Abstract
Signal transduction in response to ligand recognition by T cell receptors regulates T cell fate within and beyond the thymus. Herein we examine the involvement of the CD4 molecule in the regulation of T helper cell survival. T helper cells that lack CD4 expression are prone to apoptosis and show diminished survival after adoptive transfer to irradiated recipients. The helper lineage in CD4−/− animals shows a higher than normal apparent rate of cell division and is also enriched for cells exhibiting a memory cell phenotype. Thus the data point to a necessary role for CD4 in the regulation of T helper cell survival and homeostasis.
The thymus has a dominant and essential role in establishing TCR diversity. One of its major contributions is to allow V(D)J rearrangements to occur at the genomic T cell receptor (TCR) loci of immature T cell precursors (1, 2). Another critical function is the imposition of a stringent selection for cells expressing TCRs that engage their peptide/MHC ligands with appropriate kinetics (3–5). As a consequence of this selection, the population of cells that emerges from the thymus is substantially enriched for cells that are likely to be beneficial to the host during immune responses.
In addition to selection in the thymus, there is evidence that the nascent T cell repertoire comes under selective pressure in the periphery (6–11). Much of this evidence comes from adoptive transfer experiments that trace the fate of T cells in hosts with defective peptide/MHC ligand expression (7, 10). In some of these experiments, T cell numbers markedly decline, implicating the TCR and peptide/MHC engagement in the provision of signals that are critical for the survival of T cells. There are also data indicating that the requirement for ligand-dependent TCR signaling may be more important for naive than memory T cells (12, 13).
Through its capacity to bind MHC class II molecules and recruit p56lck to the vicinity of antigen-bound TCRs, CD4 can substantially improve the reactivity of class II-restricted thymocytes and T cells (14, 15). The significance of this coreceptor function is particularly apparent in the thymus, where loss of CD4 expression results in a substantial, yet not insurmountable, developmental block (16, 17). It is also evident in immune responses that are similarly disabled by CD4 deficiency (18, 19). Herein we show that, in addition to its crucial role during thymocyte development and immune responses, CD4 is also critically involved in promoting the survival of T helper cells. We find that the lack of CD4 expression significantly increases the likelihood of T helper cell apoptosis and leads to a dramatic depletion of naive phenotype cells from the T helper cell population. The data make clear an essential function for CD4 in peripheral selection of the T helper cell repertoire.
Materials and Methods
Mice, Antibodies, and Reagents.
C57BL/6, MRL/MpJ, B6.Ly5.1, B6.RAG-1−/−, B6.CD4−/−, Eμ-bcl-2–25, and B6.β2m−/− mice were obtained from the Jackson Laboratory or Charles River Breeding Laboratories or were bred in-house in the Parnassus Heights Barrier Facility. MRL/lpr CD4−/− mice were provided by David Wofsy (Univ. of California, San Francisco). Mice expressing a Tva transgene controlled by CD4 regulatory elements (cd4-TVA) have been described.† In CD4+/+ cd4-TVA transgenic mice, Tva, the receptor for subgroup A avian sarcoma and leukosis viruses (21), is expressed on all thymocytes and T cells in a fashion that follows endogenous CD4 expression. Tva is thus a marker for the CD4 lineage, similar to other previously described markers (22, 23).
Subgroup A avian sarcoma and leukosis viruses envelope-Ig fusion protein (SUA-rIgG), used to detect Tva, was as described (24). Fluorescent antibodies and secondary reagents were from PharMingen, Caltag, Becton Dickinson, Southern Biotechnology Associates, R&D Systems, and Jackson ImmunoResearch. Carboxyfluorescein succinimidyl ester (CFSE) was purchased from Molecular Probes.
Flow Cytometry.
Single-cell suspensions were incubated with antibodies for 30 min at 4°C in PBS/0.3% BSA/0.01% NaN3. The cells were washed once between staining steps and three times before analysis. Cells were analyzed with a Becton Dickinson FACScan and CELLQUEST software. Anti-BrdUrd staining was as described (25, 26). IFN-γ staining was performed on cells sorted for Tva and high or low CD62L expression with a MoFlo (Cytomation, Fort Collins, CO) cell sorter. After sorting, the cells were activated with phorbol 12-myristate 13-acetate (50 ng/ml) and ionomycin (2 μg/ml) for 4 h, with brefeldin A (10 μg/ml) present during the last 2 h. The cells were then fixed in 4% paraformaldehyde, washed, permeabilized in 0.5% saponin in PBS/1% FCS, and stained with FITC-coupled anti-IFN-γ in the permeabilization buffer. The cells were washed twice in the same buffer before analysis with the FACScan. Apoptotic cells were detected by flow cytometry with annexin V-coupled to FITC according to the manufacturer's instructions.
Intrathymic Injections.
Injections were performed as described (27). Briefly, 4- to 5-week-old mice were anesthetized with avertin (tribromoethanol; Sigma). An incision was made in the sternum to reveal the thymus, and approximately 20 μl of a FITC solution (1 mg/ml) in PBS was injected into each thymic lobe. Pooled cells from the spleen, mesenteric, inguinal, brachial, and cervical lymph nodes were analyzed by flow cytometry 24 h after injection for the presence of FITC-labeled recent thymic emigrants.
Cell Transfers and CFSE Labeling.
CD4−/− males that were homozygous for the cd4-TVA transgene were mated to either B6.Ly5.1+ or CD4−/− Ly5.1− females, and fetal liver tissue was obtained by dissection of embryos on gestational day 14. Cell suspensions were prepared in IMDM supplemented with 20% FCS by passing tissue through a 22-gauge needle followed by a 25-gauge needle. Fetal liver preparations from each genotype were pooled and combined in the indicated mixtures. Approximately 4 × 106 cells were then injected into the tail veins of irradiated (1000 rads; 1 rad = 0.01 Gy) 6-week-old C57BL/6 mice. Transplanted animals received water containing neomycin (1.1%) and polymixin B sulfate (850 U/ml) until analysis 6–8 weeks after transplantation.
Single-cell suspensions of pooled lymph nodes and spleen were prepared from C57BL/6 or CD4−/− donors, washed in PBS, and incubated in 3.3 μM CFSE in PBS for 8 min at room temperature (28). At the end of the incubation, FCS was added to quench the CFSE, and cells were washed twice in RPMI medium containing 15% FCS. The labeled cells were then injected into the tail veins of sex-matched 8- to 12-week-old RAG-1−/− mice (1 × 107 or 5 × 107 cells per recipient). For transfers of purified T cells, single-cell suspensions were sorted by negative selection with anti-B220, anti-MHC class II, and anti-CD8 microbeads and LS+ magnetic columns (Miltenyi Biotec, Auburn, CA). Wild-type and CD4-deficient cells were then mixed at a 1:1 ratio and 5 × 106 total CD4 lineage cells were transferred into RAG-1−/− recipients as above.
Results
Reduced Numbers of T Cells in CD4−/−β2m−/− Mice.
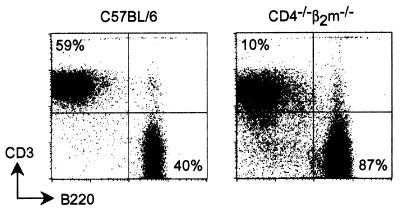
During analysis of the phenotype of CD4-deficient mice, we had generated CD4−/−β2m−/− doubly deficient animals (29). Mice lacking CD4 alone and the double null mice contained mature CD4 lineage cells that were capable of helper cell responses despite the absence of CD4 (18, 19, 29, 30). These cells normally make up ≈10% of the peripheral T cell pool in CD4−/− mice. Similar to mice that lack MHC class II or CD8 expression (31, 32), mice lacking CD4 or β2m alone had an approximately normal amount of total T cells in their peripheral lymphoid tissues (16, 17, 33). In contrast, the CD4−/−β2m−/− double null mice had many fewer mature αβ T cells than wild-type mice (Fig. 1). Lymph nodes from C57BL/6 control animals had an average of 62.4 ± 5.4% T cells, but those of CD4−/−β2m−/− animals contained only 12.8 ± 2.8% T cells (n = 8 mice per group). The same relative proportions of T cells were observed in both young and old (>6 months) mice. Although the absence of CD4 impairs positive selection of the CD4 lineage in the thymus (16, 17), wild-type T helper cells are capable of extensive proliferation and small numbers can readily expand to reconstitute peripheral compartments (34). We therefore sought a better understanding of how the absence of CD4 impaired the representation of peripheral T helper cells in CD4−/− and CD4−/−β2m−/− animals.
Figure 1.
Reduced frequency of peripheral T cells in CD4−/−β2m−/− mice. Lymph node cells from C57BL/6 and CD4−/−β2m−/− mice were analyzed by flow cytometry with anti-B220 and anti-CD3ɛ antibodies. Data are representative of 10 animals of each genotype examined in three experiments.
Reduced Positive Selection but Efficient Thymic Egress in the Absence of CD4.
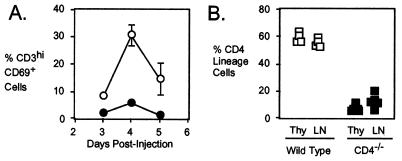
To determine the magnitude of the effect of CD4 on the efficiency of positive selection, CD4−/−β2m−/− and CD4-expressing β2m−/− animals were injected with BrdUrd to pulse-label a cohort of dividing cortical thymocytes. The development of these cells into the CD4 lineage was then followed by flow cytometry over the next 5 days, by using up-regulation of CD3 and CD69 as markers of positive selection. β2m-deficient animals were used so that the selection of cells on MHC class II molecules could be examined in the absence of MHC class I-dependent selection. As shown in Fig. 2A, the percentage of labeled positively selected thymocytes (CD3hiCD69+ cells) was typically 5-fold lower, as measured at day 4 in the absence of CD4 (5.9 ± 1.0% labeled cells), than in the presence of CD4 (30.6 ± 3.9% labeled cells). Similar results were obtained when mature thymocytes were identified by up-regulation of CD2 (35) rather than CD69 (data not shown). These kinetic data indicate that positive selection of CD4 lineage thymocytes is about 5-fold less efficient in the absence of CD4, consistent with the steady-state representation of mature T helper lineage cells in CD4-expressing vs. CD4−/− thymuses (16, 17).
Figure 2.
Decreased positive selection but efficient thymic egress of CD4 lineage cells in the absence of CD4. (A). A cohort of dividing immature thymocytes was labeled by injecting 5- to 7-week-old CD4-expressing β2m−/− (○) or CD4−/−β2m−/− (●) mice with BrdUrd (administered i.p. as two 1-mg doses of BrdUrd, given 4 h apart). The percentages of BrdUrd+ cells that express high levels of CD3 and CD69 as indicated are shown. Each point represents three animals of each strain and are representative of four experiments. (B) Thymocytes in 4- to 5-week-old CD4−/− or wild-type mice were labeled intrathymically by injection of FITC. The percentage of cells is shown that were CD8− in the total CD3hi thymocyte population and also in the FITC-labeled (i.e., recently migrated from the thymus) peripheral CD3+ T cell population in wild-type (open blocks) or CD4−/− (solid blocks) animals 24 h after injection. Each point represents one animal and data are representative of three experiments.
To examine the possibility that loss of CD4 might impair the efficiency of emigration from the thymus, we used an intrathymic FITC injection procedure (27) to compare the export of CD4 lineage cells from the thymuses of CD4−/− and wild-type mice. As shown in Fig. 2B, FITC-labeled CD8− T cells (i.e., those that include primarily the CD4 lineage) could be detected in the peripheral lymphoid organs of both genotypes of mice 24 h after FITC injection. In the CD4−/− animals, the representation of these recent thymic emigrant cells was, if anything, higher than the representation of the precursor (CD3hiCD8−) population in the thymus. Similar results were found when animals were fed water containing BrdUrd and then monitored for the accumulation of weakly BrdUrd-labeled cells in their peripheral lymphoid tissues (data not shown), a method that also allows for recent thymic emigrants to be identified (26). From these data, we conclude that the loss of CD4 does not appear to impair the rate of export of CD4 lineage cells from the thymus to the periphery.
The above results show that, in the absence of CD4, the output of CD4 lineage cells from the thymus is about 20% of normal due to inefficient positive selection. Because of the proliferative capacity of CD4 lineage T cells (34), a reduction in output seemed insufficient to account for the low numbers of these cells in the periphery. We therefore hypothesized that an additional peripheral defect accounted for decreased representation of CD4 lineage cells in CD4−/− or CD4−/−β2m−/− mice. To test this hypothesis, we established radiation chimeras in which recipient mice received a roughly equal mixture of fetal liver from CD4-expressing and CD4-deficient embryos. In these chimeras, wild-type donor cells were distinguished by expression of Ly5.1, and CD4 lineage cells from wild-type and mutant donors by the expression of a transgenic CD4 lineage-specific reporter (cd4-TVA, a chicken Tva cDNA controlled by CD4 regulatory elements; Fig. 3A). With this combination of surface molecules, the relative representation of CD4−/−- vs. CD4-expressing T helper lineage cells could be determined in the thymuses and periphery of chimeric animals.
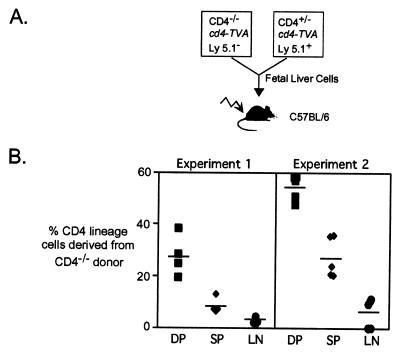
Figure 3.
Diminished representation of CD4-deficient T helper cells in mixed fetal liver chimeras. (A) Fetal liver (embryonic day 14) cells from CD4+/−Tva+ Ly5.1+ and CD4−/−Tva+ Ly5.1− animals were mixed at a ratio of 70/30 (experiment 1) or 40/60 (experiment 2) and injected into lethally irradiated C57BL/6 hosts. Recipients were analyzed 6–8 weeks later by flow cytometry. Donor-derived CD4 lineage thymocytes and lymphocytes were identified by expression of Tva; CD4 genotype was inferred by expression of Ly5.1. (B) The percentage of thymocytes is shown that were Tva+ CD8+ Ly5.1− (DP, ■) or Tva+ CD8−Ly5.1− (SP, ⧫) or lymph node cells that were Tva+CD8− Ly5.1− (LN, ●). Data are representative of three experiments.
Two conclusions can be drawn from analysis of the chimeric mice. First, in agreement with the observations above, CD4−/− cells were 3- to 4-fold less frequent than CD4-expressing cells at the single-positive stage of development than at the double-positive stage (Fig. 3B). The magnitude of this reduction in positive selection was close to that expected from the kinetic data described above. Second, the chimeras revealed a marked reduction in the frequency of CD4 lineage cells between the single-positive thymocyte compartment and the periphery, which was unaffected by the input ratio of CD4−/−- to CD4-expressing cells and typically accounted for a 2- to 3-fold decrease in the cellularity of the CD4 lineage (Fig. 3B). Because we could find no evidence for a defect in exit of CD4−/− cells from the thymus, the data suggest that the absence of CD4 impairs some aspect of peripheral T cell homeostasis, with a defect in T cell proliferation or survival being likely candidates.
Increased Apoptosis in CD4−/− Cells and Failure to Thrive after Adoptive Transfer.
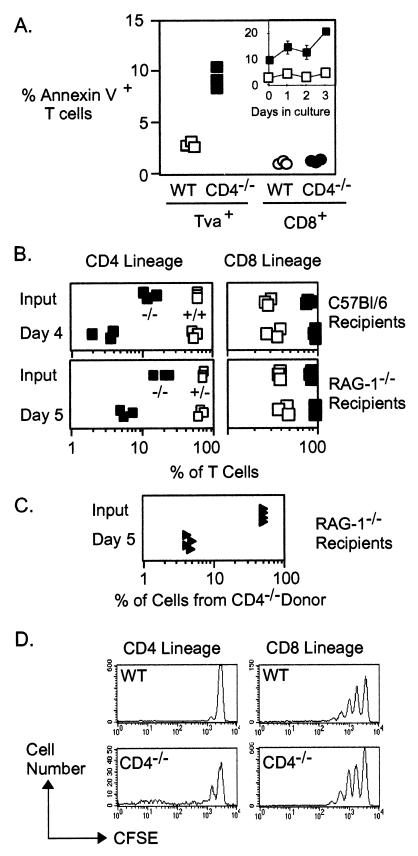
With flow cytometry with annexin V-FITC, we attempted to determine whether the loss of CD4 expression conferred an enhanced tendency for apoptosis on T helper cells. As shown in Fig. 4A, the frequency of apoptotic cells within fresh ex vivo CD4 lineage cells was consistently higher in the absence of CD4 than in its presence. No difference was found in the frequency of apoptotic CD8+ T cells from the same mice. An elevated frequency of apoptotic cells was also a persistent characteristic of CD4 lineage cells from CD4−/− mice when they were placed in culture for 4 days in the absence of stimulation (Fig. 4A Inset). These data indicate that the absence of CD4 has a significant negative effect on the survival of mature CD4 lineage T cells.
Figure 4.
Impaired survival of CD4 lineage cells from CD4−/− mice. (A) Pooled lymph node cells from CD4+/−Tva+ (open symbols) or CD4−/−Tva+ (solid symbols) mice were stained with annexin V, anti-TCRβ and SUA-rIgG or anti-CD8. The cells were analyzed by flow cytometry using a live cell forward/side scatter gate. (Left) Percentage of scatter-gated TCRβ+Tva+ cells that stained with annexin V. (Right) TCRβ+CD8+ cells. Cells from three animals of each genotype were analyzed, and data are representative of five experiments. (Inset) Representation of annexin V-positive CD4 lineage cells from CD4-expressing (□) and CD4−/− mice (filled squares) over time following in vitro culture in the absence of intentional stimulation. (B) Pooled lymph node and spleen cells from wild-type (open squares) or CD4−/− (■) donors were labeled with 0.1 μM CFSE. Approximately 3 or 5 × 107 labeled cells were then injected into the tail veins of nonirradiated sex-matched C57BL/6 (Upper) or RAG-1−/− (Lower) mice, respectively. The percentage of CD4 (Left) and CD8 (Right) lineage CFSE+ T cells found in the recipient lymph nodes (Upper) or spleens (Lower) at the indicated times after transfer is shown. Each symbol represents one recipient from an individual donor animal. (C) Approximately 2.5 × 106 purified CD8− T cells from wild-type animals were mixed with an equal number of such cells from CD4−/− animals and injected into the tail veins of RAG-1−/− recipients. The percentage of total T cells derived from the CD4−/− donor is shown, and each point represents one recipient animal. (D) Representative CFSE fluorescence profiles of CD4 lineage (Left) or CD8 lineage T cells (Right) in RAG-1−/− recipients 5 days after transfer of wild-type (Upper) or CD4−/− (Lower) cells. Data are representative of three experiments.
T cells from CD4-expressing or CD4−/− mice were also tested for their capacity to repopulate peripheral compartments after transfer into C57BL/6 or alymphoid (RAG-1−/−) recipients (Fig 4B). CD8+ T cells from both types of mice were indistinguishable in their capacity for efficient repopulation of both types of recipients. By contrast, CD4 lineage cells from CD4−/− mice but not from CD4-expressing mice were markedly deficient in this capacity and decreased in representation by 3- to 5-fold over 4–5 days. Similar results were also obtained with chimeras that had been repopulated with a mixture of both CD4-expressing and CD4-deficient T cells (Fig. 4C). As shown in Fig. 4D, CFSE-labeled CD4 lineage cells from both types of mice retained a largely undiluted level of fluorescence during these short-term transfers, indicating that very little proliferation had occurred. Thus, the reduced representation of CD4-lineage T cells in the recipient mice appeared to be a consequence of impaired survival rather than a failure to proliferate. CD4-deficient CD4 lineage cells could be detected in the chimeras 24 h after transfer at near-input levels, indicating that the failure to thrive after transfer was not due to a failure to engraft (data not shown). These data are therefore consistent with the in vitro apoptosis data (Fig. 4A) and support a necessary role for CD4 in the survival of T helper cells in vivo.
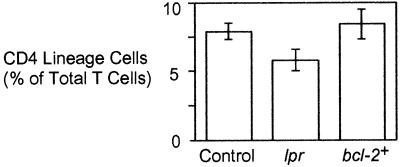
Apoptosis of T cells can be potentiated by signals from members of the tumor necrosis family receptor family. In particular, activation-induced T cell death involves Fas-associated death domain protein (FADD)-dependent signals from CD95 (Fas) (36, 37). To determine whether the apoptosis of CD4-deficient T helper cells required CD95 signaling, we examined the representation of CD4 lineage cells in CD4−/− mice that were homozygous for the lpr mutation and, therefore, lacked functional CD95 (38). As shown in Fig. 5, the absence of CD95 afforded no apparent decrease in apoptosis or increase in survival of CD4-deficient CD4 lineage T cells. Such data suggest that apoptosis of CD4-deficient T helper cells is not dependent on CD95 signaling.
Figure 5.
Effect of Fas deficiency and ectopic bcl-2 expression on the representation of CD4 lineage cells in CD4−/− mice. Pooled lymph node cells from 3- to 5-week-old animals as indicated were examined by flow cytometry for expression of TCRβ and CD8. The average percentage of total TCRβ+ cells that were CD8− in mice (n ≥ 3) of each strain is shown.
Finally, we also examined whether the survival of CD4-deficient T helper cells could be enhanced by enforced expression of the antiapoptotic protein bcl-2. Ectopic bcl-2 expression, as in Eμ-bcl-2–25 transgenic mice, can protect lymphocytes from various forms of apoptosis (39) and can prolong the survival of T cells that respond to exogenous antigens (37). bcl-2 can also at least partially suppress some mutations that impair cell survival (40, 41). Nonetheless, compared with nontransgenic littermates, CD4−/− mice carrying the Eμ-bcl-2–25 transgene showed no detectable increase in the frequency of peripheral CD4 lineage T cells (Fig. 5). Thus, the survival defect caused by loss of CD4 could not be alleviated by the level of bcl-2 expression achieved in peripheral T cells of Eμ-bcl-2–25 transgenic mice.
Enrichment for Memory T Helper Cells in the Absence of CD4.
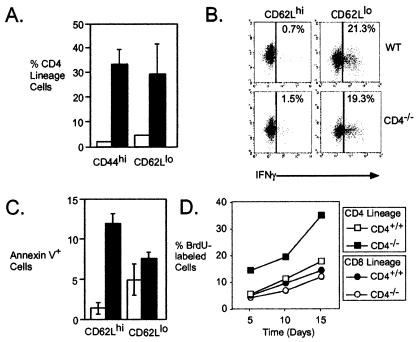
Flow cytometry analysis of the steady-state CD4 lineage in CD4−/− mice showed significant enrichment for cells with a surface phenotype that is normally typical of memory T cells (Fig. 6A). Nearly 35% of CD4 lineage cells from CD4−/− mice expressed high levels of CD44, compared with only 2% of the lineage in CD4-expressing mice. A similar enrichment was seen in the fraction of cells expressing low levels of CD62L (29% CD4−/− vs. 5% CD4+/+). As a control, we found no difference in the relative expression of any of these markers on CD8+ T cells from the same animals (data not shown). Consistent with their cell surface phenotype, we found that CD62Llo CD4 lineage cells from CD4−/− mice exhibited rapid production of IFN-γ, typical of memory T helper cells (42, 43). As shown in Fig. 6B, 20% of CD62Llo Tva-expressing cells produced IFN-γ after brief in vitro activation with phorbol ester and ionomycin. This frequency of cytokine-producing cells was very similar to that observed for CD62Llo Tva-expressing cells from C57BL/6 mice and clearly distinct from the low frequency of cytokine-positive CD62L+ (naïve) cells from either type of mouse (Fig. 6B). Interestingly, we noted a much higher frequency of apoptotic cells among naive than among memory CD4−/− T helper cells (Fig. 6C).
Figure 6.
Frequency and phenotype of naive and memory T cells in the CD4 lineage of CD4−/− mice. (A) Pooled lymph node cells were examined by flow cytometry for expression of CD8, CD4, or Tva and the indicated markers. The percentage of live CD4 lineage T cells (CD4+, wild-type, open bars; Tva+, CD4−/−, solid bars) as indicated is shown. Data from three mice of each genotype are shown and representative of three other experiments. (B) Pooled lymph node and spleen cells were sorted by flow cytometry for Tva and CD62L. Cells were then activated for 4 h with phorbol 12-myristate 13-acetate and ionomycin, fixed, and stained for IFN-γ expression. Data are the cytokine expression in CD62Lhi (Left) or CD62Llo (Right) CD4 lineage cells from wild-type or CD4−/− mice, as indicated. The y axes represent an irrelevant (unstained) parameter. Data are representative of two experiments. (C) Pooled lymph node cells from wild-type (open bars) or CD4−/−Tva+ (solid bars) mice were stained for expression of CD4 (or Tva), CD62L and annexin V. The average percentage of CD62Lhi (Left) or CD62Llo (Right) CD4 lineage cells that stained positively with annexin V. Data from three mice of each genotype are represented by each bar, and data are representative of three experiments. (D) Increased turnover of CD4-deficient CD4 lineage T cells. Wild-type and CD4−/− mice were given BrdUrd (BrdU) in their drinking water. CD4 and CD8 lineage T cells from the mice were then analyzed by flow cytometry as indicated for incorporation of BrdUrd (indicative of prior DNA synthesis during cell division). CD4 lineage cells from CD4−/−β2m−/− mice analyzed in the same experiment showed a similarly high rate of BrdUrd incorporation (data not shown).
Acquisition of a memory cell phenotype has been recently described as an attribute of T cells that undergo successive rounds of proliferation after placement in T cell-deficient environments (44, 45). To determine whether this type of phenomenon might be part of the CD4−/− phenotype, we used a continuous BrdUrd-labeling procedure to look for evidence of increased cell division among CD4−/− T helper cells. As shown in Fig. 6D, the absence of CD4 correlated with increased incorporation of BrdUrd in CD4 lineage cells over a 15-day period. In other experiments, we found that thymectomy incurred no substantial decrease in the relative representation of CD4 lineage cells in CD4−/− or CD4−/−β2m−/− double-deficient mice (data not shown). Thus, a high rate of cell division appears to balance the increased apoptosis observed in CD4-deficient T helper cells. The outgrowth of cells that exhibit a memory phenotype is a likely consequence of the increased proliferation, which serves to maintain CD4 lineage representation at a stable but nonetheless subnormal level.
Discussion
Small numbers of wild-type CD4 lineage cells can readily repopulate the T helper cell compartment of T lymphopenic hosts (34). As shown herein, CD4-deficient T helper cells are obviously defective in this respect. This observation is made clear by the dominance of the CD8 lineage in mice that lack CD4, by the reduced number of total T cells in CD4−/−β2m−/− mice and by the failure of CD4-deficient T helper cells to thrive after adoptive transfer to RAG-1−/− hosts. Furthermore, a defect in peripheral T cell homeostasis in the absence of CD4 was also readily apparent from the low ratio of CD4−/−- to CD4-expressing cells in the secondary lymphoid tissues of mixed bone-marrow chimeras. The most likely explanation for the observed deficiency is that the survival of T helper cells was impaired in the absence of CD4, consistent with data showing diminished CD4+ T cell survival when the CD4-MHC class II interaction is inhibited (46).
In contrast to normal CD4+ T cells, the surface phenotype of a large proportion of CD4−/− T helper cells resembled that of memory rather than naive T cells. Increased representation of the memory phenotype could reflect enrichment for cells that have been involved in overt immune responses. The BrdUrd incorporation data (Fig. 6D) would fit with this observation because memory cells enter the cell cycle more frequently than their naive precursors (26). It is also possible, however, that some of the cells may have acquired their memory phenotype as a consequence of homeostatic proliferation in response to a perceived T cell void (44, 45). In this context, the high rate of cell division in the CD4-deficient T helper cell population would be consistent with homeostatic proliferation still being possible despite the absence of CD4. The fact that T helper lineage cells do not accumulate to normal levels, however, indicates that such proliferation must be insufficient to compensate for the increased occurrence of apoptosis. Alternatively, the proliferation itself may impose the stress that induces apoptosis. Additional experiments are required to clarify the extent to which CD4 deficiency might impair the capacity for successive homeostatic cell divisions by T cells and also to determine whether the increased prevalence of the memory phenotype reflects selection for bona fide antigen-experienced cells.
Although it is possible that interactions between CD4 and non-MHC-encoded ligands [e.g., IL-16 (47) or gp17/GCDFP-15 (48)] may potentiate T helper cell survival under some circumstances, the most common effects of such non-MHC class II-dependent CD4 ligation appear to be antiproliferative (49) or proapoptotic (50, 51). By contrast, several reports have demonstrated the significance of ligand-dependent TCR signaling for T cell homeostasis (6–9, 11), with a key role emerging for such signaling during proliferation in T cell-deficient environments (20, 52). It seems probable, therefore, that the main impact of CD4 in enhancing the representation of T helper cells is to improve the effectiveness of immunological synapse formation and the transduction of prosurvival signals from TCRs engaged by peptide/MHC complexes. Interestingly, apoptotic cells were significantly less frequent in the population of CD4-deficient T helper cells that had a memory rather than naive phenotype, suggesting that transition to the memory state may decrease the CD4 dependency of a cell. This last interpretation would be consistent with reports showing differences in the degree to which memory and naive T cells depend on contact with MHC ligands for survival (12, 13).
The mechanism by which naive T cells are protected from cell death by TCR/coreceptor signaling is unclear. The analysis conducted herein indicates that inadequate signaling caused by coreceptor deficiency involves a form of cell death that is CD95-independent and yet insensitive to ectopic bcl-2 expression. In these respects, the cell death appears to be superficially distinct from other forms of activation-induced cell death and passive cell death (36, 37). Further experiments are required to determine whether these characteristics are a general property of apoptosis caused by ineffective TCR/coreceptor signaling and also to determine exactly how the TCR/coreceptor propagates its survival effect.
One obvious concern about the design of the experiments described in this paper is that they involve observations and manipulations of peripheral phenotypes for cells that have undergone thymic development in the absence of CD4. Although it is possible that some of the phenotypes we have observed are the result of abnormal thymopoiesis, preliminary observations (Q.W., J.S., and N.K., unpublished results) on cells that have lost CD4 expression as a postthymic event appear to be consistent with the data presented herein and support the thesis that loss of CD4 function disturbs T helper cell homeostasis. Furthermore, despite their failure to differentiate into Th2 T helper cells (18, 19), CD4-deficient T helper cells can become effective Th1 T helper cells and make useful contributions to immune responses (29). They also express TCRs that are similar in their recognition properties to those of normal CD4+ T cells.† For these reasons, we believe that our observations are relevant to the function of CD4 in normal CD4+ T cell homeostasis.
In conclusion, the expression of CD4 potentiates the survival of naive T helper cells, but its absence imposes selection for cells with a memory phenotype. Defective survival in the absence of CD4 is likely a consequence of inadequate signaling by TCRs as they engage their extrathymic MHC/peptide ligands. Thus, in addition to revealing the involvement of CD4 in survival signaling by TCRs, the data support a role for ligand discrimination by TCRs in the selection of T cells beyond the thymus.
Acknowledgments
We thank the reviewers, Jason Cyster, J. Mike McCune, and members of the Killeen laboratory for helpful discussions and/or comments on the manuscript; Richard Locksley for use of the MoFlo cell sorter; Kurt Zingler for provision of the SUA-rIgG fusion protein; and Debbie Fowell for helpful advice. This work was supported by grants from the National Institutes of Health (Grant AI39506), the University of California AIDS Research Program ( Grant R99-SF-080) and the University of California, San Francisco Howard Hughes Medical Institute Research Resources Program.
Abbreviations
- TCR
T cell receptor
- SUA-rIgG
subgroup A avian sarcoma and leukosis viruses envelope-Ig fusion protein
- CFSE
carboxyfluorescein succinimidyl ester
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Q.W., L. Malherbe, K. Zingler, D. R. Littman, N. Glaichenhaus, and N.K., unpublished work.
References
- 1.Haks M C, Oosterwegel M A, Blom B, Spits H M, Kruisbeek A M. Semin Immunol. 1999;11:23–37. doi: 10.1006/smim.1998.0153. [DOI] [PubMed] [Google Scholar]
- 2.Fehling H J, von Boehmer H. Curr Opin Immunol. 1997;9:263–275. doi: 10.1016/s0952-7915(97)80146-x. [DOI] [PubMed] [Google Scholar]
- 3.Hsu B L, Evavold B D, Allen P M. J Exp Med. 1995;181:805–810. doi: 10.1084/jem.181.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alam S M, Travers P J, Wung J L, Nasholds W, Redpath S, Jameson S C, Gascoigne N R. Nature (London) 1996;381:616–620. doi: 10.1038/381616a0. [DOI] [PubMed] [Google Scholar]
- 5.Williams C B, Engle D L, Kersh G J, White J M, Allen P M. J Exp Med. 1999;189:1531–1544. doi: 10.1084/jem.189.10.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeda S, Rodewald H R, Arakawa H, Bluethmann H, Shimizu T. Immunity. 1996;5:217–228. doi: 10.1016/s1074-7613(00)80317-9. [DOI] [PubMed] [Google Scholar]
- 7.Tanchot C, Lemonnier F A, Perarnau B, Freitas A A, Rocha B. Science. 1997;276:2057–2062. doi: 10.1126/science.276.5321.2057. [DOI] [PubMed] [Google Scholar]
- 8.Brocker T. J Exp Med. 1997;186:1223–1232. doi: 10.1084/jem.186.8.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markiewicz M A, Girao C, Opferman J T, Sun J, Hu Q, Agulnik A A, Bishop C E, Thompson C B, Ashton-Rickardt P G. Proc Natl Acad Sci USA. 1998;95:3065–3070. doi: 10.1073/pnas.95.6.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zinkernagel R M, Althage A. Proc Natl Acad Sci USA. 1999;96:8092–8097. doi: 10.1073/pnas.96.14.8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freitas A A, Rocha B. Curr Opin Immunol. 1999;11:152–156. doi: 10.1016/s0952-7915(99)80026-0. [DOI] [PubMed] [Google Scholar]
- 12.Murali-Krishna K, Lau L L, Sambhara S, Lemonnier F, Altman J, Ahmed R. Science. 1999;286:1377–1381. doi: 10.1126/science.286.5443.1377. [DOI] [PubMed] [Google Scholar]
- 13.Swain S L, Hu H, Huston G. Science. 1999;286:1381–1383. doi: 10.1126/science.286.5443.1381. [DOI] [PubMed] [Google Scholar]
- 14.Marrack P, Endres R, Shimonkevitz R, Zlotnik A, Dialynas D, Fitch F, Kappler J. J Exp Med. 1983;158:1077–1091. doi: 10.1084/jem.158.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glaichenhaus N, Shastri N, Littman D R, Turner J M. Cell. 1991;64:511–520. doi: 10.1016/0092-8674(91)90235-q. [DOI] [PubMed] [Google Scholar]
- 16.Rahemtulla A, Fung-Leung W P, Schilham M W, Kundig T M, Sambhara S R, Narendran A, Arabian A, Wakeham A, Paige C J, Zinkernagel R M, et al. Nature (London) 1991;353:180–184. doi: 10.1038/353180a0. [DOI] [PubMed] [Google Scholar]
- 17.Killeen N, Sawada S, Littman D R. EMBO J. 1993;12:1547–1553. doi: 10.1002/j.1460-2075.1993.tb05798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown D R, Moskowitz N H, Killeen N, Reiner S L. J Exp Med. 1997;186:101–107. doi: 10.1084/jem.186.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fowell D J, Magram J, Turck C W, Killeen N, Locksley R M. Immunity. 1997;6:559–569. doi: 10.1016/s1074-7613(00)80344-1. [DOI] [PubMed] [Google Scholar]
- 20.Seddon B, Legname G, Tomlinson P, Zamoyska R. Science. 2000;290:127–131. doi: 10.1126/science.290.5489.127. [DOI] [PubMed] [Google Scholar]
- 21.Bates P, Rong L, Varmus H E, Young J A, Crittenden L B. J Virol. 1998;72:2505–2508. doi: 10.1128/jvi.72.3.2505-2508.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawada S, Scarborough J D, Killeen N, Littman D R. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 23.Chan S, Correia-Neves M, Dierich A, Benoist C, Mathis D. J Exp Med. 1998;188:2321–2333. doi: 10.1084/jem.188.12.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zingler K, Young J A. J Virol. 1996;70:7510–7516. doi: 10.1128/jvi.70.11.7510-7516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carayon P, Bord A. J Immunol Methods. 1992;147:225–230. doi: 10.1016/s0022-1759(12)80012-3. [DOI] [PubMed] [Google Scholar]
- 26.Tough D F, Sprent J. J Exp Med. 1994;179:1127–1135. doi: 10.1084/jem.179.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donovan J, Brown P. In: Current Protocols in Immunology. Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W, editors. Vol. 1. New York: Wiley; 1994. pp. 1.6.1–1.6.10. [Google Scholar]
- 28.Lyons A B, Parish C R. J Immunol Methods. 1994;171:131–137. doi: 10.1016/0022-1759(94)90236-4. [DOI] [PubMed] [Google Scholar]
- 29.Locksley R M, Reiner S L, Hatam F, Littman D R, Killeen N. Science. 1993;261:1448–1451. doi: 10.1126/science.8367726. [DOI] [PubMed] [Google Scholar]
- 30.Rahemtulla A, Kundig T M, Narendran A, Bachmann M F, Julius M, Paige C J, Ohashi P S, Zinkernagel R M, Mak T W. Eur J Immunol. 1994;24:2213–2218. doi: 10.1002/eji.1830240942. [DOI] [PubMed] [Google Scholar]
- 31.Grusby M J, Johnson R S, Papaioannou V E, Glimcher L H. Science. 1991;253:1417–1420. doi: 10.1126/science.1910207. [DOI] [PubMed] [Google Scholar]
- 32.Fung-Leung W P, Schilham M W, Rahemtulla A, Kundig T M, Vollenweider M, Potter J, van Ewijk W, Mak T W. Cell. 1991;65:443–449. doi: 10.1016/0092-8674(91)90462-8. [DOI] [PubMed] [Google Scholar]
- 33.Zijlstra M, Bix M, Simister N E, Loring J M, Raulet D H, Jaenisch R. Nature (London) 1990;344:742–746. doi: 10.1038/344742a0. [DOI] [PubMed] [Google Scholar]
- 34.Rocha B, Dautigny N, Pereira P. Eur J Immunol. 1989;19:905–911. doi: 10.1002/eji.1830190518. [DOI] [PubMed] [Google Scholar]
- 35.Teh S J, Killeen N, Tarakhovsky A, Littman D R, Teh H S. Blood. 1997;89:1308–1318. [PubMed] [Google Scholar]
- 36.Van Parijs L, Abbas A K. Science. 1998;280:243–248. doi: 10.1126/science.280.5361.243. [DOI] [PubMed] [Google Scholar]
- 37.Van Parijs L, Peterson D A, Abbas A K. Immunity. 1998;8:265–274. doi: 10.1016/s1074-7613(00)80478-1. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe-Fukunaga R, Brannan C I, Copeland N G, Jenkins N A, Nagata S. Nature (London) 1992;356:314–317. doi: 10.1038/356314a0. [DOI] [PubMed] [Google Scholar]
- 39.Strasser A, Harris A W, Cory S. Cell. 1991;67:889–899. doi: 10.1016/0092-8674(91)90362-3. [DOI] [PubMed] [Google Scholar]
- 40.Akashi K, Kondo M, von Freeden-Jeffry U, Murray R, Weissman I L. Cell. 1997;89:1033–1041. doi: 10.1016/s0092-8674(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 41.Costello P S, Cleverley S C, Galandrini R, Henning S W, Cantrell D A. J Exp Med. 2000;192:77–86. doi: 10.1084/jem.192.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Croft M, Bradley L M, Swain S L. J Immunol. 1994;152:2675–2685. [PubMed] [Google Scholar]
- 43.Rogers P R, Dubey C, Swain S L. J Immunol. 2000;164:2338–2346. doi: 10.4049/jimmunol.164.5.2338. [DOI] [PubMed] [Google Scholar]
- 44.Goldrath A W, Bogatzki L Y, Bevan M J. J Exp Med. 2000;192:557–564. doi: 10.1084/jem.192.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murali-Krishna K, Ahmed R. J Immunol. 2000;165:1733–1737. doi: 10.4049/jimmunol.165.4.1733. [DOI] [PubMed] [Google Scholar]
- 46.Maroto R, Shen X, Konig R. J Immunol. 1999;162:5973–5980. [PubMed] [Google Scholar]
- 47.Center D M, Kornfeld H, Cruikshank W W. Immunol Today. 1996;17:476–481. doi: 10.1016/0167-5699(96)10052-i. [DOI] [PubMed] [Google Scholar]
- 48.Autiero M, Cammarota G, Friedlein A, Zulauf M, Chiappetta G, Dragone V, Guardiola J. Eur J Immunol. 1995;25:1461–1464. doi: 10.1002/eji.1830250550. [DOI] [PubMed] [Google Scholar]
- 49.Cruikshank W W, Lim K, Theodore A C, Cook J, Fine G, Weller P F, Center D M. J Immunol. 1996;157:5240–5248. [PubMed] [Google Scholar]
- 50.Newell M K, Haughn L J, Maroun C R, Julius M H. Nature (London) 1990;347:286–289. doi: 10.1038/347286a0. [DOI] [PubMed] [Google Scholar]
- 51.Somma F, Tuosto L, Montani M S, Di Somma M M, Cundari E, Piccolella E. J Immunol. 2000;164:5078–5087. doi: 10.4049/jimmunol.164.10.5078. [DOI] [PubMed] [Google Scholar]
- 52.Clarke S R, Rudensky A Y. J Immunol. 2000;165:2458–2464. doi: 10.4049/jimmunol.165.5.2458. [DOI] [PubMed] [Google Scholar]