Abstract
Background
Oil palm is the world's most productive oil-food crop despite yielding well below its theoretical maximum. This maximum could be approached with the introduction of elite F1 varieties. The development of such elite lines has thus far been prevented by difficulties in generating homozygous parental types for F1 generation.
Results
Here we present the first high-throughput screen to identify spontaneously-formed haploid (H) and doubled haploid (DH) palms. We secured over 1,000 Hs and one DH from genetically diverse material and derived further DH/mixoploid palms from Hs using colchicine. We demonstrated viability of pollen from H plants and expect to generate 100% homogeneous F1 seed from intercrosses between DH/mixoploids once they develop female inflorescences.
Conclusions
This study has generated genetically diverse H/DH palms from which parental clones can be selected in sufficient numbers to enable the commercial-scale breeding of F1 varieties. The anticipated step increase in productivity may help to relieve pressure to extend palm cultivation, and limit further expansion into biodiverse rainforest.
Background
Success of early F1 hybrid maize varieties exemplifies the advantages of heterosis [1]. The use of doubled haploids as parents for F1 variety production fully exploits this phenomenon and has enabled substantial yield improvements in several crops [2,3]. This strategy was outlined with the first DH crop variety [4] and has led to H/DH production systems being described for > 250 species [5]. However, few of these protocols generate the large numbers of Hs/DHs needed for commercial breeding, with just three methods (androgenesis, wide crossing, gynogenesis [6]) routinely adopted for H/DH production in only 30 species [5]. The most important of these methods in widespread use in commercial breeding is the generation of haploids in maize via pollination with a haploid inducing line such as a 'Stock 6' derivative. Desire for a more generic H/DH production system to improve agricultural yields is increasing as population growth, climate change, biofuel demand and other land-use pressures intensify. Clearly, in any species the production of F1 varieties depends not only on the production of homozygous lines to act as parents, but also it requires an efficient method to intercross the parents. This latter procedure is relatively simple in species with an outcrossing breeding system, like maize or oil palm, compared with those with an inbreeding system like rice or wheat. Production of F1 hybrids has been achieved successfully in this category of crops (for example hybrid rice in China) but often requires a male sterility system.
Annually, oil palm (Elaeis guineensis) yields eight to ten times more oil per hectare than rapeseed or soybean [7,8] and in 2008 generated 38.9 million tonnes of oil worldwide [9]. The area assigned to the crop expanded ~1.7 fold between 1997 (8.7 M ha) and 2007 (14.6 M ha) [9] with further increases forecast. Over this same period global production of palm oil increased ~2.2 fold from 18 to 38.9 Mt y-1. Thus, yield increases have been achieved predominantly by expansion of cultivated area and not through yield enhancement. This trend raises concerns over the ecological impact of felling rainforest to accommodate oil palm cultivation [10,11] and has stimulated debate over strategies to limit further agricultural expansion [12-14]. One option explored here is to use market forces to help address the problem. If F1 varieties could increase yields sufficiently to exceed demand, commodity prices would fall. This would discourage clear felling and simultaneously incentivise early replacement of existing plantations with high-yielding varieties. Feasibility of the approach clearly relies on the ability to gain marked improvements in yield. Current yields of oil palm (generally 4-10.5 t ha-1) [15,16] are much lower than the most conservative estimates of the crop's potential (17 t ha-1 [14] to 60 t ha-1 [16]). Indeed, yields per hectare in the two largest producer countries (Indonesia and Malaysia) have remained static for 30 years [9]. It should be noted, however, that in both these countries there are examples of selected varieties with much higher yields, with the highest yields from commercial breeding trials already exceeding 10 t ha-1.
To date, a H/DH-derived F1 breeding approach has been precluded by the repeated failure to secure H/DHs via anther or microspore culture [17] and successful generation of H/DHs in oil palm is unreported in the literature. The report of a spontaneous H in the related coconut palm [18] and in other species [19] nevertheless gave hope that spontaneous Hs may also occur in oil palm. However, the characteristically rare occurrence of spontaneous H/DHs necessitates development of an effective high-throughput screening system. Phenotypic characteristics of H/DH (slow growth, altered flowering phenology, smaller stomata and smaller organs [5]) could be used for diagnosis but are difficult to score qualitatively on a large scale and require plants of a reasonable size. An alternative strategy is to seek undefined atypical phenotypic features that may arise from reduced cell size and/or the hemizygous state of haploid individuals (homozygous for DHs) and that are manifest at the seedling stage when high-throughput visual assessment is more plausible. A more directed approach is also possible. Spontaneous H/DH seedlings are often associated with aberrant germination features, such as twin embryos from the same carpel [20], providing a defined feature for phenotypic selection. Here, we combined a large-scale visual survey for undefined atypical palm seedling phenotypes coupled with active selection for seeds with twin embryos to assemble a sub-population of seedlings enriched for H/DHs.
Results
Over two years, we performed two large-scale screens for morphological 'off-types' among oil palm seedlings generated by the Bah Lias Research Station, Indonesia. The first screen utilised 10,900,000 seedlings from a wide range of crosses and identified 3,854 morphological 'off-types' (H/DH candidates), of which 53 had twin embryos and 3,801 were phenotypically abnormal (Figure 1). The second screen of approximately 10,000,000 seedlings from commercial seed production activities and approximately 1,000,000 seedlings from breeding experiments generated 5,704 H/DH candidates, of which 5,601 were phenotypically abnormal and 103 had twin embryos. More than 2,000 of these seedlings (including all those with twin embryos) were transferred to the nursery prior to further screening. Although Hs could be identified relatively easily on the basis of their reduced genome size, we initially wished to target the more difficult, but more valuable DHs to circumvent the need for chromosome doubling. For the second level screen, we exploited the fact that Hs and DHs would be either hemi- or homozygous across all loci; thus individuals exhibiting heterozygosity at any locus could be discarded. Applying this logic, we performed a sequential screen using 9-15 microsatellite markers (Table 1) on all individuals and found 117 seedlings that exhibited a single allele across all loci (Table 2). These individuals were retained as candidate H/DH, and subsequent flow cytometry of leaf samples identified 83 as H, and 34 as diploid (Table 2). The haploid status of six palms was further confirmed by cytological examination of intact cells from root squashes. Each contained the expected 16 chromosomes (Figure 2).
Figure 1.
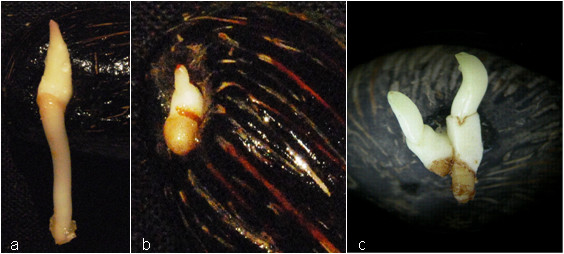
Seed germination morphology for H/DH identification. a: normal; b: abnormal; c: twin embryo.
Table 1.
Microsatellite primer pairs used to identify homozygous DH or hemizygous H candidates in the initial molecular screen.
| No. | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| 1 | GAGATTACAAAGTCCAAACC | TCAAAATTAAGAAAGTATGC |
| 2 | ACGCATGCAGCTAGCTTTTC | CGCGTGAAAGATATGAATCAAC |
| 3 | CACGCACGCAGTTTATTCTT | GGATGTATGCTTTACCTCCGAAT |
| 4 | CCCCTTTTGCTTCCCTATTT | CTCCTTTTCCCCATCACAGA |
| 5 | GACACAAGCAAAAACAAAAGCA | ATTCTGAAAGGAGGGGGAAA |
| 6 | ATATGTGTGGGTGTGCGTGT | TGCCTCTGGTTGTTAGTCTGG |
| 7 | TCTCTCTCTCTCTCTCTATGTGTGTGT | TGGCAATCAGCACACATTCT |
| 8 | GCAGCTCTTTCCACACCTCT | TGTGGTCTCCTGAGGAAGATG |
| 9 | TTTTCCCCATCACAGAATTG | CCCCTTTTGCTTCCCTATTT |
| 10 | TAGCCGCACTCCCACGAAGC | CCAGAATCATCAGACTCGGACAG |
| 11 | AGCTCTCATGCAAGTAAC | TTCAACATACCGTCTGTA |
| 12 | CCTTCAAGCAAAGATACC | GGCACCAAACACAGTAA |
| 13 | GTAGCTTGAACCTGAAA | AGAACCACCGGAGTTAC |
| 14 | GCTCGTTTTTGTTTAGGTGA | TTTTCTCCATAGTCCGTTAC |
| 15 | CCTCGGGTTATCCTTTTTACC | TGGCTGGCTTCGGTCTTAG |
Markers 10-15 obtained from Billotte et al. [27].
Table 2.
Results of ploidy analysis by flow cytometry of 117 candidate H/DH palms identified as both morphologically atypical and homozygous for the markers listed in Table 1.
| Candidate | DNA sample code | No. markers used | Ploidy |
|---|---|---|---|
| 50-Mix5-7 | 11260406301 | 9 | x |
| 50-03060367C | 07280501801 | 15 | x |
| 50-03060260C-2 | 07280501901 | 15 | x |
| 53-03080954C-2 | 09270500101 | 10 | x |
| 53-03090761C-5 | 09280504501 | 10 | x |
| BATCH 51;03060318C;1 | 060728_0010_01_a | 15 | x |
| BATCH 53;03090761C;5 | 060728_0018_01_a | 15 | x |
| 0623/172;05095508C;1 | 060728_0021_01_a | 15 | x |
| BATCH 50;03060260C;2 | 060728_0027_01_a | 15 | x |
| 0611/32;05050248C;1 | 060728_0032_01_a | 15 | x |
| 0611/16;05050228C;1 | 060728_0034_01_a | 15 | x |
| BATCH 53;03080954C;2 | 060728_0035_01_a | 15 | x |
| 06 412;04059061B;3 | 060728_0050_01_a | 14 | 2x |
| 0628/152;05100720C;1 | 060729_0021_01_a | 15 | x |
| 0628/185;05100351C;1 | 060729_0063_01_a | 15 | x |
| BATCH 51;03060626C;1 | 060729_0127_02_a | 15 | x |
| BATCH 67;0409034MC;2 | 060729_0130_02_a | 14 | 2x |
| BATCH 67;0409034MC;4 | 060729_0131_02_a | 15 | 2x |
| BATCH 67;0409034MC;15 | 060729_0132_02_a | 15 | 2x |
| BATCH 65;0409034MC;7 | 060729_0134_02_a | 15 | 2x |
| BATCH 65;0409034MC;35 | 060729_0138_02_a | 15 | 2x |
| BATCH 65;0409034MC;56 | 060729_0139_02_a | 15 | 2x |
| BATCH 65;0409034MC;50 | 060729_0141_02_a | 15 | 2x |
| BATCH 65;0409034MC;47 | 060729_0142_02_a | 15 | 2x |
| 0628/53;05090595C;1 | 060731_0043_01_a | 15 | x |
| 0627/125;05090717C;2 | 060731_0065_01_a | 15 | x |
| 0627/12;05080220C;1 | 060731_0080_01_a | 15 | x |
| 0627/6;05080095C;1 | 060731_0086_01_a | 14 | x |
| 0631/Normal;05039033B;31 | 060731_0265_01_a | 14 | x |
| 64-0409021MC-34 | 02130604301 | 15 | 2x |
| 64-0410040MC-1 | 02130604801 | 15 | 2x |
| 51-03060626C | 02130605301 | 15 | x |
| 64-0410040MC-20 | 02140600401 | 15 | 2x |
| 64-0410040MC-16 | 02140600801 | 15 | 2x |
| 65-0409021MC-2 | 02140601001 | 15 | 2x |
| 06 412B-04059061B-3 | 02170605501 | 15 | 2x |
| 06 412B-04129091B | 02170605801 | 15 | 2x |
| 0550-15/05010827C | 02200602401 | 15 | x |
| 0550-17/05010442C-1 | 02200602601 | 15 | x |
| 0550-23/05020059C | 02200603101 | 15 | x |
| 0550-33/05020568C | 02200603401 | 15 | x |
| 0550-36/05020420C-2 | 02200603701 | 15 | x |
| 0550-40/05010880C | 02200607501 | 14 | x |
| 0551-36/05020511C | 02200607601 | 15 | x |
| 0551-32/05020361C-1 | 02210600401 | 15 | x |
| 0552-4/05010836C-2 | 02210600901 | 15 | x |
| 0552-38/05020501C | 02210603101 | 14 | x |
| 0552-39/05020415C | 02210603201 | 15 | x |
| 0552-31/05020858C | 02210603701 | 15 | x |
| 0552-91/05020375C | 02210603901 | 15 | x |
| 0552-111/05020626C | 02210607201 | 15 | x |
| 0552-128/05020558C-1 | 02210607701 | 15 | x |
| 0601-35/05020946C | 02210608201 | 15 | x |
| 0601-42/05030201C-6 | 02210609501 | 15 | x |
| 0601-51/05030224C-2 | 02220600201 | 15 | x |
| 0607-21/05040317C-3 | 02220601801 | 14 | x |
| 0606-32/05040240C | 02220606201 | 13 | x |
| 0601-77/05020961C | 02230600701 | 15 | x |
| 0601-62/05030147C | 02230601401 | 15 | x |
| 0601-54/05030462C | 02230601901 | 15 | x |
| 0551-21/05020271C-1 | 02200605801 | 14 | x |
| 0601-9/05020843C-2 | 02230603101 | 15 | x |
| 0602-17/05020631C-1 | 02230605501 | 15 | x |
| 0607-111/05040970C-1 | 03010600201 | 15 | x |
| 0607-81/05040578C-1 | 03010600501 | 15 | x |
| 0607-73/05040573C-1 | 03010605101 | 15 | x |
| 0607-89/05040748C-3 | 03010605501 | 15 | x |
| 0607-102/05050016C-2 | 03010606601 | 15 | x |
| 0608-15/05040519C-3 | 03010606901 | 15 | x |
| 0608-45/05041003C-1 | 03150603401 | 15 | x |
| 0610-60/05041024C-2 | 03150604401 | 15 | x |
| 0610-124/05055039C-1 | 03150604601 | 15 | x |
| 0609-54/05050089C-2 | 03150604701 | 15 | x |
| 0610-41/05050352C-1 | 03150606701 | 15 | x |
| 0609-58/05050255C-1 | 03220600201 | 15 | x |
| 0610-82/05050099C-2 | 03220601401 | 15 | x |
| 0610-77/05050353C-1 | 03220602701 | 15 | x |
| 0610-121/05055090C-1 | 03220603301 | 15 | x |
| 0610-81/05050099C-1 | 03220605901 | 15 | x |
| 0609-100/05055311C-1 | 03290600301 | 15 | x |
| 0610-11/05040938C-1 | 03290601101 | 15 | x |
| 0610-68/05050376C-3 | 03290602001 | 15 | x |
| 0610-58/05050344C-1 | 03290602201 | 15 | x |
| 0610-73/05050594C-3 | 03290603301 | 15 | x |
| 0611-84/05050714C-4 | 03290605001 | 15 | x |
| 0611-70/05050223C-1 | 03290606701 | 15 | x |
| 0611-73/05050351C-1 | 03290608001 | 15 | x |
| 0610-67/05050376C-2 | 04050600501 | 15 | x |
| 0610-40/05050102C-2 | 04050600901 | 15 | x |
| 0611-99/05050544C-1 | 04050602601 | 15 | x |
| 0611-110/05055011C-1 | 04050603601 | 15 | x |
| 0612-2/05050017C-1 | 04050609101 | 15 | x |
| 0612-70/05050530C-1 | 04050609201 | 15 | x |
| 0612-76/05050512C-1 | 04050610301 | 15 | x |
| 0611-109/05055144C-1 | 04120600101 | 15 | x |
| 0611-31/05050220C-1 | 04120600601 | 15 | x |
| 0611-38/05050284C-4 | 04120600901 | 15 | x |
| 0611-40/05050171C-1 | 04120601101 | 14 | x |
| 0612-80/05050713C-1 | 04120603101 | 15 | x |
| 65-0409034 MC-66 | 060829_0001_02_a | 15 | 2x |
| 65-0409034 MC-68 | 060829_0002_02_a | 15 | 2x |
| 65-0409034 MC-72 | 060829_0003_02_a | 14 | 2x |
| 65-0409034 MC-111 | 060829_0005_02_a | 15 | 2x |
| 65-0409034 MC-94 | 060829_0011_02_a | 14 | 2x |
| 65-0409034 MC-120 | 060829_0012_02_a | 15 | 2x |
| 65-0409034 MC-144 | 060829_0013_02_a | 15 | 2x |
| 65-0409034 MC-133 | 060829_0015_02_a | 15 | 2x |
| 65-0409034 MC-187 | 060829_0020_02_a | 15 | 2x |
| 65-0409034 MC-193 | 060829_0021_02_a | 14 | 2x |
| 65-0409034 MC-199 | 060829_0023_02_a | 15 | 2x |
| 65-0409034 MC-135 | 060829_0025_02_a | 15 | 2x |
| 65-0409034 MC-114 | 060829_0026_02_a | 13 | 2x |
| 65-0409034 MC-147 | 060829_0027_02_a | 15 | 2x |
| 65-0409034 MC-36 B | 060829_0030_02_a | 15 | 2x |
| 65-0409034 MC-39 A | 060829_0031_02_a | 15 | 2x |
| 65-0409034 MC-73 A | 060829_0034_02_a | 15 | 2x |
| 65-0409034 MC-71 A | 060829_0035_02_a | 14 | 2x |
Note: in this initial round, no DH was found. The DH (0644-219/05049582C) was detected in a subsequent batch.
Figure 2.
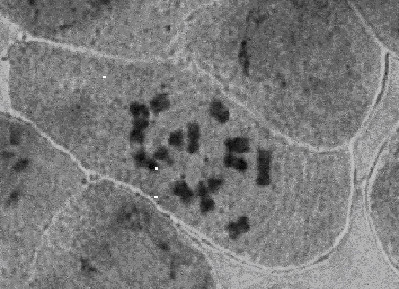
Chromosome spread of a haploid root cell from oil palm containing 16 C-metaphase chromosomes.
A larger-scale survey for heterozygosity was then performed using 97 additional microsatellites (Table 3) to confirm absolute hemizygosity of Hs and identify 'false' candidate DHs showing any heterozygosity. All Hs produced single-allele peak profiles across all microsatellites, thereby discounting fixed heterozygosity via locus duplication for all markers used. All diploids were heterozygous at several loci and so discarded. However, one diploid (0644-219/05049582C) identified from a later screen (see below) was homozygous across all 36 mapped loci found to be heterozygous in the maternal parent (palm number BL013/12-06). Taking account of linkage between mapped markers, the probability of such an individual occurring by chance following selfing was 8.72 × 10-8 (see Methods). This palm was therefore deemed a spontaneous DH.
Table 3.
Microsatellite markers (described by Billotte et al. [27]) used for a larger-scale survey for hemizygosity of Hs and homozygosity of DH candidates previously identified by the morphological screen, microsatellite pre-screen (15 markers) and flow cytometry screen.
| No. | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| 16 | GACCTTTGTCAGCATACTTGGTGTG | GCAGGCCTGAAATCCCAAAT |
| 17 | ATGCATGTGATTTTATTAGGTGAGA | CGACCCTCAGTCAATCAGTAAG |
| 18 | AAGCTAGCGACCTATGATTTTAGA | AAACAAGTAATGTGCATAACCTTTC |
| 19 | CCCACCACCCCTAGCTTCTC | ACCCCGGTCCAAATAAAATC |
| 20 | AGAGAGAGAGAGTGCGTATG | GTCCCTGTGGCTGCTGTTTC |
| 21 | GGGTAGCAAACCTTGTATTA | ACTTCCATTGTCTCATTATTCT |
| 22 | CGAGGCCCAAAAACATTCAC | GGTCCCGATCCCGTCTACTG |
| 23 | TTGCGGCCCATCGTAATC | TCCCTGCAGTGTCCCTCTTT |
| 24 | AGGGAATTGGAAGAAAAGAAAG | TCCTGAGCTGGGGTGGTC |
| 25 | AGCAAGAGCAAGAGCAGAACT | CTTGGGGGCTTCGCTATC |
| 26 | TAGCCATGCCGCCACCACTT | CAATCCATTAGCGTGCCCTTCT |
| 27 | CTTACCCCGCCTCCTCTCCT | CGAAATGCCCTTCCTTTACACTA |
| 28 | CCTTATATCGCACGGGTTCC | TTCTTGGGGTCTCGCTACGG |
| 29 | GCAAGATGCAATGGAGTTCA | CAAACCGCAGCAAGTCAGA |
| 30 | GCAAAATTCAAAGAAAACTTA | CTGACAGTGCAGAAAATGTTATAGT |
| 31 | CGTTCATCCCACCACCTTTC | GCTGCGAGGCCACTGATAC |
| 32 | GAATGTGGCTGTAAATGCTGAGTG | AAGCCGCATGGACAACTCTAGTAA |
| 33 | ACATTCCCTCTATTATTCTCAC | GTTTTGTTTGGTATGCTTGT |
| 34 | AAGCCAACTTCACAGATATGTTGAT | ATGAGCCTAACAAAGCACATTCTAA |
| 35 | AGTGAGGTATGGTTGATTAGGA | TATTGATAGCATTTGGGATTAG |
| 36 | CTCCGATGGTCAAGTCAGA | AAATGGGGAAGGCAATAGTG |
| 37 | GCCGTTCAAGTCAATTAGAC | TTTGGGAGCAAGCATTATCA |
| 38 | TGCTTCTTGTCCTTGATACA | CCACGTCTACGAAATGATAA |
| 39 | CACCACATGAAGCAAGCAGT | CCTACCACAACCCCAGTCTC |
| 40 | TTTTATTTTCCCTCTCTTTTGA | ATTGCGTCTCTTTCCATTGA |
| 41 | CATATGGCGCACAGGCAC | GCAATACAAGAGCACCCAAAT |
| 42 | AGTTGGTTTGCTGATTTG | TGTTGCTTCTTTGATTTTC |
| 43 | GCTGAAGATGAAATTGATGTA | TTCAGGTCCACTTTCATTTA |
| 44 | ATGACCTAAAAATAAAATCTCAT | ACAGATCATGCTTGCTCACA |
| 45 | GGTGCAAGAGAGGAGGAATG | TTTGGTAGTCGGGCGTTTTA |
| 46 | GTTTGGCTTTGGACATG | TCCATCACAGGAGGTATAG |
| 47 | TGTTTTGTTTCGTGCATGTG | GGCTGACATGCAACACTAAC |
| 48 | CGGTTTTGTCGCATCTATG | GTCGTCAGGGAACAACAGT |
| 49 | CAATCATTGGCGAGAGA | CGTCACCTTTCAGGATATG |
| 50 | GAGCATGACGCAAACAAAGG | GCAACATGTTTGATGCATTAATAGTC |
| 51 | TCCAAGTAGCAAATGATGAC | TGCCCTGAAACCCTTGA |
| 52 | GAAGGGGCATTGGATTT | TACCTATTACAGCGAGAGTG |
| 53 | AACACTCCAGAAGCCAGGTC | GGTTTAGGTATTGGAACTGATAGAC |
| 54 | GATCCCAATGGTAAAGACT | AAGCCTCAAAAGAAGACC |
| 55 | TGTGGTTTGAGGCATCTTCT | GCCCACCAAAAGAAAGTAGT |
| 56 | TAGCCGCACTCCCACGAAGC | CCAGAATCATCAGACTCGGACAG |
| 57 | TCAAAGAGCCGCACAACAAG | ACTTTGCTGCTTGGTGACTTA |
| 58 | GGGGATGAGTTTGTTTGTTC | CCTGCTTGGCGAGATGA |
| 59 | TCTAATGCTCCCAAGGTACA | GGCTTGGTCCACGATCTT |
| 60 | AGCTCTCATGCAAGTAAC | TTCAACATACCGTCTGTA |
| 61 | TCCTCACTGCTCCTCTAATC | ACTCCCTATGGACCTTAGTC |
| 62 | AGGGAGGCGAACGAGAAACA | CGACTGCTGATGGGGAAGAG |
| 63 | CTACGGACTCACACCTATAT | ATGGTTCATCAATGAGATC |
| 64 | GTGAGCGATTGAGGGGTGTG | GGGGCTTGATTGAGTATTTCCA |
| 65 | AGGGCAAGTCATGTTTC | TATAAGGGCGAGGTATT |
| 66 | GAAGCCTGAGACCGCATAGA | TTCGGTGATGAAGATTGAAG |
| 67 | TTTCTTATGGCAATCACACG | GGAGGGCAGGAACAAAAAGT |
| 68 | GTTTATCATTTTGGGGTCAG | CGGTGTCCCTCAGGATGTA |
| 69 | CATGCACGTAAAGAAAGTGT | CCAAATGCACCCTAAGA |
| 70 | AATCCAAGTGGCCTACAG | CATGGCTTTGCTCAGTCA |
| 71 | TGTAGGTGGTGGTTAGG | TGTCAGACCCACCATTA |
| 72 | AGCAAGACACCATGTAGTC | GACACGTGGGATCTAGAC |
| 73 | AAAAGCCGATAGTGGGAACA | ATGCTGAGAGGTGGAAAATAGAG |
| 74 | GTCCATGTGCATAAGAGAG | CTCTTGGCATTTCAGATAC |
| 75 | AGCCAATGAAGGATAAAGG | CAAGCTAAAACCCCTAATC |
| 76 | CAATTCCAGCGTCACTATAG | AGTGGCAGTGGAAAAACAGT |
| 77 | GGGCTTTCATTTTCCACTAT | GCTCAACCTCATCCACAC |
| 78 | GACAGCTCGTGATGTAGA | GTTCTTGGCCGCTATAT |
| 79 | ACTTGTAAACCCTCTTCTCA | GTTTCATTACTTGGCTTCTG |
| 80 | CCTTCAAGCAAAGATACC | GGCACCAAACACAGTAA |
| 81 | CCACTGCTTCAAATTTACTAG | GCGTCCAAAACATAAATCAC |
| 82 | GGGAGAGGAAAAAATAGAG | CCTCCCTGAGACTGAGAAG |
| 83 | AGCAGGGCAAGAGCAATACT | TTCAGCAGCAGGAAACATC |
| 84 | GCCTATCCCCTGAACTATCT | TGCACATACCAGCAACAGAG |
| 85 | CATCAGAGCCTTCAAACTAC | AGCCTGAATTGCCTCTC |
| 86 | ATTCATTGCCATTCCCTTCA | TTGTCCCCTCTGTTCACTCA |
| 87 | ATTGCAGAGATGATGAGAAG | GAGATGCTGACAATGGTAGA |
| 88 | TCTCCCAAATCACTAGAC | ATCTGCAAGGCATATTC |
| 89 | ACGTTTTGGCAACTCTC | ACTCCCCTCTTTGACAT |
| 90 | TCCACTCTGGCAACTCC | AAGGATGGGCTTTGTAGT |
| 91 | TTTAGAGGACAAGGAGATAAG | CGACCGTGTCAAGAGTG |
| 92 | AGCAAAATGGCAAAGGAGAG | GGTGTGTGCTATGGAAGATCATAGT |
| 93 | GTAGCTTGAACCTGAAA | AGAACCACCGGAGTTAC |
| 94 | AAGCCACCAGGATCATC | GTCATTGCCACCTCTAACT |
| 95 | TTACTTGCTAAGCTCTCTAGC | TGGCTGTTTAATCTGTCTG |
| 96 | TCTATATTTGGTTGGCTTGA | ACTCATTTCAATCTCAGTGTC |
| 97 | TGCTACGTGCTGAAATA | ATTTCAGGTTCGCTTCA |
| 98 | CCTCCACTTCTCTTCATCTT | CTTCCTCAAGCTCAAACAAT |
| 99 | GATGTTGCCGCTGTTTG | CATCCCATTTCCCTCTT |
| 100 | ATGCTCCACCAAGTTTA | CACATCCTAGCATCATTG |
| 101 | AAGCAATATAGGTTCAGTTC | TCATTTTCTAATTCCAAACAAG |
| 102 | GCTCGTTTTTGTTTAGGTGA | TTTTCTCCATAGTCCGTTAC |
| 103 | CAGCACACAAATGACAT | CACCTTTCCTTTTTGTC |
| 104 | CCTATTCCTTACCTTTCTGT | GACTTACTATCTTGGCTCAC |
| 105 | CCTTGCATTCCACTATT | AGTTCTCAAGCCTCACA |
| 106 | CCTCCTTTGGAATTATG | GTGTTTGATGGGACATACA |
| 107 | ATTGGAGAGCACTTGGATAG | TTCTCTTCCTTCTCACTTGT |
| 108 | AGCCAGATGGAAATACAC | GTGCGATAAAGAGGAGAGT |
| 109 | TAGTTTTCCCATCACAGAGT | ACAATATTTAGACCTTCCATGAG |
| 110 | GTGCAGATGCAGATTATATG | CCTTTAGAATTGCCGTATC |
| 111 | ACAATAACCTGAGACAACAAGAAAC | ATACATCCCCTCCCCTCTCT |
| 112 | GAACTTGGCGTGTAACT | TGGTAGGTCTATTTGAGAGT |
These initial screens collectively revealed 83 spontaneous Hs but no DHs (although one DH was discovered subsequently), with the undirected phenotypic 'off-type' selection proving substantially more effective than screening for twin embryos. This result suggests that our method could be used to secure large numbers of Hs but is less able to isolate DHs at useful frequencies. This finding, when coupled with the routine nature of H chromosome doubling in other crops [21], suggested the most promising route for commercial DH production lay in the isolation of Hs followed by somatic doubling. In subsequent screening of abnormal seedlings, high-throughput flow cytometry therefore replaced molecular analysis for candidate H identification. Haploid identity was then supported using at least 15 microsatellite markers. Plants identified as diploid by flow cytometry continued to be screened for DHs as above. Using this amended screening procedure, we have identified over 1,100 H palms from approximately 60 million seedlings (to July 2009).
To have maximum utility this H/DH material should encompass as much genetic diversity from within the breeding germplasm as possible. A Principal Coordinates Analysis performed on H profiles using 28 microsatellite loci showed the first two axes accounted for 58% of the detected variation. While most Hs had a strong affinity to commercial duras, Hs have also been generated from pisifera types and overall variability amongst Hs encompassed that seen for the entire commercial palm material (Figure 3).
Figure 3.
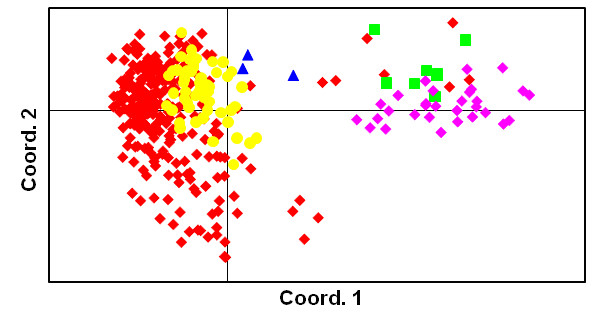
Principal Coordinates Analysis Plot of 95 diploid and 27 haploid palms based on 28 microsatellites. Red diamonds: haploids; green squares: commercial pisiferas; blue triangles: commercial teneras; yellow diamonds: commercial duras; purple diamonds: Ghanaian wild material. Microsatellite data in Table 7.
Effort then focussed on the creation of DHs from this rich germplasm of H genotypes (Figure 4). The most direct route to obtain DHs is to use chemical application to induce chromosome doubling. We applied a range of treatments to 50 H seedlings and screened leaves of the recovered material for evidence of chromosome doubling. Flow cytometry revealed that 48 seedlings contained substantial diploid sectors in their leaves; one palm was 100% doubled after exposure to10 mM colchicine (Figure 5) and 100 ppm GA3. To date, 16 H genotypes have produced pollen. This finding demonstrates scope for securing fertile gametes from diploid inflorescences or inflorescence sectors for DH or F1 production. Indeed, seed set using pollen from DH material has now been achieved (data not shown). Whilst further optimization work is required, our results when combined with experience in other crops [21] suggest routine production of fertile DH oil palm lines will be a relatively simple task.
Figure 4.
Selection of haploid oil palm plants growing in a nursery.
Figure 5.
Doubled haploid palm.
Discussion and Conclusions
The simple high-throughput phenotypic-genotypic seedling selection system used here provides a fourth practical approach to supplement androgenesis, wide crossing and gynogenesis [6] and has potential for many crops where H/DH production remains elusive. The prospect of adopting a similar untargeted approach more widely seems both plausible and attractive, and may be possible without experienced operators, especially as sophisticated phenomic screening systems [22] become more accessible.
In the case of oil palm, the efficacy of our H screening combined with the demonstrated ability to create DH palms, opens the way for the development of 100% true-breeding parental clones for F1 variety breeding. Thereafter, it is hoped that the potential genetic gain available from oil palm F1 hybrids will match that in other crops. If such a gain is achieved it could be beneficial in several ways. First, high-yielding F1 palms are likely to accelerate replacement of palms in existing plantations and cause a step-increase in production. Secondly, this breeding strategy provides greater flexibility for breeders to respond rapidly to emergent threats (e.g. climate change). Thirdly, using palm oil and its associated wastes for energy generation [7] could substantially reduce carbon-based emissions currently associated with the palm oil lifecycle [23]. Fourthly, DH oil palms could be exploited in combination with transgenic techniques that are now available for this crop [24]. Looking forward, the clear challenge is to maintain and improve oil palm productivity in the face of a changing climate sufficient to keep pace with growing demand [25]. However, it is important to point out that breeding is simply one stage in a long process from plantation to the eventual processed product and the economic realities of this international industry will finally determine the impact of any novel technology on the global agricultural system for this crop.
The provision here of a system for haploid-based F1 hybrid breeding in oil palm represents the first technological breakthrough likely to lead to step improvements in yield for this crop, and can also be applied to other crops recalcitrant to in vitro based H/DH systems. This methodology, in particular the application of high-throughput flow cytometry, has recently been applied successfully to two other tropical crops, namely rubber (Hevea brasiliensis L.) and cocoa (Theobroma cacao L.) (Nasution et al. unpublished).
Methods
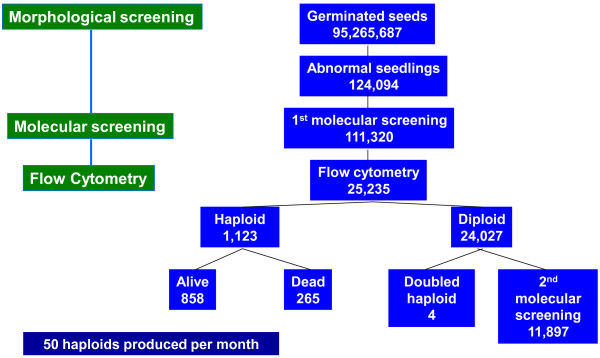
Hs and DHs were identified using three methods: a morphological screen; homozygosity/hemizygosity assessment; and ploidy level measurement. Initial screens emphasized identification of candidate DHs where seedling morphology screening was followed by homozygosity/hemizygosity assessment using microsatellites. H/DHs were then distinguished by flow cytometry and DHs subjected to an extensive homozygosity screen (Figure 6). As spontaneous DH frequency was low, later screens emphasized H recovery where the morphological screen was followed by flow cytometry; homozygosity of candidate Hs was thereafter confirmed with microsatellites.
Figure 6.
Summary of stages for identification of haploid and doubled haploid palm.
Seed morphological screen
For seed storage, mesocarps were removed from freshly harvested seed, and seeds air-dried at ambient temperature (24 h). Seeds were thereafter stored at 25°C with 15-18% moisture content. To induce germination, stored seeds were re-hydrated over 3 d to 18-20% moisture content, followed by 38-40°C incubation (40-60 d). Seeds were then re-hydrated for a further 5 d to >22% moisture content, and air-dried at ambient temperature (4 h). Seeds were germinated at ambient temperature (7 d to 3 months after treatment) and examined for atypical germination morphology (Figure 1).
Molecular pre-screen to exclude heterozygotes
DNA was isolated from leaf tissue using DNeasy 96 Plant Kit (Qiagen, UK). Initial heterozygosity screens used 15 microsatellites (Table 1) yielding alleles readily distinguished by agarose gel electrophoresis (Figure 7). 10 μl PCR mixes comprised 1.0 μl 10× NH4 buffer (Bioline), 0.3 μl MgCl2 (10 mM), 0.4 μl dNTPs (10 mM), 0.2 μl each primer (10 mM), 1-5 ng DNA and 1U Taq polymerase (Bioline). Thermocycling conditions: 2 min at 94°C followed by 35 cycles of 94°C for 30 s, 52-58°C for 30 s and 72°C for 45 s, with a final extension of 72°C for 7 min. Candidates presenting two allelic bands after fractionation by (2-3% w/v metaphor) agarose gel electrophoresis were discarded.
Figure 7.
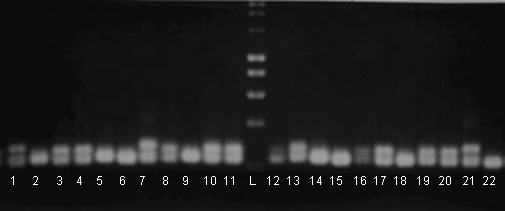
PCR amplicons generated by microsatellite marker 10 fractionated in 2% w/v agarose. Lanes 1-11 & 12-20: candidate H/DH palm plants; lane L: HyperladderI (Bioline, UK); lane 21: heterozygote control; lane 22: homozygote control. Candidates in lanes 1, 3, 4, 7, 8, 10, 11, 13, 16, 17, 19, 20 were deemed heterozygous and discarded.
Extended molecular screen
Candidate DHs and some Hs were subjected to an extensive assay for heterozygosity using 97 fluorescently-labelled microsatellites (Table 3) with 150 seedlings of normal phenotype and 24 heterozygous tenera palms as controls. PCR conditions were as described above and resultant products were fractionated on an ABI3730XL capillary sequencer (Applied Biosystems, USA) by Macrogen Inc (Korea). Allele size was determined (Genemapper v4.0) against a GS400HD standard. Individuals with two alleles at any locus were discarded.
DH candidate verification
To verify DH candidate 0644-219/05049582C we screened 212 microsatellites (Table 4) for heterozygosity in the maternal parent (BL013/12-06). 10 μl PCR mixes comprising: 5 μl BioMix™(Bioline, UK), 0.05 μl forward primer plus M13 adaptor (10 μM), 0.2 μl labelled M13(-29) (10 μM) (Sigma Genosys, UK), 0.2 μl reverse primer (10 μM) and 5-10 ng DNA were subjected to: 2 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 52°C, 45 s at 72°C, with a final extension of 72°C for 7 min. Amplicons were surveyed for heterozygosity by high-resolution melt (HRM) analysis according to Croxford et al. [26] using the candidate as the reference comparator. Samples with amplicons variable between the maternal parent and candidate DH were fractionated by capillary electrophoresis as above. 48 markers identified as heterozygous in the maternal parent (Table 5) were applied to the DH candidate to assess homozygosity.
Table 4.
Microsatellite markers used to screen for heterozygosity on the maternal parent (palm BL013/12-06) of DH candidate palm (0644-219/05049582C).
| No | Marker | Forward Primer (5'-3') | Reverse Primer (5'-3') |
|---|---|---|---|
| 1 | VS1 | GAGATTACAAAGTCCAAACC | TCAAAATTAAGAAAGTATGC |
| 2 | OPSSR 3 | ACGCATGCAGCTAGCTTTTC | CGCGTGAAAGATATGAATCAAC |
| 3 | OPSSR 7 | CACGCACGCAGTTTATTCTT | GGATGTATGCTTTACCTCCGAAT |
| 4 | OPSSR 8 | CCCCTTTTGCTTCCCTATTT | CTCCTTTTCCCCATCACAGA |
| 5 | OPSSR 9 | GACACAAGCAAAAACAAAAGCA | ATTCTGAAAGGAGGGGGAAA |
| 6 | OPSSR 14 | ATATGTGTGGGTGTGCGTGT | TGCCTCTGGTTGTTAGTCTGG |
| 7 | OPSSR 19 | TCTCTCTCTCTCTCTCTATGTGTGTGT | TGGCAATCAGCACACATTCT |
| 8 | OPSSR 29 | GCAGCTCTTTCCACACCTCT | TGTGGTCTCCTGAGGAAGATG |
| 9 | OPSSR 30 | TTTTCCCCATCACAGAATTG | CCCCTTTTGCTTCCCTATTT |
| 10 | OPSSR32 | GAACAAAACGGGAAGAAGCA | CCTCAAATGGGAGAAACCAG |
| 11 | mEgUWA07 | CGGATAGAGGCAGCAAGACT | CTCGGGTTGTTTAACCCATT |
| 12 | mEgUWA44 | TTGAGACGTCGTTCCTTTCC | AGCGGAGACCCAATAATCCT |
| 13 | mEgUWA50 | CCTGCAACTGCAAATGAGAC | TCCAGACACAAACTACACACACC |
| 14 | mEgCIR0037 | Published by Billotte et al. [27] | |
| 15 | mEgCIR0055 | Published by Billotte et al. [27] | |
| 16 | mEgCIR0059 | Published by Billotte et al. [27] | |
| 17 | mEgCIR0067 | Published by Billotte et al. [28] | |
| 18 | mEgCIR0074 | Published by Billotte et al. [27] | |
| 19 | mEgCIR0146 | Published by Billotte et al. [27] | |
| 20 | mEgCIR0163 | Published by Billotte et al. [27] | |
| 21 | mEgCIR0173 | Published by Billotte et al. [27] | |
| 22 | mEgCIR0177 | Published by Billotte et al. [27] | |
| 23 | mEgCIR0192 | Published by Billotte et al. [27] | |
| 24 | mEgCIR0195 | Published by Billotte et al. [27] | |
| 25 | mEgCIR0243 | Published by Billotte et al. [27] | |
| 26 | mEgCIR0246 | Published by Billotte et al. [27] | |
| 27 | mEgCIR0257 | Published by Billotte et al. [27] | |
| 28 | mEgCIR0268 | Published by Billotte et al. [27] | |
| 29 | mEgCIR0328 | Published by Billotte et al. [27] | |
| 30 | mEgCIR0359 | Published by Billotte et al. [27] | |
| 31 | mEgCIR0366 | Published by Billotte et al. [27] | |
| 32 | mEgCIR0369 | Published by Billotte et al. [27] | |
| 33 | mEgCIR0380 | Published by Billotte et al. [27] | |
| 34 | mEgCIR0399 | Published by Billotte et al. [27] | |
| 35 | mEgCIR0408 | Published by Billotte et al. [27] | |
| 36 | mEgCIR0409 | Published by Billotte et al. [27] | |
| 37 | mEgCIR0425 | Published by Billotte et al. [27] | |
| 38 | mEgCIR0433 | Published by Billotte et al. [27] | |
| 39 | mEgCIR0439 | Published by Billotte et al. [27] | |
| 40 | mEgCIR0445 | Published by Billotte et al. [27] | |
| 41 | mEgCIR0446 | Published by Billotte et al. [27] | |
| 42 | mEgCIR0465 | Published by Billotte et al. [27] | |
| 43 | mEgCIR0521 | Published by Billotte et al. [27] | |
| 44 | mEgCIR0551 | Published by Billotte et al. [27] | |
| 45 | mEgCIR0555 | Published by Billotte et al. [27] | |
| 46 | mEgCIR0588 | Published by Billotte et al. [27] | |
| 47 | mEgCIR0772 | Published by Billotte et al. [27] | |
| 48 | mEgCIR0773 | Published by Billotte et al. [27] | |
| 49 | mEgCIR0774 | Published by Billotte et al. [27] | |
| 50 | mEgCIR0775 | Published by Billotte et al. [27] | |
| 51 | mEgCIR0778 | Published by Billotte et al. [27] | |
| 52 | mEgCIR0779 | Published by Billotte et al. [27] | |
| 53 | mEgCIR0781 | Published by Billotte et al. [27] | |
| 54 | mEgCIR0786 | Published by Billotte et al. [27] | |
| 55 | mEgCIR0787 | Published by Billotte et al. [27] | |
| 56 | mEgCIR0788 | Published by Billotte et al. [27] | |
| 57 | mEgCIR0790 | Published by Billotte et al. [27] | |
| 58 | mEgCIR0793 | Published by Billotte et al. [27] | |
| 59 | mEgCIR0800 | Published by Billotte et al. [27] | |
| 60 | mEgCIR0801 | Published by Billotte et al. [27] | |
| 61 | mEgCIR0802 | Published by Billotte et al. [27] | |
| 62 | mEgCIR0803 | Published by Billotte et al. [27] | |
| 63 | mEgCIR0804 | Published by Billotte et al. [27] | |
| 64 | mEgCIR0825 | Published by Billotte et al. [27] | |
| 65 | mEgCIR0827 | Published by Billotte et al. [27] | |
| 66 | mEgCIR0844 | Published by Billotte et al. [27] | |
| 67 | mEgCIR0874 | Published by Billotte et al. [27] | |
| 68 | mEgCIR0878 | Published by Billotte et al. [27] | |
| 69 | mEgCIR0882 | Published by Billotte et al. [27] | |
| 70 | mEgCIR0886 | Published by Billotte et al. [27] | |
| 71 | mEgCIR0894 | Published by Billotte et al. [27] | |
| 72 | mEgCIR0905 | Published by Billotte et al. [27] | |
| 73 | mEgCIR0906 | Published by Billotte et al. [27] | |
| 74 | mEgCIR0910 | Published by Billotte et al. [27] | |
| 75 | mEgCIR0912 | Published by Billotte et al. [27] | |
| 76 | mEgCIR1729 | Published by Billotte et al. [27] | |
| 77 | mEgCIR1740 | Published by Billotte et al. [27] | |
| 78 | mEgCIR1753 | Published by Billotte et al. [27] | |
| 79 | mEgCIR1773 | Published by Billotte et al. [27] | |
| 80 | mEgCIR1917 | Published by Billotte et al. [27] | |
| 81 | mEgCIR1977 | Published by Billotte et al. [27] | |
| 82 | mEgCIR1996 | Published by Billotte et al. [27] | |
| 83 | mEgCIR2110 | Published by Billotte et al. [27] | |
| 84 | mEgCIR2144 | Published by Billotte et al. [27] | |
| 85 | mEgCIR2149 | Published by Billotte et al. [27] | |
| 86 | mEgCIR2188 | Published by Billotte et al. [27] | |
| 87 | mEgCIR2212 | Published by Billotte et al. [27] | |
| 88 | mEgCIR2215 | Published by Billotte et al. [27] | |
| 89 | mEgCIR2380 | Published by Billotte et al. [27] | |
| 90 | mEgCIR2387 | Published by Billotte et al. [27] | |
| 91 | mEgCIR2414 | Published by Billotte et al. [27] | |
| 92 | mEgCIR2417 | Published by Billotte et al. [27] | |
| 93 | mEgCIR2422 | Published by Billotte et al. [27] | |
| 94 | mEgCIR2423 | Published by Billotte et al. [27] | |
| 95 | mEgCIR2427 | Published by Billotte et al. [27] | |
| 96 | mEgCIR2436 | Published by Billotte et al. [27] | |
| 97 | mEgCIR2440 | Published by Billotte et al. [27] | |
| 98 | mEgCIR2492 | Published by Billotte et al. [27] | |
| 99 | mEgCIR2518 | Published by Billotte et al. [27] | |
| 100 | mEgCIR2525 | Published by Billotte et al. [27] | |
| 101 | mEgCIR2569 | Published by Billotte et al. [27] | |
| 102 | mEgCIR2575 | Published by Billotte et al. [27] | |
| 103 | mEgCIR2577 | Published by Billotte et al. [27] | |
| 104 | mEgCIR2590 | Published by Billotte et al. [27] | |
| 105 | mEgCIR2595 | Published by Billotte et al. [27] | |
| 106 | mEgCIR2600 | Published by Billotte et al. [27] | |
| 107 | mEgCIR2621 | Published by Billotte et al. [27] | |
| 108 | mEgCIR2628 | Published by Billotte et al. [27] | |
| 109 | mEgCIR2763 | Published by Billotte et al. [27] | |
| 110 | mEgCIR2813 | Published by Billotte et al. [27] | |
| 111 | mEgCIR2860 | Published by Billotte et al. [27] | |
| 112 | mEgCIR2887 | Published by Billotte et al. [27] | |
| 113 | mEgCIR2893 | Published by Billotte et al. [27] | |
| 114 | mEgCIR3040 | Published by Billotte et al. [27] | |
| 115 | mEgCIR3111 | Published by Billotte et al. [27] | |
| 116 | mEgCIR3160 | Published by Billotte et al. [27] | |
| 117 | mEgCIR3194 | Published by Billotte et al. [27] | |
| 118 | mEgCIR3213 | Published by Billotte et al. [27] | |
| 119 | mEgCIR3232 | Published by Billotte et al. [27] | |
| 120 | mEgCIR3295 | Published by Billotte et al. [27] | |
| 121 | mEgCIR3296 | Published by Billotte et al. [27] | |
| 122 | mEgCIR3297 | Published by Billotte et al. [27] | |
| 123 | mEgCIR3298 | Published by Billotte et al. [27] | |
| 124 | mEgCIR3300 | Published by Billotte et al. [27] | |
| 125 | mEgCIR3301 | Published by Billotte et al. [27] | |
| 126 | mEgCIR3305 | Published by Billotte et al. [27] | |
| 127 | mEgCIR3307 | Published by Billotte et al. [27] | |
| 128 | mEgCIR3310 | Published by Billotte et al. [27] | |
| 129 | mEgCIR3311 | Published by Billotte et al. [27] | |
| 130 | mEgCIR3316 | Published by Billotte et al. [27] | |
| 131 | mEgCIR3321 | Published by Billotte et al. [27] | |
| 132 | mEgCIR3328 | Published by Billotte et al. [27] | |
| 133 | mEgCIR3350 | Published by Billotte et al. [27] | |
| 134 | mEgCIR3384 | Published by Billotte et al. [27] | |
| 135 | mEgCIR3389 | Published by Billotte et al. [27] | |
| 136 | mEgCIR3399 | Published by Billotte et al. [27] | |
| 137 | mEgCIR3400 | Published by Billotte et al. [27] | |
| 138 | mEgCIR3402 | Published by Billotte et al. [27] | |
| 139 | mEgCIR3427 | Published by Billotte et al. [27] | |
| 140 | mEgCIR3428 | Published by Billotte et al. [27] | |
| 141 | mEgCIR3433 | Published by Billotte et al. [27] | |
| 142 | mEgCIR3439 | Published by Billotte et al. [27] | |
| 143 | mEgCIR3477 | Published by Billotte et al. [27] | |
| 144 | mEgCIR3519 | Published by Billotte et al. [27] | |
| 145 | mEgCIR3526 | Published by Billotte et al. [27] | |
| 146 | mEgCIR3533 | Published by Billotte et al. [27] | |
| 147 | mEgCIR3534 | Published by Billotte et al. [27] | |
| 148 | mEgCIR3535 | Published by Billotte et al. [27] | |
| 149 | mEgCIR3538 | Published by Billotte et al. [27] | |
| 150 | mEgCIR3543 | Published by Billotte et al. [27] | |
| 151 | mEgCIR3544 | Published by Billotte et al. [27] | |
| 152 | mEgCIR3546 | Published by Billotte et al. [27] | |
| 153 | mEgCIR3555 | Published by Billotte et al. [27] | |
| 154 | mEgCIR3557 | Published by Billotte et al. [27] | |
| 155 | mEgCIR3563 | Published by Billotte et al. [27] | |
| 156 | mEgCIR3567 | Published by Billotte et al. [27] | |
| 157 | mEgCIR3569 | Published by Billotte et al. [27] | |
| 158 | mEgCIR3574 | Published by Billotte et al. [27] | |
| 159 | mEgCIR3587 | Published by Billotte et al. [27] | |
| 160 | mEgCIR3590 | Published by Billotte et al. [27] | |
| 161 | mEgCIR3592 | Published by Billotte et al. [27] | |
| 162 | mEgCIR3593 | Published by Billotte et al. [27] | |
| 163 | mEgCIR3607 | Published by Billotte et al. [27] | |
| 164 | mEgCIR3622 | Published by Billotte et al. [27] | |
| 165 | mEgCIR3633 | Published by Billotte et al. [27] | |
| 166 | mEgCIR3639 | Published by Billotte et al. [27] | |
| 167 | mEgCIR3643 | Published by Billotte et al. [27] | |
| 168 | mEgCIR3649 | Published by Billotte et al. [27] | |
| 169 | mEgCIR3653 | Published by Billotte et al. [27] | |
| 170 | mEgCIR3655 | Published by Billotte et al. [27] | |
| 171 | mEgCIR3663 | Published by Billotte et al. [27] | |
| 172 | mEgCIR3668 | Published by Billotte et al. [27] | |
| 173 | mEgCIR3672 | Published by Billotte et al. [27] | |
| 174 | mEgCIR3683 | Published by Billotte et al. [27] | |
| 175 | mEgCIR3684 | Published by Billotte et al. [27] | |
| 176 | mEgCIR3691 | Published by Billotte et al. [27] | |
| 177 | mEgCIR3693 | Published by Billotte et al. [27] | |
| 178 | mEgCIR3696 | Published by Billotte et al. [27] | |
| 179 | mEgCIR3698 | Published by Billotte et al. [27] | |
| 180 | mEgCIR3705 | Published by Billotte et al. [27] | |
| 181 | mEgCIR3711 | Published by Billotte et al. [27] | |
| 182 | mEgCIR3716 | Published by Billotte et al. [27] | |
| 183 | mEgCIR3718 | Published by Billotte et al. [27] | |
| 184 | mEgCIR3722 | Published by Billotte et al. [27] | |
| 185 | mEgCIR3727 | Published by Billotte et al. [27] | |
| 186 | mEgCIR3728 | Published by Billotte et al. [27] | |
| 187 | mEgCIR3732 | Published by Billotte et al. [27] | |
| 188 | mEgCIR3737 | Published by Billotte et al. [27] | |
| 189 | mEgCIR3739 | Published by Billotte et al. [27] | |
| 190 | mEgCIR3745 | Published by Billotte et al. [27] | |
| 191 | mEgCIR3747 | Published by Billotte et al. [27] | |
| 192 | mEgCIR3750 | Published by Billotte et al. [27] | |
| 193 | mEgCIR3755 | Published by Billotte et al. [27] | |
| 194 | mEgCIR3766 | Published by Billotte et al. [27] | |
| 195 | mEgCIR3769 | Published by Billotte et al. [27] | |
| 196 | mEgCIR3775 | Published by Billotte et al. [27] | |
| 197 | mEgCIR3782 | Published by Billotte et al. [27] | |
| 198 | mEgCIR3785 | Published by Billotte et al. [27] | |
| 199 | mEgCIR3787 | Published by Billotte et al. [27] | |
| 200 | mEgCIR3788 | Published by Billotte et al. [27] | |
| 201 | mEgCIR3792 | Published by Billotte et al. [27] | |
| 202 | mEgCIR3807 | Published by Billotte et al. [27] | |
| 203 | mEgCIR3808 | Published by Billotte et al. [27] | |
| 204 | mEgCIR3809 | Published by Billotte et al. [27] | |
| 205 | mEgCIR3813 | Published by Billotte et al. [27] | |
| 206 | mEgCIR3819 | Published by Billotte et al. [27] | |
| 207 | mEgCIR3825 | Published by Billotte et al. [27] | |
| 208 | mEgCIR3826 | Published by Billotte et al. [27] | |
| 209 | mEgCIR3828 | Published by Billotte et al. [27] | |
| 210 | mEgCIR3847 | Published by Billotte et al. [27] | |
| 211 | mEgCIR3850 | Published by Billotte et al. [27] | |
| 212 | mEgCIR3869 | Published by Billotte et al. [27] | |
Table 5.
Markers shown to be heterozygous in the maternal parent (palm BL013/12-06) and homozygous in the DH candidate (0644-219/05049582C).
| No | Marker | Linkage Group |
|---|---|---|
| 1 | mEgCIR0268 | 1 |
| 2 | mEgCIR0874 | 1 |
| 3 | mEgCIR3847 | 1 |
| 4 | mEgCIR2149 | 2 |
| 5 | mEgCIR2518 | 3 |
| 6 | mEgCIR0425 | 3 |
| 7 | mEgCIR3544 | 3 |
| 8 | mEgCIR3716 | 4 |
| 9 | mEgCIR1917 | 4 |
| 10 | mEgCIR3535 | 4 |
| 11 | mEgCIR3310 | 4 |
| 12 | mEgCIR3705 | 4 |
| 13 | mEgCIR3477 | 4 |
| 14 | mEgCIR0059 | 4 |
| 15 | mEgCIR3557 | 4 |
| 16 | mEgCIR2813 | 5 |
| 17 | mEgCIR3543 | 6 |
| 18 | mEgCIR0195 | 6 |
| 19 | mEgCIR0894 | 7 |
| 20 | mEgCIR0905b | 7 |
| 21 | mEgCIR0774 | 8 |
| 22 | mEgCIR2440 | 8 |
| 23 | mEgCIR0825 | 10 |
| 24 | mEgCIR3826 | 10 |
| 25 | mEgCIR0788 | 10 |
| 26 | mEgCIR2628 | 10 |
| 27 | mEgCIR0146 | 10 |
| 28 | mEgCIR0878 | 11 |
| 29 | mEgCIR1773 | 12 |
| 30 | mEgCIR3311 | 12 |
| 31 | mEgCIR0779 | 14 |
| 32 | mEgCIR0588 | 14 |
| 33 | mEgCIR3737 | 15 |
| 34 | mEgCIR3850 | 15 |
| 35 | mEgCIR3639 | 16 |
| 36 | mEgCIR0905a | 16 |
| 37 | mEgCIR3739 | unlinked |
| 38 | mEgCIR3160 | unmapped |
| 39 | mEgCIR3360 | unmapped |
| 40 | mEgCIR0801 | unmapped |
| 41 | mEgCIR2577 | unmapped |
| 42 | OPSSR14 | unmapped |
| 43 | OPSSR30 | unmapped |
| 44 | OPSSR32 | unmapped |
| 45 | mEgUWA44 | unmapped |
| 46 | mEgUWA50 | unmapped |
| 47 | mEgUWA07 | unmapped |
| 48 | VS1 | unmapped |
Linkage group assigned according to Billotte et al. [27].
DH candidate 0644-219/05049582C was found to be homozygous across all 48 loci that were heterozygous in its maternal parent. Of these 48 loci, 36 have been mapped by Billotte et al. [27] (Table 5). We first considered the probability of obtaining the observed homozygosity levels via independent assortment using only the unlinked markers from this group. For unlinked loci, the probability of homozygous offspring arising by independent assortment is 0.5 per locus. Given that heterozygous loci were secured from 14 of the 16 linkage groups, with the addition of a further unlinked (unassigned) marker, the probability of these markers all becoming homozygous by chance is therefore: P = 0.515 = 0.000030517578125.
This figure was further reduced by the inclusion of the remaining 21 markers that had been assigned a map position [27]. Here, linkage was accommodated by multiplying by 1-(distance in cM/100). Thus the inclusion of a new marker 10 cM from an existing marker would mean multiplying the cumulative total by 1- (10/100) = 1-0.1 = 0.9 (rather than 0.5 for an unlinked marker). This reduced the probability as follows:
Flow Cytometry
Newly matured leaflets or radicles from candidate H/DH palms were subjected to flow cytometry according to Anumaganathan & Earle [29] to establish ploidy level. Commercial tenera palms were included as diploid controls. For high-throughput mass screening, tissue samples were bulked at a rate of five individual tissue samples per bulk. Bulked samples (about 0.5 cm2 for radicles and 1 cm2 for leaf material (per each individual) were sliced by chopping with a sharp clean razor-blade (20-30 chops), in a plastic 9 cm diameter Petri dish containing 1.5 ml of cold (5°C) CyStain® UV Ploidy solution (Partec, Germany) modified by addition of 6.48 mM dithiothreitol (DTT) and 1% (v/v) polyvinylpyrrolidone (PVP-40) (Sigma-Aldrich, USA). The addition of DTT and PVP-40 were found to reduce background counts ('noise') in output histograms of particle fluorescence in the analyte.
Confirmation of Hs by chromosome squashes
Harvested roots were pre-treated in iced water (24 h), then fixed in 3:1 v/v alcohol: glacial acetic acid at 4°C (24 h). They were then rinsed in water, softened in 1N HCl (20 min), rinsed in water (2 min) and stained in saturated aceto-orcein (1 min). The root tip was then squashed, mounted onto a glass slide, and examined using a compound photomicroscope.
Principal Coordinates Analysis
The genetic affinity of 270 Hs was compared with 95 representative diploids (Table 6) using 28 microsatellites (Table 7) by Principal Coordinates Analysis (PCoA). The PCoA was constructed using GenAlEx v6 [30]. Genetic distance option 'codominant-genotypic' was applied, where pairwise, individual-by-individual (N × N) genetic distances are calculated for codominant data. For a single-locus analysis, with i-th, j-th, k-th and l-th different alleles, a set of squared distances is defined as d2(ii, ii) = 0, d2(ij, ij) = 0, d2(ii, ij) = 1, d2(ij, ik) = 1, d2(ij, kl) = 2, d2(ii, jk) = 3, and d2(ii, jj) = 4. The algorithm used in GenAlEx is based on Orloci [31] using distance matrix with standardization (by dividing the distance inputs by the square root of n-1). Here, Hs were treated as the DHs they were assumed to generate; thus genotypes were homozygous not hemizygous.
Table 6.
Identification codes, oil palm type and ploidy level of oil palm genotypes used in the Principal Coordinates Analysis
| No | Label no in PCO | Sample name in PCO | Palm Id | Ploidy level |
|---|---|---|---|---|
| 1 | 1 | haploid | 05020271_0001 | x |
| 2 | 2 | haploid | 05050099_0001 | x |
| 3 | 3 | haploid | 05050099_0002 | x |
| 4 | 4 | haploid | 05020961_0001 | x |
| 5 | 5 | haploid | 05020511_0001 | x |
| 6 | 6 | haploid | 05020946_0001 | x |
| 7 | 8 | haploid | 05030147_0001 | x |
| 8 | 9 | haploid | 05030462_0001 | x |
| 9 | 10 | haploid | 05020420_0002 | x |
| 10 | 11 | haploid | 05020361_0001 | x |
| 11 | 12 | haploid | 05030060_0001 | x |
| 12 | 13 | haploid | 05020558_0001 | x |
| 13 | 14 | haploid | 05020631_0001 | x |
| 14 | 15 | haploid | 05040748_0003 | x |
| 15 | 16 | haploid | 05030308_0001 | x |
| 16 | 18 | haploid | 05080318_0003 | x |
| 17 | 19 | haploid | 06020186_0001 | x |
| 18 | 20 | haploid | 05110212_0001 | x |
| 19 | 21 | haploid | 05120555_0001 | x |
| 20 | 22 | haploid | 06011022_0001 | x |
| 21 | 23 | haploid | 05020059_0001 | x |
| 22 | 24 | haploid | 06020320_0004 | x |
| 23 | 25 | haploid | 06020571_0004 | x |
| 24 | 26 | haploid | 06020381_0001 | x |
| 25 | 27 | haploid | 05060119_0001 | x |
| 26 | 28 | haploid | 05090172_0001 | x |
| 27 | 30 | haploid | 05100321_0001 | x |
| 28 | 31 | haploid | 06010670_0006 | x |
| 29 | 32 | haploid | 06010842_0004 | x |
| 30 | 33 | haploid | 05050228_0001 | x |
| 31 | 34 | haploid | 05110260_0001 | x |
| 32 | 35 | haploid | 05110260_0002 | x |
| 33 | 36 | haploid | 05110162_0001 | x |
| 34 | 37 | haploid | 05101030_0001 | x |
| 35 | 38 | haploid | 05040273_0001 | x |
| 36 | 39 | haploid | 05110003_0001 | x |
| 37 | 40 | haploid | 05120002_0001 | x |
| 38 | 41 | haploid | 05080095_0001 | x |
| 39 | 43 | haploid | 06110122_0002 | x |
| 40 | 44 | haploid | 05110716_0001 | x |
| 41 | 45 | haploid | 05010836_0001 | x |
| 42 | 46 | haploid | 05120155_0001 | x |
| 43 | 47 | haploid | 05110875_0001 | x |
| 44 | 48 | haploid | 05070553_0001 | x |
| 45 | 49 | haploid | 05070466_0001 | x |
| 46 | 50 | haploid | 06010650_0001 | x |
| 47 | 51 | haploid | 05110718_0001 | x |
| 48 | 52 | haploid | 05110496_0001 | x |
| 49 | 53 | haploid | 06010107_0001 | x |
| 50 | 54 | haploid | 05120429_0002 | x |
| 51 | 55 | haploid | 06010953_0001 | x |
| 52 | 56 | haploid | 05030686_0001 | x |
| 53 | 57 | haploid | 05060107_0001 | x |
| 54 | 58 | haploid | 05030791_0001 | x |
| 55 | 59 | haploid | 05080585_0001 | x |
| 56 | 60 | haploid | 05020375_0001 | x |
| 57 | 61 | haploid | 05121048_0001 | x |
| 58 | 62 | haploid | 05055090_0001 | x |
| 59 | 63 | haploid | 05121004_0002 | x |
| 60 | 64 | haploid | 06030064_0001 | x |
| 61 | 65 | haploid | 05121061_0004 | x |
| 62 | 66 | haploid | 05060276_0001 | x |
| 63 | 67 | haploid | 05100988_0001 | x |
| 64 | 68 | haploid | 05060315_0001 | x |
| 65 | 69 | haploid | 06030324_0003 | x |
| 66 | 70 | haploid | 05080506_0001 | x |
| 67 | 71 | haploid | 06010813_0001 | x |
| 68 | 72 | haploid | 05110881_0001 | x |
| 69 | 73 | haploid | 05100717_0001 | x |
| 70 | 74 | haploid | 06020169_0009 | x |
| 71 | 75 | haploid | 05110134_0001 | x |
| 72 | 76 | haploid | 05030196_0001 | x |
| 73 | 77 | haploid | 05050220_0001 | x |
| 74 | 78 | haploid | 06011195_0001 | x |
| 75 | 79 | haploid | 05120725_0001 | x |
| 76 | 80 | haploid | 05100510_0001 | x |
| 77 | 81 | haploid | 05060624_0001 | x |
| 78 | 82 | haploid | 05060712_0001 | x |
| 79 | 83 | haploid | 05030150_0001 | x |
| 80 | 84 | haploid | 06030180_0001 | x |
| 81 | 85 | haploid | 06020915_0001 | x |
| 82 | 86 | haploid | 05101150_0003 | x |
| 83 | 87 | haploid | 05101152_0001 | x |
| 84 | 88 | haploid | 05020415_0001 | x |
| 85 | 89 | haploid | 05040029_0002 | x |
| 86 | 90 | haploid | 05040035_0003 | x |
| 87 | 91 | haploid | 06020573_0001 | x |
| 88 | 93 | haploid | 05121112_0008 | x |
| 89 | 94 | haploid | 05090078_0001 | x |
| 90 | 95 | haploid | 05060495_0001 | x |
| 91 | 96 | haploid | 05070484_0001 | x |
| 92 | 97 | haploid | 06020455_0001 | x |
| 93 | 98 | haploid | 05075185_0001 | x |
| 94 | 99 | haploid | 05090522_0004 | x |
| 95 | 100 | haploid | 06020625_0002 | x |
| 96 | 101 | haploid | 05100812_0002 | x |
| 97 | 102 | haploid | 05100862_0001 | x |
| 98 | 103 | haploid | 05030224_0002 | x |
| 99 | 104 | haploid | 05040439_0001 | x |
| 100 | 105 | haploid | 05040317_0003 | x |
| 101 | 106 | haploid | 05080030_0001 | x |
| 102 | 107 | haploid | 05070703_0003 | x |
| 103 | 108 | haploid | 05080485_0001 | x |
| 104 | 109 | haploid | 05110470_0002 | x |
| 105 | 110 | haploid | 05100423_0001 | x |
| 106 | 111 | haploid | 05110423_0001 | x |
| 107 | 112 | haploid | 05080362_0003 | x |
| 108 | 113 | haploid | 05110625_0001 | x |
| 109 | 114 | haploid | 05120719_0001 | x |
| 110 | 115 | haploid | 05121073_0002 | x |
| 111 | 116 | haploid | 06050726_0002 | x |
| 112 | 117 | haploid | 06060063_0001 | x |
| 113 | 119 | haploid | 06121220_0001 | x |
| 114 | 120 | haploid | 06080516_0001 | x |
| 115 | 121 | haploid | 06090505_0002 | x |
| 116 | 122 | haploid | 06090407_0004 | x |
| 117 | 123 | haploid | 06051133_0002 | x |
| 118 | 124 | haploid | 06060740_0031 | x |
| 119 | 125 | haploid | 06060740_0077 | x |
| 120 | 126 | haploid | 06060740_0090 | x |
| 121 | 127 | haploid | 06120178_0001 | x |
| 122 | 128 | haploid | 06090960_0003 | x |
| 123 | 129 | haploid | 06090657_0001 | x |
| 124 | 130 | haploid | 06120377_0001 | x |
| 125 | 131 | haploid | 06070208_0001 | x |
| 126 | 132 | haploid | 07010308_0001 | x |
| 127 | 133 | haploid | 06121125_0001 | x |
| 128 | 134 | haploid | 06121125_0002 A | x |
| 129 | 135 | haploid | 06121125_0002 B | x |
| 130 | 136 | haploid | 06019052_0005 | x |
| 131 | 137 | haploid | 06129197_0001 | x |
| 132 | 138 | haploid | 06079077_0001 | x |
| 133 | 139 | haploid | 07019130_0003 | x |
| 134 | 140 | haploid | 06075474_0001 | x |
| 135 | 141 | haploid | 06075474_0003 | x |
| 136 | 142 | haploid | 06075544_0001 | x |
| 137 | 143 | haploid | 06045801_0001 | x |
| 138 | 144 | haploid | 06065285_0001 | x |
| 139 | 145 | haploid | 06081027_0001 | x |
| 140 | 146 | haploid | 06090264_0001 | x |
| 141 | 147 | haploid | 06090264_0002 | x |
| 142 | 148 | haploid | 06070430_0001 | x |
| 143 | 149 | haploid | 06090861_0001 | x |
| 144 | 150 | haploid | 06051245_0001 | x |
| 145 | 151 | haploid | 06070716_0001 | x |
| 146 | 152 | haploid | 06051468_0001 | x |
| 147 | 153 | haploid | 06075617_0001 | x |
| 148 | 154 | haploid | 06040273_0001 | x |
| 149 | 155 | haploid | 06080584_0001 | x |
| 150 | 156 | haploid | 06070825_0001 | x |
| 151 | 158 | haploid | 06110390_0015 | x |
| 152 | 159 | haploid | 06031385_0001 | x |
| 153 | 160 | haploid | 06045657_0001 | x |
| 154 | 161 | haploid | 06110204_0008 | x |
| 155 | 162 | haploid | 06050161_0001 | x |
| 156 | 163 | haploid | 06071068_0010 | x |
| 157 | 164 | haploid | 06100785_0002 | x |
| 158 | 165 | haploid | 06010987_0028 | x |
| 159 | 166 | haploid | 07010166_0001 | x |
| 160 | 167 | haploid | 06100730_0001 | x |
| 161 | 168 | haploid | 06080681_0001 | x |
| 162 | 169 | haploid | 06080532_0005 | x |
| 163 | 170 | haploid | 06040024_0001 | x |
| 164 | 172 | haploid | 06080217_0010 | x |
| 165 | 173 | haploid | 06120975_0001 | x |
| 166 | 174 | haploid | 06070581_0002 | x |
| 167 | 175 | haploid | 06060477_0001 | x |
| 168 | 176 | haploid | 06120852_0001 | x |
| 169 | 177 | haploid | 06091392_0001 | x |
| 170 | 178 | haploid | 06060344_0001 | x |
| 171 | 179 | haploid | 06090211_0001 | x |
| 172 | 180 | haploid | 06100858_0001 | x |
| 173 | 181 | haploid | 06080272_0007 | x |
| 174 | 182 | haploid | 06050493_0004 | x |
| 175 | 183 | haploid | 06101033_0002 | x |
| 176 | 184 | haploid | 06081043_0001 | x |
| 177 | 185 | haploid | 07011057_0001 | x |
| 178 | 186 | haploid | 06070921_0001 | x |
| 179 | 187 | haploid | 06111210_0002 | x |
| 180 | 188 | haploid | 06121495_0001 | x |
| 181 | 189 | haploid | 06110610_0001 | x |
| 182 | 190 | haploid | 06090772_0001 | x |
| 183 | 191 | haploid | 06090318_0002 | x |
| 184 | 192 | haploid | 06121313_0001 | x |
| 185 | 193 | haploid | 06085027_0001 | x |
| 186 | 194 | haploid | 06090109_0001 | x |
| 187 | 195 | haploid | 06080157_0001 | x |
| 188 | 196 | haploid | 06121316_0001 | x |
| 189 | 197 | haploid | 06110900_0001 | x |
| 190 | 198 | haploid | 06070228_0002 | x |
| 191 | 199 | haploid | 06101174_0001 | x |
| 192 | 200 | haploid | 06060805_0001 | x |
| 193 | 201 | haploid | 06085063_0001 | x |
| 194 | 202 | haploid | 06101037_0001 | x |
| 195 | 203 | haploid | 06110444_0002 | x |
| 196 | 204 | haploid | 06101487_0001 | x |
| 197 | 205 | haploid | 06100937_0001 | x |
| 198 | 206 | haploid | 06090820_0002 | x |
| 199 | 207 | haploid | 06070039_0001 | x |
| 200 | 208 | haploid | 06070772_0001 | x |
| 201 | 209 | haploid | 07011408_0001 | x |
| 202 | 210 | haploid | 07011408_0002 | x |
| 203 | 211 | haploid | 06100319_0001 | x |
| 204 | 212 | haploid | 06070468_0001 | x |
| 205 | 213 | haploid | 06121385_0002 | x |
| 206 | 214 | haploid | 06100537_0001 | x |
| 207 | 215 | haploid | 06120726_0001 | x |
| 208 | 216 | haploid | 06070883_0001 | x |
| 209 | 217 | haploid | 06040041_0001 | x |
| 210 | 218 | haploid | 06100263_0001 | x |
| 211 | 219 | haploid | 06040043_0009 | x |
| 212 | 220 | haploid | 06101232_0001 | x |
| 213 | 221 | haploid | 06060189_0003 | x |
| 214 | 222 | haploid | 06091275_0002 | x |
| 215 | 223 | haploid | 06060097_0001 | x |
| 216 | 224 | haploid | 06100873_0001 | x |
| 217 | 225 | haploid | 06050038_0001 | x |
| 218 | 226 | haploid | 06100025_0001 | x |
| 219 | 227 | haploid | 06100940_0002 | x |
| 220 | 228 | haploid | 06040800_0001 | x |
| 221 | 229 | haploid | 06071007_0002 | x |
| 222 | 230 | haploid | 06020043_0026 | x |
| 223 | 231 | haploid | 06060811_0153 | x |
| 224 | 232 | haploid | 06080751_0001 | x |
| 225 | 233 | haploid | 06050178_0068 | x |
| 226 | 234 | haploid | 06040287_0001 | x |
| 227 | 236 | haploid | 06101496_0001 | x |
| 228 | 237 | haploid | 06040643_0001 | x |
| 229 | 238 | haploid | 06045788_0003 | x |
| 230 | 239 | haploid | 06050326_0001 | x |
| 231 | 240 | haploid | 06080649_0002 | x |
| 232 | 241 | haploid | 06080649_0003 | x |
| 233 | 242 | haploid | 06080601_0001 | x |
| 234 | 243 | haploid | 06101247_0001 | x |
| 235 | 244 | haploid | 06111271_0001 | x |
| 236 | 245 | haploid | 06090337_0001 | x |
| 237 | 246 | haploid | 06050125_0002 | x |
| 238 | 247 | haploid | 06050331_0001 | x |
| 239 | 248 | haploid | 06060728_0002 | x |
| 240 | 249 | haploid | 06080109_0001 | x |
| 241 | 250 | haploid | 06101048_0001 | x |
| 242 | 251 | haploid | 06051077_0001 | x |
| 243 | 253 | haploid | 06041067_0003 | x |
| 244 | 254 | haploid | 06040302_0002 | x |
| 245 | 255 | haploid | 06110121_0001 | x |
| 246 | 256 | haploid | 06090845_0001 | x |
| 247 | 257 | haploid | 06060375_0001 | x |
| 248 | 258 | haploid | 06070494_0001 | x |
| 249 | 259 | haploid | 06040938_0003 | x |
| 250 | 260 | haploid | 06081010_0001 | x |
| 251 | 261 | haploid | 06070415_0003 | x |
| 252 | 263 | haploid | 07010776_0001 | x |
| 253 | 264 | haploid | 06120890_0001 | x |
| 254 | 265 | haploid | 06120316_0001 | x |
| 255 | 266 | haploid | 06121413_0001 | x |
| 256 | 267 | haploid | 06090247_0001 | x |
| 257 | 268 | haploid | 06090247_0002 | x |
| 258 | 269 | haploid | 06090801_0001 | x |
| 259 | 270 | haploid | 06041160_0002 | x |
| 260 | 271 | haploid | 06031248_0001 | x |
| 261 | 272 | haploid | 07010075_0001 | x |
| 262 | 273 | haploid | 07011039_0001 | x |
| 263 | 274 | haploid | 06041232_0001 | x |
| 264 | 275 | haploid | 06101271_0002 | x |
| 265 | 276 | haploid | 06060506_0001 | x |
| 266 | 277 | haploid | 06080566_0001 | x |
| 267 | 278 | haploid | 06060124_0001 | x |
| 268 | 279 | haploid | 07020168_0001 | x |
| 269 | 281 | haploid | 06090909_0002 | x |
| 270 | 282 | haploid | 06080869_0001 | x |
| 271 | 1 | commercial pisifera | BL605/39-04 | 2x |
| 272 | 2 | commercial pisifera | BL607/91-10 | 2x |
| 273 | 3 | commercial pisifera | BL612/84-05 | 2x |
| 274 | 4 | commercial pisifera | BL1120/75-07 | 2x |
| 275 | 5 | commercial pisifera | BL143/04-10 | 2x |
| 276 | 6 | commercial pisifera | BL147/21-05 | 2x |
| 277 | 7 | commercial pisifera | BL148/05-08 | 2x |
| 278 | 8 | commercial pisifera | BL158/A2-13 | 2x |
| 279 | 1 | commercial tenera | BL10452/207-02 | 2x |
| 280 | 2 | commercial tenera | BL10323/104-06 | 2x |
| 281 | 3 | commercial tenera | BL1177/184-09 | 2x |
| 282 | 1 | commercial dura | BL10887/08-22 | 2x |
| 283 | 2 | commercial dura | BL10885/08-27 | 2x |
| 284 | 3 | commercial dura | BL1221/51-14 | 2x |
| 285 | 4 | commercial dura | BL1222/32-02 | 2x |
| 286 | 5 | commercial dura | BL1224/14-19 | 2x |
| 287 | 6 | commercial dura | BL1231/02-01 | 2x |
| 288 | 7 | commercial dura | BL1235/14-01 | 2x |
| 289 | 8 | commercial dura | BL1125/03-02 | 2x |
| 290 | 9 | commercial dura | BL1124/17-09 | 2x |
| 291 | 10 | commercial dura | BL1136/01-02 | 2x |
| 292 | 11 | commercial dura | BL10868/12-10 | 2x |
| 293 | 12 | commercial dura | BL10868/12-11 | 2x |
| 294 | 13 | commercial dura | BL10868/12-13 | 2x |
| 295 | 14 | commercial dura | BL10879/08-06 | 2x |
| 296 | 15 | commercial dura | BL10879/08-07 | 2x |
| 297 | 16 | commercial dura | BL10879/08-09 | 2x |
| 298 | 17 | commercial dura | BL10883/04-06 | 2x |
| 299 | 18 | commercial dura | BL10883/04-08 | 2x |
| 300 | 19 | commercial dura | BL10883/04-09 | 2x |
| 301 | 20 | commercial dura | BL10883/05-06 | 2x |
| 302 | 21 | commercial dura | BL10891/04-23 | 2x |
| 303 | 22 | commercial dura | BL10891/04-24 | 2x |
| 304 | 23 | commercial dura | BL10891/05-22 | 2x |
| 305 | 24 | commercial dura | BL10891/05-23 | 2x |
| 306 | 25 | commercial dura | BL10873/52-18 | 2x |
| 307 | 26 | commercial dura | BL10873/52-19 | 2x |
| 308 | 27 | commercial dura | BL10873/52-21 | 2x |
| 309 | 28 | commercial dura | BL10873/53-19 | 2x |
| 310 | 29 | commercial dura | BL1229/48-15 | 2x |
| 311 | 30 | commercial dura | BL1230/42-15 | 2x |
| 312 | 31 | commercial dura | A1122/04-01 | 2x |
| 313 | 32 | commercial dura | A1122/12-05 | 2x |
| 314 | 33 | commercial dura | A1122/12-08 | 2x |
| 315 | 34 | commercial dura | A1122/36-02 | 2x |
| 316 | 35 | commercial dura | A1123/01-02 | 2x |
| 317 | 36 | commercial dura | A1123/01-06 | 2x |
| 318 | 37 | commercial dura | A1123/01-07 | 2x |
| 319 | 38 | commercial dura | A1123/01-12 | 2x |
| 320 | 39 | commercial dura | A1130/02-02 | 2x |
| 321 | 40 | commercial dura | A1130/02-06 | 2x |
| 322 | 41 | commercial dura | A1130/02-10 | 2x |
| 323 | 42 | commercial dura | A1130/02-16 | 2x |
| 324 | 43 | commercial dura | A1127/08-16 | 2x |
| 325 | 44 | commercial dura | A1127/08-06 | 2x |
| 326 | 45 | commercial dura | A1127/05-11 | 2x |
| 327 | 46 | commercial dura | A1127/05-03 | 2x |
| 328 | 47 | commercial dura | B1134/35-09 | 2x |
| 329 | 48 | commercial dura | B1133/07-10 | 2x |
| 330 | 49 | commercial dura | B1136/21-11 | 2x |
| 331 | 50 | commercial dura | B1136/21-12 | 2x |
| 332 | 51 | commercial dura | C1128/07-14 | 2x |
| 333 | 52 | commercial dura | C1121/13-08 | 2x |
| 334 | 53 | commercial dura | BL11508/111-1 | 2x |
| 335 | 54 | commercial dura | BL11396/11-21 | 2x |
| 336 | 1 | Ghana wild | K31-1/GHANA/1-1 | 2x |
| 337 | 2 | Ghana wild | K31-1/GHANA/41-498 | 2x |
| 338 | 3 | Ghana wild | K31-1/GHANA/39-875 | 2x |
| 339 | 4 | Ghana wild | K31-1/GHANA/31-430 | 2x |
| 340 | 5 | Ghana wild | K31-1/GHANA/26-629 | 2x |
| 341 | 6 | Ghana wild | K31-1/GHANA/24-1164 | 2x |
| 342 | 7 | Ghana wild | K31-1/GHANA/56-1185 | 2x |
| 343 | 8 | Ghana wild | K31-1/GHANA/29-1087 | 2x |
| 344 | 9 | Ghana wild | K31-1/GHANA/38-1193 | 2x |
| 345 | 10 | Ghana wild | K31-1/GHANA/43-994 | 2x |
| 346 | 11 | Ghana wild | K31-1/GHANA/8-1100 | 2x |
| 347 | 12 | Ghana wild | K31-1/GHANA/11-1192 | 2x |
| 348 | 13 | Ghana wild | K31-1/GHANA/35-1190 | 2x |
| 349 | 14 | Ghana wild | K31-1/GHANA/3-46 | 2x |
| 350 | 15 | Ghana wild | K31-1/GHANA/5-102 | 2x |
| 351 | 16 | Ghana wild | K31-1/GHANA/7-121 | 2x |
| 352 | 17 | Ghana wild | K31-1/GHANA/12-239 | 2x |
| 353 | 18 | Ghana wild | K31-1/GHANA/14-350 | 2x |
| 354 | 19 | Ghana wild | K31-1/GHANA/18-368 | 2x |
| 355 | 20 | Ghana wild | K31-1/GHANA/19-245 | 2x |
| 356 | 21 | Ghana wild | K31-1/GHANA/21-1180 | 2x |
| 357 | 22 | Ghana wild | K31-1/GHANA/32-1141 | 2x |
| 358 | 23 | Ghana wild | K31-1/GHANA/37-1124 | 2x |
| 359 | 24 | Ghana wild | K31-1/GHANA/45-448 | 2x |
| 360 | 25 | Ghana wild | K31-1/GHANA/47-1175 | 2x |
| 361 | 26 | Ghana wild | K31-1/GHANA/50-1037 | 2x |
| 362 | 27 | Ghana wild | K31-1/GHANA/52-547 | 2x |
| 363 | 28 | Ghana wild | K31-1/GHANA/53-1167 | 2x |
| 364 | 29 | Ghana wild | K31-1/GHANA/54-1196 | 2x |
| 365 | 30 | Ghana wild | K31-1/GHANA/57-1153 | 2x |
Table 7.
Primer pairs used in the Principal Coordinates Analysis to compare the genetic diversity and affinities of Hs compared with a representative sample of commercial and wild diploid palms (listed in Table 6).
| No | Primer | Forward (5'-3') | Reverse (5'-3') |
|---|---|---|---|
| 1 | 1996 | CACTGGGGTCATCTTCATCT | TCGTTCTCTTTCCTTTTGTC |
| 2 | 2215 | GAACTTGGCGTGTAACT | TGGTAGGTCTATTTGAGAGT |
| 3 | 2427 | GAAGGGGCATTGGATTT | CAGGTGACCAAGTGTAAT |
| 4 | 2569 | TAGCCGCACTCCCACGAAGC | CCAGAATCATCAGACTCGGACAG |
| 5 | 2595 | TCAAAGAGCCGCACAACAAG | ACTTTGCTGCTTGGTGACTTA |
| 6 | 2600 | GGGGATGAGTTTGTTTGTTC | GGCAACATGAAGGTAAG |
| 7 | 3282 | GTAACAGCATCCACACTAAC | GCAGGACAGGAGTAATGAGT |
| 8 | 3298 | GACTACCGTATTGCGTTCAG | TTTATCAGGAGTTTTTGTTTGAGAG |
| 9 | 3311 | AATCCAAGTGGCCTACAG | TCCCTACAATAGCCATCTC |
| 10 | 3321 | CAAGGAGGAGCAGGTGAG | TACGGCCTCGGTTCTACAC |
| 11 | 3399 | AGCCAATGAAGGATAAAGG | CCACTTAGAGGTAAAACAACAG |
| 12 | 3400 | CAATTCCAGCGTFAFTATAG | AGTGGCAGTGGAAAAACAGT |
| 13 | 3433 | GGTTCAATGGCATACAT | ACTCCCCTCTTTGACAT |
| 14 | 3538 | TCAAGCCACATCCTAACTAC | CTCATAGCCTTTGTTGTGT |
| 15 | 3544 | AGCAGGGCAAGAGCAATACT | TTCAGCAGCAGGAAACATC |
| 16 | 3546 | GCCTATCCCCTGAACTATCT | TGCACATACCAGCAACAGAG |
| 17 | 3574 | AGAGACCCTATTTGCTTGAT | GACAAAGAGCTTGTCACAC |
| 18 | 3711 | GTCTCATGTGGCTACCTCTC | GCTAGGTGAAAAATAAAGTT |
| 19 | 3819 | CCTCCTTTGGAATTATG | GTGTTTGATGGGACATACA |
| 20 | 219 | TTTGCTCGGCGGATACAT | GGAGGGCAGGAACAAAAAGT |
| 21 | 257 | GCAGCTAGTCACCTGAAC | GACGAGACTGGAAAGATG |
| 22 | 782 | CGTTCATCCCACCACCTTTC | GCTGCGAGGCCACTGATAC |
| 23 | 783 | GAATGTGGCTGTAAATGCTGAGTG | AAGCCGCATGGACAACTCTAGTAA |
| 24 | 882 | TTGATCTTAGACATAACATACTGTA | AAAGCGCGTAATCTCATAGT |
| 25 | 894 | TGCTTCTTGTCCTTGATACA | CCACGTCTACGAAATGATAA |
| 26 | 3213 | GCTCTTTGTATTTCCTGGTTC | AGCAGCAAACCCTACTAACT |
| 27 | 3691 | GCATCATTGGACTATCATACC | TTGTGAACCAGGGAACTATC |
| 28 | vs1 | GAGATTACAAAGTCCAAACC | TCAAAATTAAGAAAGTATGC |
All primers except VS1 were taken from Billotte et al. [27].
Colchicine treatment
Roots of confirmed haploid seedlings were washed and immersed in 2.5, 5.0, 7.5, or 10 mM aqueous colchicine for 5 h. Seedlings were then rinsed with water and planted (2:1:1 v/v compost, sand and soil).
Cross-fertilization using pollen from H plants
A developing male inflorescence of a confirmed H at the PMC stage was treated with 2.5 mM colchicine via injection into the spathe. This treatment was repeated at weekly intervals. The resultant pollen (0.03 g) was applied to a targeted section of the female inflorescence of a diploid dura palm. The inflorescence was then bagged to prevent inadvertent wind pollination.
In addition, some untreated H plants contained up to 30% fully stained pollen using Fluorescein diacetate (FDA) that was presumed to be viable. Pollen from these plants and from palms with apparently inviable pollen (unstained) was applied to targeted sections of a female inflorescence of diploid dura palms in the same way as above.
Competing interests
JMD, MJW, AEC and CSF have received research funding from BioHybrids International Ltd; SN, SW, ACS, DM and YA are employed fully or in part by Sumatra Bioscience; BPF is contracted to BioHybrids International Ltd; PDSC is Managing Director of BioHybrids International Ltd.
Authors' contributions
JMD, PDSC, SN and MJW conceived the project. SN, BPF and ACS supervised the phenotypic screen and flow cytometry. MJW supervised the molecular analysis conducted by SW, AEC, CSF and YA, and the cytology conducted by DM. JMD and MJW wrote the manuscript and all authors discussed the results and commented on the manuscript.
Contributor Information
Jim M Dunwell, Email: j.m.dunwell@reading.ac.uk.
Mike J Wilkinson, Email: jjw@aber.ac.uk.
Stephen Nelson, Email: stephen.nelson@sumatrabioscience.com.
Sri Wening, Email: sri.wening@sumatrabioscience.com.
Andrew C Sitorus, Email: andrew.sitorus@sumatrabioscience.com.
Devi Mienanti, Email: devimienanti@yahoo.com.
Yuzer Alfiko, Email: yuzer.alfiko@sumatrabioscience.com.
Adam E Croxford, Email: aoc@aber.ac.uk.
Caroline S Ford, Email: csf@aber.ac.uk.
Brian P Forster, Email: brianforster@biohybrids.co.uk.
Peter DS Caligari, Email: pcaligari@utalca.cl.
Acknowledgements
This work was funded by Sumatra Bioscience as part of their R&D programme in oil palm. The authors are grateful for the assistance of all staff of the Breeding Department and Seed Production Unit at Bah Lias Research Station, Indonesia.
References
- Duvick DN. Biotechnology in the 1930s: the development of hybrid maize. Nature Rev Genet. 2001;2:69–73. doi: 10.1038/35047587. [DOI] [PubMed] [Google Scholar]
- Horie T, Shiraiwa T, Homma K, Katsura K, Maeda S, Yoshida H. Can yields of lowland rice resume the increases that they showed in the 1980s? Plant Prod Sci. 2005;8:259–274. doi: 10.1626/pps.8.259. [DOI] [Google Scholar]
- Forster BP, Thomas WTB. Doubled haploids in genetics and plant breeding. Plant Breeding Rev. 2005;25:57–88. [Google Scholar]
- Cook RR. A haploid Marglobe tomato. J Hered. 1936;27:433–435. [Google Scholar]
- Maluszynski M, Kasha KJ, Szarejko I. In: Doubled Haploid Production in Crop Plants: A Manual. Maluszynski M, Kasha KJ, Forster BP, Szarejko I, editor. Dordrecht: Kluwer; 2003. Published protocols for other crop plant species; pp. 309–335. [Google Scholar]
- Dunwell JM. Haploids in flowering plants: origins and exploitation. Plant Biotech J. 2010;8:377–424. doi: 10.1111/j.1467-7652.2009.00498.x. [DOI] [PubMed] [Google Scholar]
- Kelly-Yong TL, Lee KT, Mohamed AR, Bhatia S. Potential of hydrogen from oil palm biomass as a source of renewable energy worldwide. Energ Policy. 2007;35:5692–5701. doi: 10.1016/j.enpol.2007.06.017. [DOI] [Google Scholar]
- Basiron Y. Palm oil production through sustainable plantations. Eur J Lipid Sci Technol. 2007;109:289–295. doi: 10.1002/ejlt.200600223. [DOI] [Google Scholar]
- Food and Agriculture Organization of the United Nations. Agricultural Production Statistics. http://faostat.fao.org/
- Stone R. Ecology - Can palm oil plantations come clean? Science. 2007;317:1491. doi: 10.1126/science.317.5844.1491. [DOI] [PubMed] [Google Scholar]
- Wilcove DS, Koh LP. Addressing the threats to biodiversity from oil-palm agriculture. Biodivers Conserv. 2010;19:999–1007. doi: 10.1007/s10531-009-9760-x. [DOI] [Google Scholar]
- Koh LP, Wilcove DS. Cashing in palm oil for conservation. Nature. 2007;448:993–994. doi: 10.1038/448993a. [DOI] [PubMed] [Google Scholar]
- Clements R, Posa MRC. Conservationists could slip up in oil-palm enterprise. Nature. 2007;449:403. doi: 10.1038/449403d. [DOI] [PubMed] [Google Scholar]
- Venter O, Meijaard E, Wilson K. Strategies and alliances needed to protect forest from palm-oil industry. Nature. 2008;451:16. doi: 10.1038/451016a. [DOI] [PubMed] [Google Scholar]
- Corley RHV. Potential productivity of tropical perennial crops. Exp Agric. 1983;19:217–237. doi: 10.1017/S0014479700022742. [DOI] [Google Scholar]
- Murphy DJ. Oil palm: future prospects for yield and quality improvements. Lipid Technology. 2009;21:257–260. doi: 10.1002/lite.200900067. [DOI] [Google Scholar]
- Jones LH. Prospects for biotechnology in oil palm (Elaeis guineensis) and coconut (Cocos nucifera) improvement. Biotech Genet Engin Rev. 1989;7:281–296. [Google Scholar]
- Whitehead RA, Chapman GP. Twinning and haploidy in Cocos nucifera. Nature. 1962;195:1228–1229. doi: 10.1038/1951228a0. [DOI] [Google Scholar]
- Bouvier L, Zhang YX, Lespinasse Y. Two methods of haploidization in pear, Pyrus communis L.: greenhouse seedling selection and in situ parthenogenesis induced by irradiated pollen. Theor Appl Genet. 1993;87:229–232. doi: 10.1007/BF00223769. [DOI] [PubMed] [Google Scholar]
- Germana MA. In: Advances in Haploid Production in Higher Plants. Touraev A, Forster BP, Jain SM, editor. The Netherlands: Springer-Verlag; 2009. Haploids and doubled haploids in fruit trees; pp. 241–263. full_text. [Google Scholar]
- Lim W, Earle ED. Enhanced recovery of doubled haploid lines from parthenogenetic plants of melon. Plant Cell Tiss Org. 2009;98:351–356. doi: 10.1007/s11240-009-9563-5. [DOI] [Google Scholar]
- Finkel E. With 'phenomics,' plant scientists hope to shift plant breeding into overdrive. Science. 2009;325:380–381. doi: 10.1126/science.325_380. [DOI] [PubMed] [Google Scholar]
- Reijnders L, Huijbregts MAJ. Palm oil and the emission of carbon-based greenhouse gases. J Clean Product. 2008;16:477–482. doi: 10.1016/j.jclepro.2006.07.054. [DOI] [Google Scholar]
- Parveez GKA. Biolistic mediated production of transgenic oil palm. Method Mol Biol. 2009;477:301–320. doi: 10.1007/978-1-60327-517-0_23. full_text. [DOI] [PubMed] [Google Scholar]
- Persson UM, Azar C. Preserving the world's tropical forests - a price on carbon may not do. Environ Sci Technol. 2010;44:210–215. doi: 10.1021/es902629x. [DOI] [PubMed] [Google Scholar]
- Croxford AE, Rogers T, Caligari PDS, Wilkinson MJ. High-resolution melt analysis to identify and map sequence-tagged site anchor points onto linkage maps: a white lupin (Lupinus albus) map as an exemplar. New Phytol. 2008;180:594–607. doi: 10.1111/j.1469-8137.2008.02588.x. [DOI] [PubMed] [Google Scholar]
- Billotte N, Marseillac N, Risterucci AM, Adon B, Brottier P, Baurens FC, Singh R, Herran A, Asmady H, Billot C, Amblard P, Durand-Gasselin T, Courtois B, Asmono D, Cheah SC, Rohde W, Ritter E, Charrier A. Microsatellite-based high density linkage map in oil palm (Elaeis guineensis Jacq.) Theor Appl Genet. 2005;110:754–765. doi: 10.1007/s00122-004-1901-8. [DOI] [PubMed] [Google Scholar]
- Billotte N, Risterucci AM, Barcelos E, Noyer JL, Amblard P, Baurens FC. Development, characterisation, and across-taxa utility of oil palm (Elaeis guineensis Jacq.) microsatellite markers. Genome. 2001;44:413–425. doi: 10.1139/gen-44-3-413. [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED. Estimation of nuclear DNA content of plants by flow cytometry. Plant Mol Biol Rep. 1991;9:221–231. [Google Scholar]
- Peakall R, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1111/j.1471-8286.2005.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orloci L. Multivariate analysis in vegetation research. 2. Boston: The Hague; 1978. [Google Scholar]