Abstract
Background
Ionic aluminum (mainly Al3+) is rhizotoxic and can be present in acid soils at concentrations high enough to inhibit root growth. Many forest tree species grow naturally in acid soils and often tolerate high concentrations of Al. Previously, we have shown that aspen (Populus tremula) releases citrate and oxalate from roots in response to Al exposure. To obtain further insights into the root responses of aspen to Al, we investigated root gene expression at Al conditions that inhibit root growth.
Results
Treatment of the aspen roots with 500 μM Al induced a strong inhibition of root growth within 6 h of exposure time. The root growth subsequently recovered, reaching growth rates comparable to that of control plants. Changes in gene expression were determined after 6 h, 2 d, and 10 d of Al exposure. Replicated transcriptome analyses using the Affymetrix poplar genome array revealed a total of 175 significantly up-regulated and 69 down-regulated genes, of which 70% could be annotated based on Arabidopsis genome resources. Between 6 h and 2 d, the number of responsive genes strongly decreased from 202 to 26, and then the number of changes remained low. The responses after 6 h were characterized by genes involved in cell wall modification, ion transport, and oxidative stress. Two genes with prolonged induction were closely related to the Arabidopsis Al tolerance genes ALS3 (for Al sensitive 3) and MATE (for multidrug and toxin efflux protein, mediating citrate efflux). Patterns of expression in different plant organs and in response to Al indicated that the two aspen genes are homologs of the Arabidopsis ALS3 and MATE.
Conclusion
Exposure of aspen roots to Al results in a rapid inhibition of root growth and a large change in root gene expression. The subsequent root growth recovery and the concomitant reduction in the number of responsive genes presumably reflect the success of the roots in activating Al tolerance mechanisms. The aspen genes ALS3 and MATE may be important components of these mechanisms.
Background
Acid soils are prevalent in many regions of the world and present a range of stresses to plants. One of the major stresses caused by these soils is aluminum (Al), which is solubilized by the acidity into the soil solution. Soluble Al exists in its most toxic form as Al3+, which can inhibit root growth in many plant species at micromolar concentrations. The resulting reduced and damaged root system limits the capacity of plants to uptake water and nutrients, and increases their susceptibility to other stresses.
The mechanisms by which Al inhibits root growth are not well understood, despite extensive physiological investigations. The root apex is the most sensitive part of the root to Al because it is the site of cell division and cell elongation [1,2]. Since Al is so reactive, it can interact with multiple structures in the apoplasm and symplasm of root cells. In the cell wall, Al primarily binds to the pectin matrix and thereby alters the physical properties of the cell wall [3,4]. In the symplasm, sites of Al interaction include membrane constituents, ion channels, metabolic enzymes, components of signaling pathways, members of the cytoskeleton, and the DNA [3,5]. Although some of the resulting cellular alterations have been proposed to affect cell division or cell elongation, a recent study conducted in Arabidopsis thaliana indicates that it is not Al toxicity that is directly responsible for the inhibition of root growth. Genetic and biochemical evidence suggest that the cells of the root apex have an ATR-controlled mechanism to monitor Al-induced DNA damage [6]. In plants exposed to Al, this mechanism activates blockage of cell cycle progression and thus root growth. This active response of roots to Al may not protect individual plants, but it may help to safeguard plant populations by preventing the passage of damaged DNA to subsequent plant generations [6].
Plant species vary considerably in their degree of Al tolerance, and even genotypes within the same plant species vary in their ability to cope with Al. The mechanisms providing enhanced Al tolerance can be classified into external and internal mechanisms [5,7]. The best-documented external mechanism is the efflux of organic acid anions, such as malate, citrate, and oxalate, from the roots in response to Al exposure [8]. These organic acid anions effectively chelate Al and thereby detoxify Al in the rhizosphere. Other proposed external mechanisms involve secretion of proteins [9], root-mediated increase of the rhizosphere pH [10], and masking Al binding sites at the cell wall [11,12]. Proposed internal tolerance mechanisms include chelation of Al by organic acid anions or phenolic compounds and sequestration of Al in the vacuole [5].
The genes responsible for the Al-induced efflux of malate and citrate have been recently isolated and demonstrated to represent major genes for Al tolerance in several plant species [8]. The genes responsible for the efflux of malate belong to the ALMT (for Al-activated malate transporter) gene family [13-15] and those involved in the efflux of citrate to the MATE (for multidrug and toxin efflux protein) gene family [16-20]. All these genes encode membrane proteins, consistent with their role in facilitating the efflux of organic acid anions. Additional genes with putative roles in Al tolerance have been identified in Al-sensitive mutants of rice (Oryza sativa) and Arabidopsis. The rice mutants star1 (for sensitive to Al rhizotoxicity 1) and star2 were found to be mutated in genes encoding two proteins that form together an ATP-binding cassette (ABC) transporter [21]. This complex mediates the transport of UDP-glucose to the cell wall, where the molecule is believed to play a role in masking Al binding sites. Similarly to star1 and star2, the Al-sensitive mutants als1 (for Al sensitive 1) and als3 of Arabidopsis are mutated in genes encoding ABC transporter-like proteins [22,23]. Although the substrate of these proteins is not known, the mutant phenotypes and patterns of gene expression have led to the proposal that the two proteins transport and sequester Al to confer Al tolerance. ALS1 is believed to be involved in the intracellular transport of Al to the vacuole [22], whereas ALS3 appears to be necessary for the long-distance transport of Al to the aerial parts of the plant [23].
Further insight into the molecular mechanisms involved in Al toxicity and tolerance come from gene expression analyses. Genome-wide transcriptome analyses in roots of Arabidopsis have revealed a number of cellular processes that are altered in response to Al exposure. Examples are cell wall modification, protein metabolism, transport processes, and oxidative stress [24,25]. In maize (Zea mays), wheat (Triticum aestivum), and Medicago truncatula, gene expression was analyzed in plant lines with contrasting levels of Al tolerance [26-28]. These studies have led to the identification of several candidate genes for Al tolerance, including previously identified genes encoding organic acid efflux transporters, genes controlling levels of reactive oxygen species (ROS), as well as genes involved in pectin modification and immobilization of Al by phosphate.
Forest trees generally tolerate high concentrations of Al [29]. For example, seedlings of Norway spruce (Picea abies) and birch (Betula pendula) did not show any reduction in root growth at Al concentrations below 0.3 and 3 mM, respectively [30,31]. In contrast, Al concentrations as low as 50 μM tend to affect the root growth of Arabidopsis and several crop plants (e.g. [24,28,32]). Since many forest tree species grow naturally in acid soils, it appears likely that such species have developed adaptive mechanisms that enable them to tolerate high Al conditions. Analyses of the root responses to Al in forest trees may thus broaden our understanding of Al tolerance mechanisms in plants.
In a previous study, we used clonal aspen (Populus tremula, clone Birmensdorf) to investigate Al-induced efflux of organic acid anions from roots [33]. The results showed that Al concentrations ≥ 200 μM induced the efflux of citrate, while Al concentrations ≥ 500 μM enhanced the efflux of oxalate. At these concentrations, Al did not cause any visible symptoms at the root tips, indicating that the aspen clone examined tolerates high concentrations of Al. Using the same aspen clone, we investigated temporal patterns of root gene expression under Al stress. Changes in gene expression were assessed by application of the Affymetrix poplar genome array. The expression of selected genes was further analyzed by reverse-transcription PCR.
Results
Effect of Al on root growth and callose formation
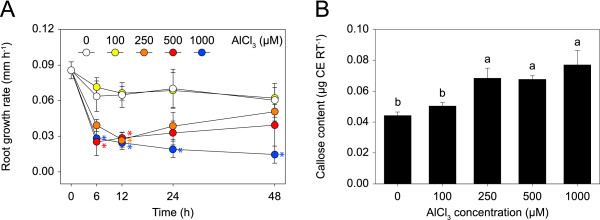
To determine plant treatment conditions suitable for gene expression profiling, we examined the effect of Al on root growth. Clonal aspen was treated with either no Al or increasing concentrations of Al up to 1000 μM for 2 d in solution culture (Fig. 1). Exposure of the roots to 100 μM Al did not affect their growth (Fig. 2A). In contrast, 250, 500, and 1000 μM Al caused a rapid and strong inhibition of root growth, with a reduction in growth rate of ≥ 40% observed after 6 h. During prolonged Al exposure, the root growth of plants treated with 250 and 500 μM Al partially recovered, while that of plants treated with 1000 μM further decreased, although not significantly (p > 0.1). As a further indicator of Al toxicity, we quantified callose, which accumulates in many plant species upon Al exposure [3]. The content of callose in the root tips increased with Al in the medium, with a significant increase observed at Al concentrations ≥ 250 μM (Fig. 2B).
Figure 1.
Solution culture of clonal aspen (Populus tremula). A Solution cultures were established in modified Magenta boxes. B Root growth was photographically monitored. Glass slides with a 5 μm grid were used as a reference to measure root growth.
Figure 2.
Effect of Al on root growth and callose formation in aspen. A Time series of the root growth rates of plants treated with no or increasing concentrations of Al for 2 d. Asterisks indicate significant differences between treated and control plants (repeated measures ANOVA; p < 0.05). B Callose content in root tips (RT) after 2 d of Al treatment. Callose content is expressed as a curdlan equivalent (CE). The means and standard error values from four independent roots are shown. Different letters indicate significant differences between treatments (ANOVA; p < 0.05).
Root growth of plants used for gene expression profiling
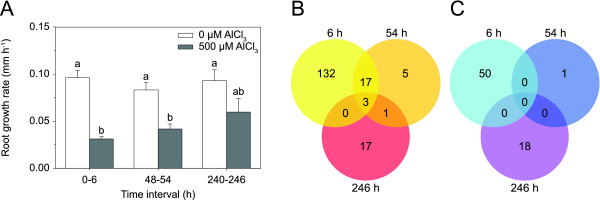
Based on the results of the dose-response experiment, plants were treated with 500 μM Al for gene expression profiling. Treatment with 500 μM was preferred over 1000 μM, which tended to cause necrotic lesions at the root tips. The time points selected were 6 h, 54 h (designated as 2 d), and 246 h (10 d). The 6 h time point marked the rapid Al-induced inhibition of root growth. The 2 and 10 d time points represented the period of root growth recovery. Since some plant genes are regulated by a diurnal rhythm and circadian clock, the exact duration of the treatments were designed such that the roots could be sampled at the same time each. During the 10 d treatment, the culture medium was exchanged every 2 d to maintain a constant Al stress. Consistent with the results of the dose-response experiment, Al induced a rapid inhibition of root growth (68% reduction at 6 h; Fig. 3A). The growth of roots treated with Al for 2 and 10 d gradually recovered (50% reduction at 2 d; 36% reduction at 10 d). Al concentrations of the culture medium decreased only slightly during the 2 d treatment periods (on average by 36.7 ± 13.9 μM), and the pH remained constant (pH 4.0 ± 0.1). Therefore, the increase in root growth could not be explained by either a possible decrease of Al in the culture medium due to the uptake of Al by the plants, or altered Al speciation due to pH change.
Figure 3.
Aspen genes with significantly altered expression in Al-treated root tips compared to control root tips. A Root growth rates of plants used for the gene expression profiling. Roots were treated with 500 μM Al for 6 h, 54 h, and 246 h. During the 246 h treatment, the culture medium was exchanged every 2 d to maintain a constant Al stress. Means and standard error values from four independent roots are shown. Different letters indicate significant differences between treatments (ANOVA; p < 0.05). Number of up-regulated genes (B) and down-regulated genes (C) after 6 h, 54 h, and 246 h of Al treatment (≥ 2-fold change in expression).
Changes in gene expression
Gene expression profiles were determined using the Affymetrix poplar genome array, interrogating over 56,000 transcripts and gene predictions. To detect genes that are significantly regulated by Al, we employed an approach that allowed an estimation of the false discovery rate (FDR) in multiple testing. The q-value, which is a positive FDR analogue of the p-value [34], was set to 0.15. As a further criterion, we used a two-fold change cut-off. By these criteria and after removing redundant probe sets, a total of 244 genes were differentially expressed. Treatment of the roots for 6 h resulted in the up-regulation of 152 genes and the down-regulation of 50 genes (Fig. 3B; C). These numbers decreased significantly when the roots were treated for 2 d (26 genes up-regulated, 1 down-regulated). Of the induced genes at this time point, 20 were also induced after 6 h. Treatment of the roots for 10 d yielded a similar low number of responsive genes (21 genes up-regulated, 18 down-regulated). Three genes were induced across all three time points.
Validation of microarray data
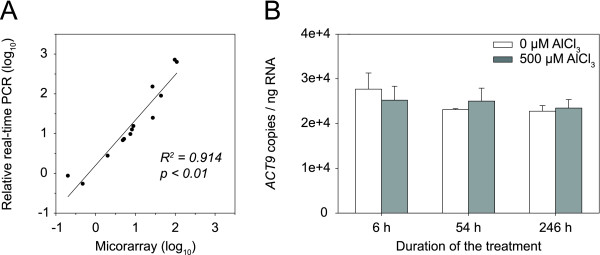
The microarray data were independently verified by real-time reverse transcription PCR (qRT-PCR). Eight genes were analyzed, displaying a wide range of expression profiles. Transcripts were quantified relative to the actin 9 (ACT9) gene, which was isolated and sequenced in this study. The qRT-PCR analyses were performed with RNA from the microarray experiment (18 samples) and with RNA from an independent Al treatment experiment (2 samples). Six genes were tested with the RNA from both experiments. A significant correlation was found between the microarray and qRT-PCR data (R2 = 0.91; p < 0.01; Fig. 4A). The magnitude of the relative changes in transcript abundance did not differ greatly between the two techniques. Exceptions were two strongly induced genes encoding a basic pathogenesis-related protein and a family 3 sulfate transporter: expression differences measured by qRT-PCR were 10 times greater than those measured by microarrays, probably due to the fact that qRT-PCR has a wider dynamic range than microarrays [35]. To examine the validity of using ACT9 as a reference gene, absolute qRT-PCR was performed. The analysis showed that the expression of the gene did not change significantly during the 10 d treatment (p > 0.6; Fig. 4B).
Figure 4.
Validation of microarray data. A Relationship between microarray and qRT-PCR data. Transcript levels of eight differentially expressed genes were quantified by qRT-PCR relative to ACT9. The fold changes in gene expression in response to Al treatment were log10 transformed. The microarray data were plotted against the qRT-PCR data. B Expression levels of ACT9 in the root samples used for microarray experiments. ACT9 transcript levels did not significantly change during the 10 d Al treatment (ANOVA; p > 0.6). The means and standard error values from three independent samples are shown.
Annotation and functional categorization of Al-responsive genes
BLASTX searches against the Arabidopsis protein database allowed 172 of the responsive genes to be annotated (expectation (E)-value ≤ 1 × 10-10). Additional 29 genes had matches to Arabidopsis genes with unknown functions, and 44 genes had no detectable similarity to Arabidopsis proteins (E-value ≥ 1 × 10-4). A complete list of the responsive genes is presented in additional file 1.
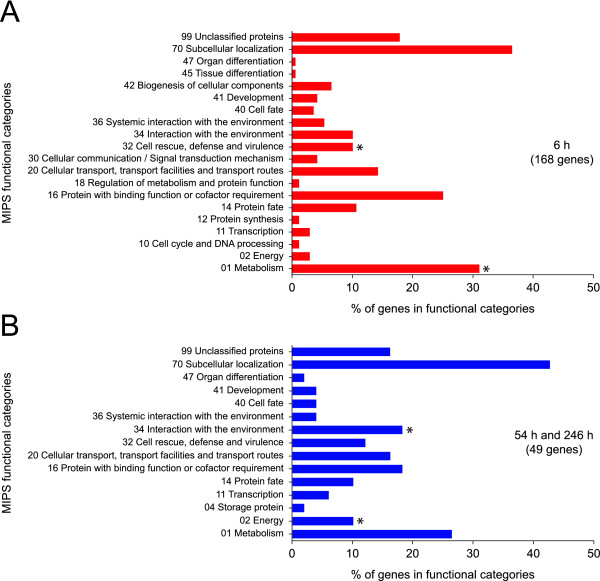
To identify biological processes, the genes with known functions were classified using the Munich Information Center for Protein Sequences (MIPS) functional catalogue [36]. The genes present on the microarray were also classified to allow identification of categories whose genes were over-represented compared to the genes present on the microarray. These analyses were carried out for the genes regulated after 6 h and for the combined set of genes regulated after 2 and 10 d. Top-level categories and subcategories with an enrichment p-value < 0.05 and a FDR < 0.05 were regarded as pertinent to the time points examined. By these criteria, genes assigned to the top-level categories 'metabolism' and 'cell rescue and defense' were enriched after 6 h (Fig. 5A; additional file 2). Within the 'metabolism' category, the genes related to 'carbohydrate metabolism' were enriched and included several genes involved in cell wall modification (additional file 2). Additional enriched genes related to cell wall modification were assigned to the top-level category 'biogenesis of cellular components'. The top-level category 'cell rescue and defense' included a number of enriched genes related to 'oxidative stress response' and 'detoxification'. An additional major group of enriched genes had functions in 'ion transport'. Smaller groups of enriched genes were related to 'cell death' and 'plant signaling molecules'. Genes implicated in 'ion transport' and 'carbohydrate metabolism' were also enriched during prolonged Al exposure, but their number was small (additional file 2). Additional genes enriched during prolonged Al exposure were assigned to the top-level categories 'energy' and 'interaction with the environment' (Fig. 5B).
Figure 5.
Functional categorization of the differentially expressed aspen genes using the Munich Information Center for Protein Sequences (MIPS) functional catalogue. Categories of the differentially expressed genes after 6 h of Al exposure (A), and the combined set of genes differentially expressed after 2 and 10 d of Al exposure (B). Categories whose members were enriched are indicated by asterisks (p < 0.05, FDR < 0.05).
Genes related to cell wall modification
All the genes recorded that are related to cell wall modification were regulated after 6 h of Al exposure (Table 1). Nine of these genes have functions in the modification of pectin. A pectin methylesterase gene, two pectate lyase genes, and two galacturonosyltransferase genes were down-regulated. Of the three pectin methylesterase inhibitor genes identified, two were down-regulated and the other induced. Additional genes encoded proteins that target cellulose and xyloglucan. The three endo-1,4-β-glucanase and three xyloglucan endotransglucosylase/hydrolase genes recorded showed contrasting patterns of up- and down-regulated expression.
Table 1.
Selected genes differentially expressed in Al-treated aspen roots.
| Fold change | ||||||
|---|---|---|---|---|---|---|
| Affymetrix probe set IDa | Gene model nameb | AGI n°c | Annotation of best hit in Arabidopsis thaliana genomed | 6 h | 2 d | 10 d |
| Cell walle | ||||||
| PtpAffx.21218.1.S1_at | eugene3.00280166 | AT3G13750 | Beta-galactosidase 1 (BGAL1; GH35) | 0.2 | 0.5ns | 0.5ns |
| PtpAffx.122394.1.S1_at | gw1.I.26.1 | AT5G64570 | Beta-xylosidase 4 (XYL4; GH3) | 0.2 | 0.8ns | 1.3ns |
| PtpAffx.212738.1.S1_at | estExt_fgenesh4_pg.C_LG_XV0425 | AT5G62620 | Galactosyltransferase family protein | 2.1 | 1.0ns | 1.2ns |
| Ptp.7955.1.S1_at | gw1.VIII.37.1 | AT1G24170 | Galacturonosyltransferase-like 8 (GATL8; GT9) | 0.5 | 0.7ns | 1.0ns |
| PtpAffx.208384.1.S1_at | gw1.VIII.37.1 | AT1G70090 | Galacturonosyltransferase-like 9 (GATL9; GT8) | 0.5 | 0.8ns | 1.1ns |
| PtpAffx.209224.1.S1_at | fgenesh4_pg.C_LG_X001601 | AT1G65610 | Endo-1,4-beta-glucanase Korrigan 2 (KOR2; GH9) | 2.1 | 1.3ns | 1.0ns |
| PtpAffx.207811.1.S1_at | estExt_fgenesh4_pg.C_LG_VIII0680 | AT1G65610 | Endo-1,4-beta-glucanase Korrigan 2 (KOR2; GH9) | 2.9 | 1.3ns | 1.0ns |
| Ptp.4073.1.S1_s_at | estExt_fgenesh4_pg.C_LG_XIV0665 | AT4G02290 | Endo-1,4-beta-glucanase 17 (GH9) | 0.2 | 0.5ns | 1.0ns |
| PtpAffx.116752.1.A1_at | gw1.VIII.287.1 | AT3G26380 | Glycosyl hydrolase family protein 27 (GH27) | 2.0 | 1.6 | 1.4ns |
| PtpAffx.20309.1.S1_at | estExt_fgenesh4_pg.C_LG_X2099 | AT5G04500 | Glycosyl transferase family protein 64 (GT64) | 5.2 | 3.8 | 1.7ns |
| Ptp.4642.1.S1_at | estExt_fgenesh4_pg.C_LG_XI1340 | AT1G30620 | UDP-D-xylose 4-epimerase 1 (UXE1) | 2.2 | 1.7 | 1.1ns |
| PtpAffx.31211.1.A1_at | eugene3.00140929 | AT1G05560 | UDP-glucose transferase 1 (UGT1; GT1) | 6.4 | 0.8ns | 1.5ns |
| Ptp.160.1.S1_x_at | fgenesh4_pm.C_LG_II000873 | AT3G62830 | UDP-glucuronic acid decarboxylase 2 (UXS2) | 2.2 | 1.2ns | 1.3ns |
| PtpAffx.119179.1.A1_at | gw1.1681.2.1 | AT5G48070 | Xyloglucan endotransglucosylase/hydrolase 20 (XTH20; GH16) | 4.3 | 1.6ns | 1.7ns |
| Ptp.2467.1.A1_x_at | gw1.XIX.2748.1 | AT4G03210 | Xyloglucan endotransglucosylase/hydrolase 9 (XTH9; GH16) | 0.3 | 0.6ns | 1.1ns |
| Ptp.3050.1.S1_s_at | estExt_Genewise1_v1.C_LG_XIV2162 | AT1G10550 | Xyloglucan:xyloglucosyltransferase 33 (XTH33; GH16) | 0.3 | 0.8ns | 0.9ns |
| Ptp.4810.1.A1_s_at | estExt_Genewise1_v1.C_LG_III0932 | AT1G04680 | Pectate lyase family protein | 0.5 | 0.8ns | 0.9ns |
| PtpAffx.1316.2.S1_s_at | eugene3.00010425 | AT1G04680 | Pectate lyase family protein | 0.2 | 1.0ns | 0.9ns |
| PtpAffx.9932.3.S1_a_at | eugene3.00030462 | AT1G53830 | Pectin methylesterase 2 (PME2) | 0.4 | 0.9ns | 0.8ns |
| PtpAffx.9932.2.A1_s_at | estExt_fgenesh4_pm.C_290002 | AT3G14310 | Pectin methylesterase inhibitor 3 (PMEI3) | 0.5 | 1.0ns | 1.0ns |
| Ptp.7635.1.S1_at | eugene3.00140717 | AT1G02810 | Pectin methylesterase inhibitor 7 (PMEI7) | 4.0 | 1.5ns | 0.9ns |
| PtpAffx.207505.1.S1_at | gw1.VIII.1476.1 | AT3G10720 | Pectin methylesterase inhibitor 25 (PMEI25) | 0.3 | 0.5ns | 1.0ns |
| Ptp.3290.1.S1_at | gw1.VII.2504.1 | AT5G08200 | Peptidoglycan-binding LysM domain-containing protein | 2.2 | 1.2ns | 1.1ns |
| Ptp.2725.1.S1_at | gw1.X.2924.1 | AT5G62150 | Peptidoglycan-binding LysM domain-containing protein | 3.1 | 1.2ns | 1.2ns |
| PtpAffx.208179.1.S1_at | eugene3.00081504 | AT2G23770 | Peptidoglycan-binding LysM domain-containing protein | 2.3 | 1.5ns | 1.1ns |
| Transport | ||||||
| Ion transport | ||||||
| Ptp.6087.1.S1_at | eugene3.97260001 | AT5G01600 | Ferritin 1 (FER1) | 0.3ns | 0.4ns | 0.3 |
| PtpAffx.595.4.S1_s_at | estExt_fgenesh4_pg.C_1470038 | AT2G24520 | H+-ATPase 5 (HA5) | 2.3 | 1.5ns | 1.6ns |
| PtpAffx.208738.1.S1_s_at | estExt_fgenesh4_pm.C_LG_X0276 | AT5G64560 | Mg transporter CorA-like family protein (MRS2-2) | 4.8 | 2.2ns | 1.4ns |
| PtpAffx.46328.1.A1_at | gw1.I.4154.1 | AT3G19640 | Mg transporter CorA-like family protein (MRS2-3) | 3.8 | 2.8 | 1.3ns |
| PtpAffx.204370.1.S1_at | fgenesh4_pg.C_LG_IX000025 | AT5G44370 | Phosphate transporter 4;6 (PHT4;6) | 4.3 | 1.8ns | 1.1ns |
| PtpAffx.249.377.A1_at | fgenesh4_pm.C_LG_V000517 | AT3G51895 | Sulfate transmembrane transporter 3;1 (SULTR3;1) | 2.4 | 1.3ns | 1.0ns |
| PtpAffx.63924.1.S1_at | eugene3.01570002 | AT5G19600 | Sulfate transmembrane transporter 3;5 (SULTR3;5) | 26.8 | 109.2 | 3.2ns |
| PtpAffx.46298.1.S1_at | estExt_fgenesh4_pg.C_LG_VIII0032 | AT5G55630 | Two pore K+ channel 1 (TPK1) | 2.1 | 1.3ns | 1.1ns |
| Transport facilities | ||||||
| PtpAffx.119032.1.S1_s_at | gw1.XVI.2587.1 | AT2G37330 | Aluminum sensitive 3 (ALS3) | 44.4 | 27.5 | 5.2 |
| Ptp.5171.1.S1_at | gw1.VI.655.1 | AT5G03910 | ABC transporter homolog 12 (ATH12) | 2.5 | 1.4ns | 1.1ns |
| PtpAffx.204839.1.S1_at | gw1.IX.3299.1 | AT3G08040 | MATE (FRD3) | 8.9 | 4.2ns | 1.7ns |
| Ptp.2869.1.A1_at | gw1.I.5916.1 | AT1G30690 | SEC14 cytosolic factor family protein/phosphoglyceride transfer family protein | 0.3 | 0.8ns | 0.9ns |
| Oxidative stress responsef | ||||||
| Ptp.2903.1.A1_s_at | gw1.XII.485.1 | AT3G22370 | Alternative oxidase 1A (AOX1A) | 8.6 | 1.5ns | 3.6ns |
| PtpAffx.56141.1.A1_at | grail3.0007029701 | AT5G20230 | Blue copper binding protein (BCB) | 2.4 | 1.4ns | 2.4ns |
| PtpAffx.153878.1.A1_at | gw1.XV.2559.1 | AT5G51100 | Fe superoxide dismutase (FSD2) | 1.4ns | 1.5ns | 2.8 |
| PtpAffx.134361.1.A1_s_at | eugene3.00031141 | AT1G64500 | Glutaredoxin family protein | 0.4 | 0.7 | 0.4ns |
| PtpAffx.2286.4.S1_a_at | - | AT1G17180 | Glutathione S-transferase tau 25 (GSTU25) | 0.4 | 0.5ns | 1.9ns |
| PtpAffx.23427.1.S1_s_at | estExt_fgenesh4_pg.C_LG_VIII1530 | AT2G29420 | Glutathione S-transferase tau 7 (GSTU7) | 2.9 | 1.0ns | 1.5ns |
| PtpAffx.29337.1.A1_at | eugene3.00030584 | AT5G67400 | Peroxidase 73 (P73) | 0.5 | 1.0ns | 1.0ns |
| PtpAffx.36879.1.A1_s_at | gw1.VII.698.1 | AT5G24070 | Peroxidase family protein | 0.5 | 0.7ns | 0.8ns |
| PtpAffx.43372.1.A1_at | fgenesh4_pg.C_LG_XVI000455 | AT5G06720 | Peroxidase, putative | 3.1 | 1.4ns | 0.9ns |
| PtpAffx.54628.1.S1_at | estExt_fgenesh4_pg.C_LG_XVI1240 | AT5G05340 | Peroxidase, putative | 13.3 | 1.7ns | 1.1ns |
| PtpAffx.55376.1.S1_at | fgenesh4_pg.C_LG_XIV000840 | AT5G05340 | Peroxidase, putative | 2.0ns | 0.4ns | 0.1 |
aAffymetrix probe set identifier (ID) of the GeneChip Poplar Genome Array 61 K (Affymetrix).
bPreferred gene model name attributed by Poparray v1.2 http://aspendb.uga.edu/poparray.
cArabidopsis genome identifier (AGI n°).
dAnnotation of best hit in Arabidopsis genome with a E-value cut-off of < 0.05.
eC-compound and carbohydrate metabolism and cell wall.
fOxidative stress response and detoxification.
nsnot significantly up- or down-regulated.
Genes related to ion transport
The regulated genes encoding ion transporters included both cation and anion transporters. With the exception of a ferritin gene, all the ion transporters were induced. A two-pore K+ channel gene was closely related to the Arabidopsis gene TPK1, which is a key regulator of K homeostasis [37]. Two genes encoded CorA-like Mg transporters, belonging to a class of transmembrane proteins that are suggested to function in the uptake and intracellular transport of Mg [38]. The anion transporters were composed of a family 4 phosphate transporter and two family 3 sulfate transporter genes. The encoded family 4 phosphate transporter was related to the Arabidopsis protein PHT4;6, proposed to be involved in the biosynthesis of cell wall polysaccharides [39]. One of the encoded sulfate transporters was related to the Arabidopsis SULTR3;5, a protein functioning in the root-to-shoot transport of sulfate [40].
Genes related to oxidative stress
Three of the regulated genes related to oxidative stress belong to the network of genes that control levels of ROS [41]. An alternative oxidase and a Fe superoxide dismutase gene were up-regulated, while a glutaredoxin gene was down-regulated. Additional genes encoded tau-type glutathione S-transferases and peroxidases. Individual members of these gene families showed contrasting patterns of up- and down-regulated expression.
Identification of putative Al tolerance genes
Among the regulated genes encoding transport facilitators, there were two genes that may play a role in Al tolerance. One was related to the Arabidopsis Al tolerance gene ALS3 [23]. The other belonged to the MATE gene family and was related to the Arabidopsis citrate efflux transporter gene MATE [18] and to FRD3 (for ferric reductase defective 3), which encodes a citrate transporter responsible for loading Fe into the xylem [42]. Both aspen genes were induced: ALS3 was up-regulated at all three time points, and MATE at 6 h and 2 d.
To examine whether the genes identified by microarrays are indeed genes related to the Arabidopsis ALS3 and MATE, RT-PCR was performed and the PCR products were sequenced. In the case of ALS3, the entire coding sequence was isolated. The predicted protein shared 79% sequence identity with the Arabidopsis ALS3 [23] and 71% with STAR2 of rice [21]. All of these genes encode the transmembrane domain of ABC transporters and lack the ATP-binding cassette domain. In the case of MATE, approximately 80% was isolated. The predicted protein shared 60% sequence identity with the Arabidopsis MATE [18] and 62% with FRD3 [43]. Similar degrees of sequence identities were found between the aspen MATE and MATE of sorghum (54%) [16], barley (54%) [17], and maize (56%) [20].
Expression of ALS3 and MATE
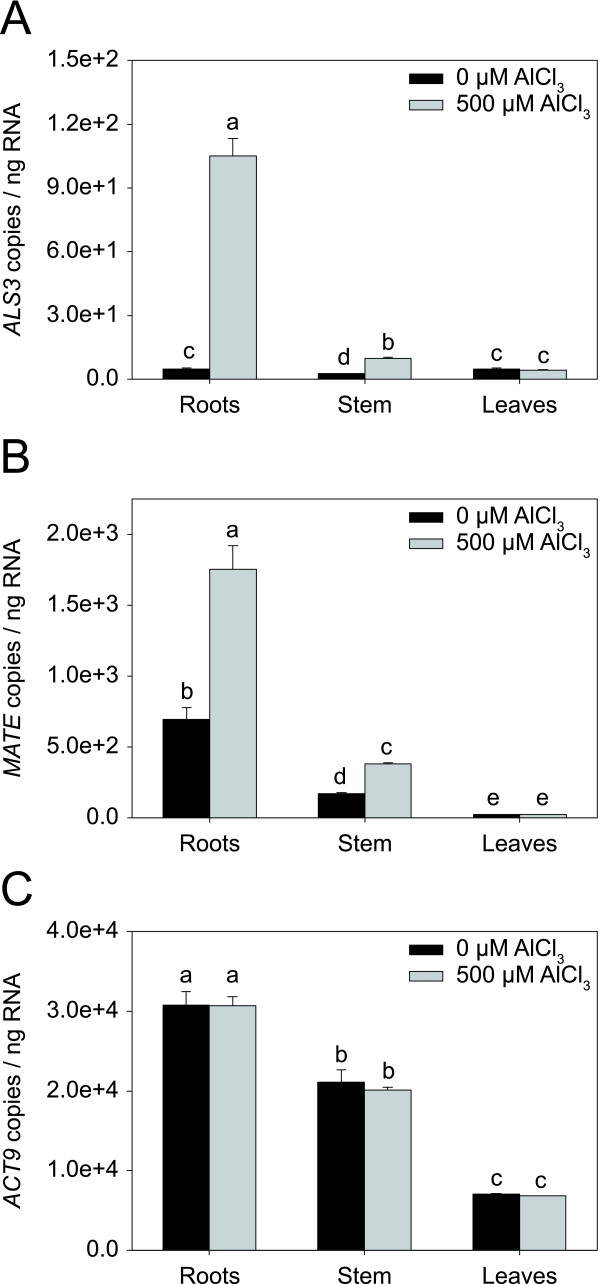
Each of the three Arabidopsis genes ALS3, MATE, and FRD3 are characterized by a distinct pattern of expression. Under non-stressed conditions, ALS3 is expressed in the phloem throughout the plant and in the root epidermis [23]. MATE and FRD3 are primarily expressed in the root [18,43]. Exposure to Al strongly induces ALS3 and MATE in the root, but not FRD3 [18,23]. To examine whether the expression of the aspen ALS3 and MATE is coherent with that of the Arabidopsis ALS3 and MATE, absolute qRT-PCR was carried out with RNA isolated from root, stem, and leaf tissue of a separate set of plants treated with either no Al or 500 μM Al for 2 d. The results of the analysis showed that ALS3 is expressed in all three tissues under non-stress conditions with little differences among the tissues (Fig. 6A). In contrast, MATE was more strongly expressed in the root than in the stem and leaves (Fig. 6B). Treatment of the plants with Al led to an induction of both genes (Fig. 6A; B). ALS3 was strongly induced in the root (22.3-fold) and to a lower extent in the stem (3.9-fold), while MATE was induced both in the root (2.5-fold) and in the stem (2.3-fold). The expression levels of ACT9 did not differ between the treatments, but did between the tissues, where they were highest in the root (Fig. 6C). However, the differences in ACT9 expression in the different tissues did not explain the tissue-specific expression of ALS3 and MATE, as was shown by plotting transcript levels relative to ACT9 (data not shown). Taken together, expression patterns of the two aspen genes were highly coherent with that of the Arabidopsis ALS3 and MATE.
Figure 6.
Expression levels of ALS3, MATE, and ACT9 in different aspen tissues. Expression levels of ALS3 (A), MATE (B), and ACT9 (C) in root, stem, and leaf tissue of plants treated with no or 500 μM Al for 2 d. Transcript levels were quantified by absolute qRT-PCR. The means and standard error values from three independent samples are shown. Different letters indicate significant differences between treatments and tissues (ANOVA; p < 0.05).
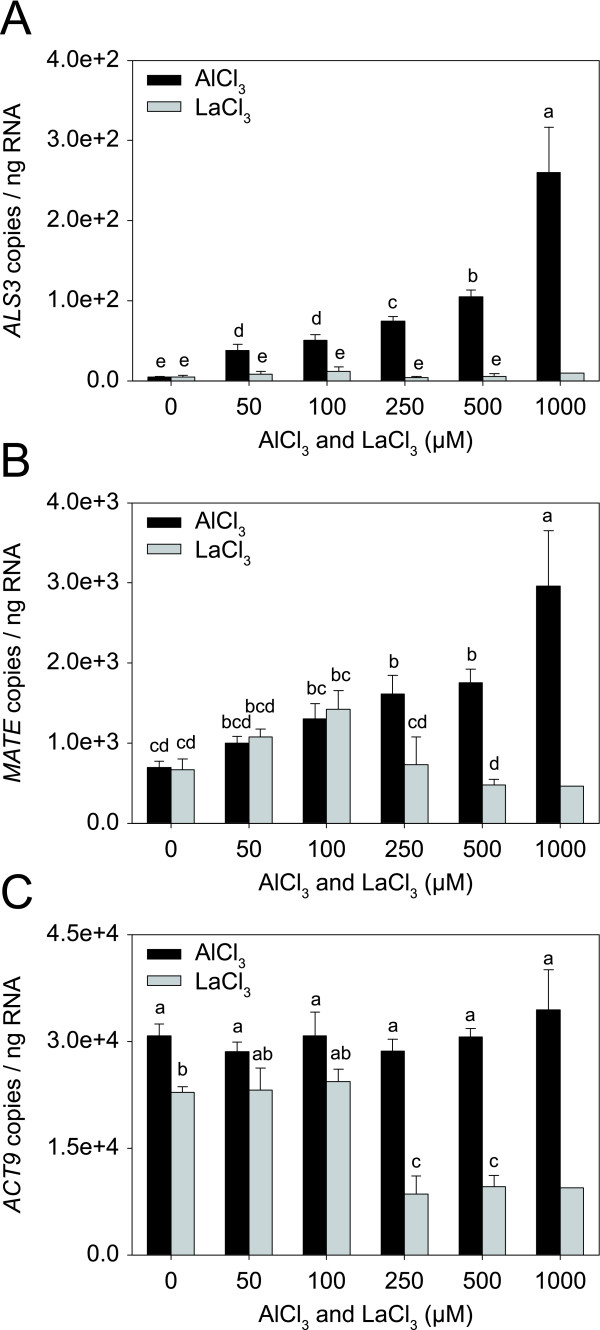
To examine whether Al induces the two genes in a concentration-dependent way, a separate set of plants were treated with increasing concentrations of Al up to 1000 μM Al. Plants were also treated with increasing concentrations of lanthanum (La), which has chemical properties similar to those of Al and is known to inhibit root growth [44]. Root growth measurements showed that La inhibited root growth in a similar way to Al, but the inhibition was stronger (data not shown). Absolute qRT-PCR showed that the expression of both genes increased with Al in the medium (Fig. 7A; B). In contrast, exposure of the roots to La induced MATE, but not ALS3 (Fig. 7A; B). Based on levels of ACT9 expression, it appeared that La concentrations ≥ 250 μM, affected transcription (Fig. 7C).
Figure 7.
Expression levels of ALS3, MATE, and ACT9 in aspen roots treated with Al and La. Expression levels of ALS3 (A), MATE (B) and ACT9 (C) in the root tips of plants treated with no or increasing Al and La concentrations for 2 d. Transcript levels were quantified by absolute qRT-PCR. The means and standard error values from three independent samples are shown, except for the roots treated with 1000 μM La. Different letters indicate significant differences between treatments (ANOVA; p < 0.05).
Discussion
In this study, we analyzed temporal patterns of root growth and root gene expression in aspen under Al stress. Two phases of root growth could be distinguished: a rapid Al-induced growth inhibition (within 6 h) and a subsequent growth recovery. From our analysis of gene expression at three time points, we found a pronounced decrease in the number of responsive genes from 202 to 27 between 6 h and 2 d of Al exposure. The number of changes then remained low. Similar patterns of root growth and gene expression were observed in an Al-tolerant line of Medicago truncatula [28]. The root growth of this line largely recovered within 2 d of Al exposure, while that of an Al-sensitive line remained inhibited. The number of responsive genes was found to decrease in both lines between 12 h and 2 d of Al exposure, but the reduction was stronger in the Al-tolerant line (3.3-fold) than in the Al-sensitive line (1.5-fold). These patterns may reflect the success of roots in activating Al tolerance mechanisms and the restoration of the transcriptome to a prestress program, and in the case of the Al-sensitive line of Medicago the failure to establish an adaptive response.
Consistent with the strong Al-induced root growth inhibition at the 6 h time point, a number of differentially expressed genes can be associated with toxic effects of Al. Our enrichment analysis identified cell wall modification, ion transport, and oxidative stress as major biological processes operating at this time point. The regulation of cell wall-related genes was not surprising because the cell wall is considered to be a major site of Al toxicity [4]. Physiological studies show consistently that a large portion of the Al absorbed by roots is localized to the apoplasm [5]. Several studies indicate that the Al bound to the apoplasm can make the cell wall more rigid, thus reducing its extensibility, which is required for normal cell extension [45,46]. Plants are believed to employ several different mechanisms to loosen the cell wall during cell extension. Proposed cell wall loosening agents include expansin, xyloglucan endotransglucosylase/hydrolase (XHT), endo-1,4-β-glucanase, and hydroxyl radicals [47]. In this study, we recorded several genes encoding XHTs and endo-1,4-β-glucanases, with some of the genes being up-regulated and others being down-regulated. These contrasting responses may reflect the different enzymatic functions that members of the XHT and endo-1,4-β-glucanase protein families can fulfill [47]. Although the exact function of the XHT and endo-1,4-β-glucanase genes identified is not known, it appears likely that these genes play a role in minimizing the toxic effects of Al on the cell wall. A number of physiological studies demonstrate that Al strongly interacts with membrane channel proteins, resulting in the disruption of the uptake and homeostasis of cations, such as Ca, Mg, and K [3]. In our study, we recorded up-regulation of a two-pore K+ channel gene and two CorA-like Mg transporter genes, indicating that Al interferes with K and Mg homeostasis in aspen roots. Mg transporters have so far not been found to be Al inducible. Nevertheless, two lines of evidence indicate that Mg transporters play an important role in Al toxicity. The activity of a CorA-like Mg transporter of Arabidopsis was shown to be blocked by micromolar concentrations of Al [38]. In addition, the same CorA-like Mg transporter alleviated Al toxicity when overexpressed in planta [48]. Although oxidative stress is commonly regarded to be a major component of Al toxicity, we recorded only a small number of genes belonging to the ROS network of genes. A Fe superoxide dismutase and a mitochondrial alternative oxidase gene were up-regulated. While Fe superoxide dismutases are responsible for ROS scavenging [49], alternative oxidases serve to lower mitochondrial ROS formation [50]. Induction of the alternative oxidase is entirely in agreement with the finding that Al can induce ROS formation in mitochondria [51].
Based on information from Arabidopsis and crop plants, three differentially expressed aspen genes may play a role in Al tolerance mechanisms. Two genes were closely related to the Arabidopsis Al tolerance genes ALS3 and MATE. Patterns of expression in different plant organs and in response to Al strongly suggest that the two aspen genes are homologs of the Arabidopsis ALS3 and MATE. The exact function of the Arabidopsis ALS3 is not known. Mutant als3 seedlings grown in Al-containing medium exhibit a severe arrest of root growth and inhibited leaf expansion [52]. Based on this phenotype and patterns of tissue-specific expression, ALS3 has been proposed to mediate transport of Al away from sensitive root tissues to aerial parts of the plant for sequestration or exudation [23]. The Arabidopsis ALS3 is expressed in the phloem throughout the plant and is strongly induced by Al in the root cortex [23]. Our aspen ALS3 gene was expressed in the root, stem, and leaves, and was strongly induced by Al in the root. In addition, the aspen ALS3 was inducible by Al, but not by La. This is consistent with the finding that the Arabidopsis mutant als3 is not affected by La [52]. Based on information from Arabidopsis, it appears likely that the aspen gene identified is functioning in internal Al tolerance. The predicted aspen ALS3 also shared significant sequence identity with STAR2 of rice, which has been suggested to be involved in masking Al binding sites at the cell wall. However, since STAR2 is expressed in the root only [21], it seems unlikely that the aspen gene is a homolog of STAR2. The second putative aspen Al tolerance gene belongs to the MATE gene family. Members of this gene family mediate the release of citrate into the rhizosphere and have been demonstrated to represent major genes of Al tolerance in several plant species [8]. The aspen MATE was primarily expressed in the root and was inducible by Al, a pattern comparable to that of the Arabidopsis MATE and MATE of other plant species [16-20]. Previously, we have shown that aspen releases citrate and oxalate from roots in response to Al exposure. Therefore, it is possible that the aspen MATE is involved in the release of citrate and that this mechanism is regulated at least in part at the transcriptional level. An additional aspen gene that may play a role in Al tolerance encodes a pectin methylesterase. This gene was down-regulated early in the response to Al. Pectin methylesterases demethylate pectin and thereby generate free pectin carboxylic groups. The degree of pectin demethylation largely determines the negative charge the pectin matrix carries and thus the amount of Al that can bind to the cell wall. In maize and rice, the degree of pectin methylation has been linked to genotypic differences in Al tolerance. It was demonstrated that the root tips of Al-sensitive lines had a lower degree of pectin methylation and that larger amounts of Al were bound to the cell wall when compared with Al-tolerant lines [11,12]. Consistent with this, higher levels of pectin methylesterase expression were observed in an Al-sensitive maize line than in an Al-tolerant line [26]. Down-regulation of the aspen pectin methylesterase gene thus may serve to reduce Al binding sites at the pectin matrix, and consequently to limit accumulation of Al in the apoplasm.
Conclusion
This study shows that aspen roots respond to Al exposure with a rapid inhibition of root growth and a large change in gene expression. This early response to Al was characterized by genes involved in cell wall modification, ion transport, and oxidative stress. The subsequent root growth recovery and the concomitant reduction in responsive genes strongly suggest that aspen roots are capable to activate Al tolerance mechanisms when exposed to Al. Based on information from Arabidopsis and other plant species, it appears likely that ALS3, MATE, and possibly a pectin methylesterase gene are important components of the Al tolerance mechanisms in aspen. These genes and genes with unknown function provide candidates for further studies to elucidate the molecular basis of Al tolerance in aspen.
Methods
Plant material
Experiments were performed with in vitro propagated plants of the aspen (Populus tremula L.) clone Birmensdorf [33]. The plants were maintained in Magenta vessels (GA-7) on 80 ml of 1/2 MS medium (Murashige and Skoog), supplemented with 1% sucrose and solidified with 0.8% agar. The plants were cultivated in a greenhouse and multiplied every 6-8 weeks.
Solution culture experiments
Aspen roots were treated with Al and La in solution culture prepared in modified Magenta vessels (GA-7). Teflon-racks, placed on the floor of the vessels, supported a 1190 μm nylon mesh (Sefar) as substrate for the plants and two glass slides with a 5 μm grid (N°5, Boraglas) used as a reference to measure root growth. The modified Magenta vessels were sterilized by autoclaving, and filled with 120 ml of autoclaved 1/6 MS solution (pH 4.0) containing 50 nM indol-3-butyric acid (IBA) to induce and synchronize root formation [53]. In each vessel, four cuttings with 3-4 internodes and 1-2 apical leaves were inserted into the mesh so that the roots could form between the wall of the vessel and the glass slide (slotted 1 cm away of the vessel wall). After one day, the nutrient solution was replaced with fresh 1/6 MS medium (pH 4.0) without IBA. The cuttings were incubated without aeration in a climate chamber maintained at 20 ± 2°C with a 16 h light/8 h darkness period (Osram Dulux L 36W/840 color white, Osram). The nutrient solution was replaced twice a week. After 20 d of incubation, after the cuttings had formed 5-15 adventitious roots, the nutrient solution was replaced with treatment solution composed of autoclaved 1/6 MS medium (pH 4.0), supplemented with increasing concentrations of filter-sterilized AlCl3 up to 1000 μM (dissolved in 1/6 MS). The pH of the Al treatment solutions was adjusted prior to Al addition with filter-sterilized base (1 M KOH) at amounts empirically determined to ensure that the final pH was 4.0. Treatment experiments with LaCl3 were identical to those carried out with AlCl3.
The root growth was monitored photographically prior to the treatment (2 d) and during the entire treatment at 12 h intervals and during the first day of the treatment at 6 h intervals. We used a Canon EOS 400 D digital camera fitted with a Canon macro lens EF 100 mm focused on the 5 μm grid of the glass slides. Files were transferred to an Apple MacBookPro, and the pictures were cropped and normalized using the grid on the glass slide with IMAGEJ 1.36b for Macintosh http://rsb.info.nih.gov/ij/. The normalized pictures were used to measure the increase in root length during the particular time intervals. The root growth rate was estimated by dividing each increment by the time elapsed.
Following these treatments, the roots were separated from the shoots and were rinsed in 1/6 MS pH 4.0. The first centimeter of each root was collected, and all the roots processed per plant were transferred to a sterile 1.5 ml tube. The pooled leaves and the stem were collected separately. All the tissues were frozen in liquid nitrogen and stored at -80°C until RNA extraction.
Quantification of callose
Callose was quantified in the first centimeter of the roots essentially as described by Köhle et al. [54]. Ethanol-fixed root tips (four per plant) were blotted dry and homogenized in 0.5 ml of 1 M NaOH with two steel beads in a Retsch MM 200 mixing mill for 3 min. The homogenate was supplemented with 0.5 ml of 1 M NaOH and incubated at 80°C for 30 min. Following centrifugation, 200 μl of the supernatant were mixed with 400 μl of 0.1% (w/v) aniline blue and 1 M glycine NaOH buffer (pH 9.5), and incubated at 50°C for 20 min. Callose was quantified fluorometrically at 400 nm excitation and at 512-521 nm emission wavelength (FluroLog FL3-22, Jobin Yvon), using curdlan as a reference.
Treatment of plants used for gene expression profiling
Gene expression profiles were determined in a single set of clonal plants grown simultaneously to produce RNA. The plants were treated with no or 500 μM AlCl3 for 6 h, 54 h, and 246 h. For each treatment period, three solution cultures were established to allow three independent plants per treatment to be analyzed. Following treatment, the roots were processed as described above and stored at -80°C until RNA extraction. To control the Al concentrations in the nutrient solutions, total Al was quantified by inductively coupled plasma optical emission (ICP-OES; Optima 7300 DV, Perkin Elmer, Wellesley, MA). Prior to analysis, the samples were acidified with nitric acid at 1.5% (v/v).
RNA isolation
Total RNA from the root tips, stems and leaves were isolated with the Agilent Total RNA Isolation Mini-Kit (Agilent Technologies) according to the manufacturer's instructions. Steps 10 and 11 of the protocol (Agilent Technologies, 2005) were modified as follows: step 10 was carried out twice with 400 μl of wash solution, and step 11 with 400 μl and centrifugation for 1 min, followed by an additional centrifugation for 1 min to ensure that the membrane of the column was completely dry. The concentration of total RNA was determined with a Nanodrop ND-1000 spectrophotometer and the integrity of the RNA was determined with the Eukaryote Total RNA Nano Assay (Agilent Technologies) using the 2100 Agilent Bioanalyzer. Only RNAs with a 260 nm/280 nm ratio between 1.8 and 2.1 and a RNA integrity number between 7 and 10 were processed further.
Microarray analyses
Microarray analyses were carried out with the Affymetrix GeneChip poplar genome array, which was designed based on sequence information from different poplar species. Synthesis of cDNA, cRNA labeling, and hybridization to the GeneChip poplar genome array were essentially performed as described in the Affymetrix GeneChip Expression Analysis Technical Manual (2005). The cDNA synthesis was performed with 2 μg of total RNA, and the quality of the labeled cRNA was determined using Bioanalyzer 2100.
After hybridization and scanning, probe cell intensities were calculated with the Affymetrix Microarray Analysis Suite (MAS version 5.0) [55]. The Robust Multichip Average (RMA) summary algorithm [56] as implemented in GeneSpring GX 7.3 (Agilent Technologies Inc.) was used to generate and normalize raw gene expression data from probe intensities. Genespring was also used to filter out normalized expression values when not showing present calls in all replicate measurements of at least one condition. To identify genes whose expression differed upon Al treatment at each time point, a Student's t-test was performed in R http://www.r-project.org. To reduce the number of true discoveries that include false positives in multiple and simultaneous statistical tests, a positive false discovery rate (FDR), called q, was estimated for each gene. Q-values were calculated with the QVALUE software (implemented in R) with the p-values (p < 0.05, obtained from the t-test) as input and the bootstrap robust settings [34]. Genes were considered to be differentially expressed when (1) the transcript abundance in Al-treated plants was significantly different from that of control plants as determined by the Student's t-test, (2) the calculated q was ≤ 0.15, and (3) the change in expression between treated and control plants was at least two-fold. Probe-sets matching the same gene model of the black cottonwood genome sequence http://www.phytozome.net/poplar were declared as redundant. Microarray data are available in the Gene Omnibus Database as accessions GSE19297.
To annotate the expressed transcripts, we performed BLASTX searches against the Arabidopsis protein database (TAIR; http://www.arabidopsis.org). The best match was reported, and the resulting list of Arabidopsis identifiers was applied to the Functional Catalogue developed by MIPS http://mips.gsf.de/proj/funcatDB to identify biological processes. The MIPS singular enrichment tool was used to identify categories whose members were statistically over-represented compared to the genes present on the microarray. The frequency of genes of a given category on the microarray was calculated as the ratio of the number of genes of this category divided by the total number of genes on the microarray, and the frequency of regulated genes of a given category was calculated as the ratio of the number of regulated genes of this category divided by the total number of regulated genes. Because differentially expressed genes were annotated based on information from Arabidopsis, we only considered genes of the microarray representing putative homologs of Arabidopsis genes (15,216 genes). Our significance test to search for enriched categories was based on the hypergeometric distribution and is identical to the corresponding one-tailed version of Fisher's exact test, calculating the probability of observing data as extreme or more extreme if the null hypothesis is true. Two strategies to correct for multiple testing were used. The first was Bonferroni's correction, which is a conservative way to control the family wise error rate, and the second a FDR. Functional categories with a p-value < 0.05 and a FDR < 0.05 were regarded to be enriched.
Real-time reverse transcription PCR
Real-time reverse transcription PCR (qRT-PCR) was performed with primers matching aspen sequences that were obtained as follows. Sequence information on the Affymetrix probes was used to design primers, which amplified as long fragments as possible. These primers were applied in RT-PCR and the resulting RT-PCR products were directly sequenced. The aspen sequences obtained were then used to design new primers, which amplified short fragments suitable for qRT-PCR (90-114 bp). When possible, one of the two primers was designed across an exon-exon junction using information from the black cottonwood genome sequence. Primers were designed with the Primer3 software [57]. Sequences of the primers are given in additional file 3.
To validate the results of the microarray analysis, we quantified the expression of eight genes relative to ACT9. RT was performed with 200 ng of total RNA using the QuantiTect Reverse Transcription Kit (Qiagen) under conditions recommended by the manufacturer. PCR was performed with the power SYBR green PCR master mix (Applied Biosystems) in a reaction volume of 25 μl containing 5 μl of diluted cDNA. Cycling was carried out on an ABI 7500 Fast real-time cycler (Applied Biosystems) with the following cycling profile: 10 min activation of AmpliTaq Gold Polymerase at 95°C, 45 cycles of 15 sec denaturation at 95°C, 30 sec annealing at 60°C, 30 sec extension at 72°C, followed by a dissociation step of 15 sec at 95°C, 15 sec at 60°C, and 15 sec at 95°C to detect primer dimers and non-specific amplification products. For each primer pair, we determined the PCR efficiency and the dynamic range of PCR by plotting the threshold cycle (Ct) values generated over a range of dilutions against the log input of cDNA amount. To obtain accurate results, only primer pairs yielding PCR efficiencies of 90-110% (slope of regression between -3.2 and -3.5) were considered [58]. To quantify the transcripts, each cDNA of three biological replicates was diluted in duplicate and used in duplicate PCR. The relative abundance of each transcript was estimated using the ΔΔCt method [59].
The expression of selected genes (ALS3, MATE, and ACT9) was further quantified by absolute qRT-PCR. Copy numbers of the transcripts were calculated from standard curves that were obtained as follows. Single-stranded sense olignonucleotides specifying amplicons of the selected genes were synthesized (Operon; for sequences see additional file 4). Information from the manufacturer was used to calculate the copy number of the oligonucleotides present in 1 μl of 10 mM Tris-HCl pH 8.5 (Qiagen). Serial dilutions of the stocks were carried out in duplicate, and dilutions in the range from 102-108 copies were used in duplicate PCR to generate standard curves. The standard curve was obtained by plotting the logs of the calculated copy number against Ct. The copy numbers of unknown samples were calculated from the regression line. Each cDNA was diluted and run in duplicate, and the transcript copy number was expressed per nanogram of total RNA.
Authors' contributions
NG carried out the molecular and plant studies, participated in the microarray data analysis, and drafted the manuscript. MGK carried out the screening of the microarrays, SZ performed the statistical analysis of the microarray data, and MM performed the enrichment analysis. TL quantified the callose and participated in the plant studies. IB, LB, and JPM participated in the design of the study and improved the quality of the manuscript. CS coordinated the study and wrote the final manuscript. All authors have read and approved the final manuscript.
Supplementary Material
List of Al-responsive genes in root tips of aspen.
Lists of MIPS categories whose genes were significantly enriched (p < 0.05; FDR < 0.05).
Primers used to perform real-time reverse transcription PCR in aspen.
Sequences of single-stranded sense oligonucleotides specifying amplicons of ACT9, ALS3, and MATE of aspen.
Contributor Information
Nadine Grisel, Email: nadine.grisel@wsl.ch.
Stefan Zoller, Email: stefan.zoller@env.ethz.ch.
Marzanna Künzli-Gontarczyk, Email: marzanna.kuenzli@fgcz.ethz.ch.
Thomas Lampart, Email: thomas.lampart@dualsystems.com.
Martin Münsterkötter, Email: m.muensterkoetter@helmholtz-muenchen.de.
Ivano Brunner, Email: ivano.brunner@wsl.ch.
Lucien Bovet, Email: lucien.bovet@pmintl.com.
Jean-Pierre Métraux, Email: jean-pierre.metraux@unifr.ch.
Christoph Sperisen, Email: christoph.sperisen@wsl.ch.
Acknowledgements
We thank Daniela Steiner for her help in our laboratory and in propagating aspen. We also thank Enrico Martinoia for helpful discussions, Ulrich Wagner for suggestions about the design and data analysis of test microarray experiments, and Silvia Dingwall for improving the English. The work was partially founded by the State Secretariat for Education and Research (SER; grant C04.0259 to CS, LB, IB) through the COST E38 Action 'Woody root processes' and by the European Network of Excellence EVOLTREE supporting courses for NG.
References
- Ryan PR, Ditomaso JM, Kochian LV. Aluminium toxicity in roots: an investigation of spatial sensitivity and the role of the root cap. Journal of Experimental Botany. 1993;44:437–446. doi: 10.1093/jxb/44.2.437. [DOI] [Google Scholar]
- Sivaguru M, Horst WJ. The distal part of the transition zone is the most aluminum-sensitive apical root zone of maize. Plant Physiology. 1998;116:155–163. doi: 10.1104/pp.116.1.155. [DOI] [Google Scholar]
- Kochian LV, Piñeros MA, Hoekenga OA. The physiology, genetics and molecular biology of plant aluminum resistance and toxicity. Plant and Soil. 2005;274:175–195. doi: 10.1007/s11104-004-1158-7. [DOI] [Google Scholar]
- Horst WJ, Wang Y, Eticha D. The role of the root apoplast in aluminium-induced inhibition of root elongation and in aluminium resistance of plants: a review. Annals of Botany. 2010;106:185–197. doi: 10.1093/aob/mcq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF. Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. International Review of Cytology. 2007;264:225–252. doi: 10.1016/S0074-7696(07)64005-4. full_text. [DOI] [PubMed] [Google Scholar]
- Rounds MA, Larsen PB. Aluminum-dependent root-growth inhibition in Arabidopsis results from AtATR-regulated cell-cycle arrest. Current Biology. 2008;18:1495–1500. doi: 10.1016/j.cub.2008.08.050. [DOI] [PubMed] [Google Scholar]
- Kochian LV, Hoekenga OA, Piñeros MA. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annual Review of Plant Biology. 2004;55:459–493. doi: 10.1146/annurev.arplant.55.031903.141655. [DOI] [PubMed] [Google Scholar]
- Delhaize E, Gruber BD, Ryan PR. The roles of organic anion permeases in aluminium resistance and mineral nutrition. FEBS Letters. 2007;581:2255–2262. doi: 10.1016/j.febslet.2007.03.057. [DOI] [PubMed] [Google Scholar]
- Basu U, Basu A, Taylor GJ. Differential exudation of polypeptides by roots of aluminum-resistant and aluminum-sensitive cultivars of Triticum aestivum L. in response to aluminum stress. Plant Physiology. 1994;106:151–158. doi: 10.1104/pp.106.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt J, Larsen PB, Howell SH, Kochian LV. Aluminum resistance in the Arabidopsis mutant alr-104 is caused by an aluminum-induced increase in rhizosphere pH. Plant Physiology. 1998;117:19–27. doi: 10.1104/pp.117.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eticha D, Stass A, Horst WJ. Cell-wall pectin and its degree of methylation in the maize root-apex: significance for genotypic differences in aluminium resistance. Plant, Cell and Environment. 2005;28:1410–1420. doi: 10.1111/j.1365-3040.2005.01375.x. [DOI] [Google Scholar]
- Yang JL, Li YY, Zhang YJ, Zhang SS, Wu YR, Wu P, Zheng SJ. Cell wall polysaccharides are specifically involved in the exclusion of aluminum from the rice root apex. Plant Physiology. 2008;146:602–611. doi: 10.1104/pp.107.111989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Delhaize E, Matsumoto H. A wheat gene encoding an aluminum-activated malate transporter. Plant Journal. 2004;37:645–653. doi: 10.1111/j.1365-313X.2003.01991.x. [DOI] [PubMed] [Google Scholar]
- Hoekenga OA, Maron LG, Piñeros MA, Cançado GMA, Shaff J, Kobayashi Y, Ryan PR, Dong B, Delhaize E, Sasaki T. et al. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:9738–9743. doi: 10.1073/pnas.0602868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligaba A, Katsuhara M, Ryan PR, Shibasaka M, Matsumoto H. The BnALMT1 and BnALMT2 genes from rape encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiology. 2006;142:1294–1303. doi: 10.1104/pp.106.085233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes JV, Liu J, Guimarães CT, Lana UGP, Alves VMC, Wang YH, Schaffert RE, Hoekenga OA, Piñeros MA, Shaff JE. et al. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nature Genetics. 2007;39:1156–1161. doi: 10.1038/ng2074. [DOI] [PubMed] [Google Scholar]
- Furukawa J, Yamaji N, Wang H, Mitani N, Murata Y, Sato K, Katsuhara M, Takeda K, Ma JF. An aluminum-activated citrate transporter in barley. Plant and Cell Physiology. 2007;48:1081–1091. doi: 10.1093/pcp/pcm091. [DOI] [PubMed] [Google Scholar]
- Liu J, Magalhaes JV, Shaff J, Kochian LV. Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant Journal. 2009;57:389–399. doi: 10.1111/j.1365-313X.2008.03696.x. [DOI] [PubMed] [Google Scholar]
- Ryan PR, Raman H, Gupta S, Horst WJ, Delhaize E. A second mechanism for aluminum resistance in wheat relies on the constitutive efflux of citrate from roots. Plant Physiology. 2009;149:340–351. doi: 10.1104/pp.108.129155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron LG, Piñeros MA, Guimarães CT, Magalhaes JV, Pleiman JK, Mao C, Shaff J, Belicuas SNJ, Kochian LV. Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. Plant Journal. 2010;61:728–740. doi: 10.1111/j.1365-313X.2009.04103.x. [DOI] [PubMed] [Google Scholar]
- Huang CF, Yamaji N, Mitani N, Yano M, Nagamura Y, Ma JF. A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell. 2009;21:655–667. doi: 10.1105/tpc.108.064543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PB, Cancel J, Rounds M, Ochoa V. Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta. 2007;225:1447–1458. doi: 10.1007/s00425-006-0452-4. [DOI] [PubMed] [Google Scholar]
- Larsen PB, Geisler MJB, Jones CA, Williams KM, Cancel JD. ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant Journal. 2005;41:353–363. doi: 10.1111/j.1365-313X.2004.02306.x. [DOI] [PubMed] [Google Scholar]
- Kumari M, Taylor GJ, Deyholos MK. Transcriptomic responses to aluminum stress in roots of Arabidopsis thaliana. Molecular Genetics and Genomics. 2008;279:339–357. doi: 10.1007/s00438-007-0316-z. [DOI] [PubMed] [Google Scholar]
- Goodwin SB, Sutter TR. Microarray analysis of Arabidopsis genome response to aluminum stress. Biologia Plantarum. 2009;53:85–99. doi: 10.1007/s10535-009-0012-4. [DOI] [Google Scholar]
- Maron LG, Kirst M, Mao C, Milner MJ, Menossi M, Kochian LV. Transcriptional profiling of aluminum toxicity and tolerance responses in maize roots. New Phytologist. 2008;179:116–128. doi: 10.1111/j.1469-8137.2008.02440.x. [DOI] [PubMed] [Google Scholar]
- Houde M, Diallo AO. Identification of genes and pathways associated with aluminum stress and tolerance using transcriptome profiling of wheat near-isogenic lines. BMC Genomics. 2008;9:400. doi: 10.1186/1471-2164-9-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran D, Sharopova N, VandenBosch KA, Garvin DF, Samac DA. Physiological and molecular characterization of aluminum resistance in Medicago truncatula. BMC Plant Biology. 2008;8:89. doi: 10.1186/1471-2229-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaedle M, Thornton FC, Raynal DJ, Tepper HB. Response of tree seedlings to aluminum. Tree Physiology. 1989;5:337–356. doi: 10.1093/treephys/5.3.337. [DOI] [PubMed] [Google Scholar]
- Göransson A, Eldhuset TD. Effects of aluminium on root growth and nutrient uptake of Betula pendula seedlings. Physiologia Plantarum. 1987;69:193–199. doi: 10.1111/j.1399-3054.1987.tb04275.x. [DOI] [Google Scholar]
- Göransson A, Eldhuset TD. Effects of aluminium on root growth and nutrient uptake of small Picea abies and Pinus sylvestris plants. Trees-Structure and Function. 1991;5(3):136–142. doi: 10.1007/BF00204334. [DOI] [Google Scholar]
- Brauer D. Rapid inhibition of root growth in wheat associated with aluminum uptake as followed by changes in morin fluorescence. Journal of Plant Nutrition. 2001;24:1243–1253. doi: 10.1081/PLN-100106979. [DOI] [Google Scholar]
- Qin R, Hirano Y, Brunner I. Exudation of organic acid anions from poplar roots after exposure to Al, Cu and Zn. Tree Physiology. 2007;27:313–320. doi: 10.1093/treephys/27.2.313. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D. Quantification using real-time PCR technology: applications and limitations. Trends in Molecular Medicine. 2002;8:257–260. doi: 10.1016/S1471-4914(02)02355-9. [DOI] [PubMed] [Google Scholar]
- Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Güldener U, Mannhaupt G, Münsterkötter M, Mewes HW. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Research. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert A, Isayenkov S, Voelker C, Czempinski K, Maathuis FJM. The two-pore channel TPK1 gene encodes the vacuolar K+ conductance and plays a role in K+ homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10726–10731. doi: 10.1073/pnas.0702595104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Tutone AF, Drummond RSM, Gardner RC, Luan S. A novel family of magnesium transport genes in Arabidopsis. Plant Cell. 2001;13:2761–2775. doi: 10.1105/tpc.13.12.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubero B, Nakagawa Y, Jiang XY, Miura KJ, Li F, Raghothama KG, Bressan RA, Hasegawa PM, Pardo JM. The phosphate transporter PHT4;6 is a determinant of salt tolerance that is localized to the Golgi apparatus of Arabidopsis. Molecular Plant. 2009;2:535–552. doi: 10.1093/mp/ssp013. [DOI] [PubMed] [Google Scholar]
- Kataoka T, Hayashi N, Yamaya T, Takahashi H. Root-to-shoot transport of sulfate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiology. 2004;136:4198–4204. doi: 10.1104/pp.104.045625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends in Plant Science. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Durrett TP, Gassmann W, Rogers EE. The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiology. 2007;144:197–205. doi: 10.1104/pp.107.097162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EE, Guerinot ML. FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis. Plant Cell. 2002;14:1787–1799. doi: 10.1105/tpc.001495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinraide TB, Ryan PR, Kochian LV. Interactive effects of Al3+, H+, and other cations on root elongation considered in terms of cell-surface electrical potential. Plant Physiology. 1992;99:1461–1468. doi: 10.1104/pp.99.4.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi A, Matsumoto H. Changes in cell-wall properties of wheat (Triticum aestivum) roots during aluminum-induced growth inhibition. Physiologia Plantarum. 2001;112:353–358. doi: 10.1034/j.1399-3054.2001.1120308.x. [DOI] [PubMed] [Google Scholar]
- Jones DL, Blancaflor EB, Kochian LV, Gilroy S. Spatial coordination of aluminium uptake, production of reactive oxygen species, callose production and wall rigidification in maize roots. Plant, Cell and Environment. 2006;29:1309–1318. doi: 10.1111/j.1365-3040.2006.01509.x. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Growth of the plant cell wall. Nature Reviews Molecular Cell Biology. 2005;6:850–861. doi: 10.1038/nrm1746. [DOI] [PubMed] [Google Scholar]
- Deng W, Luo K, Li D, Zheng X, Wei X, Smith W, Thammina C, Lu L, Li Y, Pei Y. Overexpression of an Arabidopsis magnesium transport gene, AtMGT1, in Nicotiana benthamiana confers Al tolerance. Journal of Experimental Botany. 2006;57:4235–4243. doi: 10.1093/jxb/erl201. [DOI] [PubMed] [Google Scholar]
- Alscher RG, Erturk N, Heath LS. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. Journal of Experimental Botany. 2002;53:1331–1341. doi: 10.1093/jexbot/53.372.1331. [DOI] [PubMed] [Google Scholar]
- Maxwell DP, Wang Y, McIntosh L. The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:8271–8276. doi: 10.1073/pnas.96.14.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Kobayashi Y, Devi SR, Rikiishi S, Matsumoto H. Aluminum toxicity is associated with mitochondrial dysfunction and the production of reactive oxygen species in plant cells. Plant Physiology. 2002;128:63–72. doi: 10.1104/pp.010417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PB, Kochian LV, Howell SH. Al inhibits both shoot development and root growth in als3, an Al-sensitive Arabidopsis mutant. Plant Physiology. 1997;114:1207–1214. doi: 10.1104/pp.114.4.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pythoud F, Buchala AJ. The fate of vitamin D3 and indolylbutyric acid applied to cuttings of Populus tremula L. during adventitious root formation. Plant, Cell and Environment. 1989;12:489–494. doi: 10.1111/j.1365-3040.1989.tb02121.x. [DOI] [Google Scholar]
- Köhle H, Jeblick W, Poten F, Blaschek W, Kauss H. Chitosan-elicited callose synthesis in soybean cells as a Ca2+-dependent process. Plant Physiology. 1985;77:544–551. doi: 10.1104/pp.77.3.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–1592. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA. Preprocessing of oligonucleotide array data. Nature Biotechnology. 2004;22:656–658. doi: 10.1038/nbt0604-656b. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. In: Bioinformatics Methods and Protocols: Methods in Molecular Biology. Misener S, Krawetz SA, editor. Totowa NJ: Humana Press; 2000. Primer3 on the WWW for general users and for biologist programmers; pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nature Protocols. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCTmethod. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of Al-responsive genes in root tips of aspen.
Lists of MIPS categories whose genes were significantly enriched (p < 0.05; FDR < 0.05).
Primers used to perform real-time reverse transcription PCR in aspen.
Sequences of single-stranded sense oligonucleotides specifying amplicons of ACT9, ALS3, and MATE of aspen.