Abstract
Background
Glycine soja is a wild relative of soybean that has purple flowers. No flower color variant of Glycine soja has been found in the natural habitat.
Results
B09121, an accession with light purple flowers, was discovered in southern Japan. Genetic analysis revealed that the gene responsible for the light purple flowers was allelic to the W1 locus encoding flavonoid 3'5'-hydroxylase (F3'5'H). The new allele was designated as w1-lp. The dominance relationship of the locus was W1 >w1-lp >w1. One F2 plant and four F3 plants with purple flowers were generated in the cross between B09121 and a Clark near-isogenic line with w1 allele. Flower petals of B09121 contained lower amounts of four major anthocyanins (malvidin 3,5-di-O-glucoside, petunidin 3,5-di-O-glucoside, delphinidin 3,5-di-O-glucoside and delphinidin 3-O-glucoside) common in purple flowers and contained small amounts of the 5'-unsubstituted versions of the above anthocyanins, peonidin 3,5-di-O-glucoside, cyanidin 3,5-di-O-glucoside and cyanidin 3-O-glucoside, suggesting that F3'5'H activity was reduced and flavonoid 3'-hydroxylase activity was increased. F3'5'H cDNAs were cloned from Clark and B09121 by RT-PCR. The cDNA of B09121 had a unique base substitution resulting in the substitution of valine with methionine at amino acid position 210. The base substitution was ascertained by dCAPS analysis. The polymorphism associated with the dCAPS markers co-segregated with flower color in the F2 population. F3 progeny test, and dCAPS and indel analyses suggested that the plants with purple flowers might be due to intragenic recombination and that the 65 bp insertion responsible for gene dysfunction might have been eliminated in such plants.
Conclusions
B09121 may be the first example of a flower color variant found in nature. The light purple flower was controlled by a new allele of the W1 locus encoding F3'5'H. The flower petals contained unique anthocyanins not found in soybean and G. soja. B09121 may be a useful tool for studies of the structural and functional properties of F3'5'H genes as well as investigations on the role of flower color in relation to adaptation of G. soja to natural habitats.
Background
Soybean (Glycine max (L.) Merr.) is believed to have been domesticated in north-eastern China from its wild relative, Glycine soja Sieb. & Zucc. [1]. Glycine soja is native throughout China, the adjacent area of Russia, Korea, Japan and Taiwan [1]. Flower color of G. soja is almost exclusively purple; by contrast, 33% (5,544 out of 16,855) of the soybean accessions in the USDA Soybean Germplasm Collections have white flowers (Dr. R.L. Nelson, personal communication 2006). The reason why G. soja almost lacks flower color variants is uncertain [2]. A few white-flowered G. soja accessions were reported in a Chinese germplasm collection, but these had high 100-seed weight, strongly suggesting recent outcrossing with G. max [2]. One white-flowered plant (PI 424008C) was found in 1998 among the progeny of a purple-flowered G. soja accession (PI 424008A) that was originally introduced from South Korea in 1976 [2]. Genetic analysis indicated that the white flower was caused by a recessive allele at the W1 locus similar to the white-flowered soybeans [2].
In soybean, six genes (W1, W2, W3, W4, Wm and Wp) primarily control flower color and two genes (T and Td) control pubescence color [3,4]. The hydroxylation pattern of B-ring in flavonoids plays an important role in the coloration of seed coats, flower and pubescence of soybeans. The B-ring of flavonoids can be hydroxylated at either the 3' position leading to the production of cyanidin-based pigments, or at both the 3' and 5' positions to produce delphinidin-based pigments. Two key enzymes involved in this pathway are flavonoid 3'-hydroxylase (F3'H) and flavonoid 3'5'-hydroxylase (F3'5'H) which are both microsomal cytochrome P450 dependent monooxygenases that require NADPH as a co-factor [5]. Chromatographic experiments suggested that T and W1 loci are responsible for the formation of flavonoids with 3', 4' and 3', 4', 5' B-ring hydroxylation patterns, respectively [6-8]. Hence, T and W1 are presumed to encode F3'H and F3'5'H, respectively. The F3'H cDNA was cloned and characterized from a pair of near-isogenic lines (NILs) for the T locus, To7B (TT, tawny pubescence) and To7G (tt, gray pubescence) [9]. Sequence analysis revealed that they differed by a single-base deletion of C in the coding region of To7G. The deletion generated a truncated polypeptide lacking the GGEK consensus sequence of F3'H gene and the heme-binding domain, resulting in non-functional protein.
The W1 gene has a pleiotropic effect on flower and hypocotyl color: soybean cultivars with purple/white flowers have purple/green hypocotyls. The soybean F3'5'H gene was cloned from NILs for W1 and confirmed that W1 encodes F3'5'H and that the gene of white-flowered NILs contained a 65 bp insertion in the coding region [10]. In addition to the F3'5'H protein, a cytochrome b5 is required for full activity of F3'5'H in petunia, and mutation in cytochrome b5 results in a reduction in F3'5'H activity and alteration of anthocyanin amount and composition [11].
The flavonoids in flower petals of soybean have been analyzed [12,13]. The primary components of anthocyanin in purple-flowered cultivars were malvidin 3,5-di-O-glucoside, petunidin 3,5-di-O-glucoside, delphinidin 3,5-di-O-glucoside and delphinidin 3-O-glucoside. In addition, eight flavonol glycosides, kaempferol 3-O-gentiobioside, kaempferol 3-O-rutinoside, kaempferol 3-O-glucoside, kaempferol 3-O-glycoside, kaempferol 3-O-rhamnosylgentiobioside, kaempferol 7-O-glucoside, kaempferol 7-O-diglucoside and quercetin 3-O-gentiobioside, and one dihydroflavonol, aromadendrin 3-O-glucoside were identified [12,13]. The sugar component of kaempferol 3-O-glycoside could not be determined. No anthocyanins were detected in Clark NILs for w1 and w4, Clark-w1 and Clark-w4, and only a trace amount was detected in Clark-W3w4. The amount of flavonols and dihydroflavonol in NILs for w1 or w4 was largely similar to the NILs with purple flowers suggesting that W1 and W4 affect only anthocyanin biosynthesis. However, the flavonoid composition in flower petals of G. soja has not been analyzed so far.
Yasuda discovered B09121, a flower color variant of G. soja, at a slope between a paddy field and a ditch in Karatsu, Saga Prefecture (southern Japan) in 2002 (unpublished result) (Figure 1). Its banner petals have a pale pinkish hue and a pronounced light purple pigmentation that originates from the base of the petals and spreads in streaks towards the petal margins. We designated the flower color as light purple. Considering its growth habit, small seed size and unique flower color, it is unlikely that the flower color of B09121 was derived from outcrossing with soybean. To our knowledge, B09121 is the first example of a flower color variant of G. soja found in nature. The first objective of this study was to determine the genetic basis of purple flower color by crossing experiments. The second objective was to analyze the flavonoids in flower petals of G. soja accessions. The third objective was to clone and characterize a gene responsible for light purple flowers.
Figure 1.
Glycine soja line B09121 with light purple flowers discovered in southern Japan.
Methods
Genetic analysis
A Canadian soybean cultivar Harosoy (W1W1 W2W2 w3w3 W4W4 WmWm WpWp tt) with purple flowers and gray pubescence and a NIL of a US soybean cultivar Clark for W1 gene, Clark-w1 (L63-2373, w1w1 W2W2 w3w3 W4W4 WmWm WpWp TT) with white flowers and tawny pubescence were crossed with B09121 having light purple flowers and tawny pubescence in 2005. Flowers of Harosoy and Clark-w1 were emasculated one day before opening and pollinated with B09121. Hybridity of the F1 plants was ascertained by tawny pubescence color in crossing with Harosoy and by spindly growth habit in crossing with Clark-w1. Seeds of L63-2373 were provided by the USDA Soybean Germplasm Collection. The NIL was produced by crossing Clark with T139 and backcrossing the progeny to Clark up to BC6 [14].
A total of 120 to 130 F2 seeds derived from the crossing with Harosoy and Clark-w1 were planted in field (low-humic andosols) on June 13 in 2007 at the National Institute of Crop Science, Tsukuba, Japan (36°06'N, 140°05'E). A bulk of 30 seeds each of 36 F3 families derived from the cross with Clark-w1 were planted at the same location on June 8 in 2008. N, P and K were applied at 3.0, 4.4 and 8.3 g m-2, respectively. Flower color was scored in individual F2 and F3 plants.
Analysis of flavonoids
Clark, B09121 and four additional G. soja accessions were used for flavonoid analysis of flower petals. Seeds of Japanese accessions with purple flowers Kokaigawa-1 collected in Ibaraki Prefecture and COL/AOMORI/1983/NASU-2 collected in Aomori Prefecture were obtained from Ms. N. Mihashi and the NIAS Genebank, respectively. Seeds of a South Korean G. soja line with purple flowers (PI 424008A) and its derivative mutant line with white flowers (PI 424008C) were obtained from the USDA Soybean Germplasm Collection. Seeds of the G. soja lines were planted in vermiculite in a glasshouse on June 16 in 2006, and transplanted on June 26 to pots (24 cm diameter) filled with 10 kg soil (low-humic andosols). Fertilizers were applied at rates similar to the field plantings.
Banner petals were collected with forceps at the day of opening. Two 200 mg samples of banner petals were collected in 2 ml of MeOH containing 0.1% (v/v) HCl for anthocyanin analysis. Two 200 mg samples in 2 ml of absolute MeOH were also collected for the determination of flavonol and dihydroflavonol. High performance liquid chromatography (HPLC) of anthocyanins, flavonols and dihydroflavonol was performed following previously described protocols [12]. The 2 ml extracts were filtered through disposable filtration units (Maishoridisc H-13-5, Tosoh, Japan) and 10 μl from each sample was subjected to HPLC analysis. The amount of flavonoids was estimated from the pertinent peak area in the HPLC chromatogram (detection wavelength of anthocyanins = 530 nm; flavonols= 351 nm; dihydroflavonols = 290 nm). The peak area was subjected to analysis of variance using Statistica software (StatSoft, Inc. Tulsa, OK).
Three additional anthocyanins were characterized by direct HPLC comparison with authentic specimens, i.e., chrysanthemin (cyanidin 3-O-glucoside) from the autumn leaves of Acer species [15], cyanin (cyanidin 3,5-di-O-glucoside) from the flowers of Dahlia variabilis (Willd.) Desf. [16] and peonin (peonidin 3,5-di-O-glucoside) from the flowers of Paeonia albiflora Pall. var. hortensis [17].
RNA extraction and cDNA cloning
Total RNA was extracted from banner flower petals (100 mg) of Clark, Clark-w1 and B09121 using the TRIZOL Reagent (Invitrogen) according to the manufacturer's instructions. cDNA was synthesized by reverse transcription of 5 μg of total RNA using the Superscript III First-Strand Synthesis System (Invitrogen) and an oligo(dT) primer according to the manufacturer's instructions. The full-length cDNA was cloned from Clark and B09121, using a pair of PCR primers (5'-AACTAGCAAATTAATTAGCTT and 5'-CAACCCAAACATTACTTAT) and end-to-end PCR. The PCR mixture contained 0.5 μg of cDNA, 10 pmol of each primer, 10 pmol of nucleotides and 1 unit of ExTaq in 1 × ExTaq Buffer supplied by the manufacturer (Takara) in a total volume of 50 μl. A 5 min denaturation at 94°C was followed by 30 cycles of 30 sec denaturation at 94°C, 1 min annealing at 58°C and 1 min extension at 72°C. A final 7 min extension at 72°C completed the program. The PCR was performed in an Applied Biosystems 9700 thermal cycler. The ~1.8 kbp PCR product was cloned into pCR 2.1 vector (Invitrogen) and sequenced.
DNA sequencing
Nucleotide sequences of both strands were determined with the BigDye terminator cycle method using an ABI3100 Genetic Analyzer (Applied Biosystems). Nucleotide sequences and the putative amino acid translations were analyzed with the BLAST program [18]. Primers for sequencing the internal regions of F3'5'H genes were 5'-TGTGGTGGTGGAAATGT, 5'-CTATAGAAAGCACCCTTCAA, 5'-GCCTCATCCCTCTTGTTACA and 5'-TTTGAGACGGCGATCCCTTC.
dCAPS and indel analyses
Genomic DNA of Clark, Clark-w1, B09121 and 36 F2 plants that were used for F3 progeny tests was isolated from trifoliolate leaves by CTAB [19]. A pair of PCR primers (5'-GTCTAACGAGTTCAAGGCCAT, 5'-CAACTTGGCCAAAAAGGGTAT) was designed to detect a single-base substitution at nucleotide number 653 that is unique to B09121. The first primer contains a nucleotide C that is mismatched with its target DNA to artificially create a restriction site of NcoI (CCATGG) in Clark (Figure 2). The base substitution within the restriction site would result in presence/absence of the restriction site in the amplified product to generate a polymorphism. The PCR mixture contained 30 ng of genomic DNA, 5 pmol of each primer, 10 pmol of nucleotides and 1 unit of ExTaq in 1 × ExTaq Buffer supplied by the manufacturer (Takara) in a total volume of 25 μl. After an initial 30 sec denaturation at 94°C, there were 30 cycles of 30 sec denaturation at 94°C, 1 min annealing at 56°C and 1 min extension at 72°C. A final 7 min extension at 72°C completed the program. The amplified products were digested with NcoI, and the digests were separated on an 8% nondenaturing polyacrylamide gel in 1 × TBE buffer (90 mM Tris-borate, 2 mM EDTA, pH 8.0). After electrophoresis, the gel was stained with ethidium bromide and the DNA fragments were visualized under UV light.
Figure 2.
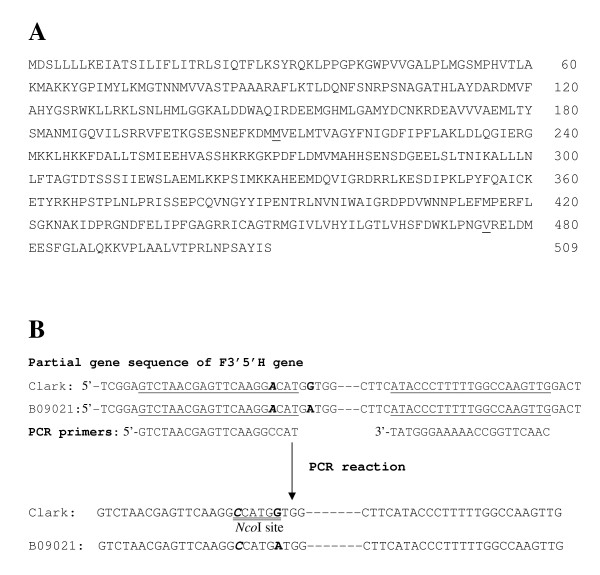
Amino acid sequence of F3'5'H gene from Glycine soja line B09121 with light purple flowers and schematic diagram of dCAPS analysis to detect a base substitution found in B09121. (A) Amino acid sequence of F3'5'H gene from Glycine soja line B09121 with light purple flowers. Amino acid position 210 (underlined) was methionine (valine in Clark) and amino acid position 475 (underlined) was valine (glutamic acid in Clark). (B) Schematic diagram of dCAPS analysis to detect a single-base substitution found in B09121 (shown in bold). NcoI site (CCATGG, double underlined) was artificially introduced in the PCR products of Clark using a forward primer with a mismatched base C that is exhibited in bold italic. Annealing sites of PCR primers are underlined. PCR product is digested by NcoI only in Clark generating size differences of 18 bp.
A pair of indel PCR primers (5'-TTTTGAGCTTATTCCATTTGG, 5'-TGAATATTCGAACCCAACCA) was designed to identify the 65 bp insertion in F3'5'H gene of soybean lines with w1 allele based on the previous report [10]. The PCR profile and electrophoresis conditions were identical with the dCAPS analysis except that annealing was conducted at 59°C.
Semi-quantitative RT-PCR analysis
Semi-quantitative RT-PCR was conducted by reverse-transcription of 5 μg of total RNA using the Superscript III First-Strand Synthesis System and an oligo d(T) primer according to the manufacturer's instruction. To test the transcription level of the F3'5'H gene, PCR reactions were carried out in a volume of 25 μl, using 125 ng of cDNA. The initial 30 sec denaturation at 94°C was followed by 26 cycles of 30 sec denaturation at 94°C, 1 min annealing at 59°C and 1 min extension at 72°C. A final 7 min extension at 72°C completed the program. The primers were 5'-GACGCTGAGGATATTCAACC and 5'-AGAAATCTGTGAGGTCACGA. A soybean actin gene was used as a control. The initial 30 sec denaturation at 94°C was followed by 20 cycles of 30 sec denaturation at 94°C, 1 min annealing at 56°C and 1 min extension at 72°C. A final 7 min extension at 72°C completed the program. The primers were 5'-CTGGGGATGGTGTCAGCCACAC and 5'-CACCGAACTTTCTCTCGGAAGGTG. PCR products were loaded on a 1.2% agarose gel, stained by ethidium bromide and visualized under UV light.
Accession Numbers
Sequence data from this article have been deposited with the DDBJ Data Libraries under accession nos. AB540111 (Clark) and AB540112 (B09121).
Results
Genetic analysis
F1 plants produced by a cross between the purple-flowered cultivar Harosoy and B09121 had purple flowers, indicating that purple flower was dominant to light purple flower (Table 1). The F2 plants segregated into 3 purple: 1 light purple flower suggesting involvement of a single gene.
Table 1.
Segregation of flower color in F1 plants and F2 population derived from a cross between a soybean cultivar Harosoy with purple flowers and B09121, a Glycine soja accession with light purple flowers, and segregation of flower color in F1 plants, and F2 and F3 populations derived from a cross between a soybean near-isogenic line Clark-w1 with white flowers and B09121 in Tsukuba, Japan.
| Generation | Year | Number of plants | Expected ratio | χ2 value | Probability (P value) |
|||
|---|---|---|---|---|---|---|---|---|
| Total | Purple | Light purple | White | |||||
| B09121 (B) | 2007 | 10 | - | 10 | - | - | - | - |
| Harosoy (H) | 2007 | 10 | 10 | - | - | - | - | - |
| H × B F1 | 2007 | 3 | 3 | - | - | - | - | - |
| H × B F2 | 2007 | 119 | 91 | 28 | - | 3:1 | 0.14 | 0.71 |
| Clark-w1 (w1) | 2007 | 10 | - | - | 10 | - | - | - |
| w1 × B F1 | 2007 | 3 | - | 3 | - | - | - | - |
| w1 × B F2 | 2007 | 124 | 1 | 98 | 25 | 3:1a | 1.43 | 0.23 |
| w1 × B F3 (purple)b | 2008 | 69 | 51 | 18 | - | 3:1 | 0.04 | 0.83 |
aPlants with light purple flowers: plants with white flowers.
bA line derived from an F2 plant with purple flowers.
F1 plants produced from a cross between Clark-w1 and B09121 had light purple flowers (Table 1). Segregation of the F2 plants fitted a 3 light purple: 1 white ratio except one plant that had purple flowers. The results suggested that the W1 locus was responsible for light purple flowers and that light purple flower was dominant to white flower. Six F3 families derived from F2 plants with white flowers produced only plants with white flowers (Table 2). Thirty F3 families derived from F2 plants with light purple flowers produced twenty-one families segregating for flower color and nine families fixed for light purple flowers; the segregation pattern fit a ratio of 2 segregating lines: 1 line fixed for light purple flowers (Table 2). Among the twenty-one segregating families, seventeen families segregated into light purple and white flowers, whereas four families each contained one plant with purple flowers in addition to light purple and white flowers. The frequency of plants with light purple and white flowers fitted 3:1 ratio in the twenty-one segregating families. The frequency of purple-flowered plants was similar across generations; 0.81% (1/124 plants) in F2 and 0.95% (4/421 plants) in F3. Progeny of an F2 plant with purple flowers segregated at a ratio of 3 purple: 1 light purple flower (Table 1). Results of the F3 progeny tests supported the hypothesis that light purple flower was controlled by a new allele at the W1 locus that was dominant to the w1 allele. We designated the allele as w1-lp (light purple). The gene symbol was approved by the Soybean Genetics Committee. Dominance relationship of the locus was W1 >w1-lp >w1.
Table 2.
Segregation of flower color in F3 lines derived from a cross between a soybean near-isogenic line Clark-w1 with white flowers and B09121, a Glycine soja accession with light purple flowers in Tsukuba, Japan in 2008.
| Line | Number of lines | Expected ratio | χ2 value | Probability (P value) |
|||
|---|---|---|---|---|---|---|---|
| Total | Fixed for light purple | Segregating | Fixed for white | ||||
| F3 (light purple)a | 30 | 9 | 21b | - | 1:2 | 0.15 | 0.70 |
| F3 (white)c | 6 | - | - | 6 | - | - | - |
aF3 lines derived from F2 plants with light purple flowers.
bFour lines each contained one plant with purple flowers.
cF3 lines derived from F2 plants with white flowers.
Flavonoid analysis
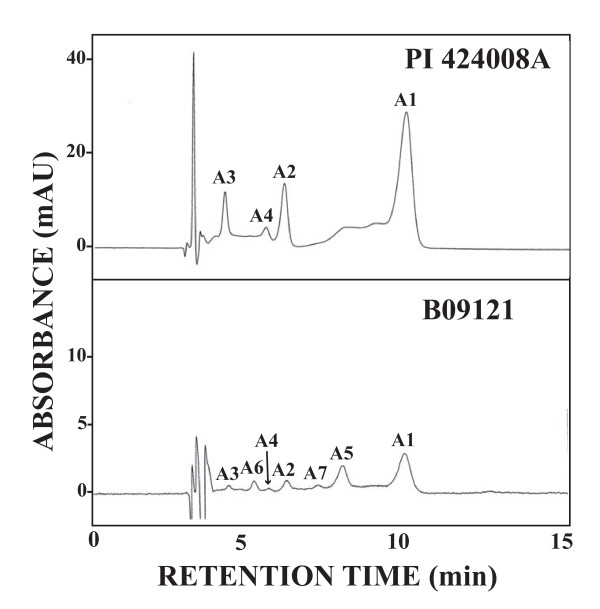
The amount of anthocyanins estimated from peak area by HPLC analysis is presented in Table 3. HPLC chromatograms for anthocyanins in PI 424008A and B09121 are presented in Figure 3. In the purple-flowered G. soja lines, four HPLC peaks, viz., A1 (malvidin 3,5-di-O-glucoside), A2 (petunidin 3,5-di-O-glucoside), A3 (delphinidin 3,5-di-O-glucoside) and A4 (delphinidin 3-O-glucoside), were found similar to Clark. The amount of malvidin 3,5-di-O-glucoside in purple-flowered G. soja lines was 78 to 252% higher than Clark. The amounts of the other three anthocyanins were not substantially different from Clark.
Table 3.
Mean and standard errors (× 103) for anthocyanin contents by HPLC analysis of flower petals from soybean cv, Clark and five G. soja accessions in 2006 at Tsukuba, Japan.
| Line name | A1a | A2 | A3 | A4 | A5 | A6 | A7 | Total |
|---|---|---|---|---|---|---|---|---|
| Clark | 766 ± 186 | 282 ± 20 | 399 ± 45 | 191 ± 10 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 1,638 ± 260 |
| COL/AOMORI/1983/NASU-2 | 2,700 ± 769 | 396 ± 106 | 315 ± 61 | 201 ± 23 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 3,613 ± 959 |
| Kokaigawa-1 | 1,927 ± 438 | 340 ± 20 | 374 ± 17 | 195 ± 11 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 2,836 ± 486 |
| PI 424008A | 1,365 ± 228 | 350 ± 24 | 366 ± 9 | 196 ± 21 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 2,276 ± 174 |
| PI 424008C | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 |
| B09121 | 317 ± 4 | 172 ± 4 | 147 ± 2 | 116 ± 2 | 287 ± 19 | 160 ± 1 | 142 ± 18 | 1,342 ± 33 |
| LSD0.05 | 1,384 | 166 | 116 | 52 | 29 | 2 | 27 | 1,662 |
aA1: malvidin 3,5-di-O-glucoside, A2: petunidin 3,5-di-O-glucoside, A3: delphinidin 3,5-di-O-glucoside, A4: delphinidin 3-O-glucoside, A5: peonidin 3,5-di-O-glucoside, A6: cyanidin 3,5-di-O-glucoside, A7: cyanidin 3-O-glucoside.
Figure 3.
HPLC chromatogram of anthocyanins extracted from flower petals of PI 424008A (upper panel) and B09121 (lower panel). 200 mg of banner petals was extracted with 2 ml of MeOH containing 0.1% HCl. Eluents: MeCN/HOAc/H2O/H3BO3 (6:8:83:3). Flow-rate: 1.0 ml/min. Injection: 10 μl. Detection: 530 nm, A1 = malvidin 3,5-di-O-glucoside, A2 = petunidin 3,5-di-O-glucoside, A3 = delphinidin 3,5-di-O-glucoside, A4 = delphinidin 3-O-glucoside, A5 = peonidin 3,5-di-O-glucoside, A6 = cyanidin 3,5-di-O-glucoside, A7 = cyanidin 3-O-glucoside.
In contrast, flower petals of white-flowered line PI 424008C contained no anthocyanins. Further, the flower petals of B09121 contained about half the amount of A1 to A4 compared with Clark. The HPLC chromatogram of B09121 exhibited three additional anthocyanin peaks, A5 to A7, that were not found in soybeans or other G. soja lines (Figure 3). Based on the comparison of retention time with authentic specimens, peonin, cyanin and chrysanthemin, A5, A6 and A7 were estimated as peonidin 3,5-di-O-glucoside, cyanidin 3,5-di-O-glucoside and cyanidin 3-O-glucoside, respectively.
Eight peaks, F1 to F8, corresponding to flavonol glycosides, were detected by HPLC analysis: F1 (kaempferol 3-O-gentiobioside), F2 (kaempferol 3-O-rutinoside), F3 (kaempferol 3-O-glucoside), F4 (kaempferol 3-O-glycoside), F5 (kaempferol 3-O-rhamnosyl-(1→2)-[glucosyl-(1→6)-galactoside]), F6 (quercetin 3-O-gentiobioside), F7 (kaempferol 7-O-glucoside) and F8 (kaempferol 7-O-diglucoside). The sugar component of F4 could not be determined. Though F5 has been partially characterized as kaempferol 3-O-rhamnosylgentiobioside [12], our data indicates that it is actually kaempferol 3-O-rhamnopyranosyl-(1→2)-[glucopyranosyl-(1→6)-galactopyranoside] by 1H and 13C NMR survey. The pertinant 1H and 13C NMR data of F5 are as follows:
1H NMR (600 MHz, pyridine-d5): δ 13.43 (1 H, s, 5-OH), 8.61 (2 H, d, J = 8.8 Hz, H-2',6'), 7.30 (2 H, d, J = 8.9 Hz, H-3',5'), 6.80 (1 H, d, J = 2.1 Hz, H-8), 6.72 (1 H, d, J = 2.0 Hz, H-6), 6.39 (1 H, d, J = 7.7 Hz, galactosyl H-1), 6.34 (1 H, s, rhamnosyl H-1), 4.73 (1 H, d, J = 7.8 Hz, glucosyl H-1), 1.53 (3 H, d, J = 6.2 Hz, rhamnosyl CH3). 13C NMR (125 MHz, pyridine-d5): (kaempferol) δ 157.4 (C-2), 134.2 (C-3), 178.7 (C-4), 163.0 (C-5), 99.7 (C-6), 165.7 (C-7), 94.5 (C-8), 157.6 (C-9), 105.5 (C-10), 122.3 (C-1'), 131.9 (C-2',6'), 116.2 (C-3',5'), 161.6 (C-4'); (galactose) δ 100.7 (C-1), 76.5 (C-2), 75.7 (C-3), 69.5 (C-4), 74.9 (C-5), 68.3 (C-6); (glucose) δ 104.9 (C-1), 75.0 (C-2), 78.1 (C-3), 71.4 (C-4), 78.3 (C-5), 62.3 (C-6); (rhamnose) δ 102.5 (C-1), 72.9 (C-2), 72.7 (C-3), 74.2 (C-4), 69.9 (C-5), 18.3 (C-6).
The amounts of flavonol glycosides estimated by peak areas in HPLC analysis are presented in Table 4. PI 424008A and PI 424008C contained all eight of the flavonol glycosides found in Clark. COL/AOMORI/1983/NASU-2 lacked F4 and Kokaigawa-1 lacked F8. B09121 was devoid of F4 and F7. However, the total amount of flavonol glycosides was not significantly different among the Clark and G. soja lines included in this report.
Table 4.
Mean and standard errors (× 103) of flavonol contents by HPLC analysis of flower petals from soybean cv, Clark and five G. soja accessions in 2006 at Tsukuba, Japan.
| Line name | F1a | F2 | F3 | F4 | F5 | F6 | F7 | F8 | Total |
|---|---|---|---|---|---|---|---|---|---|
| Clark | 7,581 ± 391 | 792 ± 160 | 221 ± 78 | 300 ± 24 | 323 ± 45 | 162 ± 36 | 88 ± 60 | 144 ± 35 | 9,612 ± 829 |
| COL/AOMORI/1983/NASU-2 | 11,097 ± 357 | 816 ± 40 | 256 ± 7 | 0 ± 0 | 724 ± 40 | 177 ± 17 | 36 ± 2 | 240 ± 1 | 13,346 ± 463 |
| Kokaigawa-1 | 8,812 ± 611 | 56 ± 3 | 159 ± 13 | 327 ± 27 | 215 ± 17 | 207 ± 35 | 29 ± 6 | 0 ± 0 | 9,805 ± 712 |
| PI 424008A | 14,138 ± 4,800 | 1,091 ± 328 | 339 ± 102 | 624 ± 226 | 643 ± 196 | 290 ± 124 | 156 ± 53 | 199 ± 65 | 17,481 ± 5,896 |
| PI 424008C | 14,025 ± 637 | 774 ± 3 | 321 ± 20 | 533 ± 5 | 585 ± 36 | 116 ± 3 | 29 ± 1 | 131 ± 28 | 16,513 ± 686 |
| B09121 | 13,465 ± 1,026 | 776 ± 92 | 279 ± 27 | 0 ± 0 | 745 ± 61 | 376 ± 7 | 0 ± 0 | 228 ± 27 | 15,868 ± 1,240 |
| LSD0.05 | nsb | 562 | ns | 340 | 323 | ns | ns | 124 | ns |
aF1: kaempferol 3-O-gentiobioside, F2: kaempferol 3-O-rutinoside, F3: kaempferol 3-O-glucoside, F4: kaempferol 3-O-glycoside, F5: kaempferol 3-O-rhamnosyl-(1→2)-[glucosyl-(1→6)-galactoside], F6: quercetin 3-O-gentiobioside, F7: kaempferol 7-O-glucoside, F8: kaempferol 7-O-diglucoside.
bnot significant at 5% level by analysis of variance
One peak (F9) corresponding to dihydroflavonol (aromadendrin 3-O-glucoside) was found by HPLC analysis in Clark and all of the G. soja lines. The amount of aromadendrin 3-O-glucoside estimated by peak area in HPLC analysis is presented in Table 5. The G. soja lines had 33 to 155% more aromadendrin 3-O-glucoside than Clark.
Table 5.
Mean and standard errors (× 103) of dihydroflavonol contents by HPLC analysis of flower petals from soybean cv, Clark and five G. soja accessions in 2006 at Tsukuba, Japan.
| Line name | F9a |
|---|---|
| Clark | 789 ± 54 |
| COL/AOMORI/1983/NASU-2 | 1,286 ± 38 |
| Kokaigawa-1 | 1,046 ± 53 |
| PI 424008A | 1,574 ± 382 |
| PI 424008C | 1,547 ± 91 |
| B09121 | 2,010 ± 94 |
| LSD0.05 | 612 |
aF9: aromadendrin 3-O-glucoside
cDNA cloning
Four F3'5'H cDNA clones each from Clark and B09121 were sequenced from the 5' and 3' ends. The cDNA sequence of both lines consisted of 1657 nucleotides encoding 509 amino acids (Figure 2). Compared with Clark, five nucleotides were substituted in B09121 at nucleotide positions 653, 1084, 1366, 1449 and 1534 in the coding region. Identical base substitutions at nucleotide positions 1084, 1449 and 1534 were also observed in cultivar Chin-Ren-Woo-Dou with purple flowers (GenBank accession number AY117551). Substitution at nucleotide positions 1084, 1366 and 1534 had no effects on amino acid sequences. In contrast, the base substitution at nucleotide positions 653 and 1449 changed amino acids from valine to methionine (amino acid position 210) and from glutamic acid to valine (amino acid position 475), respectively (Figure 2). The latter amino acid substitution was also found in Chin-Ren-Woo-Dou, whereas the former was unique to B09121.
dCAPS and indel analysis
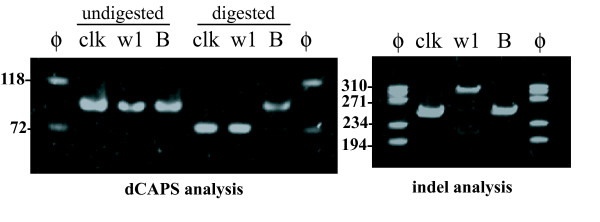
The PCR reaction for dCAPS analysis produced bands with expected size of about 100 bp in Clark, Clark-w1 and B09121 (Figure 4). NcoI digested the bands of Clark and Clark-w1 and shortened the bands by 18 bp. In contrast, the band from B09121 was largely undigested. In addition, a faint band with approximately the same size was also observed among the digested bands in B09121. Similar results were obtained in dCAPS analysis using a different set of PCR primers and a different restriction enzyme (HphI) (data not shown). The PCR reaction for indel analysis produced shorter bands (255 bp) in Clark and B09121, and a longer band in Clark-w1 (310 bp) due to the 65 bp insertion (Figure 4). In addition, a faint band was also observed in Clark-w1 that was approximately the same size as the shorter bands in Clark and B09121.
Figure 4.
Results of dCAPS and indel analysis of F3'5'H gene in a soybean cultivar Clark, a near-isogenic line of Clark with white flowers, Clark-w1 and a Glycine soja line with light purple flowers, B09121. (Left panel) Results of dCAPS analysis. PCR products amplified with dCAPS primers were digested by NcoI and the digests were separated on an 8% polyacrylamide gel. ϕ, molecular marker ϕx174/HaeIII; clk, Clark; w1, Clark-w1; B, B09121. The migration of size markers is shown to the left of the gel. (Right panel) Results of indel analysis. PCR products amplified with indel primers were separated on an 8% polyacrylamide gel. ϕ, molecular marker ϕx174/HaeIII; clk, Clark; w1, Clark-w1; B, B09121.
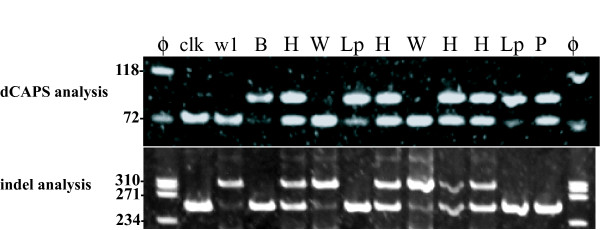
In dCAPS analysis of the F2 population, plants with w1w1 genotype (white flower) had only shorter bands, whereas plants with w1-lpw1-lp genotype (light purple flower) had longer bands with faint bands similar to B09121 (Figure 5). Heterozygous plants with the w1-lpw1 genotype (light purple flower) had both bands at similar band intensity. In indel analysis, plants with w1-lpw1-lp genotype had only shorter bands, whereas plants with w1w1 genotype had longer bands and faint shorter bands. Heterozygous plants with w1-lpw1 genotype had both bands at similar band intensity. Thus, dCAPS and indel markers co-segregated in plants with white and light purple flowers. In contrast, the F2 plant with purple flowers had the two bands with similar intensities in dCAPS analysis and only a shorter band in indel analysis.
Figure 5.
dCAPS and indel analyses of an F2 population derived from a cross between a soybean near-isogenic line with white flowers, Clark-w1 and a Glycine soja line with light purple flowers, B09121. (Upper panel) Results of dCAPS analysis. PCR products with dCAPS primers were digested with NcoI and the digests were separated on an 8% polyacrylamide gel. ϕ, molecular marker ϕx174/HaeIII; clk, Clark; w1, Clark-w1; B, B09121; W, F2 plants homozygous for w1 allele (white flower); Lp, F2 plants homozygous for w1-lp allele (light purple flower); H, heterozygous F2 plants (light purple flower); P, an F2 plant with purple flowers. The migration of size markers is shown to the left of the gel. (Lower panel) Results of indel analysis. PCR products amplified with indel primers were separated on an 8% polyacrylamide gel.
Semi-quantitative RT-PCR analysis
Results of semi-quantitative RT-PCR suggested that the transcript level of the F3'5'H gene was not substantially different among lines (data not shown).
Discussion
Flower color of G. soja is almost exclusively purple, whereas about 30% of soybean cultivars have white flowers. The reason why G. soja almost lacks flower color variants is uncertain [2]. In 1998, researchers found a white-flowered variant of PI 424008A, a USDA accession of G. soja with purple flowers that was originally introduced from South Korea in 1976 [2]. The mutation may have occurred during propagation at USDA. To our knowledge, B09121 is the first example of flower color variant found in the natural habitat.
Genetic analysis suggested that light purple flower is controlled by a new allele at the W1 locus, w1-lp. Dominance relationship of the locus was W1 >w1-lp >w1. Interestingly, one F2 plant and four F3 plants with purple flowers were generated in the cross with Clark-w1. Considering the fact that purple-flowered plants were produced from heterozygous plants (w1-lp w1) in both F2 and F3 generations and that frequency of purple-flowered plants was similar (about 1%) across generations, the purple flower color may have been produced by a crossover in the W1 gene instead of seed contamination or out-crossing. The purple-flowered F2 plant produced F3 plants with purple and light purple flowers at a 3:1 ratio; this suggests that the region including the 65 bp insertion was eliminated from the genome. The dCAPS and indel analyses indicated that the base substitution was heterozygous but the indel region was homozygous without the 65 bp insertion in the purple-flowered F2 plant. The results further supported elimination of the insertion that is responsible for gene dysfunction from the plant by intragenic recombination. It remains to be investigated whether the existence of tandem repeats derived from the insertion is responsible for the high frequency of intragenic recombination.
Sequence analysis of F3'5'H cDNA from Clark and B09121 indicated that two amino acids (amino acid numbers 210 and 475) were substituted. The former substitution has not been observed in soybean cultivars examined to date. It may be responsible for light purple flower and unique anthocyanin composition. However, no catalytic domains have been assigned to the region. The spontaneous mutation leading to flower color change may not have affected amount of F3'5'H gene transcripts, based on the results from semi-quantitative RT-PCR analyses.
Flavonoids in flower petals of G. soja with purple flowers were generally similar to those of soybean cultivars with purple flowers. Flower petals of PI 424008C with white flowers had no anthocyanin but contained comparable amounts of flavonol glycosides and dihydroflavonols. It is consistent with the result from Clark-w1, a soybean NIL with white flowers, whose flower petals contained no anthocyanin although it had levels of flavonol glycosides and dihydroflavonol similar to Clark [12]. The present results confirmed that W1 solely controls anthocyanin biosynthesis in G. soja.
Purple flowers of soybean and G. soja contain four major anthocyanins with 3'4'5'-substituted form, malvidin 3,5-di-O-glucoside, petunidin 3,5-di-O-glucoside, delphinidin 3,5-di-O-glucoside and delphinidin 3-O-glucoside. In contrast, flower petals of B09121 contained lower amounts of the four major anthocyanins. In addition, they contained small amounts of the 5'-unsubstituted versions of the above anthocyanins, peonidin 3,5-di-O-glucoside, cyanidin 3,5-di-O-glucoside and cyanidin 3-O-glucoside. It appears that F3'5'H activity was reduced and F3'H activity was increased in flower petals of B09121. Flower petals of soybean and G. soja contain large amounts of kaempferols and very small amounts of quercetins [12,13], suggesting that F3'H activity may be very low. Further, alleles at the T locus encoding F3'H did not affect 3'-hydroxylation of flavonols in flower petals [12,13]. Therefore, it is unlikely that the F3'H gene might be responsible for the anthocyanin alteration. Alternatively, mutation in the F3'5'H gene possibly led to a reduction in F3'5'H activity and an increase in F3'H activity. In petunia, a cytochrome b5 is required for full activity of F3'5'H. A mutation in cytochrome b5 reduced 3'4'5'-substituted anthocyanins and increased 3'4'-substituted anthocyanins [11]. It is possible that the amino acid substitution might directly affect the amount and composition of anthocyanins or interact with cytochrome b5. Functional analysis using yeast recombinant assays may be necessary to identify the amino acid substitution that led to light purple flower and unique anthocyanin composition, to investigate the association with cytochrome b5, and to verify whether the amino acid substitution generated F3'H activity. Transformation experiments using a soybean line with w1 allele may be necessary to verify the function of the F3'5'H gene from B09121. B09121 may be the first example of a flower color variant of G. soja found in the natural habitat. It may be a useful tool for studies of the structural and functional properties of F3'5'H genes as well as investigations on the role of flower color in relation to adaptation of G. soja to natural habitats.
Conclusions
This study is the first report of a flower color variant of wild soybean G. soja discovered in nature. Genetic analysis revealed that light purple flower of the accession B09121 was controlled by a new allele of W1 locus encoding F3'5'H. The new allele was designated as w1-lp. The dominance relationship of the locus was W1 >w1-lp >w1. In crossing experiments, purple-flowered plants were generated in the cross between B09121 and a soybean near-isogenic line with w1 allele. F3 progeny test, and dCAPS and indel analyses suggested that the plants with purple flowers might be due to intragenic recombination. Flower petals of B09121 contained lower amounts of four major anthocyanins common in purple flowers and contained small amounts of the 5'-unsubstituted versions of the above anthocyanins that are absent in soybeans and other G. soja accessions. The results suggested that F3'5'H activity was reduced and flavonoid 3'-hydroxylase activity was increased in the flower petals. The cDNA of B09121 had a unique base substitution resulting in the substitution of valine with methionine. B09121 may be a useful tool for studies of the structural and functional properties of F3'5'H genes as well as investigations on the role of flower color in relation to adaptation of G. soja to natural habitats.
Authors' contributions
RT carried out genetic analysis. RT, JGD and HM participated in the molecular genetic studies. KY discovered a G. soja line with unique flower color. TI carried out chemical analysis of flavonoids. JGD critically revised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Ryoji Takahashi, Email: masako@affrc.go.jp.
Joseph G Dubouzet, Email: jgd111660@gmail.com.
Hisakazu Matsumura, Email: hisaca@affrc.go.jp.
Kentaro Yasuda, Email: kentaroy@akita-pu.ac.jp.
Tsukasa Iwashina, Email: iwashina@kahaku.go.jp.
Acknowledgements
We thank Dr. R.L. Nelson at USDA/ARS Univ. of Illinois for supplying the seeds of Clark isolines and G. soja lines. We are grateful to Ms. N. Mihashi and the NIAS Genebank for providing seeds of the G. soja accessions, Dr. Junichi Kitajima (Showa Pharmaceutical University) for NMR measurement, and Dr. R.G. Palmer (Iowa State University) for advice.
References
- Hymowitz T. In: Soybeans: Improvement, Production, and Uses, Agron. Monogr. 16. 3. Boerma HR, Specht JE, editor. ASA, CSSA, and SSSA, Madison, WI; 2004. Speciation and cytogenetics; pp. 97–136. [Google Scholar]
- Chen Y, Nelson RL. Identification and characterization of a white-flowered wild soybean plant. Crop Sci. 2004;44:339–342. [Google Scholar]
- Palmer RG, Pfeiffer TW, Buss GR, Kilen TC. In: Soybeans: Improvement, Production, and Uses, Agron. Monogr. 16. 3. Boerma HR, Specht JE, editor. ASA, CSSA, and SSSA, Madison, WI; 2004. Qualitative genetics; pp. 137–233. [Google Scholar]
- Takahashi R, Matsumura H, Oyoo ME, Khan NA. Genetic and linkage analysis of purple-blue flower in soybean. J Hered. 2008;99:593–597. doi: 10.1093/jhered/esn041. [DOI] [PubMed] [Google Scholar]
- Forkmann G. Flavonoids as flower pigments: The formation of the natural spectrum and its extension by genetic engineering. Plant Breed. 1991;106:1–26. doi: 10.1111/j.1439-0523.1991.tb00474.x. [DOI] [Google Scholar]
- Buttery BR, Buzzell RI. Varietal differences in leaf flavonoids of soybeans. Crop Sci. 1973;13:103–106. [Google Scholar]
- Buzzell RI, Buttery BR. Genetics of black pigmentation of soybean seed coats/hila. Soybean Genet Newsl. 1982;9:26–29. [Google Scholar]
- Iwashina T, Benitez ER, Takahashi R. Analysis of flavonoids in pubescence of soybean near-isogenic lines for pubescence color loci. J Hered. 2006;97:438–443. doi: 10.1093/jhered/esl027. [DOI] [PubMed] [Google Scholar]
- Toda K, Yang D, Yamanaka N, Watanabe S, Harada K, Takahashi R. A single-base deletion in soybean flavonoid 3'-hydroxylase gene is associated with gray pubescence color. Plant Mol Biol. 2002;50:187–196. doi: 10.1023/A:1016087221334. [DOI] [PubMed] [Google Scholar]
- Zabala G, Vodkin LO. A rearrangement resulting in small tandem repeats in the F3'5'H genes of white flower genotypes is associated with the soybean W1 locus. Plant Genome (A Supplement to Crop Science) 2007;47:S113–S124. [Google Scholar]
- de Vetten N, ter Horst J, van Schaik HP, de Boer A, Mol J, Koes R. A cytochrome b5 is required for full activity of flavonoid 3', 5'-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc Natl Acad Sci USA. 1999;96:778–783. doi: 10.1073/pnas.96.2.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwashina T, Githiri SM, Benitez ER, Takemura T, Kitajima J, Takahashi R. Analysis of flavonoids in flower petals of soybean near-isogenic lines for flower and pubescence color genes. J Hered. 2007;98:250–257. doi: 10.1093/jhered/esm012. [DOI] [PubMed] [Google Scholar]
- Iwashina T, Oyoo ME, Khan NA, Matsumura H, Takahashi R. Analysis of flavonoids in flower petals of soybean flower color variants. Crop Sci. 2008;48:1918–1924. doi: 10.2135/cropsci2008.01.0031. [DOI] [Google Scholar]
- Bernard RL, Nelson RL, Cremeens CR. USDA soybean genetic collection: Isoline collection. Soybean Genet Newsl. 1991;18:27–57. [Google Scholar]
- Hattori S, Hayashi K. Über Anthocyane II. Über die Farbstoffe aus den rotten Herbstblättern von einigen Acer-Arten. Acta Phytochim. 1937;10:129–138. [Google Scholar]
- Hayashi K. Vereinfachte Darstellungs-methode der Anthocyanpräparate aus Dahlienblüten. Bot Mag Tokyo. 1933;47:394–399. [Google Scholar]
- Hayashi K. Studien über Anthocyane, IV. Isolierung des Paeonins aus den Blüten von Paeonia albiflora var. hortensis. Acta Phytochim. 1939;11:80–90. [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]