterized Arabidopsis PFT and PGGT1 with respect to substrate specificity and catalysis. These studies were aimed at answering the following questions: 1) What distinguishes plant CaaX prenyltransferases from animal and fungal prenyltransferases and what gives them their unique substrate specificities? 2) Do the substrate specificities of Arabidopsis PFT and PGGT1 potentially explain the phenotypes of era1 and ggb mutants? The results reported here indicate that Arabidopsis PFT exhibits less specificity for the terminal position of the CaaX motif than PFT enzymes from metazoans and yeast and Arabidopsis PGGT1 exhibits greater specificity for CaaX motifs with leucine in the terminal position than PGGT1 enzymes from metazoans and yeast. These results potentially explain the phenotypes of era1 and ggb mutants. Moreover, we show that different CaaX substrates exhibit differences in reactivity rather than differences in affinity in the presence of Arabidopsis PFT and PGGT1.
Results
Recombinant Arabidopsis PFT is more specific for isoprenoid substrates than PGGT1, whereas PGGT1 is more specific for CaaX substrates
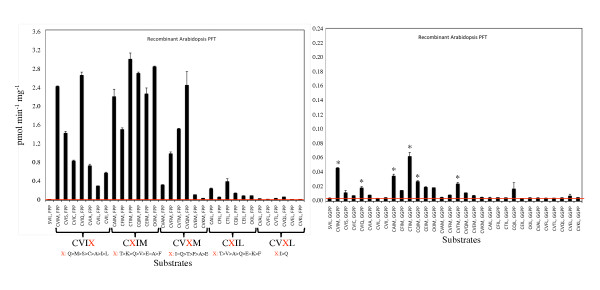
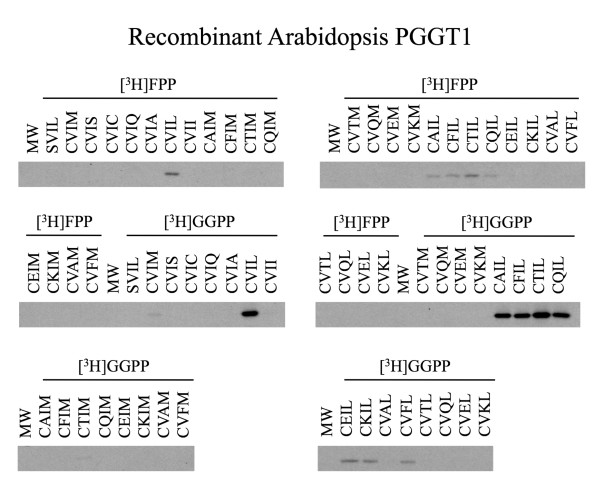
To functionally characterize Arabidopsis PFT and PGGT1, we co-expressed the PLP and ERA1 coding sequences in E. coli using the pETDuet-1 vector (PLP was expressed with an amino terminal FLAG tag and ERA1 was expressed with an amino terminal 6 × His tag). We also co-expressed the PLP and GGB coding sequences in E. coli (PLP was expressed with an amino terminal FLAG tag and GGB was expressed with an amino terminal 6 × His tag). IPTG-inducible PFT and PGGT1 activities were detected in E. coli extracts and analyzed for substrate specificity using [1-3H]FPP, [1-3H]GGPP, and 32 distinct GFP-BD-CaaX protein substrates, which were generated by site-directed mutagenesis of the GFP-BD-CaaX constructs recently reported by Gerber et al. (each protein substrate consists of GFP fused to the carboxyl terminal basic domain of the rice CaM61 protein and one of 32 different CaaX motifs) [50]. As shown in Figures 2 and 3, recombinant Arabidopsis PFT exhibited modest selectively for the terminal amino acid of the Ca1a2X motif. GFP-BD-CaaX substrates with glutamine, methionine, serine, cysteine, alanine, isoleucine, and even leucine (in descending order) were appreciably farnesylated by Arabidopsis PFT. As previously reported, PFT exhibited low selectivity for the a1 position of the Ca1a2X motif, consistent with the observation that the a1 position is solvent exposed and not constrained by active site amino acids [51-55]. In contrast, PFT exhibited high selectivity for the a2 position, with charged amino acids (basic as well as acidic) strongly excluded. GFP-BD-CaaX substrates that were efficiently farnesylated were also weakly geranylgeranylated by Arabidopsis PFT, but GFP-BD-CaaX farnesylation was 50-fold greater than geranylgeranylation (i.e., the y-axes in the two graphs of Figure 2 are not the same).
Figure 2.
Substrate specificity of recombinant Arabidopsis PFT. Quantitative filter assay data are shown for recombinant Arabidopsis PFT in the presence of [1-3H]FPP, [1-3H]GGPP, and 32 distinct GFP-BD-CaaX substrates. The CaaX substrates are grouped with the variable amino acid indicated in red. Asterisks indicate PFT-catalyzed protein geranylgeranylation detectable after 4 days by SDS-PAGE and fluorography. The red line indicates background. The standard error of the mean is shown.
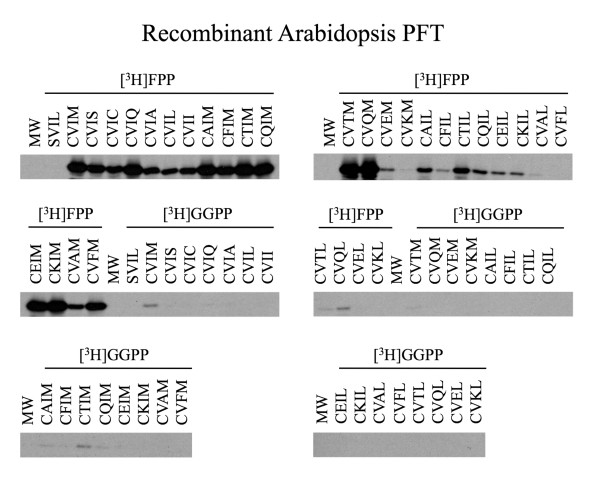
Figure 3.
Radiofluorograms of prenylation assays performed in the presence of recombinant Arabidopsis PFT. Radiofluorograms corresponding to the quantitative filter assay data in Figure 2 are shown.
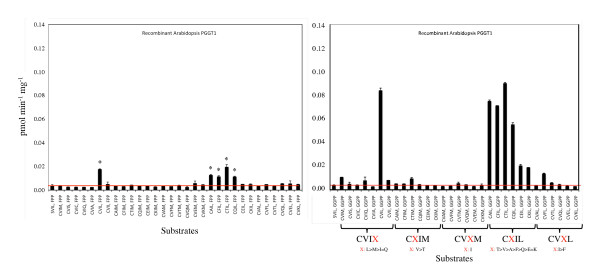
As shown in Figures 4 and 5, recombinant Arabidopsis PGGT1 exhibited high selectivity for the terminal amino acid of the Ca1a2X motif. Only GFP-BD-CaaX substrates ending in leucine were significantly prenylated by this enzyme (not even CVII, which is a good substrate for mammalian PGGT1, was appreciably prenylated by Arabidopsis PGGT1). As with PFT, PGGT1 exhibited low selectivity for the a1 position of the Ca1a2X motif, consistent with the observation that the a1 position is solvent exposed and not constrained by active site amino acids. On the other hand, PGGT1 exhibited extremely high selectivity for the a2 position, and only GFP-BD-CaaX substrates with a hydrophobic amino acid at the a2 position (CVIL and CVFL) were prenylated. GFP-BD-CaaX substrates that were efficiently geranylgeranylated were also farnesylated by PGGT1. Indeed, GFP-BD-CaaX geranylgeranylation was only 4-fold greater than farnesylation in the presence of recombinant Arabidopsis PGGT1.
Figure 4.
Substrate specificity of recombinant Arabidopsis PGGT I. Quantitative filter assay data are shown for recombinant Arabidopsis PGGT I in the presence of [1-3H]FPP, [1-3H]GGPP, and 32 distinct CaaX substrates. The CaaX substrates are grouped with the variable amino acid indicated in red. Asterisks indicate PGGT1-catalyzed protein farnesylation detectable after 4 days by SDS-PAGE and fluorography. The red line indicates background. The standard error of the mean is shown.
Figure 5.
Radiofluorograms of prenylation assays performed in the presence of recombinant Arabidopsis PGGT1. Radiofluorograms corresponding to the quantitative filter assay data in Figure 4 are shown.
Purified recombinant PFT is more active than purified recombinant PGGT1
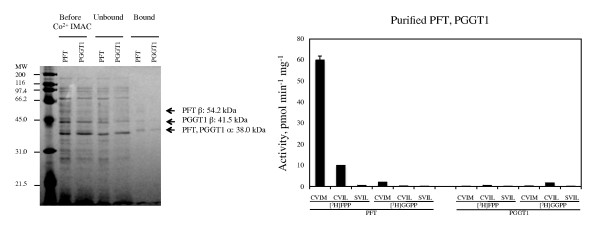
The next step in the characterization of Arabidopsis PFT and PGGT1 was to examine the activity and substrate specificity of purified recombinant enzymes. The enzymes described above were purified by immobilized metal affinity chromatography (IMAC) using Talon®; Co2+-based resin. As shown in Figure 6, IMAC-purified enzymes (80-90% pure) were assayed and found to exhibit the same substrate specificities as described above (PFT exhibits low specificity for the terminal amino acid of the Ca1a2X motif, whereas PGGT1 is highly selective for GFP-BD-CaaX substrates ending in leucine). Comparing purified recombinant PFT and PGGT1 allowed us to make the following conclusion: purified recombinant PFT is 30- to 100-fold more active than purified recombinant PGGT1. This is not due to errors in the expressed sequences, nor is it likely to be due to differential effects of the FLAG tag on the alpha subunit or the 6 × His-tags on the two β-subunits because the amino termini of prenyltransferase α- and β-subunits are solvent exposed and not involved in the formation or stabilization of active heterodimers [56,57]. Moreover, plant extracts (tobacco BY2 as well as Arabidopsis extracts) consistently exhibit 30-100 fold higher PFT activity compared with PGGT1 activity [12].
Figure 6.
Activity and substrate specificity of IMAC-purified recombinant Arabidopsis PFT and PGGT I. Left Panel: E. coli extracts before Co2+-IMAC, unbound proteins, and bound (IMAC-purified) proteins are shown. An E. coli protein at 42 kDa, which co-migrates with the PGGT I β-subunit is present at low levels in the purified PFT and PGGT I samples. Right Panel: quantitative activity data for purified PFT and PGGT I. The standard error of the mean is shown.
Kinetic Analysis of Recombinant Arabidopsis PFT and PGGT1
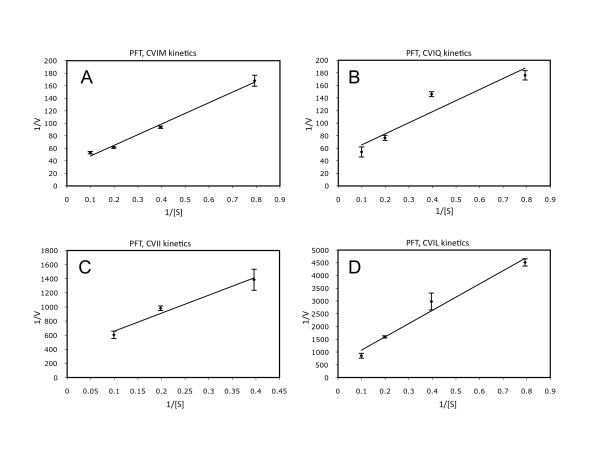
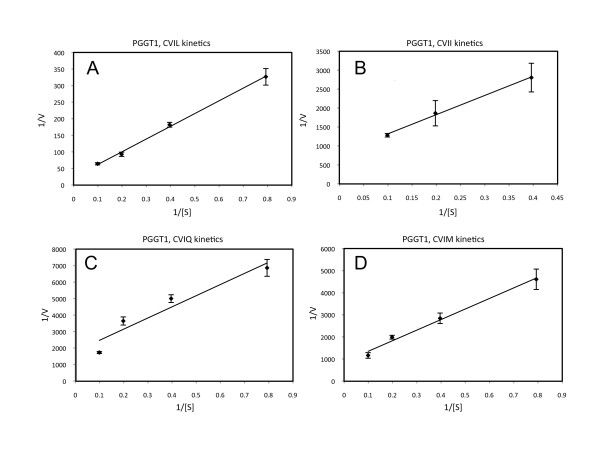
Purified recombinant Arabidopsis PFT and PGGT1 were subjected to kinetic analyses under Michaelis-Menten conditions (product formation was linear with time and substrate conversion was less than 10%). The results of these experiments were interpreted by Lineweaver-Burk analysis and are shown in Figures 7 and 8. The catalytic constants (kcat/Km) shown in Table 1 for Arabidopsis PFT confirm the results shown in Figures 2 and 3. Both data sets demonstrate the following substrate preferences for Arabidopsis PFT (normalized to 1.0 for GFP-BD-CVIQ): GFP-BD-CVIQ (1.0), GFP-BD-CVIM (0.60), GFP-BD-CVII (0.21) and GFP-BD-CVIL (0.06). Moreover, the results in Table 1 show that, while different CaaX substrates (CVIQ, CVIM, CVII, and CVIL) have similar Km values, they have markedly different kcat values in the presence of Arabidopsis PFT. Thus, PFT substrate specificities reflect differences in catalytic turnover rate rather than differences in binding affinity. The catalytic constants (kcat/Km) for Arabidopsis PGGT1, which are shown in Table 2, confirm the results shown in Figures 4 and 5. Both data sets demonstrate the following substrate preferences for Arabidopsis PGGT1 (normalized to 1.0 for GFP-BD-CVIL): GFP-BD-CVIL (1.0), GFP-BD-CVII (0.14), GFP-BD-CVIM (0.07) and GFP-BD-CVIQ (0.07). Moreover, while different CaaX substrates have slightly different Km values, they have dramatically different kcat values. Indeed, the significantly higher kcat value for GFP-BD-CVIL is the primary determinant of substrate specificity for Arabidopsis PGGT1. While these kcat values can be compared, they are nevertheless low, suggesting that only a fraction of the purified PGGT1 protein was catalytically active.
Figure 7.
Lineweaver-Burk plots for purified PFT. A, GFP-BD-CVIM and [1-3H]FPP were used as substrates. B, GFP-BD-CVIQ and [1-3H]FPP were used as substrates. C, GFP-BD-CVII and [1-3H]FPP were used as substrates. D, GFP-BD-CVIL and [1-3H]FPP were used as substrates. The standard error of the mean is shown.
Figure 8.
Lineweaver-Burk plots for purified PGGT1. A, GFP-BD-CVIL and [1-3H]GGPP were used as substrates. B, GFP-BD-CVII and [1-3H]GGPP were used as substrates. C, GFP-BD-CVIQ and [1-3H]GGPP were used as substrates. D, GFP-BD-CVIM and [1-3H]GGPP were used as substrates. The standard error of the mean is shown.
Table 1.
Kinetic constants for recombinant Arabidopsis PFT*
| PFT substrate |
Km (μM) | Specific Activity (pmol min-1 mg-1) |
kcat (hr-1) | kcat/Km (μM-1 hr-1) |
n |
|---|---|---|---|---|---|
| GFP-BD-CVIM | 5.4 +/- 0.6 | 5200 +/- 700 | 28.8 +/- 4.1 | 5.3 +/- 1.0 | 8 |
| GFP-BD-CVIQ | 5.5 +/- 0.9 | 8900 +/- 1900 | 49.1 +/- 10.5 | 8.9 +/- 2.4 | 8 |
| GFP-BD-CVII | 6.9 +/- 1.0 | 2300 +/- 700 | 12.8 +/- 3.7 | 1.9 +/- 0.6 | 8 |
| GFP-BD-CVIL | 7.0 +/- 1.5 | 670 +/- 140 | 3.7 +/- 0.8 | 0.5 +/- 0.2 | 8 |
Standard errors are given.
*PFT assays were performed in the presence of [1-3H]farnesyl diphosphate.
Table 2.
Kinetic constants for recombinant Arabidopsis PGGT1*
| PGGT1 substrate |
Km (μM) | Specific Activity (pmol min-1 mg-1) |
kcat (hr-1) | kcat/Km (μM-1 hr-1) |
n |
|---|---|---|---|---|---|
| GFP-BD-CVIL | 17.2 +/- 4.4 | 110 +/- 30 | 0.50 +/- 0.12 | 0.029 +/- 0.010 | 7 |
| GFP-BD-CVII | 19.0 +/-14.4 | 17 +/- 15 | 0.08 +/- 0.07 | 0.004 +/- 0.005 | 4 |
| GFP-BD-CVIQ | 3.5 +/- 0.6 | 1.7 +/- 0.2 | 0.008 +/- 0.001 | 0.002 +/- 0.001 | 8 |
| GFP-BD-CVIM | 5.8 +/- 1.0 | 2.6 +/- 0.3 | 0.012 +/- 0.001 | 0.002 +/- 0.001 | 8 |
Standard errors are given.
*PGGT1 assays were performed in the presence of [1-3H]geranylgeranyl diphosphate.
Purified recombinant Arabidopsis PFT and PGGT1 were also analyzed with respect to isoprenyl diphosphate specificity. As shown in Table 3, Km values for FPP and GGPP are an order of magnitude lower than Km values for CaaX substrates in the presence of recombinant Arabidopsis PFT and PGGT1. However, as with CaaX substrates, the different specificities of Arabidopsis PFT and PGGT1 for isoprenyl diphosphates cannot be explained by differences in Km. Thus, the preferences of PFT and PGGT1 for different isoprenyl diphosphate substrates is primarily determined by reactivity rather than binding affinity.
Table 3.
Km values (in μM) for isoprenyl diphosphates in the presence of Arabidopsis PFT and PGGT1*
| Prenyltransferase | FPP | GGPP | n |
|---|---|---|---|
| PFT | 0.2 +/- 0.1 | 0.7 +/- 0.3 | 7 |
| PGGT1 | 0.6 +/- 0.5 | 0.8 +/- 0.1 | 7 |
Standard errors are given.
*PFT assays were performed in the presence of GFP-BD-CVIQ and PGGT1 assays were performed in the presence of GFP-BD-CVIL.
Discussion
In this report, it is shown that recombinant Arabidopsis PFT exhibits broad specificity for CaaX substrates with Gln, Met, Ser, Cys, Ala, Ile, or Leu in the terminal 'X' position, whereas PGGT1 exhibits strict specificity for CaaX substrates ending in Leu. Both PFT and PGGT1 exhibit little or no specificity for the a1 position of the Ca1a2X motif, which is consistent with previous observations using mammalian prenyltransferases that the a1 position is solvent exposed and not constrained by active site amino acids [51-55]. In contrast, both prenyltransferases exhibit specificity for the a2 position of the Ca1a2X motif. While the mechanism for CaaX specificity remains unknown for Arabidopsis PFT and PGGT1, it is clear that the substrate specificities of both prenyltransferases reflect differences in catalytic turnover rates rather than differences in Km values (Tables 1 and 2). This finding suggests that, while binding affinities of GFP-BD-CVIQ, GFP-BD-CVIM, GFP-BD-CVII, and GFP-BD-CVIL to the active sites of PFT and PGGT1 are similar, the terminal amino acid of the CaaX motif dramatically affects catalysis. Moreover, the data described above provide an explanation for era1 and ggb phenotypes. PFT prenylates a wide range of CaaX substrates and compensates almost fully for loss of PGGT1 in ggb plants. However, PGGT1 specifically prenylates CaaL substrates and only partially compensates for loss of PFT in era1 plants. These biochemical differences potentially account for the mild phenotype of ggb mutants and the dramatic phenotype of era1 mutants.
The different isoprenoid specificities of Arabidopsis PFT and PGGT1 cannot be explained by differences in Km. Indeed, the Km for FPP was only slightly lower than that for GGPP in the presence of recombinant Arabidopsis PFT, despite the fact that CaaX farnesylation was 50-fold greater than geranylgeranylation in the presence of this enzyme (Figures 2 and 3). Moreover, the Km values for FPP and GGPP were almost identical in the presence of recombinant Arabidopsis PGGT1, despite the fact that PGGT1 catalyzed CaaX geranylgeranylation 4-fold more efficiently than farnesylation. Thus, the primary determinant of isoprenoid substrate specificity is reactivity rather than binding affinity.
The results in Figure 6 raise an interesting question. Given the higher specific activity and lower CaaX substrate specificity of PFT, why are CaaX substrates with leucine in the terminal position predominantly geranylgeranylated rather than farnesylated in planta [12,50]? The results in Figure 6 suggest that a CAIL (or CVIL) protein should be predominantly farnesylated in planta because farnesylation of CAIL (or CVIL) by Arabidopsis PFT is approximately 20% as efficient as farnesylation of CAIM (or CVIM), which greatly exceeds the efficiency of CAIL (or CVIL) geranylgeranylation by PGGT1. We propose that PFT is regulated in planta, perhaps by post-translational modifications or protein-protein interactions, to reduce recognition and farnesylation of CaaX substrates with leucine in the terminal position. Nevertheless, it is likely that CaaX substrates with leucine in the terminal position are, to some extent, farnesylated by PFT and that these aberrantly farnesylated proteins retain full or partial function. This explains why ggb mutants, which were expected to exhibit severe meristem and tip-growth defects due to loss of ROP function, do not exhibit these phenotypes [12,22,23].
We propose that PGGT1 activity is higher in planta than the purified recombinant, E. coli-expressed PGGT1 activity we have characterized. The activity observed with purified recombinant PGGT1 was low, suggesting that only a portion of the purified PGGT1 enzyme was active. Despite this, the relative kcat/Km values obtained for different CaaX substrates in the presence of PGGT1 were consistent with the results shown in Figures 4 and 5.
Conclusions
In this report, recombinant Arabidopsis PFT is shown to prenylate CaaX substrates with little specificity for the terminal amino acid. In contrast, recombinant Arabidopsis PGGT1 is shown to exclusively prenylate CaaX substrates with leucine in the terminal position. These different substrate specificities provide a straightforward explanation for the phenotypes of era1 and ggb mutant plants. In addition, substrate specificities for PFT and PGGT1 are shown to reflect differences in catalytic turnover rates rather than differences in substrate binding.
Methods
RNA isolation
Total RNA was isolated from wild type Arabidopsis plants (ecotype Col-0) using TRIzol® Reagent according to the manufacturer's instructions (Invitrogen/Life Technologies Corp., Carlsbad, CA).
PFT and PGGT1 expression constructs
The coding sequences of the PLP (At3g59380), ERA1 (At5g40280), and GGB (At2g39550) genes were amplified by reverse-transcriptase-PCR using 0.5 μg of total RNA, 5 pmol of forward primer, 5 pmol of reverse primer, and the Platinum Quantitative RT-PCR Thermoscript One-Step System (Invitrogen/Life Technologies Corp., Carlsbad, CA). RT-PCR conditions included a 20-min reverse transcription step at 50°C, followed by a 5-min pre-soak at 95°C, and 25-35 cycles of the following PCR program: 95°C, 30 sec; 55°C, 30 sec; 72°C, 90 sec. A post-soak was performed at 72°C for 7 min to ensure complete product synthesis. RT-PCR products were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining. The primers used for RT-PCR were as follows: PLP-CDS-forward: 5'-cac gga tcc acc atg gat tac aag gat gac gac gat aag aat ttc gac gag acc gtg cca-3'; PLP-CDS-reverse: 5'-cac gga tcc tca aat tgc tgc cac tgt aat ctt g-3'; ERA1-CDS-forward: 5'-cac gga tcc acc atg cac cac cat cac cat cac cca gta gta acc cgc ttg att-3'; ERA1-CDS-reverse: 5'-cac gga tcc tca tgc tgc ttt aaa gaa gaa ctc-3'; GGB-CDS-forward: 5'-cac gga tcc acc atg cat cat cat cat cat cat tca gag acc gcc gtg tca atc-3'; GGB-CDS-reverse: 5'-cac gga tcc tca aat tcc cgg ggc tgc aag aa-3'. RT-PCR products were confirmed by DNA sequence analysis and ligated into the BamH1 (PLP) or BglII (ERA1, GGB) sites of the pETDuet-1 vector (EMD Biosciences, Gibbstown, NJ).
GFP-BD-CaaX mutagenesis
GFP-BD-CaaX expression constructs [50] were mutagenized using the QuikChange II site-directed mutagenesis kit from Stratagene (La Jolla, CA) to generate the following CaaX motifs: CVIM, CVIS, CVIC, CVIQ, CVIA, CVIL, CVII, CAIM, CFIM, CTIM, CQIM, CEIM, CKIM, CVAM, CVFM, CVTM, CVQM, CVEM, CVKM CAIL, CFIL, CTIL, CQIL CEIL, CKIL, CVAL, CVFL, CVTL, CVQL, CVEL, CVKL, SVIL. The following primers were used for mutagenesis of the GFP-BD-CVIM template: CVII-F: 5'- cgt ggc cag aag tgc gtg atc atc taa cgg gat ccc gcc -3'; CVII-R: 5'- ggc ggg atc ccg tta gat gat cac gca ctt ctg gcc acg -3'; CVIS-F: 5'- cgt ggc cag aag tgc gtg atc tcg taa cgg gat ccc gcc -3'; CVIS-R: 5'- ggc ggg atc ccg tta cga gat cac gca ctt ctg gcc acg -3'; CVIC-F: 5'- cgt ggc cag aag tgc gtg atc tgc taa cgg gat ccc gcc -3'; CVIC-R: 5'- ggc ggg atc ccg tta gca gat cac gca ctt ctg gcc acg -3'; CAIM-F: 5'- cgt ggc cag aag tgc gcg atc atg taa cgg gat ccc gcc -3'; CAIM-R: 5'- ggc ggg atc ccg tta cat gat cgc gca ctt ctg gcc acg -3'; CFIM-F: 5'- cgt ggc cag aag tgc ttt atc atg taa cgg gat ccc gcc -3'; CFIM-R: 5'- ggc ggg atc ccg tta cat gat aaa gca ctt ctg gcc acg -3'; CTIM-F: 5'- cgt ggc cag aag tgc acg atc atg taa cgg gat ccc gcc -3'; CTIM-R: 5'- ggc ggg atc ccg tta cat gat cgt gca ctt ctg gcc acg -3'; CQIM-F: 5'- cgt ggc cag aag tgc cag atc atg taa cgg gat ccc gcc -3'; CQIM-R: 5'- ggc ggg atc ccg tta cat gat ctg gca ctt ctg gcc acg -3'; CEIM-F: 5'- cgt ggc cag aag tgc gag atc atg taa cgg gat ccc gcc -3'; CEIM-R: 5'- ggc ggg atc ccg tta cat gat ctc gca ctt ctg gcc acg -3'; CKIM-F: 5'- cgt ggc cag aag tgc aag atc atg taa cgg gat ccc gcc -3'; CKIM-R: 5'- ggc ggg atc ccg tta cat gat ctt gca ctt ctg gcc acg -3'; CVAM-F: 5'- cgt ggc cag aag tgc gtg gcc atg taa cgg gat ccc gcc -3'; CVAM-R: 5'- ggc ggg atc ccg tta cat ggc cac gca ctt ctg gcc acg -3'; CVFM-F: 5'- cgt ggc cag aag tgc gtg ttc atg taa cgg gat ccc gcc -3'; CVFM-R: 5'- ggc ggg atc ccg tta cat gaa cac gca ctt ctg gcc acg -3'; CVTM-F: 5'- cgt ggc cag aag tgc gtg acc atg taa cgg gat ccc gcc -3'; CVTM-R: 5'- ggc ggg atc ccg tta cat ggt cac gca ctt ctg gcc acg -3'; CVQM-F: 5'- cgt ggc cag aag tgc gtg cag atg taa cgg gat ccc gcc -3'; CVQM-R: 5'- ggc ggg atc ccg tta cat ctg cac gca ctt ctg gcc acg -3'; CVEM-F: 5'- cgt ggc cag aag tgc gtg gag atg taa cgg gat ccc gcc -3'; CVEM-R: 5'- ggc ggg atc ccg tta cat ctc cac gca ctt ctg gcc acg -3'; CVKM-F: 5'- cgt ggc cag aag tgc gtg aag atg taa cgg gat ccc gcc -3'; CVKM-R: 5'- ggc ggg atc ccg tta cat ctt cac gca ctt ctg gcc acg -3'. The following primers were used for mutagenesis of the GFP-BD-CVIL template: CVIQ-F: 5'- cgt ggc cag aag tgc gtg atc cag taa cgg gat ccc gcc -3'; CVIQ-R: 5'- ggc ggg atc ccg tta ctg gat cac gca ctt ctg gcc acg -3'; CVIA-F: 5'- cgt ggc cag aag tgc gtg atc gcg taa cgg gat ccc gcc -3'; CVIA-R: 5'- ggc ggg atc ccg tta cgc gat cac gca ctt ctg gcc acg -3'; CAIL-F: 5'- cgt ggc cag aag tgc gcg atc ctg taa cgg gat ccc gcc -3'; CAIL-R: 5'- ggc ggg atc ccg tta cag gat cgc gca ctt ctg gcc acg -3'; CFIL-F: 5'- cgt ggc cag aag tgc ttt atc ctg taa cgg gat ccc gcc -3'; CFIL-R: 5'- ggc ggg atc ccg tta cag gat aaa gca ctt ctg gcc acg -3'; CTIL-F: 5'- cgt ggc cag aag tgc acg atc ctg taa cgg gat ccc gcc -3'; CTIL-R: 5'- ggc ggg atc ccg tta cag gat cgt gca ctt ctg gcc acg -3'; CQIL-F: 5'- cgt ggc cag aag tgc cag atc ctg taa cgg gat ccc gcc -3'; CQIL-R: 5'- ggc ggg atc ccg tta cag gat ctg gca ctt ctg gcc acg -3'; CEIL-F: 5'- cgt ggc cag aag tgc gag atc ctg taa cgg gat ccc gcc -3'; CEIL-R: 5'- ggc ggg atc ccg tta cag gat ctc gca ctt ctg gcc acg -3'; CKIL-F: 5'- cgt ggc cag aag tgc aag atc ctg taa cgg gat ccc gcc -3'; CKIL-R: 5'- ggc ggg atc ccg tta cag gat ctt gca ctt ctg gcc acg -3'; CVAL-F: 5'- cgt ggc cag aag tgc gtg gcc ctg taa cgg gat ccc gcc -3'; CVAL-R: 5'- ggc ggg atc ccg tta cag ggc cac gca ctt ctg gcc acg -3'; CVFL-F: 5'- cgt ggc cag aag tgc gtg ttc ctg taa cgg gat ccc gcc -3'; CVFL-R: 5'- ggc ggg atc ccg tta cag gaa cac gca ctt ctg gcc acg -3'; CVTL-F: 5'- cgt ggc cag aag tgc gtg acc ctg taa cgg gat ccc gcc -3'; CVTL-R: 5'- ggc ggg atc ccg tta cag ggt cac gca ctt ctg gcc acg -3'; CVQL-F: 5'- cgt ggc cag aag tgc gtg cag ctg taa cgg gat ccc gcc -3'; CVQL-R: 5'- ggc ggg atc ccg tta cag ctg cac gca ctt ctg gcc acg -3'; CVEL-F: 5'- cgt ggc cag aag tgc gtg gag ctg taa cgg gat ccc gcc -3'; CVEL-R: 5'- ggc ggg atc ccg tta cag ctc cac gca ctt ctg gcc acg -3'; CVKL-F: 5'- cgt ggc cag aag tgc gtg aag ctg taa cgg gat ccc gcc -3'; CVKL-R: 5'- ggc ggg atc ccg tta cag ctt cac gca ctt ctg gcc acg -3'. All GFP-BD-CaaX constructs were confirmed by DNA sequence analysis.
Protein expression and purification
Cultures of E. coli Rosetta cells (EMD Biosciences, Gibbstown, NJ) containing PFT, PGGT1, or GFP-BD-CaaX expression constructs were grown to log phase (A600 = 0.7-0.8) in Luria Broth containing 100 μg ml-1 ampicillin. Recombinant protein expression was then induced with 1 mM IPTG (PFT and PGGT1) or 0.1% L-arabinose (GFP-BD-CaaX proteins) at 20°C for 16 hr. After centrifugation at 16,000g for 2 min, cell pellets were resuspended in 1 ml of STE buffer (150 mM NaCl, 10 mM Tris-HCl, pH 7.5, 1 mM EDTA) containing Complete Protease Inhibitors (Roche Diagnostics, Indianapolis, IN) and cells were disrupted by vigorous agitation in the presence of glass beads. Cell extracts were cleared by centrifugation at 16,000g for 2 min at 4°C. Protein purification was accomplished at 4°C in three steps: 1) PD-10 gel filtration chromatography in 50 mM sodium phosphate, pH 7.0, 0.3 M NaCl [to remove EDTA], 2) immobilized metal affinity chromatography [IMAC] using Talon® Co2+-based resin (Clontech, Mountain View, CA) according to the manufacturer's instructions, and 3) PD-10 gel filtration chromatography in 50 mM sodium phosphate, pH 7.0, 0.3 M NaCl [to remove imidazole, which inhibits protein prenylation].
In vitro prenylation assays
In vitro prenylation reactions contained protein prenyltransferase (100 μg of E. coli extract containing recombinant PFT or PGGT1, 0.005 μg of purified PFT, or 0.5 μg of purified PGGT1), 5-40 μg of GFP-BD-CaaX protein, and either [1-3H]farnesyl diphosphate (26.2 Ci/mmol, Perkin Elmer, Waltham, MA) or [1-3H]geranylgeranyl diphosphate (19.5 Ci/mmol, Perkin Elmer, Waltham, MA) in 125 μl of 50 mM Hepes (pH 7.5), 20 mM MgCl2, 5 mM DTT, 0.1% Zwittergent, and 5 mM ZnCl2. Reactions were incubated at 30°C for 30 min, after which two 50-μl portions were terminated in 950 μl of 1 M ethanolic HCl. Precipitated proteins were collected on GF/A glass fiber filters (Whatman, Piscataway, NJ), washed with 10 ml of 95% ethanol, and quantified by liquid scintillation using BioSafe II cocktail (RPI Corporation, Mt. Prospect, IL). A 25 μl portion of each reaction was resolved by SDS-PAGE and prenylated proteins were visualized using Amplify fluorographic reagent (GE Healthcare, Piscataway, NJ) and Kodak AR5 film (Eastman Kodak, Rochester, NY).
Authors' contributions
MA performed site-directed mutagenesis on GFP-BD-CaaX proteins, protein expression and purification (i.e., PFT, PGGT1 and GFP-BD-CaaX proteins), and the kinetic assays described in Figures 7 and 8 and Tables 1, 2 and 3. DHH created the PLP and PLP + GGB expression constructs. DNC designed all experiments, created the PLP + ERA1 expression construct and performed the experiments shown in Figures 2, 3, 4, 5 and 6. All authors read and approved the final manuscript.
Contributor Information
Michelle Andrews, Email: andrmich@isu.edu.
David H Huizinga, Email: DHHuizinga@dow.com.
Dring N Crowell, Email: crowdrin@isu.edu.
Acknowledgements
This work was supported by NSF grant MCB-0900962 to DNC and by NIH Grant P20RR16454 from the INBRE program of the National Center for Research Resources, which provided funds for the Molecular Research Core Facility at Idaho State University. The authors thank Dr. Caryn Evilia for critical reading of the manuscript.
References
- Crowell DN. Functional implications of protein isoprenylation in plants. Prog Lipid Res. 2000;39(5):393–408. doi: 10.1016/S0163-7827(00)00010-2. [DOI] [PubMed] [Google Scholar]
- Clarke S. Protein isoprenylation and methylation at carboxyl-terminal cysteine residues. Annu Rev Biochem. 1992;61:355–386. doi: 10.1146/annurev.bi.61.070192.002035. [DOI] [PubMed] [Google Scholar]
- Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- Crowell DN, Huizinga DH. Protein isoprenylation: the fat of the matter. Trends Plant Sci. 2009;14(3):163–170. doi: 10.1016/j.tplants.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Hartman HL, Hicks KA, Fierke CA. Peptide specificity of protein prenyltransferases is determined mainly by reactivity rather than binding affinity. Biochemistry. 2005;44(46):15314–15324. doi: 10.1021/bi0509503. [DOI] [PubMed] [Google Scholar]
- Qian D, Zhou D, Ju R, Cramer CL, Yang Z. Protein farnesyltransferase in plants: molecular characterization and involvement in cell cycle control. Plant Cell. 1996;8(12):2381–2394. doi: 10.1105/tpc.8.12.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Cramer CL, Watson JC. Protein farnesyltransferase in plants. Molecular cloning and expression of a homolog of the beta subunit from the garden pea. Plant Physiol. 1993;101(2):667–674. doi: 10.1104/pp.101.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loraine AE, Yalovsky S, Fabry S, Gruissem W. Tomato Rab1A homologs as molecular tools for studying rab geranylgeranyl transferase in plant cells. Plant Physiol. 1996;110(4):1337–1347. doi: 10.1104/pp.110.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S, Loraine AE, Gruissem W. Specific prenylation of tomato Rab proteins by geranylgeranyl type-II transferase requires a conserved cysteine-cysteine motif. Plant Physiol. 1996;110(4):1349–1359. doi: 10.1104/pp.110.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldelari D, Sternberg H, Rodríguez-Concepción M, Gruissem W, Yalovsky S. Efficient prenylation by a plant geranylgeranyltransferase-I requires a functional CaaL box motif and a proximal polybasic domain. Plant Physiol. 2001;126(4):1416–1429. doi: 10.1104/pp.126.4.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P. A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science. 1996;273(5279):1239–1241. doi: 10.1126/science.273.5279.1239. [DOI] [PubMed] [Google Scholar]
- Johnson CD, Chary SN, Chernoff EA, Zeng Q, Running MP, Crowell DN. Protein geranylgeranyltransferase I is involved in specific aspects of abscisic acid and auxin signaling in Arabidopsis. Plant Physiol. 2005;139(2):722–733. doi: 10.1104/pp.105.065045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei ZM, Ghassemian M, Kwak CM, McCourt P, Schroeder JI. Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science. 1998;282(5387):287–290. doi: 10.1126/science.282.5387.287. [DOI] [PubMed] [Google Scholar]
- Running MP, Lavy M, Sternberg H, Galichet A, Gruissem W, Hake S, Ori N, Yalovsky S. Enlarged meristems and delayed growth in plp mutants result from lack of CaaX prenyltransferases. Proc Natl Acad Sci USA. 2004;101(20):7815–7820. doi: 10.1073/pnas.0402385101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GJ, Murata Y, Chu SP, Nafisi M, Schroeder JI. Hypersensitivity of abscisic acid-induced cytosolic calcium increases in the Arabidopsis farnesyltransferase mutant era1-2. Plant Cell. 2002;14(7):1649–1662. doi: 10.1105/tpc.010448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Sarkar SF, Bonetta D, McCourt P. The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J. 2003;34(1):67–75. doi: 10.1046/j.1365-313X.2003.01707.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ying J, Kuzma M, Chalifoux M, Sample A, McArthur C, Uchacz T, Sarvas C, Wan J, Dennis DT. et al. Molecular tailoring of farnesylation for plant drought tolerance and yield protection. Plant J. 2005;43(3):413–424. doi: 10.1111/j.1365-313X.2005.02463.x. [DOI] [PubMed] [Google Scholar]
- Bonetta D, Bayliss P, Sun S, Sage T, McCourt P. Farnesylation is involved in meristem organization in Arabidopsis. Planta. 2000;211(2):182–190. doi: 10.1007/s004250000283. [DOI] [PubMed] [Google Scholar]
- Yalovsky S, Kulukian A, Rodríguez-Concepción M, Young CA, Gruissem W. Functional requirement of plant farnesyltransferase during development in arabidopsis. Plant Cell. 2000;12(8):1267–1278. doi: 10.1105/tpc.12.8.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelhoffer EC, Medrano LJ, Meyerowitz EM. Cloning of the Arabidopsis WIGGUM gene identifies a role for farnesylation in meristem development. Proc Natl Acad Sci USA. 2000;97(13):7633–7638. doi: 10.1073/pnas.130189397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Running MP, Fletcher JC, Meyerowitz EM. The WIGGUM gene is required for proper regulation of floral meristem size in Arabidopsis. Development. 1998;125(14):2545–2553. doi: 10.1242/dev.125.14.2545. [DOI] [PubMed] [Google Scholar]
- Li H, Shen JJ, Zheng ZL, Lin Y, Yang Z. The Rop GTPase switch controls multiple developmental processes in Arabidopsis. Plant Physiol. 2001;126(2):670–684. doi: 10.1104/pp.126.2.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemichez E, Wu Y, Sanchez JP, Mettouchi A, Mathur J, Chua NH. Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Genes & Development. 2001;15(14):1808–1816. doi: 10.1101/gad.900401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt RE, Ainley WM, Nagao RT, Conner TW, Key JL. Expression of the Arabidopsis AtAux2-11 auxin-responsive gene in transgenic plants. Plant Mol Biol. 1993;22(5):731–749. doi: 10.1007/BF00027361. [DOI] [PubMed] [Google Scholar]
- Trusov Y, Rookes JE, Tilbrook K, Chakravorty D, Mason MG, Anderson D, Chen JG, Jones AM, Botella JR. Heterotrimeric G protein gamma subunits provide functional selectivity in Gbetagamma dimer signaling in Arabidopsis. Plant Cell. 2007;19(4):1235–1250. doi: 10.1105/tpc.107.050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Concepción M, Yalovsky S, Zik M, Fromm H, Gruissem W. The prenylation status of a novel plant calmodulin directs plasma membrane or nuclear localization of the protein. EMBO J. 1999;18(7):1996–2007. doi: 10.1093/emboj/18.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Xin H, Dong A, Sun C, Cao K. A novel calmodulin-like protein gene in rice which has an unusual prolonged C-terminal sequence carrying a putative prenylation site. DNA Res. 1999;6(3):179–181. doi: 10.1093/dnares/6.3.179. [DOI] [PubMed] [Google Scholar]
- Dykema PE, Sipes PR, Marie A, Biermann BJ, Crowell DN, Randall SK. A new class of proteins capable of binding transition metals. Plant Mol Biol. 1999;41(1):139–150. doi: 10.1023/A:1006367609556. [DOI] [PubMed] [Google Scholar]
- Zhu JK, Bressan RA, Hasegawa PM. Isoprenylation of the plant molecular chaperone ANJ1 facilitates membrane association and function at high temperature. Proc Natl Acad Sci USA. 1993;90(18):8557–8561. doi: 10.1073/pnas.90.18.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Yamaguchi Y, Koizumi N, Sano H. Functional characterization of a heavy metal binding protein CdI19 from Arabidopsis. Plant J. 2002;32(2):165–173. doi: 10.1046/j.1365-313X.2002.01412.x. [DOI] [PubMed] [Google Scholar]
- Galichet A, Hoyerova K, Kaminek M, Gruissem W. Farnesylation directs AtIPT3 subcellular localization and modulates cytokinin Biosynthesis in Arabidopsis. Plant Physiol. 2008;146:1155–1164. doi: 10.1104/pp.107.107425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerlin A, Bach TJ. Effects of mevinolin on cell cycle progression and viability of tobacco BY-2 cells. Plant J. 1998;14:65–74. doi: 10.1046/j.1365-313X.1998.00095.x. [DOI] [PubMed] [Google Scholar]
- Hemmerlin A, Fischt I, Bach TJ. Differential interaction of branch-specific inhibitors of isoprenoid biosynthesis with cell cycle progression in tobacco BY-2 cells. Physiol Plant. 2000;110:343–350. [Google Scholar]
- Bracha K, Lavy M, Yalovsky S. The Arabidopsis AtSTE24 is a CAAX protease with broad substrate specificity. J Biol Chem. 2002;277(33):29856–29864. doi: 10.1074/jbc.M202916200. [DOI] [PubMed] [Google Scholar]
- Cadiñanos J, Varela I, Mandel DA, Schmidt WK, Díaz-Perales A, López-Otín C, Freije JM. AtFACE-2, a functional prenylated protein protease from Arabidopsis thaliana related to mammalian Ras-converting enzymes. J Biol Chem. 2003;278(43):42091–42097. doi: 10.1074/jbc.M306700200. [DOI] [PubMed] [Google Scholar]
- Boyartchuk VL, Ashby MN, Rine J. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis.[see comment] Science. 1997;275(5307):1796–1800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- Bergo MO, Leung GK, Ambroziak P, Otto JC, Casey PJ, Young SG. Targeted inactivation of the isoprenylcysteine carboxyl methyltransferase gene causes mislocalization of K-Ras in mammalian cells. J Biol Chem. 2000;275(23):17605–17610. doi: 10.1074/jbc.C000079200. [DOI] [PubMed] [Google Scholar]
- Hancock JF, Cadwallader K, Marshal CJ. Methylation and proteolysis are essential for efficient membrane binding of prenylated p21K-ras(B) EMBO J. 1991;10:641–646. doi: 10.1002/j.1460-2075.1991.tb07992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish CA, Rando RR. Isoprenylation/methylation of proteins enhances membrane association by a hydrophobic mechanism. Biochemistry. 1996;35(26):8473–8477. doi: 10.1021/bi960603g. [DOI] [PubMed] [Google Scholar]
- Parish CA, Smrcka AV, Rando RR. The role of G protein methylation in the function of a geranylgeranylated beta gamma isoform. Biochemistry. 1996;35(23):7499–7505. doi: 10.1021/bi960271f. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Concepción M, Toledo-Ortiz G, Yalovsky S, Caldelari D, Gruissem W. Carboxyl-methylation of prenylated calmodulin CaM53 is required for efficient plasma membrane targeting of the protein. Plant J. 2000;24(6):775–784. doi: 10.1046/j.1365-313x.2000.00924.x. [DOI] [PubMed] [Google Scholar]
- Sapperstein S, Berkower C, Michaelis S. Nucleotide sequence of the yeast STE14 gene, which encodes farnesylcysteine carboxyl methyltransferase, and demonstration of its essential role in a-factor export. Mol Cell Biol. 1994;14(2):1438–1449. doi: 10.1128/mcb.14.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell DN, Sen SE, Randall SK. Prenylcysteine alpha-carboxyl methyltransferase in suspension-cultured tobacco cells. Plant Physiol. 1998;118(1):115–123. doi: 10.1104/pp.118.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chary SN, Bultema RL, Packard CE, Crowell DN. Prenylcysteine alpha-carboxyl methyltransferase expression and function in Arabidopsis thaliana. Plant J. 2002;32(5):735–747. doi: 10.1046/j.1365-313X.2002.01463.x. [DOI] [PubMed] [Google Scholar]
- Crowell DN, Kennedy M. Identification and functional expression in yeast of a prenylcysteine alpha-carboxyl methyltransferase gene from Arabidopsis thaliana. Plant Mol Biol. 2001;45(4):469–476. doi: 10.1023/A:1010671202925. [DOI] [PubMed] [Google Scholar]
- Huizinga DH, Omosegbon O, Omery B, Crowell DN. Isoprenylcysteine methylation and demethylation regulate abscisic acid signaling in Arabidopsis. Plant Cell. 2008;20(10):2714–2728. doi: 10.1105/tpc.107.053389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem AK, Bultema RL, Crowell DN. Prenylcysteine methylesterase in Arabidopsis thaliana. Gene. 2006;380(2):159–166. doi: 10.1016/j.gene.2006.05.023. [DOI] [PubMed] [Google Scholar]
- Armstrong SA, Hannah VC, Goldstein JL, Brown MS. CAAX geranylgeranyl transferase transfers farnesyl as efficiently as geranylgeranyl to RhoB. J Biol Chem. 1995;270(14):7864–7868. doi: 10.1074/jbc.270.14.7864. [DOI] [PubMed] [Google Scholar]
- Trueblood CE, Ohya Y, Rine J. Genetic evidence for in vivo cross-specificity of the CaaX-box protein prenyltransferases farnesyltransferase and geranylgeranyltransferase-I in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13(7):4260–4275. doi: 10.1128/mcb.13.7.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber E, Hemmerlin A, Hartmann M, Heintz D, Hartmann MA, Mutterer J, Rodríguez-Concepción M, Boronat A, Van Dorsselaer A, Rohmer M. et al. The plastidial 2-C-methyl-D-erythritol 4-phosphate pathway provides the isoprenyl moiety for protein geranylgeranylation in tobacco BY-2 cells. Plant Cell. 2009;21(1):285–300. doi: 10.1105/tpc.108.063248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss Y, Stradley SJ, Gierasch LM, Brown MS, Goldstein JL. Sequence requirement for peptide recognition by rat brain p21ras protein farnesyltransferase. Proc Natl Acad Sci USA. 1991;88(3):732–736. doi: 10.1073/pnas.88.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane KT, Beese LS. Thematic review series: lipid posttranslational modifications. Structural biology of protein farnesyltransferase and geranylgeranyltransferase type I. J Lipid Res. 2006;47(4):681–699. doi: 10.1194/jlr.R600002-JLR200. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Reid TS, Terry KL, Casey PJ, Beese LS. Structure of mammalian protein geranylgeranyltransferase type-I. EMBO J. 2003;22(22):5963–5974. doi: 10.1093/emboj/cdg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland CL, Windsor WT, Syto R, Wang L, Bond R, Wu Z, Schwartz J, Le HV, Beese LS, Weber PC. Crystal structure of farnesyl protein transferase complexed with a CaaX peptide and farnesyl diphosphate analogue. Biochemistry. 1998;37(47):16601–16611. doi: 10.1021/bi981197z. [DOI] [PubMed] [Google Scholar]
- Long SB, Casey PJ, Beese LS. The basis for K-Ras4B binding specificity to protein farnesyltransferase revealed by 2 A resolution ternary complex structures. Structure. 2000;8(2):209–222. doi: 10.1016/S0969-2126(00)00096-4. [DOI] [PubMed] [Google Scholar]
- Long SB, Casey PJ, Beese LS. Cocrystal structure of protein farnesyltransferase complexed with a farnesyl diphosphate substrate. Biochemistry. 1998;37(27):9612–9618. doi: 10.1021/bi980708e. [DOI] [PubMed] [Google Scholar]
- Park HW, Boduluri SR, Moomaw JF, Casey PJ, Beese LS. Crystal structure of protein farnesyltransferase at 2.25 angstrom resolution.[see comment][erratum appears in Science 1997 Apr 4;276(5309):21] Science. 1997;275(5307):1800–1804. doi: 10.1126/science.275.5307.1800. [DOI] [PubMed] [Google Scholar]