Abstract
Cyclosporine A (CsA) is a substrate for cytochrome P450 3A and the efflux transporter P-glycoprotein (P-gp; ABCB1), both abundantly expressed in the kidney. We retrospectively investigated the role of polymorphisms in CYP3A4, CYP3A5 and ABCB1 kidney graft donors on recipient renal function and subsequent graft loss, in a long-term cohort of recipients transplanted between 1990 and 2005. DNA from 227 donors and clinical data from the respective 259 recipients were analysed. Graft loss was significantly associated with the donor ABCB1 variant haplotype (1236T/2677T/3435T vs others haplotype: HR=9.346; 95%CI (2.278–38.461); p=0.0019) and with acute rejection episodes (HR=3.077; 95%CI (1.213–7.812); p=0.0178). The variant haplotype was also associated with a greater decrease in renal function (homozygotes for TTT −3.047 mL.min−1/y; heterozygotes for TTT −4.435 mL.min−1/y; others −2.186 mL.min−1/y; p=0.0240). ABCB1 polymorphisms in donors influence long-term graft outcome, favouring the decrease in renal function and graft loss in transplant recipients receiving CsA.
Keywords: Adult; Aged; Analysis of Variance; Cohort Studies; Cyclosporine; therapeutic use; Cytochrome P-450 CYP3A; genetics; Female; Follow-Up Studies; Genotype; Graft Rejection; drug therapy; genetics; Haplotypes; Humans; Immunosuppressive Agents; therapeutic use; Kaplan-Meiers Estimate; Kidney; physiology; Kidney Function Tests; Kidney Transplantation; physiology; Male; Middle Aged; P-Glycoprotein; genetics; Polymorphism, Genetic; genetics; physiology; Retrospective Studies; Risk Factors; Tissue Donors; statistics & numerical data; Young Adult
Keywords: Kidney transplantation, cyclosporine A, pharmacogenetics, graft loss, P-glycoprotein, CYP3As
Introduction
Cyclosporine A (CsA) is a calcineurin inhibitor which has considerably improved graft outcome in solid organ transplantation. However, its clinical benefits are balanced by its side effects, mainly chronic nephrotoxicity which remains a major problem in all types of solid organ transplantation (1). CsA is a substrate of the efflux transporter P-glycoprotein (P-gp; ABCB1 gene) extruding xenobiotics from the cell (2). The most frequent polymorphisms of ABCB1 are located in exons 12 (1236 C>T), 21 (2677 G>T) and 26 (3435 C>T). There have been few studies investigating the consequences of ABCB1 polymorphisms on the efficacy or toxicity of immunosuppressive therapy in renal transplantation. Crettol et al. described higher cyclosporine concentrations in the lymphocytes of carriers of the variant ABCB1 3435T allele (3) suggesting that P-gp could influence CsA activity independently of its effect on the drug bioavailability or clearance. However, only a weak association was found between the recipient’s ABCB1 haplotype gathering the exon 12, 21 and 26 alleles and acute graft rejection, and no association with graft loss (4). The impact of the silent 3435 C>T SNP on the pharmacokinetics of CsA is controversial (5). It has been argued that ABCB1 mRNA stability would be lower when the 3435-T allele is present at exon 26 (6), or that P-gp conformation would be altered as a result of this silent mutation, the rarity of the corresponding codon affecting the timing of cotranslational folding and insertion of P-gp into the membrane (7). Since P-gp is highly expressed in renal proximal tubule epithelial cells (8), its activity in the kidney graft, which carries the donor’s and not the recipient’s genome, may contribute to CsA toxicity. This was confirmed by Hauser et al. (n=97), who showed that the donor’s but not the recipient’s ABCB1 3435 variant genotype was associated with CsA nephrotoxicity (OR, 13.4; 95% CI, 1.2 to 148; p=0.034) (9). On the other hand, CsA is a substrate for CYP3A enzymes. Among them CYP3A5 is highly polymorphic: the CYP3A5*3 allele results in an alternative splice site in the pre-mRNA leading to a truncated enzyme (10); CYP3A5*1 is the wild-type allele, associated with enzyme activity. It was shown that CYP3A4 kidney expression is much lower than that of CYP3A5 (when expressed) and that the oxidative metabolism of CsA in human renal microsomes is strongly dependent to CYP3A5 activity (11–13). It is thus possible that the donor’s status for CYP3A5 could also have a role in the in situ accumulation, hence local toxicity, of CsA.
The goal of this study was to investigate the influence of the donor’s polymorphisms in CYP3A and ABCB1 on graft loss in a large retrospective cohort with a long-term follow-up of renal transplant recipients given CsA.
Results
Clinical data
Clinical and demographic characteristics of the 259 recipients who fulfilled the inclusion and exclusion criteria are reported in Table 1. In particular, we included recipients for whom there was donor DNA available from an original set of 473 renal transplants, performed at Limoges University Hospital between 1990 and 2006 (inclusive). Patients’ follow up was 72 months on average (ranging from 8 to 190 months). The grafts came from 227 cadaveric donors with age range of 12–73 years (39±14) and sex ratio of 155/72 (M/F). A total of 20 graft losses (7.7% of recipients) were observed in this cohort within the period considered (Table 1).
Table 1.
Patients’ characteristics
| Recipients | |
|---|---|
| n | 259 |
| Age (min-max) | 54±14 years (19–78) |
| Sex ratio M/F | 162/97 |
| Cold ischemia time (min-max) | 1195±347 min (303–2580) |
| Number of HLA mismatches (min-max) | 3.6±1.2 (0–6) |
| Follow-up period (min-max) | 72.0±46.2 months (8.1–190.0) |
| Acute rejection: n (%) with ≥ 1 episode | 80 (30.9%) |
| Graft loss: n (%) | 20 (7.7%) |
| Time between transplantation and graft loss (min-max) | 78.1±41.1 months (13.8–161.6) |
| Delta creatinine clearance per year (ml.min−1/year) | −3.39±6.07 |
Parameters are expressed as mean±SD
Genotype/haplotype distribution and association with graft loss
Genotyping results were in the Hardy-Weinberg equilibrium and similar to those reported in the literature (14) (Table 2). As previously described, a strong LD was observed between exons 12, 21, 26 SNPs of ABCB1 (D′12–26=0.69, r212–26=0.33; D′12–21=0.92, r212–21=0.84; D′21–26=0.72, r221–26=0.37). The most probable haplotype could not be inferred in 6 patients because the probability of the 2 haplotypes proposed was close to 50%. Of the 8 haplotypes identified, the 3 most frequent ones were CGC (40.9%), TTT (36.2%) and CGT (14.1%), the other five representing 8.8% of patients’ haplotypes.
Table 2.
Frequency and distribution of the studied polymorphisms in donors (n=227)
| Gene | Polymorphisms | Allelic frequencies (variant allele) | Genotype distributions | ||
|---|---|---|---|---|---|
| wt/wt | wt/m | m/m | |||
| ABCB1 | 1236 C>T | 0.427 | 75/227 | 110/227 | 42/227 |
| 2677 G>T | 0.427 | 75/227 | 110/227 | 42/227 | |
| 3435 C>T | 0.509 | 59/227 | 105/227 | 63/227 | |
| CYP3A4 | −392 A>G (CYP3A4*1B) | 0.037 | 210/227 | 17/227 | 0/227 |
| CYP3A5 | 6986 A>G (CYP3A5*3) | 0.923 | 0/227 | 35/227 | 192/227 |
wt: wild type, m: mutated
Univariate analysis showed that patients who received their graft from donors carrying the variant ABCB1 1236 TT, 2677TT and 3435TT genotypes had an increased risk of graft loss (Table 3) compared to patients whose donors were carriers of the wild-type genotypes (CC1236, GG2677 and CC3435).
Table 3.
Univariate analysis (Cox Model) of the influence of the different variables studied on graft loss.
| Variable | Category2 | Hazard ratio | CI 95% | p |
|---|---|---|---|---|
| 6986 A>G (CYP3A5*3) | AG vs GG | 2.437 | 0.867–6.850 | 0.0910 |
| −392 A>G (CYP3A4*1B) | AG vs AA | 0.913 | 0.121–6.896 | 0.9299 |
| ABCB1 1236 C>T | TT vs CC | 8.403 | 1.733–41.667 | 0.0083 |
| CT vs CC | 1.825 | 0.705–4.717 | 0.2149 | |
| ABCB1 2677 G>T | TT vs GG | 10.417 | 2.105–52.631 | 0.0041 |
| GT vs GG | 2.174 | 0.829–5.682 | 0.1143 | |
| ABCB1 3435 C>T | TT vs CC | 3.921 | 1.059–14.493 | 0.0407 |
| CT vs CC | 1.211 | 0.313–4.695 | 0.7820 | |
| ABCB1 haplotype 1236 C>T/2677 G>T/3435 C>T | TTT vs CGC1 | 2.616 | 1.373–4.986 | 0.0035 |
| Age recipient | per year increase | 0.995 | 0.965–1.026 | 0.7496 |
| Age donor | per year increase | 1.030 | 0.995–1.067 | 0.1254 |
| Sex recipient | Female vs Male | 0.806 | 0.329–1.977 | 0.6398 |
| Sex donor | Female vs Male | 0.885 | 0.360–2.178 | 0.7906 |
| Acute rejection | ≥1 vs 0 | 2.756 | 1.095–6.933 | 0.0321 |
| Cold ischemia time | per min. increase | 0.999 | 0.998–1.001 | 0.3218 |
| Total HLA mismatch | per mismatch increase | 0.885 | 0.629–1.245 | 0.4851 |
Estimation of the risk when carrying the mutated allele vs. the wild type allele calculated with THESIAS
For the most frequent category of the variable, taken as reference, HR=1.
Haplotype analysis revealed that patients who received their graft from a donor carrying the TTT haplotype had an increased risk of graft loss compared to those whose grafts came from a CGC haplotype carrier (HR=2.616; 95%CI (1.373–4.986); p=0.0035) (Table 3). Graft loss episodes occurred in 6/33 (18%) of the recipients from donors homozygous for the TTT haplotype, 11/123 (9%) of those with donors heterozygous for TTT and 3/97 (3%) of hosts of another haplotype.
No significant association was found between the donor’s CYP3A4*1B or CYP3A5 genotype and graft loss (Table 3).
Association of non genetic factors with graft loss
Univariate analysis of non genetic variables revealed a strong association between occurrence of acute rejection (AR ≥ 1 episode) and graft loss (HR=2.756; 95%CI (1.095–6.933); p=0.0321) (Table 3). On the other hand, no significant association between graft loss and recipients’ sex or age, donor’s sex or age, cold ischemia time or number of HLA mismatches was found (Table 3).
Independent risk factors for graft loss: multivariate analysis
Only the donor’s ABCB1 G2677T SNP and acute rejection remained significant in the final multivariate Cox model (Table 4). Patients who received their graft from a donor carrying the 2677TT or the G2677T genotype had a graft loss hazard ratio of 12.0 and 4.7, respectively as compared to those transplanted from carriers of the GG2677 genotype. Simultaneously, patients who had at least one episode of acute rejection had a graft loss hazard ratio of 3.1 as compared to those who did not experience acute rejection.
Table 4.
Multivariate analysis (Cox Model) for risk of graft loss (taking in account either the three ABCB1 genotypes separately or the corresponding haplotype).
| Variable | Category | Adjusted Hazard ratio | Adjusted CI 95% | p |
|---|---|---|---|---|
| ABCB1 2677 G>T | TT vs GG | 12.048 | 2.392–58.823 | 0.0025 |
| GT vs GG | 4.673 | 1.020–21.276 | 0.0471 | |
| Acute rejection | ≥1 vs 0 | 3.077 | 1.213–7.812 | 0.0178 |
| ABCB1 haplotype | TTT/TTT vs other/other | 9.346 | 2.278–38.461 | 0.0019 |
| TTT/other vs other/other | 3.322 | 0.918–12.048 | 0.0671 | |
| Acute rejection | ≥1 vs 0 | 2.849 | 1.129–7.194 | 0.0266 |
In the final Cox model including the donor’s ABCB1 haplotype instead of the ABCB1 SNPs, again only the ABCB1 haplotype and acute rejection remained significant. Patients who received their graft from a donor homozygous or heterozygous for the ABCB1 variant (TTT) haplotype had a graft loss hazard ratio of 9.3 and 3.3, respectively, as compared to those transplanted from carriers of other haplotype combinations (Table 4). Simultaneously, patients who had had at least one acute rejection episode had a graft loss hazard ratio of 2.8 as compared to those with no acute rejection episode.
Survival analysis further confirmed that the cumulative incidence of graft loss was significantly higher when donors carried the ABCB1 TTT haplotype (p=0.0028) (Figure 1).
Figure 1.
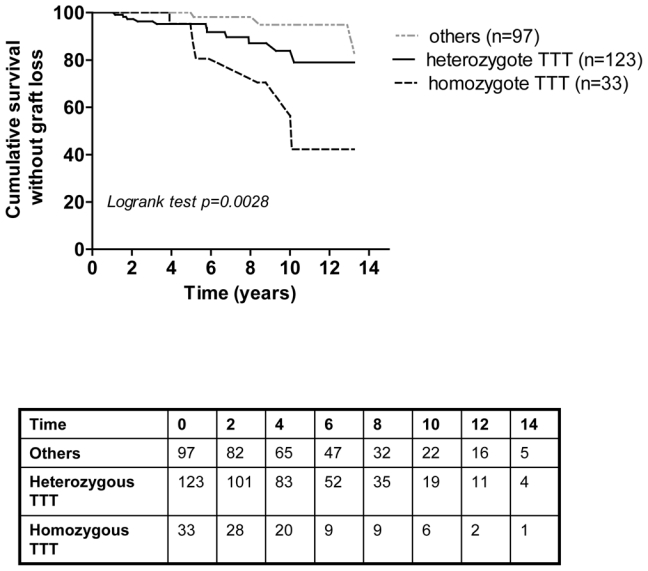
Kaplan-Meier curves of cumulative survival without graft loss for patients transplanted from donor homozygous carriers, heterozygous or non-carriers of the ABCB1 TTT haplotype.
Association of ABCB1 haplotype with renal function
A significantly more important decrease of creatinine clearance over the follow-up period (ΔCrCl/y) was observed when donors were carriers of the variant TTT haplotype (homozygotes for TTT: μΔCrCl/y =−3.047 ± 4.008 mL.min−1 per year, heterozygotes for TTT: μΔCrCl/y =−4.435 ± 6.803 mL.min−1 per year and others: μΔCrCl/y =−2.186 ± 5.583 mL.min−1 per year; p=0.0240). Similar results were obtained when regression analysis of creatinine clearance measured annually was performed and the average slope was compared between the different haplotype groups (data not shown).
Discussion
Based on the data collected retrospectively in 259 renal transplant patients receiving CsA with a mean follow-up of 72 months (range: 8 to 190 months), we found that the ABCB1 1236T, 2677T and 3435T variant alleles and the corresponding (1236T-2677T-3435T) variant haplotype in graft donors were associated with a higher risk of graft loss. Multivariate analysis considering this haplotype, the CYP3A5*3 and CYP3A4*1B polymorphisms recipients’ demographic characteristics, donors’ age and sex, graft cold ischemia time, number of HLA mismatches, and occurrence of acute rejection identified the ABCB1 variant haplotype and acute rejection as the only significant predictors of long-term graft survival. Moreover, the decrease in renal function over the follow-up period (estimated as delta creatinine clearance per year) was more pronounced when the donor was carrier of the ABCB1 TTT haplotype.
This study was based on a long-term collection of donors’ DNA samples by the Immunology laboratory of Limoges University Hospital and was based on the hypothesis that polymorphisms of the donor’s P-gp or CYP3A proteins (reflecting their local activity in the kidney graft) could influence CsA long-term nephrotoxicity and finally graft loss. P-gp acts as a transport pump actively excreting CsA from the epithelial tubular cells into urine. As it is abundantly expressed in these cells, normal P-gp activity in the graft may protect them from CsA toxicity.
Consistently, Hauser et al., 2004 showed in 97 kidney transplant patients that approximately 40% of kidney recipients transplanted from donors homozygous for the ABCB1 3435T variant allele developed CsA nephrotoxicity within 2.5 years post-transplantation, compared to only 10% of patients transplanted from donors carrying the heterozygous (C3435T) or the reference (CC3435) genotype (9). In addition, Joy et al. showed using immunohistochemistry that kidney P-gp expression was lower in 39 renal transplant patients exhibiting structural signs of nephrotoxicity to calcineurin inhibitors (CNI) than in 32 controls (15). Finally, a recent study by Naesens et al. showed in patients on tacrolimus that the donor’s and recipient’s ABCB1 TT 3435 C>T variant alleles were associated with an increased risk of chronic allograft damage (16). This group further showed there was no association with tacrolimus exposure suggesting that this outcome is likely to be related to accumulation of tacrolimus in epithelial cells. The authors hypothesized that the recipient’s polymorphism is significantly linked to the outcome due to important epithelial chimerism after renal transplantation. Although no association between ABCB1 polymorphism and graft survival was found at 3 years, they suggested that a longer follow-up would be necessary to observe the progression from chronic histological damage to renal graft loss. Our retrospective study provides convincing evidence of the importance of the donor’s ABCB1 genotype on graft loss in renal transplant patients on CsA with long-term follow-up. Moreover, the analysis of the evolution of renal function showed that when the donor carried the ABCB1 TTT haplotype creatinine clearance declined more rapidly, leading to a shorter time to graft loss. It is noteworthy that we did not observe a gradual change of renal function with haplotype copy number. A possible explanation is that creatinine clearance is a continuous variable subject to high variability, and that the number of TTT homozygotes was too low as compared to heterozygotes to observe a potential additive effect.
Cattaneo et al. recently found that recipients carriers of the T allelic variants of exons 21 and 26 had an increased risk for CsA-related adverse events, including delayed graft function and lower GFR (17). In the present study, the analysis of recipient DNA polymorphisms was performed on 9 graft loss episodes only, because 11 recipients with graft loss died before the beginning of this retrospective study, i.e. before we could collect their DNA (these patients were amongst those transplanted farthest back in time and graft loss may have shortened their survival). We found no association between ABCB1 recipient haplotype and graft loss (data not shown) but the low number of events makes any conclusion difficult to drawn.
In addition to ABCB1 polymorphism, multivariate analysis identified acute rejection as another, already well-known (18–20), independent factor of graft loss, however with lower hazard ratio values than the ABCB1 polymorphisms or haplotype.
There are some limitations to this study. First, there is a lack of an independent validation group. It is rare to find recipients with long-term follow-up data for whom donor DNA is available as can be seen by the paucity of studies in this domain (21, 22). When such data exist, there is a short retrospective follow-up of recipients (9, 16). Despite our efforts and contacts with colleagues, we have been unable to find an independent database with a follow-up period as long as for our recipients.
Secondly, we missed data on immunosuppressant blood concentrations, in particular CsA, due to the impossibility to reliably retrieve these data retrospectively over such a long period of time. Third, precise histological analysis of explanted grafts is missing. Out of the 28 patients who lost their graft, 8 were excluded from the analysis owing to graft loss attributable to clear external causes. The 20 patients remaining had a graft biopsy at one point in time during the follow-up period. Seventeen of these biopsies allowed histological analysis of the graft and most (12/17) showed histological damages compatible with CsA toxicity. Unfortunately, case-control comparisons on histological findings in the whole cohort could not be undertaken because biopsies were not performed in a majority of patients. Finally, it is evident that graft loss is the result of multiple other factors, the study of which was beyond the goal of the present work that specifically aimed to investigate the role of genetic factors from the donor in the occurrence of graft loss, owing to its retrospective design that made it possible to study this rare and late occurring event.
In conclusion, we found that the ABCB1 variant haplotype of graft donors is associated with a significantly higher risk of graft loss in kidney transplant recipients. It suggests that genetic variations in ABCB1 are predictive of graft outcomes through P-gp influence on local CsA concentration. Our study provides additional and complementary evidence to the results of Naesens et al. who showed that in patients receiving calcineurin inhibitors ABCB1 polymorphisms are predictive of the renal graft deterioration independently of graft quality at transplantation (16). Owing to developments in genetic technology for clinical use, it can be proposed that genotyping for ABCB1 be included as part of systematic genotyping of the donor in routine practice and that the donor ABCB1 haplotype be taken in account in the choice or the doses of the immunosuppressants.
Methods
Patient population
The clinical data-on-file from renal graft recipients between 1990 and 2005, routinely followed as outpatients at Limoges University hospital were retrospectively studied. The ethics committee of Limoges hospital approved the protocol. Informed consent was obtained from each living patient, while French Health Authorities have waived the requirement for consent for deceased patients and donors. The following inclusion criteria were used: recipient age > 18 years; kidney graft from a cadaveric donor (following brain death); patients treated with CsA (SANDIMMUN® or NEORAL®); functional graft after more than one year posttransplantation; donor’s DNA available. The exclusion criteria were: patient age < 18 years; pregnancy; combined kidney and pancreas, heart or liver transplantation; patient treated with tacrolimus or an mTOR inhibitor; graft survival < 1 year so as to exclude precocious graft loss, which is not due to nephrotoxicity. Eight patients were excluded from the analysis because they lost their graft as a consequence of a clearly identified cause: recurrence of the primary kidney disease or de novo glomerulopathy (4), myeloma (1), lymphoma (1), uterus cancer (1), and septicaemia (1). Patients who died with a functional graft were excluded at the time of death (death-censored graft survival). For each patient, the following clinical data were recorded from the medical file by the same nephrologist (JPR) before the genetic study was carried out: date of birth, sex, HLA mismatches between donor and recipient, duration of cold ischemia, induction therapy, immunosuppressive drug regimens, treated acute rejection episodes within the period of follow-up (acute rejection proven by histological examination of a biopsy or treated as acute rejection without biopsy confirmation), creatinine clearance each year and graft survival. The creatinine clearance was estimated for each patient using the Cockcroft and Gault formula. Patients’ decrease in renal function over time was estimated by subtracting the last creatinine clearance value available to the value at one year posttransplantation divided by follow-up duration (delta creatinine clearance per year). The only data available for donors was age and sex.
Donors’ DNA bank
Each donor’s genomic DNA was extracted from lymph nodes or spleen lymphocytes by salting out extraction as previously described (23) and banked by the Immunology laboratory of Limoges University Hospital as part of routine practice.
Identification of genotypes
Donor’s genotypes for CYP3A4 −392 A>G (CYP3A4*1B allele, rs2740574), CYP3A5 6986 A>G (CYP3A5*3 allele, rs776746) SNPs, and ABCB1 1236 C>T (exon 12, rs1128503), 2677 G>T (exon 21, rs2032582) and 3435 C>T (exon 26, rs1045642) SNPs were determined using TaqMan allelic discrimination assays on an ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Courtaboeuf, France). Primers and probes were purchased from Applied-Biosystems as validated custom or DME assays.
Statistical analyses
Deviations from the Hardy-Weinberg equilibrium were studied using the Fisher’s exact test. The effect of gene polymorphisms (SNPs or haplotypes) on graft loss was investigated using the Cox proportional hazard model. P values less than 0.05 were considered significant and 95% confidence intervals were calculated when relevant. For SNPs and haplotypes association analyses, the most frequent allele was considered as the reference. For multivariate analysis, the significance of variables in the final model was tested by a backward stepwise process using the likelihood ratio to evaluate the effect of omitting variables.
After studying the effect of each polymorphism independently, the association of haplotypes with graft loss was analyzed using the THESIAS program (http://genecanvas.ecgene.net) (24), based on the maximum likelihood model described by Tregouët et al. and linked to the s.e.m. algorithm. This software was used to estimate pairwise linkage disequilibrium (LD) between ABCB1 polymorphisms. The extent of disequilibrium was expressed in terms of D′ and r2.
The most probable haplotypes were inferred for each patient using the PHASE V2.0 program (25) and for the significant polymorphisms, time-to-event data were estimated using Kaplan-Meier analysis for patient with or without the polymorphism of interest, and groups were compared using the log-rank test.
One way analysis of variance (ANOVA) was performed to test the influence of significant polymorphisms on delta creatinine clearance per year.
All statistical analyses were performed using Statview 5.0 (SAS Institute Inc, Cary, NC, USA), except when stated otherwise.
Acknowledgments
We thank the CHU Limoges and the University of Limoges for their support. We gratefully thank J. H. Comte for his contribution and Jana Stojanova for manuscript editing.
References
- 1.Myers BD, Ross J, Newton L, Luetscher J, Perlroth M. Cyclosporine-associated chronic nephropathy. N Engl J Med. 1984;311:699–705. doi: 10.1056/NEJM198409133111103. [DOI] [PubMed] [Google Scholar]
- 2.Sakaeda T, Nakamura T, Okumura K. Pharmacogenetics of MDR1 and its impact on the pharmacokinetics and pharmacodynamics of drugs. Pharmacogenomics. 2003;4:397–410. doi: 10.1517/phgs.4.4.397.22747. [DOI] [PubMed] [Google Scholar]
- 3.Crettol S, et al. Influence of ABCB1 genetic polymorphisms on cyclosporine intracellular concentration in transplant recipients. Pharmacogenet Genomics. 2008;18:307–15. doi: 10.1097/FPC.0b013e3282f7046f. [DOI] [PubMed] [Google Scholar]
- 4.Bandur S, Petrasek J, Hribova P, Novotna E, Brabcova I, Viklicky O. Haplotypic structure of ABCB1/MDR1 gene modifies the risk of the acute allograft rejection in renal transplant recipients. Transplantation. 2008;86:1206–13. doi: 10.1097/TP.0b013e318187c4d1. [DOI] [PubMed] [Google Scholar]
- 5.Leschziner GD, Andrew T, Pirmohamed M, Johnson MR. ABCB1 genotype and PGP expression, function and therapeutic drug response: a critical review and recommendations for future research. Pharmacogenomics J. 2007;7:154–79. doi: 10.1038/sj.tpj.6500413. [DOI] [PubMed] [Google Scholar]
- 6.Wang D, Johnson AD, Papp AC, Kroetz DL, Sadee W. Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharmacogenet Genomics. 2005;15:693–704. [PubMed] [Google Scholar]
- 7.Kimchi-Sarfaty C, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–8. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 8.Thiebaut F, Tsuruo T, Hamada H, Gottesman MM, Pastan I, Willingham MC. Cellular localization of the multidrug-resistance gene product P-glycoprotein in normal human tissues. Proc Natl Acad Sci U S A. 1987;84:7735–8. doi: 10.1073/pnas.84.21.7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hauser IA, et al. ABCB1 genotype of the donor but not of the recipient is a major risk factor for cyclosporine-related nephrotoxicity after renal transplantation. J Am Soc Nephrol. 2005;16:1501–11. doi: 10.1681/ASN.2004100882. [DOI] [PubMed] [Google Scholar]
- 10.Kuehl P, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–91. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 11.Haehner BD, et al. Bimodal distribution of renal cytochrome P450 3A activity in humans. Mol Pharmacol. 1996;50:52–9. [PubMed] [Google Scholar]
- 12.Dai Y, et al. In vitro metabolism of cyclosporine A by human kidney CYP3A5. Biochem Pharmacol. 2004;68:1889–902. doi: 10.1016/j.bcp.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 13.Dai Y, et al. Effect of CYP3A5 polymorphism on tacrolimus metabolic clearance in vitro. Drug Metab Dispos. 2006;34:836–47. doi: 10.1124/dmd.105.008680. [DOI] [PubMed] [Google Scholar]
- 14.Anglicheau D, et al. CYP3A5 and MDR1 genetic polymorphisms and cyclosporine pharmacokinetics after renal transplantation. Clin Pharmacol Ther. 2004;75:422–33. doi: 10.1016/j.clpt.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 15.Joy MS, Nickeleit V, Hogan SL, Thompson BD, Finn WF. Calcineurin inhibitor-induced nephrotoxicity and renal expression of P-glycoprotein. Pharmacotherapy. 2005;25:779–89. doi: 10.1592/phco.2005.25.6.779. [DOI] [PubMed] [Google Scholar]
- 16.Naesens M, Lerut E, de Jonge H, Van Damme B, Vanrenterghem Y, Kuypers DR. Donor age and renal P-glycoprotein expression associate with chronic histological damage in renal allografts. J Am Soc Nephrol. 2009;20:2468–80. doi: 10.1681/ASN.2009020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cattaneo D, et al. ABCB1 genotypes predict cyclosporine-related adverse events and kidney allograft outcome. J Am Soc Nephrol. 2009;20:1404–15. doi: 10.1681/ASN.2008080819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cosio FG, et al. Impact of acute rejection and early allograft function on renal allograft survival. Transplantation. 1997;63:1611–5. doi: 10.1097/00007890-199706150-00013. [DOI] [PubMed] [Google Scholar]
- 19.Lindholm A, Ohlman S, Albrechtsen D, Tufveson G, Persson H, Persson NH. The impact of acute rejection episodes on long-term graft function and outcome in 1347 primary renal transplants treated by 3 cyclosporine regimens. Transplantation. 1993;56:307–15. doi: 10.1097/00007890-199308000-00010. [DOI] [PubMed] [Google Scholar]
- 20.Matas AJ, Gillingham KJ, Payne WD, Najarian JS. The impact of an acute rejection episode on long-term renal allograft survival (t1/2) Transplantation. 1994;57:857–9. doi: 10.1097/00007890-199403270-00015. [DOI] [PubMed] [Google Scholar]
- 21.Kreutz R, et al. CYP3A5 genotype is associated with longer patient survival after kidney transplantation and long-term treatment with cyclosporine. Pharmacogenomics J. 2008;8:416–22. doi: 10.1038/sj.tpj.6500488. [DOI] [PubMed] [Google Scholar]
- 22.Varagunam M, Yaqoob MM, Dohler B, Opelz G. C3 polymorphisms and allograft outcome in renal transplantation. N Engl J Med. 2009;360:874–80. doi: 10.1056/NEJMoa0801861. [DOI] [PubMed] [Google Scholar]
- 23.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tregouet DA, Tiret L. Cox proportional hazards survival regression in haplotype-based association analysis using the Stochastic-EM algorithm. Eur J Hum Genet. 2004;12:971–4. doi: 10.1038/sj.ejhg.5201238. [DOI] [PubMed] [Google Scholar]
- 25.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–9. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]


